Abstract
When nanoparticles (NPs) are exposed to the biological environment, their surfaces become covered with proteins and biomolecules (e.g. lipids). Here, we report that this protein coating, or corona, reduces the targeting capability of surface engineered NPs by screening the active sites of the targeting ligands.
Nanoparticles (NPs) are promising materials for the targeted delivery of therapeutic drugs to the desired site in the human body.1 One strategy to obtain a high targeting yield is to functionalize the surface of the NPs with targeting ligands (e.g. antibodies or aptamers) that enhance NP binding to receptors on the target cells and facilitate NP uptake by receptor-mediated endocytosis.2, 3 In vitro results generally confirm the high capability of functionalized NPs for targeting to the desired cells. However, lower targeting yields and an unfavorable biodistribution (i.e., NP accumulation in the liver and spleen instead of the desired tumor tissue) is often observed in vivo.4
The discrepancy between the in vitro and in vivo results is due, in part, to the adsorption of proteins and other biomolecules to the NP’s surface upon exposure to the biological medium in vivo.1, 5, 6 In contrast, the NP surface remains nearly pristine in the presence of the serum-free medium that is often used for in vitro studies of NP uptake.1 Therefore, cells in in vitro experiments that employ serum-free medium interact with the original NP surface, whereas cells in vivo interact with the protein coating, which is called the protein corona.1, 5, 6 The presence of the protein corona on the NP, which is strongly related to the physicochemical properties of the NP and protein sources,7–9 is reported to alter the biodistribution, cellular uptake mechanism, and intracellular location of the NPs in vivo.4, 10–13 In addition to these changes in biological phenomena, the protein corona may compromise the targeting efficiency of NPs that are functionalized with targeting ligands via a non-biological mechanism. Specifically, the protein corona surrounding the NP is hypothesized to hinder interactions between the NP’s ligands and their targets on the cell surface,4 though this mechanism has not been directly tested. Thus, the main aim of this work is to investigate whether the protein corona inhibits NP ligand binding to reactive moieties on a surface.
To permit separating the protein corona’s effects on biological phenomena (i.e., uptake mechanism) from its potential ability to limit access to the targeting ligand, we selected a copper-free click reaction between NPs functionalized with a strained cycloalkyne, bicyclononyne (BCN) and an azide on a silicon substrate as the model targeting reaction (Scheme 1). We prepared fluorescent silica NPs with diameters of 75 nm using the Stöber method (see SI for details),14 and then conjugated BCN targeting ligands (synthesized as reported,15 Scheme S1) to the NP surface (Scheme S2). Silicon substrates were modified with an azide-terminated self-assembled monolayer (SAM) by reaction with 11-azidoundecyltrimethoxysilane (Scheme S3). The presence of the azides on the substrates was confirmed by X-ray photoelectron spectroscopy (XPS) (Figure S1).
Scheme 1.
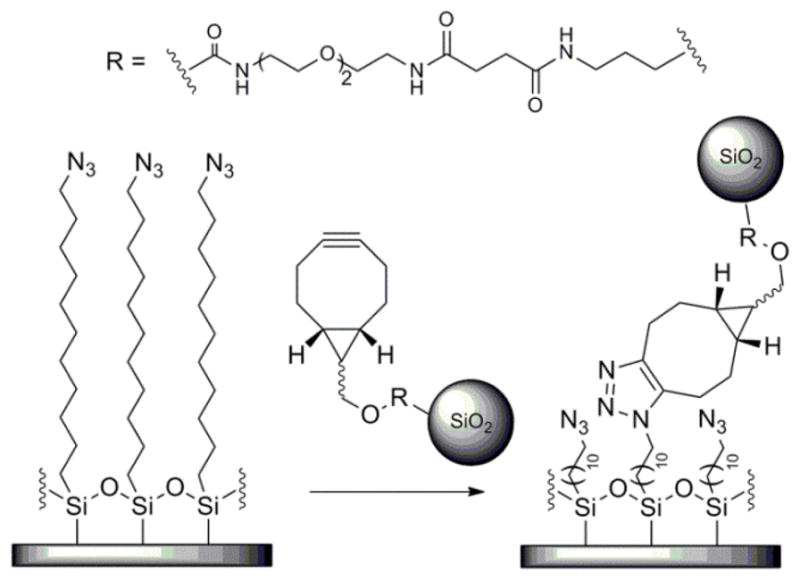
A copper-free click reaction between the BCN moieties on the NPs and the azides on the modified silicon substrate was selected as the model targeting reaction.
