Abstract
Src family kinases (SFKs) regulate the completion of cytokinesis through signal transduction pathways that lead to the Rab11-dependent phosphorylation of ERK and its localization to the midbody of cytokinetic cells. We find that UNC119a, a known activator of SFKs, plays essential roles in this signaling pathway. UNC119a localizes to the centrosome in interphase cells and begins to translocate from the spindle pole to the spindle midzone after the onset of mitosis; it then localizes to the intercellular bridge in telophase cells and to the midbody in cytokinetic cells. We show that the midbody localization of UNC119a is dependent on Rab11, and that knocking down UNC119a inhibits the Rab11-dependent phosphorylation and midbody localization of ERK and cytokinesis. Moreover, we demonstrate that UNC119a interacts with a Src family kinase, Fyn and is required for the activation of this kinase. These results suggest that UNC119a plays a key role in the Fyn signal transduction pathway, which regulates the completion of cytokinesis via Rab11.
Keywords: cytokinesis, UNC119, SFKs signaling, Fyn kinase, Rab11, cytokinesis failure
Introduction
During cytokinesis, the final stage of cell division, a cell containing two duplicated nuclei physically separates into two daughter cells. This process requires the orchestrated interaction of various cellular activities, including signal transductions, the reorganization of the cytoskeleton and the transportation and fusion of membrane components to the dividing site (for a review, see refs. 1–4). Src family kinases (SFKs) transduce signals from the extracellular environment to intracellular signal transduction pathways and, hence, are involved in various cellular functions such as cell proliferation and differentiation, organization of the cytoskeleton, adhesion to the extracellular matrix, motility and migration (for a review, see refs. 5–6).
Several reports have suggested a possible role of SFK signaling in cytokinesis; however, the underlying mechanism remains to be elucidated.7-9 Recently, Kasahara et al.10 reported that SFK signaling leads to the phosphorylation and midbody localization of ERK, and that this process is required for the completion of cytokinesis in HeLa cells. In this report, the phosphorylation and midbody localization of ERK was shown to be Rab11-dependent.10 These analyses and previous reports addressing the role of Rab11 in cytokinesis11-14 suggest that SFK signaling is transmitted to Rab11 for the completion of cytokinesis.
unc (uncoordinated) 119 was first identified in Caenorhabiditis elegans mutants that showed defects in locomotion, feeding and chemosensation.15 Independently, Higashide et al. reported the cloning of the human ortholog of unc119 (HRG4) by screening a human retinal cDNA library.16 Since these initial findings, homologs of this gene have been identified in a wide range of organisms, from protists to mammals, including Naegleria gruberi,17Trypanosome bruci,18 fruit flies,19 zebra fish,20 mice21 and rats.22 Previous studies have proposed various functions for UNC119. UNC119 has been reported to be involved in the maintenance of the nervous system in C. elegans,15 synaptic functions in photoreceptor cells,21-28 the signal transduction in immune cells as a Src family kinase activator,29-31 endosome recycling,32 the uptake of bacteria and endocytosis33,34 and protein trafficking in sensory neurons.35,36 Although it has been reported that two UNC119 isoforms, UNC119a and UNC119b (~60% identical in amino acid sequences), exist in mammals, most studies have focused on UNC119a. Recently, it was reported that UNC119b functions in the transport of proteins to the primary cilium.37
Several lines of evidence suggest that UNC119a may be involved in the SFK signal transduction pathway that controls cytokinesis: (1) UNC119a is a known activator of different SFKs (Fyn,30,31 Lyn,29 Lck30); (2) SFK signaling that determines the completion of cytokinesis is dependent on Rab1110; and (3) UNC119a interacts with and activates Rab11.32 Based on this hypothesis, we tested whether UNC119a functions in the SFK signal transduction that leads to the completion of cytokinesis. In this paper, we report that UNC119a localizes to the midbody of dividing animal cells in a Rab11-dependent manner and plays essential roles in transmitting Fyn kinase signals to Rab11, suggesting that UNC119a plays an essential role in the completion of cytokinesis.
