Abstract
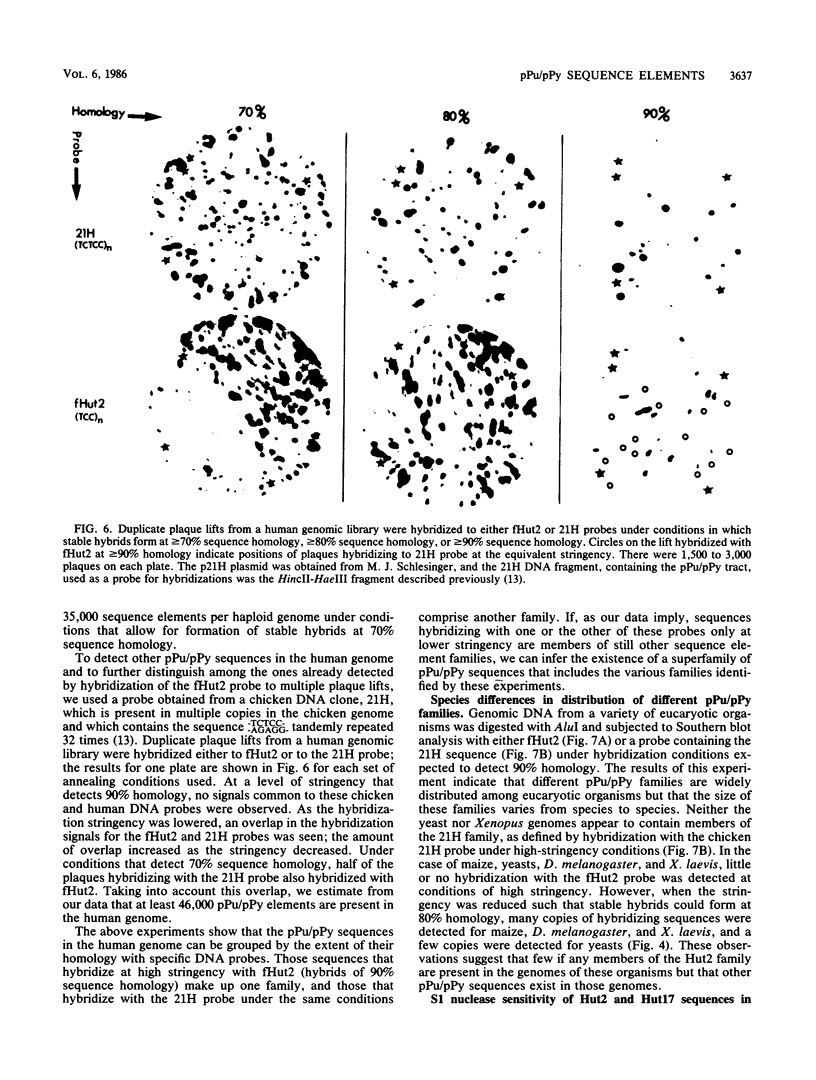
We previously have shown that homologs of the outer domain segment of the inverted repeat termini (IVR-OD) of the sea urchin TU transposons are conserved among multiple eucaryotic species, including humans. We report here that two cloned human DNA IVR-OD homologs, Hut2 and Hut17, consist of a series of tandem repeats of the trimer AGG/TCC, forming segments (313 and 221 base pairs in length, respectively) of polypurine/polypyrimidine (pPu/pPy or "Puppy") asymmetry in the two DNA strands; these are punctuated at certain sites with variant trimers, which are different for the two clones. Sequences homologous to the Hut2 pPu/pPy tract exist at multiple sites in the DNA of a wide variety of eucaryotes. Hybridization of human DNA with a Hut2 probe or with a previously described chicken DNA pPu/pPy sequence indicates that pPu/pPy sequences can be grouped into families distinguishable by the extent of their homology with each probe at different hybridization stringencies. Moreover, particular pPu/pPy tracts show species-specific differences in their distribution. Both the Hut2 and Hut17 pPu/pPy tracts are cleaved by S1 nuclease when tested on supercoiled plasmids. Most if not all of the 313-base-pair Hut2 pPu/pPy tract is also sensitive to S1 in its native location in HeLa cell chromatin, indicating that the sequence contains conformational information that can be expressed in vivo. This view is supported by evidence that exogenously derived Hut2 pPu/pPy tracts introduced into mouse L cells and integrated in chromatin can assume an S1-sensitive conformation.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bell G. I., Selby M. J., Rutter W. J. The highly polymorphic region near the human insulin gene is composed of simple tandemly repeating sequences. Nature. 1982 Jan 7;295(5844):31–35. doi: 10.1038/295031a0. [DOI] [PubMed] [Google Scholar]
- Bennett K. L., Hill R. E., Pietras D. F., Woodworth-Gutai M., Kane-Haas C., Houston J. M., Heath J. K., Hastie N. D. Most highly repeated dispersed DNA families in the mouse genome. Mol Cell Biol. 1984 Aug;4(8):1561–1571. doi: 10.1128/mcb.4.8.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Sederoff R. R., Paterson M. C. Distribution of polypyrimidine . polypurine segments in DNA from diverse organisms. Eur J Biochem. 1979 Jul;98(1):301–307. doi: 10.1111/j.1432-1033.1979.tb13189.x. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Davidson E. H. Gene regulation for higher cells: a theory. Science. 1969 Jul 25;165(3891):349–357. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- Cantor C. R., Efstratiadis A. Possible structures of homopurine-homopyrimidine S1-hypersensitive sites. Nucleic Acids Res. 1984 Nov 12;12(21):8059–8072. doi: 10.1093/nar/12.21.8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophe D., Cabrer B., Bacolla A., Targovnik H., Pohl V., Vassart G. An unusually long poly(purine)-poly(pyrimidine) sequence is located upstream from the human thyroglobulin gene. Nucleic Acids Res. 1985 Jul 25;13(14):5127–5144. doi: 10.1093/nar/13.14.5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. B., Effron K., Rechavi G., Ben-Neriah Y., Zakut R., Givol D. Simple DNA sequences in homologous flanking regions near immunoglobulin VH genes: a role in gene interaction? Nucleic Acids Res. 1982 Jun 11;10(11):3353–3370. doi: 10.1093/nar/10.11.