Abstract
The fetal Alz-50 reactive clone 1 (FAC1) protein exhibits altered expression and subcellular localization during neuronal development and neurodegenerative diseases such as Alzheimer's disease. Using the yeast two-hybrid screen, the human orthologue of Keap1 (hKeap1) was identified as a FAC1 interacting protein. Keap1 is an important regulator of the oxidative stress response pathway through its interaction with the Nrf family of transcription factors. An interaction between full-length FAC1 and hKeap1 proteins has been demonstrated, and the FAC1 binding domain of hKeap1 has been identified as the Kelch repeats. In addition, FAC1 colocalizes with endogenous Keap1 within the cytoplasm of PT67 cells. Exogenously introduced eGFP:hKeap1 fusion protein redistributed FAC1 to colocalize with eGFP: hKeap1 in perinuclear, spherical structures. The interaction between FAC1 and hKeap1 is reduced by competition with the Nrf2 protein. However, competition by Nrf2 for hKeap1 is reduced by diethylmaleate (DEM), a known disrupter of the Nrf2:Keap1 interaction. DEM does not affect the ability of FAC1 to bind hKeap1 in our assay. These results suggest that hKeap1 regulates FAC1 in addition to its known role in control of Nrf2. Furthermore, the observed competition between FAC1 and Nrf2 for binding hKeap1 indicates that the interplay between these three proteins has important implications for neuronal response to oxidative stress.
Fetal ALZ-50 Reactive Clone 1 (FAC1)1 is a transcriptional regulator with enhanced expression patterns in both developing and degenerating neurons (1– 4). FAC1 transcript and protein levels have been shown to be much higher in the developing fetal brain compared to the adult brain (5). Similarly, FAC1 is elevated in response to nerve growth factor induced differentiation of PC12 cells (6). FAC1 protein expression is enhanced during several neurodegenerative diseases. For instance, FAC1 protein is elevated in affected brain regions of patients with Alzheimer's disease (AD) (4). Furthermore, β-amyloid treatment of PC12 cells, an in vitro model of AD, increases FAC1 transcript and protein levels analogously to that observed in AD (7).
Changes in FAC1 protein levels are accompanied by altered subcellular localization in both development and disease. During cortical development FAC1 is localized to the cytoplasm and elongating processes of immature, migrating neurons (5); however, FAC1 is predominantly nuclear in mature, differentiated neurons and in the adult brain (5). In AD, FAC1 localizes to dystrophic neurites associated with subsets of β-amyloid-containing plaques, a major hallmark of the disease (4). FAC1 is also found in the nucleus and cytoplasm of motor neurons and white matter tracts in the spinal cords of victims of amyotrophic lateral sclerosis (ALS) (3). These data suggest that FAC1 subcellular localization is altered in response to degenerative stimuli and that this alteration mirrors changes seen in developing neurons.
Such changes in FAC1 subcellular localization would be expected to affect the function of FAC1 as a transcriptional regulator (8). FAC1 has been shown to recognize the DNA sequence CACAACAC and repress transcription through this site in a phosphorylation-dependent manner (1, 8). The presence of this DNA element in the regulatory regions of the amyloid precursor protein, presenilin 1, and Cu/Zn superoxide dismutase 1 suggests that FAC1 may regulate expression of these genes, which have been implicated in neurodegeneration (9– 11). Because nuclear FAC1 has been shown to repress transcription, removal of FAC1 from the nucleus in response to neurodegenerative or developmental stimuli would be expected to alter FAC1-regulated gene expression.
Because subcellular distribution is often regulated by protein:protein interaction, we sought to identify FAC1-interacting proteins using the dihybrid yeast screen to gain insight into the mechanisms determining the alterations in FAC1 localization seen in disease and development. This approach yielded several previously identified nuclear proteins that bind FAC1 and alter FAC1 transcriptional regulation, including the Myc-associated zinc finger protein as we have previously reported (12). Here we report the identification of the human orthologue [KIAA0132 (13)] of the murine Kelch-like Ech-associated protein (Keap1) (14) as a FAC1 binding protein. Keap1 has been shown to regulate the subcellular distribution of another transcription factor, Nrf2 (14). In response to oxidative damage, Keap1 releases Nrf2, which translocates to the nucleus where it activates promoters containing the antioxidant response element (ARE) (15, 16). Thus, Keap1 and hKeap1 are strong candidates for regulation of FAC1 subcellular localization. Here we demonstrate that FAC1 interacts with hKeap1 through a similar domain as Nrf2 and that Nrf2 and FAC1 compete for hKeap1 binding. We also find that FAC1 colocalizes with Keap1 in the cytoplasm and that increased expression of hKeap1 redistributes FAC1 protein. Taken together, these findings suggest a role for hKeap1 in regulating FAC1 function and subcellular localization.
Experimental Procedures
Two-Hybrid Yeast Screen
(a) Strains and Media
The strain of Saccharomyces cerevisiae used for the two-hybrid yeast screen was previously described by Vojtek et al. (17). The reporter strain used was L40 [MATa,trp1,leu2,LYS2∷lexA-HIS3,URA3∷lexA-lacZ (Sternglanz, Weintraub, and Hollenburg, unpublished data)]. Yeast cells were grown in rich medium YPDA (1% yeast extract, 2% bactopeptone, 2% glucose, and 0.1 μg/mL adenine) or in synthetic medium lacking amino acids for which the yeast cells are auxotrophic due to the presence of a particular plasmid marker.
(b) Plasmids
The LexA:FAC1(438–810) plasmid encoded amino acids 438–810 of the human FAC1 cDNA. This sequence was cloned in frame with the LexA coding sequence in plasmid pBTM116 (18, 19). Amino acids 438–810 were used as a fusion with the LexA binding domain as bait because this fusion did not activate expression of the HIS3 or LACZ reporter genes alone or in the presence of the library vector alone. Unfortunately, fusion between LexA and residues 1–437 of FAC1 activated HIS3 and LACZ expression in the absence of library vector, making it inappropriate for use in this screen. For prey, a human fetal brain cDNA library was constructed in the plasmid pACT2 (Clontech). pACT2 is a high-copy yeast vector that contains the coding region for the transcriptional activation domain of the herpes virus VP16 transactivator as well as the leu2 marker gene coding for a gene product needed for leucine biosynthesis (17).
(c) Library Screen
The screen was performed as previously described by Jordan et al. (22). L40 cells were transformed with LexA:FAC1(438–810). The plasmid was maintained by auxotrophy for the TRP1 marker. L40 containing LexA:FAC1(438–810) was then prepared for electroporation of the library. Two hundred micrograms of human fetal brain cDNA library cloned into the pACT2 plasmid was electroporated into 50 μL aliquots of L40 containing pLexA-FAC1(438–810). Electroporated cells were plated onto media lacking Ura, Lys, Trp, Leu, and His. Growth on His(−) plates indicates an interaction between FAC1(438–810) and the protein coded by the unknown cDNA. Colonies were then assayed for β-galactosidase activity to ensure that the interaction between FAC1(438– 810) and the protein coded by the unknown cDNA was not specific for the heterologous promoter driving His3 expression. The pACT2 plasmid containing the unknown cDNA was then isolated by the “smash and grab” method (17), and the cDNA insert was sequenced.
GST Fusion Affinity Column Chromatography
cDNAs coding for FAC1(438–810), FAC1(610–810), FAC1(401– 500), and FAC1(501–610) were cloned into pGEX-5x-1 (Pharmacia) in frame with the glutathione-S-transferase gene (GST). Fusion proteins were produced as described in Jordan et al. (20). Briefly, the proteins were induced with IPTG for 3 h, bacteria were lysed, and protein was separated from cellular debris by centrifugation. Using the bacterial extracts, the GST fusion proteins were purified by the specific interaction between GST and glutathione immobilized on Sepharose beads (Pharmacia). At this point the protein: Sepharose column was used as an affinity column for the fusion protein it contained.
In Vitro Transcription/Translation
In vitro transcription reactions were performed by cloning hKeap1 into the Hind III Xba I site of pcDNA3.1+, which contains a T7 promoter at the 5′ end. An initiator methionine sequence was cloned into the Hind III site for translation initiation. pcDNA3.1+ hKeap1 was transcribed and translated in the presence of [35S]methionine (NEN DuPont) using the TNT T7 Coupled Reticulocyte Lysate in vitro Transcription/Translation System (Promega). [35S]Methionine at 0.9 mCi/mL was used for these reactions.
