Abstract
The sixth step in the lipid A biosynthetic pathway involves phosphorylation of the tetraacyldisaccharide-1-phosphate (DSMP) intermediate by the cytosol-facing inner membrane kinase LpxK, a member of the P-loop containing nucleoside triphosphate (NTP) hydrolase superfamily. We report the kinetic characterization of LpxK from Aquifex aeolicus and the crystal structures of LpxK in complex with ATP in a pre-catalytic binding state, the ATP analog AMP-PCP in the closed catalytically competent conformation, and a chloride anion revealing an inhibitory conformation of the nucleotide-binding P-loop. We demonstrate that LpxK activity in vitro requires the presence of a detergent micelle and formation of a ternary LpxK-ATP/Mg2+-DSMP complex. Using steady-state kinetics, we have identified crucial active site residues, leading to the proposal that the interaction of D99 with H261 acts to increase the pKa of the imidazole moiety, which in turn serves as the catalytic base to deprotonate the 4'-hydroxyl of the DSMP substrate. The fact that an analogous mechanism has not yet been observed for other P-loop kinases highlights LpxK as a distinct member of the P-loop kinase family, a notion that is also reflected through its localization at the membrane, lipid substrate, and overall structure.
Gram-negative bacteria differentiate themselves from their Gram-positive counterparts by the presence of an outer membrane, the outer leaflet of which is composed of the lipid-anchored complex carbohydrate known as lipopolysaccharide (LPS). The lipid portion of LPS is an acylated glucosamine disaccharide known as lipid A, which even without the presence of the immunogenic O-antigen can elicit a mammalian inflammatory response through activation of the macrophage Toll-like receptor 4 and myeloid differentiation protein 2 complex (TLR4-MD2) (1, 2). Nine enzymatic steps make up the constitutive pathway of lipid A biosynthesis in Escherichia coli, and because lipid A is essential for the viability of the vast majority of Gram-negative bacteria, the pathway stands as an attractive target for the development of novel antimicrobials (3).
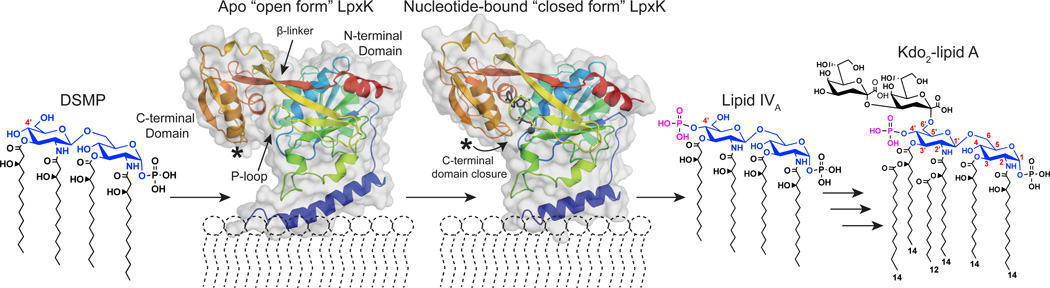
The sixth step of the lipid A biosynthetic pathway is the phosphorylation of the 4'-hydroxyl group of tetraacyldisaccharide-1-phosphate (DSMP) as catalyzed by LpxK, a divergent member of the P-loop containing nucleoside triphosphate (NTP) hydrolase superfamily (Pfam02606, CL0023), which resides on the cytosolic face of the inner membrane (Scheme 1) (4–7). The active site Walker A (P-loop) and Walker B (Mg2+-binding) motifs are common to all P-loop kinase family members (8). Recent structural characterization of LpxK from Aquifex aeolicus revealed a two α/β/α domain topology in which the second α/β/α domain, a substructure unique to LpxK, was implicated in nucleotide binding through a hinge motion about its base (Scheme 1) (9). Further analysis led to the conclusion that the hydrophobic lower face of the N-terminal helix may be responsible for membrane association, assisted by charge-charge interactions of surrounding basic residues with the anionic phospholipids of the membrane. Despite some differences regarding the presence of N- versus O-linked acyl chains and chain lengths of Aquifex DSMP (10, 11), A. aeolicus LpxK can readily phosphorylate the E. coli-derived DSMP substrate (9).
Scheme 1.
The reaction catalyzed by LpxK in Kdo2-lipid A biosynthesis. LpxK is responsible for the phosphorylation (purple) of the 4′-hydroxyl of tetraacyldisaccharide-1-phosphate (DSMP). Positions on the glucosamine backbone (blue) of DSMP are labeled (red). The crystal structures of apo A. aeolicus LpxK (PDB: 4EHX) and ADP/Mg2+-bound LpxK (PDB: 4EHY) in their predicted membrane orientations are included colored from N- to C-terminus (blue to red). ADP is shown as black sticks and Mg2+ is shown as a black sphere in the “closed” form. The N-terminal helix (blue cartoon) is implicated in membrane binding through a hydrophobic lower face assisted by surrounding basic residues of the N-terminal core domain. In its “closed” catalytically competent form, the C-terminal domain of LpxK undergoes a hinge motion to close around the nucleotide substrate upon binding (indicated with curved arrow).
A handful of P-loop kinases have been kinetically characterized, however, determination of a conserved catalytic mechanism has remained elusive, possibly due to the fact that P-loop kinases have evolved to act upon a highly diverse array of substrates (4, 12). Structural and kinetic characterization of several P-loop kinases have suggested direct nucleophilic attack by the acceptor substrate, either assisted or unassisted by an aspartate/glutamate catalytic base of the Walker B motif (12–14). A distribution of positively charged residues in the active site, especially the conserved P-loop lysine, may assist in shielding the negative charges generated during phosphate transfer. For a few kinases and sulfotransferases outside of the P-loop kinase family, a histidine residue has been implicated in acting as a general base catalyst (15, 16), presenting another mechanism by which base-mediated phosphate transfer may occur.
We present herein the kinetic characterization of purified LpxK and show that the enzyme catalyzes phosphate transfer through formation of a ternary complex at the membrane interface. Determination of steady-state kinetic parameters for a variety of active site point mutants and additional crystal structures of AMP-PCP-bound LpxK, ATP-bound LpxK in the open conformation (Scheme 1), and LpxK with an alternative P-loop conformation leads to the proposal of an overall catalytic mechanism. The detailed kinetic and structural characterization of LpxK reported here bestows further insight into the action of this unique P-loop kinase and provides additional groundwork for the continued development of LpxK into a viable antibiotic target.
Experimental Procedures
Cloning and expression of LpxK constructs
All primers, plasmids, and strains referenced can be found in Tables S1 and S2. Wild type A. aeolicus LpxK was generated by growth of C41(DE3) cultures expressing the construct pRPE7 and purified as previously described (9, 17). Purified LpxK was stored in a buffer containing ~0.5 % (w/v) dodecyl maltoside (DDM) (Anatrace, Maumee, OH), 750 mM NaCl, 20 % (v/v) glycerol, and 50 mM HEPES pH 8.0. Quikchange mutagenesis (Stratagene, La Jolla, CA) was employed to generate point mutants S49A, Y74A, D99A, D99N, D99E, E100A, E100Q, E100D, D138N, D139N, D260A, and H261A using the primer pairs listed in Table S1 and resulting in the plasmids listed in Table S2. All constructs were validated by sequencing with primers prT7F and prT7R. Plasmids containing alanine point mutants for K51, T52, S53, D138, and D139 had been constructed in previous work (9). To generate partially purified LpxK point mutants, the plasmids were transformed into C41(DE3), expressed, and solubilized from membranes as previously described (9, 18).
Assay and kinetic characterization of LpxK activity
The lipid assay components 32P-radiolabeled DSMP and non-radioactive DSMP were prepared as previously described (9). The standard assay conditions included 50 µM 32P-DSMP (10,000 cpm/nmol), 5 mM ATP, 5 mM MgCl2, 50 mM Tris pH 8.5, 0.5 % (w/v) Triton X-100 (Thermo Scientific, Rockford, IL), 1 mg/mL BSA (Sigma-Aldrich, St. Louis, MO), 0.1 M NaCl, and A. aeolicus LpxK at 30 °C (9). Typically, LpxK was first diluted in 0.5 % (w/v) Triton X-100, 0.5 M NaCl, and 50 mM Tris buffer before being diluted 5-fold (4 µL into 16 µL) into the assay to begin the reaction. 4 µL aliquots from the reaction mixtures were spotted onto 10 cm tall thin-layer chromatography (TLC) plates (EMD Chemicals, Gibbstown, NJ), developed in a chloroform/methanol/water/acetate (25:15:4:2) (v:v:v:v) tank system, exposed to 35 cm × 43 cm Molecular Dynamics PhosphorImager screens, and scanned on a Storm 840 phosphorimager (GE Healthcare, Waukesha, WI).
