Abstract
Motivation: Fragmented RNA immunoprecipitation combined with RNA sequencing enabled the unbiased study of RNA epigenome at a near single-base resolution; however, unique features of this new type of data call for novel computational techniques.
Result: Through examining the connections of RNA epigenome sequencing data with two well-studied data types, ChIP-Seq and RNA-Seq, we unveiled the salient characteristics of this new data type. The computational strategies were discussed accordingly, and a novel data processing pipeline was proposed that combines several existing tools with a newly developed exome-based approach ‘exomePeak’ for detecting, representing and visualizing the post-transcriptional RNA modification sites on the transcriptome.
Availability: The MATLAB package ‘exomePeak’ and additional details are available at http://compgenomics.utsa.edu/exomePeak/.
Contact: yufei.huang@utsa.edu or jmeng@mit.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Despite the unprecedented prosperity in epigenetics studies of DNA and histone modifications with next-generation sequencing data (Bernstein et al., 2007; Thurman et al., 2012), the RNA epigenetics remains a largely uncharted territory (He, 2010) and has not benefitted as much from the advancement in sequencing technology until lately. Two recent studies of transcriptome-wide mRNA m6A methylation (Dominissini et al., 2012; Meyer et al., 2012) proposed a new powerful protocol (differently named as ‘m6A-Seq’ and ‘MeRIP-Seq’), where mRNA is fragmented before the immunoprecipitation with anti-m6A antibody, and the immunoprecipitated and input control fragments are then sequenced for reconstructing transcriptome-wide m6A methylation sites. This protocol in theory enabled the transcriptome-wide unbiased study of a repertoire of >100 known post-transcriptional RNA modifications (Cantara et al., 2011) at a near single-base resolution, provided the corresponding antibody is available. To unify the nomenclatures, we call this protocol ‘FRIP-Seq’, which stands for ‘Fragmented RNA ImmunoPrecipitation Sequencing’. FRIP-Seq is different from RIP-Seq (Zhao et al., 2010), where full-length RNA is subjected to immunoprecipitation for detection of protein–RNA interaction. In contrast, the immunoprecipitation of fragmented RNA in FRIP-Seq enables the precise prediction of RNA modification sites on the transcriptome.
From a technological perspective, FRIP-Seq can be considered a marriage of two relatively well-studied techniques: ChIP-Seq (Kidder et al., 2011) and RNA-Seq (Garber et al., 2011). This new technique brings a host of new computational challenges yet to be adequately addressed. Next, we discuss briefly the best practice for FRIP-Seq data analysis.
Mapping and Filtering Short Reads: As FRIP-Seq sequences mRNA, spliced aligners that allow reads to span exon–exon junctions should be implemented. One important issue is the widespread repetitive elements (Treangen and Salzberg, 2012) in a broad range of species (∼50% of the human genome) that can lead to multi-reads (reads that could be mapped to multiple genomic locations) and ambiguities in alignment. Of the various existing strategies, the simplest yet effective way is to exclude the multi-reads completely from the analysis. (See Supplementary Material for detailed discussion.)
Fragment Length and Shifting Size: Currently, the most popular RNA sequencing protocol (unstranded RNA library and single-end sequencing) produces two shifted peaks on the ‘+’ and ‘-’ strands with a distance equal to the fragment length, and this pattern is also observed in FRIP-Seq. To correctly predict the precise methylation sites, reads need to be shifted by half of the fragment length or extended to the full length towards the 3′-end. In case that the fragment length is unknown, it may be estimated from the bimodal pattern (Zhang et al., 2008) or the cross-strand correlation (Kharchenko et al., 2008).
Peak Calling, Sequencing Bias and Control Sample: The detection of interaction sites has been formulated as the peak detection problem in ChIP-Seq (Micsinai et al., 2012; Wilbanks and Facciotti, 2010). Different from the mild sequencing bias in ChIP-Seq owing to nucleosome loss around the transcription starting sites, FRIP-Seq suffers from the depletion at both 5′- and 3′-end as a result of RNA fragmentation, considerable variations of expression levels for different genes, and most importantly, the positional bias on the gene locus caused by different isoform transcripts. Although ChIP-Seq peak calling can be conducted in the absence of a control sample by estimating the background from the neighborhood genomic regions, FRIP-Seq peak calling does require a paired input control sample of preferably fragmented RNAs before immunoprecipitation as supposed to an immunoglobulin G control sample.
Peak Annotation, Gene and Isoform Transcripts: The association between detected RNA modification sites and the specific mRNA transcripts can be highly problematic. Recent study shows that with an average of 10–12 isoforms per gene, most genes tend to express multiple isoforms simultaneously (Djebali et al., 2012). Since the sequencing read length is mostly <100bp, isoform quantification can be difficult, not to mention calling peaks for each individual isoform transcript. Nevertheless, an mRNA methylation site may be uniquely associated with a transcript when the site spans across the nearest exon(s) that uniquely belong to that transcript.