GST-Pulldown Assays
In vitro transcribed and translated protein was added to GST affinity columns in binding buffer [150 mM NaCl, 20 mM Tris (pH 8), 1 mM EDTA (pH 8.0), 125 mM PMSF, 2 μg/mL pepstatin A, 1 μg/mL benzamidine, and 1 μg/mL leupeptin]. The affinity columns were then washed five times with 10 bed volumes of NTEN [150 mM NaCl, 20 mM Tris (pH 8), 1 mM EDTA, 0.5% NP40, 125 μM PMSF, 2 μg/mL pepstatin A, and 1 μg/mL leupeptin]. Proteins retained on the column were eluted by boiling the columns in SDS–PAGE sample buffer. The eluted proteins were then electrophoresed on a 10% SDS-polyacrylamide gel. For the in vitro translated proteins, the gels were dried and exposed to X-ray film.
Transfection/Cell Lines
The murine fibroblast cell line, PT67, was maintained at 37 °C under 5% CO2 using Dulbecco's modified Eagles' media (DMEM) supplemented with 10% fetal calf serum, 10 units/mL penicillin, 10 μg/mL streptomycin, and 2 mM L-glutamate. All transfections were done using the Fugene 6 transfection reagent (Roche) following the manufacturer's specifications. The day before transfection, cells were split to 20% confluence. Equal quantities of DNA were used in each transfection with a total of 6 μg of DNA for each 10 cm plate of cells. The cells were allowed to grow for 48 h following transfection and either harvested for immunoblotting or fixed for immunofluorescent staining.
Protein Extracts and Immunoblotting
Transfected PT67 cell protein extracts were prepared by detergent lysis. Cells were scraped in PBS and pelleted at 1000 rpm in a clinical centrifuge for 5 min. Pellets were resuspended in detergent lysis buffer [0.1% NP-40, 10 mM Tris (pH 8.0), 10 mM MgCl2, 15 mM NaCl, 0.5 mM PMSF, 2 μg/mL pepstatin A, and 1 μg/mL leupeptin] and incubated on ice for 15 min. Debris was removed by centrifugation at 1400g for 5 min. The supernatant was saved and assayed for protein concentration via the Bio-Rad protein assay. Equal amounts of protein were loaded onto a 4–12% NuPAGE gradient gel (Invitrogen) and fractionated by size via electrophoresis for 1 h at 100 V.
The proteins were transferred from the NuPAGE gel to PVDF by electrophoresis and blocked in 2% bovine serum albumin in TBS [10 mM Tris (pH 8.0), 150 mM NaCl]. Monoclonal M2 antibody (Kodak, IBI) and the polyclonal Keap1 E20 antibody (Santacruz) were used at 1:1000 in TBS with 0.1% Tween 20 (TBST) overnight at 4 °C. The blot was washed three times in TBS for 15 min. M2 antibody was detected with goat anti-mouse IgG–HRP secondary antibody (1:500, Jackson Laboratories), and Keap1 antibody was bound by donkey anti-goat–HRP (1:500, Jackson laboratories). The secondary antibody was washed in TBST three times for 20 min. The antibody was then visualized using enhanced chemiluminescence (ECL) (Renaissance, NEN Life Science Products, Inc.).
Immunofluorescent Laser Confocal Microscopy
Fixed cells on coverslips were permeabilized and blocked in 0.1% Triton X-100 and 0.2% bovine serum albumin in PBS. After washing, the cells were incubated at 4 °C overnight with biotinylated M2 antibody (Sigma), which recognizes the FLAG epitope tag on the amino terminus of exogenously expressed FAC1 diluted 1:100 in normal antibody diluent (Scytek, Logan, UT). Cells were washed three times in PBS 0.1% Tween-20 (PBST) and incubated with strepavidin-Cy5 (1:400) in blocking buffer (Tyramide Signal Amplification System, Perkin-Elmer). When indicated, endogenous Keap1 was detected by incubation with the anti-Keap1 polyclonal antibody (E-20, Santa Cruz). Cells were washed three times in PBST and incubated with donkey anti-goat–FITC (1:200, Jackson Immunologicals). Cells were then incubated with phalloidin conjugated to TRITC (Sigma) at 2 μg/mL and 10 μM DAPI for 30 min. Cells were washed three times with PBST, mounted on slides with gelvatol (21), and analyzed by laser confocal microscopy using a four-laser 2100 Radiance (Bio-Rad). This confocal microscope has an argon laser exciting at 488 for FITC, a green He/Ne laser exciting at 543 for TRITC, a red diode laser exciting at 637 for Cy5, and a blue diode laser exciting at 405 for DAPI. The emission filter used for FITC labeling and GFP detection was 515 ± 30, the filter used for TRITC was 590 ± 70, the filter used for Cy5 was 660 LP, and the filter used for DAPI was 476 ± 48.
Colocalization of each fluor was determined using image analysis software (Metamorph 6.1; Universal Imaging Corp.). Each image was segmented by selecting pixels above a constant threshold with intensity values indicative of true fluorescence (intensity values = 158–255; >60% of background). The integrated intensity of pixels within the region of overlap is expressed as a percentage of pixels that had the same brightness value and spatial location.
Results
FAC1 Interacts with hKeap1 in Vivo As Assayed by the Dihybrid Yeast Screen
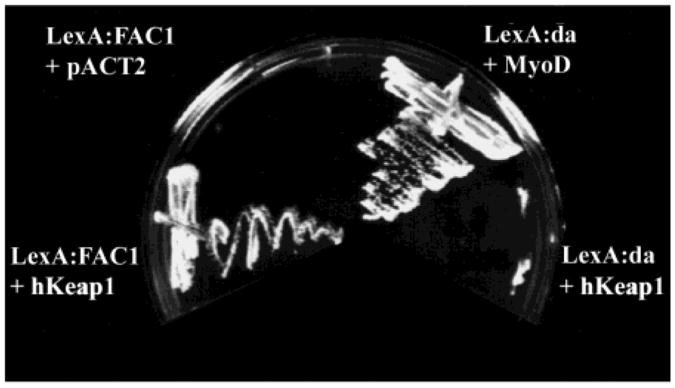
Because both transcriptional regulation and changes in subcellular localization are often regulated through protein:protein interaction, we hypothesized that FAC1 function was regulated by interaction with other cellular proteins. A two-hybrid yeast screen was used to identify proteins that may function in such a capacity(12, 17, 22). Amino acids 438–810 of FAC1 were fused to the LexA DNA binding domain for use as the “bait”. Amino acids 1–437 were removed as these residues resulted in activation of the reporter genes in the absence of the cDNA library. A human fetal brain cDNA library cloned in frame with the VP16 acidic activation domain was used as the “prey”. Interaction between FAC1 and a protein of interest reconstitutes a functional transcriptional activator by bringing the acidic domain and DNA binding domain into the same vicinity of promoters containing the LexA binding site. Such sites are located in the promoter regions of the HIS3 gene, which encodes a protein required for histidine synthesis, and the LacZ gene, which converts Xgal to a blue metabolite. As a control, FAC1(438–810) was introduced into yeast with the acidic domain alone to rule out interaction between the bait and the VP16 activation domain. This did not produce histidine-synthesizing yeast (Figure 1, Lex:FAC1 +pACT2), suggesting it would be appropriate for use in the screen.
Figure 1.
FAC1 interacts with hKeap1 in vivo as assayed by the dihybrid yeast screen. Four cultures of the L40 yeast strain were transformed with (1) pLexA:FAC1(438–810) and hKeap1 (amino acids 42–624 as identified in the screen), (2) pLexA:FAC1 and pACT2 (library vector only), (3) LexA:da and MyoD (positive control for functioning interactions), or (4) LexA:da and hKeap1 (control for nonspecific interaction with LexA or an unrelated protein). Colonies were streaked on plates lacking the amino acid histidine on which only yeast containing interacting fusion proteins will grow.