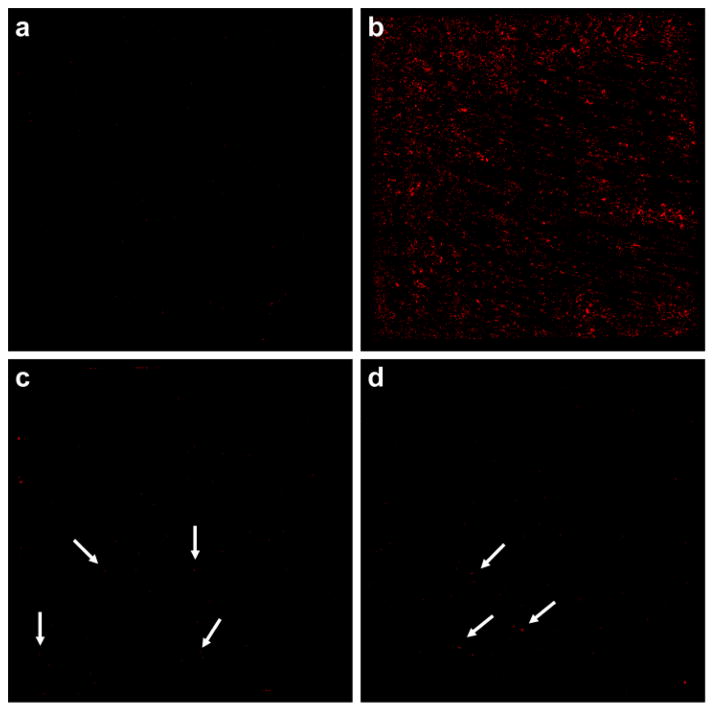
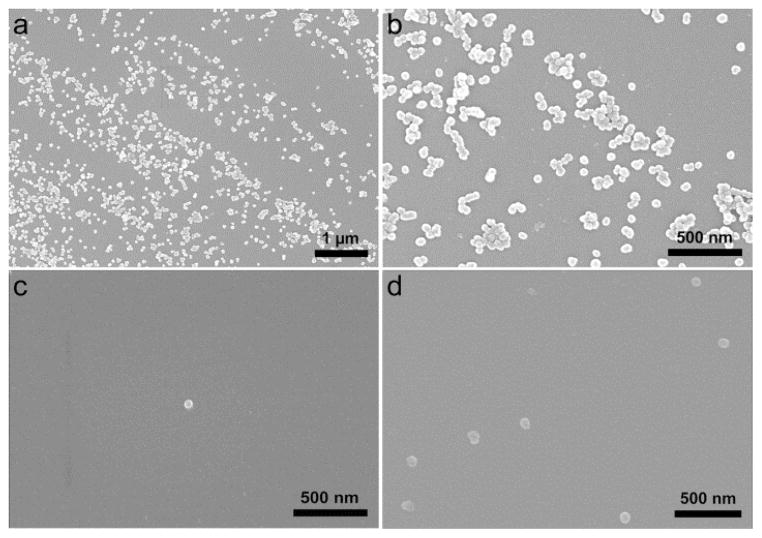
We evaluated whether BCN-NP conjugation to the azide-functionalized substrates was reduced by the protein corona that results from exposing the BCN-functionalized silica NPs (BCN-NPs) to biological fluids. We compared the conjugation of pristine BCN-NPs to those of BCN-NPs exposed to mediums that mimic in vitro culture conditions (i.e., medium with 10% serum) and the biological fluids present in vivo (i.e., 100% serum). Pristine BCN-NPs and BCN-NPs exposed to medium with 10% serum (10% serum corona BCN-NPs) or 100% serum (100% serum corona BCN-NPs) were incubated with azide-functionalized substrates for 90 min in phosphate buffered saline (PBS), and then conjugation was assessed with fluorescence microscopy and SEM. Control experiments in which pristine BCN-NPs were incubated with azide-free substrates confirmed that non-specific BCN-NP binding to the substrate was insignificant (Figure 1a). The fluorescence microscopy images show a high number of the pristine BCN-NPs conjugated to the azide-functionalized substrate (Figure 1b). In contrast, fluorescence microscopy showed few 10% or 100% serum corona BCN-NPs had attached to the azide-functionalized substrates (Figure 1c and d). Quantitative analysis indicated that the number of conjugated NPs, and therefore the targeting efficiencies for the 10% and 100% serum corona BCN-NPs were lower than that of the pristine BCN-NPs by 94 and 99%, respectively. SEM imaging confirmed the fluorescence microscopy results. The SEM images show numerous pristine BCN-NPs, but very few 10% or 100% serum corona BCN-NPs conjugated to the azide-functionalized substrates (Figures 2). These findings indicate the protein corona inhibits the NP’s targeting capability.
