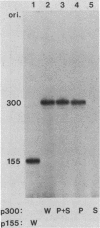
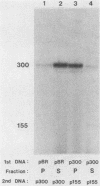
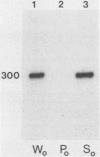
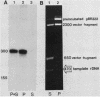
Abstract
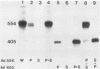
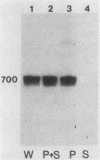
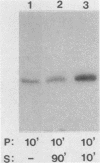
RNA synthesis in eucaryotes takes place on template molecules that are activated by stably associating with limiting transcription factors. In this paper we demonstrate that such stable transcription complexes can be specifically sedimented from in vitro transcription reaction mixtures by mild centrifugation. This occurs with stable complexes of genes transcribed by all three classes of eucaryotic RNA polymerase and with S-100 as well as whole-cell extracts. However, the transcriptional capacity of the isolated complex differs for the three polymerase classes. The pelleted ribosomal DNA (polymerase I) complex contains all the factors necessary for transcription, each purified 25- to 50-fold, whereas the pelleted adenovirus major late promoter (polymerase II) complex lacks a factor that remains in the supernatant. In the case of 5S DNA (polymerase III), a necessary factor associates slowly with the sedimentable complex. Notably, the interactions responsible for this rapid sedimentation are specific for DNA molecules in stable complexes, suggesting that the in vitro sedimentable complex mirrors the in vivo structural organization of active genes.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackerman S., Bunick D., Zandomeni R., Weinmann R. RNA polymerase II ternary transcription complexes generated in vitro. Nucleic Acids Res. 1983 Sep 10;11(17):6041–6064. doi: 10.1093/nar/11.17.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenmeier E. H., Brown D. D., Jordan E. A nuclear extract of Xenopus laevis oocytes that accurately transcribes 5S RNA genes. Cell. 1978 Nov;15(3):1077–1086. doi: 10.1016/0092-8674(78)90291-x. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Wormington W. M., Brown D. D. Stable transcription complexes of Xenopus 5S RNA genes: a means to maintain the differentiated state. Cell. 1982 Feb;28(2):413–421. doi: 10.1016/0092-8674(82)90359-2. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brown D. D. The role of stable complexes that repress and activate eucaryotic genes. Cell. 1984 Jun;37(2):359–365. doi: 10.1016/0092-8674(84)90366-0. [DOI] [PubMed] [Google Scholar]
- Cizewski V., Sollner-Webb B. A stable transcription complex directs mouse ribosomal RNA synthesis by RNA polymerase I. Nucleic Acids Res. 1983 Oct 25;11(20):7043–7056. doi: 10.1093/nar/11.20.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A. H., Reudelhuber T. L., Garrard W. T. Varigated chromatin structures of mouse ribosomal RNA genes. J Mol Biol. 1983 Jun 15;167(1):133–155. doi: 10.1016/s0022-2836(83)80038-2. [DOI] [PubMed] [Google Scholar]
- Davison B. L., Egly J. M., Mulvihill E. R., Chambon P. Formation of stable preinitiation complexes between eukaryotic class B transcription factors and promoter sequences. Nature. 1983 Feb 24;301(5902):680–686. doi: 10.1038/301680a0. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J., Bloomer L. S. Assembly of transcriptionally active 5S RNA gene chromatin in vitro. Cell. 1982 Apr;28(4):781–791. doi: 10.1016/0092-8674(82)90057-5. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lassar A. B., Martin P. L., Roeder R. G. Transcription of class III genes: formation of preinitiation complexes. Science. 1983 Nov 18;222(4625):740–748. doi: 10.1126/science.6356356. [DOI] [PubMed] [Google Scholar]
- Manley J. L., Fire A., Cano A., Sharp P. A., Gefter M. L. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesfeld R., Arnheim N. Identification of the in vivo and in vitro origin of transcription in human rDNA. Nucleic Acids Res. 1982 Jul 10;10(13):3933–3949. doi: 10.1093/nar/10.13.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K. G., Sollner-Webb B. Transcription of mouse rRNA genes by RNA polymerase I: in vitro and in vivo initiation and processing sites. Cell. 1981 Nov;27(1 Pt 2):165–174. doi: 10.1016/0092-8674(81)90370-6. [DOI] [PubMed] [Google Scholar]
- Mishima Y., Financsek I., Kominami R., Muramatsu M. Fractionation and reconstitution of factors required for accurate transcription of mammalian ribosomal RNA genes: identification of a species-dependent initiation factor. Nucleic Acids Res. 1982 Nov 11;10(21):6659–6670. doi: 10.1093/nar/10.21.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey J. H. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal Biochem. 1981 Nov 1;117(2):307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- Pardoll D. M., Vogelstein B. Sequence analysis of nuclear matrix associated DNA from rat liver. Exp Cell Res. 1980 Aug;128(2):466–470. doi: 10.1016/0014-4827(80)90083-x. [DOI] [PubMed] [Google Scholar]
- Robinson S. I., Nelkin B. D., Vogelstein B. The ovalbumin gene is associated with the nuclear matrix of chicken oviduct cells. Cell. 1982 Jan;28(1):99–106. doi: 10.1016/0092-8674(82)90379-8. [DOI] [PubMed] [Google Scholar]
- Rose S. M., Garrard W. T. Differentiation-dependent chromatin alterations precede and accompany transcription of immunoglobulin light chain genes. J Biol Chem. 1984 Jul 10;259(13):8534–8544. [PubMed] [Google Scholar]
- Sakonju S., Brown D. D. Contact points between a positive transcription factor and the Xenopus 5S RNA gene. Cell. 1982 Dec;31(2 Pt 1):395–405. doi: 10.1016/0092-8674(82)90133-7. [DOI] [PubMed] [Google Scholar]
- Schaack J., Sharp S., Dingermann T., Söll D. Transcription of eukaryotic tRNA genes in vitro. II. Formation of stable complexes. J Biol Chem. 1983 Feb 25;258(4):2447–2453. [PubMed] [Google Scholar]
- Schwartz L. B., Sklar V. E., Jaehning J. A., Weinmann R., Roeder R. G. Isolation and partial characterization of the multiple forms of deoxyribonucleic acid-dependent ribonucleic acid polymerase in the mouse myeloma, MOPC 315. J Biol Chem. 1974 Sep 25;249(18):5889–5897. [PubMed] [Google Scholar]
- Tolunay H. E., Yang L., Anderson W. F., Safer B. Isolation of an active transcription initiation complex from HeLa cell-free extract. Proc Natl Acad Sci U S A. 1984 Oct;81(19):5916–5920. doi: 10.1073/pnas.81.19.5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandelt C., Grummt I. Formation of stable preinitiation complexes is a prerequisite for ribosomal DNA transcription in vitro. Nucleic Acids Res. 1983 Jun 11;11(11):3795–3809. doi: 10.1093/nar/11.11.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson J. A., Miller K. G., Sollner-Webb B. Dinucleotide primers facilitate convenient identification of the mouse ribosomal DNA transcription initiation site. A general method for analysis of transcription by RNA polymerases I and III. J Biol Chem. 1983 Nov 25;258(22):13919–13928. [PubMed] [Google Scholar]
- Wingender E., Shi X. P., Houpert A., Seifart K. H. Isolation of a transcription complex for ribosomal 5S RNA. EMBO J. 1984 Aug;3(8):1761–1768. doi: 10.1002/j.1460-2075.1984.tb02043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]