Of the 8 × 105 colonies screened, 88 were positive for histidine synthesis, suggesting an interaction between FAC1-(438–810) and the protein produced by the cDNA present in the library vector. A secondary screen for LacZ expression yielded 44 LACZ positive clones for further consideration. True interacting proteins were further delineated by isolation of the cDNA library vector, amplification and purification of the plasmid in bacteria, followed by fresh transformation with LexA:FAC1(438–810) into the L40 yeast strain. The library vectors were also introduced by themselves into yeast to ensure that the proteins they encoded were not activating transcription by directly binding the promoter, bypassing the need for interaction with the LexA:FAC1(438–810) fusion protein (data not shown). Of the 44 remaining clones only 11 retained the ability to induce growth on histidine-deficient plates. These 11 clones were sequenced, compared to Genbank, and found to contain five unique sequences. One of these was the myc-associated zinc finger protein, ZF87/MAZ, which we have previously reported (12). Three others await further investigation. The final clone (isolated twice) encoded amino acids 42–624 of the predicted sequence for the EST KIAA0132 (13). This human cDNA shows 94% identity with a recently isolated murine protein Keap1 (14). We will refer to the identified clone (KIAA1032) as hKeap1 throughout the paper to differentiate between it and the murine orthologue (Keap1). Figure 1 demonstrates that yeast expressing both LexA:FAC1(438–810) and VP16:hKeap1 fusion proteins grow on plates lacking histidine, suggesting that interaction between FAC1 and hKeap1 brings the VP16 activation domain into the vicinity of the LexA DNA binding domain and induces expression of the HIS3 gene product (Figure 1, Lex:FAC1 + hKeap1). LexA:FAC1(438–810) was introduced with pACT2, which expresses the VP16 activation domain (VP16) to show that the interaction requires the presence of the hKeap1 polypeptide. To demonstrate that the hKeap1 is not binding to the LexA DNA binding domain and is specific for FAC1, VP16:hKeap1 was cotransformed with a plasmid expressing a LexA:daughter-less fusion protein (Figure 1, Lex:da + hKeap1). The failure of yeast expressing Lex:da and hKeap1 fusion proteins to grow on histidine-lacking plates suggests that hKeap1 does not interact with LexA or daughterless protein. Finally, as a positive control LexA:da was cotransformed with a plasmid expressing a known daughterless interacting protein, MyoD: VP16 (Figure 1, Lex:da + MyoD). Growth of yeast expressing LexA:da and MyoD:VP16 on histidine-deficient plates indicates an interaction between these two proteins. These data suggest that the carboxy-terminal domain of FAC1 interacts with the hKeap1 protein when coexpressed in yeast.
The hKeap1 protein is a 624 amino acid protein encoding two domains of known function: a BTB/POZ domain and double-glycine repeats (DGR, also known as the Kelch-like domain). Both of these domains are characteristic of the drosophila Kelch protein, which has numerous homologues including c-elegans spe-26, vaccinia virus A55R, and several human genes: ENC1, Mayven, and NRP/B (23– 27). The BTB/POZ domain is so named because it is contained in the drosophila genes broad complex, tramtrack, and bric á brac (BTB) and several pox virus genes that have zinc fingers (POZ) (28– 30). This domain is believed to function as a protein:protein interaction domain and is found in combination with two other classic protein motifs: zinc finger DNA binding motifs or Kelch-like domains (28– 30). BTB/POZ containing proteins with a DNA binding domain would be expected to function in the nucleus to regulate gene expression.
Expression of FAC1 mRNA and protein has been found in multiple tissues with the highest levels being in kidney, muscle, and liver (5, 12). By Northern analysis, the ∼3000 base pair hKeap1 transcript was present in a variety of tissues with highest levels in skeletal muscle (data not shown). The wide range of tissue expression for hKeap1 mRNA is similar to that reported for the mouse orthologue Keap1 (14).
hKeap1 Interacts with a Region of FAC1 Containing a Putative PEST Domain
To demonstrate a specific interaction between FAC1 and hKeap1, we used a panel of FAC1 mutants in a GST pull-down assay. In vitro translated (IVT) hKeap1 (amino acids 42–624) was incubated with affinity columns containing glutathione:Sephadex beads and either GST or GST:FAC1 fusion proteins (Figure 2A). The GST: FAC1 fusion proteins were a panel of deletion mutants containing specific functional domains of FAC1 to identify motifs involved in the interaction with hKeap1. The same amount of IVT hKeap1 was loaded onto each column, and an aliquot was saved and loaded in the first lane of the gel (Figure 2B, lane 1). Although no detectable IVT hKeap1 bound to GST alone (Figure 2B, lane 2), significant binding to GST:FAC1 (438–810) (Figure 2B, lane 3) was observed, confirming the results of the two-hybrid screen. GST:FAC1-(501–610) (Figure 2B, lane 6), which contains a putative PEST domain, was sufficient for binding hKeap1 as well as or better than the larger domain used in the original dihybrid screen (438–810). IVT hKeap1 does not significantly interact with either GST:FAC1(611–810), which contains putative nuclear localization and nuclear export sequences, or GST:FAC1(401–500) (Figure 2B, lanes 4 and 5) within this batch column experiment. Equal amounts of fusion protein were present on each of the columns as verified by Ponceau S staining (data not shown). These data suggest that hKeap1 interacts with FAC1 in vitro specifically through a domain containing the putative PEST domain.
Figure 2.
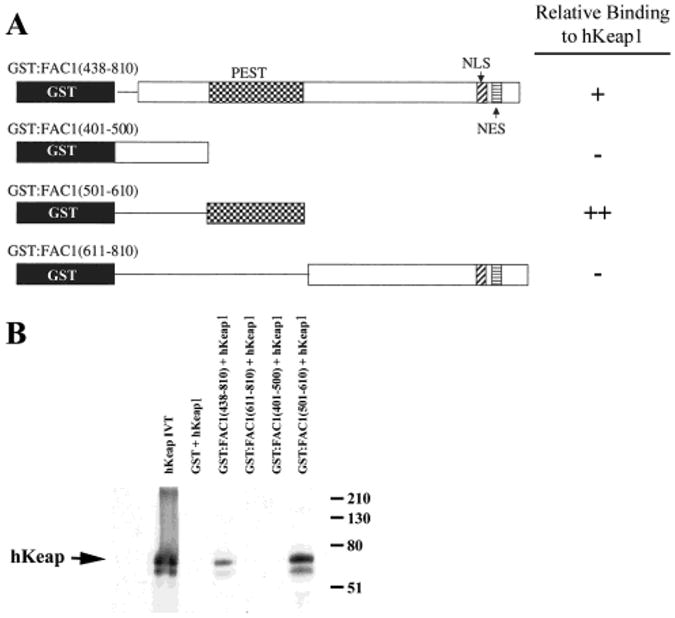
hKeap1 interacts with a region of FAC1 containing a putative PEST domain. (A) GST:FAC1 fusion proteins shown here were used to make affinity columns. GST is shown in black, whereas the remainder contains the indicated FAC1 domains. FAC1 deletion mutants containing the putative PEST domain, nuclear localization sequence (NLS), and a nuclear export sequence (NES) are shown. Relative binding of each FAC1 domain to hKeap1 is indicated on the right. In (B) 35S-labeled, in vitro translated hKeap1 (42–624) (hKeap1; lane 1) was incubated with the following affinity columns: GST (lane 2), GST:FAC1(438–810) (lane 3), GST:FAC1(611–810) (lane 4), GST:FAC1(401–500) (lane 5), and GST:FAC1(501–610). Column-bound proteins were fractionated by size on a 10% SDS–polyacrylamide gel, which was transferred to PVDF. The autoradiograph of the transferred gel is shown in (B).
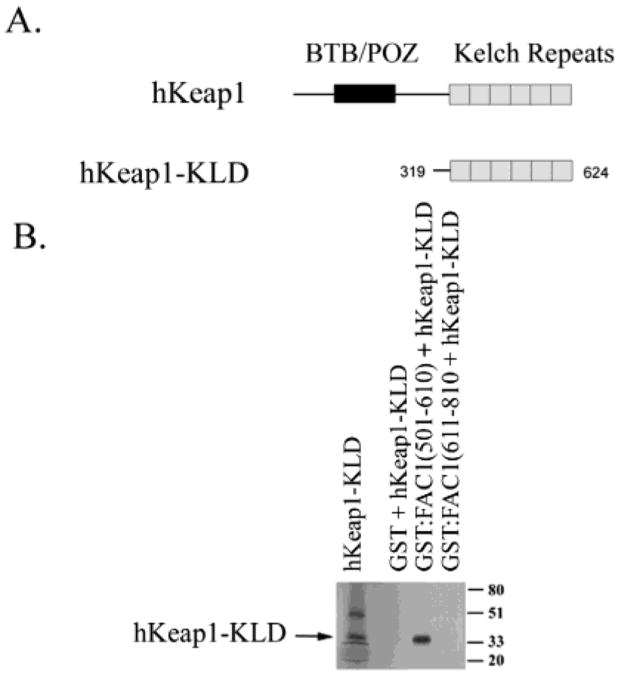
FAC1 Binds the Kelch-like Domain of hKeap1
The region of hKeap1 important for binding the PEST domain of FAC1 was also determined by GST pull-down experiments. There are two conserved domains with hKeap1, the BTB/POZ domain (Figure 3A), which is known to be involved in dimerization of Keap1, and the DGR, Kelch repeats within the C terminus of hKeap1, which are protein-binding domains required for Keap1 binding to actin cytoskeleton (31, 32). The carboxy-terminal half of hKeap1 lacking the BTB/POZ domain (hKeap1-KLD) was sufficient for binding to GST:FAC1(501–610) but not to GST:FAC1(611–810) (Figure 3B). These results suggest that the PEST domain of FAC1 binds directly to the Kelch repeats of hKeap1.