In order to assess the pH dependence for wild-type enzyme, the D99A point mutant, and the H261A point mutant, LpxK was assayed in the presence of a three-component buffer system consisting of 100 mM sodium acetate, 50 mM bis-Tris, and 50 mM Tris of pH 5 through 9.5 replacing the usual Tris buffer. The enzyme concentration in the assay was varied (between 0.3 and 3 nM for the wild type enzyme) to keep conversion within the linear range. Enzyme, 100-fold concentrated with respect to the final assay condition, was first diluted 20-fold into 0.5 % (w/v) Triton X-100, 0.5 M NaCl, and 50 mM pH buffer, and then 5-fold into the assay. The resulting curve was fitted to equation 1 in order to assign pKa and pKb using Kaleidegraph (Synergy Software, Reading, PA) as previously described (19). In equation 1, v is the rate of the reaction, C is the pH-independent rate, [H] is the hydrogen ion concentration, and Ka and Kb are the ionization constants of the acid and base species of LpxK.
| (Eq. 1) |
In order to determine the detergent dependence of LpxK activity, the assay was carried out in the standard conditions with Triton X-100 varied from 0.05 to 62 mM. We were unable to assay LpxK at lower detergent concentrations because of the necessity of detergent to solubilize the DSMP substrate in its stock solution. The enzyme concentration was varied between 0.5 and 10 nM to keep conversion within the linear range.
To assess which metal cations support LpxK activity, enzyme concentrated at 270 nM was incubated in the standard enzyme dilution buffer in 1 mM EDTA for 1 hr at 4 °C. This solution was diluted 20-fold into dilution buffer containing 10 mM CaCl2, CoCl2, CuCl2, FeCl2, MgCl2, MnCl2, NiCl2, or ZnCl2 and incubated for 1 hr at 4 °C. This enzyme solution was next diluted 5-fold into the standard assay without MgCl2. Conversion of DSMP to lipid IVA was determined by TLC after 8 minutes. To determine whether excess Mg2+ inhibited LpxK activity, the enzyme was assayed at between 0.3 and 8 nM in the standard assay condition with 5 mM ATP and increasing amounts of MgCl2 ranging from 0.015 to 128 mM.
To examine whether excess salt inhibited LpxK activity, the standard assay was performed in the presence of 0 to 800 mM NaCl or KBr. Enzyme first diluted to 5 nM in 0.5 % (w/v) Triton X-100 and 50 mM Tris pH 8.5 buffer was immediately diluted 5-fold into the assay and specific activity assessed.
Determination of apparent steady-state kinetic parameters
The apparent kinetic parameters for the ATP/Mg2+ complex were determined by performing the standard assay at a fixed concentration of 50 µM DSMP while varying the amount of ATP and MgCl2 from 0.3 to 10 mM. The LpxK concentration in these assays varied between 0.3 and 2 nM. Conversely, to determine the apparent kinetic parameters for DSMP, the ATP/MgCl2 concentration was fixed at 5 mM while the lipid concentration varied from 1.56 to 100 µM. Both curves were fit to the Michaelis-Menten equation using KadeidaGraph (Synergy Software, Reading, PA) in order to determine apparent KM and kcat with respect to each substrate. For determination of apparent kinetic parameters of the solubilized point mutants, ATP/MgCl2 concentration was varied from 0.3 to 15 mM at a fixed 50 µM DSMP concentration and again fit to the Michaelis-Menten equation. Enzyme concentration was varied to capture the linear range of activity for each mutant.
To analyze LpxK activity through bi-substrate kinetics, the enzyme was assayed with ATP/MgCl2 concentration varied from 0.3 to 10 mM at four fixed concentrations of DSMP (2.5, 5, 15, and 50 µM). The data was fit to equations describing either a ping-pong (Eq. 2) or sequential (Eq. 3) mechanism as has been described previously, using PRISM (GraphPad Software, La Jolla, CA) (20). A and B represent the concentration of the two substrates, KMa is the KM for substrate A, KMb is the KM for substrate B, Kia is the dissociation constant for substrate A, v is the reaction velocity, and Vm is the maximal velocity.
| (Eq. 2) |
| (Eq. 3) |
Crystallization and structure determination of LpxK in complex with AMP-PCP, ATP, and with an alternate P-loop conformation
Crystals were grown in 24-well trays (Hampton Research, Aliso Viejo, CA) using the sitting drop vapor diffusion method. All conditions included 2-methyl-2,4-pentanediol (MPD) and HEPES (Qiagen, Valencia, CA), with a 700 µL well volume and 10 µL drop volume incubated at 20 °C. All crystals were harvested after growing for one month and were immediately flash frozen in liquid nitrogen after being looped. Following collection, all data were reduced and scaled using HKL-2000 (21) and the structure solved by molecular replacement using PHASER within the PHENIX software suite (22). The model was manually rebuilt in COOT (23) between rounds of iterative maximum-likelihood refinement in PHENIX, which included TLS options. Using an electron-density acceptance criterion of d ≥ 3 σ (d) in the Fo – Fc difference electron-density map, ligands (AMP-PCP, ATP, chloride, HEPES, MPD, or Glycerol) were added to the respective models. The overall structures were validated using MOLPROBITY (24) and imaged in PyMOL (25). Data collection and refinement statistics can be found in Table 1.
Table 1.
Data Collection and Refinement Statistics
| AMP-PCP LpxK | ATP LpxK | Compact P-loop LpxK |
|
|---|---|---|---|
| Data Collection | |||
| Space group | P212121 | P212121 | P212121 |
| Unit Cell a,b,c (Å) | 66.1, 75.4, 104.6 | 62.7, 68.1, 105.9 | 62.1, 68.5, 107.6 |
| Wavlength (Å) | 1.0 | 1.0 | 1.0 |
| Resolution (Å) | 50.0–2.1 (2.14–2.10)* | 50.0–2.2 (2.24–2.20) | 50.0–2.2 (2.24–2.20) |
| Rmerge | 0.059 (0.391) | 0.073 (0.420) | 0.046 (0.451) |
| I/σ | 41.3 (2.8) | 16.5 (2.3) | 37.7 (2.6) |
| Completeness (%) | 92.3 (62.8) | 97.1 (98.4) | 97.4 (83.4) |
| Redundancy | 4.4 (3.1) | 4.0 (3.6) | 5.5 (4.2) |
| Reflections/unique | 129,472/29,108 | 92,699/23,000 | 128,849/23,573 |
| Refinement | |||
| Rwork/Rfree (%) | 16.7/19.0 | 16.2/21.0 | 19.3/22.9 |
| Number of atoms (Avg. B-factor, Å2) | |||
| Protein | 2,527 (50.4) | 2,585 (30.3) | 2,555 (71.7) |
| Water | 55 (41.7) | 139 (28.6) | 73 (53.6) |
| Glycerol, MPD, Hepes | 18 (89.4) | 29 (44.3) | 39 (96.3) |
| AMP-PCP | 31 (32.1) | - | - |
| ATP | - | 31 (20.6) | - |
| Chloride | - | - | 1 (60.5) |
| Ramachandran Plot | |||
| Favored/allowed/outlier (%) | 98.0/2.0/0.0 | 97.8/2.2/0.0 | 97.7/2.3/0.0 |
| Rms deviations | |||
| Bond length (Å) | 0.014 | 0.01 | 0.01 |
| Bond angles (°) | 1.48 | 1.16 | 1.15 |
Values parenthesis indicate highest resolution shell
Rod-shaped crystals (0.2 mm × 0.05 mm × 0.05 mm) resulted when LpxK was incubated with AMP-PCP (Sigma-Aldrich, St. Louis, MO). The drop contained four parts of a reservoir solution consisting of 50 % (v/v) MPD and 0.1 M HEPES pH 7.5, and one part protein solution containing 13 mg/mL LpxK, 4.3 mM AMP-PCP, 1 mM EDTA, 0.5 % (w/v) DDM, 540 mM NaCl, 14 % (v/v) glycerol, and 35 mM HEPES pH 8.0. Similar crystals resulted after a round of microseeding where 1 µL of a seed stock (one crystal dissolved in 0.5 mL of reservoir solution) was added to the drop. The harvested crystal diffracted to 2.1 Å and belonged to the P212121 space group (a = 66.1 Å, b = 75.4 Å, c = 104.6 Å). Data were collected at a single wavelength (1.0 Å) at the Southeast Regional Collaborative Access Team (SERCAT) 22-ID line at the Advanced Photon Source (APS, Argonne National Laboratory). The structure was solved by molecular replacement using the previously reported ADP/Mg2+ LpxK structure (PDB: 4EHY) as the search model with all ligands removed, and AMP-PCP was subsequently added to the model.
Rod-shaped crystals (0.25 mm × 0.05 mm × 0.05 mm) resulted when LpxK was incubated with ATP. The drop contained seventeen parts of a reservoir solution consisting of 60 % (v/v) MPD and 0.1 M HEPES pH 7.5, and three parts protein solution containing 7.4 mg/mL LpxK, 10 mM ATP, 1 mM EDTA, 0.35 % (w/v) DDM, 700 mM NaCl, 18.5 % (v/v) glycerol, and 45 mM HEPES pH 8.0. A harvested crystal diffracted to 2.2 Å and data were collected at a single wavelength (1.0 Å) at the SERCAT BM-line with a P212121 space group (a = 62.7 Å, b = 68.1 Å, c = 105.9 Å). The structure was solved using the previously reported apo LpxK structure (PDB: 4EHX) as the molecular replacement search model and ATP was subsequently added to the structure.
