Abstract
The properties of the human macrophage galactose receptor have been investigated. Specificity for N-acetylgalactosamine (GalNAc) residues with exposed 3- and 4-hydroxyl groups explains virtually all of the results obtained from a recently expanded array of synthetic glycans and is consistent with a model for the structure of the binding site. This simple interaction is sufficient to explain the ability of the receptor to bind to tumor-cell glycans bearing Tn and sialyl-Tn antigens, but not to more elaborate O-linked glycans that predominate on normal cells. This specificity also allows for binding of parasite glycans and screening of an array of bacterial outer membrane oligosaccharides confirms that the receptor binds to a subset of these structures with appropriately exposed GalNAc residues. A key feature of the receptor is the clustering of binding sites in the extracellular portion of the protein, which retains the trimeric structure observed in the cell membrane. Chemical crosslinking, gel filtration, circular dichroism analysis and differential scanning calorimetry demonstrate that this trimeric structure of the receptor is stabilized by an α-helical coiled coil that extends from the surface of the membrane to the globular carbohydrate-recognition domains. The helical neck domains form independent trimerization domains. Taken together, these results indicate that the macrophage galactose receptor shares many of the features of serum mannose-binding protein, in which clusters of monosaccharide-binding sites serve as detectors for a simple epitope that is not common on endogenous cell surface glycans but that is abundant on the surfaces of tumor cells and certain pathogens.
Keywords: Carbohydrate-recognition domain, Galactose receptor, Glycan array, Glycan-binding receptor, Lectin
Introduction
Carbohydrate-binding receptors in mammals interact with both endogenous glycans, such as those on cell surfaces and circulating glycoproteins, and oligosaccharides and polysaccharides on the surfaces of micro-organisms. Recognition of endogenous glycans provides a mechanism for trafficking of glycoproteins within and between cells as well as cell–cell adhesion and signaling, while binding of sugars on the surfaces of viruses, bacteria, fungi and parasites provides a mechanism of pathogen recognition in the innate immune system (Robinson et al. 2006; Vasta et al. 2007; Geijtenbeek and Gringhuis 2009). These different functions are often mediated by structurally related receptors, many of which contain modular carbohydrate-recognition domains (CRDs) that fall into more than a dozen different structural families (Taylor and Drickamer 2011). The largest and most diverse of the families of receptors contain Ca2+-dependent CRDs that share a common folded structure and are designated C-type CRDs (Weis et al. 1998).
Discrimination of different sugar structures by C-type CRDs can be viewed as occurring at multiple levels, each of which imposes increasing specificity on the interaction. Initial discrimination between different classes of monosaccharides is achieved in a primary sugar-binding site that is constructed around a conserved Ca2+-binding site, in which the orientation of adjacent hydroxyl groups of the sugar determines the coordination geometry with the Ca2+. Most binding sites involve interaction with the 3- and 4-hydroxyl groups and bind selectively either to mannose-type sugars, including mannose, glucose and GlcNAc, or to galactose-type sugars, including galactose and GalNAc (Weis and Drickamer 1996). Further specificity for individual monosaccharides results from local contacts with residues near the Ca2+ site. Oligosaccharide recognition is achieved through interactions further away on the surface of the CRD, often through bumps and protrusions that form extended or secondary binding sites (Taylor and Drickamer 2009).
Beyond the recognition of individual glycans through these mechanisms, higher order specificity is often achieved by placing multiple CRDs in groups, either as a string of CRDs in a single polypeptide or as a cluster of CRDs in an oligomer of receptor polypeptides (Dam and Brewer 2008; Coombs et al. 2010). From the organization of the relatively few receptors that have been examined in molecular detail, two general paradigms for oligomer organization have emerged. In one mechanism, first described in serum mannose-binding protein, CRDs with simple primary-binding sites that bind mannose-type sugars are placed at a fixed, wide spacing to allow high avidity, multivalent interaction with dense arrays of mannose and GlcNAc found on bacterial and fungal surfaces (Weis and Drickamer 1994; Weis et al. 1998). The dendritic cell surface receptor DC-SIGN (dendritic cell-specific intercellular adhesion molecule-3 grabbing nonintegrin) interacts with viral surfaces by a somewhat different mechanism, in which mammalian-type high-mannose oligosaccharides are bound selectively through extended-binding sites in the CRDs, which are then flexibly positioned in the receptor oligomer to accommodate the organization of glycans on the viral surface (Feinberg et al. 2009; Menon et al. 2009).
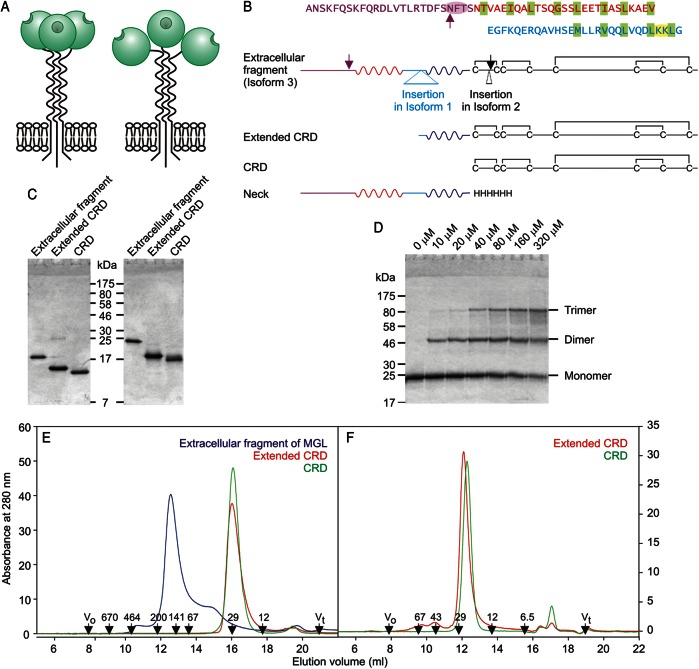
The macrophage galactose receptor (macrophage galactose lectin (MGL)) is a transmembrane cell surface receptor on macrophages and dendritic cells (Kawasaki et al. 1986; Suzuki et al. 1996). Although MGL has been reported to bind to a variety of galactose- and GalNAc-containing ligands, earlier glycan array analysis suggested that the primary targets are terminal GalNAc residues in the Tn antigen, α-GalNAc-Ser/Thr (van Vliet et al. 2005; Napoletano et al. 2007; Saeland et al. 2007). The Tn antigen, particularly on mucin MUC1, is associated with tumor cell surfaces (Ju et al. 2008). Binding of tumor antigens can lead to endocytosis (Higashi et al. 2002; Napoletano et al. 2007). MGL on dendritic cells binding to Tn antigen is associated with antigen presentation and regulation of activation of immune cells (Denda-Nagai et al. 2010; Singh et al. 2011; Napoletano et al. 2012). A further target is the LacdiNAc disaccharide, GalNAcβ1-4GlcNAc, which is abundant in parasitic helminthes (van Vliet et al. 2005; van Die and Cummings 2010) and which is present on pituitary hormones, although in this case it typically bears a 4-sulfate group (Mi et al. 2008). In addition, MGL binding to the mucin-like domain of filoviruses such as Zaire virus provides a mechanism for entry into macrophages and dendritic cells (Takada et al. 2004). Chemical crosslinking indicates that MGL is a homo-trimer (Iida et al. 1999), but the arrangement of CRDs in the receptor has not been investigated. MGL is formed from a single type of subunit, which is a Type II transmembrane polypeptide in which a C-terminal CRD is separated from the membrane anchor by an extended neck domain (Figure 1A).
Fig. 1.