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E. H., Britten R. J. Regulation of gene expression: possible role of repetitive sequences. Science. 1979 Jun 8;204(4397):1052–1059. doi: 10.1126/science.451548. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Jacobs H. T., Britten R. J. Very short repeats and coordinate induction of genes. Nature. 1983 Feb 10;301(5900):468–470. doi: 10.1038/301468a0. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Posakony J. W. Repetitive sequence transcripts in development. Nature. 1982 Jun 24;297(5868):633–635. doi: 10.1038/297633a0. [DOI] [PubMed] [Google Scholar]
- Davis M. M., Kim S. K., Hood L. E. DNA sequences mediating class switching in alpha-immunoglobulins. Science. 1980 Sep 19;209(4463):1360–1365. doi: 10.1126/science.6774415. [DOI] [PubMed] [Google Scholar]
- Dybvig K., Clark C. D., Aliperti G., Schlesinger M. J. A chicken repetitive DNA sequence that is highly sensitive to single-strand specific endonucleases. Nucleic Acids Res. 1983 Dec 10;11(23):8495–8508. doi: 10.1093/nar/11.23.8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson B. M., Felsenfeld G. Specific factor conferring nuclease hypersensitivity at the 5' end of the chicken adult beta-globin gene. Proc Natl Acad Sci U S A. 1984 Jan;81(1):95–99. doi: 10.1073/pnas.81.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson B. M., Lewis C. D., Felsenfeld G. Interaction of specific nuclear factors with the nuclease-hypersensitive region of the chicken adult beta-globin gene: nature of the binding domain. Cell. 1985 May;41(1):21–30. doi: 10.1016/0092-8674(85)90057-1. [DOI] [PubMed] [Google Scholar]
- Evans T., Schon E., Gora-Maslak G., Patterson J., Efstratiadis A. S1-hypersensitive sites in eukaryotic promoter regions. Nucleic Acids Res. 1984 Nov 12;12(21):8043–8058. doi: 10.1093/nar/12.21.8043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finer M. H., Fodor E. J., Boedtker H., Doty P. Endonuclease S1-sensitive site in chicken pro-alpha 2(I) collagen 5' flanking gene region. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1659–1663. doi: 10.1073/pnas.81.6.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch E. F., Lawn R. M., Maniatis T. Molecular cloning and characterization of the human beta-like globin gene cluster. Cell. 1980 Apr;19(4):959–972. doi: 10.1016/0092-8674(80)90087-2. [DOI] [PubMed] [Google Scholar]
- Gilliam A. C., Shen A., Richards J. E., Blattner F. R., Mushinski J. F., Tucker P. W. Illegitimate recombination generates a class switch from C mu to C delta in an IgD-secreting plasmacytoma. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4164–4168. doi: 10.1073/pnas.81.13.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies S. D., Folsom V., Tonegawa S. Cell type-specific enhancer element associated with a mouse MHC gene, E beta. Nature. 1984 Aug 16;310(5978):594–597. doi: 10.1038/310594a0. [DOI] [PubMed] [Google Scholar]
- Glikin G. C., Gargiulo G., Rena-Descalzi L., Worcel A. Escherichia coli single-strand binding protein stabilizes specific denatured sites in superhelical DNA. Nature. 1983 Jun 30;303(5920):770–774. doi: 10.1038/303770a0. [DOI] [PubMed] [Google Scholar]
