Abstract
We have conducted a screen to identify developmentally regulated enhancers that drive tissue-specific Gal4 expression in zebrafish. We obtained 63 stable transgenic lines with expression patterns in embryonic or adult zebrafish. The use of a newly identified minimal promoter from the medaka edar locus resulted in a relatively unbiased set of expression patterns representing many tissue types derived from all germ layers. Subsequent detailed characterization of selected lines showed strong and reproducible Gal4-driven GFP expression in diverse tissues, including neurons from the central and peripheral nervous systems, pigment cells, erythrocytes, and peridermal cells. By screening adults for GFP expression, we also isolated lines expressed in tissues of the adult zebrafish, including scales, fin rays, and joints. The new and efficient minimal promoter and large number of transactivating driver-lines we identified will provide the zebrafish community with a useful resource for further enhancer trap screening, as well as precise investigation of tissue-specific processes in vivo.
Introduction
Enhancer detection through random vector insertion into a metazoan genome was pioneered in Drosophila using a vector in which a minimal promoter is positioned in front of a reporter protein to identify cell-type specific markers.1 After random insertion of the reporter construct, transgenic animals were screened for expression within specified cells or tissues during development. The particular regulation of the transgene suggests control of the promoter by local cis-regulatory enhancers in a tissue- or cell-specific manner. In addition to the expression pattern supporting the possible role for the adjacent gene in the tissue of interest, the transgenic lines serve as markers for future studies. The extensive use of this technique has led to the generation of thousands of transgenic lines in Drosophila,2–4 which have contributed substantially to our understanding of the function of cis-regulatory networks in metazoan development.5–7
In exchange for a reporter protein, the addition of the Gal4 transcriptional activator from yeast strengthened the use of such enhancer screens by further allowing tissue-specific manipulation of embryos in vivo.8 When Gal4 is expressed in the tissue of interest, it facilitates the transcription of genes directly downstream of the Upstream Activating Sequence (UAS). In this way, it is possible to achieve spatial and temporal control over the expression of effector constructs fused to a UAS cassette. By simply crossing a stable transgenic line expressing the Gal4 driver to a line expressing the UAS:effector transgene, the effector construct will be activated only in the cell type and at the time when the Gal4 protein is present. Not only do these lines allow for more detailed observation of developmental processes in vivo, but the driver lines in particular are a powerful tool for real-time manipulation of development through the targeted expression of effector constructs such as endogenous genes,9 subcellular or physiological markers,10 ablation or synaptic antagonist toxins,11,12 photoswitchable fluorophores,13 or optico-genetic molecules that can be used for controlling neuronal activity.14
Although broad-based enhancer screens have been widely applied to many nonvertebrate model systems,15–19 they have only been moderately used in mouse due to the difficulties of large-scale transgenesis and embryonic expression screening in mammals.20–22 However, the use of enhancer detection in zebrafish has expanded dramatically in recent years, due to the fish's transparency, external development, and amenability to high-throughput screening.23–25 These advantages have long been employed for systematic genetic screens,26–28 but advances in retroviral29,30 and transposon31 based transgenesis techniques have enabled the efficient and large-scale detection of enhancer regions in the zebrafish genome,23,25,32 and the identification of transactivating Gal4-driver lines for targeted transgene expression in a tissue-specific manner.11,33–36
The initial enhancer detection screens in fish were performed to optimize transposon-based transgenesis techniques with simple reporter transgenes in medaka37 and zebrafish.25,38 In spite of the successful and continuing use of transposon-based methodologies, the first large-scale enhancer screen using a fluorescent reporter was carried out with a murine leukemia retrovirus (MLV) that was engineered for zebrafish transgenesis.23 With this method, about 95 distinct transgenic YFP-expressing lines were obtained with a diverse set of tissue-specific and ubiquitous expression patterns.23 However, because of the specialized training, equipment, and permitting that is required to work with MLV, it is not used as commonly as transposon-based methods.
Transposons have been an essential tool of Drosophila genetics for decades, but the use of transposons in zebrafish has been limited, until recently, by the lack of transposable elements in the zebrafish genome.39 The distinct advantages of transposon-based systems for enhancer detection and routine transgenesis such as their simplicity and efficiency, resulted in the development of diverse transposon vectors (e.g., Sleeping Beauty,40,41 Tc3,42 AC-DS,43 and Tol244) that allowed for high frequency chromosomal integration in germ cells. The Tol2 system in particular, which is derived from an endogenous medaka transposable element, has dramatically increased the efficiency and ease of both making transgenic lines and mapping their integration sites.
Here we present the results of our Tol2 transposon-based Gal4-driver line screen with a minimal promoter derived from the medaka edar locus, which demonstrates tissue-specific expression in lines encompassing a diverse set of cell types in embryonic, larval, and adult zebrafish. Our lines will serve as an important supplement to the growing driver line collection in the zebrafish community and will strengthen the position of the zebrafish as an excellent model for studying fundamental developmental processes through in vivo imaging and tissue-specific genetic manipulations. Additionally, the use of this promoter with refined Gal4 UAS constructs will aid future enhancer screens in the zebrafish.