Results
UNC119a localizes to the midbody of cytokinetic cells
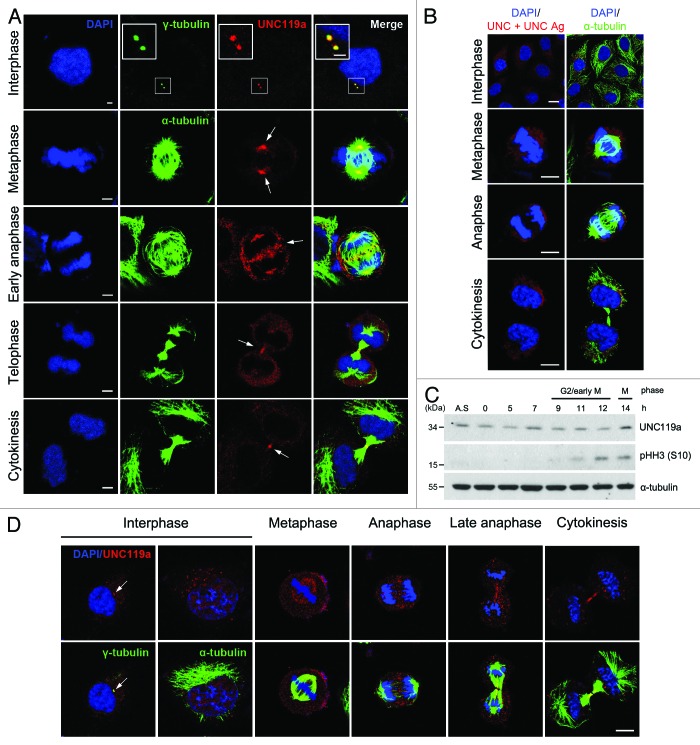
To examine the distribution of UNC119a in mammalian cells, we fixed HeLa cells with cold methanol and immunostained the cells with an anti-UNC119a antibody (UNC-Ab) raised against mouse UNC119a (Fig. 1; see “Materials and Methods” for details). In interphase cells, UNC119a was concentrated in one or two distinct spots. Double immunostaining of cells with UNC-Ab and monoclonal γ-tubulin-antibody (Ab) indicated that UNC119a was concentrated at the centrosomes (Fig. 1A). In metaphase cells, UNC119a was mainly found at the spindle poles and in some of the spindle fibers. Many punctate UNC119a spots were also found throughout the cytoplasm. In early anaphase cells, UNC119a was concentrated at the spindle poles, as in metaphase cells, and at the spindle midzone in numerous spots. In late anaphase cells, UNC119a was not observed at the spindle poles but was concentrated at the intercellular bridge of the cells. In telophase and cytokinetic cells, UNC119a was concentrated at the intercellular bridge and midbody of the cells (Fig. 1A). Pre-incubation of UNC-Ab with GST (glutathione-s-transferase)-tagged UNC119a completely inhibited the binding of the Ab in the fixed cells, suggesting that UNC-Ab binding was specific (Fig. 1B). Western blotting experiments with cell extracts prepared at different stages of the cell cycle showed no significant changes in the cellular content of UNC119a during the cell cycle (Fig. 1C). This cell cycle-dependent localization of UNC119a was also observed in five different cell types fixed using the same method (three human cell lines, HEK293, U2OS and htRPE-1; and two mouse cell lines, NIH3T3 and MEF) (Fig. S1).
Figure 1. Cell cycle-dependent localization of UNC119a. Asynchronously growing HeLa cells were fixed in cold methanol (A and B) or paraformaldehyde (D). Fixed cells were double immunostained with UNC-Ab and monoclonal α-tubulin-Ab (or monoclonal γ-tubulin-Ab). The binding of the antibodies was visualized using Alexa 546-conjugated goat anti-rabbit IgG (red) and Alexa 488-conjugated goat anti-mouse IgG (green), respectively. The nuclei and chromosomes were stained with DAPI. (A) Cell cycle-dependent localization of UNC119a. White arrows show regions of highly concentrated UNC119a. Bars: 5 μm, in interphase; 10 μm, in other images. (B) UNC-Ab specifically recognizes endogenous UNC119a. Fixed HeLa cells were double immunostained with monoclonal α-tubulin-Ab and UNC-Ab pre-incubated with GST-UNC119a (UNC + UNC Ag). Pre-incubation of UNC-Ab with the antigen completely inhibited the binding of UNC-Ab to the endogenous protein. Bars, 20 μm. (C) The cellular content of UNC119 does not change during the cell cycle. HeLa cells were synchronized at G1/S using a double thymidine block. Synchronized cells were released into fresh medium, collected at the indicated times and lysed with RIPA buffer. Western blotting was performed with three antibodies: UNC-Ab for UNC119a; anti-phosphor-histone H3 (Ser 10)-Ab, pHH3 (S10), as a mitosis marker; and monoclonal α-tubulin-Ab as a loading control. A.S., asynchronous cell extracts. (D) Cell cycle-dependent localization of UNC119a in paraformaldehyde-fixed cells. Asynchronously growing HeLa cells were fixed with 4% paraformaldehyde and permeabilized in 0.2% Triton X-100 in PBS. Double immunostaining was performed as described above. White arrows indicate the centrosomes. Bar, 20 μm.