Boxy rod-shaped crystals (0.15 mm × 0.1 mm × 0.1 mm) resulted from a drop which contained three parts of a reservoir solution consisting of 40 % (v/v) MPD and 0.1 M HEPES pH 7.5, and one part protein solution containing 8.3 mg/mL LpxK, 4 mM methyl 2-acetamido-2-deoxy-β-D-glucopyranoside (26), 0.35 % (w/v) DDM, 625 mM NaCl, 17 % (v/v) glycerol, and 45 mM HEPES pH 8.0. A harvested crystal diffracted to 2.2 Å and data were collected at a single wavelength (1.0 Å) at the SERCAT BM-line with a P212121 space group (a = 62.1 Å, b = 68.5 Å, c = 107.6 Å). The structure was solved using the apo LpxK structure (PDB: 4EHX) as the molecular replacement search model and spherical active site density refined well as a chloride ion.
Results
pH, detergent, and metal dependence of LpxK activity
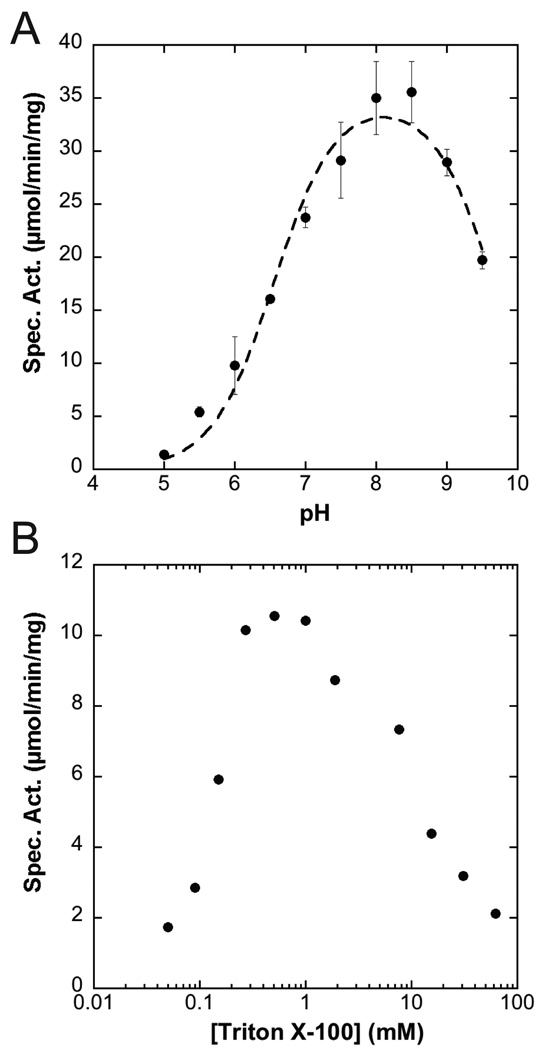
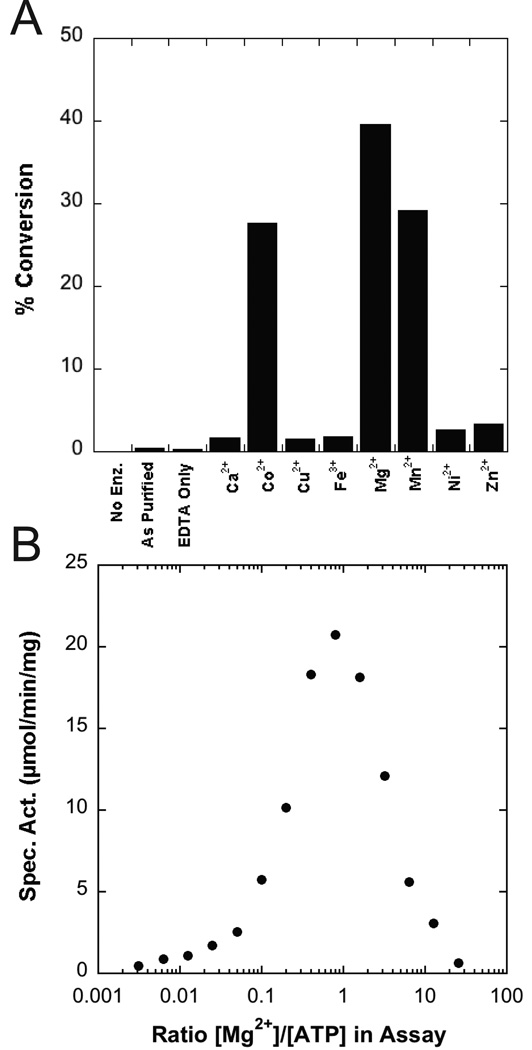
The A. aeolicus LpxK assay was performed in triplicate in the presence of a triple-buffer system at various pH values in order to determine the effect of pH on overall activity. LpxK was found to tolerate a broad pH optimum with a pKa of 6.6 ± 0.1 and pKb of 9.7 ± 0.1 (Fig. 1A). Inactivation of the enzyme at low pH was reversible as this enzyme could be re-diluted into a buffer with a pH of 8 to restore full activity (data not shown). LpxK was also assayed in the presence of increasing amounts of detergent (0.05 to 62 mM) while keeping the DSMP concentration fixed at 50 µM in order to determine the effects of increasing micelle surface area on activity (27). LpxK achieved maximal activity as the amount of Triton X-100 in the assay overcame the critical micelle concentration (~0.2 mM), followed by decay in activity as the micelle surface area increased with the amount of detergent (Fig 1B). In order to assess the metal dependence of kinase activity, LpxK was incubated with EDTA, then diluted into buffer containing select cations, and further diluted into buffer containing the rest of the reaction components. Activity was stimulated by the presence of Mg2+, Co2+, and Mn2+ while significantly less phosphoryl transfer was observed when LpxK was incubated with Ca2+, Ni2+, Cu2+, Zn2+, or Fe3+ (Fig. 2A). Almost no activity was observed for enzyme as purified without the addition of any metals. In order to determine whether excess Mg2+ in the assay condition was inhibitory to activity, LpxK was assayed in the standard assay condition in the presence of 0.015 to 128 mM of the cation. Optimal activity was observed at an equimolar ratio of ATP to Mg2+, with a decrease in activity at higher amounts of the divalent cation (Fig. 2B).
Fig. 1.
pH and detergent dependence of LpxK activity. (A) A. aeolicus LpxK was assayed in triplicate in a triple-buffer system (100 mM sodium acetate, 50 mM Tris, 50 mM bis-Tris) at the indicated pH values and specific activity assessed. A pKa of 6.6 ± 0.1 and pKb of 9.7 ± 0.1 were calculated based on the data. (B) The enzyme was also assayed in the standard assay conditions in the presence 0.05 to 62 mM Triton X-100 and activity assessed. Both the detergent requirement and reduced activity with increased micellular surface area are indicative that LpxK-catalyzed phosphorylation of DSMP occurs at the micelle surface.
Fig. 2.
Metal dependence of LpxK activity. (A) Purified LpxK, which was first incubated with EDTA, was then diluted into assay buffer containing 2 mM ATP and the indicated cation, and reaction was quenched by spotting on a TLC plate after 8 minutes. (B) The standard assay was performed in the presence of 5 mM ATP, and Mg2+ ranging from 0.015 to 128 mM. The cation is necessary for activity, but becomes inhibitory at high concentrations with respect to ATP.
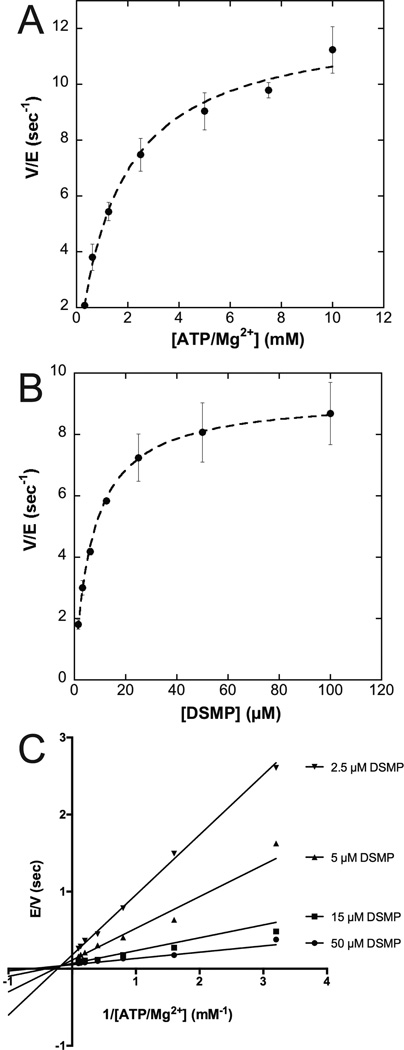
Apparent steady-state kinetic parameters for LpxK activity support the formation of a ternary LpxK-ATP/Mg2+-DSMP complex
Due to the surface dilution effects of the mixed micelle assay, all kinetic parameters are deemed “apparent” (Fig. 1B) (27). Velocity of the A. aeolicus LpxK phosphoryl transfer reaction was assessed for various concentrations of ATP and Mg2+ held in a 1:1 ratio, at a fixed concentration of DSMP. The KM for the nucleotide/cation complex was determined to be 1.6 ± 0.2 mM with a kcat of 12.3 ± 0.4 sec−1 (Fig. 3A). The kinetic parameters for the lipid substrate DSMP were also determined by varying the lipid concentration in the assay at saturating amounts of ATP/Mg2+, yielding a KM of 7.0 ± 0.3 µM and a kcat of 9.2 ± 0.1 sec−1 (Fig. 3B).
