Abstract
“Explosive” adaptive radiations on islands remain one of the most puzzling evolutionary phenomena and the evolutionary genetic processes behind such radiations remain unclear. Rapid morphological and ecological evolution during island radiations suggests that many genes may be under fairly strong selection, although this remains untested. Here, we report that during a rapid recent diversification in the Hawaiian endemic plant genus Schiedea (Caryophyllaceae), 5 in 36 studied genes evolved under positive selection. Positively selected genes are involved in defence mechanisms, photosynthesis, and reproduction. Comparison with eight mainland plant groups demonstrates both the relaxation of purifying selection and more widespread positive selection in Hawaiian Schiedea. This provides compelling evidence that adaptive evolution of protein-coding genes may play a significant role during island adaptive radiations.
Keywords: adaptive radiation, Hawaiian Islands, positive selection, relaxation of purifying selection, Schiedea
Introduction
Studies of adaptive radiations of species on oceanic islands have yielded important insights into our understanding of evolution ever since the pioneering works of Darwin (1845, 1859) and Wallace (1881). Although island adaptive radiations may be viewed as extreme examples of evolutionary diversification, it is thought that major adaptive radiations in the history of our planet have followed the same evolutionary processes as island endemic radiations, so islands may be viewed as evolutionary laboratories one can use to understand the general processes of adaptation and speciation (Emerson 2002; Gavrilets and Losos 2009).
The most dramatic “bursts” of adaptive radiation often occur within confined geographical regions (e.g., oceanic islands or large lakes; cf. Schluter 2000). This might seem surprising given that natural selection is more effective in large populations, while island habitats are always limited in area and island populations are thus of limited size. In addition, many island species are thought to have been formed via the colonization of new islands or habitats (Hollocher and Williamson 1996) and such colonization events should lead to a drastic reduction in the effective population size. Low levels of genetic variability caused by the small size of island populations have been reported for island species—and in particular for endemic ones (Frankham 1997; Eldridge et al. 1999; Dixon et al. 2011)—although exceptions do occur, especially among species with high dispersal abilities (Bonhomme et al. 1989; Frankham 1997). The small population sizes of island species may render selection less effective in the face of stronger genetic drift (Ohta 2002) and the genetic processes that control the rapid morphological and ecological evolution during island adaptive radiations remain unclear.
Many groups of animals and plants have radiated extensively in the Hawaiian archipelago (Wagner and Funk 1995). The Hawaiian Islands were created by the interplay of an active volcanic hot spot and continental drift. New islands are created by volcanic activity at the eastern end of the archipelago, whereas older islands are eroding away at the western end as continental drift moves them away from the volcanic hotspot (Fleischer et al. 1998). The linear sequence of the archipelago, known ages of the individual islands and the richness of endemic flora and fauna make Hawaii one of the best locations to study adaptive radiations.
The genus Schiedea (Caryophyllaceae: Alsinoideae) is one of the largest plant adaptive radiations on the Hawaiian archipelago, comprising 34 endemic palaeopolyploid species (Kapralov et al. 2009) of perennial herbs, vines, and shrubs (Wagner et al. 2005). Schiedea species from contrasting environments (e.g., rainforest vs. coastal cliffs) are dramatically different from each other in morphology and many physiological traits (Wagner et al. 2005), which may require differences in gene expression and/or alterations in coding sequences. We demonstrate that all the morphological and ecological diversity in Schiedea has evolved surprisingly rapidly, as interspecific divergence at silent sites of nuclear genes in the genus does not exceed 4%. Low molecular divergence together with species biogeography suggests that most extant Schiedea species evolved in less than a couple of million years. This suggests that many genes might have been under fairly strong positive selection during the radiation of Schiedea.
Only few previous studies have investigated the action of selection at the molecular level during island adaptive radiations (Barrier et al. 2001; Remington and Purugganan 2002). They found some increase in the ratio of nonsynonymous (dN) to synonymous (dS) substitution rates in the Hawaiian silversword alliance, which may reflect a slight relaxation of purifying selection in small island populations, but no compelling evidence of positive selection was reported. Here, we report the analysis of positive and purifying selection during a rapid recent adaptive radiation in Schiedea and compare it with the mainland plant groups.
Materials and Methods
Polymerase Chain Reaction Amplification and Sequencing of Schiedea Genes
The data set is comprised of partial sequences of 36 nuclear genes from up to 27 species of the island endemic genus Schiedea (supplementary tables S1 and S2, Supplementary Material online). Plant samples used in this study (supplementary table S2, Supplementary Material online) have been described previously (Kapralov and Filatov 2006). Genomic DNA was isolated from fresh leaf material using magnetic beads-based Plant DNA Charge Switch Kit (Invitrogen) in accordance with manufacturer’s protocol. Given that Schiedea is not a model group and no cDNA or genomic sequences are available, we sequenced 1,000 clones from a Schiedea globosa cDNA library (Atanassov I and Filatov DA, unpublished data) and used them as queries for blast-searches against the GenBank database. Thus, after removing duplicates and genes with unknown functions, we were left with approximately 150 nonredundant sequences representing protein-coding genes with known functions. Polymerase chain reaction (PCR) primers were designed for these genes and tested across all species. PCR amplification of all studied genes was conducted using BioMix Red (Bioline) with the following PCR conditions: one cycle of 94 °C, 2.5 min, 55 °C, 30 s, 72 °C, 3.5 min followed by 37 cycles of 93 °C, 20 s, 53 °C, 30 s, 72 °C, 2.5 min. For 36 genes (supplementary tables S1 and S3, Supplementary Material online), clear single band PCR products were obtained for 20 or more Schiedea species. Primers for these genes are listed in the supplementary table S4, Supplementary Material online. The PCR products were separated on 1% agarose gels and extracted from the gels using Qiagen gel extraction kit. The relatively short length of the obtained PCR products (<1,200 base pairs) allowed us to use the same primers for both PCR and sequencing. Sequencing was performed using ABI BigDye v3.1 system on an ABI3700 automated sequencing machine. Sequence chromatograms were checked and corrected, and the contigs were assembled and aligned using ProSeq3 software (Filatov 2009). All polymorphic sites were checked against the original sequence chromatograms and problematic regions were resequenced; the sequences obtained were compared with S. globosa cDNA sequence and homologs from GenBank, and open reading frame (ORF) integrity was confirmed for protein-coding sequences.