As it has been reported that UNC119 is associated with membrane vesicles,22,26,32 we also examined the distribution of UNC119a in cells fixed using a different method that well preserved the staining of the membrane vesicle-associated protein, p23038,39 (Fig. S2A). In these experiments, where cells were fixed with paraformaldehyde, permeabilized with Triton X-100 (see “Materials and Methods”) and then immunostained with UNC-Ab, we also observed that UNC119a was concentrated in different regions of the cells depending on the cell cycle stage (Fig. 1D).
Although the results of these two sets of experiments were basically identical, there were some minor, but reproducible, differences depending on the applied fixation method. In interphase cells fixed with cold methanol, UNC119a was distinctly concentrated at the centrosomes (Fig. 1A). However, in interphase cells fixed with paraformaldehyde, UNC119a was observed as many punctate spots in the cytoplasm and at the centrosomes (Fig. 1D). In metaphase cells fixed with paraformaldehyde, UNC119a was detected as numerous spots along spindle fibers. Although some of the UNC119a spots were concentrated at the spindle poles, the localization to the poles was not as distinct as it was in methanol-fixed cells (see metaphase cells in Fig. 1A and D). Concentration of UNC119a at the intercellular bridges of cytokinetic cells was evident in cells fixed with either method (Fig. 1A and D); however, concentration of UNC119a at the midbody was not observed in paraformaldehyde-fixed cells (Fig. 1D). Instead, there was a clear gap in the middle of the intercellular bridges, corresponding to the presumed site of the midbody (Fig. 1D). It has been reported that the midbody is surrounded by electron-dense material of an unknown nature,40-42 suggesting that the paraformaldehyde fixation and permeabilization procedure was not sufficient to allow UNC-Ab to penetrate the surrounding material. We obtained essentially identical immunostaining results when U2OS cells were fixed with paraformaldehyde and permeabilized as described above (data not shown). Alternatively, we observed that UNC119a was concentrated at centrosomes and midbodies when GFP-UNC119a was expressed in U2OS cells (Fig. S2B). These results showed that the localization of UNC119a is regulated during the cell cycle in these cells and suggested that this process is associated with vesicle-transport during the cell cycle.
Different regions of UNC119a are required for the cell cycle-regulated localization of UNC119a
To identify domains that function in the cell-cycle regulated localization of UNC119a, we constructed five deletion mutants of human UNC119a and expressed the GFP-tagged deletion mutants in U2OS cells (Fig. S3). We first identified domains involved in the centrosomal localization of UNC119a by fixing transfected cells with cold methanol 12 h after transfection and immunostaining the cells with monoclonal γ-tubulin-Ab. When the GFP-tagged C-terminal domain of UNC119a (amino acids 121–240, GFP-C) was expressed, concentration of GFP-C at the centrosome was observed in 73.3% of the cells expressing the fusion protein (Fig. S3). Centrosomal localization of a truncation mutant including amino acids 60–240 (GFP-MC) and that of wild type UNC119a (GFP-T) was observed in 39% and 30% of cells expressing the fusion proteins, respectively (Fig. S3). Although western blotting results indicated that all GFP-tagged mutant UNC119a proteins were synthesized in similar amounts in the transfected cells (data not shown), the distribution of amino acids 1–59 (GFP-N) or that of amino acids 60–120 (GFP-M) was difficult to detect in cells fixed with cold methanol. These results suggested that the C-terminal half of UNC119 (amino acids 121–240) was required for the centrosomal localization of the protein and that the N-terminal half of UNC119a was likely associated with vesicles (see above).