Fig. 3.
Apparent kinetic parameters and bi-substrate kinetic analysis of LpxK. (A) A. aeolicus LpxK was assayed at a fixed 50 µM concentration of DSMP and varied amounts of ATP/Mg2+ ranging from 0.3 to 10 mM. The apparent KM for ATP/Mg2+ was determined to be 1.6 ± 0.2 mM with a kcat of 12.3 ± 0.4 sec−1. (B) The concentration of ATP and Mg2+ in the assay was fixed at 5 mM while the concentration of DSMP was varied between 1.56 and 100 µM. The apparent KM for DSMP was determined to be 7.0 ± 0.3 µM with a kcat of 9.2 ± 0.1 sec−1. (C) In this series of assays, the concentration of ATP/Mg2+ was varied between 0.3 and 10 mM for four fixed concentrations of the DSMP substrate (2.5, 5, 15 and 50 µM). The data was fit by a non-linear least-squares method to a sequential mechanism.
The LpxK reaction was further analyzed via bi-substrate kinetics to determine whether the reaction proceeds through a ping-pong type mechanism (transient existence of a phospho-enzyme intermediate) or a sequential mechanism (formation of a ternary complex) (20). Lineweaver-Burk plots constructed for varied ATP/MgCl2 concentrations (held at a 1:1 ratio) at four fixed concentrations of the lipid substrate were overlaid to reveal intersecting lines, indicating the necessity of the formation of a ternary complex for catalysis (Fig. 3C). The global fit of the data for a sequential mechanism was favored over a ping-pong type mechanism with a Vm of 18.0 ± 0.7 sec−1, a KMa of 1.0 ± 0.2 mM, a KMb of 7 ± 1 µM, and a Kia of 3.1 ± 0.8 mM, where substrate A is ATP/Mg2+ and substrate B is DSMP.
Crystal structure of AMP-PCP bound LpxK
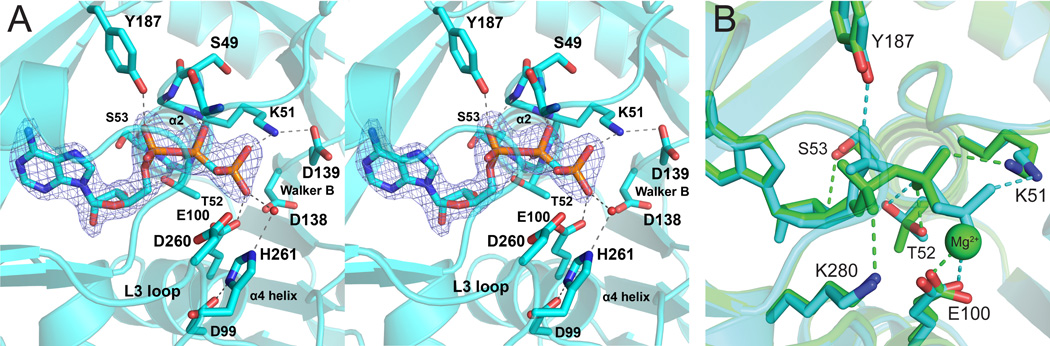
In order to identify the pre-catalytic position of the ATP γ-phosphate in LpxK, co-crystals in complex with the non-hydrolizable analog AMP-PCP were generated and the structure solved via molecular replacement (Table 1). The AMP-PCP containing structure shows the analog bound to the enzyme in the closed form (Scheme 1), with the β-phosphate sitting at the base of helix α2 (Fig. 4A). The AMP-PCP structure overlays well with the structure previously determined for post-catalytic ADP/Mg2+-bound LpxK structure with three differences (9) (Fig. 4B). First, the position of the α-phosphate is shifted slightly between the two structures, the ADP/Mg2+ α-phosphate favoring an interaction with the side chain of K280 while the AMP-PCP favors hydrogen bonding to Y187. Secondly, the β-phosphate of AMP-PCP resides in a position occupied by the cation in the ADP/Mg2+ structure, participating in hydrogen bonding interactions with E100 of the α4 helix and K51 of the P-loop motif. Third, T52 forms a hydrogen bond with a ribose hydroxyl, which is bound to S53 in the ADP/Mg2+ structure. In both the ADP/Mg2+ and AMP-PCP crystal structures, the side chain of D99 is bonded to H261 of the L3 loop, reminiscent of the charge relay system of serine proteases (28) (Fig. 4A). The γ-phosphate of AMP-PCP is well-positioned for transfer as it is directly adjacent to the putative DSMP-binding pocket on the underside of the N-terminal domain.
Fig. 4.
Active site of AMP-PCP bound to LpxK. (A) Active site stereo view reveals AMP-PCP bound in the “closed” enzyme conformation between the C-terminal domain and β–linker. Waters (red spheres) and putative hydrogen bonds (dashed lines) are indicated. Simulated annealing omit electron density for AMP-PCP (mesh) was calculated with coefficients Fo – Fc, contoured at 4 σ. (B) Overlay of AMP-PCP (cyan) and ADP-Mg2+ (PDB: 4EHY) (green) LpxK structures. Side chains and hydrogen bonds (dashed lines) are colored accordingly. The Mg2+ ion of the ADP/Mg2+ structure is bound at the same site as the γ–phosphate of AMP-PCP.
Kinetic parameter perturbations of LpxK active site point mutants
To gain insight into the LpxK catalytic mechanism, point mutants of various conserved active site residues informed by LpxK structures with bound nucleotide were generated and partially purified through solubilization from membranes of the expression strain. All point mutants expressed to similar levels and behaved as wild type throughout the partial purification as judged by SDS-PAGE (data not shown). Apparent kinetic parameters for ATP/Mg2+ were determined for the wild type enzyme along with each point mutant in order to assess whether these residues have a catalytic role and/or assist in binding the nucleotide/cation substrate. The results are displayed in Table 2.
Table 2.
Kinetic parameters of LpxK point mutants with respect to ATP/Mg2+
| LpxK Mutant | App. KM (mM) | Fold KM increase |
App. kcat (sec−1) | Fold kcat decrease |
|---|---|---|---|---|
| WT | 1.2 ± 0.2 | 1.0 | 3.9 ± 0.2 | 1.0 |
| S49A | 2.7 ± 0.3 | 2.3 | 3.9 ± 0.1 | 1.0 |
| K51A | 1.5 ± 0.4 | 1.3 | (1.3 ± 0.1)×10−3 | 3000 |
| T52A | 2.2 ± 0.3 | 1.8 | (1.3 ± 0.1)×10−3 | 3000 |
| S53A | 6 ± 1 | 5 | (4.7 ± 0.6)×10−1 | 8.3 |
| Y74A | 1.7 ± 0.3 | 1.4 | (2.2 ± 0.1)×10−2 | 180 |
| D99A | 2.8 ± 0.5 | 2.3 | (1.5 ± 0.1)×10−3 | 2600 |
| D99N | 1.1 ± 0.2 | 1.0 | (3.5 ± 0.2)×10−4 | 11000 |
| D99E | 2.3 ± 0.2 | 1.9 | (5.0 ± 0.2)×10−2 | 78 |
| E100A | 1.9 ± 0.5 | 1.6 | (2.9 ± 0.2)×10−3 | 1300 |
| E100Q | 2.9 ± 0.4 | 2.4 | (4.8 ± 0.2)×10−4 | 8100 |
| E100D | 2.0 ± 0.3 | 1.7 | (2.7 ± 0.1)×10−3 | 1400 |
| D138A | 4.0 ± 0.6 | 3.3 | (8.3 ± 0.4)×10−4 | 4700 |
| D138N | 3.2 ± 0.7 | 2.7 | (5.7 ± 0.5)×10−2 | 68 |
| D139A | 5 ± 1 | 4 | (4.8 ± 0.5)×10−4 | 8100 |
| D139N | 1.4 ± 0.3 | 1.2 | (7.6 ± 0.5)×10−3 | 510 |
| D260A | 1.6 ± 0.3 | 1.3 | (7.3 ± 0.4)×10−2 | 53 |
| H261A | 1.3 ± 0.2 | 1.1 | (4.6 ± 0.2)×10−3 | 850 |
Alanine mutants of Walker A motif residues S49A and S53A, implicated in ATP binding through interaction with the β-phosphate and a ribose hydroxyl (Fig. 4) had little relative effect on kcat while showing an increased KM value for the nucleotide/cation complex of 2.3 and 5-fold above wild type, respectively. The K51A mutant had a much more significant effect on kcat (3000-fold), while displaying only a slight ATP/Mg2+ KM increase. The T52A mutant also led to a large decrease in turnover (3000-fold) and a modest ATP/Mg2+ KM increase (1.8-fold).