Figure 3.
FAC1 binds the Kelch-like domain of hKeap1. (A) hKeap1 has two conserved motifs, the BTB/POZ domain and the Kelch-like repeat domain (KLD). In (B) 35S-labeled, in vitro translated hKeap1-KLD (lane 1) was incubated with the following affinity columns: GST (lane 2), GST:FAC1(501–610) (lane 3), and GST:FAC1(611–810) (lane 4). Column-bound proteins were fractionated by size on a 10% SDS–polyacrylamide gel, which was transferred to PVDF. The autoradiograph of the transferred gel is shown in (B).
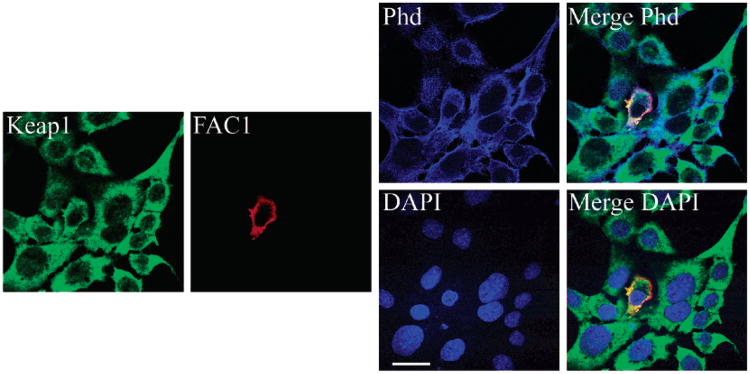
FAC1 Colocalizes with Endogenous Keap1 and Actin in Mouse Fibroblasts
The Kelch-like domain, present in Keap1 and hKeap1, mediates the interaction between many DGR-containing proteins and actin microfilaments. Because FAC1 also binds to the DGR domain, we assessed FAC1 and Keap1 association with actin. Using quadruple-label immunofluorescent confocal microscopy, we assessed localization of transfected, epitope-tagged FAC1 with endogenous Keap1, filamentous-actin (f-actin), and nuclei in PT67 murine fibroblasts. We used the PT67 cell line as it does not express FAC1 (1), but does express Keap1 (the murine orthologue of hKeap1). Forty-eight hours after transfection, cells were fixed and immunostained for exogenous FAC1 using M2 antibody specific for the FLAG epitope and endogenous Keap1 using anti-Keap1 antibody (E-20; Santa Cruz Biotechnology). Polymerized actin (f-actin) was visualized using TRITC-conjugated phalloidin (Phd), and nuclei were stained with DAPI. In untransfected cells, Keap1 exhibited colocalization with a subset of f-actin and was observed predominantly in the cytoplasm (Figure 4; Keap1, green; phalloidin, blue; Merge Phd). Quantification of the colocalization revealed that 55.3% of the f-actin colocalized with Keap1, whereas 69.9% of the Keap1 colocalizaed with f-actin. Immunostaining for transfected FAC1 colocalized with immunostaining for endogenous Keap1 (Figure 4; Keap1, green; FAC1, red; phalloidin, blue; three fluors together appear white in Merge Phd). Quantification of the colocalization between FAC1 and Keap1 demonstrated that 87.3% of FAC1 colocalized with Keap1 in the eFAC1 transfected cell, whereas only 71.6% of Keap1 colocalized with FAC1 in the FAC1 transfected cell. FAC1 and Keap1 further colocalized with a subset of f-actin (Figure 4, Merge Phd); 71.5% of FAC1 and 69.9% of Keap1 colocalized with f-actin. Neither FAC1 nor Keap1 was detectable within the nucleus (Figure 4; DAPI, blue; Merge DAPI; overlapping red and green fluors appear yellow and do not overlap with blue; <3% of Keap1 or FAC1 colocalized with DAPI). These results indicate that f-actin-associated Keap1 also interacts with FAC1.
Figure 4.
FAC1 colocalizes with endogenous Keap1 and actin in mouse fibroblasts. PT67 cells were transfected with epitope tagged FAC1. Quadruple-label immunofluorescent confocal microscopy for endogenous murine Keap1 (green), FAC1 (red), actin via phalloidin (Phd, blue), and nuclear DNA via DAPI (DAPI, blue) demonstrated colocalization between FAC1 and Keap1 (Merge Phd and Merge DAPI). FAC1 and Keap1 also colocalized with actin (Merge Phd; green, red, and blue colocalization appears white). However, neither FAC1 nor Keap1 colocalizes with DAPI (FAC1 and Keap1 appear yellow to orange, indicating colocalization of the red and green, but not the blue). Bar= 20 μM.
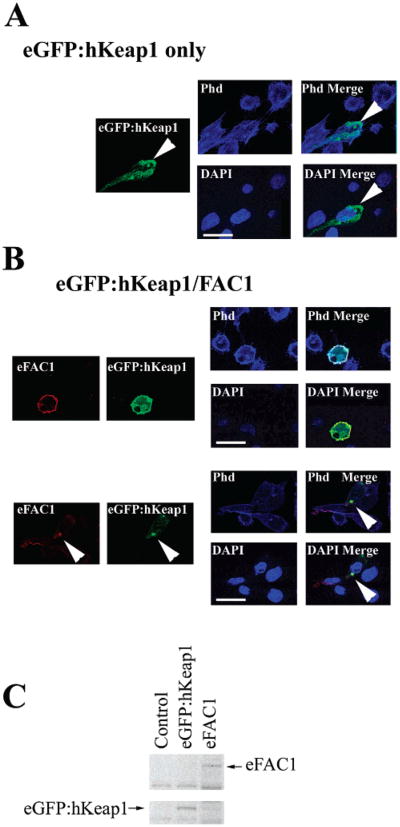
Coexpression of hKeap1 with FAC1 Redistributes FAC1 within the Cytoplasm
To further demonstrate that FAC1 colocalizes with the human Keap1 protein (hKeap1), we generated a plasmid that expressed hKeap1 fused to green fluorescent protein (eGFP:hKeap1). PT67 cells were transfected with eGFP:hKeap1, FAC1, or eGFP:hKeap1 and FAC1 together. Western blot analysis confirmed the expression of the exogenous proteins in PT67 (Figure 5C). Using immunofluorescent laser confocal microscopy, FAC1 alone exhibited the same staining observed in Figure 4. Overexpressed eGFP:hKeap1 localized to a fraction of f-actin as seen with endogenous Keap1 (Figure 5A). When quantified, we found 95.6% of the exogenously expressed eGFP:hKeap1 colocalized with 66.9% of the f-actin. However, eGFP: hKeap1 also exhibited a localization unique to cells overexpressing eGFP:hKeap1, which is observed as several large spherical structures adjacent to the nucleus (Figure 5A; eGFP: hKeap1 only; eGFP:hKeap1, green; arrowhead). By laser confocal microscopy, these structures were confirmed to be outside the nuclear compartment (Figure 5A; arrowhead; Merge DAPI, <0.07% colocalization between eGFP:hKeap1 and DAPI). Furthermore, the spherical structures did not colocalize with actin (Figure 5A: eGFP:hKeap1, green; phalloidin, blue; Merge Phd). In cells coexpressing FAC1 and eGFP:hKeap1, 99.2% of FAC1 colocalized with eGFP: hKeap1 by immunofluorescent laser confocal microscopy in the cytoplasm, where it colocalized with 95.5% of the f-actin (Figure 5B; eGFP:Keap1, green; FAC1, red; phalloidin, blue; Merge Phd) and in the perinuclear spherical structures (Figure 5B,C; eGFP:hKeap1, green; FAC1, red). Importantly, FAC1 does not localize to perinuclear spherical structures in the absence of excess hKeap1 (Figure 4), suggesting that it is the overexpression of hKeap1 that causes FAC1 to localize to these structures. These data support a role for hKeap1 in localization of FAC1 when it resides in the cytoplasm, suggesting that FAC1 cytoplasmic localization may be determined by its interaction with hKeap1 or its family members.
