Abstract
Post-translational modifications of histone proteins lie at the heart of the epigenetic landscape in the cell’s nucleus and the precise regulation of gene expression. A myriad of studies have showed that several histone-modifying enzymes are controlled by modulatory protein partner subunits and other post-transcriptional modifications deposited in the vicinity of the targeted site. All together, these mechanisms create an intricate network of interactions that regulate enzymatic activities and ultimately control the site-specific deposition of covalent modifications. In this Point-of-View, we discuss our evolving understanding on the assembly and architecture of histone H3 Lys-4 (H3K4) methyltransferase COMPASS complexes and the techniques that progressively allowed us to better define the molecular basis of complex formation and function. We further briefly discuss some of the challenges lying ahead and additional approaches required to understand mechanistic details for the function of such complexes.
Keywords: H3K4 methylation, chromatin modification, COMPASS, SET, structural biology, RbBP5, WDR5, ASH2
Introduction
In the eukaryotic cell’s nucleus, chromatin is the host of several complex regulatory processes associated with DNA transcription, replication, and repair. Studies in the past 20 years have shown that several evolutionary conserved protein assemblies regulate these events, including the displacement of histone octamers as well as the covalent modifications of histone tails. These same studies highlighted that intricate networks of intermolecular interactions at the core of these protein assemblies often determine their subunits’ activities. However, owing to the exquisite requirement of each individual subunit to stably interact with other components of the complex, our understanding of the biochemical and structural determinants underlying the assembly of these protein complexes has lagged behind. Here we provide a brief historical perspective on identifying the structural elements controlling the formation of the histone H3K4 methylase Set1/COMPASS and its human homolog MLL1/COMPASS-like complexes, and we briefly present the challenges that lie ahead to fully characterize their function.
What is COMPASS
In the early 2000s, five laboratories independently characterized the protein SET1,1-5 a protein sharing high sequence homology with trithorax, an important regulator of homeotic genes in drosophila.6,7 Initial purifications by the Shilatifard laboratory revealed that at least seven different polypeptides formed a complex associated with SET1 (COMPASS).4 While the substrate of SET1 was still obscure at the time, the presence of a Su(var)3–9, Enhancer of Zeste and Trithorax (SET) domain in its C-terminus suggested that SET1 was able to methylate lysine residues. Shortly after, it was demonstrated that Set1/COMPASS was capable of methylating Lys-4 of histone H3, suggesting that SET1 was a genuine histone H3 Lys-4 (H3K4) methyltransferase.8,9 Combined with genome-wide studies, these findings attracted a great deal of attention since H3K4 mono- and di-methylation are now commonly known to be enriched in transcriptional enhancer elements at active and potentially active genes, respectively, while trimethylation of the same residue is a mark associated with the transcriptional start sites of actively transcribed genes.10,11
Additional analysis of the COMPASS complex revealed that several of its protein subunits (Cps) were evolutionary conserved. Cps60 (also referred to as Bre2) shared high sequence homology with drosophila ASH2, a trithorax protein required for the controls of genes important for disc patterning in flies12 (Table 1). The mammalian homolog of Cps40 (also referred to as Spp1), Cfp1, was found to be homologous to a protein now known to bind unmethylated CpG islands13,14 and to prevent spurious H3K4 trimethylation in embryonic stem cells.15 Initially identified as interacting proteins in a yeast two-hybrid system,16 the WD40-motif containing proteins Cps50 and Cps30 (also referred to as Swd1 and Swd3) were found to share high sequence homology with RBQ-3, a protein identified to bind Retinoblastoma (Rb),17 and a 7-bladed β-propeller protein named BIG-3. RBQ-3 and BIG-3 are now commonly referred as RbBP5 and WDR5, respectively. Another predicted β-propeller protein, Cps35 (also referred to as Swd2), was found to be an essential gene in budding yeast and link H3K4 methylation to H2B ubiquitylation. It was later established that Cps35 is homologous to WDR82 in mammals.18,19 One of the smallest subunit of the complex, Cps25 (also referred to as Sdc1), shared high sequence homology with DPY30 which was initially found to play a role in dosage compensation of gene expression on the X-chromosome in C. elegans20,21 (Table 1). Finally, Roguev et al. identified Shg1 as a subunit of COMPASS, however no mammalian homologs of this protein has been identified thus far8 and the loss of Shg1 has no effect on COMPASS stability or the pattern of H3K4 methylation. Along with studies on mammalian COMPASS-like complexes,22-27 these findings illustrated the complexity of these multi-subunit protein machineries and established the foundation to perform detailed molecular analysis on the role of each subunit in the assembly and function of COMPASS.