Both alanine and asparagine mutants of the Walker B carboxylate-containing residues D138 and D139 were generated. The alanine mutants of both residues had a significantly decreased kcat (4700 and 8100-fold compared to wild type, respectively) along with increases in ATP/Mg2+ KM (3.3 and 4-fold). The asparagine mutants of these residues showed some restoration of activity when compared to the alanine mutants (only a 68 and 510-fold loss in activity of D138N and D139N with respect to wild type). The KM for the nucleotide/cation complex is recovered as well, to 2.7-fold higher than WT for the D138N mutant and near-WT levels for D139N.
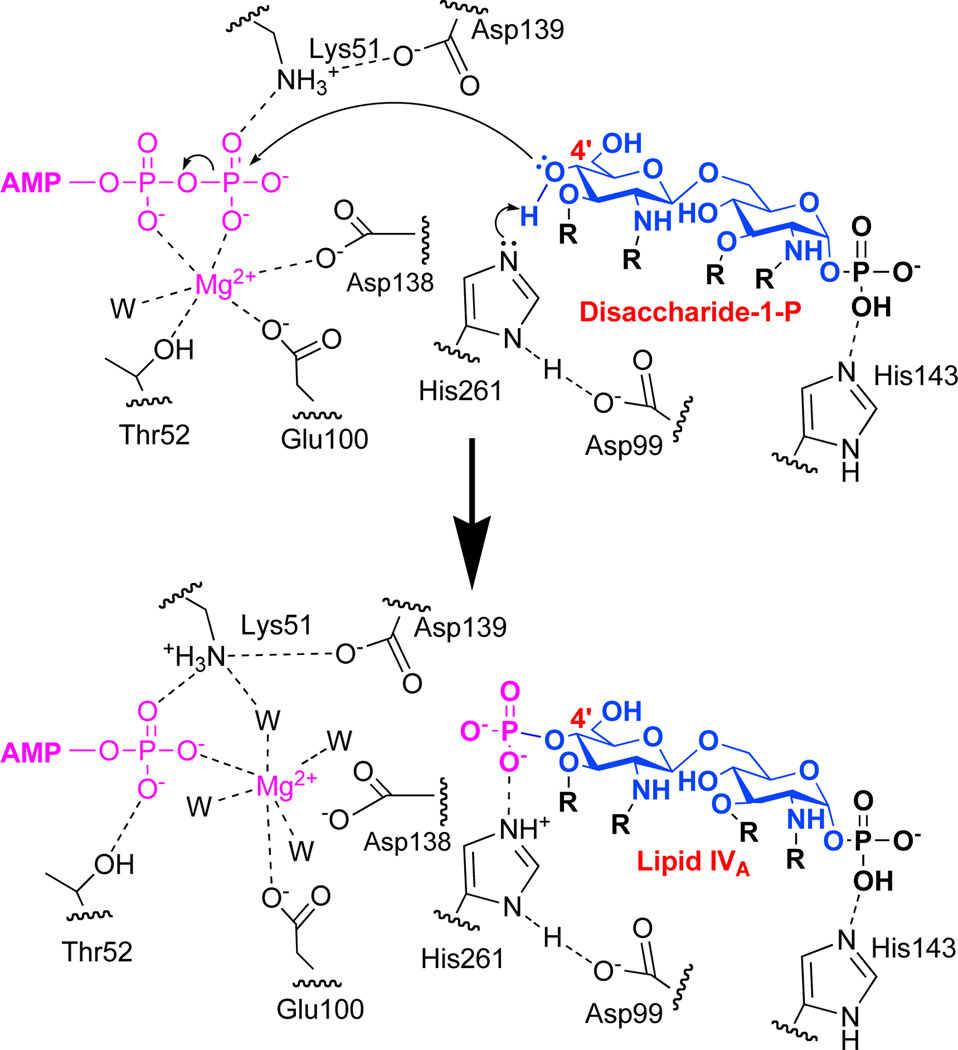
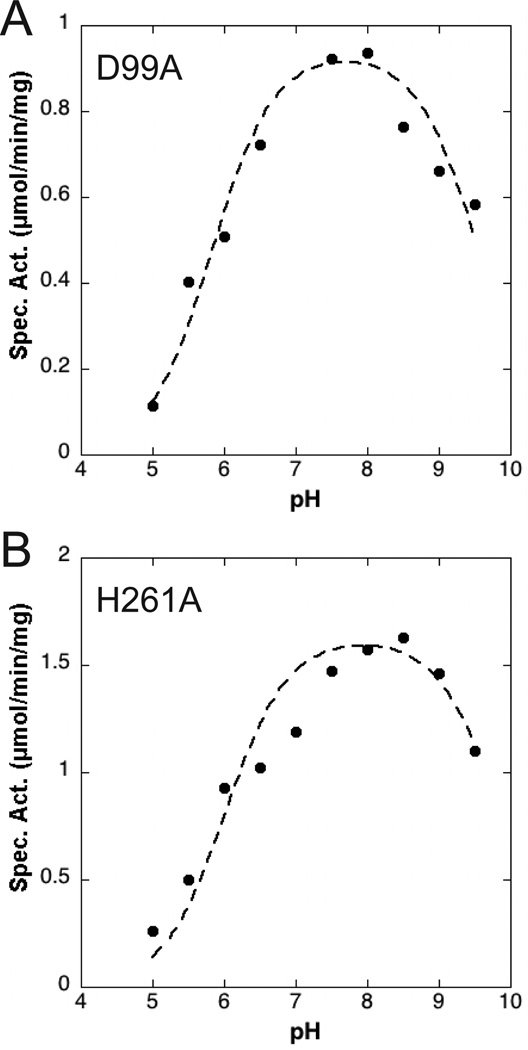
Conserved carboxylate-containing residues of the α4 helix were also deemed to be critical for activity. D99, whose side chain is bonded to H261 of the L3 loop in the closed conformation of LpxK (Fig. 4A), was mutated to alanine, asparagine, and glutamate. The alanine mutant of this residue resulted in a 2600-fold decrease in kcat and a 2.3-fold increase in ATP/Mg2+ KM. Interestingly, the asparagine mutant of this same residue displayed a further decreased kcat (11000-fold) with a full restoration of KM for the nucleotide/cation. The glutamate mutant of D99 results in only a 78-fold decrease in velocity with a 1.9-fold increase in KM. Because the proper ionization state of this residue seems crucial to efficient turnover, the pH-dependence of the D99A point mutant was assessed, revealing a shift in pKa from 6.6 ± 0.1 to 5.8 ± 0.1 with respect to the wild-type enzyme while leaving the pKb undisturbed at 9.6 ± 0.1 (Fig. 5A). It is important to note that due to potential chemical rescue with acetate found in the pH buffer, the apparent pKa and pKb for this mutant may be slightly altered from the true values. The E100 residue was mutated to alanine, glutamine, and aspartate, which resulted in 1300, 8100, and 1400-fold decreases in kcat respectively. The ATP/Mg2+ KM for the E100 mutants is increased 1.6, 2.4, and 1.7-fold for the alanine, glutamine, and asparate mutants respectively. The pattern from the D99 mutants was preserved, where the carboxamide mutant showed a more significantly decreased kcat when compared to the alanine mutant, but is partially recovered with the restoration of the carboxylate functional group with E100D.
Fig. 5.
pH-dependence of active site point mutants. (A) The D99A LpxK mutant was assayed in a triple-buffer system (100 mM sodium acetate, 50 mM Tris, 50 mM bis-Tris) at the indicated pH values and specific activity assessed. A pKa of 5.8 ± 0.1 and pKb of 9.6 ± 0.1 were calculated based on the data. (B) In the same manner, the pH dependence of the H261A mutant was analyzed resulting in pKa and pKb values of 6.0 ± 0.1 and 9.9 ± 0.2, respectively.
Further alanine point mutants were generated for other residues which occupy the active site. Y74 lines the putative DSMP-binding pocket, but is close enough to the active site to participate in catalysis. The alanine point mutant of Y74 resulted in a 1.4-fold increase in ATP/Mg2+ KM and a 180-fold decrease in kcat. An alanine mutant of D260 of loop L3 (Fig. 4A) displays a slightly increased nucleotide/cation KM (1.3-fold) and a 53-fold decreased kcat parameter. H261, a residue of loop L3 that is hydrogen bonded to D99 of helix α4, has little effect on the nucleotide/cation KM but shows an 850-fold decreased kcat when mutated to alanine, indicating its importance for kinase activity. The pH-dependence the H261A mutant was also assessed due to its contact with the critical D99 residue. The pKa and pKb were calculated to be 6.0 ± 0.1 and 9.9 ± 0.2, revealing a similar pattern as the D99A mutant where the pKa is downshifted with respect to wild type (Fig. 5B).