Figure 5.
Coexpression of hKeap1 with FAC1 redistributes FAC1 within the cytoplasm. PT67 murine fibroblasts were transfected with eGFP:hKeap1 alone (A) or with eGFP:hKeap1 and epitope tagged FAC1 (B). Transfected cells were stained for FAC1 by immunohistochemistry, actin by TRITC conjugated phalloidin, and DNA by DAPI and analyzed by quadruple-label immunofluorescent laser confocal microscopy. For panels A and B, FAC1 is shown in red, eGFP:hKeap1 is shown in green, actin is labeled with phalloidin and shown as blue in the Phd panel, and DAPI labeling nuclei are shown as blue in the DAPI panel. Panel B demonstrates FAC1 colocalization with eGFP:hKeap1. In panel C, western analysis of cells transfected with eGFP:hKeap1 (lane 2) and epitope-tagged FAC1 (eFAC1; lane 3) or untransfected (lane 1) demonstrates the levels of eGFP:hKeap1 and eFAC1 upon transfection. Bar = 20 μM.
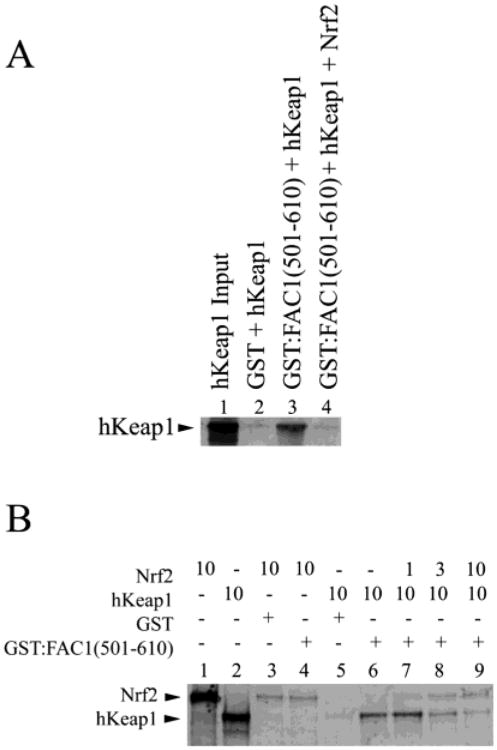
FAC1 Binding to hKeap1 Is Dramatically Reduced by Competition with NRF2
Like FAC1, the Nrf2 protein is retained in the cytoplasm by Keap1, and this repression of Nrf2 activity is mediated through a direct interaction with the Kelch domains of Keap1. Because the interaction between FAC1 and hKeap1 is mediated through the conserved Kelch repeat domain of hKeap1, we hypothesized that Nrf2 would compete with FAC1 for hKeap1 binding. To test this hypothesis, we assessed the ability of Nrf2 to compete for hKeap1 binding to GST:FAC1(501–610) in an in vitro GST pull-down assay. As shown for hKeap1(42– 624), there was significant binding between full-length hKeap1 and GST:FAC1(501–610) (Figure 6A, lane 3; Figure 6B, lane 6); however, the addition of Nrf2 (Figure 6A, lane 4) reduced the level of hKeap1 bound to FAC1 to background levels (Figure 6A, lanes 2 and 4). In vitro translated Nrf2 (Figure 6B, lane 1) does not bind directly to GST:FAC1 (501–610) above background binding to GST alone (Figure 6B, lanes 3 and 4). When increasing amounts of Nrf2 were added, hKeap1 binding to FAC1 was reduced in a dose-dependent fashion (Figure 6B, lanes 7–9). Equivalent amounts of Nrf2 outcompete FAC1 for hKeap1 binding even in the presence of excess FAC1 (Figure 6B, lane 9). The amount of Nrf2 retained on the GST:FAC1-(501–610) columns in the presence or absence of hKeap1 is similar to that observed with GST alone (Figure 6B, lanes 3 and 7–9) and therefore considered to be background. These findings suggest that Nrf2 has a higher affinity for hKeap1 than FAC1 under the conditions used in our assay. Furthermore, Nrf2 and FAC1 are potentially competitors for binding the Kelch domains of hKeap1.
Figure 6.
FAC1 binding to hKeap1 is dramatically reduced by competition with NRF2. In (A), 35S-labeled hKeap1 (lane 1) was incubated with GST (lane 2) or GST:FAC1(501–610) in the presence (lane 4) or absence (lane 3) of in vitro translated Nrf2. Bound proteins were electrophoresed on a 10% SDS–polyacryl-amide gel, transferred to PVDF, and visualized by autoradiography. In (B), 35S-labeled, in vitro translated Nrf2 (lane 1) was incubated with GST affinity column (lane 3) or GST:FAC1(501–610) (lane 4). Shown also is the 35S-labeled, in vitro translated hKeap1 (lane 2), which is in equal amounts to Nrf2 (lane 1). A constant amount of hKeap1 (10 μL) was added to each reaction indicated. GST and hKeap1 alone (lane 5) did not interact. GST:FAC1(501–610) bound hKeap1 (lane 6) as shown in Figure 2. However, when Nrf2 was added to the GST:FAC1(501–610) binding reaction (lanes 7–9), we saw a reduction in bound hKeap1 in a dose-dependent manner. Nrf2 retention on GST:FAC1(501–610) was the same as GST control (lanes 7–9 compared to lane 3). At equimolar amounts of hKeap1 and Nrf2, >90% of the hKeap1 no longer bound FAC1.
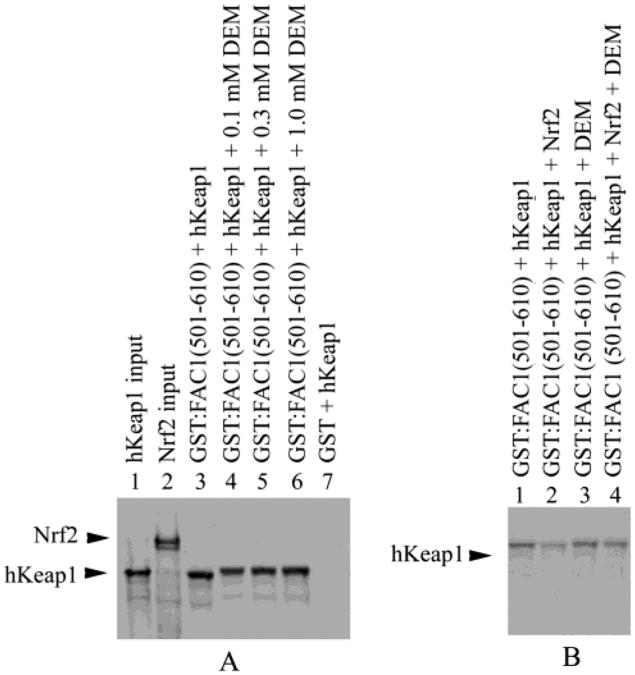
Nrf2 Does Not Outcompete FAC1 for hKeap1 as Efficiently in the Presence of Diethylmaleate (DEM)
In several models of oxidative stress, Nrf2 binding to hKeap1 is abrogated, at least in part, by oxidation of specific cysteine residues (46). To determine if disruption of the Nrf2:hKeap1 interaction by known chemical inducers alters the ability of Nrf2 to outcompete FAC1 for hKeap1 binding, we repeated our in vitro binding assays in the presence of DEM, a molecule known to disrupt Nrf2:Keap1 interaction. As above, hKeap1 bound GST:FAC1(501–610) (Figure 7A, lane 3). Addition of increasing concentrations of DEM did not reduce binding of hKeap1 to the GST:FAC1(501–610) (Figure 7A, lanes 4–6). DEM did, however, slightly alter the mobility of hKeap1 in the non-denaturing gel. As in Figure 6, hKeap1 did not show appreciable binding to GST column matrix alone (Figure 7A, lane 7). As in Figure 6A,B, addition of Nrf2 to the hKeap1 and GST:FAC1(501–610) reaction mixture markedly reduced hKeap1 binding to the GST:FAC1(501–610) (Figure 7B, lanes 1 and 2). Addition of DEM to the GST:FAC1(501–610) and hKeap1 binding reaction did not significantly alter retention of hKeap1 on the column as in Figure 7A (Figure 7B, lane 3). However, including DEM with the Nrf2, hKeap1, and GST:FAC1-(501–610) binding reaction reduced the ability of Nrf2 to compete for hKeap1 with FAC1(501–610) (Figure 7B, lane 4). These findings suggest that DEM interferes with the ability of Nrf2 to compete for hKeap1 binding without altering binding between hKeap1 and FAC1.