To identify domains involved in the midbody localization of UNC119a, we incubated cells for 48 h after transfection to obtain a sufficient number of cells undergoing cell division and fixed the cells with paraformaldehyde as the above results suggested that the N-domain (amino acids 1–59) and M-domain (amino acids 60–120) of UNC119a were associated with vesicles. The cells were then immunostained with monoclonal α-tubulin-Ab, and the cytokinetic cells expressing GFP were scored. Concentration of GFP-N and of GFP-M at the midbody was observed in 82.4% and 60% of the cells, respectively (Fig. S3). Fusion of the two domains (GFP-NM) had no additive effect on the midbody localization (53%). Only 6.3% of the cells showed midbody localization when GFP-C was expressed (Fig. S3). Together, these results suggested that amino acids 1–120 and 121–240 of UNC119a were each strongly involved in the midbody localization and centrosomal localization of UNC119a, and that the midbody localization of UNC119a might be related to vesicle transport.
UNC119a is required for the completion of cytokinesis
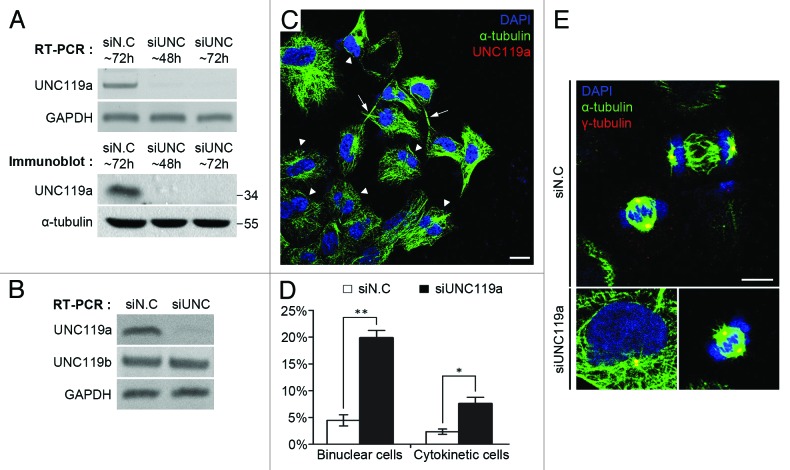
Because UNC119a was concentrated at the intercellular bridge and midbody in cytokinetic cells, we next tested whether UNC119a was involved in cytokinesis by treating HeLa cells with siRNAs targeting human UNC119a (Fig. 2). RT-PCR and western blotting experiments showed that the siRNA treatment effectively decreased the cellular concentration of UNC119a (Fig. 2A and B) but had no effect on the cellular concentration of UNC119b mRNA (Fig. 2B). To test the effect of UNC119a siRNA on cell division, siRNA-treated HeLa cells were fixed and double immunostained with UNC-Ab and monoclonal α-tubulin-Ab. UNC119a siRNA treatment significantly increased the percentages of bi-nucleated cells and cytokinetic cells containing a long intercellular bridge with a centrally located midbody (Fig. 2C). In control cells, the percentages of bi-nucleated cells and cytokinetic cells were approximately 4.47% and 2.36%, respectively, while in UNC119a siRNA-transfected cells, the percentages increased to 19.86% and 7.58%, respectively (Fig. 2D). The siRNA treatment had no detectable effect either on the organization of cytoplasmic microtubules in interphase cells or the formation of spindle fibers in mitotic cells (Fig. 2E). These results suggested that UNC119a is required for the completion of cytokinesis, but not for either the organization of cytoplasmic microtubules or the formation of spindle fibers. Identical results were obtained when similar experiments were performed in htRPE-1 cells (data not shown).