Sequence Data for Mainland Groups
Homologs of genes analyzed in Schiedea were retrieved using BLAST (Altschul et al. 1990) from Genbank (http://www.ncbi.nlm.nih.gov) for eight mainland plant groups (families Asteraceae, Fabaceae, and Poaceae; tribe Cichorieae; and genera Citrus, Helianthus, Populus, and Solanum). Schiedea globosa genes with introns removed were blast-searched against the Genbank expressed sequence tag (EST) database with searches limited to one selected species from each of the eight mainland groups. The sequences obtained were aligned with Schiedea sequences and trimmed to the same length as the analyzed Schiedea sequences. These trimmed sequences from mainland groups were used as queries for new BLAST searches narrowed to species from the respective mainland groups with sufficient numbers of available ESTs. The data sets obtained were aligned using ClustalW and manually checked; ambiguous sites and gaps were removed before further analyses. Sequences that diverged from the queries substantially more than expected from average species divergence were not included into alignments as they are likely to be paralogs. Including more species inevitably reduced the number of genes in the analysis, as not all genes are available for all species. Subject to this trade-off, 22, 21, and 28 species were included in the analysis of families Asteraceae, Fabaceae, and Poaceae, respectively, whereas from 6 to 8 species were sampled from the tribe Cichorieae and genera Citrus, Helianthus, Populus, and Solanum (supplementary table S5, Supplementary Material online, for species names and GenBank accession numbers). Six of the mainland groups were represented by herbaceous plants (Asteraceae, Cichorieae, Fabaceae, Helianthus, Poaceae, and Solanum) and two—by shrubs and trees (Citrus and Populus); and the sizes of corresponding data sets varied from 11 to 21 genes, as not all genes we sequenced in Schiedea are sequenced in other groups (supplementary table S5, Supplementary Material online). Both number of sequences in a data set and their divergence affect performance of phylogeny based tests for positive selection (Anisimova et al. 2001). Thus, ideal groups for comparison with Schiedea would be ones with similar species number and interspecific divergence. We could not find data sets of over 20 closely related plant species with available sequences homologous to obtained Schiedea genes. Hence, we used three data sets of similar size to Schiedea one consisting of species from families Asteraceae, Fabaceae, and Poaceae, which are much more diverged than Schiedea species. There is less statistical power to find positive selection in a less diverged group (Schiedea) compared with more diverged ones (Anisimova et al. 2001), that makes these comparisons more conservative. Also we used five data sets of six to eight closely related plant species (tribe Cichorieae; and genera Citrus, Helianthus, Populus, and Solanum). These data sets were compared with the subset comprised of the seven most diverged Schiedea species to minimize differences in interspecific divergence and species numbers per data set.
Data Analyses
Alignments of obtained sequences were created manually or using ClustalW with default parameter values (Larkin et al. 2007). All alignments were unambiguous, which is not surprising given the low divergence between Schiedea species as well as within the groups of mainland plants. Coding region assignment, intron splicing, and gene concatenation were performed in ProSeq3 (Filatov 2009). Average pairwise divergence per nucleotide for silent and nonsilent sites as well as preliminary gene trees using the neighbor-joining algorithm were obtained using Mega v.4 (Tamura et al. 2007). The fully resolved phylogenies of Schiedea and mainland groups were reconstructed using silent mutations (noncoding and the third positions of exons) and indels manually recoded as single mutations in a concatenated data set of all sequenced genes. The statistical selection of best-fit models of nucleotide substitution was done using jModelTest (Posada and Crandall 1998; Posada 2008). The phylogenies were reconstructed by Bayesian analysis implemented in MrBayes 3.1.2 (Ronquist and Huelsenbeck 2003) using a GTR+Γ model of DNA sequence evolution. Potential topological conflicts between the phylogenetic trees obtained using concatenated data sets and individual genes were assessed with approximately unbiased (AU) and weighted Shimodaira–Hasegawa (WSH) tests (Shimodaira 2002) performed in the Treefinder package (Jobb et al. 2004). The fully resolved phylogenies of Schiedea and the mainland groups obtained using concatenated data sets were in agreement with previously published ones (Wagner et al. 2005) and showed no significant topological conflict with individual genes at the 5% level or below in AU and WSH tests. These fully resolved phylogenies of Schiedea and the mainland groups were used as input trees for analyses of individual genes using PAML, sitewise likelihood-ratio (SLR), and FitModel (discussed later) to increase analytical power, which would be compromised if we used less resolved phylogenies obtained using individual genes (Anisimova et al. 2001). When some species were missing for particular genes, these species were manually pruned from the input tree.
Molecular adaptation can be inferred from the comparison of the rates of nonsynonymous (changing amino acid protein sequence, dN) and synonymous (resulting in no change at the protein level, dS) mutations. Under neutrality the two rates are expected to be equal (dN/dS ∼ 1), whereas purifying or adaptive selection is expected to reduce (dN/dS < 1) or inflate (dN/dS > 1) this ratio, respectively. To infer molecular adaptation, we used three programs (PAML, SLR, and FitModel) calculating dN/dS ratio within phylogenetic framework. First, we used the program codeml in the PAML 4.0 package (Yang 2007) to estimate the nonsynonymous divergence (dN), synonymous divergence (dS), and their ratio (dN/dS) in the model M0, which allows for a single dN/dS value throughout the whole phylogenetic tree. We tested for the presence of positive selection in Schiedea and in the mainland data sets by comparing a pair of models, M8 versus M8a, implemented in PAML 4.0 (Yang 2007). These are nested models and their relative fit to the data can be compared in a likelihood ratio test (LRT). Both M8 and M8a models allow the dN/dS ratio to vary between 0 and 1, which takes into account those codons that evolve under purifying selection as well as codons evolving neutrally. In addition, the M8 model also allows for a class of codons with dN/dS > 1 that accommodates codons under positive selection. In the M8a model, the positively selected codons are forced to have dN/dS = 1. The M8/M8a LRT specifically tests whether dN/dS for codons falling into the positively selected class is significantly larger than unity (Swanson et al. 2003). The significance of the LRTs was calculated assuming that twice the difference in the log of maximum likelihood between the two models is distributed as a chi-squared distribution with the degrees of freedom (df) given by the difference in the numbers of parameters in the two nested models. It has been argued that for M8a–M8 comparisons the appropriate test would use a 50:50 mixture of df = 0 and df = 1 (Swanson et al. 2003); thus, our approach is conservative. The P values of the LRT tests were corrected using the false discovery rate (FDR) procedure (Benjamini and Hochberg 1995). Codon distributions were extracted from the model M8 outputs and basic descriptive statistics were calculated in Excel. Groups were compared using the nonparametric Kruskal–Wallis test, which does not make assumptions about normality and is designed for conditions when both nominal and measurement variables are present.