Figure 7.
Nrf2 does not outcompete FAC1 for hKeap1 as efficiently in the presence of diethylmaleate (DEM). In (A), 35S-labeled hKeap1 (lane 1) was incubated with GST (lane 7) or GST: FAC1(501–610) in the presence of increasing concentrations of DEM (lane 4–6) or in the absence of DEM (lane 3). Bound proteins were electrophoresed on a 10% SDS–polyacrylamide gel, transferred to PVDF, and visualized by autoradiography. Also shown is the in vitro translated 35S-labeled Nrf2 used in part B (lane 2). In (B), 35S-labeled, in vitro translated hKeap1 alone (lanes 1 and 3) or equal amounts of 35S-labled in vitro translated hKeap1 and Nrf2 (lanes 2 and 4) were incubated with GST:FAC1(501–610) in the absence (lanes 1 and 2) or presence of 0.2 μM DEM (lanes 3 and 4). GST:FAC1(501–610) bound hKeap1 (lane 1) as shown in Figures 2, 6A, and 7A. However, when Nrf2 was added to the GST:FAC1(501–610) binding reaction (lane 2), we saw a reduction in bound hKeap1 as seen in Figure 6. In the presence of 0.2 μM DEM, hKeap1 binding to GST:FAC1(501–610) was not altered (lane 3) as shown in Figure 7A. However, Nrf2 was not able to compete as efficiently with GST:FAC1(501–610) for hKeap1 binding in the presence of 0.2 μM DEM (lane 4).
Discussion
By employing the yeast two-hybrid assay to identify proteins that regulate FAC1 localization and activity, we have isolated a human orthologue of the Keap1 protein (hKeap1). Keap1 belongs to a growing family of proteins that are characterized by similar primary structure consisting of an NH2-terminal BTB/POZ dimerization domain followed by six Kelch repeats. We confirmed the interaction between FAC1 and hKeap1 in vitro and determined that the putative PEST domain of FAC1 is required for interaction with hKeap1. The PEST domain has been implicated in targeting proteins for proteosomal degradation. Because hKeap1 interacts directly with the FAC1 PEST containing domain, it is possible that hKeap1 regulates FAC1 proteins' stability. In support of this role, hKeap1 has been reported to regulate proteosome degradation of Nrf2 (15, 33). Further investigation is warranted to determine the functional significance of interactions between FAC1 and hKeap1.
We have further demonstrated that the Kelch-like domain of hKeap1 is sufficient for interaction with FAC1. Because this hKeap1 domain is also important for interaction with actin and Nrf2, it is plausible that this domain will localize FAC1 to actin-containing structures in the cytoplasm. Because the FAC1 protein is capable of regulating gene expression and is found in the nucleus of adult neurons, its localization to the cytoplasm would impact FAC1 transcriptional activity. This is interesting in light of the alteration of FAC1 localization to the neurites and cytoplasm of degenerating neurons of AD and ALS as well as differentiating neurons in developing cortex (3, 4). Our studies utilizing immunofluorescent laser confocal microscopy confirm that FAC1 colocalizes with endogenous Keap1 and f-actin (Figure 5), suggesting that hKeap1 can bind FAC1 and actin simultaneously. Furthermore, overexpression of hKeap1 is capable of altering FAC1 intracellular localization to perinuclear spherical structures in the cytoplasm. Although these perinuclear structures may be artifacts of hKeap1 overexpression or indicate a previously unidentified facet of hKeap1 function independent of actin association, the fact that FAC1 localizes to such structures only in the presence of hKeap1 suggests that hKeap1 can dictate FAC1 subcellular distribution in the cytoplasm. These findings suggest that hKeap1 may regulate FAC1 localization in tissues where both are expressed.
The hKeap1 protein is the first predominantly cytoplasmic protein identified to interact with FAC1, which suggests new roles for FAC1 involving cytoskeletal changes and oxidative stress. The Kelch repeat domain within Keap1 and hKeap1, which is responsible for FAC1 interaction, is believed to be a β-propeller structure on the basis of its homology to the fungal galactose oxidase protein (32, 34). For many of the Kelch family members, the function of the β-propeller is to associate with cytoskeletal structures including actin filaments, nuclear matrix, and adhesion junctions (35). Although some discrepancy exists about whether Keap1 associates directly with actin (36), Keap1 and actin have been reported to co-immunoprecipitate (37). Furthermore, GFP-Keap1 has been reported to colocalize with actin filaments and to concentrate in a perinuclear region in NIH 3T3 cells (37) similar to our observations of eGFP:hKeap1 in PT67 cells. Our findings support an association between f-actin and hKeap1 in PT67 cells.
Investigation into FAC1 has revealed an association between this protein and neuronal development and response to disease. While hKeap1 is expressed in the brain, but further investigation is needed to determine what role hKeap1 may play in the central nervous system. However, several related proteins with Kelch domains and homologies to hKeap1 have been reported to have important neuronal functions. Both Actinfilin and Mayven are Kelch repeat containing proteins that bind to f-actin and are expressed in neurons (27, 38). Another Kelch-containing protein, NRP/B, is involved in neurite outgrowth and neuronal differentiation (26). Finally, mutations in Gigaxonin, another protein containing BTB/POZ and Kelch domains, results in giant axonal neuropathy (GAN), a disease associated with disorganization of cytoskeletal intermediate filaments within neurons (39). Further experimentation is warranted to determine which of these Kelch repeat containing proteins contribute to regulation of FAC1 localization and function in the central nervous system during development and neurodegenerative disease.
Among Kelch-containing proteins, Keap1 is unique in its regulation of the Nrf2 transcription factor, which transactivates the antioxidant response element (ARE). Genes with the ARE in their promoter include glutathione S-transferase, NAD(P)H quinone reductase, and other proteins necessary to reduce cellular toxicity in response to reactive oxygen species (ROS) (40– 45). Keap1 functions as a molecular sensor for oxidative stress through electrophile-induced disruption of its BTB/POZ domain homodimerization (46). Abrogation of Keap1 dimerization disrupts the Nrf2:Keap1 interaction, allowing Nrf2 to enter the nucleus and activate transcription of target genes containing the ARE (47). Both Nrf2 and FAC1 bind to hKeap1 through the Kelch repeats [Figure 3(14)]. Our findings indicate that Nrf2 competes for binding to hKeap1 with FAC1 in a dose-dependent manner (Figure 6). Because binding of Nrf2 and FAC1 to hKeap1 is mutually exclusive and Nrf2 interaction with hKeap1 is disrupted by oxidative stress, we further investigated the impact of chemical disrupters of the Nrf2:hKeap1 interaction. Using DEM as a known chemical disrupter of Nrf2:Keap1 interaction, we demonstrate here that DEM does not alter the ability of FAC1 to bind hKeap1, but does reduce the ability of Nrf2 to compete with FAC1 for hKeap1. These findings have interesting implications for the interplay between FAC1, hKeap1, and Nrf2 in neurons responding to oxidative stress.
Oxidative damage has been implicated as a major contributor to pathogenesis in several neurodegenerative diseases including AD and ALS (48– 51). Thus, hKeap1 is likely to be oxidized in these disease states and therefore free of Nrf2. Furthermore, FAC1 has been shown to exhibit cytoplasmic localization in neurons of affected brain regions in AD and ALS (3, 4). Taken together, these findings suggest that Keap1 regulation of FAC1 and Nrf2 may play an important role in determining neuronal response to oxidative damage. It has been previously shown that Nrf2 protects against neuronal death in response to oxidative stressors including H2O2 and glutamate (52, 56). Interestingly, β-amyloid treatment, which has been reported to induce formation of ROS (49), induces redistribution of FAC1 in the cytoplasm (R. Bowser, unpublished observations). These findings suggest that Keap1 may bind Nrf2 and FAC1 in a reciprocal fashion: Nrf2 under reducing conditions and FAC1 under oxidizing conditions. Further investigation into the interplay between these proteins is warranted to understand their role in determining neuronal viability in response to oxidative stress and neurodegenerative stimuli. Understanding the role of FAC1, Keap1, and Nrf2 in neuronal response to oxidative stress could lead to novel neuroprotective strategies for neurodegenerative diseases.