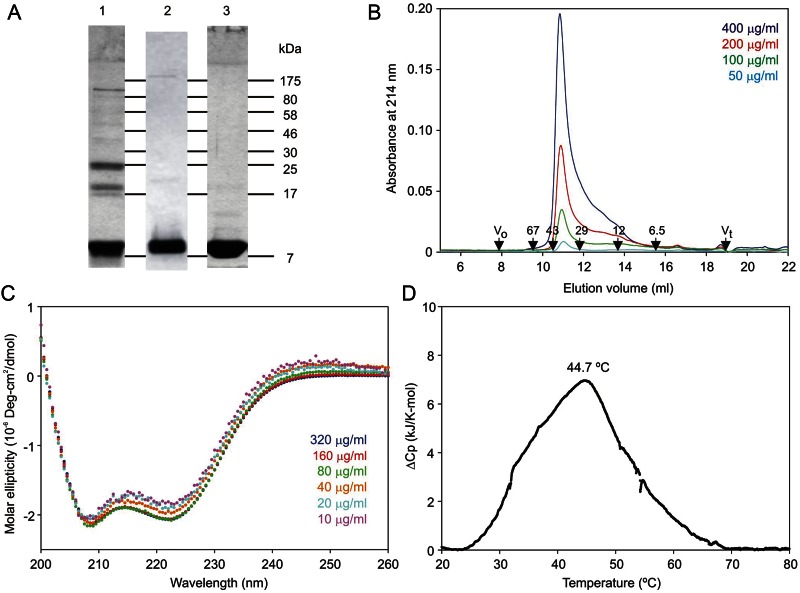
Organization of the MGL polypeptide. (A) A summary of two potential views of MGL organization based on paradigms from mannose-binding protein (left), in which CRDs interact extensively and DC-SIGN (right), in which the neck regions mediate oligomer formation independently of the CRDs. (B) Linear diagrams of the MGL polypeptide based on the most abundant isoform, which is designated isoform 3. Additional sequences found in some cDNAs from alternatively spliced mRNA molecules are present in isoforms 1 and 2. Regions expressed are shown, along with the sequence of the neck region divided into four sub-regions, which are color-coded to correspond to the sequence above. Two of the sub-regions contain heptad repeat patterns of nonpolar aliphatic amino acid side chains highlighted with green shading. Glycosylation sites are noted with arrows. The disulfide bonding pattern in the CRD is also indicated. (C) SDS–PAGE (17.5% gel) of purified fragments of MGL. Left panel, samples without reducing agent. Right panel, samples reduced with 2-mercaptoethanol. Gel was stained with Coomassie blue. (D) SDS–PAGE (10% gel) of the extracellular fragment of MGL subjected to chemical crosslinking with bis-sulfosuccinimidylsuberate. Concentrations of cross-linking reagent are indicated at the top of each well. Gel was stained with Coomassie blue. (E) Gel-filtration analysis of fragments of MGL on a Superdex S200 column. (F) Gel-filtration analysis of CRD and extended CRD of MGL on a Superdex S75 column. In (E and F), elution positions of standards are indicated in kDa at the bottom.
In the present work, physical and chemical methods have been used to show that the extracellular portion of MGL is a trimer that is stabilized primarily by an almost entirely helical coiled-coil neck region and that the CRDs form relatively independent domains. The CRDs show specificity for GalNAc residues with exposed 3- and 4-hydroxyl groups. As in the case of serum mannose-binding protein, this combination of selective binding to a simple monosaccharide epitope and appropriate clustering of binding sites is sufficient to make the receptor a detector for nonself cell surfaces.
Results and discussion
Characterization of the extracellular portion of MGL
Three isoforms of human MGL, originating from alternative splicing in the extracellular region, have been described (Higashi et al. 2002). Compared with isoform 2 (National Center for Biotechnology Infomation accession number NP_006335) originally cloned from macrophages (Suzuki et al. 1996), a form cloned from dendritic cells, which we designate isoform 3, has a deletion of three amino acids in the N-terminal portion of the CRD (Figure 1B). A further form, designated isoform 1 (accession number NP_878910), was cloned from immature dendritic cells and has an additional insertion of 27 amino acids in the middle of the neck domain (Valladeau et al. 2001). Screening of tissue cDNA libraries with polymerase chain reaction primers reveals that the most common form of mRNA encodes isoform 3.
Portions of MGL isoform 3 corresponding to the whole extracellular region, as well as subfragments representing the minimal globular CRD and the CRD with the C-terminal portion of the neck, were expressed using a bacterial system in which the protein was renatured from inclusion bodies and the resulting protein was purified using affinity chromatography on galactose-Sepharose. Additional purification to homogeneity was achieved by ion exchange chromatography (Figure 1C). Cross-linking analysis of the extracellular fragment revealed a clear progression from the monomeric polypeptide to a trimer of crosslinked polypeptides at increasing reagent concentrations (Figure 1D). These results are consistent with cross-linking experiments conducted in cells and suggest that trimer formation is mediated by regions in the extracellular portion of the protein. Gel-filtration analysis revealed an asymmetrical peak, the shape of which suggests the presence of a dissociating oligomer (Figure 1E). Based on simple molecular weight calibration of the column, a molecular weight of 137 kDa was obtained for the main species present, compared with a predicted value of 77.1 kDa (3 × 25.7 kDa) for three polypeptides. In combination, these results are consistent with the presence of a dissociating trimer that forms an asymmetrical structure due to the presence of an extended neck or stalk domain together with the CRDs. The presence of some dissociation for the isolated extracellular fragment probably reflects the absence of the membrane anchor, which would stabilize the oligomer by holding the necks in an orientation that favors interactions in a parallel coiled coil.
Binding properties of MGL
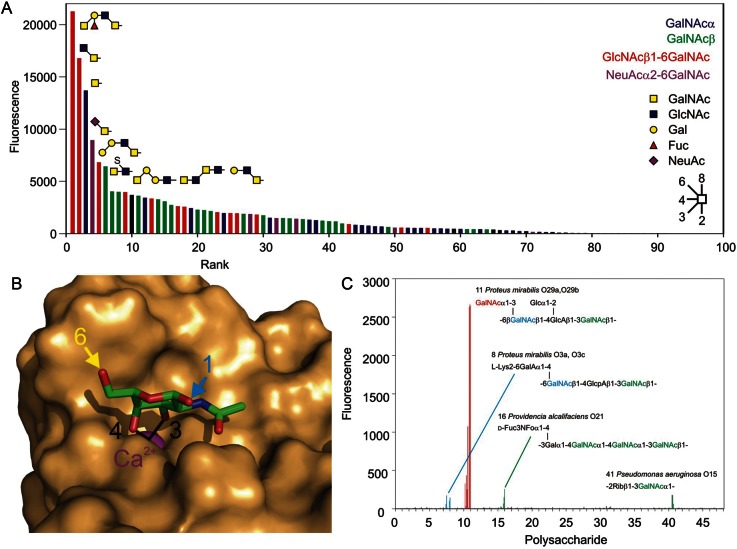
Previous studies of the binding specificity of MGL employed artificial oligomers obtained with Fc fusion domains (van Vliet et al. 2005). In order to verify the relevance of these results to the natural trimeric form of the receptor and to take advantage of the recently enlarged glycan array developed by the Consortium for Functional Glycomics, a fluorescently labeled preparation of the extracellular fragment of MGL was used to probe the array (Figure 2A). Binding to a large number of oligosaccharides is observed. All but one of the 63 glycans with signals at 2% or higher compared with the maximal signal share a common feature, which is the presence of a GalNAc residue with exposed 3- and 4-hydroxyl groups. Of the next 12 glycans in rank order of binding, a further seven meet this criterion and of the remaining 535 glycans on the array, which score at <0.5% of the maximal value, only six glycans meet this criterion. A strict requirement for this pair of exposed hydroxyl groups is consistent with the proposed structure of the ligand complex based on the structure of the closely related asialoglycoprotein receptor and the CRD from mannose-binding protein which has been modified to mimic the specificity for GalNAc (Kolatkar et al. 1998; Feinberg et al. 2000). Binding requires that the 3- and 4-hydroxyl groups ligate to the conserved Ca2+ residue in the CRD (Figure 2B). Additional contacts with the 2-acetamido group render binding specific for GalNAc over galactose.
Fig. 2.
Glycan array and pathogen array analysis of MGL-binding specificity. (A) Synthetic glycan array. Results are arranged in rank order based on decreasing signal observed on the glycan array probed with fluorescently labeled extracellular fragment of MGL at 10 μg/mL. Similar results were obtained at 1 and 90 μg/mL. The complete glycan array results are provided in Supplementary data, Table S1. All signals shown in color result from binding of glycans containing GalNAc with free 3- and 4-hydroxyl groups. In the top 65 glycans, the single exception to this rule is a strong signal for a simple α-linked fucose residue (Position 7). No studies with MGL binding to earlier versions of the glycan array have revealed binding to this sugar and none of the 170 other glycans on the array that contain α-linked fucose but lack GalNAc give signals above background, suggesting that this signal does not reflect an alternative binding specificity for glycans. (B) A model of the binding site of MGL based on the structure of the CRD of mannose-binding protein modified with portions of the MGL sequence so that it binds GalNAc (Feinberg et al. 2000). This figure was prepared with PyMol based on Protein Data Bank entry 1fih. The sequences of the model protein and human MGL are compared in Supplementary data, Figure S1. (C) Bacterial O-specific polysaccharide array. Each oligosaccharide is represented five times printed directly and five times printed after derivatization with a disubstituted oxamine linker, each at 0.03, 0.06, 0.125, 0.25 and 0.5 mg/mL. Red, GalNAc residues in terminal positions on branches; blue, 6-substituted GalNAc in the backbone sequence; green, 3- or 4-substituted GalNAc in the backbone sequence. A complete list of the bacterial sources of the lipopolysaccharides and their O-specific polysaccharide core structures is provided in Supplementary data, Table S2.