Second, we used the SLR program, which implements SLR method for detecting nonneutral evolution, a statistical test that can identify sites under positive selection even when the strength of selection is low (Massingham and Goldman 2005). The SLR test consists of performing a likelihood-ratio test on a sitewise basis, testing the null model (neutrality dN/dS = 1) against an alternative model (dN/dS ≠ 1). SLR method is a test of whether a given site has undergone selection, and the test statistic summarizes the strength of the evidence for selection rather than the strength of the selection itself (Massingham and Goldman 2005). The SLR program implements Hochberg’s step-up procedure (Hochberg 1988) to correct P values for multiple tests.
Third, we applied switching Markov modulated codon models as implemented in the program FitModel (Guindon et al. 2004) to genes shown to be under positive selection by PAML and SLR. Unlike site model, switching models allow each codon site to change the selective regime, and thus be affected by different selective pressures at different time points. This is accomplished by using an additional Markov process to describe the switches between selection regimes at any individual site. We used model M2a (same as in PAML) both with switches of selective pressure over time (M2a + S1) and without switches (M2a) and the LRTs were performed to test whether switches of selective pressure over time occurred (M2a vs. M2a + S1). Sites with episodes under positive selection were detected a posteriori using the Bayesian approach (Guindon et al. 2004). The same input files with sequence alignment and species phylogeny were used for PAML, SLR, and FitModel.
Results
Analysis of Selection in Schiedea
To test whether rapid diversification and adaptation to contrasting conditions in Schiedea was accompanied by widespread adaptation at the molecular level, we obtained partial sequences of 36 nuclear genes from up to 27 species of the island endemic genus Schiedea (supplementary tables S1 and S2, Supplementary Material online). With a total alignment length of approximately 24 kb, our data set represents by far the largest multigenic study of substitution rates in rapidly radiating island endemics. The sequenced genes are involved in a wide range of metabolic pathways and biological functions (supplementary table S3, Supplementary Material online) and have not been chosen in any specific way with respect to their function. Interspecific divergence in Schiedea averaged across all genes and species was quite low: 2% for noncoding regions and synonymous sites and 0.5% for nonsynonymous sites (supplementary table S1, Supplementary Material online). The largest pairwise silent divergence between Schiedea species averaged across all genes did not exceed 4%, demonstrating that all the morphological and phenotypic diversity in this endemic genus evolved in a very short period of time. Although the divergence between Schiedea species is relatively low, it is still at least an order of magnitude higher than intraspecific polymorphism (Filatov and Burke 2004; Wallace et al. 2009; Dixon et al. 2011).
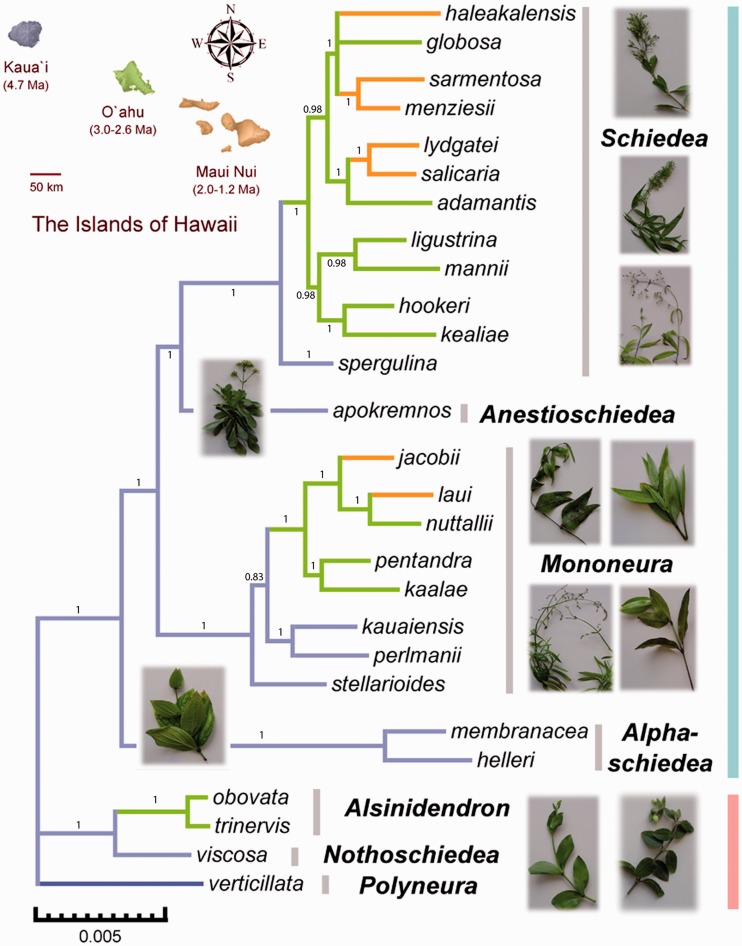
We reconstructed a fully resolved phylogeny of Schiedea from a concatenated data set of all 36 nuclear genes, using Bayesian inference as implemented in MrBayes 3.1.2 (Ronquist and Huelsenbeck 2003). The phylogeny we obtained (fig. 1) was similar to the previously published Schiedea phylogenies based on morphology and internal/external transcribed spacers (ITS/ETS) sequence data (Wagner et al. 2005) and three nuclear genes (Willyard et al. 2011) but was better resolved and supported. Potential topological conflicts between Schiedea phylogenies obtained using the concatenated data set and those obtained from individual genes were assessed using AU and weighted Shimodaira–Hasegawa tests (Shimodaira 2002). None of these tests revealed any significant topological conflict and the phylogeny based on the concatenated data set was therefore used in subsequent tests for positive selection in individual Schiedea genes.
Fig. 1.
Unrooted tree of 27 Schiedea species representing perennial herbs, vines, and shrubs occurring in a range of contrasting environments. The phylogeny was estimated using a concatenated alignment of 36 nuclear genes using MrBayes (Ronquist and Huelsenbeck 2003); posterior probabilities are shown next to branches. All recognized sections are monophyletic and indicated by section names in bold and gray bars on the right of species names. Branches are color-coded according to the islands where each species is found (see map of central Hawaii, top left). Images of representative species are placed near the section names. Blue and pink bars to the right of the plant photographs indicate the borders of the former genera Schiedea and Alsinidendron, respectively, before they were merged by Wagner et al. (2005).