Table 1. Budding yeast COMPASS subunits and their corresponding homologs in mammals.
| Budding yeast | Mammals |
|---|---|
| Set1 |
SET1A/B, MLL1–4 |
| Cps60 (Bre2) |
ASH2L |
| Cps50 (Swd1) |
RbBP5 (RBQ-3) |
| Cps40 (Spp1) |
CFP1 (CXXC1) |
| Cps35 (Swd2) |
WDR82 |
| Cps30 (Swd3) |
WDR5 (BIG-3) |
| Cps25 (Sdc1) |
DPY30 |
| Shg1 | - |
Subunit Deletion: The First Approach to Dissect Protein Complex Formation
Budding yeast is a powerful model system to study evolutionary conserved protein complexes.28 In fact, non-lethal homozygous deletion strains are commercially available and can be tailored, when combined with tandem affinity purification (TAP) tagging approaches, to verify protein complex formation. Using a TAP-tagging approach in combination with strains harboring genetic deletion of COMPASS components, Roguev et al. determined that Cps30 and Cps50 remained bound upon deletion of S. cerevisiae (Sc) SET1 but lost interaction with the other COMPASS subunits, suggesting that these two β-propeller domain containing proteins formed a heterodimer.8 Subsequent studies confirmed that this heterodimer was critical for maintaining global levels of H3K4 mono- di- and tri-methylation in vivo,24,29-31 demonstrating the importance of the global organization of the ScSET1 complex for its function. Similar to the Cps30-Cps50 heterodimer, deletion of ScSET1 resulted in the dissociation of a Cps60-Cps25 heterodimer,8 thus supporting previous genome wide yeast two-hybrid studies showing that Cps60 and Cps25 stably associated in budding yeast.16,32 These in vivo findings were also recently supported by three independent studies with the human homologs, showing that ASH2L (Cps60) and DPY30 (Cps25) interact independently of MLL1.33-35 Finally, recent studies in yeast COMPASS demonstrated that histone H2B monoubiquitination regulates the incorporation of Cps35 in COMPASS.36 Overall, these initial deletion studies provided the first insights into the assembly of COMPASS.
Protein Complex Reconstitution: A second Challenging Step
Studying protein complex assembly by homozygous deletion in budding yeast, although informative, can be hampered by compensatory mechanisms or the instability of the dissociated proteins. This is exemplified by the deletion of Cps50 and Cps30 which decreases the overall level of ScSET1.2,29,31,37 To tackle these problems, in vitro reconstitution of protein complexes with purified components from a homologous or heterologous system, such as baculovirus-infected insect cells, have proven to be well-suited approaches.
In human, the first lines of evidence assigning roles of MLL1 regulatory subunits originated from the Roeder laboratory.23 Using recombinant baculovirus expressing a FLAG-tagged WDR5, Roeder and coworkers reconstituted and purified the WDR5-ASH2L-RbBP5-MLL1 complex. In addition, using combinations of recombinant baculoviruses, they determined the hierarchical organization of the MLL1 complex, showing that RbBP5 directly interacts with both ASH2L and WDR5. The Cosgrove group further delineated the MLL1 complex assembly using analytical ultracentrifugation analysis. By iterative addition of each subunit, they confirmed the directionality of the interactions as MLL1-WDR5-RbBP5-ASH2L-DPY30 (MLL1-WRAD).35 Utilizing sequence alignment, isothermal titration calorimetry, and GST pull-down experiments with proteins harboring deletions, the Cosgrove and Kingston laboratories showed that WDR5 bound to a region preceding the SET domain of MLL1, which they coined as WDR5 INteracting (WIN) motif.38 Using a similar line of experiments, the Wilson and Couture groups identified a region of RbBP5 bound by WDR5 [referred to as WDR5 Binding Motif (WBM)].39,40 Mutations of WBM resulted in the loss of stimulation of MLL1 methyltransferase activity, probably through the disruption of the complex. Overall, by combining several biochemical approaches, these studies provided important information of the regions of each member allowing the assembly of COMPASS or COMPASS-like complexes. However the precise molecular determinants defining the interactions between the Cps subunits were still elusive.
