Abstract
The transcriptional activator CLOCK is a histone acetyltransferase that is required for the circadian expression of many genes. Asher et al. (2008) and Nakahata et al. (2008) now demonstrate that the NAD+-dependent enzyme SIRT1 functions as a histone deacetylase that counteracts the activity of CLOCK. These results broaden our understanding of the impact of cellular metabolism on the circadian system.
The discovery that the transcriptional activator CLOCK functions as a histone acetyltransferase (HAT) (Doi et al., 2006) has implicated chromatin remodeling in the circadian regulation of gene expression. In this sense, the existence of a histone deacetylase (HDAC) that counteracts the HAT activity of CLOCK could perhaps have been anticipated. However, not all deacetylases achieve their goal in the same way, and this is in part why the work by Asher et al. (2008) and Nakahata et al. (2008) presented in this issue is of such compelling interest. Both groups show that the HDAC SIRT1 is involved in regulating the amplitude of circadian clock-controlled gene expression. Mammalian SIRT proteins (homologs of Saccharomyces mating type cassette regulator Sir2p) use energy stored in nicotinamide adenine dinucleotide (NAD+) to catalyze the removal of the acetyl group from substrates (Sauve et al., 2006). Given that cellular NAD+ levels are coupled to metabolic activity, the identification of SIRT1 as a histone deacetylase that counteracts CLOCK function brings this protein with its extensive connections, both to aging and energy metabolism, into the circadian fold.
Analysis of the molecular basis of the circadian rhythm in the fruit fly Drosophila, the fungus Neurospora, mice, and human cells in culture has revealed at its core a common architecture, a transcription-translation feedback loop in which a heterodimeric transcription factor, whose parts interact via PAS domains, drives the expression of proteins that turn down the activity of their heterodimeric activator. In mammalian cells, the transcriptional activator complex is composed of CLOCK and BMAL1, which drive expression of the negative regulators, two PER and two CRY proteins. Targets of CLOCK-mediated HAT activity include both histones H3 and H4 (Doi et al., 2006) and BMAL1 (Hirayama et al., 2007), all of which display cycles in acetylation. PER2 is now reported to be cyclically acetylated, a modification that may also be dependent on CLOCK (Asher et al., 2008). Importantly, PER2 (Asher et al., 2008) and possibly BMAL1 (Nakahata et al., 2008) appear more stable when acetylated. Both the acetylase CLOCK and the deacetylase SIRT1 associate with BMAL1 as well as with each other, so in this light SIRT1 may be a point through which changes in cellular energy metabolism impact the functioning of the clock. Indeed, SIRT1 activity also cycles (peaking in the early evening), confirming the potential for a rhythmic input to the circadian clock.
Because mice homozygous for SIRT1 deletions fail to thrive, analysis of the functional significance of SIRT1 required ingenuity. To address this problem, both groups made imaginative use of mouse embryonic fibroblasts from knockout mice as well as dominant-negative and catalytically dead proteins and potent inhibitors. Through these analyses, the rhythm in SIRT1 activity is credited with driving the rhythms in BMAL1, H3, and PER2 acetylation. Moreover, SIRT1 activity influences the amplitude of a variety of molecular rhythms in clock gene and clock-controlled gene expression including that of Bmal1, Per2, Dbp, Rorγ, and Cry1. Significantly though, neither the cycling of SIRT1 nor its activity is required for the core circadian rhythm per se. Although this might seem unexpected at first glance, it is perhaps not entirely a surprise: circadian rhythms are noted for the degree to which they are compensated against changes in temperature, nutrition, or metabolism.