Acknowledgments
We thank Dr. Michael Freeman for the kind gift of full-length KIAA0132, the human orthologue of Keap1. We also thank Andy Shih for the kind gift of full-length Nrf2. We appreciate technical assistance from Courtney Wilson.
Footnotes
This project was supported by NIH Grants NS41202, AG13208, and NS10572 to K.J.-S. and NS42902 to R.B.
Abbreviations: AD, Alzheimer's disease; ALS, amyotrophic lateral sclerosis; ARE, antioxidant response element; BTB, broad complex, tramtrack, and bric á brac; DEM, diethylmaleate; DGR, double-glycine repeats; FAC1, fetal Alz-50 reactive clone 1; GST, glutathione-S-transferase gene; GFP, green fluorescent protein; IVT, in vitro translated; KLD, Kelch-like domain; Keap1, Kelch-like Ech-associated protein; hKeap1, human Keap1; Phd, phalloidin; PBS, phosphatebuffered saline; POZ, pox virus genes that have zinc fingers; TBS, Tris-buffered saline.
References
- 1.Jordan-Sciutto KL, Dragich JM, Bowser R. DNA binding activity of the fetal Alz-50 clone 1 (FAC1) protein is enhanced by phosphorylation. Biochem Biophys Res Commun. 1999;260:785–789. doi: 10.1006/bbrc.1999.0986. [DOI] [PubMed] [Google Scholar]
- 2.Styren SD, Bowser R, Dekosky ST. Expression of fetal ALZ-50 reactive clone 1 (FAC1) in dentate gyrus following entorhinal cortex lesion. J Comp Neurol. 1997;386:555–561. [PubMed] [Google Scholar]
- 3.Mu X, Springer JE, Bowser R. FAC1 expression and localization in motor neurons of developing, adult, and amyotrophic lateral sclerosis spinal cord. Exp Neurol. 1997;146:17–24. doi: 10.1006/exnr.1997.6508. [DOI] [PubMed] [Google Scholar]
- 4.Schoonover S, Davies P, Bowser R. Immunolocalization and redistribution of the FAC1 protein in Alzheimer's disease. J Neuropathol Exp Neurol. 1996;55:444–455. doi: 10.1097/00005072-199604000-00007. [DOI] [PubMed] [Google Scholar]
- 5.Bowser R, Giambrone A, Davies P. FAC1, a novel gene identified with the monoclonal antibody Alz50, is develop-mentally regulated in human brain. Dev Neurosci. 1995;17:20–37. doi: 10.1159/000111270. [DOI] [PubMed] [Google Scholar]
- 6.Rhodes J, Lutka FA, Jordan-Sciutto KL, Bowser R. Altered expression and distribution of FAC1 during NGF-induced neurite outgrowth of PC12 cells. Neuroreport. 2003;14:449–452. doi: 10.1097/00001756-200303030-00030. [DOI] [PubMed] [Google Scholar]
- 7.Jordan-Sciutto K, Rhodes J, Bowser R. Altered subcellular distribution of transcriptional regulators in response to Abeta peptide and during Alzheimer's disease. Mech Ageing Dev. 2001;123:11–20. doi: 10.1016/s0047-6374(01)00334-7. [DOI] [PubMed] [Google Scholar]
- 8.Jordan-Sciutto KL, Dragich JM, Rhodes JL, Bowser R. Fetal Alz-50 clone 1, a novel zinc finger protein, binds a specific DNA sequence and acts as a transcriptional regulator. J Biol Chem. 1999;274:35262–35268. doi: 10.1074/jbc.274.49.35262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salbaum JM, Weidemann A, Lemaire HG, Masters CL, Beyreuther K. The promoter of Alzheimer's disease amyloid A4 precursor gene. EMBO J. 1988;7:2807–2813. doi: 10.1002/j.1460-2075.1988.tb03136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitsuda N, Roses AD, Vitek MP. Transcriptional regulation of the mouse presenilin-1 gene. J Biol Chem. 1997;272:23489–23497. doi: 10.1074/jbc.272.38.23489. [DOI] [PubMed] [Google Scholar]
- 11.Kim HT, Kim YH, Nam JW, Lee HJ, Rho HM, Jung G. Study of 5′-flanking region of human Cu/Zn superoxide dismutase. Biochem Biophys Res Commun. 1994;201:1526–1533. doi: 10.1006/bbrc.1994.1877. [DOI] [PubMed] [Google Scholar]
- 12.Jordan-Sciutto KL, Dragich JM, Caltagarone J, Hall DJ, Bowser R. Fetal Alz-50 clone 1 (FAC1) protein interacts with the Myc-associated zinc finger protein (ZF87/MAZ) and alters its transcriptional activity. Biochemistry. 2000;39:3206–3215. doi: 10.1021/bi992211q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagase T, Seki N, Tanaka A, Ishikawa K, Nomura N. Prediction of the coding sequences of unidentified human genes. IV. The coding sequences of 40 new genes (KIAA0121-KIAA0160) deduced by analysis of cDNA clones from human cell line KG-1. DNA Res. 1995;2:167–174. 199–210. doi: 10.1093/dnares/2.4.167. [DOI] [PubMed] [Google Scholar]
- 14.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McMahon M, Itoh K, Yamamoto M, Hayes JD. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J Biol Chem. 2003;278:21592–21600. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- 16.Nguyen T, Huang HC, Pickett CB. Transcriptional regulation of the antioxidant response element. Activation by Nrf2 and repression by MafK. J Biol Chem. 2000;275:15466–15473. doi: 10.1074/jbc.M000361200. [DOI] [PubMed] [Google Scholar]
- 17.Vojtek AB, Hollenberg SM, Cooper JA. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 18.Chien CT, Bartel PL, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 20.Jordan KL, Haas AR, Logan TJ, Hall DJ. Detailed analysis of the basic domain of the E2F1 transcription factor indicates that it is unique among bHLH proteins. Oncogene. 1994;9:1177–1185. [PubMed] [Google Scholar]
- 21.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. In: Current Protocols in Molecular Biology. Janssen K, editor. Wiley; New York: 1994. pp. 12.11.1–12.11.8. [Google Scholar]
- 22.Jordan KL, Evans DL, Steelman S, Hall DJ. Isolation of two novel cDNAs whose products associate with the amino terminus of the E2F1 transcription factor. Biochemistry. 1996;35:12320–12328. doi: 10.1021/bi9611927. [DOI] [PubMed] [Google Scholar]
- 23.Varkey JP, Muhlrad PJ, Minniti AN, Do B, Ward S. The Caenorhabditis elegans spe-26 gene is necessary to form spermatids and encodes a protein similar to the actin-associated proteins kelch and scruin. Genes Dev. 1995;9:1074–1086. doi: 10.1101/gad.9.9.1074. [DOI] [PubMed] [Google Scholar]
- 24.Goebel SJ, Johnson GP, Perkus ME, Davis SW, Winslow JP, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–266. 517–563. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 25.Hernandez MC, Andres-Barquin PJ, Kuo WL, Israel MA. Assignment of the ectodermal-neural cortex 1 gene (ENC1) to human chromosome band 5q13 by in situ hybridization. Cytogenet Cell Genet. 1999;87:89–90. doi: 10.1159/000015398. [DOI] [PubMed] [Google Scholar]
- 26.Kim TA, Lim J, Ota S, Raja S, Rogers R, Rivnay B, Avraham H, Avraham S. NRP/B, a novel nuclear matrix protein, associates with p110(RB) and is involved in neuronal differentiation. J Cell Biol. 1998;141:553–566. doi: 10.1083/jcb.141.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soltysik-Espanola M, Rogers RA, Jiang S, Kim TA, Gaedigk R, White RA, Avraham H, Avraham S. Characterization of Mayven, a novel actin-binding protein predominantly expressed in brain. Mol Biol Cell. 1999;10:2361–2375. doi: 10.1091/mbc.10.7.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Albagli O, Dhordain P, Deweindt C, Lecocq G, Leprince D. The BTB/POZ domain: a new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Diff. 1995;6:1193–1198. [PubMed] [Google Scholar]