X-ray Crystallography Helps Identifying Structural Elements Underpinning COMPASS Assembly
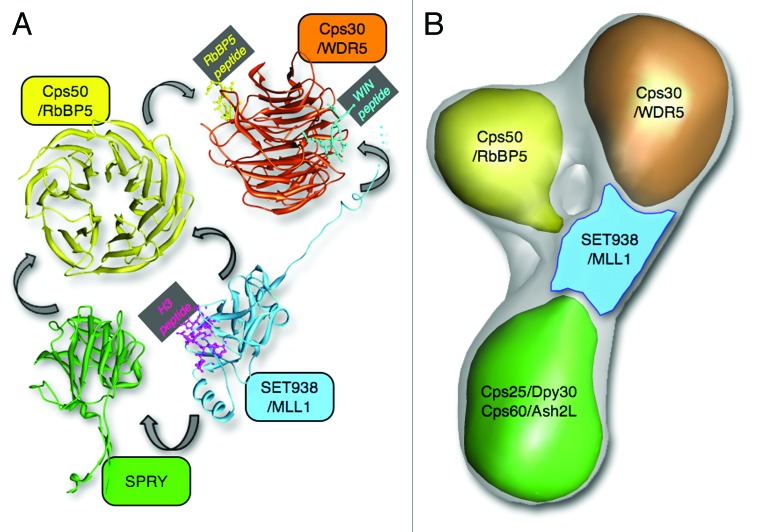
X-ray crystallography has traditionally been an invaluable method for obtaining high-resolution snapshots of macromolecular complexes and for delineating the structural principles of their assembly. Coupled with site-directed mutagenesis and binding assays, X-ray crystallography has also provided crucial mechanistic insights into the molecular basis for the formation of mammalian COMPASS-like complexes. Interestingly, the crystal structure of the MLL1 catalytic SET domain in complex with the product cofactor S-Adenosyl-L-homocysteine (AdoHcy) and a histone H3 peptide revealed that the MLL1 catalytic domain has a more open active site when compared with the structure of other SET domain methyltransferases which do not require the presence of protein cofactors for optimal activity (Fig. 1A).41 This open conformation, which might possibly impair optimal recognition of the substrate, gave rise to the hypothesis that members of the WRAD complex are required to orient regions involved in the binding of the peptide substrates or to position active site residues in an orientation amenable for catalysis.
Figure 1. COMPASS structure and network of interactions. (A) Crystal structure models of Cps domains. Note that SET938 was co-crystallized with an N-terminal H3 peptide (pdb:2W5Z), while WDR5 was co-crystallized with RbBP5 and WIN (SET1) peptides (pdb:2XL2). The model shown for the WD40 domain of Cps50/RbBP5 is from the homologous domain of WDR5 (pdb:2XL2). The SPRY domain crystal structure shown is the one determined from human ASH2L (pdb:3TOJ). The arrows indicate interactions between Cps subunits as revealed by biochemical and structural studies. (B) Cryo-EM 3D map of yeast Set1-Cps50-Cps30- Cps60-Cps25 complex showing the relative location of Cps subunits as suggested by EM and biochemical studies.