We analyzed selection in the sequenced protein-coding genes using a phylogeny-based maximum likelihood approach based on a comparison of nonsynonymous (i.e., changing the protein sequence, dN) and synonymous (resulting in no change at the protein level, dS) substitution rates (Yang 2007). Under neutrality, the two rates are expected to be equal (dN/dS ∼ 1), whereas purifying or adaptive selection is expected to reduce (dN/ dS < 1) or inflate (dN/ dS > 1) this ratio, respectively. The average dN/dS ratio among 36 nuclear genes of the island genus Schiedea was 0.39 (ranging from 0.03 to 1.44; supplementary table S6, Supplementary Material online), which is relatively high, compared with other studied groups (e.g., Drosophila [Drosophila 12 Genomes Consortium 2007]). This may be due either to somewhat relaxed purifying selection or to widespread positive selection in Schiedea. To test for positive selection in individual genes, we employed a LRT that compares the fit of two nested models, the more general model allowing for positive selection and more restrictive model that assumes only purifying selection or neutral evolution. The signal of positive selection was detectable by PAML in 10 of the 36 genes (P < 0.05; M8/M8a LRT; Yang 2007), and remained significant in five of these genes after correction for multiple tests using the FDR (supplementary table S6, Supplementary Material online). Thus, at a 5% FDR, at least 5 in 36 of the studied protein-coding genes were shown to be under positive selection in Schiedea (supplementary table S6, Supplementary Material online), which gives conservative estimate of proportion of genes under positive selection in Schiedea within the binomial confidence interval from 5% to 30%. This is likely to be an underestimate given that we only analyzed an average of approximately 30% of the length of the protein-coding regions in each gene (supplementary table S1, Supplementary Material online), and hence could have missed regions under positive selection.
Results of analyses performed with the SLR program (Massingham and Goldman 2005) showed presence of codons under positive selection in nine genes; genes evolving under positive selection were the same as shown by PAML before FDR with exception of the glyceraldehyde-3-P dehydrogenase gene, which was shown under selection by PAML before FDR but not by SLR.
LRTs M2a versus M2a + S1 were performed using the FitModel program (Guindon et al. 2004) to compare the model that allows the site-specific selection process to vary along lineages of a phylogenetic tree with one that does not take into account switches between selection patterns. The model that allows selection process to vary along lineages provided significantly better fit to the data for two Schiedea genes under positive selection, CBP5 and putative cell division protein genes with P values of 0.009 and 0.002, respectively. Positively selected substitutions in the CBP5 gene were either associated with branches leading to species of the Mononeura section or all branches except of Mononeura, whereas positively selected substitutions in the putative cell division protein gene appeared across all phylogeny.
Selection in the Mainland Plant Groups
To compare Schiedea to mainland plants, we compiled three data sets for herbaceous plants from the families Asteraceae, Fabaceae, and Poaceae, which were of comparable size with the Schiedea data set. In addition, we compiled five small data sets consisting of six to eight closely related species from the tribe Cichorieae and genera Citrus, Helianthus, Populus, and Solanum and compared them with the subset of seven Schiedea species. These data sets consist of homologs of the genes we sequenced in Schiedea (supplementary table S5, Supplementary Material online). The fully resolved Bayesian phylogenies of the concatenated data sets for each mainland group are shown in supplementary figures S1–S8, Supplementary Material online. No topological conflict was detected between the genes, and the phylogenies matched the current classification in each group with minor differences which likely aroused due to different sampling designs and common usage of chloroplast genes in published phylogenies but not in our ones (Nicolosi et al. 2000; Doyle and Luckow 2003; Hamzeh and Dayanandan 2004; Weese and Bohs 2007; Panero and Funk 2008; Schneider et al. 2009; Kane et al. 2011; Morrone et al. 2011).
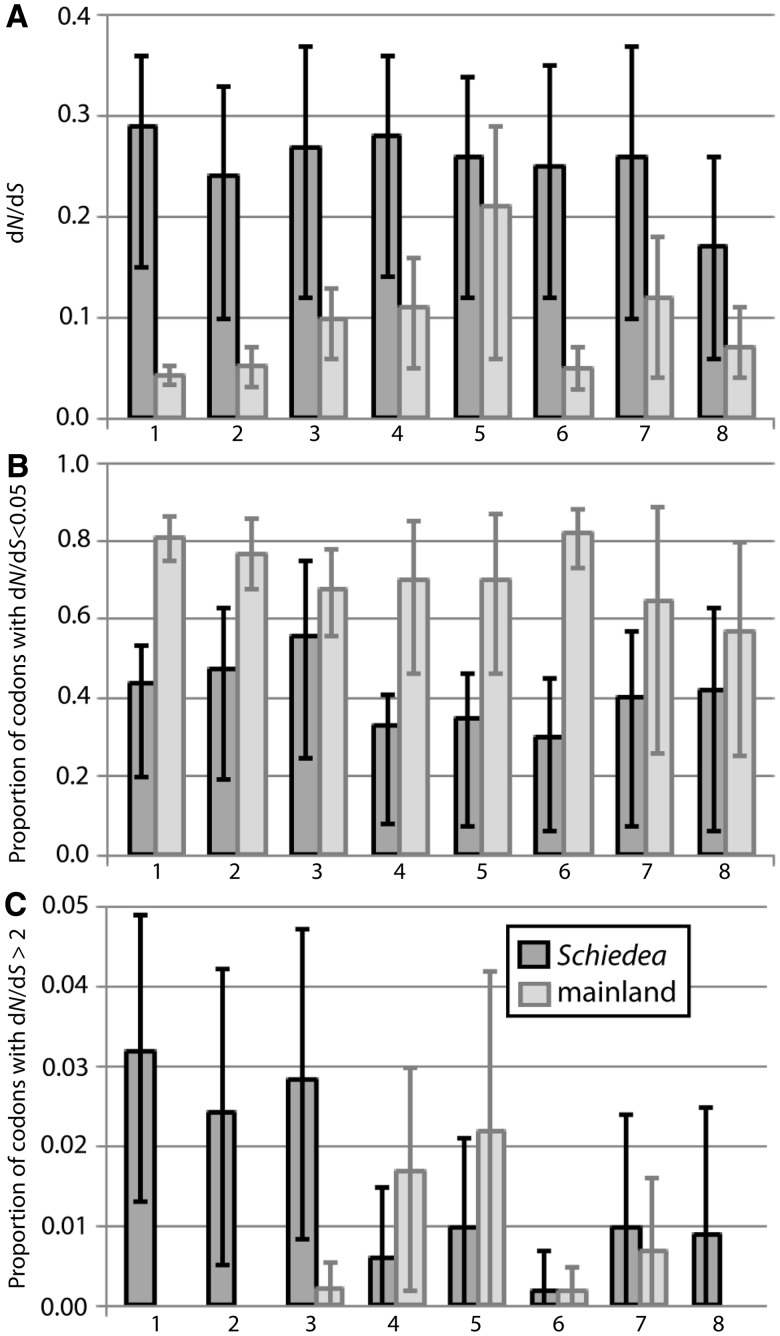
Average dN/dS ratios of all mainland groups were lower than in Schiedea (fig. 2A; supplementary table S6, Supplementary Material online), supporting the view that purifying selection is relaxed and/or there is more positive selection in island Schiedea, compared with the mainland groups. These differences were significant when the data set of 27 Schiedea species was compared with family-level mainland groups with over 20 species per group (P < 0.05, the Kruskal–Wallis test; fig. 2A), but not significant when the subset of seven Schiedea species was compared with genus-level mainland groups of 6–8 species (with exception of Cichorieae; fig. 2A). To test for positive selection in the mainland groups, we used the same models as in the analyses of Schiedea. Higher divergence in the mainland groups (supplementary table S6, Supplementary Material online), compared with Schiedea should lead to more power to detect positive selection in the mainland groups (Anisimova et al. 2001); hence, this comparison is conservative, if it is used to address the question whether there is more positive selection in Schiedea, compared with mainland groups. Nevertheless, with the exception of one gene in Helianthus, no genes were identified as evolving under positive selection in the mainland groups by PAML (after FDR correction, see supplementary table S6, Supplementary Material online). Another approach, SLR, showed no codons under positive selection in the mainland groups with exception of one gene in Helianthus and one gene in Populus (1-aminocyclopropane oxidase and mannitol dehydrogenase, respectively). The lack of positive selection cannot be due to lower power of the analysis in mainland groups given that species divergence in the mainland groups was higher than in Schiedea (supplementary table S6, Supplementary Material online).