- Goodbourn S., Zinn K., Maniatis T. Human beta-interferon gene expression is regulated by an inducible enhancer element. Cell. 1985 Jun;41(2):509–520. doi: 10.1016/s0092-8674(85)80024-6. [DOI] [PubMed] [Google Scholar]
- Harris S. E., Mansson P. E., Tully D. B., Burkhart B. Seminal vesicle secretion IV gene: allelic difference due to a series of 20-base-pair direct tandem repeats within an intron. Proc Natl Acad Sci U S A. 1983 Nov;80(21):6460–6464. doi: 10.1073/pnas.80.21.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilig R., Muraskowsky R., Mandel J. L. The ovalbumin gene family. The 5' end region of the X and Y genes. J Mol Biol. 1982 Mar 25;156(1):1–19. doi: 10.1016/0022-2836(82)90455-7. [DOI] [PubMed] [Google Scholar]
- Heller M., Flemington E., Kieff E., Deininger P. Repeat arrays in cellular DNA related to the Epstein-Barr virus IR3 repeat. Mol Cell Biol. 1985 Mar;5(3):457–465. doi: 10.1128/mcb.5.3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller M., Henderson A., Kieff E. Repeat array in Epstein-Barr virus DNA is related to cell DNA sequences interspersed on human chromosomes. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5916–5920. doi: 10.1073/pnas.79.19.5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel C. C. Homocopolymer sequences in the spacer of a sea urchin histone gene repeat are sensitive to S1 nuclease. Nature. 1982 Feb 25;295(5851):714–716. doi: 10.1038/295714a0. [DOI] [PubMed] [Google Scholar]
- Hoffman-Liebermann B., Liebermann D., Kedes L. H., Cohen S. N. TU elements: a heterogeneous family of modularly structured eucaryotic transposons. Mol Cell Biol. 1985 May;5(5):991–1001. doi: 10.1128/mcb.5.5.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Htun H., Lund E., Dahlberg J. E. Human U1 RNA genes contain an unusually sensitive nuclease S1 cleavage site within the conserved 3' flanking region. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7288–7292. doi: 10.1073/pnas.81.23.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinek W. R., Toomey T. P., Leinwand L., Duncan C. H., Biro P. A., Choudary P. V., Weissman S. M., Rubin C. M., Houck C. M., Deininger P. L. Ubiquitous, interspersed repeated sequences in mammalian genomes. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1398–1402. doi: 10.1073/pnas.77.3.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao F. T. Human genome structure. Int Rev Cytol. 1985;96:51–88. doi: 10.1016/s0074-7696(08)60594-x. [DOI] [PubMed] [Google Scholar]
- Kedes L. H. Histone genes and histone messengers. Annu Rev Biochem. 1979;48:837–870. doi: 10.1146/annurev.bi.48.070179.004201. [DOI] [PubMed] [Google Scholar]