Among the 75 glycans on the array that contain GalNAc residues with exposed 3- and 4-hydroxyl groups, other parts of the glycan structures do not appear to correlate with the apparent strength of binding. Only two oligosaccharides bind better than a simple GalNAc residue. One of these is the next simplest structure, the GlcNAcβ1-6GalNAc disaccharide that forms the O-linked glycan Core 6, and the top-ranked glycan contains two different GalNAc residues, either of which could be accommodated in the binding site. In roughly half the glycans, the GalNAc residue is exposed as a nonreducing terminal residue while in the remainder it is derivatized on the 6-hydroxyl group. In many cases, the 6-linked sugar is GlcNAc, but in others sialic acid or sulfate is attached at the 6 position. In the proposed ligand-binding complexes, the 6-hydroxyl group is projected away from the binding site (Figure 2B), which is consistent with the finding that substitution at this position is tolerated. Similar binding to carbohydrates modified in the 6 position has been reported for hepatic asialoglycoprotein receptors, although these receptors bind to galactose-terminated ligands as well (Park and Baenziger 2004; Coombs et al. 2006). Reference to the model in combination with the glycan array results serves to highlight the fact that MGL does not just bind to GalNAc residues in nonreducing terminal positions, since the requirement is for unmodified 3- and 4-hydroxyl groups.
In the glycans in which GalNAc is the nonreducing terminal residue, there is an approximately equal mixture of ligands with α- and β-linked GalNAc. This observation is consistent with the accessibility of the anomeric carbon of the GalNAc residue in the binding site observed in the structural model (Figure 2B), so that attachment to hydroxyl groups in either configuration is possible. There is no clear pattern to the rank ordering of binding and the presence or absence of specific structural features beyond the GalNAc residue. Given that the simple mono- and disaccharides bind better than other structures that contain these motifs, the primary effect of various elaborations is to reduce binding, suggesting that in larger glycans, access of the key hydroxyl groups to the binding site is reduced as a result of steric hindrance.
The combination of a strict requirement for exposed 3- and 4-hydroxyl groups on GalNAc and a preference for smaller glycans containing this motif explains the ability of MGL to bind to the Tn antigen, consisting of GalNAc linked to serine or threonine residues. The sialyl-Tn structure, NeuAcα2-6GalNAc, as well as the Core 6 structure also binds. The Tn, sialyl-Tn and Core 6 structures are all rare on normal cell surfaces but particularly the first two of these glycans are commonly present on cell surface mucins of carcinomas and other tumor cells (Ju et al. 2008). In contrast, binding to the T antigen or Core 1 structure, consisting of Galβ1-3GalNAc linked to serine or threonine residues, is not observed, since the 3-linked galactose would block binding to the GalNAc residue. Thus, the simple specificity deduced from the array explains the ability of MGL to distinguish normal from tumor cells and is not necessary to invoke an extended oligosaccharide-binding site to explain the ability of MGL to bind to tumor cells.
The binding specificity observed using the natural trimer and the expanded synthetic glycan array are consistent with previous studies using earlier versions of the array containing more limited sets of glycans and with competition studies (van Vliet et al. 2005). The glycans on the current array are presented as oligosaccharides directly linked to coated glass slides, while the previously reported array employed biotinylated oligosaccharides presented on streptavidin-coated plastic wells. Thus, the current results demonstrate that the observed specificity is platform-independent as well as testing a large number of glycans that do and do not contain the binding motif. The finding that similar glycans are bound regardless of whether the CRDs are presented in the natural trimer used here or as part of Fc fusion proteins used in earlier studies indicates that binding specificity for individual oligosaccharides arises from contacts with individual CRDs.
Binding of MGL to nonself glycans
The array of synthetic glycans generated by the Consortium for Functional Glycomics contains only a limited number of sugar combinations commonly found on bacterial pathogens, although it does contain glycans with terminal GalNAcβ1-4GlcNAc (LacdiNAc) disaccharides that are commonly found on helminthes. To investigate the ability of MGL to interact with nonself glycans on micro-organisms, the same preparation of labeled, trimeric extracellular fragment was used to probe a prototype pathogen array, which contains 48 terminal oligosaccharide structures from bacterial outer membrane lipopolysaccharides. The results reveal a strong signal with a single oligosaccharide and much weaker signals for three other oligosaccharides (Figure 2C). These results are fully consistent with the proposed specificity based on the synthetic array. The strong signal arises from binding to the terminal GalNAc residues present on the repeated branches of the oligosaccharide from Proteus mirabilis O29a/O29b. The weaker signal for the oligosaccharide from P. mirabilis O3a/O3c reflects the presence of two GalNAc residues in the backbone, one of which would be a potential ligand if branches were absent from the 4 position in some copies of the repeat unit. The other GalNAc residue in this oligosaccharide as well as the GalNAc residues in the backbone sequences of Providencia alcalifaciens O21 and Pseudomonas aeruginosa O15 would not be expected to bind to MGL, since they are substituted at the 3- or 4-position. However, some copies of the oligosaccharide will have a single terminal GalNAc residue, so there will be a limited number of sites for binding to MGL. The frequency of such residues, at most one per oligosaccharide molecule, is much less than for the frequency of branches in the P. mirabilis O3a/O3c oligosaccharide, leading to weaker binding. There are other oligosaccharides on the array that contain backbone GalNAc residues, but these occur at only one position in longer repeats, while GalNAc makes up between 50 and 75% of the residues in the three oligosaccharides that show weak binding, resulting in a predicted higher frequency of exposed terminal GalNAc residues in these cases.
The strong selectivity of human MGL for GalNAc contrasts with the comparable binding of both galactose- and GalNAc-terminated structures by the single rat ortholog of human MGL (Iobst and Drickamer 1996; Coombs et al. 2006). Of the two mouse orthologs, one shows binding to both galactose and GalNAc, with preferential binding to GalNAc and the other is largely specific for oligosaccharides containing Lewisa or Lewisx epitopes (Tsuiji et al. 2002; Singh et al. 2009). Such variation in the binding specificity of glycan-binding receptors between species seems to be more common for receptors that bind pathogens, such as DC-SIGN, compared with those that bind endogenous ligands, such as the selectins (Powlesland et al. 2006).
Cluster of binding sites generated by trimer formation in the neck region
The confirmation that the binding epitope is relatively simple is analogous to the finding that mannose-binding protein binds to terminal sugar residues that have free 3- and 4-hydroxyl groups with the stereochemistry found in mannose and GlcNAc. In the same way that mannose-binding protein binds to bacterial and fungal walls because of the presence of high concentrations of these target sugars, it seems likely that MGL binds to pathogens and tumor cells largely as a result of the presence of high concentrations of appropriately exposed GalNAc rather than a more specific structure.
Based on the similar ways that MGL and mannose-binding protein each bind a simple epitope common on target cells but rare on endogenous cells, the relative positioning of binding sites in MGL may be an important determinant of biological targets as it is in MBP. The structural basis for formation of the oligomers was investigated by comparing the extracellular fragment of MGL and CRD-containing fragments (Figure 1E and F). Gel-filtration analysis of the CRD and the extended CRD yielded molecular weight values corresponding closely to the expected values for globular polypeptides of 18 and 20 kDa, respectively. These results, combined with the earlier analysis of the full extracellular fragment of the polypeptide, indicate that the neck region of the protein is required for oligomer formation.
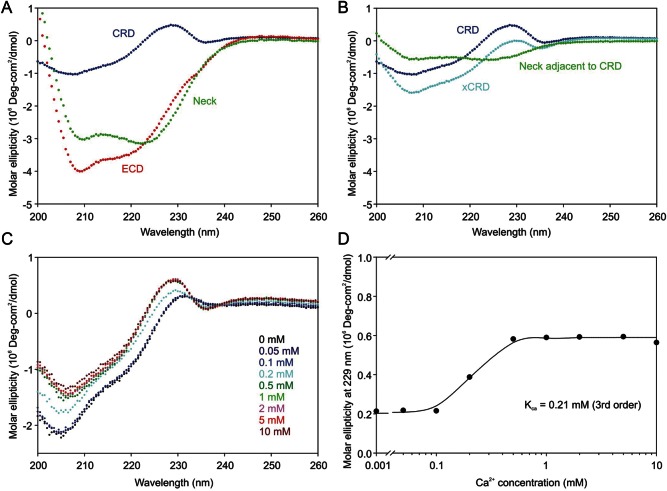
N-terminal to the first cysteine residue that defines the minimal CRD (Figure 1B), the sequence of much of the neck contains aliphatic amino acid side chains arranged in heptad repeats that are characteristic of α-helical coiled coils. However, the heptad repeat pattern is not evident in the N-terminal 20 amino acid residues of the neck, and there is an interruption in the middle of the neck. In order to investigate the conformation of the neck domain in the extracellular fragment of MGL by circular dichroism analysis, it was necessary to subtract the spectrum of the CRD from larger fragments (Figure 3A and B). An unusual feature of the CD spectrum is a region of positive ellipticity at 229 nm followed by a negative inflection at 235 nm. Signals at this wavelength are generally associated with amino acid side chains, particularly the indole ring of tryptophan, interacting with backbone amides (Liang and Chakrabarti 1982; Woody 1994). A key conserved tryptophan residue in C-type CRDs lies under the sugar-binding Ca2+ site, packed against an unusual cis-proline residue that is flanked by two of the ligands for the Ca2+ (Ng et al. 1998; Ng and Weis 1998). In mannose-binding protein, the configuration of this proline residue changes in the absence of Ca2+ and it moves away from the tryptophan residue (Ng et al. 1998; Ng and Weis 1998). Titration of the CRD with Ca2+ revealed a change in the signal at 229 nm which occurs with a midpoint for the transition at 0.2 mM Ca2+ (Figure 3C and D), which corresponds to the concentration range for Ca2+ binding as assessed by the Ca2+-dependence of ligand binding to C-type CRDs (Loeb and Drickamer 1988; Weis et al. 1991). The sensitivity of the CD signal in this region of the spectrum is thus consistent with a model for the structure of the CRD from MGL based on the CRD of mannose-binding protein, in which the CD signal of the conserved tryptophan is sensitive to the position and conformation of the proline residue.