Figure 1.
Fluorescence microscopy images of 5 mm by 5 mm silicon substrates after incubation with pristine BCN-NPs and those coated with a protein corona. (a) Little non-specific binding of pristine BCN-NPs to the azide-free substrate occurred. (b) Numerous pristine BCN-NPs were conjugated to the azide-functionalized substrate. (c and d) Few 10% (c) or 100% (d) corona BCN-NPs were visible on the azide-functionalized substrates. Arrows designate individual NPs.
Figure 2.
SEM images of silicon substrates modified with azide-terminated SAMs after incubation with (a and b) pristine BCN-functionalized NPs, (c) 10% serum corona BCN-NPs, and (d) 100% serum corona BCN-NPs.
Characterization of the BCN-NPs in terms of size and zeta potential indicated that exposing the BCN-NPs to medium containing 10% or 100% serum increased their size, but only slightly decreased their negative charge (Table S1). The increase in size upon exposure to serum-containing media reflects formation of the protein corona as well as larger protein-NP complexes.16 The slight decrease in negative charge is due to screening of the negatively charged surface of the silica NP by the protein corona, and should not drastically alter the interactions between the substrate and the BCN-NPs in these experiments. Although NP uptake by cells is very size-dependent,8 we do not expect that the approximately two-fold increase in BCN-NP diameter that occurred after exposure to 10% serum medium is the primary cause of the ~94% reduction in targeting efficiency we observed under these conditions. Therefore, the protein corona-induced inhibition of NP targeting capability we observed in this system was mainly caused by screening of the interactions between the NP’s targeting ligands and their reactive partners on the substrate.
Next, we used liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS) to assess the compositions of the protein coronas on the BCN-NPs that resulted from exposure to mediums that mimic the in vitro (10% serum) or in vivo (100% serum) environments. A spectral counting method was used to normalize the protein-specific spectral counts detected with LC-MS/MS (SpC) with respect to the molecular weight of protein k (MW) and the total spectral counts detected according to Eq. S1. This method yields the normalized percentage of spectral counts for protein k (NpSpCk)5.
The most abundant proteins detected in the coronas on the 10% and 100% serum corona BCN-NPs are listed in Table 1. A more comprehensive list that includes the less abundant proteins detected is provided in Table S1. Evaluation of the normalized abundances of proteins within a specific molecular weight range (Figure S2) indicated that ≥88% of the proteins in the coronas on the BCN-NPs exposed to either serum-containing medium had a molecular weight below 30 kDa. Despite their relatively low molecular weights, these proteins established a shell that significantly reduced the BCN targeting ligand’s ability to react with the azides on the surface. Although Table 1 and Table S1 show that the proportions of the different proteins within each of the two coronas differed, the proportions of proteins with a specific molecular weight range, and the reduction in NP conjugation were very similar. Thus, the protein’s molecular weight is the main factor correlating its propensity to adsorb to the NP and obstruct its targeting ligands.
Table 1.
Protein composition of representative hard corona formed on BCN-NPs incubated in 10% and 100% FBS, as identi ed by LC-MS/MS.