Fig. 2.
The three leftmost pairs of columns represent comparisons between the data set of 27 Hawaiian Schiedea species (dark gray bars) and three mainland plant groups represented by over 20 species each (Poaceae, Asteraceae, and Fabaceae; light gray); the following five pairs of columns represent comparisons between the subset of seven Schiedea species (dark gray bars) and five mainland plant groups represented by 6–8 species each (Helianthus, Populus, Cichorieae, Citrus, and Solanum; light gray). (A) dN/dS estimated assuming a single ratio for all codons and branches of a tree (M0 model) and averaged over all studied genes; (B) proportions of codons under strong purifying selection (with dN/dS < 0.05), calculated using model M8 implemented in PAML; (C) proportions of codons under strong positive selection (with dN/dS > 2), calculated as in (B). Each of eight pairs of columns represents data averaged across 20, 14, 13, 19, 15, 14, 12, and 10 genes, respectively. Differences between Schiedea and three family-level mainland groups (Poaceae, Asteraceae, and Fabaceae) were significant (P < 0.05; the Kruskal–Wallis test), with the exception of the comparison of purifying selection between Schiedea and Fabaceae (B-3). Differences between Schiedea and the genus-level mainland groups (Helianthus, Populus, Cichorieae, Citrus, and Solanum) were not significant (P > 0.05; the Kruskal–Wallis test), with the exception of the comparisons for average dN/dS and purifying selection between Schiedea and Cichorieae (A-6 and B-6). All presented data are back-transformed from log transformation performed to calculate 95% confidence interval shown by error bars. Original values are presented in the supplementary table S6, Supplementary Material online.
The model M8 allows the dN/dS ratio to vary between 0 and 1, which takes into account those codons that evolve under purifying selection as well as codons evolving neutrally. In addition, it also allows a class of codons with dN/dS >1 that accommodates codons under positive selection.We extracted data on dN/dS ratios and the size of the classes from the M8 output for each gene and compared the proportion of codons under strong purifying selection (dN/dS < 0.05) and strong positive selection (dN/dS >2) in island Schiedea and the mainland plant groups. We found a lower proportion of codons under strong purifying selection in Schiedea than in mainland groups; these differences being significant in comparisons between Schiedea and Poaceae, Asteraceae, and Cichorieae, P < 0.05, the Kruskal–Wallis test; fig. 2B). The proportion of codons under strong positive selection was higher in Schiedea compared with all mainland groups with exception of Helianthus and Populus (fig. 2C). This difference was statistically significant for large mainland groups (P < 0.05, the Kruskal–Wallis test; fig. 2C), but not for small ones (fig. 2C).
Thus, strong purifying selection is somewhat relaxed and positive selection is more common in island Schiedea than in the most of studied mainland plant groups. This supports the view that rapid morphological evolution in Schiedea is driven by fairly strong positive selection, as might be expected for an island radiation. Population sizes on islands are often small, so the dynamics of mutations with weak effect are dominated by drift and only mutations of strong effect are “visible” to selection. Indeed, the dN/dS ratios for positively selected Schiedea genes are quite high (supplementary table S6, Supplementary Material online).
Discussion
Our estimates of relatively low interspecific divergence in Schiedea confirms that remarkable morphological diversity of leaf shapes, reproductive syndromes, and growth forms have evolved recently and rather rapidly in Schiedea (Wagner et al. 2005). During an island adaptive radiation, Schiedea species not only underwent dramatic morphological evolution but also colonized a range of habitats from shady understory of rainforest to desert-like sun-exposed oceanic cliffs (Wagner et al. 2005). It is not known whether such rapid phenotypic and ecological diversification predominantly occurs due to strong selection at only a few genes, or if many genes are involved. It is also not clear whether it is mainly due to the evolution of gene expression and regulatory regions, or due to selection for amino acid replacements in the protein-coding regions.