Crystal structure of ATP bound to open form LpxK
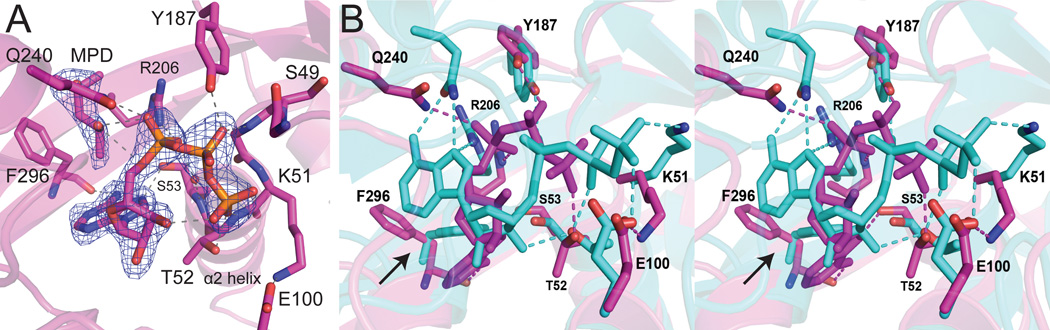
In at attempt to obtain an ATP-bound structure, LpxK was incubated with EDTA and ATP in the MPD condition. Rod-shaped crystals were obtained which surprisingly showed ATP bound in the open form of the enzyme (Scheme 1) (Fig. 6A). This ATP does not occupy the same binding site as AMP-PCP, likely because in the open form, the C-terminal domain is not closed around the nucleotide. Instead, ATP is found in a “bent” conformation, which may represent the binding state of the nucleotide before closure of the C-terminal domain. A well-ordered MPD molecule is also found bound in this pocket. The conformation of ATP consists of the α-phosphate positioned at the base of helix α2, the site normally occupied by the β-phosphate in both the ATP/Mg2+ (9) and AMP-PCP bound structures.
Fig. 6.
Active sites of ATP-bound A. aeolicus LpxK in the “open form”. (A) Simulated annealing omit electron density for ATP and MPD (mesh) in the ATP-bound structure was calculated using coefficients Fo – Fc, contoured at 4 σ. The “bent” ATP molecule is bound to the “open”, normally apo conformation of the enzyme. Dashed lines indicate hydrogen bonds. (B) Stereo view of the overlay of AMP-PCP (cyan) and ATP (magenta) bound LpxK structures reveals a shift in nucleotide binding upon domain closure. Side chains and hydrogen bonds are colored accordingly. Side chain rotation of F296 between the two structures is indicated with an arrow.
Comparisons of the AMP-PCP and ATP bound LpxK structures reveal that besides the large difference in position of the C-terminal domain (Scheme 1), specific residues show significant shifts (Fig. 6B). Q240 trades a hydrogen bond with the α-phosphate of ATP for the adenosine ring of AMP-PCP. The F296 side chain is rotated 180° to mediate a pi-stacking interaction with this same adenosine. Other side chains (Y187, S53, R206, and T52) retain the same relative positioning but trade binding partners between the two structures. Y187 participates in a hydrogen bond with the β-phosphate in the ATP structure, but switches to the α-phosphate of AMP-PCP. S53 is bound to an adenosine nitrogen atom of ATP, but contacts a ribose hydroxyl in the AMP-PCP structure. R206 trades a bond with the α-phosphate of ADP for the adenosine ring, and T52 binds the γ-phosphate of ATP only to switch to the β-phosphate and a ribose hydroxyl in the AMP-PCP structure.
Crystal structure of a “compact” P-loop conformation
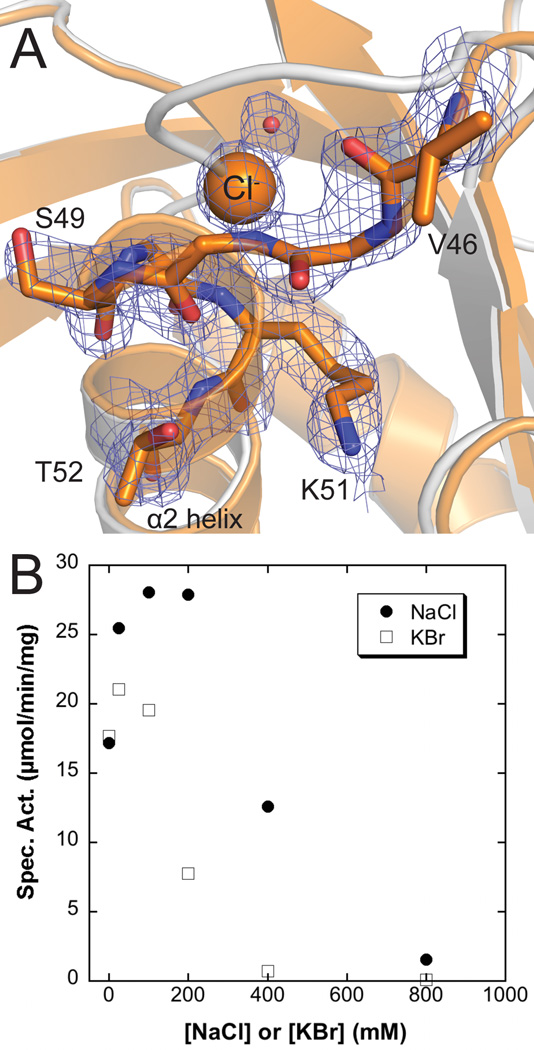
In the same drop condition which had produced apo LpxK crystals (9) except for a slightly higher salt concentration, boxy rod-shaped crystals of the same space group and unit cell dimensions appeared, which when the structure was solved were observed to have an unaltered overall appearance except for the position of the P-loop (Fig. 7A). In these crystals, the P-loop is “compact”, with the residues G48, S49, and G50 of the P-loop shifted to complete another turn of the α2 helix. Pinning down these residues at the base of the helix is spherical density which, given the high chloride concentration in the drop (~160 mM) and the dipole properties of helices, was assigned as a chloride ion. This structure may explain weak positive difference density seen in the apo LpxK structure (PDB: 4EHX), as the P-loop may exist in a semi-dynamic state alternating between these two conformations (9). In order to determine the effect of salt on activity, the assay was performed in the presence of increasing NaCl and KBr (Fig. 7B). Though both salts initially stimulated activity up to concentrations of 50–100 mM, excess salt was found to become inhibitory.
Fig. 7.
Structural basis for anion inhibition of LpxK (A) The “compact” P-loop conformation along with a chloride ion (orange sphere) and water molecule (red sphere) likely display an inactive P-loop conformer. Electron density calculated using coefficients 2Fo – Fc contoured at 2 σ is shown (mesh). To note the P-loop shift, the apo LpxK cartoon is overlaid (gray) (PDB: 4EHX) (B) Salt dependence of LpxK activity. The standard assay was performed in the presence of 0 to 0.8 M NaCl or KBr. Though having some salt (either NaCl or KBr) in the assay condition is stimulatory, excess salt becomes inhibitory. This result may represent the biochemical consequence of the “compact” P-loop conformation observed with a chloride ion obstructing the active site.
Discussion
LpxK catalyzes the sixth step in the lipid A biosynthetic pathway, and its activity is essential to bacterial viability (29). The crystal structures of A. aeolicus LpxK in both apo and ADP/Mg2+-bound forms have been previously reported, revealing a significant domain rearrangement upon nucleotide binding (Scheme 1) (9). In order to better understand the catalytic mechanism of LpxK and the molecular basis for phosphoryl transfer, we have performed detailed kinetic characterization of the kinase from Aquifex aeolicus and solved additional crystal structures of LpxK bound to AMP-PCP and ATP, as well as an open form of LpxK with an unusual “compact” P-loop conformation.
Biochemical characteristics of LpxK differentiate this membrane-bound kinase from its cytosolic relatives
We kinetically characterized LpxK in order to understand how membrane-associated kinases function at the lipid interface. LpxK’s lipid kinase activity requires the presence of detergents at concentrations above CMC and is susceptible to surface-dilution (Fig. 1B). The observed surface-dilution effect supports the notion that catalysis is occurring at the membrane interface (27). This notion correlates with previously published crystal structures of LpxK that revealed the presence of an extended, hydrophobic N-terminal helix, which we proposed functions to associate LpxK with the membrane.
Apart from the requirement of detergent, LpxK displays a bell-shaped, pH rate profile that maximizes at slightly basic pH (Fig. 1A), distinguishing it from other P-loop kinases which function best in an acidic environment (30). Due to the limitations of pH vs. activity assays, we have been cautious not to over-interpret the pKa and pKb values obtained as they may not represent a single ionizable group or take into account pH-dependence of the substrate KM values (31). However, the restoration of activity upon return to neutral pH after incubation and low pH suggests that protonation of necessary active site residues for substrate binding and/or catalysis may reversibly inactivate the enzyme. LpxK can utilize magnesium, cobalt, or manganese as metal cofactors, albeit Mg2+ is likely the preferred divalent cation (Fig. 2A). Excess magnesium is inhibitory, as has been described for other kinases, perhaps due to the existence of a second inhibitory cation binding site as has been described in many protein kinases (Fig. 2B) (32). For LpxK, the previously observed putative “post-catalytic” binding site of Mg2+ may be inhibitory, effectively trapping the ADP product in the active site (Fig. 4B) (9).