Figure 2. UNC119a is required for the completion of cytokinesis. (A and B). HeLa cells were treated with a mixture of UNC119a siRNAs (see “Materials and Methods”). Total RNAs and cell extracts were prepared from the transfected cells at the indicated times after siRNA treatment. RT-PCR was performed with primers specific for UNC119a, UNC119b or Glyceraldehyde 3-phosphate dehydrogenase (GAPDH). siN.C, siRNA negative control. siUNC, mixture of ds-RNAs specific for UNC119a. (A) Effects of UNC119a siRNA treatment. Top: RT-PCR. Bottom: western blotting with UNC-Ab and monoclonal α-tubulin-Ab. (B) UNC119a siRNA treatment specifically decreases UNC119a mRNA. (C) Increased appearance of bi-nucleated cells and cytokinetic cells with elongated intercellular bridges in UNC119a siRNA transfected cells. Cells treated with the UNC119a siRNA mixture were fixed with cold methanol at 72 h after transfection and double-immunostained with UNC-Ab and monoclonal α-tubulin-Ab as described in Figure 1. DAPI staining was used to identify nuclei. Triangles indicate bi-nucleated cells. Arrows indicate cytokinetic cells with elongated intercellular bridges. Bar, 20 μm. (D) UNC119a siRNA treatment increases the number of bi-nucleated cells and cytokinetic cells with long intercellular bridges. Graphs represent the mean ± standard deviation (S.D.) of three independent experiments. siN.C: bi-nucleated cells, 4.47 ± 1.03%; cytokinetic cells, 2.36 ± 0.49%. n = 1,000: experiment 1, 300; experiment 2, 300; experiment 3, 400. siUNC: bi-nucleated cells, 19.86 ± 1.39%; cytokinetic cells, 7.58 ± 1.19%. n = 1000: experiment 1, 300; experiment 2, 300; experiment 3, 400. ***p < 0.001; *p < 0.05. (E) UNC119 siRNA treatment has no effect on either the organization of cytoplasmic microtubules in interphase cells or the formation of spindle fibers in mitotic cells (merged images). UNC119a siRNA-transfected cells were fixed with cold methanol and double immunostained with monoclonal α-tubulin-Ab (green) and polyclonal γ-tubulin-Ab (red; yellow, merge). Nuclei and chromosomes were stained with DAPI. Bar, 20 μm.
UNC119a interacts with Rab11 in a cell cycle-dependent manner
A tendency of bi-nucleated cells and cytokinetic cells that exhibit an elongated intercellular bridge with a midbody has been observed in cells that are defective in abscission.10,11,43-46 It has also been reported that Rab11, a small GTPase, is localized at the intercellular bridge and midbody in cytokinetic cells and functions in abscission via transporting vesicles to the midbody.11 Moreover, an interaction between Rab11 and UNC119a has been reported in the transportation of Lck, a member of the SFKs, to the plasma membrane during T-cell activation.32 Based on these observations, we examined the possible interaction of UNC119a with Rab11 in proliferating HeLa cells.
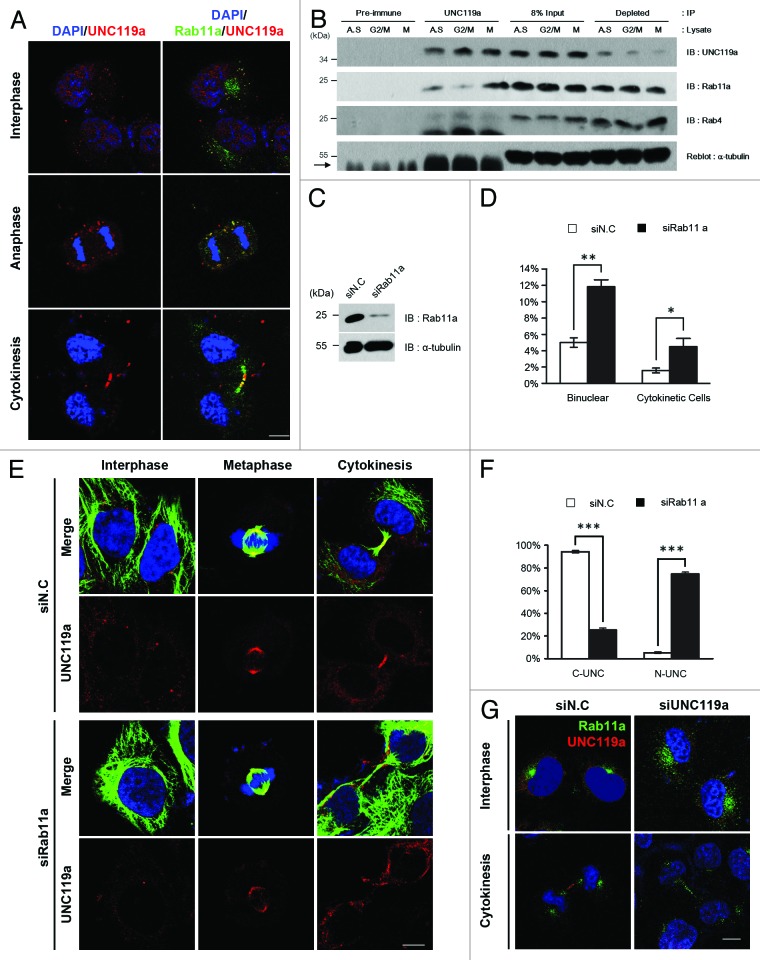
In interphase cells fixed with paraformaldehyde and double immunostained with monoclonal Rab11a-Ab and UNC-Ab, both Rab11a and UNC119a were found throughout the cytoplasm of the cells in numerous spots. However, co-localization of UNC119a and Rab11a at these spots was not evident (Fig. 3A). In anaphase cells, both Rab11a and UNC119a were found as punctate dots, and co-localization of the two proteins was observed in most of the spots. In telophase and cytokinetic cells, the two proteins were present at the intercellular bridge and the midbody (Fig. 3A). These results showed that UNC119a and Rab11a co-localize in dividing cells and suggested that the co-localization of the two proteins is a regulated process during the cell cycle.