There are multiple examples demonstrating the importance of gene expression in adaptive evolution and speciation (reviewed in Hoekstra and Coyne 2007; Wray 2007). Studies of selection at the molecular level in crop species also often point toward the importance of cis-regulatory changes. For example, the dramatic evolution of candelabra-like teosinte into pole-like maize (the development of strong apical dominance) was demonstrated to be due to strong selection in the cis-regulatory region of the TB1 gene that led to much higher expression of that gene in maize, compared with teosinte (White and Doebley 1998; Wang et al. 1999). However, it is not clear whether the rapid morphological and ecological evolution during island adaptive radiations is mainly driven by selection on cis-regulatory sequences, or in the protein-coding genes. Because of the difficulties of working with nonmodel island species, there are only a few reports devoted to the analysis of the genetic bases of adaptation in island radiations (Barrier et al. 2001; Remington and Purugganan 2002; Abzhanov et al. 2004, 2006) and all these reports point toward the importance of cis-regulatory changes and evolution of gene expression. However, the previous studies focused on only a few genes and the wider survey of genes undertaken in our study does reveal the acceleration of the rate of protein evolution in the island Schiedea, compared with mainland species. After FDR correction, at least 5 in 36 of studied protein-coding genes of Schiedea were shown to be under positive selection
Similar phylogeny-based analyses of positive selection in animals revealed that 878 out of 8,510 (binomial confidence interval is 9.7–11.0%) protein-coding genes in six species of Drosophila, and 400 out of 16,529 (binomial confidence interval is 2.2–2.7%) in six mammalian genomes, evolved under positive selection (Drosophila 12 Genomes Consortium 2007; Kosiol et al. 2008; Larracuente et al. 2008). The analyzed Drosophila and mammalian genomes cover phylogenies that are more than an order of magnitude deeper than that of Schiedea (Drosophila 12 Genomes Consortium 2007; Kosiol et al. 2008). If positive selection is not persistent and acts only occasionally in the phylogeny, then with longer divergence times, more genes are expected to have undergone positive selection, compared with a shallower phylogeny of species that diverged only recently. In addition, greater divergence between the species provides more power to detect positive selection using the phylogeny-based maximum likelihood approach we used, and species divergence in these groups is nearly optimal for detecting positively selected genes (Anisimova et al. 2001). Another issue when analyzing highly diverged lineages is high proportion of false-positives produced by PAML due to misalignment of sequences that was demonstrated recently on the data set of 12 Drosophila genomes (Markova-Raina and Petrov 2011). Although number of genes detected to be under positive selection might decrease after removing misaligned regions from analysis of highly diverged groups (Markova-Raina and Petrov 2011), this will not affect our findings in Schiedea where alignments were unambiguous due to low divergence and absence of indels in the analyzed regions of exons. Thus, the estimate of at least 5% to 30% of Schiedea genes evolving under positive selection is thus surprisingly high given the recent divergence in the genus. However, selection may act differently in plants and animals. For example, many plant genes are expressed in the haploid gametophyte, whereas only a few animal genes are expressed in gametes (Borg et al. 2009), which may lead to inherent differences in selective pressures between the two kingdoms. Nevertheless, the comparison with mainland plant groups revealed significantly more positive selection in Schiedea, which may reflect the acceleration of adaptive evolution in protein-coding genes during the rapid morphological and ecological radiation in this Hawaiian endemic genus. Colonization of island habitats and expansion into unoccupied niches might be among the conditions that make widespread positive selection possible.
The ubiquity of positive selection in Schiedea genes might seem surprising given positive selection is associated with a substantial cost, often expressed in terms of genetic load. In the classic paper on genetic load Haldane (1957) argued that a reasonable estimate to the limit for selection is approximately 1 substitution per 300 generations. Our analysis of selection in Schiedea spans over 2–3 My, or at least a million generations. According to Haldane, this would be sufficiently long time for selection to fix substitutions in over 3,000 genes. Assuming that Schiedea genome contains approximately 30,000 genes, our estimate of 5–30% of genes under positive selection is compatible with Haldane’s limits for selection. Furthermore, the original Haldane’s view of the cost of positive selection is now considered too extreme (Turner and Williamson 1968; Nunney 2003), as much of adaptation can occur via soft or polygenic selection (Pritchard et al. 2010) that does not require elimination of all individuals that do not possess the advantageous allele.
Three of the five genes identified to evolve under positive selection are potentially involved in plant defence mechanisms, whereas photosynthesis and reproduction were represented by one gene each (supplementary tables S3 and S6, Supplementary Material online). Schiedea ubiquitin activating enzyme is similar to Arabidopsis thaliana AT2G30110 gene, which encodes an enzyme, involved in the first step in conjugating multiple ubiquitins to proteins targeted for degradation and plays a role in plant defence signaling. Schiedea glycosyl hydrolase gene is similar to A. thaliana glycosyl hydrolases family 17 protein (AT1G11820) which is involved in metabolic processes of carbohydrates and possibly in defense of plants against pathogens (Dong et al. 1991). Schiedea putative cell division protein is similar to the cell division protein of Bacillus cereus and putative allergen protein from sugar beet (Fowler et al. 2000). Finding that three in five of Schiedea genes under positive selection are potentially involved in defence response is in agreement with previous results from genome-wide studies of various organisms showing that a category of defence-related genes is enriched with genes targeted by positive selection (Drosophila 12 Genomes Consortium 2007; Petersen et al. 2007).
Schiedea gene for NHL repeat-containing protein is similar to Arabidopsis thaliana gene AT5G14890 that is expressed in the flowers. Expression of this gene has been shown to accompany pollen germination and tube growth in Arabidopsis (Schmid et al. 2005; Wang et al. 2008). Flower development and reproduction are areas, which experienced strong diversification and directional selection during Schiedea evolution (Wagner et al. 2005).
Schiedea CBP5 gene is similar to A. thaliana gene AT3G27690, which encodes the light-harvesting chlorophyll a/b-binding (LHC) proteins that constitute the antenna system of the photosynthetic apparatus (Jansson 1999). This is the second photosynthetic gene reported to be under positive selection in Schiedea after the chloroplast gene rbcL encoding the large subunit of the Rubisco enzyme (Kapralov and Filatov 2006, 2007). Adaptive radiation in Schiedea included diversification into niches with contrasting light regimes, such as shadowy understory of wet forest and oceanic cliffs exposed to direct sun, and species from different habitats differ in photosynthetic parameters (Wagner et al. 2005).
Although we detect positive selection in 5 out of 36 nuclear genes in Schiedea, no significant signal of adaptation was found in a DNA polymorphism analysis in a subset of these genes (Gossmann et al. 2010). The difference between the results of the two studies may be due to timing of selection in Schiedea—if selection occurred predominantly during the early evolution of the genus, then this may not be detectable in polymorphism data without a sufficiently diverged outgroup, but can be detected by the phylogeny-based approach employed here. Alternatively, the discrepancy may be due to different sensitivities of the analyses in the two studies. Indeed, out of 10 different plant groups analyzed by Gossman et al. (2010), Schiedea was the only genus that showed some signs of adaptive evolution (positive, but not significant alpha). The Schiedea data set was the smallest in the earlier study and with a larger data set the signal of adaptive evolution in this genus might become detectable with polymorphism-based analyses.
Analysis of the fossil record suggests that many major groups of organisms have evolved very rapidly (e.g., flowering plants) and adaptive radiations on islands provide us with a way to analyze the evolutionary processes during such rapid evolutionary events. Unfortunately, the analysis of island radiations is often hampered at the population genetic and phylogenetic level: the lack of intraspecific DNA polymorphism and molecular divergence between the species makes it necessary to sequence multiple genes to uncover enough molecular variation for further analysis. However, improvements in molecular techniques (in particular, the availability of high-throughput sequencing) provides hope that more studies devoted to the genetic basis of adaptation during island radiations will soon be published. Undoubtedly, such studies will significantly improve our understanding of the evolutionary processes that act during the relatively short but critically important periods of rapid morphological and ecological change.