- 29.Bardwell VJ, Treisman R. The POZ domain: a conserved protein-protein interaction motif. Gene Dev. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- 30.Chen W, Zollman S, Couderc JL, Laski FA. The BTB domain of bric a brac mediates dimerization in vitro. Mol Cell Biol. 1995;15:3424–3429. doi: 10.1128/mcb.15.6.3424. retraction in Chen, W., Zollman, S., Couderc, J. L., and Laski, F. A. (1997) Mol. Cell. Biol. 17, 6772; PMID: 9380040. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Robinson DN, Cooley L. Drosophila kelch is an oligomeric ring canal actin organizer. J Cell Biol. 1997;138:799–810. doi: 10.1083/jcb.138.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams J, Kelso R, Cooley L. The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol. 2000;10:17–24. doi: 10.1016/s0962-8924(99)01673-6. [DOI] [PubMed] [Google Scholar]
- 33.Sekhar KR, Yan XX, Freeman ML. Nrf2 degradation by the ubiquitin proteasome pathway is inhibited by KIAA0132, the human homolog to INrf2. Oncogene. 2002;21:6829–6834. doi: 10.1038/sj.onc.1205905. [DOI] [PubMed] [Google Scholar]
- 34.Bork P, Doolittle RF. Drosophila kelch motif is derived from a common enzyme fold. J Mol Biol. 1994;236:1277–1282. doi: 10.1016/0022-2836(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 35.Velichkova M, Guttman J, Warren C, Eng L, Kline K, Vogl AW, Hasson T. A human homologue of Drosophila kelch associates with myosin-VIIa in specialized adhesion junctions. Cell Motil Cytoskeleton. 2002;51:147–64. doi: 10.1002/cm.10025. [DOI] [PubMed] [Google Scholar]
- 36.T'Jampens D, Devriendt L, De Corte V, Vandekerckhove J, Gettemans J. Selected BTB/POZ-kelch proteins bind ATP. FEBS Lett. 2002;516:20–26. doi: 10.1016/s0014-5793(02)02456-0. [DOI] [PubMed] [Google Scholar]
- 37.Kang MI, Kobayashi A, Wakabayashi N, Kim SG, Yamamoto M. Scaffolding of Keap1 to the actin cyto-skeleton controls the function of Nrf2 as key regulator of cytoprotective phase 2 genes. Proc Natl Acad Sci USA. 2004;101:2046–2051. doi: 10.1073/pnas.0308347100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Y, Derin R, Petralia RS, Li M. Actinfilin, a brain-specific actin-binding protein in postsynaptic density. J Biol Chem. 2002;277:30495–30501. doi: 10.1074/jbc.M202076200. [DOI] [PubMed] [Google Scholar]
- 39.Bomont P, Cavalier L, Blondeau F, Ben Hamida C, Belal S, Tazir M, Demir E, Topaloglu H, Korinthenberg R, Tuysuz B, Landrieu P, Hentati F, Koenig M. The gene encoding gigaxonin, a new member of the cytoskeletal BTB/kelch repeat family, is mutated in giant axonal neuropathy. Nat Genet. 2000;26:370–374. doi: 10.1038/81701. see comment. [DOI] [PubMed] [Google Scholar]
- 40.Hayes JD, McMahon M. Molecular basis for the contribution of the antioxidant responsive element to cancer chemoprevention. Cancer Lett. 2001;174:103–113. doi: 10.1016/s0304-3835(01)00695-4. [DOI] [PubMed] [Google Scholar]
- 41.Rushmore TH, King RG, Paulson KE, Pickett CB. Regulation of glutathione S-transferase Ya subunit gene expression: identification of a unique xenobiotic-responsive element controlling inducible expression by planar aromatic compounds. Proc Natl Acad Sci USA. 1990;87:3826–3830. doi: 10.1073/pnas.87.10.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Favreau LV, Pickett CB. The rat quinone reductase antioxidant response element. Identification of the nucleotide sequence required for basal and inducible activity and detection of antioxidant response element-binding proteins in hepatoma and non-hepatoma cell lines. J Biol Chem. 1995;270:24468–24474. doi: 10.1074/jbc.270.41.24468. [DOI] [PubMed] [Google Scholar]
- 43.Favreau LV, Pickett CB. Transcriptional regulation of the rat NAD(P)H:quinone reductase gene. Characterization of a DNA-protein interaction at the antioxidant responsive element and induction by 12-O-tetradecanoylphorbol 13-acetate. J Biol Chem. 1993;268:19875–19881. [PubMed] [Google Scholar]
- 44.Rozen F, Nguyen T, Pickett CB. Isolation and characterization of a human glutathione S-transferase Ha1 subunit gene. Arch Biochem Biophys. 1992;292:589–593. doi: 10.1016/0003-9861(92)90035-u. [DOI] [PubMed] [Google Scholar]
- 45.Rushmore TH, Morton MR, Pickett CB. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J Biol Chem. 1991;266:11632–11639. [PubMed] [Google Scholar]
- 46.Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci USA. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zipper LM, Mulcahy RT. The Keap1 BTB/POZ dimerization function is required to sequester Nrf2 in cytoplasm. J Biol Chem. 2002;277:36544–36552. doi: 10.1074/jbc.M206530200. [DOI] [PubMed] [Google Scholar]
- 48.Benzi G, Moretti A. Are reactive oxygen species involved in Alzheimer's disease? Neurobiol Aging. 1995;16:661–674. doi: 10.1016/0197-4580(95)00066-n. see comment. [DOI] [PubMed] [Google Scholar]
- 49.Butterfield DA. Amyloid beta-peptide [1-42]-associated free radical-induced oxidative stress and neurodegeneration in Alzheimer's disease brain: mechanisms and consequences. Curr Med Chem. 2003;10:2651–2659. doi: 10.2174/0929867033456422. [DOI] [PubMed] [Google Scholar]
- 50.Simpson EP, Yen AA, Appel SH. Oxidative stress: a common denominator in the pathogenesis of amyotrophic lateral sclerosis. Curr Opin Rheumatol. 2003;15:730–736. doi: 10.1097/00002281-200311000-00008. [DOI] [PubMed] [Google Scholar]
- 51.Carri MT, Ferri A, Cozzolino M, Calabrese L, Rotilio G. Neurodegeneration in amyotrophic lateral sclerosis: the role of oxidative stress and altered homeostasis of metals. Brain Res Bull. 2003;61:365–374. doi: 10.1016/s0361-9230(03)00179-5. [DOI] [PubMed] [Google Scholar]
- 52.Johnson DA, Andrews GK, Xu W, Johnson JA. Activation of the antioxidant response element in primary cortical neuronal cultures derived from transgenic reporter mice. J Neurochem. 2002;81:1233–1241. doi: 10.1046/j.1471-4159.2002.00913.x. [DOI] [PubMed] [Google Scholar]
- 53.Lee JM, Calkins MJ, Chan K, Kan YW, Johnson JA. Identification of the NF-E2-related factor-2-dependent genes conferring protection against oxidative stress in primary cortical astrocytes using oligonucleotide microarray analysis. J Biol Chem. 2003;278:12029–12038. doi: 10.1074/jbc.M211558200. [DOI] [PubMed] [Google Scholar]
- 54.Lee JM, Shih AY, Murphy TH, Johnson JA. NF-E2-related factor-2 mediates neuroprotection against mitochondrial complex I inhibitors and increased concentration of intracellular calcium in primary cortical neurons. J Biol Chem. 2003;278:37948–37956. doi: 10.1074/jbc.M305204200. [DOI] [PubMed] [Google Scholar]
- 55.Shih AY, Johnson DA, Wong G, Kraft AD, Jiang L, Erb H, Johnson JA, Murphy TH. Coordinate regulation of glutathione biosynthesis and release by Nrf2-expressing glia potently protects neurons from oxidative stress. J Neurosci. 2003;23:3394–3406. doi: 10.1523/JNEUROSCI.23-08-03394.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kraft AD, Johnson DA, Johnson JA. Nuclear factor E2-related factor 2-dependent antioxidant response element activation by tert-butylhydroquinone and sulforaphane occurring preferentially in astrocytes conditions neurons against oxidative insult. J Neurosci. 2004;24:1101–1112. doi: 10.1523/JNEUROSCI.3817-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]