Figure 3. UNC119a co-localizes and interacts with Rab11a. (A) Co-localization of UNC119a and Rab11a. HeLa cells were fixed with paraformaldehyde, permeabilized and double-immunostained with UNC-Ab and monoclonal Rab11a-Ab. The binding of the antibodies was visualized using Alexa 546-conjugated goat anti-rabbit IgG (red) and Alexa 488-conjugated goat anti-mouse IgG (green), respectively. Nuclei and chromosomes were stained with DAPI. Left: DAPI/UNC119a. Right: DAPI/UNC119a/Rab11a. Bar, 20 μm. (B) UNC119a and Rab11a interaction is regulated during the cell cycle. Immunoprecipitation with UNC-Ab (or pre-immune serum) was performed using HeLa cell extracts prepared from asynchronously growing cells (A.S.), G2/M-arrested cells (G2/M) and mitotic cells (M) (“Materials and Methods”). Western blotting was performed with UNC-Ab, monoclonal Rab11a-Ab and monoclonal Rab4-Ab. Pre-immune, immunoprecipitation with rabbit pre-immune serum; UNC119a, immunoprecipitation with UNC-Ab; 8% Input, an 8% volume of each immunoprecipitation was included as a loading control; Depleted, 8% volume of recovered supernatant of each immunoprecipitation. Reblotting of the top membrane with monoclonal α-tubulin-Ab shows that UNC-Ab specifically precipitates UNC119a. Arrow indicates IgG heavy chain. (C) Rab11a siRNA depletes endogenous Rab11a. HeLa cells were transfected with Rab11a siRNAs, cell lysates were prepared after 72 h, and western blotting was conducted with monoclonal Rab11a-Ab and α-tubulin-Ab. siN.C, control; siRab11a, Rab11a siRNA. (D) Rab11a siRNA treatment inhibits cytokinesis. HeLa cells were transfected with Rab11a siRNA and, after 72 h, fixed with paraformaldehyde. The numbers of bi-nucleated cells and cytokinetic cells were counted. Graphs represent the mean ± SD of three independent experiments. siN.C: bi-nucleated cells, 5 ± 0.58%; cytokinetic cells, 1.58 ± 0.3%. n = 1,000: experiment 1, 300; experiment 2, 300; experiment 3, 400. siRab11a: bi-nucleated cells, 12.22 ± 0.72%; cytokinetic cells, 5.22 ± 0.89%. n = 1600: experiment 1, 600; experiment 2, 400; experiment 3, 600. **p < 0.01; *p < 0.05. (E) Rab11a siRNA treatment inhibits the localization of UNC119a to the intercellular bridge and midbody but has no effect on the centrosome/spindle pole localization. Rab11a siRNA-treated HeLa cells were fixed with cold methanol and double immunostained with monoclonal α-tubulin-Ab and UNC-Ab as described in Figure 1. Nuclei and chromosomes were stained with DAPI. siN.C; cells treated with control siRNA. siRab11a; cells treated with Rab11a siRNA. Bar, 20 μm. (F) Rab11a siRNA treatment inhibits the localization of UNC119a to the intercellular bridge and midbody. After the immunostaining (Fig. 3E), telophase cells and cytokinetic cells were scored and the concentration of UNC119a at the midbody (or intercellular bridge) of these cells was examined. Graphs represent the mean ± SD of three independent experiments. siN.C: cells with concentrated UNC119a (C-UNC), 94.33 ± 1.2%; no specific concentration (N-UNC), 5.33 ± 0.88%. n = 300: experiment 1, 100; experiment 2, 100; experiment 3, 100. siRab11a: C-UNC, 25.33 ± 1.76%; N-UNC, 74.67 ± 1.76%. n = 300: experiment 1, 100; experiment 2, 100; experiment 3, 100. ***p < 0.001. (G) UNC119a siRNA treatment has no effect on the localization of Rab11a. HeLa cells were transfected with a mixture of UNC119a siRNAs, as in Figure 2, then fixed, permeabilized and double immunostained, as described in Figure 3A. siN.C; cells treated with control siRNA. siUNC119a; cells treated with UNC119a siRNA. Bar, 20 μm.