Supplementary Material
Supplementary tables S1–S6 and figures S1–S8 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Stephen G. Weller for providing Schiedea leaf material; John Pannell and Roger Butlin for helpful comments on the manuscript; and Christopher Dixon for correcting the manuscript. This work was supported by the Natural Environment Research Council UK to D.A.F.
References
- Abzhanov A, Kuo WP, Hartmann C, Grant BR, Grant PR, Tabin CJ. The calmodulin pathway and evolution of elongated beak morphology in Darwin’s finches. Nature. 2006;442:563–567. doi: 10.1038/nature04843. [DOI] [PubMed] [Google Scholar]
- Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. Bmp4 and morphological variation of beaks in Darwin’s finches. Science. 2004;305:1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Bielawski JP, Yang Z. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol Biol Evol. 2001;18:1585–1592. doi: 10.1093/oxfordjournals.molbev.a003945. [DOI] [PubMed] [Google Scholar]
- Barrier M, Robichaux RH, Purugganan MD. Accelerated regulatory gene evolution in an adaptive radiation. Proc Natl Acad Sci U S A. 2001;98:10208–10213. doi: 10.1073/pnas.181257698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- Bonhomme F, Miyashita N, Boursot P, Catalan J, Moriwaki K. Genetical variation and polyphyletic origin in Japanese Mus musculus. Heredity. 1989;63:299–308. doi: 10.1038/hdy.1989.102. [DOI] [PubMed] [Google Scholar]
- Borg M, Brownfield L, Twell D. Male gametophyte development: a molecular perspective. J Exp Bot. 2009;60:1465–1478. doi: 10.1093/jxb/ern355. [DOI] [PubMed] [Google Scholar]
- Darwin CR. Journal of researches into the natural history and geology of the countries visited during the voyage of H.M.S. Beagle round the world, under the Command of Captain Fitz Roy, R.N. London: John Murray; 1845. [Google Scholar]
- Darwin CR. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. London: John Murray; 1859. [PMC free article] [PubMed] [Google Scholar]
- Dixon CJ, Kapralov MV, Filatov DA. Gene flow and species cohesion following the spread of Schiedea globosa (Caryophyllaceae) across the Hawaiian Islands. J Evol Biol. 2011;24:1–11. doi: 10.1111/j.1420-9101.2010.02128.x. [DOI] [PubMed] [Google Scholar]
- Dong X, Mindrinos M, Davis KR, Ausubel FM. Induction of Arabidopsis defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. Plant Cell. 1991;3:61–72. doi: 10.1105/tpc.3.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Luckow MA. The rest of the iceberg. Legume diversity and evolution in a phylogenetic context. Plant Phys. 2003;131:900–910. doi: 10.1104/pp.102.018150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Eldridge MDB, King JM, Loupis AK, Spencer PBS, Taylor AC, Pope LC, Hall GP. Unprecedented low levels of genetic variation and inbreeding depression in an island population of the black-footed rock-wallaby. Conserv Biol. 1999;13:531–541. [Google Scholar]
- Emerson BC. Evolution on oceanic islands: molecular phylogenetic approaches to understanding pattern and process. Mol Ecol. 2002;11:951–966. doi: 10.1046/j.1365-294x.2002.01507.x. [DOI] [PubMed] [Google Scholar]
- Filatov DA. Processing and population genetic analysis of multigenic datasets with ProSeq3 software. Bioinformatics. 2009;25:3189–3190. doi: 10.1093/bioinformatics/btp572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov DA, Burke S. DNA diversity in Hawaiian endemic plant Schiedea globosa. Heredity. 2004;92:452–458. doi: 10.1038/sj.hdy.6800440. [DOI] [PubMed] [Google Scholar]
- Fleischer RC, McIntosh CE, Tarr CL. Evolution on a volcanic conveyor belt: using phylogeographic reconstructions and K-Ar-based ages of the Hawaiian Islands to estimate molecular evolutionary rates. Mol Ecol. 1998;7:533–545. doi: 10.1046/j.1365-294x.1998.00364.x. [DOI] [PubMed] [Google Scholar]
- Fowler MR, Gartland J, Norton W, Slater A, Elliott MC, Scott NW. RS2: a sugar beet gene related to the latex allergen Hev b 5 family. J Exp Bot. 2000;51:2125–2126. doi: 10.1093/jexbot/51.353.2125. [DOI] [PubMed] [Google Scholar]
- Frankham R. Do island populations have less genetic variation than mainland populations? Heredity. 1997;78:311–327. doi: 10.1038/hdy.1997.46. [DOI] [PubMed] [Google Scholar]
- Gavrilets S, Losos JB. Adaptive radiation: contrasting theory with data. Science. 2009;323:732–737. doi: 10.1126/science.1157966. [DOI] [PubMed] [Google Scholar]
- Gossmann TI, Song BH, Windsor AJ, Mitchell-Olds T, Dixon CJ, Kapralov MV, Filatov DA, Eyre-Walker A. Genome wide analyses reveal little evidence for adaptive evolution in many plant species. Mol Biol Evol. 2010;27:1822–1832. doi: 10.1093/molbev/msq079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Rodrigo AG, Dyer KA, Huelsenbeck JP. Modeling the site-specific variation of selection patterns along lineages. Proc Natl Acad Sci U S A. 2004;101:12957–12962. doi: 10.1073/pnas.0402177101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane JBS. The cost of natural selection. Genetics. 1957;55:511–524. [Google Scholar]
- Hamzeh M, Dayanandan S. Phylogeny of Populus (Salicaceae) based on nucleotide sequences of chloroplast TRNT-TRNF region and nuclear rDNA. Am J Bot. 2004;91:1398–1408. doi: 10.3732/ajb.91.9.1398. [DOI] [PubMed] [Google Scholar]
- Hochberg Y. A sharper Bonferroni procedure for multiple tests of significance. Biometrika. 1988;75:800–802. [Google Scholar]
- Hoekstra HE, Coyne JA. The locus of evolution: evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- Hollocher H, Williamson M. Island hopping in Drosophila: patterns and processes. Philos Trans R Soc Lond B Biol Sci. 1996;351:735–743. doi: 10.1098/rstb.1996.0068. [DOI] [PubMed] [Google Scholar]
- Jansson S. A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 1999;4:236–240. doi: 10.1016/s1360-1385(99)01419-3. [DOI] [PubMed] [Google Scholar]
- Jobb G, von Haeseler A, Strimmer K. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kane NC, Barker MS, Zhan SH, Rieseberg LH. Molecular evolution across the Asteraceae: micro- and macroevolutionary processes. Mol Biol Evol. 2011;28:3225–3235. doi: 10.1093/molbev/msr166. [DOI] [PubMed] [Google Scholar]