2 METHODS
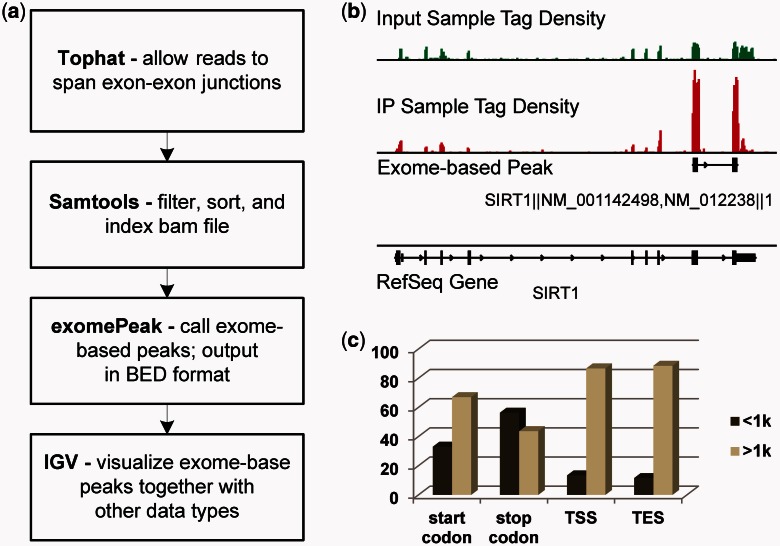
We describe in the following a pipeline for the analysis of FRIP-seq data that combines several existing tools with a novel exome-based peak calling approach (Fig. 1a). In this pipeline, short reads are first aligned to the genome assembly with Tophat (Trapnell et al., 2009); then, the resulting BAM files are filtered, sorted and indexed with Samtools (Li et al., 2009); in the last, RNA methylation sites are predicted with ‘exomePeak’, a MATLAB package that outputs exome-based peaks (the genomic locus of RNA modification sites) in the BED format to facilitate the visualization in Integrative Genomics Viewer (IGV) (Robinson et al., 2011). See the Supplementary Material for an example.
Fig. 1.
Exome-based analysis of RNA epigenome sequencing data. (a) The pipeline for FRIP-Seq processing. (b) An m6A site is found near the stop codon of Sirt1, which has two RefSeq isoform transcripts. The single RNA peak was split into two genomic loci when projected into the genome. (c) For the 3721 protein-coding genes with at least one m6A site, at least 10 kb exons, and only one RefSeq transcript, 56.3% (5195 of 9228) m6A sites fall within 1 kb distance to its stop codon on mRNA
The proposed exome-based approach can be considered as projecting transcriptome onto the genome, effectively avoiding the transcriptome heterogeneity. The advantages of exome-based analysis are mainly in two folds: first, working on exome avoids the isoform-related ambiguity, hence making all the computational operations (such as shifting, extension, smoothing, testing) straightforward. Second, it helps annotate the detected methylation sites. Despite the increasing attention on studying isoforms (Pearson, 2006), the gene convention is still at the center of most annotations and descriptions. A detected RNA modification site (on a particular transcript) needs to be associated first with a gene name before its function can be predicted. As a result, the developed ‘exomePeak’ package calls peaks on pooled exons of a specific gene so that the called peaks are automatically associated with that gene and hence its related functions.
More specifically, exomePeak first extracts and connects all the exons of a specific gene and then detects peaks using a sliding window with a conditional test that compares the means of two Poisson distributions (Przyborowski and Wilenski, 1940). The peak enrichment is considered for the IP versus control sample and for locus versus the entire gene in the IP sample. The former indicates a peak enriched in the IP sample, whereas the latter shows a locus-specific enrichment. The two P-values are then combined by the Fisher’s or Stouffer’s method. ExomePeak also swaps the IP and control samples to calculate False Discovery Rate (FDR). (See the Supplementary Material for detailed discussion.)
3 RESULTS
Our pipeline was applied to the data from (Meyer et al., 2012) and found at least one RNA m6A site on 9218 genes in HEK293T cells at a significance level of P-value = 10−5 (See the Supplementary Material for more details). We then retained only the protein-coding genes with coding length at least 10kb long and single RefSeq isoform, and then calculated the geometric distances between each detected m6A sites and four transcript landmarks of the respective gene, including the start codon, stop codon, transcription starting site and transcription ending site. Result shows that the m6A sites are enriched within 1 kb distance around the stop codons (Fig. 1c), consistent with previous studies (Dominissini et al., 2012).
Funding: The William and Ella Medical Research Foundation grant to MKR; National Institutes of Health (NIH-NCI P30CA54174) to YC; National Science Foundation (CCF-0546345) to YH; Qatar National Research Fund (09-874-3-235) to YC and YH. We thank computational support provided by the UTSA Computational Systems Biology Core Facility (NIH RCMI grant 5G12RR013646-12).
Conflict of Interest: none declared.
Supplementary Material
References
- Bernstein BE, et al. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Cantara WA, et al. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39:D195–D201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Garber M, et al. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat. Methods. 2011;8:469–477. doi: 10.1038/nmeth.1613. [DOI] [PubMed] [Google Scholar]
- He C. Grand challenge commentary: RNA epigenetics? . Nat. Chem. Biol. 2010;6:863–865. doi: 10.1038/nchembio.482. [DOI] [PubMed] [Google Scholar]
- Kharchenko PV, et al. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidder BL, et al. ChIP-Seq: technical considerations for obtaining high-quality data. Nat. Immunol. 2011;12:918–922. doi: 10.1038/ni.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micsinai M, et al. Picking ChIP-seq peak detectors for analyzing chromatin modification experiments. Nucleic Acids Res. 2012;40:e70. doi: 10.1093/nar/gks048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson H. What is a gene? Nature. 2006;441:398–401. doi: 10.1038/441398a. [DOI] [PubMed] [Google Scholar]
- Przyborowski J, Wilenski H. Homogeneity of results in testing samples from Poisson series: with an application to testing clover seed for dodder. Biometrika. 1940;31:313–323. [Google Scholar]
- Robinson JT, et al. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, et al. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat. Rev. Genet. 2012;13:36–46. doi: 10.1038/nrg3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbanks EG, Facciotti MT. Evaluation of algorithm performance in ChIP-seq peak detection. PLoS One. 2010;5:e11471. doi: 10.1371/journal.pone.0011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.