Fig. 3.
Conformations of MGL fragments analyzed by circular dichroism. Circular dichroism spectra for the entire extracellular fragment of MGL and for the CRD and extended CRD fragments were normalized on a molar basis so that difference spectra corresponding to the neck could be obtained by subtraction. (A) Calculation of difference spectrum corresponding to the entire neck domain. (B) Calculation of difference spectrum corresponding to portion of the neck adjacent to the CRD. (C) Circular dichroism spectrum of the CRD as a function of Ca2+ concentration. (D) Ca2+-dependence of circular dichroism of the CRD at 229 nm. Values for the molar ellipticity at 229 nm were fitted to a third-order binding equation using SigmaPlot.
The difference circular dichroism spectra of the CRD and the full extracellular fragment yielded a spectrum corresponding closely to >98% α-helical spectrum based on CDNN deconvolution analysis (Figure 3A). The lower trough at 222 nm compared with 208 nm is expected for a coiled coil compared with a simple helical structure (Monera et al. 1993). A similar analysis of the extended CRD compared with the CRD revealed that the region of the neck adjacent to the CRD is also largely helical even in the absence of the remainder of the neck (Figure 3B). The results confirm that, in spite of the irregularities in the heptad repeats, the neck region extends all the way to the base of the CRD defined by the first cysteine residue that is involved in disulfide bonding within the CRD.
Relationship between domains in the extracellular portion of MGL
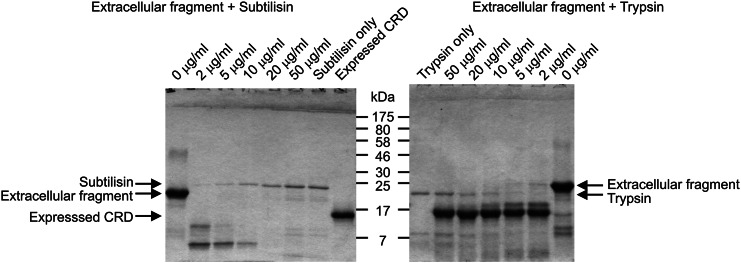
To examine the relationship between the neck domain and the CRDs, limited proteolysis was employed to probe for structurally discrete regions of MGL. In earlier studies of mannose-binding protein, subtilisin was used to define exposed regions of the polypeptide because of the relatively broad specificity of the protease (Weis et al. 1991). Digestion of MGL under similar conditions resulted in rapid digestion to low molecular weight fragments (Figure 4). Compared with the CRD of mannose-binding protein, the CRD of MGL contains an extra glycine-rich loop extending from the surface of the protein, which forms a key part of the galactose-binding site (Kolatkar et al. 1998). Thus, it seems likely that digestion by subtilisin in this extended loop structure results in degradation of the CRD and it was necessary to employ trypsin to screen for protease-sensitive sites elsewhere in the extracellular fragment of MGL. Trypsin would not be expected to cut within this loop because it lacks basic residues (Figure 4). The results reveal that the predominant cleavage results in a stable fragment corresponding to the CRD, indicating that the junction between the CRD and the neck is more protease-sensitive than other regions of the polypeptide. Alternatively, the results might reflect lower stability of the neck region, which might partially unfold and become sensitive to proteases while the CRD remains protease resistant.
Fig. 4.
Limited proteolysis of the extracellular fragment of MGL. SDS–PAGE of digestions conducted for 30 min at 37°C at increasing concentrations of protease. Gel was stained with Coomassie blue. The bacterially expressed CRD fragment of MGL was run in parallel for comparison in the final lane of the left-hand panel.
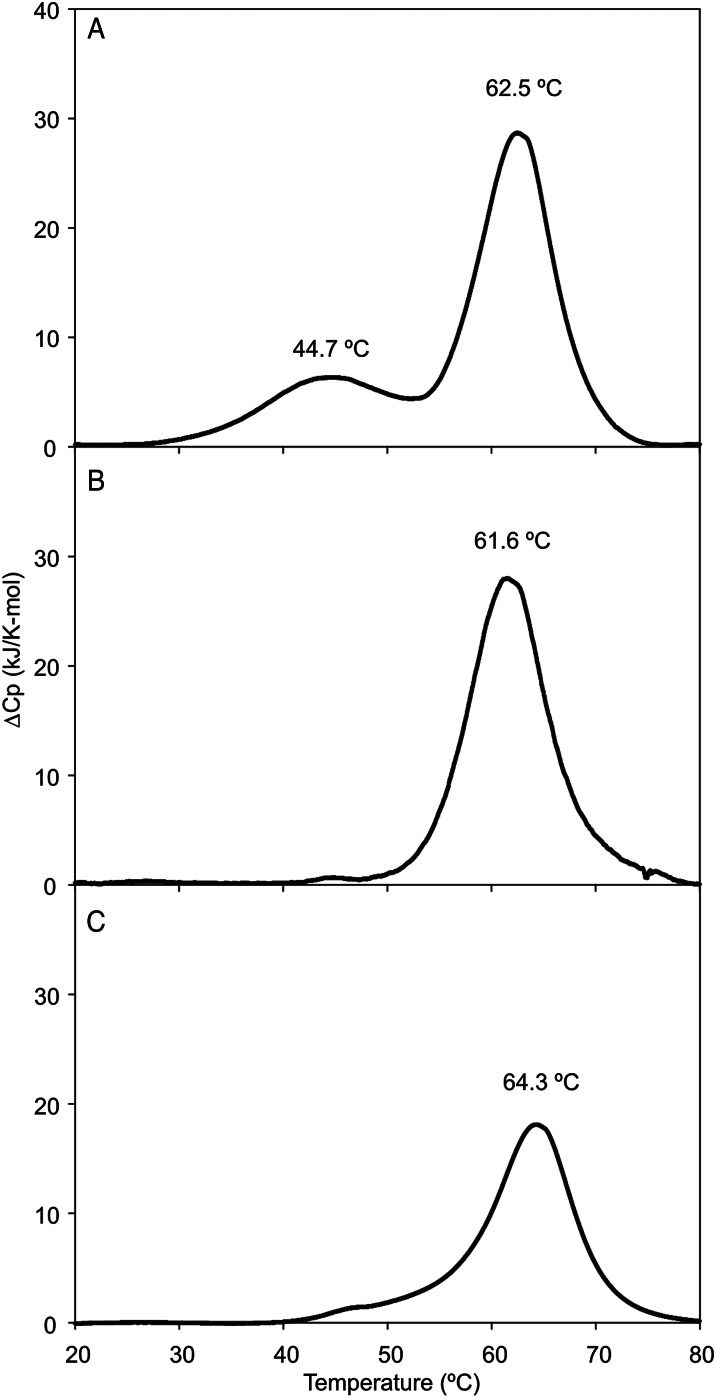
Differential scanning calorimetry was used to determine the extent to which the neck and CRD of MGL fold and function independently. Two transitions were observed upon heating of the intact extracellular fragment of MGL, one centered at 45°C and one at 62°C (Figure 5A). Based on results with other C-type CRDs, the denaturation temperature near 62°C would be expected to represent the CRD (Yu et al. 2009), a prediction that was confirmed by analysis of the CRD and the extended CRD by calorimetry (Figure 5B and C), each of which shows a single transition at approximately 62°C. Thus, the lower temperature transition was predicted to correspond to unfolding of the neck. The presence of two transitions would be consistent with the neck and CRD behaving as largely independent domains. This interpretation is supported by the fact that the CRD denatures at the same temperature in isolation as it does in the presence of the neck region.
Fig. 5.
Stability of MGL fragments analyzed by differential scanning calorimetry. Proteins were extensively dialyzed against 100 mM NaCl, 10 mM HEPES, pH 7.4 and 2.5 mM CaCl2 for calorimetry. Concentrations of proteins were (A) extracellular fragment of MGL 2.7 mg/mL; (B) extended CRD, 0.7 mg/mL; (C) CRD, 10.0 mg/mL.