To examine the possible cell cycle-dependent interaction between UNC119a and Rab11a, immunoprecipitation experiments were performed using UNC-Ab and HeLa cell extracts prepared from asynchronously growing cells, G2/M-arrested cells and cells undergoing cell division (“Materials and Methods”). Western blottings with Rab11a-Ab showed that Rab11a co-precipitated with UNC119a, and that there was a noticeable increase in the co-precipitated amount of Rab11a in the mitotic cell extract, suggesting that UNC119a and Rab11a specifically interacted in mitotic cells (Fig. 3B). The interaction between Rab11a and UNC119a was confirmed by immunoprecipitating Rab11a in mitotic cell extracts and western blotting this with UNC-Ab (Fig. S4A). These results supported the idea that UNC119a and Rab11a interact in a cell cycle-specific manner.
Western blotting with Rab4-Ab suggested a possible interaction between UNC119a and Rab4, especially in G2/M-arrested cells. However, this interaction was observed only when the film was exposed for a prolonged period (e.g., overnight). These results suggested that only a minute fraction of Rab4 interacts with UNC119a and that the Rab4-UNC119a interaction is regulated differently from Rab11a-UNC119a interaction. The interaction between UNC119a and Rab4 could not be confirmed by a reverse immunoprecipitation. Immunoprecipitaion using the Rab4-Ab and western blotting the precipitated proteins with the Ab showed that this Ab did not precipitate Rab4 (data not shown).
Midbody localization of UNC119a is dependent on Rab11a activity
We next examined the localization of UNC119a in HeLa cells treated with Rab11a siRNA to test whether Rab11a functions in the cell cycle-dependent localization of UNC119a. Rab11a siRNA treatment drastically decreased the amount of Rab11a in the treated cells (Fig. 3C) and inhibited the completion of cytokinesis as determined by the increase in both the number of bi-nucleated cells11 and the number of cytokinetic cells with long intercellular bridges (Fig. 3D). Double immunostaining cells with monoclonal α-tubulin-Ab and UNC-Ab showed that this siRNA treatment also inhibited the localization of UNC119a to the intercellular bridge and the midbody of cytokinetic cells (Fig. 3E and F). The Rab11 siRNA treatment had no effect on the concentration of UNC119a either at the centrosome of interphase cells or at the spindle poles of mitotic cells (Fig. 3E). In contrast, UNC119a siRNA treatment had no effect on the localization of Rab11a either in the pericentrosomal region in interphase cells or at the intercellular bridge of cytokinetic cells (Fig. 3G). Taken together, these results suggested that the transport of UNC119a to the intercellular bridge and midbody, possibly via associated endosomes, is dependent on Rab11a activity and is required for the completion of cytokinesis. These results also suggested that Rab11a is not involved in the centrosomal concentration of UNC119a in interphase cells. Because human cells exhibit two Rab11 isoforms, Rab11a and Rba11b, which are 91% identical in their amino acid sequences, we further examined the distribution of UNC119a in HeLa cells treated with siRNAs targeting both Rab11 isoforms. Even though the double-siRNA treatment effectively decreased the amounts of both Rab11 isoforms below the detection limit, the inhibitory effect of the double-siRNA treatment on the completion of cytokinesis and on the localization of UNC119a was almost identical to that of treatment with siRNA targeting Rab11a alone (Fig. S4B). These results suggested that Rab11a plays an important role in the midbody localization of UNC119a (see “Discussion”).
