Abstract
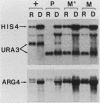
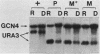
In Saccharomyces cerevisiae, many amino acid biosynthetic pathways are coregulated by a complex general control system: starvation for a single amino acid results in the derepression of amino acid biosynthetic genes in multiple pathways. Derepression of these genes is mediated by positive (GCN) and negative (GCD) regulatory genes. In this paper we describe the isolation and characterization of a previously unreported negative regulatory gene, GCD3. A gcd3 mutation is recessive to wild type, confers resistance to multiple amino acid analogs, and results in overproduction and partially constitutive elevation of mRNA levels for amino acid biosynthetic genes. Furthermore, a gcd3 mutation can overcome the derepression-deficient phenotype of mutations in the positive regulatory GCN1, GCN2, and GCN3 genes. However, the gcd3 mutation cannot overcome the derepression-deficient phenotype of a gcn4 mutation, suggesting that GCD3 acts as a negative regulator of the important GCN4 gene. Northern blot analysis confirmed this conclusion, in that the steady-state levels of GCN4 mRNA are greatly increased in a gcd3 mutant. Thus, the negative regulatory gene GCD3 plays a central role in derepression of amino acid biosynthetic genes.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Carsiotis M., Jones R. F. Cross-pathway regulation: tryptophan-mediated control of histidine and arginine biosynthetic enzymes in Neurospora crassa. J Bacteriol. 1974 Sep;119(3):889–892. doi: 10.1128/jb.119.3.889-892.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L., Carbon J. Functional expression of cloned yeast DNA in Escherichia coli: specific complementation of argininosuccinate lyase (argH) mutations. J Mol Biol. 1978 Apr 25;120(4):517–532. doi: 10.1016/0022-2836(78)90351-0. [DOI] [PubMed] [Google Scholar]
- Donahue T. F., Daves R. S., Lucchini G., Fink G. R. A short nucleotide sequence required for regulation of HIS4 by the general control system of yeast. Cell. 1983 Jan;32(1):89–98. doi: 10.1016/0092-8674(83)90499-3. [DOI] [PubMed] [Google Scholar]
- Greenberg M. L., Myers P. L., Skvirsky R. C., Greer H. New positive and negative regulators for general control of amino acid biosynthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1986 May;6(5):1820–1829. doi: 10.1128/mcb.6.5.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer H. Mechanisms of yeast genetics. Basic Life Sci. 1981;18:217–242. doi: 10.1007/978-1-4684-3980-9_14. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6442–6446. doi: 10.1073/pnas.81.20.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., Fink G. R. Positive regulation in the general amino acid control of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5374–5378. doi: 10.1073/pnas.80.17.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., Lucchini G., Fink G. R. A synthetic HIS4 regulatory element confers general amino acid control on the cytochrome c gene (CYC1) of yeast. Proc Natl Acad Sci U S A. 1985 Jan;82(2):498–502. doi: 10.1073/pnas.82.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4 protein, synthesized in vitro, binds HIS3 regulatory sequences: implications for general control of amino acid biosynthetic genes in yeast. Cell. 1985 Nov;43(1):177–188. doi: 10.1016/0092-8674(85)90022-4. [DOI] [PubMed] [Google Scholar]
- Klapholz S., Esposito R. E. A new mapping method employing a meiotic rec-mutant of yeast. Genetics. 1982 Mar;100(3):387–412. doi: 10.1093/genetics/100.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenguy F. Concerted repression of the synthesis of the arginine biosynthetic enzymes by aminoacids: a comparison between the regulatory mechanisms controlling aminoacid biosyntheses in bacteria and in yeast. Mol Gen Genet. 1979 Jan 16;169(1):85–95. doi: 10.1007/BF00267549. [DOI] [PubMed] [Google Scholar]
- Miozzari G., Niederberger P., Hütter R. Tryptophan biosynthesis in Saccharomyces cerevisiae: control of the flux through the pathway. J Bacteriol. 1978 Apr;134(1):48–59. doi: 10.1128/jb.134.1.48-59.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn M. D., Galgoci B., Greer H. Identification of AAS genes and their regulatory role in general control of amino acid biosynthesis in yeast. Proc Natl Acad Sci U S A. 1983 May;80(9):2704–2708. doi: 10.1073/pnas.80.9.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn M. D., Thireos G., Greer H. Temporal analysis of general control of amino acid biosynthesis in Saccharomyces cerevisiae: role of positive regulatory genes in initiation and maintenance of mRNA derepression. Mol Cell Biol. 1984 Mar;4(3):520–528. doi: 10.1128/mcb.4.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürch A., Miozzari J., Hütter R. Regulation of tryptophan biosynthesis in Saccharomyces cerevisiae: mode of action of 5-methyl-tryptophan and 5-methyl-tryptophan-sensitive mutants. J Bacteriol. 1974 Mar;117(3):1131–1140. doi: 10.1128/jb.117.3.1131-1140.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman S. J., Rose M., Botstein D., Fink G. R. Regulation of HIS4-lacZ fusions in Saccharomyces cerevisiae. Mol Cell Biol. 1982 Oct;2(10):1212–1219. doi: 10.1128/mcb.2.10.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skvirsky R. C., Greenberg M. L., Myers P. L., Greer H. A new negative control gene for amino acid biosynthesis in Saccharomyces cerevisiae. Curr Genet. 1986;10(7):495–501. doi: 10.1007/BF00447382. [DOI] [PubMed] [Google Scholar]
- Thireos G., Penn M. D., Greer H. 5' untranslated sequences are required for the translational control of a yeast regulatory gene. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5096–5100. doi: 10.1073/pnas.81.16.5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfner M., Yep D., Messenguy F., Fink G. R. Integration of amino acid biosynthesis into the cell cycle of Saccharomyces cerevisiae. J Mol Biol. 1975 Aug 5;96(2):273–290. doi: 10.1016/0022-2836(75)90348-4. [DOI] [PubMed] [Google Scholar]