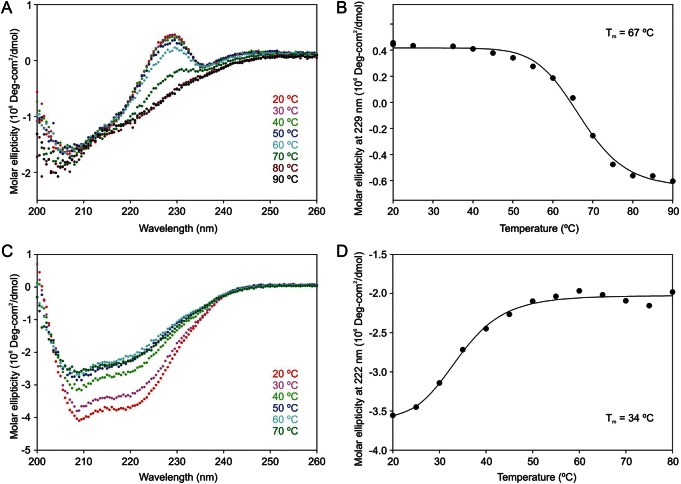
Further evidence regarding the stabilities of the two domains in the extracellular portion of MGL was obtained by examining the effect of temperature on the circular dichroism spectrum. Comparing the circular dichroism signal at 229 nm for the isolated CRD at various temperatures produced a denaturation curve with a midpoint of unfolding falling at 67°C, which is consistent with the value obtained from the calorimetry experiments (Figure 6A and B). In contrast, analysis of the signal at 222 nm for the spectrum of the intact extracellular fragment of MGL, which is dominated by the signal from the α-helical neck domain, gave a transition temperature of 34°C (Figure 6C and D). These results are consistent with the interpretation that the lower temperature transition in the calorimetry experiments corresponds to denaturation of the neck as a transition independent of the unfolding of the CRD. Corresponding to the two-step unfolding process, cooling of the denatured sample resulted in two-step renaturation, a cycle that could be repeated (data not shown). This stepwise reversibility is also consistent with the presence of independent domains.
Fig. 6.
Stability of MGL fragments analyzed by circular dichroism. (A and B) Folded conformation of the CRD (0.07 mg/mL) was assessed by comparing the molar ellipticity at 229 nm, which reflects the folded conformation of the tryptophan residue adjacent to the Ca2+ that forms the sugar-binding site (see Figure 3C and D). (C and D) Folded conformation of the neck domain (0.27 mg/mL) was assessed based on the ellipticity at 222 nm of the extracellular fragment of MGL, which is sensitive to the α-helical conformation that arises dominantly from the neck domain. Proteins were dialyzed extensively against 10 mM Tris-Cl, pH 7.4 and 2.5 mM CaCl2. Spectra represent averages of 10 scans each, taken after a 2-min stabilization period at each temperature. Data were fitted to first order transitions in order to determine the midpoint of the unfolding transitions.
The neck domain of MGL as an oligomerization domain
To complement the experiments comparing the effect of removing the neck from the CRD, the neck domain was studied in isolation. The neck polypeptide was expressed with a C-terminal His6 sequence in place of the CRD, which allowed purification on immobilized Ni2+ in the presence of urea and detection by western blotting (Figure 7A, lanes 1 and 2). The polypeptide was eluted efficiently with 100 mM imidazole. Following removal of the denaturing agent, the renatured neck polypeptide was re-purified on immobilized Ni2+. In this case, release of the protein from the resin required overnight incubation in the presence of ethylenediaminetetraacetic acid (EDTA), presumably reflecting the presence of a large cluster of histidine residues in the re-formed oligomer (Figure 7A, lane 3). Gel-filtration analysis revealed that the isolated neck polypeptide was present largely as an oligomer that eluted at an apparent molecular weight of 38 kDa (Figure 7B). As with the extracellular fragment, this value is larger than the expected molecular weight of 29.8 kDa for a trimer, which would reflect the extended nature of the polypeptide. The shape of the elution profile suggests an equilibrium with a small form, which would correspond to the monomeric neck as in the case for the full extracellular fragment of the receptor.
Fig. 7.
Purification and analysis of the neck domain of MGL. (A) SDS–PAGE (17.5% gel) of neck domain purified on immobilized Ni2+ in the presence of urea (lane 1, Coomassie blue; lane 2, detection with antibody to the His6 tag) and following further purification on immobilized Ni2+ in the absence of urea (lane 3, Coomassie blue). (B) Gel-filtration analysis of the neck domain of MGL on a Superdex S75 column. Concentrations of starting samples (100 μL) are indicated. (C) Conformation of the neck domain analyzed by circular dichroism. (D) Stability of the neck domain determined by differential scanning calorimetry. Protein concentration was 0.65 mg/mL.
The circular dichroism spectrum of the isolated neck domain is dominated by α helix (Figure 7C). As the concentration of the protein is decreased, the ratio of molar ellipticity at 222 compared with 208 decreases slightly, consistent with the suggestion that the coiled coil is partially dissociating. These results correspond to the conformation observed in the difference spectrum for the neck obtained by comparing the isolated CRD and entire extracellular fragment of the protein (Figure 3A). Thus, the neck assumes an α-helical conformation independently of the CRDs. Differential scanning calorimetry analysis of the neck domain yielded a single denaturation event centered at approximately 45°C (Figure 7D). This value closely matched the deduced value for the neck in the context of the entire extracellular portion of the polypeptide. Taken together, the gel filtration, calorimetry and circular dichroism data indicate that the neck domain of MGL functions as a coiled-coil α-helical oligomerization domain that is necessary and sufficient for formation of receptor trimers.
The finding that the neck domain is primarily responsible for trimer formation contrasts with mannose-binding protein, in which extensive interactions between the CRDs are needed to stabilize the trimer and the neck domain is too short to form a stable coiled coil on its own. The ability of the neck to form a trimer in the absence of the CRDs is more like the situation with DC-SIGN, in which the neck is a fully independent trimerization domain. The analogy with DC-SIGN, as well as the finding that the neck-CRD junction is protease sensitive, suggests that the CRDs may be flexibly positioned so that their positions can adjust to accommodate the pattern of glycans on cell surfaces.
Conclusion
The analysis presented here indicates that the mechanism of target recognition by MGL shares features with both serum mannose-binding protein and DC-SIGN. Like mannose-binding protein, MGL has a monosaccharide-specific binding site that interacts with a simple epitope, which is characteristic of nonself oligosaccharides. As with DC-SIGN, the clustering of these binding sites is achieved through an independent oligomerization domain that may provide some flexibility in the positioning of the binding sites. Interestingly, recent structural analysis of langerin indicates that it combines a different set of features, in which there are extensive interactions between the CRDs, forming a fixed cluster as in mannose-binding protein, but the individual binding sites are more extended, as in DC-SIGN, leading to more selective ligand binding (Feinberg, Powlesland et al. 2010).
Methods
Cloning of MGL fragments
The extracellular fragment of MGL was cloned from a human liver cDNA library using Advantage II polymerase (Takara, St-Germain-en-Laye, France) with flanking primers TGGCCAATTCCAAATTTCAGAGGGACCTGGTGACC (forward) and CCGGGTGGTCCCACCAAAGGCAGCTCAGTG (reverse). Products of the polymerase chain reaction were cloned into vector pCRII-TOPO (Invitrogen, Paisley, UK). Fragments for expression were amplified from this cDNA, using primers to insert into the T7 expression vector pT5T (Eisenberg et al. 1990; Iobst and Drickamer 1996). The N-terminal sequences of the expressed fragments were: (Ala)AsnSerLysPheGln for the extracellular fragment, (Ala)AlaValHisSer for the extended CRD and AlaLeuThrCysGlnVal for the CRD. For the neck domain, a reverse primer encoding the C-terminal end of the neck domain and the His6 tag resulted in a C-terminal sequence of AspLeuLysLysLeuGlyHisHisHisHisHisHis. The expression plasmids were transformed into Escherichia coli strain BL21/DE3 for production of protein.
Preparation of CRD-containing fragments of MGL
Extracellular fragment and CRD-containing fragments were produced as inclusion bodies in E. coli, extracted, refolded and purified by affinity chromatography on galactose-Sepharose (Fornstedt and Porath 1975) as previously described (Iobst and Drickamer 1996) using loading buffer consisting of 0.5 M NaCl, 25 mM Tris-Cl, pH 7.8, and 25 mM CaCl2 and eluting buffer consisting of 0.5 M NaCl, 25 mM Tris-Cl, pH 7.8, and 2.5 mM EDTA. The fragment corresponding to the extracellular portion of the protein was further purified by gel filtration on a 1 × 30 cm Superdex-200 column (GE Healthcare Life Sciences, Little Chalfont, UK) in the presence of 6 M urea, 25 mM Tris-Cl, pH 6.8, at a flow rate of 1 mL/min. Absorbance was monitored at 280 nm and on 17.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) run in the absence of reducing agent. Fractions containing monomeric fragments lacking inter-chain disulfide bonds were pooled and renatured by dialysis against 150 mM NaCl, 25 mM Tris-Cl, pH 7.8, 25 mM CaCl2. For repurification of the CRD and extended CRD, the proteins were dialyzed against 50 mM Tris-Cl, pH 7.8, and centrifuged at 100,000 × g at 4°C for 15 min before fractionation on a 1-mL MonoQ column (GE Healthcare Life Sciences). The column was eluted with a linear gradient from 0 to 0.5 M NaCl in 50 mM Tris-Cl, pH 7.8, and fractions containing purified fragments were identified by SDS–PAGE. The CRD eluted at ∼0.25 M NaCl and the extended CRD eluted at approximately 0.4 M NaCl.