Materials and Methods
Cloning of the enhancer detection construct
The promoter used to generate the TDL driver lines was derived from the immediate upstream region of the first exon of the medaka edar gene. An analysis of conserved elements in the edar locus of the zebrafish compared with medaka showed three elements sharing long stretches of identity. Whereas the medaka elements were all clustered in the 5′ upstream region, in zebrafish they were found duplicated and scattered within the first intron as well. To test the function of these elements, we isolated them from genomic DNA in three parts—one containing all elements (1140 bp), the two proximal elements (787 bp), and the most proximal element (627 bp) to the edar transcript. We cloned these putative promoters in front of Gal4 cassette in cis to 14xUAS driving GFP.54 These constructs were injected with Tol2 mRNA into single-celled embryos to facilitate transgenesis. Analysis of F1 transgenic progeny showed specific expression patterns in a broad range of tissue types. This suggests that the promoter constructs contained little specific regulation, however had sufficient information to promote expression with the specificity gained from the particular insertion site. Of the three different promoter constructs, we found no obvious difference in the types of expression or frequency of insertion/transgenesis in any of the transgenic progeny screened. The smallest proximal element showed the most robust expression and all further screening of transgenic lines used this minimal promoter element. The sequence of this proximal element is as follows:
5′TGACCTGAACCGGAACATCAGTGGGGGGACCCTTCGGTGTGGAGTTTGCATGTTCTCCCTGTGCAGGTATGGGTTCTCTCTGGGAACTCTGGCTTCCTCCCACCGTCCAAAAACATGCTTCATAGGTCAATTGGCAACTCTAAATTGTCCATAGATGTGGGTGTGAGAGTGAGTGGATGTGTGATATTTACATACACACATACATATTTGTTTTTGCAAATATTTTATTTAAAAAACATACATAGAACTCCACTAACCATTTCCAGCAGTTTGCATGCTTTGGCTCCTCCACTACATGTTATTGCATGCCTGCAAACACTAGATGGCGTGGTCGAGTCAGGCTCTGCAAACTTTTTTTTTTTTTTTTGACAGAGTTTACCTGGGACGCTTTAATCAGTTTGTCTTCCAGGGCGGAGAAAACTTCAAGGAGGAGCCTGCAGACACACCGACATGCGTCCAAACGGAGGCTGAGACACAACTTAGATTGTGTGAAGTTGAACCTTTGAGACTTGCCGGTCCGTTTGGGGATGCTACAGGAACGCCGGGTCAGAGACTGAGCGGTGACCATA3′.
Screening of F1 progeny
Mating crosses of albino mutant zebrafish were set up according to standard procedures55 and single-cell embryos were injected with 30 ng/μL of Tol2 RNA and 30 ng/μL of plasmid DNA from the enhancer trap construct in volumes of 1 nL per embryo. Capped Tol2 RNA was generated with a mMessage mMachine RNA kit (Applied Biosystems, Darmstadt Germany) according to manufacturer protocols. All injected larvae that survived up to 7 days post fertilization (dpf) were raised, and F1 progeny were screened for GFP expression every day from 24 hpf up until 7 dpf, and then once again at 30 dpf. All larvae showing GFP expression were raised and then re-identified in the F2 generation.
Mapping of insertion sites
The insertion sites were determined by thermal asymmetric interlaced (TAIL) PCR as described in Parinov et al. (2004)25 with the following modifications: 100 ng genomic DNA extracted from 20, 5 dpf larvae or caudal fin clips of adults showing GFP expression were used as template for primary PCR reactions; PCR products of secondary or tertiary PCR reactions were gel-purified (Wizard SV Gel and PCR Clean-Up System, Promega), subcloned into pGEMT-easy vector (Promega), and sequenced using the standard primers M13uni and M13rev. All sequences were analyzed for the presence of either Tol2-5′ or -3′ flanking sequences, and the cloned zebrafish genomic sequences were BLASTed to the zebrafish genome to identify the insertion site as seen in Table 2 (Ensemble release Zv9 http://www.ensembl.org/Danio_rerio/). For some lines we found two genomic positions, however it is not clear which is responsible for the GFP expression pattern.
Table 2.
Genomic Mapping of Selected Insertion Lines
| Tübingen driver line # | Expression pattern: Stage | Insertion position (LG:bp), notes |
|---|---|---|
| 6 | Primary motorneurons: 24 hpf | * |
| 13 | Muscle fibers, skin, vasculature, and unidentified cell types: 3 dpf | 23:24,886,350 intergenic, in promoter region of kans12; mutant phenotype linked to GFP expression |
| 22 | Scales, adult | |
| 40 | Trunk skeletal muscle fibers: 48 hpf | * |
| 42 | Myogenic lineage: 48 hpf | * |
| 45-1 | Epidermis: 24 hpf | * |
| 45-2 | Liver, pancreas: 4 dpf | 24:25,590,869 5′UTR of zgc:92111 (osta) |
| 64 | Trunk skeletal muscle fibers, strongest labeling in posterior of larva: 48 hpf | 18:20,649,309 exonic within synemin |
| 74 | Pectoral fin, adult | |
| 91 | Presomitic mesoderm, later in skeletal muscle in posterior of larva | 10:10,933,328 intronic of si:ch211-183l21.3 |
| 118-1 | Cells surrounding notochord (epithelial-like hexagonal shape): 48 hpf | * |
| 137 | Spinal cord neurons: 24 hpf | 10:7,973,728 intronic of LOC564899 |
| 200 | Skin, trunk skeletal muscles, notochord | 7:8,217,106 exonic within zgc:101810 |
| 201 | Cranial neural crest, branchial arches, ear: 48 hpf | 5: 16,930,847 intronic of gnb1l |
| 206 | Cranial neural crest, myogenic lineage: 24 hpf | 21:44,248,727 intergenic |
| 220-1 | Dorsal slow muscle fibers, posterior retina: 48 hpf | 9:16,204,267 intronic of si:dkey-114e9 |
| 234 | Midbrain, 24 hpf | * |
| 235-1 | CNS neurons, 24 hpf | 13:7,424,690 intronic of col13a1 |
| 237 | Trunk skeletal muscle fibers, skin, gut (strongest in posterior of larva), and pectoral fins: 48 hpf | * |
| 244-1 | Epidermis: 24 hpf | 1:34,215,682 intergenic |
| 244-3 | Vasculature: 72 hpf | * |
| 244-4 | Pectoral fins, heart valves, dorsal retina: 48 hpf | * |
| 245 | CNS neurons: 24 hpf | |
| 275 | Myogenic lineage: 48 hpf | 16:33,668,562 intergenic |
| 283 | Myogenic lineage: 24 hpf | 13:23,419,906 intronic of si:dkey-103j14.2 |
| 296-1 | Slow skeletal muscles, dermal ionocytes: 48 hpf | * |
| 296-2 | Finfold edges: 48 hpf | 5:28,460,264 intronic of si:ch211-102c2.5 |
| 296-3 | Erythrocytes: 24 hpf | 3:55,961,349 intergenic |
| 302 | Liver, pancreas: 6 dpf | 3:25,723,273 intronic of LOC100149276 (usp43) |
| 318 | Lateral line neuromasts, ENS, jaw, pharynx, tectum: 48 hpf | 25:19,091,711 intronic of met4:∼25,248, 000-25,379,000 intergenic |
| 354-1 | CNS neurons: 24 hpf | * |
| 358 | Iridophores, lateral line glia: 72 hpf Maternal/ubiquitous expression earlier |
* |
Selected lines with specific expression and minimally variegating patterns were chosen for genomic mapping of insertion sites using TAIL PCR. Some mappings resulted in a lack of clear TAIL PCR product (*). The stage in hpf (hours post fertilization) indicates the time point when the GFP expression pattern is fully developed, although GFP expression can start earlier.