Apparent kinetic parameters for both substrates were determined for Aquifex aeolicus LpxK (Fig. 3). The KM for ATP/Mg2+ was on the same order of magnitude when compared with other membrane-bound lipid kinases (33). The KM for DSMP was slightly lower than the reported KM values for KdtA and LpxL, which also utilize tetraacylated lipid substrates in the lipid A biosynthetic pathway (34, 35). It is generally accepted that P-loop kinases catalyze direct transfer of the phosphate of ATP (4, 12, 13), a property of LpxK that we investigated using steady state kinetics. Despite low homology of LpxK to other P-loop kinase family members in both sequence (outside the Walker motifs) and structure, these enzymes do share the necessity of forming a ternary complex (Fig. 3C).
Structural insights into nucleotide binding and inhibitory conformations of the LpxK P-loop
The AMP-PCP LpxK structure is consistent with the previous proposal that the pre-catalytic position of the γ-phosphate is bound to E100 of the α4 helix, and that the likely catalytic coordination of the Mg2+ ion involves T52, E100, and D138 along with the two terminal phosphates of ATP (Fig. 4A) (Scheme 2) (9). Though this structure is informative, the binding of the Mg2+ ion may further alter geometry of the nucleotide, as made evident by comparison with the ADP/Mg2+ structure (Fig. 4B). The structure of LpxK with ATP bound in the open “apo” enzyme conformation (Scheme 1) reveals a potential pre-catalytic binding site of the nucleotide prior to formation of the catalytically competent “closed” form (Fig. 6). Since there was EDTA included in the drop to prevent hydrolysis of ATP, it stands to reason that an unoccupied Mg2+ binding site could be associated with this particular conformation of the nucleotide, altering its overall geometry when the cation is present. Nevertheless, the ATP, AMP-PCP, and previously reported ADP/Mg2+-bound (9) structures may provide a glimpse of nucleotide-enzyme interactions before, during, and after catalysis has occurred.
Scheme 2.
Proposed catalytic mechanism of LpxK. H261 acts as the catalytic base which is activated by an interaction with D99 providing a proton sink. H261 abstracts a proton from the 4'-hydroxyl of DSMP which undergoes nucleophilic attack of the γ–phosphate of ATP (pink). The Mg2+ ion (pink) shifts to its post-catalysis position between E100 and the β–phosphate of ADP and becomes solvated by additional water molecules. The acyl chains (R) on the glucosamine backbone (blue) represent R-2-hydroxymyristic acid.
The third structure reported in this work shows a chloride ion pinning the P-loop onto helix α2, causing LpxK to adopt a seemingly inactive conformation and potentially demonstrating how excess salt in the assay is inhibitory (Fig. 7). Because the detergent in the assay system (Triton X-100) has a neutral head group, inhibition in high salt is likely a direct effect of the ions interacting with the protein itself and not a result of separation of LpxK from the micelle surface, which in the assay condition may be relying solely on the hydrophobic N-terminal helix for micelle attachment. Thus, this “compact” P-loop structure reveals an intriguing mode of allosteric regulation of LpxK activity and may provide important insights into the development of allosteric LpxK inhibitors.
Roles of active site residues in LpxK catalysis
Extensive point mutagenesis was performed on active site residues in order to gain insight into their roles in kinase activity (Table 2). Overall, most of the point mutants resulted in significant effects on apparent kcat, likely because even small perturbations in proper alignment of the substrates can significantly affect catalytic efficiency. Residues S49 and S53, based on the kinetic data, appear to be primarily involved in ATP binding since alanine mutants of these residues showed significantly affected KM for the nucleotide/cation with a modest decrease in relative kcat. All of the residues that are implicated in holding Mg2+ in its catalytically competent conformation (T52, E100, and D138) have elevated ATP/Mg2+ KM values when mutated to alanine, though the largest increase for this parameter is seen for D138A. The Y74A mutant had a relatively modest effect on kcat with only slight shifts in KM for nucleotide. Y74 lines the putative DSMP binding pocket and may play a greater role in lipid binding. The alanine mutant of K51, one of the most highly conserved residues of the P-loop kinase superfamily, showed little effect on nucleotide/cation apparent KM and a large decrease in kcat, highlighting its primarily catalytic role (12).
In some P-loop kinases related to LpxK, the existence of a catalytic base has been postulated to assist in phosphoryl transfer (14). The crystal structure of LpxK with bound AMP-PCP reveals several possibilities for a putative catalytic base that is responsible for the activation of the 4'-hydroxyl group of DSMP. E100 of the α4 helix, D139 of the Walker B motif, D260 of the L3 loop, and H261 of this same loop are all well-positioned within the active site near the γ-phosphate of ATP and the putative binding locale of the 4'-hydroxyl of DSMP (Fig. 4A). Of the two residues in the L3 loop, H261 appears more catalytically relevant than D260 because when the alanine mutants of these residues are compared, a loss of the imidazole ring affects catalysis much more dramatically (Table 2). Perhaps the most intriguing possibility, given the observed interaction between the carboxylate group of D99 and the imidazole ring of H261 (Fig. 4A), is the formation of an aspartate-histidine dyad, as are commonly seen in the charge relay systems found in serine proteases (28). The use of a histidine as a general base has not been previously reported for any characterized P-loop kinase family member, though a few unrelated kinases and sulfotransferases are thought to use this amino acid as a base catalyst (15, 16). In this scenario, D99 of LpxK would serve to increase the pKa of H261, allowing for deprotonation of the substrate and subsequent nucleophilic attack on the γ-phosphate of ATP (Scheme 2). The high optimal pH range for LpxK activity supports the inclusion of an activated histidine imidazole in the catalytic mechanism over a carboxylate functionality (Fig. 1A), and the downshift in pKa when the D99 carboxylate or H261 imidazole group is removed also lends credence to this assertion (Fig. 5).
Further endorsing the hypothesis that the D99-H261 dyad is critical for LpxK activity, a significant kinetic distinction is made between the acidic residues of the Walker B motif (D138 and D139) and those of the α4 helix (D99 and E100) upon mutation (Table 2, Fig. 4A). Although alanine mutants of all these residues have reduced kcat values (from 1000 to 8000-fold as compared to wild type LpxK), carboxamide mutants of D138 and D139 are more active than the corresponding alanine mutants, whereas the opposite is true for D99 and E100. This discrepancy reveals that side chain ionization of D99 and E100 must be required for efficient catalysis while it may not be necessary for D138 and D139. Because the carboxamide mutants retain some hydrogen bonding characteristics of the original carboxylates, D138 and D139 likely play a substrate-positioning role since some activity is restored in the asparagine mutants when compared to alanine. D138, as stated previously, is thought to be involved in Mg2+ coordination, and D139 could be positioned to dock part of the DSMP substrate. The D139A mutation has a significant effect on the ATP/Mg2+ KM, indicating that its hydrogen bond to K51 could be involved in maintaining active site integrity (Table 2, Fig. 4A).
In the case of D99 and E100, the alanine mutants show reduced activity, but may be somewhat compensated by an active site water molecule or nearby analogous side chain functionalities such as D260. The D99N and E100Q mutants cannot be ionized at physiological pH, and the bulky asparagine and glutamine residues block the compensatory functionalities from performing as well. As stated previously, D99 may be required to activate H261, increasing the pKa of the distal nitrogen of the imidazole ring, so that the residue can in turn deprotonate the 4′-hydroxyl of DSMP (Scheme 2). The necessity of the E100 side chain to remain a carboxylate may be attributed to the fact that unlike D138, which is also predicted to coordinate the Mg2+ cation in the pre-catalytic state, E100 must remain bound to the cation throughout the catalytic process, thus making its charge state more critical to the overall mechanism.
Conclusion
Taken together, the additional crystal structures and kinetic characterization of LpxK has led to proposal of a catalytic mechanism involving H261 as the primary catalytic base which, assisted by its interaction with D99, deprotonates the lipid substrate for nucleophilic attack. The closure of the C-terminal domain (Scheme 1) forms the active site catalytic dyad since the D99 and H261 reside on the N- and C-terminal domains respectively, and the crystal structure of ATP bound to the “open” conformation of LpxK likely highlights an intermediate step in this process. The rest of the residues in the LpxK active site position the substrates for efficient transfer, not insignificant roles given the large perturbations in reaction velocity upon mutation for the majority of these side chains. Deciphering the catalytic roles of these residues is critical to inhibitor design and evaluation, and improves the chances of successfully targeting this essential step in lipid A biosynthesis in the pursuit of novel antimicrobials. LpxK orthologs also represent a previously uncharacterized class of membrane-bound lipid kinases, and their study will further improve our understanding of common mechanisms and unique features of enzymatic catalysis at the membrane interface.