Preparation of MGL neck domain
Luria-Bertani medium (3 × 1 L) containing 50 μg/mL ampicillin was inoculated with 3 × 30 mL of an overnight culture that had been grown at 25°C. The culture was grown, with shaking, at 37°C to an OD550 of 0.8, at which point is was induced with 100 μg/mL isopropyl β-d-thiogalactopyranoside and incubated for a further 2.5 h at 37°C with shaking. Cells were harvested by centrifugation at 4000 × g at 4°C for 15 min, resuspended in 30 mL of cold 10 mM Tris-Cl, pH 7.8, buffer, and centrifuged at 10,000 × g at 4°C for 10 min. The supernatants were discarded and the pelleted cells were kept frozen at −80°C. For purification of the neck domain, the pellet was thawed and suspended in 60 mL of cold N1 buffer (0.5 M NaCl 25 mM Tris-Cl, pH 7.8) and lysed by sonication (4 bursts of 30 s duration). The lysate was centrifuged at 20,000 × g for 15 min at 4°C. Solid urea (28.8 g) was added to bring the solution to 8 M urea. Following centrifugation at 100,000 × g in a Beckman 50.2 Ti rotor for 30 min at 4°C, the supernatant was passed over a 2-mL NTA agarose column (QIAGEN, Manchester, UK) preloaded with 10 mL of 50 mM NiSO4 and equilibrated in N1 buffer at room temperature. The column was washed with 10 mL of 0.5 M NaCl, 25 mM Tris-Cl, pH 8.0, and 8 M urea with 20 mM imidazole and eluted with 3 column volumes each of N1 buffer containing 8 M urea and 50 mM, 100 and 200 mM imidazole. Following analysis on a 17.5% SDS–PAGE, fractions containing the neck domain, eluting at 100 mM imidazole, were dialyzed against N1 buffer and repurified on a 2-mL Ni2+-NTA agarose column in the absence of urea. The column was rinsed with 10 mL of N1 buffer containing 20 mM imidazole and eluted with 3 column volumes each of N1 buffer containing 50, 100 and 200 mM imidazole, followed by 6 × 1 mL of N1 buffer containing 10 mM EDTA. The column was kept overnight at 4°C and the tightly bound protein was eluted with an additional 8 × 1 mL of N1 buffer containing 10 mM EDTA. Purification was monitored by SDS–PAGE on 17.5% gels (Laemmli 1970). Western blots (Burnette 1981) were developed with mouse anti-His6 antibody (Sigma Chemical Co., Dorset, UK) followed by goat antimouse IgG coupled to alkaline phosphatase (Jackson ImmunoResearch, West Grove, PA) and developed using nitro-blue tetrazolium and 5-bromo-4-chloro-3′-indolylphosphate.
Gel-filtration analysis
Gel-filtration analysis was performed on 1 × 30 cm Superdex-75 and 200 columns (GE Healthcare Life Sciences). For extracellular and CRD fragments, columns were eluted with 10 mM Tris-Cl, pH 7.8, 100 mM NaCl and 2.5 mM EDTA at a flow rate of 0.5 mL/min and absorbance was monitored at 280 nm. In order to allow monitoring at 214 nm, EDTA was omitted from the elution buffer for samples of the neck domain fragment and the column was eluted in 0.5 M NaCl, 10 mM Tris-Cl, pH 7.8. Data were collected and processed using the UNICORN software. Elution positions of the following standard proteins (with Mr as indicated) were determined: Aprotinin (6.5 kDa), cytochrome c (12.4 kDa), bovine erythrocyte carbonic anhydrase (29 kDa), ovalbumin (43 kDa), bovine serum albumin (67 kDa), yeast alcohol dehydrogenase (141 kDa), β-amylase (200 kDa), E. coli β-galactosidase (464 kDa) and thyroglobulin, (670 kDa). The void volume was determined using dextran blue and the salt volume was determined from the elution position of salt in the sample detected by conductivity.
Glycan array analysis
The extracellular fragment of MGL (∼250 µg) was repurified on a 1-mL column of galactose-Sepharose. After washing with 5 mL of 125 mM NaCl, 100 mM Na-bicine, pH 9.0, 25 mM CaCl2, the protein was eluted in 0.5-mL aliquots with 125 mM NaCl, 100 mM Na-bicine, pH 9.0, 2.5 mM EDTA. The peak fractions adjusted to 25 mM CaCl2 and 25 µg of fluorescein isothiocyanate was added as 5 aliquots of 5 µL dissolved at 1 mg/mL in dimethyl sulfoxide. After incubation overnight at 4°C, the protein was repurified by affinity chromatography on a 1-mL column of galactose-Sepharose following the above protocol but using 125 mM NaCl, 25 mM Tris-Cl, pH 7.8, 25 mM CaCl2 loading buffer and 125 mM NaCl, 25 mM Tris-Cl, pH 7.8, 2.5 mM EDTA elution buffer. Version 5.1 of the synthetic glycan array of the Consortium for Functional Glycomics was screening by the standard protocol (Blixt et al. 2004; Feinberg, Taylor et al. 2010). Preparation of the prototype pathogen glycan array, containing bacterial polysaccharides, is summarized in Feinberg, Taylor et al. (2010), and details are available at: www.functionalglycomics.org/static/consortium/resources/timelined2p.shtml (McBride et al., in preparation). The synthetic array was screened at 1, 10 and 90 μg/mL of labeled protein and the pathogen array was screened at 90 μg/mL.
Limited proteolysis
Proteolysis with subtilisin or trypsin was performed in 0.5 M NaCl, 25 mM Tris-Cl, pH 7.8, 25 mM CaCl2. Aliquots (20 µL) of the extracellular fragment of MGL were combined with freshly prepared trypsin or subtilisin solutions and incubated 1 h at 37°C. Proteolysis was stopped by the addition of 20 µL of 2× SDS–PAGE sample buffer, followed by immediate boiling for 5 min and analysis on 17.5% polyacrylamide gels.
Crosslinking
The extracellular fragment of MGL was re-purified by affinity chromatography on a 1-mL galactose-Sepharose column that was washed with 5 mL of 125 mM NaCl, 100 mM HEPES, pH 7.8, 25 mM CaCl2 and eluted with 0.5-mL aliquots of 125 mM NaCl, 100 mM HEPES, pH 7.8, 2.5 mM EDTA. CaCl2 was added to a final concentration of 10 mM to the peak fraction, which contained 250 μg/mL protein. Aliquots (25 μL) were incubated with bis(sulfosuccinimidyl)suberate (Thermo Scientific, Hemel Hempstead, UK) for 1 h at room temperature. Reactions were stopped by adding 2 × sample buffer containing 1% 2-mercaptoethanol and were immediately analyzed on a 10% SDS–PAGE.
Circular dichroism analysis
Samples were dialyzed against 25 mM NaCl, 5 mM Tris-Cl, pH 7.8, 2.5 mM CaCl2 and analyzed at 0.1–0.2 mg/mL final concentration. For the Ca2+ titration, the sample was dialyzed against 25 mM NaCl, 5 mM Tris-Cl, pH 7.8, after which CaCl2 was added from a 1 M stock to achieve the indicated final concentrations. Circular dichroism spectra on 200 µL samples were obtained in a 1-mm quartz cell using a Chirascan spectropolarimeter (Applied Photophysics, Leatherhead, UK) at a band width of 1 nm, step size of 0.5 nm and 1 s per step. The spectra presented represent an average of ten scans. Denaturation was monitored by performing scans at intervals of 5°C, after equilibration for 2 min at each temperature. Data were fitted to a simple first-order curve using SigmaPlot. Protein concentrations were determined by the alkaline ninhydrin assay (Hirs 1967).
Differential scanning calorimetry
Samples were dialyzed extensively against 25 mM Na-HEPES, pH 7.8, 125 mM NaCl, 5 mM CaCl2 and degassed for 15 min. Analysis was performed in a Calorimetry Sciences Nano III calorimeter with a sample loop volume of 300 μL. Initial scans from 5 to 20°C were repeated until a stable signal was obtained. Baselines were calculated using a third-order polynomial equation to fit the data using SigmaPlot as previously described (Wallis et al. 2004). Protein concentrations were determined by the alkaline ninhydrin assay (Hirs 1967).