Cloning of the mitochondrial-localized construct, MLS-DsRed
The MLS-DsRed construct,55 which contains a mitochondrial targeting sequence (MLS) to target DsRed to the mitochondrial matrix, was digested with EcoRV/XhoI and inserted into vector pT2K, which contains Tol2 transposase recognition elements.32 This construct was designated pT2K-MLSDsRed. For targeted gene expression using the GAL4-UAS system, a fragment containing nine consecutive Gal4 upstream activator sequences was amplified by PCR and cloned upstream of the MLS-DsRed at SacII and SpeI sites. In cells with GAL4 expression, the mitochondrial network is marked by DsRed and visualized with the Texas Red filter set.
Imaging
Confocal imaging of live larvae was done after mounting them in 1% low-melt agarose in glass-bottom dishes on a Zeiss LSM-510 confocal microscope. Larvae from fixed fish (i.e., TDL244, 3, 22, and 74) were stained with the zn-12 (HNK1) antibody57 according to standard techniques,56 and imaged on glass slides with a LSM-510 confocal.
Results
Identification of an efficient minimal promoter and the generation of 63 Gal4 driver lines
In order to identify a new minimal promoter for enhancer detection, we cloned out two viral promoters (i.e., pTAL45 and Sv4046,47) that are commonly fused to cis-regulatory regions to drive reporter proteins in cell culture48 and in vivo.49 Given the low recovery rate of transgenic lines from these constructs (data not shown), we decided to focus our attention on endogenous promoters from teleost fish. Our previous work with the ectodysplasin receptor (edar) gene in medaka and zebrafish prompted us to evaluate the regulation of its expression.50 We observed that a 627 bp minimal promoter from this locus was not capable of independently driving expression of a reporter transgene without additional enhancer elements (data not shown, Materials and Methods). However, random integration of this construct into the genome resulted in a large number of strong and distinct expression patterns with very little nonspecific background. Thus, this minimal promoter provided an enhancer detection construct that could be used efficiently to generate tissue-specific Gal4-driver lines. We named all transgenic lines coming from this screen TDL, for Tuebingen Driver Lines.
We injected this construct along with Tol2 RNA into single-cell stage albino embryos and raised a total of 380 adult zebrafish for GFP screening of F1 progeny at 1–7 days post fertilization (dpf) and again at 30 dpf. We screened in the albino background in order to reduce the obstructing effect of melanin in larval and adult stages. We obtained a total of 63 driver lines representing all germ layers and a diverse set of tissue types (Table 1). The success rate of about 17% enhancer detection from stable integrated lines corresponds with previous Tol2 transposon-based enhancer trap screens that range from 12% to 36% transmission.25,33,36 Many founders had multiple insertions, as demonstrated by the presence of embryos with different expression patterns in the F1 clutch and subsequent segregation of the lines upon outcrossing. There was also considerable variability in the efficiency of germ line transmission in individual F0 fish, as could be seen in the number of individual F1 progeny within a clutch exhibiting a specific expression pattern. In the case of TDL296, only a single individual F1 fish was observed with the given expression pattern, but upon raising and outcrossing that individual, it was possible to maintain three stable transgenic lines from independent insertions (Table 2). In other cases, the majority of the larvae in a particular clutch exhibited a specific expression pattern, and several individuals were raised to maintain the line. However, in most cases, only between 5 and 10 F1 fish per clutch expressed the transgene in a similar way and the other larvae either showed no expression, or in rare cases the segregation of another expression pattern (e.g., TDL302 and TDL45). Besides notochord expression, which we observed in eight driver lines, the hatching gland represented the most frequent expression found in larvae of our screen.
Table 1.
Expression Patterns of Identified Insertion Lines
| Expression pattern | Total identified | Lines |
|---|---|---|
| Ubiquitous | 2 | 79, 354-2 |
| Central nervous system | 14 | 6, 71, 75, 92, 137, 210, 234, 235-1, 256, 276, 318, 323, 350, 354-1 |
| Notochord | 8 | 79, 118-1, 200, 220-2, 235-2, 276, 351, 355 |
| Hatching gland | 6 | 118-2, 220-2, 251, 276, 281, 302 |
| Rhombomeres/branchial arches | 2 | 118-3, 201 |
| Muscle: | ||
| Full myogenic lineage | 4 | 42, 206, 275, 283 |
| Skeletal muscle (trunk/tail) | 10 | 13, 40, 64, 73, 89, 91, 200, 220-1, 237, 296-1 |
| Head muscle | 1 | 276 |
| Muscle pioneers | 1 | 228 |
| Pectoral fins | 4 | 237, 244-4, 283, 297 |
| Gonad | 1 | 353 |
| Cranial neural crest | 4 | 149, 201, 206, 318 |
| Jaw | 8 | 89, 118-3, 201, 187, 215, 253, 318, 350 |
| Skin | 8 | 13, 45-1, 187, 200, 237, 244-1, 296-1, 296-2 |
| Iris | 3 | 95, 220-1, 244-4 |
| Liver/pancreas/kidney/gut | 4 | 45-2, 111, 237, 244-3 |
| Otic placode/ear | 2 | 201, 220-2 |
| Vasculature | 4 | 13, 153, 215, 244-3 |
| Heart | 1 | 244-4 |
| Blood | 1 | 296-3 |
| Cephalic mesoderm | 2 | 205, 302 |
| Lateral line | 3 | 290, 318, 358 |
| Pigment cell | 1 | 358 |
| Fin | 1 | 3, 76 |
| Scales | 1 | 22 |
GFP expression was screened from 1–7 days post fertilization (dpf) and in 30 dpf fish of progeny from founder fish injected with the TDL plasmid. The tissues in which GFP expression was observed, as well as the number of lines (and their identification number) with that expression pattern are shown.