| Protein | Composition | |
|---|---|---|
| 10% serum corona | 100% serum corona | |
| Alpha-1-antiproteinase precursor | 0.98 | 1.73 |
| Apolipoprotein E | 2.42 | 0 |
| Apolipoprotein A-I precursor | 6.05 | 5.694 |
| Hemoglobinfetal subunit beta | 18.54 | 12.35 |
| Chain A, A Novel Allosteric Mechanism In Haemoglobin | 32.25 | 47.28 |
| Apolipoprotein A-II precursor | 16.27 | 10.19 |
| Fetuin | 20.74 | 19.91 |
Altogether, this study confirms the hypothesis that the protein corona establishes a barrier that screens the interactions between the ligand and its target on a separate surface (i.e., the cell membrane) (Figure 3), thereby significantly reducing NP targeting efficiency as compared to NPs with pristine surfaces. We found that BCN-NP exposure to medium that contained as little as 10% serum, which is the typical serum concentration used for mammalian cell culture, resulted in the formation of a protein corona that significantly inhibited the model targeting reaction we studied. Our results imply that, in addition to NP clearance from the body (i.e., unfavorable biodistribution), screening of the targeting ligand by the protein corona that forms upon NP exposure to biological fluids contributes to the lower the NP targeting efficiencies attained in vivo than in vitro. Given that protein absorption to the NP surface cannot be prevented, as even modification of NPs with polyethylene glycol (PEG) reduces, but does not prevent, protein absorption,17 screening of the NP’s targeting ligand due to protein absorption must be considered when designing NPs for targeted drug delivery. Because the click chemistry reaction between BCN-NPs and the azide-functionalized surfaces used in this study enables separating the protein corona’s ability to obstruct the targeting ligand from its effects on biological phenomena, this approach may simplify assessing the extent that NPs with various surface modifications are screened by the protein corona. Of course, a cell-based platform would be required to assess the changes in biological phenomena that are induced by the protein corona, and how its composition may change as the NPs are trafficked through the endocytic compartments.18, 19 Alternatively, one might exploit the protein corona as a targeting ligand.4, 20 For example, the high vitronectin content in the protein corona of cationic lipid/DNA complexes enhances targeting to MDA-MB-435S cancer cells, a cell line derived from M14 melanoma cells, that overexpress the two major vitronectin receptors, ανβ3 and ανβ5 integrins.19 Overall, we expect that increased consideration of targeting ligand screening by the protein corona that surrounds the NP will help to improve the efficiency of NP uptake in vivo.
Figure 3.
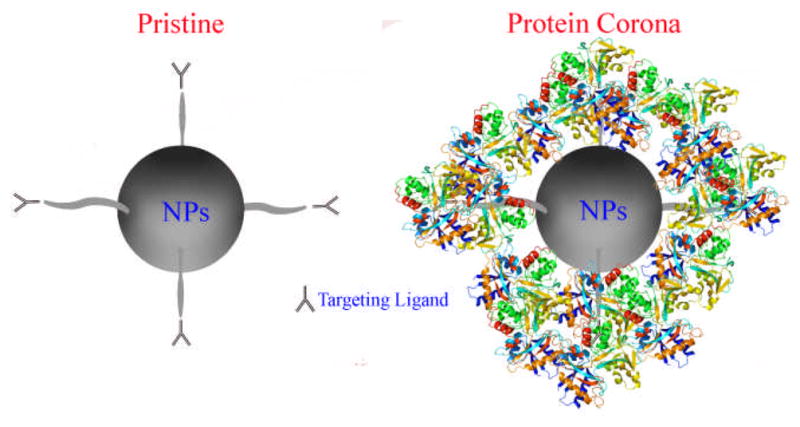
Simplified schematic of protein corona-induced screening of NP targeting ligands, which reduces targeted NP delivery. The protein corona covers the targeting ligands on the NP, preventing the ligands from binding to their targets on a separate surface (i.e., a cell).
Supplementary Material
Acknowledgments
Portions of this work were carried out in the Metabolomics Center in the Roy J. Carver Biotechnology Center, Univ. of Ill., and the Frederick Seitz Materials Research Laboratory Central Facilities, Univ. of Ill., which is partially supported by the US Department of Energy under grant DE-FG02-07ER46471. MLK holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund. JC acknowledges the support of NIH via 1R21CA152627. VM was funded by the NIH National Cancer Institute Alliance for Nanotechnology in Cancer Midwest Cancer Nanotechnology Training Center (M-CNTC) Grant. The authors thank Li Tang for assistance with NP synthesis and Qian Yin for zeta potential measurement. MM thanks Professor Kenneth S. Suslick from the Department of Chemistry at the Univ. of Ill. for the invaluable advices.