Supplementary data
Supplementary data for this article are available online at http://glycob.oxfordjournals.org/.
Funding
The Wellcome Trust (grant 093599 to M.E.T. and K.D.) and National Institutes of Health (NIH) (grant GM098791 to the Protein-Glycan Interaction Resource of the Consortium for Functional Glycomics). A.Q.-M. was recipient of a fellowship from Consejo Nacional de Ciencia y Tecnología (Mexico).
Conflict of interest
None declared.
Abbreviations
CRDs, carbohydrate-recognition domains; DC-SIGN, dendritic cell-specific intercellular adhesion molecule-3 grabbing nonintegrin; EDTA, ethylenediaminetetraacetic acid; MGL, macrophage galactose lectin; SDS–PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis.
Supplementary Material
Acknowledgements
We thank Eliot Ward for providing the initial cDNA clone of MGL and David Smith and Jamie Heimburg-Molinaro of the Protein-Glycan Interaction Resource of the Consortium for Functional Glycomics, Emory University, for performing the glycan array analyses.
References
- Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, et al. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci U S A. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. doi:10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette WN. Western blotting”: Electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981;112:195–203. doi: 10.1016/0003-2697(81)90281-5. doi:10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Coombs PJ, Harrison R, Pemberton S, Quintero-Martinez A, Parry S, Haslam SM, Dell A, Taylor ME, Drickamer K. Identification of novel contributions to high-affinity glycoprotein–receptor interactions using engineered ligands. J Mol Biol. 2010;396:685–696. doi: 10.1016/j.jmb.2009.11.073. doi:10.1016/j.jmb.2009.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs PJ, Taylor ME, Drickamer K. Two categories of mammalian galactose-binding receptors distinguished by glycan array profiling. Glycobiology. 2006;16:1C–7C. doi: 10.1093/glycob/cwj126. doi:10.1093/glycob/cwj126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dam TK, Brewer CF. Effects of clustered epitopes in multivalent ligand-receptor interactions. Biochemistry. 2008;47:8470–8476. doi: 10.1021/bi801208b. doi:10.1021/bi801208b. [DOI] [PubMed] [Google Scholar]
- Denda-Nagai K, Aida S, Saba K, Suzuki K, Moriyama S, Oo-puthinan S, Tsuiji M, Morikawa A, Kumamoto Y, Sugiura D, et al. Distribution and function of macrophage galactose-type C-type lectin 2 (MGL2/CD301b): Efficient uptake and presentation of glycosylated antigens by dendritic cells. J Biol Chem. 2010;285:19193–19204. doi: 10.1074/jbc.M110.113613. doi:10.1074/jbc.M110.113613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg SP, Evans RJ, Arend WP, Verderber E, Brewer MT, Hannum CH, Thompson RC. Primary structure and functional expression from complementary DNA of a human interleukin-1 receptor antagonist. Nature. 1990;343:341–346. doi: 10.1038/343341a0. doi:10.1038/343341a0. [DOI] [PubMed] [Google Scholar]
- Feinberg H, Powlesland AS, Taylor ME, Weis WI. Trimeric structure of langerin. J Biol Chem. 2010;285:13285–13293. doi: 10.1074/jbc.M109.086058. doi:10.1074/jbc.M109.086058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg H, Taylor ME, Razi N, McBride R, Knirel YA, Graham SA, Drickamer K, Weis WI. Structural basis for langerin recognition of diverse pathogen and mammalian glycans through a single binding site. J Mol Biol. 2010;405:1027–1039. doi: 10.1016/j.jmb.2010.11.039. doi:10.1016/j.jmb.2010.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg H, Torgersen D, Drickamer K, Weis WI. Mechanism of pH-dependent N-acetylgalactosamine binding to a function mimic of the hepatic asialoglycoprotein receptor. J Biol Chem. 2000;275:35176–35184. doi: 10.1074/jbc.M005557200. doi:10.1074/jbc.M005557200. [DOI] [PubMed] [Google Scholar]
- Feinberg H, Tso CKW, Taylor ME, Drickamer K, Weis WI. Segmented helical structure of the neck region of the glycan-binding receptor DC-SIGNR. J Mol Biol. 2009;394:613–620. doi: 10.1016/j.jmb.2009.10.006. doi:10.1016/j.jmb.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornstedt N, Porath J. Characterization studies on a new lectin found in seed of Vicia ervilia. FEBS Lett. 1975;57:187–191. doi: 10.1016/0014-5793(75)80713-7. doi:10.1016/0014-5793(75)80713-7. [DOI] [PubMed] [Google Scholar]
- Geijtenbeek TBH, Gringhuis SI. Signalling through C-type lectin receptors: Shaping immune responses. Nat Rev Immunol. 2009;9:465–479. doi: 10.1038/nri2569. doi:10.1038/nri2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi N, Fujioka K, Denda-Nagai K, Hashimoto S, Nagai S, Sato T, Fujita Y, Morikawa A, Tsuiji M, Miyata-Takeuchi M, et al. The macrophage C-type lectin specific for galactose/N-acetylgalactosamine is an endocytic receptor expressed on monocyte-derived immature dendritic cells. J Biol Chem. 2002;277:20686–20693. doi: 10.1074/jbc.M202104200. doi:10.1074/jbc.M202104200. [DOI] [PubMed] [Google Scholar]
- Hirs CHW. Detection of peptides by chemical methods. Methods Enzymol. 1967;11:325–329. doi:10.1016/S0076-6879(67)11037-9. [Google Scholar]
- Iida S, Yamamoto K, Irimura T. Interaction of human macrophage C-type lectin with O-linked N-acetylgalactosamine residues on mucin glycopeptides. J Biol Chem. 1999;274:10697–10705. doi: 10.1074/jbc.274.16.10697. doi:10.1074/jbc.274.16.10697. [DOI] [PubMed] [Google Scholar]
- Iobst ST, Drickamer K. Selective sugar binding to the carbohydrate-recognition domains of rat hepatic and macrophage asialoglycoprotein receptors. J Biol Chem. 1996;271:6686–6693. doi: 10.1074/jbc.271.12.6686. doi:10.1074/jbc.271.12.6686. [DOI] [PubMed] [Google Scholar]
- Ju T, Lanneau GS, Gautam T, Wang Y, Xia B, Stowell SR, Willard MT, Wang W, Xia JY, Zuna RE, et al. Human tumor antigens Tn and sialyl Tn arise from mutations in Cosmc. Cancer Res. 2008;68:1636–1646. doi: 10.1158/0008-5472.CAN-07-2345. [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Ii M, Kozutsumi Y, Yamashina I. Isolation and characterization of a receptor lectin specific for galactose/N-acetylgalactosamine from macrophages. Carbohydr Res. 1986;151:197–206. doi: 10.1016/s0008-6215(00)90340-9. doi:10.1016/S0008-6215(00)90340-9. [DOI] [PubMed] [Google Scholar]
- Kolatkar AR, Leung AK, Isecke R, Brossmer R, Drickamer K, Weis WI. Mechanism of N-acetylgalactosamine binding to a C-type animal lectin carbohydrate-recognition domain. J Biol Chem. 1998;273:19502–19508. doi: 10.1074/jbc.273.31.19502. doi:10.1074/jbc.273.31.19502. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. doi:10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liang JN, Chakrabarti B. Spectroscopic investigations of bovine lens crystallins 1: Circular dichrosim and intrinsic fluorescence. Biochemistry. 1982;21:1847–1852. doi: 10.1021/bi00537a022. doi:10.1021/bi00537a022. [DOI] [PubMed] [Google Scholar]
- Loeb JA, Drickamer K. Conformational changes in the chicken receptor for endocytosis of glycoproteins: Modulation of binding activity by Ca2+ and pH. J Biol Chem. 1988;263:9752–9760. [PubMed] [Google Scholar]
- Menon S, Rosenberg K, Graham SA, Ward EM, Taylor ME, Drickamer K, Leckband DE. Binding site geometry and flexibility in DC-SIGN demonstrated with surface force measurements. Proc Natl Acad Sci U S A. 2009;106:11524–11529. doi: 10.1073/pnas.0901783106. doi:10.1073/pnas.0901783106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi Y, Fiete D, Baenziger JU. Ablation of GalNAc-4-sufotransferase-1 enhances reproduction by altering the carbohydrate structures of luteinizing hormone in mice. J Clin Invest. 2008;118:1815–1824. doi: 10.1172/JCI32467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monera OD, Zhou NE, Kay CM, Hodges RS. Comparison of antiparallel and parallel two-stranded a-helical coiled coils: Design, synthesis and characterization. J Biol Chem. 1993;268:19218–19227. [PubMed] [Google Scholar]