Although we were able to identify a large number of driver lines with consistent and specific expression pattern, variegation and generational silencing were detected in many instances. Previous studies have noted general background expression with other minimal promoters such as the keratin 8 promoter,25 or the heat shock-70kD promoter,35 which could cause problems for downstream applications such as the ectopic expression of a second transgene in a tissue-specific manner. However, we have observed very little to no basal expression using the edar minimal promoter construct. In contrast, the variegation that we did observe was more often spurious GFP expression in a few individual muscle fibers or notochord cells, and the absence of GFP in a number of cells in which the transgene was normally expressed. Because of these two types of variegation (i.e., spurious expression, and patchy expression), only a subset of the most robust and specific lines were kept for subsequent detailed analysis (Table 1).
Tissue-specific expression patterns of selected driver lines
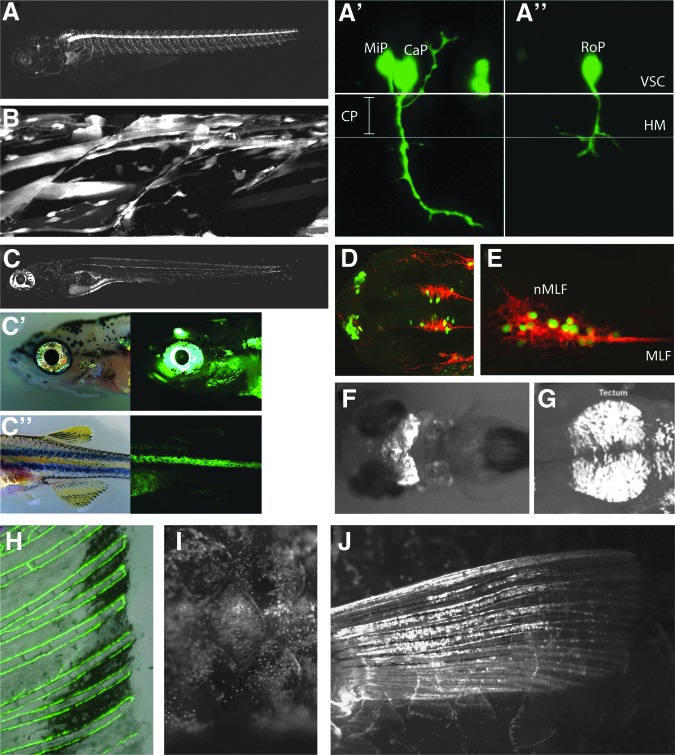
In order to more accurately describe the expression patterns of some of the transgenic lines identified in our enhancer detection screen, we performed confocal microscopy on selected lines at early larval stages and more detailed fluorescence microscopy of adult tissues (Fig. 1). Lower resolution images of a subset of the lines that were identified can be seen in Supplementary Figure S1 (Supplementary Data are available online at www.liebertpub.com/zeb). The TDL driver lines shown in Figure 1 demonstrate the typical lack of low level background expression present in most of the lines generated from the driver construct used in this study. In addition, the specificity of some lines for particular cell types can be readily observed, for example, in the case of TDL6, which labels primary motor neurons (PMN) from 24 hpf onwards (Fig. 1A). Although GFP fluorescence appears to be both strong and specific in TDL6 in all three PMN at 24 hpf (Fig. 1A′, 1A″), variegation occurs in the form of absent expression in some PMN throughout the embryo. In these cases, only two of the three primary motor neurons are labeled (Fig. 1A′), where the middle primary can be seen extending dorsally and the caudal primary migrating ventrally through the choice point past the horizontal myoseptum. In another example, neither the Middle Primary nor the Caudal Primary is labeled, but the Rostral Primary, which is the third PMN, specifically expresses GFP (Fig. 1A″). In spite of this early variegation, by 5 dpf most of the larval PMNs express GFP (Fig. 1A).
FIG. 1.
GFP expression of selected Tuebingen driver lines (TDL). (A, A′, A″) TDL6 marking primary motorneurons in larval zebrafish embryos with (A) shows a larva at 5 dpf and (A′,A″) showing detailed GFP expression at 24 hpf with middle (MiP) and caudal primary motorneuron (CaP) being labeled in A′, and the rostral primary motorneuron (RoP) being labeled in A″. (B) TDL42 labels the myogenic lineage, from myoblasts to differentiated muscle fibers. Shown is a confocal projection of two dorsal myotomes of the midtrunk of a 3 dpf larva. (C, C′, C″) TDL358 gives GFP expression in iridophores, glia of the lateral line system, and the pineal gland. (C) shows a larva at 5 dpf, with iridophores labeling being most pronounced in iridophores of the eyes and in the ventral and yolk sac stripe. Schwann cells wrapped around the lateral line nerve are visible along the horizontal myoseptum. (C′, C″) In juveniles at 30 dpf, GFP expression is most apparent in iridophores of the eyes and the first interstripe. (D) Dorsal view of a 28 hpf TDL354-line embryo combined with immunohistochemistry for the neuronal marker zn-12 (HNK1). The TDL354 line shows expression in primary neurons of the forebrain belonging to the dorso-rostral cluster (drc) and in a subset of nMLF (nucleus of the medial longitudinal fasciculus) neurons located in the ventral midbrain. (E) Lateral view of the ventral midbrain of a 28 hpf TDL235-line embryo co-stained with zn-12. Neurons belonging to the nMLF are GFP positive. (F) TDL234 shows GFP expression in the midbrain at 3 dpf. (G) In TDL318-line 28 hpf embryo, cells are labeled in the dorsal midbrain in the region of the forming tectum. (H) TDL244-3 results in GFP expression in the vasculature, here shown in the adult caudal fin. (I) TDL22 shows GFP expression in the scales of the adult zebrafish. (J) TDL74 labels the pectoral fins of the adult zebrafish.