Several co-crystal structures of WDR5 with peptides from its interacting partners, MLL1 and RbBP5, highlighted the importance of the β-propeller protein in the formation of the MLL1 complex. The crystal structure of WDR5 in complex with the WIN motif of MLL1 revealed that a conserved arginine residue (R3765) of MLL1 is positioned between two key phenylalanine residues (F133 and F263) of the WDR5 peptidyl arginine-binding cleft.42,43 Recently, the crystal structures of WDR5 in complex with different WIN regions showed that the WDR5 peptidyl arginine binding cleft enables the binding of the WIN motifs of other members of the SET1 family of methyltransferases including MLL2–4 and SET1A/B (Fig. 1A).44,45 Additional structural studies revealed that WDR5 binds RbBP5 via a V-shaped cleft located on the opposite side of the arginine binding cleft.39,40 The same studies also demonstrated that a valine-aspartate-valine (VDV) motif of RbBP5 is necessary for binding to WDR5. Identification of WDR5 peptide binding pockets confirmed the initial findings showing that WDR5 plays a pivotal role in scaffolding the MLL1 complex by bridging together MLL1 with the RbBP5-ASH2L regulatory subunits. Finally, the crystal structure of the SPRY domain of ASH2L revealed that several arginine and lysine residues form a positively charged surface important for binding a cluster of acidic residues on RbBP5 (Fig. 1A).34 Overall, these structural studies have provided detailed information regarding the specificity of MLL1 complex formation and offered a better understanding of the regulation of MLL1 by the core complex Cps members. However, given that the recent structural studies have focused on protein/peptide complexes, the full extent of the interactions underlying the formation of the WRAD complex and its interaction with SET1 members has remained unclear. Consequently, the crystal structures reported so far may have not shown the entire spectrum of interactions controlling MLL1 complex assembly. This fact is well illustrated by recent observations by the Cosgrove laboratory showing that WDR5 binds a construct encompassing the WIN and SET domains of MLL1 with ~14-fold more affinity compared with a peptide corresponding to the WIN motif.35 This finding stresses the needs to consider full-length subunits of SET1 complexes in structural studies, either using X-ray crystallography or other structure determination techniques.
Electron Microscopy and the 3D Puzzle of COMPASS Complexes
Single particle electron microscopy (EM) analysis, applied on negative stained or vitrified specimens (cryo-EM), is a powerful approach for characterizing the 3D architecture of multi-subunit complexes, albeit at typically lower resolution compared with X-ray crystallography or NMR. The application of this methodology recently started shedding light on the architecture of in vitro reconstituted yeast and human COMPASS complexes that included the methyltransferase C-terminal SET domain of Set1/MLL, Cps60/ASH2L, Cps50/RbBP5, Cps30/WDR5 and Cps25/DPY30. These subunits are all present in Set1 and MLL COMPASS complexes and were been shown to form a catalytically fully active complex.46 By combining 3D cryo-EM reconstructions with 2D negative stain analysis of individual sub-complexes, Takahashi et al. showed that yeast core COMPASS adopts a Y-shaped architecture with Cps50 and Cps30 localizing on the top two adjacent lobes and Cps60-Cps25 forming the base at the bottom (Fig. 1B).46 Fab labeling experiments localized the SET domain of Set1 in a region adjacent to a central channel at the juncture of Cps50, Cps30 and the Cps60-Cps25 module. Interestingly, the human core COMPASS-like complex revealed high architectural similarity with its yeast counterpart, further strengthening the notion that yeast is a representative model system to study the regulation of the SET1 family of methyltransferases.46 Moreover, the observed subunit arrangement is compatible with the available X-ray crystallography data, and elegantly highlights the requirement of all these subunits for the regulation of mono-, di-, and tri-methylation. It is worth noting that the position of the SET domain, immediately adjacent to each component of core COMPASS, suggests a centralized organization that reflects the importance of the SET motif in catalysis, together with complex stabilization by its closely associated subunits.