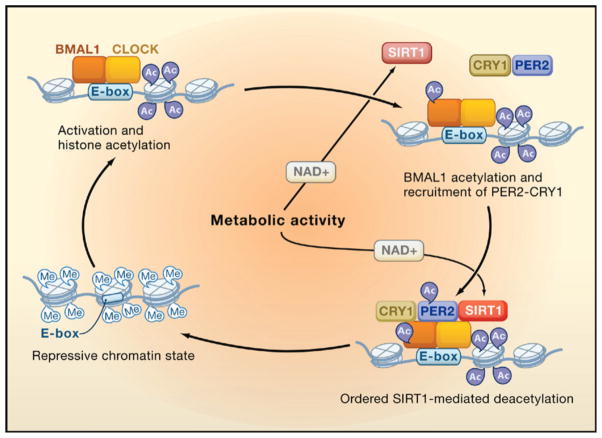
Is there a common means through which the ying and yang of CLOCK and SIRT1 might be functioning? It is perhaps too soon to tell; there are a sufficient number of differences in the details of both studies to animate journal clubs until the seasons change. For instance, an obvious difference is whether it is the amount of SIRT1 or its activity that changes with time, a distinction that may be traceable to extraction conditions, and the reports also differ in the response of gene expression to loss of SIRT1. Yet, when the dust settles it will be the commonalities between the studies that have staying power. Indeed, an attractive way to embrace the data is by assimilating it into the emerging role of chromatin structure and remodeling in facilitating clock-regulated gene expression. The first hint of this mode of regulation emerged at the beginning of the millennium with the observation that serine 10 on H3 was phosphorylated in response to light in the suprachiasmatic nucleus (SCN), a region of the hypothalamus that controls, among other things, overt circadian rhythms in locomotor activity (Crosio et al., 2000). That observation was followed by reports of rhythmic acetylation of histones, perhaps controlled by CBP/p300 (Curtis et al., 2004; Etchegaray et al., 2003), and the more recent description of CLOCK’s intrinsic HAT activity (Doi et al., 2006), the targets of which were later expanded to include BMAL1 (Hirayama et al., 2007). Apparent clock-controlled repressive effects on gene expression include histone methylation. For example, at the clock-controlled dbp locus, H3K9me2 and the binding of HP1 are rhythmic with peaks occurring during the repressive phase, supporting the notion of circadian-regulated facultative heterochromatin (Ripperger and Schibler, 2006). In contrast, Per2 appears to be repressed through methylation of histone H3 lysine 27 by the polycomb group protein EzH2 (Etchegaray et al., 2006). Integrating the action of SIRT1 into this picture suggests the simplified model depicted in Figure 1. CLOCK and BMAL1 bind to E boxes, leading to subsequent acetylation of histone H3 and H4 (presumably by both CLOCK and CBP/p300). CLOCK then acetylates BMAL1, a modification that potentiates its binding by the repressive PER/CRY complex (Hirayama et al., 2007). Once PER2 is in the complex, it too becomes acetylated (Asher et al., 2008). SIRT1 then enters (or if it is already there becomes activated), commencing the deacetylation of BMAL1, histones, and PER2. Here, metabolic activity leading to changes in NAD+ levels might also impact the cycle leading to faster or more thorough deacetylation, in effect controlling robustness. This leads to the establishment of a repressive chromatin state, which is controlled in part by histone methylation (Etchegaray et al., 2006; Ripperger and Schibler, 2006). When the repressive state needs to be undone for the next round of activation and acetylation, PER2 would become the target of SIRT1, leading to its deacetylation and destabilization. The loss of PER2 might provide a means by which SIRT1 is titrated away from the chromatin, setting the stage for a new cycle. In accord with the results published by both groups in this issue, the loss of SIRT1 in this cycle would lead to changes in the amplitude but not in rhythmicity of the circadian clock.
Figure 1. The Circadian Rhythm of Acetylation and Deacetylation.
During the activation phase, CLOCK-BMAL1 is associated with the E-box promoter element and CLOCK mediates acetylation of histone tails. BMAL1 then becomes acetylated and recruits PER2-CRY1. PER2 is acetylated and curtails the activation phase. Systematic NAD+-dependent deacetylation by SIRT of histones, BMAL1, and finally PER2 facilitates the establishment of a repressive chromatin state. For simplicity, SIRT1 is shown only when active.
References
- Asher G, Gatfield D, Stratman M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. Cell. 2008 doi: 10.1016/j.cell.2008.06.050. this issue. [DOI] [PubMed] [Google Scholar]
- Crosio C, Cermakian N, Allis CD, Sassone-Corsi P. Nat Neurosci. 2000;3:1241–1247. doi: 10.1038/81767. [DOI] [PubMed] [Google Scholar]
- Curtis AM, Seo SB, Westgate EJ, Rudic RD, Smyth EM, Chakravarti D, FitzGerald GA, McNamara P. J Biol Chem. 2004;279:7091–7097. doi: 10.1074/jbc.M311973200. [DOI] [PubMed] [Google Scholar]
- Doi M, Hirayama J, Sassone-Corsi P. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Etchegaray JP, Lee C, Wade PA, Reppert SM. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- Etchegaray JP, Yang X, DeBruyne JP, Peters AH, Weaver DR, Jenuwein T, Reppert SM. J Biol Chem. 2006;281:21209–21215. doi: 10.1074/jbc.M603722200. [DOI] [PubMed] [Google Scholar]
- Hirayama J, Sahar S, Grimaldi B, Tamaru T, Takamatsu K, Nakahata Y, Sassone-Corsi P. Nature. 2007;450:1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. Cell. 2008 doi: 10.1016/j.cell.2008.07.002. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripperger JA, Schibler U. Nat Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- Sauve AA, Wolberger C, Schramm VL, Boeke JD. Annu Rev Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]