Confocal maximum intensity projections from TDL42 show GFP in multiple cell types of the myogenic lineage, from myoblasts to differentiated muscle fibers (Fig. 1B). Line TDL358 was identified as being expressed in Schwann cells of the posterior lateral line system and in iridophores in 5 dpf larvae (Fig. 1C). This line shows similar expression in both larval (Fig. 1C) and 30 dpf fish (Fig. 1C′, 1C″). Iridescent iridophores, which are derivatives of the neural crest,51 are reflective pigment cells present on the eye and, in line TDL358, they express GFP around the eye and operculum at 30 dpf (Fig. 1C, 1C′). Iridophores are also present in the zebrafish integumentary stripes (Fig. 1C″), which strongly express GFP in this line (Fig. 1C″). Although this is the only line marking larval pigment cells, two other driver lines (i.e., TDL 290, and TDL318) also labeled Schwann cells of the lateral line system, as well as other tissues of the central nervous system (Table 1).
Expression in CNS tissues was observed in the largest class of lines we identified (Table 1), and some examples of these lines may be seen in Figure 1D–1G (i.e., TDL354, TDL, TDL235, TDL234, and TDL318, respectively). By immunohistochemistry and confocal microscopy, we further characterized two of the driver lines that showed specific expression in cells of the CNS (Fig. 1D, 1E). At 28 hpf, line TDL354 shows expression in primary neurons of the forebrain belonging to the dorso-rostral cluster (DRC) and in a subset of neurons in the nucleus of the medial longitudinal fasciculus (Fig. 1D). A lateral view of the ventral midbrain of a 28 hpf TDL235 embryo co-stained with the neuronal marker zn-12 shows that neurons belonging to the medial longitudinal fasciculus are GFP positive (Fig. 1E). At 3 dpf, TDL234 shows strong midbrain expression (Fig. 1F), and 28 hpf embryos from line TDL318 express GFP in the dorsal midbrain in the region of the forming tectum (Fig. 1G). This line additionally shows expression in dorso-rostral cluster cells, Schwann cells of the lateral line, and a population of cranial neural crest cells migrating anteriorly before 24 hpf (data not shown).
A novel aspect of our screen was that it was not limited to larval expression patterns, but because it was done in the albino background, we were able to identify a GFP signal at much later time points (Table 1). All F1 progeny were screened at 30 dpf to find driver lines with continued expression beyond larval stages or displayed new tissue-specific expression in adults. Four additional lines were identified with specific, albeit sometimes patchy expression (Table 1; Figure 1H–1J). One line (i.e., TDL244-3) had expression in the veination of the fin (Fig. 1H). This line had shown expression in the vasculature during larval stages, and this persisted in the fins of the adult. Line TDL3 showed expression in the forming joint/intraray space of the lepidotrichia of the fins (data not shown). However, there was apparent silencing of this line over several generations and so it was not kept. Line TDL22 showed expression in mosaic patches around adult scales (Fig. 1I). Finally, line TDL74, in addition to showing some early neural crest expression, labeled scleroblasts of the fins at 30 dpf (Fig. 1J).
Mapping of insertion sites identifies 23 enhancer loci
To identify the genomic integration site of selected driver lines, we re-confirmed in F2 fish the expression pattern that had been originally identified in F1 larvae. We outcrossed F1 carriers of the driver lines to albino fish and collected at least 5 individuals from each driver line for mapping by Thermal Asymmetric Interlaced PCR (TAIL-PCR)52 (see Methods). We were not able to obtain a PCR product for 12 of the lines, but we could successfully map the genomic insertion site in 17 (Table 2). Interestingly, eight of the lines mapped to intronic sites, five to intergenic sites, and three to transcribed regions (i.e., two exonic and one in the 5′UTR) (Table 2). In one case (i.e., TDL318), two insertions appeared to segregate with the transgene expression, and it was not clear if the intronic or intergenic insertion were responsible for the expression pattern observed (Table 2). In one other case (i.e., TDL283), two insertions were identified but both were determined to be intronic (Table 2). Only in two lines (i.e., TDL201 and TDL13) were mutant phenotypes observed and this was from an apparent intronic insertion in the gnb1l locus in the case of TDL201, and an intergenic insertion in the promoter region of kans12 for TDL13 (Table 2).
TDL275 can transactivate effector constructs
In order to test the ability of the TDL lines to transactivate genes under control of UAS sequences in different genomic loci, we crossed the transgenic line TDL275, which expresses GFP in progenitors and differentiated cells of the skeletal muscle lineage (Table 1), to a UAS line driving mitochondrial-localized DsRed in response to Gal4. In larvae containing both insertions, GFP and DsRed colocalize in the same cells (Fig. 2). This demonstrates that the lines we generated are able to drive UAS effector constructs in trans and could be used to activate other reporter or effector genes under control of UAS sequences.
FIG. 2.
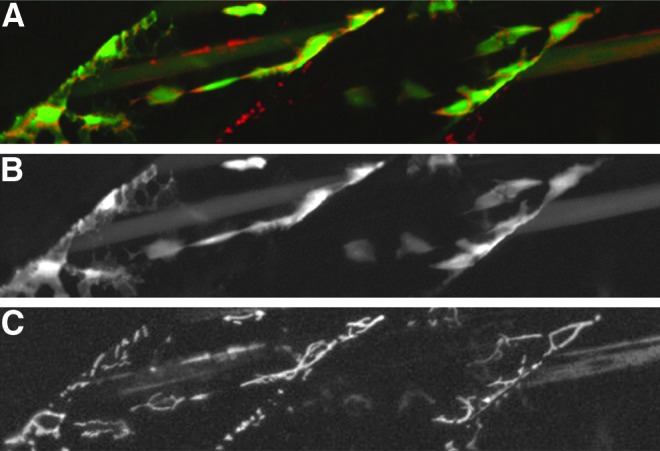
Tuebingen driver line TDL275 transactivates the effector line UAS::MLS:DsRed. (A) TDL275 expresses GFP in cells of the myogenic lineage (green) and transactivates the expression of DsRed targeted to mitochondria (red), merged, 3 dpf. (B) Single channel for GFP expression; (C) single channel for DsRed expression.