- Kuo M. T., Plunkett W. Nick-translation of metaphase chromosomes: in vitro labeling of nuclease-hypersensitive regions in chromosomes. Proc Natl Acad Sci U S A. 1985 Feb;82(3):854–858. doi: 10.1073/pnas.82.3.854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen A., Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982 Jun;29(2):609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- Lee J. S., Woodsworth M. L., Latimer L. J., Morgan A. R. Poly(pyrimidine) . poly(purine) synthetic DNAs containing 5-methylcytosine form stable triplexes at neutral pH. Nucleic Acids Res. 1984 Aug 24;12(16):6603–6614. doi: 10.1093/nar/12.16.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrman M. A., Schneider W. J., Südhof T. C., Brown M. S., Goldstein J. L., Russell D. W. Mutation in LDL receptor: Alu-Alu recombination deletes exons encoding transmembrane and cytoplasmic domains. Science. 1985 Jan 11;227(4683):140–146. doi: 10.1126/science.3155573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebermann D., Hoffman-Liebermann B., Troutt A. B., Kedes L., Cohen S. N. Sequences from sea urchin TU transposons are conserved among multiple eucaryotic species, including humans. Mol Cell Biol. 1986 Jan;6(1):218–226. doi: 10.1128/mcb.6.1.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebermann D., Hoffman-Liebermann B., Weinthal J., Childs G., Maxson R., Mauron A., Cohen S. N., Kedes L. An unusual transposon with long terminal inverted repeats in the sea urchin Strongylocentrotus purpuratus. Nature. 1983 Nov 24;306(5941):342–347. doi: 10.1038/306342a0. [DOI] [PubMed] [Google Scholar]
- Mace H. A., Pelham H. R., Travers A. A. Association of an S1 nuclease-sensitive structure with short direct repeats 5' of Drosophila heat shock genes. Nature. 1983 Aug 11;304(5926):555–557. doi: 10.1038/304555a0. [DOI] [PubMed] [Google Scholar]
- Maeda N., Bliska J. B., Smithies O. Recombination and balanced chromosome polymorphism suggested by DNA sequences 5' to the human delta-globin gene. Proc Natl Acad Sci U S A. 1983 Aug;80(16):5012–5016. doi: 10.1073/pnas.80.16.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda N., Yang F., Barnett D. R., Bowman B. H., Smithies O. Duplication within the haptoglobin Hp2 gene. Nature. 1984 May 10;309(5964):131–135. doi: 10.1038/309131a0. [DOI] [PubMed] [Google Scholar]
- Maroteaux L., Heilig R., Dupret D., Mandel J. L. Repetitive satellite-like sequences are present within or upstream from 3 avian protein-coding genes. Nucleic Acids Res. 1983 Mar 11;11(5):1227–1243. doi: 10.1093/nar/11.5.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight S. L., Gavis E. R. Expression of the herpes thymidine kinase gene in Xenopus laevis oocytes: an assay for the study of deletion mutants constructed in vitro. Nucleic Acids Res. 1980 Dec 20;8(24):5931–5948. doi: 10.1093/nar/8.24.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Miyake T., Migita K., Sakaki Y. Some KpnI family members are associated with the Alu family in the human genome. Nucleic Acids Res. 1983 Oct 11;11(19):6837–6846. doi: 10.1093/nar/11.19.6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyatake S., Yokota T., Lee F., Arai K. Structure of the chromosomal gene for murine interleukin 3. Proc Natl Acad Sci U S A. 1985 Jan;82(2):316–320. doi: 10.1073/pnas.82.2.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moos M., Gallwitz D. Structure of two human beta-actin-related processed genes one of which is located next to a simple repetitive sequence. EMBO J. 1983;2(5):757–761. doi: 10.1002/j.1460-2075.1983.tb01496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickol J. M., Felsenfeld G. DNA conformation at the 5' end of the chicken adult beta-globin gene. Cell. 1983 Dec;35(2 Pt 1):467–477. doi: 10.1016/0092-8674(83)90180-0. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Gil A., Maniatis T. The structure of the human zeta-globin gene and a closely linked, nearly identical pseudogene. Cell. 1982 Dec;31(3 Pt 2):553–563. doi: 10.1016/0092-8674(82)90311-7. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D. E., Haniford D. B., Morgan A. R. A structural basis for S1 nuclease sensitivity of double-stranded DNA. Cell. 1985 Aug;42(1):271–280. doi: 10.1016/s0092-8674(85)80122-7. [DOI] [PubMed] [Google Scholar]
- Qasba P. K., Safaya S. K. Similarity of the nucleotide sequences of rat alpha-lactalbumin and chicken lysozyme genes. Nature. 1984 Mar 22;308(5957):377–380. doi: 10.1038/308377a0. [DOI] [PubMed] [Google Scholar]
- Rich A., Nordheim A., Wang A. H. The chemistry and biology of left-handed Z-DNA. Annu Rev Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- Richards J. E., Gilliam A. C., Shen A., Tucker P. W., Blattner F. R. Unusual sequences in the murine immunoglobulin mu-delta heavy-chain region. Nature. 1983 Dec 1;306(5942):483–487. doi: 10.1038/306483a0. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Ruiz-Carrillo A. The histone H5 gene is flanked by S1 hypersensitive structures. Nucleic Acids Res. 1984 Aug 24;12(16):6473–6492. doi: 10.1093/nar/12.16.6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon E., Evans T., Welsh J., Efstratiadis A. Conformation of promoter DNA: fine mapping of S1-hypersensitive sites. Cell. 1983 Dec;35(3 Pt 2):837–848. doi: 10.1016/0092-8674(83)90116-2. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Southgate R., Ayme A., Voellmy R. Nucleotide sequence analysis of the Drosophila small heat shock gene cluster at locus 67B. J Mol Biol. 1983 Mar 25;165(1):35–57. doi: 10.1016/s0022-2836(83)80241-1. [DOI] [PubMed] [Google Scholar]
- Sures I., Lowry J., Kedes L. H. The DNA sequence of sea urchin (S. purpuratus) H2A, H2B and H3 histone coding and spacer regions. Cell. 1978 Nov;15(3):1033–1044. doi: 10.1016/0092-8674(78)90287-8. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G., Milner R. J., Gottesfeld J. M., Lerner R. A. Identifier sequences are transcribed specifically in brain. Nature. 1984 Mar 15;308(5956):237–241. doi: 10.1038/308237a0. [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Blackburn E. H. Cloning yeast telomeres on linear plasmid vectors. Cell. 1982 May;29(1):245–255. doi: 10.1016/0092-8674(82)90109-x. [DOI] [PubMed] [Google Scholar]
- Thayer R. E. An improved method for detecting foreign DNA in plasmids of Escherichia coli. Anal Biochem. 1979 Sep 15;98(1):60–63. doi: 10.1016/0003-2697(79)90705-x. [DOI] [PubMed] [Google Scholar]
- Weintraub H. A dominant role for DNA secondary structure in forming hypersensitive structures in chromatin. Cell. 1983 Apr;32(4):1191–1203. doi: 10.1016/0092-8674(83)90302-1. [DOI] [PubMed] [Google Scholar]
- Wigler M., Sweet R., Sim G. K., Wold B., Pellicer A., Lacy E., Maniatis T., Silverstein S., Axel R. Transformation of mammalian cells with genes from procaryotes and eucaryotes. Cell. 1979 Apr;16(4):777–785. doi: 10.1016/0092-8674(79)90093-x. [DOI] [PubMed] [Google Scholar]