Where to Go from Here
All of these studies have provided an initial framework for understanding the overall assembly of COMPASS complexes and the molecular basis of methylation activity. Building upon these findings, we need to expand our structural and functional understanding of COMPASS complexes by determining the crystal structures of full-length proteins alone and in complex with other COMPASS subunits. It is clear however that the application of these approaches will not be so forthcoming due to the biochemical challenges in obtaining full-length proteins and also stable complexes, both compositionally and conformationally. To this end it will be important to fully explore the use of heterologous expression systems, such as insect cells and mammalian cells, for the expression and purification of COMPASS proteins. Such protein preparation platforms will also allow the simultaneous co-expression of several Cps subunits, which might be key to obtain stable complexes for structural studies. Along the same lines, it might be necessary to probe the use of antibody fragments (Fabs), nanobodies,47 or even chemical cross-linkers like glutaraldehyde in order to limit the conformational variability of the COMPASS complex46 and render it more amenable to crystallization.
Obtaining more homogeneous preparations will also be necessary for determining cryo-EM structures at higher resolution, which will facilitate accurate docking of the X-ray structures in the EM envelopes of the full complex. Perhaps cryo-EM will be crucial for determining the structure of COMPASS in complex with the nucleosome, through which we will gain insights into the mechanisms underlying substrate recognition by the SET1 family of methyltransferases. In this direction, we also need to develop COMPASS activity assays that are based on nucleosomal preparations and not just histone peptides which, though informative, do not necessarily reflect a native substrate when isolated from the nucleosome structure.
But crystal structures or even high resolution EM densities may tell only one part of the COMPASS story, as recapitulated in static pictures of a protein entity that is by nature highly conformationally “active.” Another key to obtaining insights into dynamic aspects of COMPASS subunits and their assembly may lie in the application of more recently developed experimental approaches, such as deuterium exchange mass spectroscopy (DXMS),48 electron paramagnetic resonance (EPR) spectroscopy,49 and FRET measurements.50 These methods can accurately monitor the conformational dynamics of protein regions at a range of resolutions and permit the spatiotemporal probing of changes associated with substrate binding and dissociation. Such enabling technologies can offer remarkable information on COMPASS conformational transitions that, combined with crystallographic or EM data, will start to delineate the molecular underpinnings of COMPASS function. Altogether, these studies should guide detailed mutagenesis work to probe the role of specific regions important for both directing the assembly and also for establishing the different levels of methylation capacities built into the protein complex.
Acknowledgments
We thank Ali Shilatifard and Raymond Trievel for valuable comments on this manuscript. J.-F.C. acknowledges an Early Research award (Ontario) and a Canada Research Chair in Structural Biology and Epigenetics. J.-F.C. is also supported by a grant from the Canadian Institutes of Health Research. G.S. is supported as a University of Michigan Biological Sciences Scholar and as a Pew Scholar of Biomedical Sciences.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/24177
References
- 1.Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS, et al. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci U S A. 2002;99:8695–700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci U S A. 2002;99:90–4. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, et al. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15:3286–95. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, et al. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci U S A. 2001;98:12902–7. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nislow C, Ray E, Pillus L. SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol Biol Cell. 1997;8:2421–36. doi: 10.1091/mbc.8.12.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–8. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 7.Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 8.Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, et al. The Saccharomyces cerevisiae Set1 complex includes an ASH2 homologue and methylates histone 3 lysine 4. EMBO J. 2001;20:7137–48. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, et al. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem. 2002;277:10753–5. doi: 10.1074/jbc.C200023200. [DOI] [PubMed] [Google Scholar]