Supplementary Material
Acknowledgments
Crystallization, screening, data collection, and data processing were performed at the Duke University X-ray Crystallography Shared Resource. Diffraction data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-BM beamline at the Advanced Photon Source (APS), Argonne National Laboratory. Use of the APS was supported by the US Department of Energy, Office of Science, and the Office of Basic Energy Sciences under contract No. W-31-109-Eng-38. We would also like to thank Dr. Anthony S. Serianni for providing methyl-2-acetamido-2-deoxy-β-D-glucopyranoside and Drs. Teresa Garrett, Kelly Daughtry, and Samuel Gattis for careful review of the manuscript. Finally, we would like to thank the members of the Raetz Lab for helpful discussion. This research was funded by N.I.H. grant GM51310 to C.R.H. Raetz and P. Zhou.
This work was supported by NIH grant GM-51310 (C.R.H.R. and P.Z.).
Abbreviations
- ADP
adenosine diphosphate
- AMP-PCP
5'-adenylyl (β, γ-methylene)diphosphonate
- ATP
adenosine triphosphate
- bis-Tris
bis(2-hydroxymethyl)-imino-tris(hydroxymethyl)-methane
- CMC
critical micelle concentration
- DDM
dodecyl maltoside
- DSMP
tetraacyldisaccharide-1-phosphate
- EDTA
ethylenediaminetetraacetic acid
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- KDO
3-deoxy-d-manno-oct-2-ulosonic acid
- LPS
lipopolysaccharide
- MPD
2-methyl-2,4-pentanediol
- NTP
nucleotide triphosphate
- P-loop
glycine-rich loop implicated in phosphate binding for enzymes which utilize NTP’s
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- TLC
thin-layer chromatography
- Tris
2-amino-2-(hydroxymethyl)-1,3-propanediol
Footnotes
The coordinates and structure factors for AMP-PCP LpxK, ATP LpxK, and Compact P-loop LpxK have been deposited in the Protein Data Bank with accession codes 4ITL, 4ITM, and 4ITN respectively.
Supporting Information Available
Tables S1 and S2 describe the primers and strains used in this study. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Raetz CRH, Whitfield C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 3.Raetz CRH, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheek S, Zhang H, Grishin NV. Sequence and structure classification of kinases. J. Biol. Chem. 2002;320:855–881. doi: 10.1016/s0022-2836(02)00538-7. [DOI] [PubMed] [Google Scholar]
- 5.Ray BL, Raetz CRH. The biosynthesis of gram-negative endotoxin. A novel kinase in Escherichia coli membranes that incorporates the 4'-phosphate of lipid A. J. Biol. Chem. 1987;262:1122–1128. [PubMed] [Google Scholar]
- 6.Garrett TA, Kadrmas JL, Raetz CRH. Identification of the gene encoding the Escherichia coli lipid A 4'-kinase. Facile phosphorylation of endotoxin analogs with recombinant LpxK. J. Biol. Chem. 1997;272:21855–21864. doi: 10.1074/jbc.272.35.21855. [DOI] [PubMed] [Google Scholar]
- 7.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer ELL, Eddy SR, Bateman A, Finn RD. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emptage RP, Daughtry KD, Pemble CW, 4th, Raetz CRH. Crystal structure of LpxK, the 4'-kinase of lipid A biosynthesis and atypical P-loop kinase functioning at the membrane interface. Proc. Natl. Acad. Sci. U. S. A. 2012;109:12956–12961. doi: 10.1073/pnas.1206072109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plötz BM, Lindner B, Stetter KO, Holst O. Characterization of a novel lipid A containing D-galacturonic acid that replaces phosphate residues. The structure of the lipid a of the lipopolysaccharide from the hyperthermophilic bacterium Aquifex pyrophilus . J. Biol. Chem. 2000;275:11222–11228. doi: 10.1074/jbc.275.15.11222. [DOI] [PubMed] [Google Scholar]
- 11.Mamat U, Schmidt H, Munoz E, Lindner B, Fukase K, Hanuszkiewicz A, Wu J, Meredith TC, Woodard RW, Hilgenfeld R, Mesters JR, Holst O. WaaA of the hyperthermophilic bacterium Aquifex aeolicus is a monofunctional 3-deoxy-D-manno-oct-2-ulosonic acid transferase involved in lipopolysaccharide biosynthesis. J. Biol. Chem. 2009;284:22248–22262. doi: 10.1074/jbc.M109.033308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leipe DD, Koonin EV, Aravind L. Evolution and classification of P-loop kinases and related proteins. J. Mol. Biol. 2003;333:781–815. doi: 10.1016/j.jmb.2003.08.040. [DOI] [PubMed] [Google Scholar]
- 13.Hutter MC, Helms V. Phosphoryl transfer by a concerted reaction mechanism in UMP/CMP-kinase. Protein Sci. 2000;9:2225–2231. doi: 10.1110/ps.9.11.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charlier HA, Runquist JA, Miziorko HM. Evidence supporting catalytic roles for aspartate residues in phosphoribulokinase. Biochemistry. 1994;33:9343–9350. doi: 10.1021/bi00197a039. [DOI] [PubMed] [Google Scholar]
- 15.Forstner M, Muller A, Stolz M, Wallimann T. The active site histidines of creatine kinase. A critical role of His 61 situated on a flexible loop. Protein Sci. 1997;6:331–339. doi: 10.1002/pro.5560060208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Negishi M, Pedersen LG, Petrotchenko E, Shevtsov S, Gorokhov A, Kakuta Y, Pedersen LC. Structure and function of sulfotransferases. Arch. Biochem. Biophys. 2001;390:149–157. doi: 10.1006/abbi.2001.2368. [DOI] [PubMed] [Google Scholar]
- 17.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 18.Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 19.McClerren AL, Zhou P, Guan Z, Raetz CRH, Rudolph J. Kinetic analysis of the zinc-dependent deacetylase in the lipid A biosynthetic pathway. Biochemistry. 2005;44:1106–1113. doi: 10.1021/bi048001h. [DOI] [PubMed] [Google Scholar]
- 20.Bartling CM, Raetz CRH. Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis. Biochemistry. 2008;47:5290–5302. doi: 10.1021/bi800240r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeLano WL, Lam JW. PyMOL: A communications tool for computational models. Abstr. Pap. Am. Chem. Soc. 2005;230:U1371–U1372. [Google Scholar]
- 26.Hu XS, Zhang WH, Oliver AG, Serianni AS. Methyl 2-acetamido-2-deoxy-beta-d-glucopyranoside dihydrate and methyl 2-formamido-2-deoxy-beta-d-glucopyranoside. Acta Crystallogr. C: Cryst. Struct. Commun. 2011;67:o146–o150. doi: 10.1107/S0108270111009061. [DOI] [PubMed] [Google Scholar]
- 27.Carman GM, Deems RA, Dennis EA. Lipid signaling enzymes and surface dilution kinetics. J. Biol. Chem. 1995;270:18711–18714. doi: 10.1074/jbc.270.32.18711. [DOI] [PubMed] [Google Scholar]
- 28.Dodson G, Wlodawer A. Catalytic triads and their relatives. Trends Biochem. Sci. 1998;23:347–352. doi: 10.1016/s0968-0004(98)01254-7. [DOI] [PubMed] [Google Scholar]
- 29.Garrett TA, Que NL, Raetz CRH. Accumulation of a lipid A precursor lacking the 4'-phosphate following inactivation of the Escherichia coli lpxK gene. J. Biol. Chem. 1998;273:12457–12465. doi: 10.1074/jbc.273.20.12457. [DOI] [PubMed] [Google Scholar]
- 30.Krell T, Maclean J, Boam DJ, Cooper A, Resmini M, Brocklehurst K, Kelly SM, Price NC, Lapthron AJ, Coggins JR. Biochemical and X-ray crystallographic studies on shikimate kinase: The improtant structural role of the P-loop lysine. Protein Sci. 2001;10:1137–1149. doi: 10.1110/ps.52501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knowles JR. The intrinsic pKa-values of functional groups in enzymes: improper deductions from the pH-dependence of steady-state parameters. Crit. Rev. Biochem. 1976;4:165–173. doi: 10.3109/10409237609105457. [DOI] [PubMed] [Google Scholar]
- 32.Adams JA. Kinetic and catalytic mechanisms of protein kinases. Chem. Rev. 2001;101:2271–2290. doi: 10.1021/cr000230w. [DOI] [PubMed] [Google Scholar]
- 33.Badola P, Sanders CR., 2nd Escherichia coli diacylglycerol kinase is an evolutionarily optimized membrane enzyme and catalyzes direct phosphoryl transfer. J. Biol. Chem. 1997;272:24176–24182. doi: 10.1074/jbc.272.39.24176. [DOI] [PubMed] [Google Scholar]
- 34.Belunis CJ, Raetz CRH. Biosynthesis of endotoxins. Purification and catalytic properties of 3-deoxy-D-manno-octulosonic acid transferase from Escherichia coli . J. Biol. Chem. 1992;267:9988–9997. [PubMed] [Google Scholar]
- 35.Six DA, Carty SM, Guan Z, Raetz CRH. Purification and mutagenesis of LpxL, the lauroyltransferase of Escherichia coli lipid A biosynthesis. Biochemistry. 2008;47:8623–8637. doi: 10.1021/bi800873n. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.