- Kapralov MV, Filatov DA. Molecular adaptation during adaptive radiation in the Hawaiian endemic genus Schiedea. PLoS One. 2006;1:e8. doi: 10.1371/journal.pone.0000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapralov MV, Filatov DA. Widespread positive selection in the photosynthetic Rubisco enzyme. BMC Evol Biol. 2007;7:73. doi: 10.1186/1471-2148-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapralov MV, Stift M, Filatov DA. Evolution of genome size in Hawaiian endemic genus Schiedea (Caryophyllaceae) Tropical Plant Biol. 2009;2:77–83. [Google Scholar]
- Kosiol C, Vinar T, da Fonseca RR, Hubisz MJ, Bustamante CD, Nielsen R, Siepel A. Patterns of positive selection in six mammalian genomes. PLoS Genet. 2008;4:e1000144. doi: 10.1371/journal.pgen.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. (13 co-authors) Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Larracuente AM, Sackton TB, Greenberg AJ, Wong A, Singh ND, Sturgill D, Zhang Y, Oliver B, Clark AG. Evolution of protein-coding genes in Drosophila. Trends Genet. 2008;24:114–123. doi: 10.1016/j.tig.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Markova-Raina P, Petrov D. High sensitivity to aligner and high rate of false positives in the estimates of positive selection in the 12 Drosophila genomes. Genome Res. 2011;21:863–874. doi: 10.1101/gr.115949.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massingham T, Goldman N. Detecting amino acid sites under positive selection and purifying selection. Genetics. 2005;169:1753–1762. doi: 10.1534/genetics.104.032144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrone O, Aagesen L, Scataglini MA, Salariato DL, Denham SS, Chemisquy MA, Sede SM, Giussani LM, Kellogg EA, Zuloaga FO. Phylogeny of the Paniceae (Poaceae: Panicoideae): integrating plastid DNA sequences and morphology into a new classification. Cladistics. 2011;28:333–356. doi: 10.1111/j.1096-0031.2011.00384.x. [DOI] [PubMed] [Google Scholar]
- Nicolosi E, Deng ZN, Gentile A, La Malfa S, Continella G, Tribulato E. Citrus phylogeny and genetic origin of important species as investigated by molecular markers. Theor Appl Genet. 2000;100:1155–1166. [Google Scholar]
- Nunney L. The cost of natural selection revisited. Ann Zool Fennici. 2003;40:185–194. [Google Scholar]
- Ohta T. Near-neutrality in evolution of genes and gene regulation. Proc Natl Acad Sci U S A. 2002;99:16134–16137. doi: 10.1073/pnas.252626899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panero JL, Funk VA. The value of sampling anomalous taxa in phylogenetic studies: major clades of the Asteraceae revealed. Mol Phylogen Evol. 2008;47:757–782. doi: 10.1016/j.ympev.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Petersen L, Bollback JP, Dimmic M, Hubisz M, Nielsen R. Genes under positive selection in Escherichia coli. Genome Res. 2007;17:1336–1343. doi: 10.1101/gr.6254707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Pickrell JK, Coop G. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr Biol. 2010;20:R208–R215. doi: 10.1016/j.cub.2009.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington DL, Purugganan MD. GAI homologues in the Hawaiian silversword alliance (Asteraceae-Madiinae): molecular evolution of growth regulators in a rapidly diversifying plant lineage. Mol Biol Evol. 2002;19:1563–1574. doi: 10.1093/oxfordjournals.molbev.a004218. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Schluter D. The ecology of adaptive radiation. Oxford: Oxford University Press; 2000. [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Schneider J, Doring E, Hilu KW, Roser M. Phylogenetic structure of the grass subfamily Pooideae based on comparison of plastid matK gene-3'trnK exon and nuclear ITS sequences. Taxon. 2009;58:405–424. [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Nielsen R, Yang Q. Pervasive adaptive evolution in mammalian fertilization proteins. Mol Biol Evol. 2003;20:18–20. doi: 10.1093/oxfordjournals.molbev.a004233. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Turner JR, Williamson MH. Population size, natural selection and the genetic load. Nature. 1968;218:700. doi: 10.1038/218700a0. [DOI] [PubMed] [Google Scholar]
- Wagner WL, Funk VA, editors. Hawaiian biogeography. evolution on a hot spot archipelago. Washington: Smithsonian Institute Press; 1995. [Google Scholar]
- Wagner WL, Weller SG, Sakai A. Monograph of Schiedea (Caryophyllaceae subfam. Alsinoideae) Syst Bot Monogr. 2005;72:1–169. [Google Scholar]
- Wallace AR. Island life, or, the phenomena and causes of insular faunas and floras, including a revision and attempted solution of the problem of geological climates. London: Harper; 1881. [Google Scholar]
- Wallace LE, Weller SG, Wagner WL, Sakai AK, Nepokroeff M. Phylogeographic patterns and demographic history of Schiedea globosa (Caryophyllaceae) on the Hawaiian Islands. Am J Bot. 2009;96:958–967. doi: 10.3732/ajb.0800243. [DOI] [PubMed] [Google Scholar]
- Wang R-L, Stec A, Hey J, Lukens L, Doebley J. The limits of selection during maize domestication. Nature. 1999;398:236–239. doi: 10.1038/18435. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang W-Z, Song L-F, Zou J-J, Su Z, Wu W-H. Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Phys. 2008;148:1201–1211. doi: 10.1104/pp.108.126375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weese TL, Bohs L. A three-gene phylogeny of the genus Solanum (Solanaceae) Syst Bot. 2007;32:445–463. [Google Scholar]
- White S, Doebley J. Of genes and genomes and the origin of maize. Trends Genet. 1998;14:327–332. doi: 10.1016/s0168-9525(98)01524-8. [DOI] [PubMed] [Google Scholar]
- Willyard A, Wallace LE, Wagner WL, Weller SG, Sakai AK, Nepokroeff M. Estimating the species tree for Hawaiian Schiedea (Caryophyllaceae) from multiple loci in the presence of reticulate evolution. Mol Phylogen Evol. 2011;60:29–48. doi: 10.1016/j.ympev.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.