- 10.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 11.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LaJeunesse D, Shearn A. Trans-regulation of thoracic homeotic selector genes of the Antennapedia and bithorax complexes by the trithorax group genes: absent, small, and homeotic discs 1 and 2. Mech Dev. 1995;53:123–39. doi: 10.1016/0925-4773(95)00430-0. [DOI] [PubMed] [Google Scholar]
- 13.Xu C, Bian C, Lam R, Dong A, Min J. The structural basis for selective binding of non-methylated CpG islands by the CFP1 CXXC domain. Nat Commun. 2011;2:227. doi: 10.1038/ncomms1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JH, Skalnik DG. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J Biol Chem. 2005;280:41725–31. doi: 10.1074/jbc.M508312200. [DOI] [PubMed] [Google Scholar]
- 15.Clouaire T, Webb S, Skene P, Illingworth R, Kerr A, Andrews R, et al. Cfp1 integrates both CpG content and gene activity for accurate H3K4me3 deposition in embryonic stem cells. Genes Dev. 2012;26:1714–28. doi: 10.1101/gad.194209.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–7. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 17.Saijo M, Sakai Y, Kishino T, Niikawa N, Matsuura Y, Morino K, et al. Molecular cloning of a human protein that binds to the retinoblastoma protein and chromosomal mapping. Genomics. 1995;27:511–9. doi: 10.1006/geno.1995.1084. [DOI] [PubMed] [Google Scholar]
- 18.Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, et al. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol. 2008;28:7337–44. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JH, Skalnik DG. Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A Histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol Cell Biol. 2008;28:609–18. doi: 10.1128/MCB.01356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu DR, Chuang PT, Meyer BJ. DPY30, a nuclear protein essential early in embryogenesis for Caenorhabditis elegans dosage compensation. Development. 1995;121:3323–34. doi: 10.1242/dev.121.10.3323. [DOI] [PubMed] [Google Scholar]
- 21.Hsu DR, Meyer BJ. The dpy-30 gene encodes an essential component of the Caenorhabditis elegans dosage compensation machinery. Genetics. 1994;137:999–1018. doi: 10.1093/genetics/137.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steward MM, Lee JS, O’Donovan A, Wyatt M, Bernstein BE, Shilatifard A. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat Struct Mol Biol. 2006;13:852–4. doi: 10.1038/nsmb1131. [DOI] [PubMed] [Google Scholar]
- 23.Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13:713–9. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 24.Schneider J, Wood A, Lee JS, Schuster R, Dueker J, Maguire C, et al. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol Cell. 2005;19:849–56. doi: 10.1016/j.molcel.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 25.Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–85. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 26.Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24:5639–49. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rozenblatt-Rosen O, Rozovskaia T, Burakov D, Sedkov Y, Tillib S, Blechman J, et al. The C-terminal SET domains of ALL-1 and TRITHORAX interact with the INI1 and SNR1 proteins, components of the SWI/SNF complex. Proc Natl Acad Sci U S A. 1998;95:4152–7. doi: 10.1073/pnas.95.8.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40(Database issue):D700–5. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mersman DP, Du HN, Fingerman IM, South PF, Briggs SD. Charge-based interaction conserved within histone H3 lysine 4 (H3K4) methyltransferase complexes is needed for protein stability, histone methylation, and gene expression. J Biol Chem. 2012;287:2652–65. doi: 10.1074/jbc.M111.280867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morillon A, Karabetsou N, Nair A, Mellor J. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol Cell. 2005;18:723–34. doi: 10.1016/j.molcel.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 31.Dehé PM, Dichtl B, Schaft D, Roguev A, Pamblanco M, Lebrun R, et al. Protein interactions within the Set1 complex and their roles in the regulation of histone 3 lysine 4 methylation. J Biol Chem. 2006;281:35404–12. doi: 10.1074/jbc.M603099200. [DOI] [PubMed] [Google Scholar]