- Napoletano C, Rughetti A, Tarp MPA, Coleman J, Bennett EP, Picco G, Sale P, Denda-Hagai K, Irimura T, Mandel U, et al. Tumour associated Tn-MUC1 glycoform is internalised through the macrophage galactose C-type lectin and delivered to the HLA class I and Class II compartments in dendritic cells. Cancer Res. 2007;67:8358–8367. doi: 10.1158/0008-5472.CAN-07-1035. doi:10.1158/0008-5472.CAN-07-1035. [DOI] [PubMed] [Google Scholar]
- Napoletano C, Zizzari IG, Rughetti A, Rahimi H, Irimura T, Clausen H, Wandall HH, Belleudi F, Bellati F, Pierelli L, et al. Trageting of macrophage galactose C-type lectin (MGL) induces DC signaling and activation. Eur J Immunol. 2012;42:936–945. doi: 10.1002/eji.201142086. doi:10.1002/eji.201142086. [DOI] [PubMed] [Google Scholar]
- Ng KK-S, Park-Snyder S, Weis WI. Ca2+-dependent structural changes in C-type mannose-binding proteins. Biochemistry. 1998;37:17965–17976. doi: 10.1021/bi981972a. doi:10.1021/bi981972a. [DOI] [PubMed] [Google Scholar]
- Ng KK-S, Weis WI. Coupling of prolyl peptide bond isomerization and Ca2+ binding in a C-type mannose-binding protein. Biochemistry. 1998;37:17977–17989. doi: 10.1021/bi9819733. doi:10.1021/bi9819733. [DOI] [PubMed] [Google Scholar]
- Park EI, Baenziger JU. Closely related mammals have distinct asialoglycoprotein receptor carbohydrate specificities. J Biol Chem. 2004;279:40954–40959. doi: 10.1074/jbc.M406647200. doi:10.1074/jbc.M406647200. [DOI] [PubMed] [Google Scholar]
- Powlesland AS, Ward EM, Sadhu SK, Guo Y, Taylor ME, Drickamer K. Novel mouse homologs of human DC-SIGN: Widely divergent biochemical properties of the complete set of mouse DC-SIGN-related proteins. J Biol Chem. 2006;281:20440–20449. doi: 10.1074/jbc.M601925200. doi:10.1074/jbc.M601925200. [DOI] [PubMed] [Google Scholar]
- Robinson MJ, Sancho D, Slack EC, LeibundGut-Landmann S, Reis e Sousa C. Myeloid C-type lectins in innate immunity. Nat Immunol. 2006;7:1258–1265. doi: 10.1038/ni1417. doi:10.1038/ni1417. [DOI] [PubMed] [Google Scholar]
- Saeland E, van Vliet SJ, Backstrom M, van den Berg VC, Geijtenbeek TB, Meijer GA, van Kooyk Y. The C-type lectin MGL expressed by dendritic cells detects glycan changes on Muc1 in colon carcinoma. Cancer Immunol Immunother. 2007;56:1225–1236. doi: 10.1007/s00262-006-0274-z. doi:10.1007/s00262-006-0274-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Streng-Ouwehand I, Litjens M, Kalay H, Saeland E, Van Kooyk Y. Tumour-associated glycan modifications of antigen enhance MGL2 dependent uptake and MHC class I restricted CD8 T cell responses. Int J Cancer. 2011;128:1371–1383. doi: 10.1002/ijc.25458. doi:10.1002/ijc.25458. [DOI] [PubMed] [Google Scholar]
- Singh SK, Streng-Ouwehand I, Litjens M, Weelij DR, Garcia-Vallejo JJ, Van Vliet SJ, Saeland E, van Kooyk Y. Characterization of murine MGL1 and MGL2 C-type lectins: Distinct glycan specificities and tumor binding properties. Mol Immunol. 2009;26:1240–1249. doi: 10.1016/j.molimm.2008.11.021. doi:10.1016/j.molimm.2008.11.021. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Yamamoto K, Toyoshima S, Osawa T, Irimura T. Molecular cloning and expression of cDNA encoding human macrophage C-type lectin. J Immunol. 1996;156:128–135. [PubMed] [Google Scholar]
- Takada A, Fujioka K, Tsuiji M, Morikawa A, Higahsi N, Ebihara H, Kobasa D, Feldman H, Irimura T, Kawaoka Y. Human macrophage C-type lectin specific for galactose and N-acetylgalactosamine promotes filovirus entry. J Virol. 2004;78:2943–2947. doi: 10.1128/JVI.78.6.2943-2947.2004. doi:10.1128/JVI.78.6.2943-2947.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor ME, Drickamer K. Structural insights into what glycan arrays tell us about how glycan-binding proteins interact with their ligands. Glycobiology. 2009;19:1155–1162. doi: 10.1093/glycob/cwp076. doi:10.1093/glycob/cwp076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor ME, Drickamer K. Introduction to Glycobiology. 3rd ed. Oxford: Oxford University Press; 2011. [Google Scholar]
- Tsuiji M, Fujimori M, Ohashi Y, Higashi N, Onami TM, Hedric SM, Irimura T. Molecular cloning and characterization of a novel mouse macrophage C-type lectin, mMGL2, which has a distinct carbohydrate specificity from mMGL1. J Biol Chem. 2002;277:28892–28901. doi: 10.1074/jbc.M203774200. doi:10.1074/jbc.M203774200. [DOI] [PubMed] [Google Scholar]
- Valladeau J, Duvert-Frances V, Pin JJ, Kleijmeer MJ, Ait-Yahia S, Ravel O, Vincent C, Vega F, Jr, Helms A, Gorman D, et al. Immature human dendritic cells express asialoglycoprotein receptor isoforms for efficient receptor-mediated endocytosis. J Immunol. 2001;167:5767–5774. doi: 10.4049/jimmunol.167.10.5767. [DOI] [PubMed] [Google Scholar]
- van Die I, Cummings RD. Glycan gimmickry by parasitic helminths: A strategy for modulating the host immune response? Glycobiology. 2010;20:2–12. doi: 10.1093/glycob/cwp140. doi:10.1093/glycob/cwp140. [DOI] [PubMed] [Google Scholar]
- van Vliet SJ, van Liempt E, Saeland E, Aarnoudse CA, Appelmelk B, Irimura T, Geijtenbeek TB, Blixt O, Alvarez R, van Die I, et al. Carbohydrate profiling reveals a distinctive role for the C-type lectin MGL in the recognition of helminth parasites and tumor antigens by dendritic cells. Int Immunol. 2005;17:661–669. doi: 10.1093/intimm/dxh246. doi:10.1093/intimm/dxh246. [DOI] [PubMed] [Google Scholar]
- Vasta GR, Ahmed H, Tasumi S, Odom EW, Saito K. Biological roles of lectins in innate immunity: Molecular and structural basis for diversity in self/non-self recognition. Adv Exp Med Biol. 2007;598:389–406. doi: 10.1007/978-0-387-71767-8_27. [DOI] [PubMed] [Google Scholar]
- Wallis R, Shaw JM, Uitdehaag J, Chen C-B, Torgersen D, Drickamer K. Localization of the serine protease-binding sites in the collagen-like domain of mannose-binding protein: Indirect effects of naturally occurring mutations on protease binding and activation. J Biol Chem. 2004;278:14065–14073. doi: 10.1074/jbc.M400171200. doi:10.1074/jbc.M400171200. [DOI] [PubMed] [Google Scholar]
- Weis WI, Crichlow GV, Murthy HMK, Hendrickson WA, Drickamer K. Physical characterization and crystallization of the carbohydrate-recognition domain of a mannose-binding protein from rat. J Biol Chem. 1991;266:20678–20686. [PubMed] [Google Scholar]
- Weis WI, Drickamer K. Trimeric structure of a C-type mannose-binding protein. Structure. 1994;2:1227–1240. doi: 10.1016/S0969-2126(94)00124-3. doi:10.1016/S0969-2126(94)00124-3. [DOI] [PubMed] [Google Scholar]
- Weis WI, Drickamer K. Structural basis of lectin-carbohydrate interaction. Annu Rev Biochem. 1996;65:441–473. doi: 10.1146/annurev.bi.65.070196.002301. doi:10.1146/annurev.bi.65.070196.002301. [DOI] [PubMed] [Google Scholar]
- Weis WI, Taylor ME, Drickamer K. The C-type lectin superfamily in the immune system. Immunol Rev. 1998;163:19–34. doi: 10.1111/j.1600-065x.1998.tb01185.x. doi:10.1111/j.1600-065X.1998.tb01185.x. [DOI] [PubMed] [Google Scholar]
- Woody RW. Contributions of tryptophan side chains to the far-ultraviolet circular dichroism of proteins. Eur Biophys J. 1994;23:253–262. doi: 10.1007/BF00213575. doi:10.1007/BF00213575. [DOI] [PubMed] [Google Scholar]
- Yu QD, Oldring AP, Powlesland AS, Tso CK, Yang C, Drickamer K, Taylor M. Autonomous tetramerization domains in the glycan-binding receptors DC-SIGN and DC-SIGNR. J Mol Biol. 2009;387:1075–1080. doi: 10.1016/j.jmb.2009.02.046. doi:10.1016/j.jmb.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.