Discussion
Enhancer detection through large scale transgenesis screening provides a means to address two fundamental objectives of developmental biology: understanding the cis-regulatory control of metazoan development, and the generation of tools for the tissue-specific manipulation of ontogeny in vivo. By directly demonstrating which genomic regions are competent to drive transgene expression, random genomic integration of a reporter construct complements other primarily computational methods of predicting cis-regulatory modules. It may be difficult if not impossible in some cases to identify the actual cis-regulatory elements responsible for a particular expression pattern, since cis-regulation can occur at very large distances. However, the knowledge that a particular genomic region is able to drive specific expression is achieved without any a priori assumptions of sequence content or conservation.
Taking advantage of the efficiency of these transgenesis methods and the power of the Gal4 transactivation system, several labs have recently conducted screens to identify tissue specific driver lines in zebrafish.11,33–36,53 But because accurate transactivation requires tight temporal-spatial control, it has been important to optimize the minimal promoter that is used in these transgenic constructs. The choice of the minimal promoter may dramatically affect the amount of nonspecific background that is observed, the degree of variegation of the lines, and the tissue bias of the entire screen. It has been observed in one study, for instance, that a c-fos minimal promoter showed the lowest amount of background expression (as compared to two different length heat shock promoters), whereas the E1b promoter had a strong bias towards cranial ganglia expression.33 Likewise, in another study, greater specificity was achieved with a thymidine kinase and GATA2 promoter, than with a minimal heat shock promoter.36
While the objective of identifying cis-regulatory regions of the genome may be accomplished with a simple reporter construct, our intention here was to generate transactivating lines that may be used to drive transgene expression in larval or adult zebrafish tissues. By designing an efficient minimal promoter from the edar locus of medaka and screening the F1 progeny of 380 TDL-injected zebrafish, we identified 63 new transgenic lines displaying GFP expression in multiple tissue types (Table 1). The over-representation of some tissue types (e.g., hatching gland and notochord) in our screen did not preclude the identification of a diverse set of driver lines (Table 1). In fact, the distribution of expression patterns across various organ systems and cell types would suggest that targeted screens using this minimal promoter might be effective for many different tissue of interest to the zebrafish community.
It is worth noting that, in spite of not having screened for recessive phenotypes, we identified two apparent insertional mutants in our screen (Table 2). The apparent inefficiency of recovering mutant phenotypes (i.e., 2 out of 63 confirmed insertions) is likely the result of the screening strategy we employed. That is, having screened for consistent GFP expression in the F1 generation, we only raised and in-crossed F1 fish that had reproducible expression patterns. In all likelihood, most genomic integrations of our construct did not result in observable transgene expression and so would have been discarded. Presumably, some of those insertions would have occurred in open reading frames. However, because of our screen design, it is not possible to evaluate the efficiency of insertional mutagenesis from our construct at this time. Likewise, it will be interesting to further investigate the mechanism whereby this particular promoter minimizes background noise and allows for the identification of a broad range of tissue-specific expression patterns in the zebrafish embryo and adult.
Variegation of transgenic lines may occur in many forms, including generational silencing, spurious ectopic expression, and cell-type specific mosaicism. To varying degrees, these phenomena were all observed in some of our lines (Table 2). However, these are commonly observed properties of transgenic lines using the Gal4 system53 and thus may be general features of the Gal4 and/or Tol2 systems. Thus, the variegation we noted is not likely to be an inherent property of the minimal promoter we used in this study. The fact that variegation can cause problems for some downstream applications has encouraged several groups to improve both the transgenesis constructs used in zebrafish and the Gal4 system itself.34,36,53 As transgenesis and enhancer detection systems are continually improved, it might be advantageous for future screens to choose the minimal promoter, transposon, and Gal4-variant combinations that are best suited for the intended application.
We demonstrate here that the edar promoter from medaka can supplement the growing set of commonly used minimal promoters in the zebrafish community, due to its efficiency, low bias, and lack of background expression. This promoter construct and the new Gal4-driver lines that we describe will be a useful addition to the tool-box of other groups either wishing to conduct their own enhancer detection screens, or requiring driver lines in some of the tissues where we have found expression. All lines that were generated here and are included in Table 2 have been stored permanently as frozen sperm and are available upon request.
Supplementary Material
Acknowledgments
We would like to thank Katrin Henke, Christian Soellner, Alessandro Mongera, and Vanya Krneta-Stankic for some of the images of the driver lines presented here. This work would also not have been possible without the technical assistance of Christian Weiler and Horst Geiger, as well as the support of Christiane Nuesslein-Volhard.
Disclosure Statement
No competing financial interests exist.