- 32.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–74. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.South PF, Fingerman IM, Mersman DP, Du HN, Briggs SD. A conserved interaction between the SDI domain of Bre2 and the DPY30 domain of Sdc1 is required for histone methylation and gene expression. J Biol Chem. 2010;285:595–607. doi: 10.1074/jbc.M109.042697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y, Cao F, Wan B, Dou Y, Lei M. Structure of the SPRY domain of human ASH2L and its interactions with RbBP5 and DPY30. Cell Res. 2012;22:598–602. doi: 10.1038/cr.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel A, Dharmarajan V, Vought VE, Cosgrove MS. On the mechanism of multiple lysine methylation by the human mixed lineage leukemia protein-1 (MLL1) core complex. J Biol Chem. 2009;284:24242–56. doi: 10.1074/jbc.M109.014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee MG, Norman J, Shilatifard A, Shiekhattar R. Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomb-like protein. Cell. 2007;128:877–87. doi: 10.1016/j.cell.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 37.Mueller JE, Canze M, Bryk M. The requirements for COMPASS and Paf1 in transcriptional silencing and methylation of histone H3 in Saccharomyces cerevisiae. Genetics. 2006;173:557–67. doi: 10.1534/genetics.106.055400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patel A, Vought VE, Dharmarajan V, Cosgrove MS. A conserved arginine-containing motif crucial for the assembly and enzymatic activity of the mixed lineage leukemia protein-1 core complex. J Biol Chem. 2008;283:32162–75. doi: 10.1074/jbc.M806317200. [DOI] [PubMed] [Google Scholar]
- 39.Avdic V, Zhang P, Lanouette S, Groulx A, Tremblay V, Brunzelle J, et al. Structural and biochemical insights into MLL1 core complex assembly. Structure. 2011;19:101–8. doi: 10.1016/j.str.2010.09.022. [DOI] [PubMed] [Google Scholar]
- 40.Odho Z, Southall SM, Wilson JR. Characterization of a novel WDR5-binding site that recruits RbBP5 through a conserved motif to enhance methylation of histone H3 lysine 4 by mixed lineage leukemia protein-1. J Biol Chem. 2010;285:32967–76. doi: 10.1074/jbc.M110.159921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Southall SM, Wong PS, Odho Z, Roe SM, Wilson JR. Structural basis for the requirement of additional factors for MLL1 SET domain activity and recognition of epigenetic marks. Mol Cell. 2009;33:181–91. doi: 10.1016/j.molcel.2008.12.029. [DOI] [PubMed] [Google Scholar]
- 42.Song JJ, Kingston RE. WDR5 interacts with mixed lineage leukemia (MLL) protein via the histone H3-binding pocket. J Biol Chem. 2008;283:35258–64. doi: 10.1074/jbc.M806900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patel A, Dharmarajan V, Cosgrove MS. Structure of WDR5 bound to mixed lineage leukemia protein-1 peptide. J Biol Chem. 2008;283:32158–61. doi: 10.1074/jbc.C800164200. [DOI] [PubMed] [Google Scholar]
- 44.Zhang P, Lee H, Brunzelle JS, Couture JF. The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases. Nucleic Acids Res. 2012;40:4237–46. doi: 10.1093/nar/gkr1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dharmarajan V, Lee JH, Patel A, Skalnik DG, Cosgrove MS. Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases. J Biol Chem. 2012;287:27275–89. doi: 10.1074/jbc.M112.364125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi YH, Westfield GH, Oleskie AN, Trievel RC, Shilatifard A, Skiniotis G. Structural analysis of the core COMPASS family of histone H3K4 methylases from yeast to human. Proc Natl Acad Sci U S A. 2011;108:20526–31. doi: 10.1073/pnas.1109360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steyaert J, Kobilka BK. Nanobody stabilization of G protein-coupled receptor conformational states. Curr Opin Struct Biol. 2011;21:567–72. doi: 10.1016/j.sbi.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Englander JJ, Del Mar C, Li W, Englander SW, Kim JS, Stranz DD, et al. Protein structure change studied by hydrogen-deuterium exchange, functional labeling, and mass spectrometry. Proc Natl Acad Sci U S A. 2003;100:7057–62. doi: 10.1073/pnas.1232301100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Columbus L, Hubbell WL. A new spin on protein dynamics. Trends Biochem Sci. 2002;27:288–95. doi: 10.1016/S0968-0004(02)02095-9. [DOI] [PubMed] [Google Scholar]
- 50.Viallet PM, Vo-Dinh T. Studying 3D subdomains of proteins at the nanometer scale using fluorescence spectroscopy. Methods Mol Biol. 2005;300:165–89. doi: 10.1385/1-59259-858-7:165. [DOI] [PubMed] [Google Scholar]