Footnotes
Electronic Supplementary Information (ESI) available: Synthetic methods, substrate characterization, protein corona analysis, supplemental schemes, figures and tables. See DOI: DOI: 10.1039/b000000x/
References
- 1.Mahmoudi M, Lynch I, Ejtehadi MR, Monopoli MP, Bombelli FB, Laurent S. Chem Rev. 2011;111:5610–5637. doi: 10.1021/cr100440g. [DOI] [PubMed] [Google Scholar]
- 2.Gao X, Cui Y, Levenson RM, Chung LWK, Nie S. Nat Biotech. 2004;22:969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 3.Choi CHJ, Alabi CA, Webster P, Davis ME. Proc Natl Acad Sci. 2010;107:1235–1240. doi: 10.1073/pnas.0914140107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahon E, Salvati A, BaldelliBombelli F, Lynch I, Dawson KA. J Control Release. 2012;161:164–174. doi: 10.1016/j.jconrel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Monopoli MP, Walczyk D, Campbell A, Elia G, Lynch I, Baldelli Bombelli F, Dawson KA. J Am Chem Soc. 2011;133:2525–2534. doi: 10.1021/ja107583h. [DOI] [PubMed] [Google Scholar]
- 6.Walczyk D, Bombelli FB, Monopoli MP, Lynch I, Dawson KA. J Am Chem Soc. 2010;132:5761–5768. doi: 10.1021/ja910675v. [DOI] [PubMed] [Google Scholar]
- 7.Maiorano G, Sabella S, Sorce B, Brunetti V, Malvindi MA, Cingolani R, Pompa PP. ACS Nano. 2010;4:7481–7491. doi: 10.1021/nn101557e. [DOI] [PubMed] [Google Scholar]
- 8.Albanese A, Tang PS, Chan WCW. Annu Rev Biomed Eng. 2012;14:1–16. doi: 10.1146/annurev-bioeng-071811-150124. [DOI] [PubMed] [Google Scholar]
- 9.Deng ZJ, Liang M, Monteiro M, Toth I, Minchin RF. Nature Nanotech. 2011;6:39–44. doi: 10.1038/nnano.2010.250. [DOI] [PubMed] [Google Scholar]
- 10.Bertrand N, Leroux JC. J Control Release. 2012;161:152–163. doi: 10.1016/j.jconrel.2011.09.098. [DOI] [PubMed] [Google Scholar]
- 11.Chithrani BD, Chan WCW. Nano Lett. 2007;7:1542–1550. doi: 10.1021/nl070363y. [DOI] [PubMed] [Google Scholar]
- 12.Lesniak A, Campbell A, Monopoli MP, Lynch I, Salvati A, Dawson KA. Biomaterials. 2010;31:9511–9518. doi: 10.1016/j.biomaterials.2010.09.049. [DOI] [PubMed] [Google Scholar]
- 13.Dobrovolskaia MA, Aggarwal P, Hall JB, McNeil SE. Mol Pharm. 2008;5:487–495. doi: 10.1021/mp800032f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang L, Fan TM, Borst LB, Cheng J. ACS Nano. 2012;6:3954–3966. doi: 10.1021/nn300149c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dommerholt J, Schmidt S, Temming R, Hendriks LJA, Rutjes FPJT, van Hest JCM, Lefeber DJ, Friedl P, van Delft FL. Angew Chem Int Ed. 2010;49:9422–9425. doi: 10.1002/anie.201003761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shapero K, Fenaroli F, Lynch I, Cottell DC, Salvati A, Dawson KA. Mol BioSyst. 2011;7:371–378. doi: 10.1039/c0mb00109k. [DOI] [PubMed] [Google Scholar]
- 17.Gref R, Lück M, Quellec P, Marchand M, Dellacherie E, Harnisch S, Blunk T, Müller RH. Colloids Surf B Biointerfaces. 2000;18:301–313. doi: 10.1016/s0927-7765(99)00156-3. [DOI] [PubMed] [Google Scholar]
- 18.Lundqvist M, Stigler J, Cedervall T, Berggård T, Flanagan MB, Lynch I, Elia G, Dawson K. ACS Nano. 2011;5:7503–7509. doi: 10.1021/nn202458g. [DOI] [PubMed] [Google Scholar]
- 19.Ghavami M, Saffar S, Abd Emamy B, Peirovi A, Shokrgozar MA, Serpooshan V, Mahmoudi M. RSC Advances. 2013;3:1119–1126. [Google Scholar]
- 20.Caracciolo G, Pozzi D, Capriotti AL, Cavaliere C, Cardarelli F, Bifone A, Bardi G, Salomone F, Laganà A. Nanotechnology 2012. Santa Clara, CA: 2012. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.