References
- 1.O'Kane CJ. Gehring WJ. Detection in situ of genomic regulatory elements in Drosophila. Proc Natl Acad Sci USA. 1987;84:9123–9127. doi: 10.1073/pnas.84.24.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pfeiffer BD. Jenett A. Hammonds AS, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci USA. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayashi S. Ito K. Sado Y, et al. GETDB, a database compiling expression patterns and molecular locations of a collection of Gal4 enhancer traps. Genesis. 2002;34:58–61. doi: 10.1002/gene.10137. [DOI] [PubMed] [Google Scholar]
- 4.Han PL. Meller V. Davis RL. The Drosophila brain revisited by enhancer detection. J Neurobiol. 1996;31:88–102. doi: 10.1002/(SICI)1097-4695(199609)31:1<88::AID-NEU8>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 5.Yang MY. Armstrong JD. Vilinsky I. Strausfeld NJ. Kaiser K. Subdivision of the Drosophila mushroom bodies by enhancer-trap expression patterns. Neuron. 1995;15:45–54. doi: 10.1016/0896-6273(95)90063-2. [DOI] [PubMed] [Google Scholar]
- 6.Waddell S. Quinn WG. What can we teach Drosophila? What can they teach us? Trends Genet. 2001;17:719–726. doi: 10.1016/s0168-9525(01)02526-4. [DOI] [PubMed] [Google Scholar]
- 7.Brand AH. Dormand EL. The GAL4 system as a tool for unravelling the mysteries of the Drosophila nervous system. Curr Opin Neurobiol. 1995;5:572–578. doi: 10.1016/0959-4388(95)80061-1. [DOI] [PubMed] [Google Scholar]
- 8.Elliott DA. Brand AH. The GAL4 system: A versatile system for the expression of genes. Methods Mol Biol. 2008;420:79–95. doi: 10.1007/978-1-59745-583-1_5. [DOI] [PubMed] [Google Scholar]
- 9.Scheer N. Campos-Ortega JA. Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mech Dev. 1999;80:153–158. doi: 10.1016/s0925-4773(98)00209-3. [DOI] [PubMed] [Google Scholar]
- 10.Halpern ME. Rhee J. Goll MG, et al. Gal4/UAS transgenic tools and their application to zebrafish. Zebrafish. 2008;5:97–110. doi: 10.1089/zeb.2008.0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asakawa K. Suster ML. Mizosawa K, et al. Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc Natl Acad Sci USA. 2008;105:1255–1260. doi: 10.1073/pnas.0704963105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davison JM. Akitake CM. Goll MG, et al. Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev Biol. 2007;304:811–824. doi: 10.1016/j.ydbio.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatta K. Tsujii H. Omura T. Cell tracking using a photoconvertible fluorescent protein. Nat Protoc. 2006;1:960–967. doi: 10.1038/nprot.2006.96. [DOI] [PubMed] [Google Scholar]
- 14.Szobota S. Gotostiza P. Del Bene F, et al. Remote control of neuronal activity with a light-gated glutamate receptor. Neuron. 2007;54:535–545. doi: 10.1016/j.neuron.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 15.Bellen HJ. Ten years of enhancer detection: Lessons from the fly. Plant Cell. 1999;11:2271–2281. doi: 10.1105/tpc.11.12.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ckurshumova W. Koizuma K. Chatfield SP, et al. Tissue-specific GAL4 expression patterns as a resource enabling targeted gene expression, cell type-specific transcript profiling and gene function characterization in the Arabidopsis vascular system. Plant Cell Physiol. 2009;50:141–150. doi: 10.1093/pcp/pcn180. [DOI] [PubMed] [Google Scholar]
- 17.Bellen HJ. Wilson C. Gibson G, et al. P-element-mediated enhancer detection: A versatile method to study development in Drosophila. Genes Dev. 1989;3:1288–1300. doi: 10.1101/gad.3.9.1288. [DOI] [PubMed] [Google Scholar]
- 18.Wimmer EA. Innovations: Applications of insect transgenesis. Nat Rev Genet. 2003;4:225–232. doi: 10.1038/nrg1021. [DOI] [PubMed] [Google Scholar]
- 19.Pereira A. A transgenic perspective on plant functional genomics. Transgenic Res. 2000;9:245–260. doi: 10.1023/a:1008967916498. discussion 243. [DOI] [PubMed] [Google Scholar]
- 20.Korn R. Schoor M. Neuhaus H, et al. Enhancer trap integrations in mouse embryonic stem cells give rise to staining patterns in chimaeric embryos with a high frequency and detect endogenous genes. Mech Dev. 1992;39:95–109. doi: 10.1016/0925-4773(92)90029-j. [DOI] [PubMed] [Google Scholar]
- 21.Gossler A. Joyner AL. Rossant J. Skarnes WC. Mouse embryonic stem cells and reporter constructs to detect developmentally regulated genes. Science. 1989;244:463–465. doi: 10.1126/science.2497519. [DOI] [PubMed] [Google Scholar]
- 22.Bayer TA. Campos-Ortega JA. A transgene containing lacZ is expressed in primary sensory neurons in zebrafish. Development. 1992;115:421–426. doi: 10.1242/dev.115.2.421. [DOI] [PubMed] [Google Scholar]
- 23.Ellingsen S. Laplante MA. Konig M, et al. Large-scale enhancer detection in the zebrafish genome. Development. 2005;132:3799–3811. doi: 10.1242/dev.01951. [DOI] [PubMed] [Google Scholar]
- 24.Korzh V. Transposons as tools for enhancer trap screens in vertebrates. Genome Biol. 2007;8:S8. doi: 10.1186/gb-2007-8-s1-s8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parinov S. Kondrichin I. Korzh V. Emelyanov A. Tol2 transposon-mediated enhancer trap to identify developmentally regulated zebrafish genes in vivo. Dev Dyn. 2004;231:449–459. doi: 10.1002/dvdy.20157. [DOI] [PubMed] [Google Scholar]
- 26.Amsterdam A. Burgess S. Golling G, et al. A large-scale insertional mutagenesis screen in zebrafish. Genes Dev. 1999;13:2713–2724. doi: 10.1101/gad.13.20.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Driever W. Solnica-Krezel L. Schier AF, et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- 28.Haffter P. Granato M. Brand M, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- 29.Golling G. Amsterdam A. Sun Z, et al. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat Genet. 2002;31:135–140. doi: 10.1038/ng896. [DOI] [PubMed] [Google Scholar]
- 30.Motzer RJ. Mazumdar M. Bacik J, et al. Survival and prognostic stratification of 670 patients with advanced renal cell carcinoma. J Clin Oncol. 1999;17:2530–2540. doi: 10.1200/JCO.1999.17.8.2530. [DOI] [PubMed] [Google Scholar]
- 31.Kawakami K. Shima A. Kawakami N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc Natl Acad Sci USA. 2000;97:11403–11408. doi: 10.1073/pnas.97.21.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawakami K. takeda H. Kawakami N, et al. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 33.Scott EK. Baier H. The cellular architecture of the larval zebrafish tectum, as revealed by gal4 enhancer trap lines. Front Neural Circuits. 2009;3:13. doi: 10.3389/neuro.04.013.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Distel M. Wullimann MF. Koster RW. Optimized Gal4 genetics for permanent gene expression mapping in zebrafish. Proc Natl Acad Sci USA. 2009;106:13365–13370. doi: 10.1073/pnas.0903060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott EK. Mason L. Arrengerb AB, et al. Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nat Methods. 2007;4:323–326. doi: 10.1038/nmeth1033. [DOI] [PubMed] [Google Scholar]
- 36.Ogura E. Okuda Y. Kondoh H. Kamachi Y. Adaptation of GAL4 activators for GAL4 enhancer trapping in zebrafish. Dev Dyn. 2009;238:641–655. doi: 10.1002/dvdy.21863. [DOI] [PubMed] [Google Scholar]
- 37.Grabher C. Henrich T. Sasado T, et al. Transposon-mediated enhancer trapping in medaka. Gene. 2003;322:57–66. doi: 10.1016/j.gene.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 38.Balciunas D. Davidson AE. Sivasubbu S, et al. Enhancer trapping in zebrafish using the Sleeping Beauty transposon. BMC Genom. 2004;5:62. doi: 10.1186/1471-2164-5-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawakami K. Transposon tools and methods in zebrafish. Dev Dyn. 2005;234:244–254. doi: 10.1002/dvdy.20516. [DOI] [PubMed] [Google Scholar]
- 40.Ivics Z. Izsvak Z. Minter A. Hackett PB. Identification of functional domains and evolution of Tc1-like transposable elements. Proc Natl Acad Sci USA. 1996;93:5008–5013. doi: 10.1073/pnas.93.10.5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Izsvak Z. Ivics Z. Hackett PB. Characterization of a Tc1-like transposable element in zebrafish (Danio rerio) Mol Gen Genet. 1995;247:312–322. doi: 10.1007/BF00293199. [DOI] [PubMed] [Google Scholar]
- 42.Raz E. van Luenen HG. Schaerringer B. Plasterk RH. Driever W. Transposition of the nematode Caenorhabditis elegans Tc3 element in the zebrafish Danio rerio. Curr Biol. 1998;8:82–88. doi: 10.1016/s0960-9822(98)70038-7. [DOI] [PubMed] [Google Scholar]
- 43.Emelyanov A. Gao Y. Naqvi NI. Parinov S. Trans-kingdom transposition of the maize dissociation element. Genetics. 2006;174:1095–1104. doi: 10.1534/genetics.106.061184. , [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawakami K. Koga A. Hori H. Shima A. Excision of the tol2 transposable element of the medaka fish, Oryzias latipes, in zebrafish, Danio rerio. Gene. 1998;225:17–22. doi: 10.1016/s0378-1119(98)00537-x. [DOI] [PubMed] [Google Scholar]
- 45.Jones KA. Yamamoto KR. Tjian R. Two distinct transcription factors bind to the HSV thymidine kinase promoter in vitro. Cell. 1985;42:559–572. doi: 10.1016/0092-8674(85)90113-8. [DOI] [PubMed] [Google Scholar]
- 46.Jones RH. Moreno S. Nurse P. Jones NC. Expression of the SV40 promoter in fission yeast: Identification and characterization of an AP-1-like factor. Cell. 1988;53:659–667. doi: 10.1016/0092-8674(88)90581-8. [DOI] [PubMed] [Google Scholar]
- 47.Fromm M. Berg P. Deletion mapping of DNA regions required for SV40 early region promoter function in vivo. J Mol Appl Genet. 1982;1:457–481. [PubMed] [Google Scholar]
- 48.Cook WJ. Lin SM. DeLuca NA. Coen DM. Initiator elements and regulated expression of the herpes simplex virus thymidine kinase gene. J Virol. 1995;69:7291–7294. doi: 10.1128/jvi.69.11.7291-7294.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stuart GW. Vielkind JR. McMurray JV. Westerfield M. Stable lines of transgenic zebrafish exhibit reproducible patterns of transgene expression. Development. 1990;109:577–584. doi: 10.1242/dev.109.3.577. [DOI] [PubMed] [Google Scholar]
- 50.Harris MP. Rohner N. Schqarz H, et al. Zebrafish eda and edar mutants reveal conserved and ancestral roles of ectodysplasin signaling in vertebrates. PLoS Genet. 2008;4:e1000206. doi: 10.1371/journal.pgen.1000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lopes SS. Yang X. Muller J, et al. Leukocyte tyrosine kinase functions in pigment cell development. PLoS Genet. 2008;4:e1000026. doi: 10.1371/journal.pgen.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu YG. Whittier RF. Thermal asymmetric interlaced PCR: Automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics. 1995;25:674–681. doi: 10.1016/0888-7543(95)80010-j. [DOI] [PubMed] [Google Scholar]
- 53.Asakawa K. Kawakami K. Targeted gene expression by the Gal4-UAS system in zebrafish. Dev Growth Differ. 2008;50:391–399. doi: 10.1111/j.1440-169X.2008.01044.x. [DOI] [PubMed] [Google Scholar]
- 54.Koster RW. Fraser SE. Tracing transgene expression in living zebrafish embryos. Dev Biol. 2001;233:329–346. doi: 10.1006/dbio.2001.0242. [DOI] [PubMed] [Google Scholar]
- 55.Griparic L. Kanazawa T. van der Bliek AM. Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage. J Cell Biol. 2007;178:757–764. doi: 10.1083/jcb.200704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nüsslein-Volhard C. Dahm R. Zebrafish: A Practical Approach. 1st. Oxford University Press; 2002. [Google Scholar]
- 57.Trevarrow B. Marks DL. Kimmel CB. Organization of hindbrain segments in the zebrafish embryo. Neuron. 1990;4:669–679. doi: 10.1016/0896-6273(90)90194-k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.