UNC119a is required for the phosphorylation and transportation of ERK to the midbody
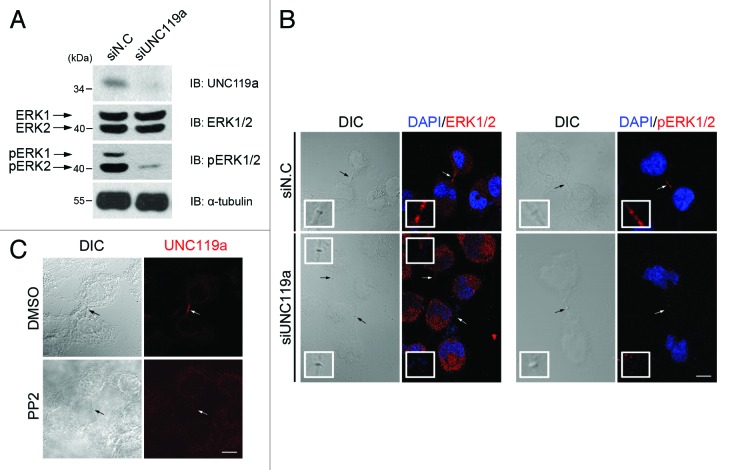
Because the above results showed that UNC119a interacts with Rab11a, we next examined whether this interaction plays a role in the Rab11-dependent phosphorylation and midbody localization of ERK during cytokinesis (Fig. 4). Western blotting experiments using UNC119a siRNA-treated HeLa cell extracts showed that depletion of UNC119a inhibited the phosphorylation of ERK1/2 but had no effect on the cellular concentrations of these proteins (Fig. 4A). In addition, the midbody localization of ERK in cytokinetic cells was abolished in UNC119a siRNA-transfected cells (Fig. 4B). Inhibition of ERK phosphorylation via UNC119a antisense RNA treatment was also reported by Gorska et al.30 during T cell activation. These results and previous reports showing the interaction of UNC119a with different SFKs29-31 suggested that UNC119a might be involved in the SFK signaling, leading to Rab11-dependent ERK phosphorylation. To test this possibility, we treated HeLa cells with PP2, an SFK inhibitor that caused cytokinetic cells with long intercellular bridges to appear (Fig. 4C), as reported by Kasahara et al.10 As expected, when we immunostained these cells with UNC-Ab, we did not observe midbody concentration of UNC119a. These results further suggested that UNC119a is involved in the SFK signal transduction, leading to completion of cytokinesis.
Figure 4. Requirement of UNC119a for the phosphorylation and midbody localization of ERK. (A) UNC119a siRNA treatment inhibits the phosphorylation of ERK. HeLa cells were treated with UNC119a siRNA (siUNC119a) or control siRNA (siN.C) and lysed with RIPA buffer. Western blotting was performed with the indicated antibodies. (B) UNC119a siRNA treatment inhibits the midbody localization of ERK. At 72 h after the siRNA treatment, cells were fixed with paraformaldehyde, permeabilized and immunostained with ERK2-Ab (left panels) or phosphor-ERK1/2-Ab (right panels). In control cells (siN.C), both antibodies bound to the intercellular bridge. Neither antibody bound to the elongated intercellular bridges in UNC119a siRNA-treated cells (siUNC119). Black arrows (DIC images) and white arrows (fluorescent images) indicate the midbody. Insets, higher magnification image of midbodies indicated by arrows. Bar, 20 μm. (C) SFK activity is required for the midbody localization of UNC119a. HeLa cells were treated with PP2 (10 μM), fixed with cold methanol and immunostained with UNC-Ab. Arrows indicate the midbody. Bar, 20 μm.
UNC119a interacts with and activates Fyn
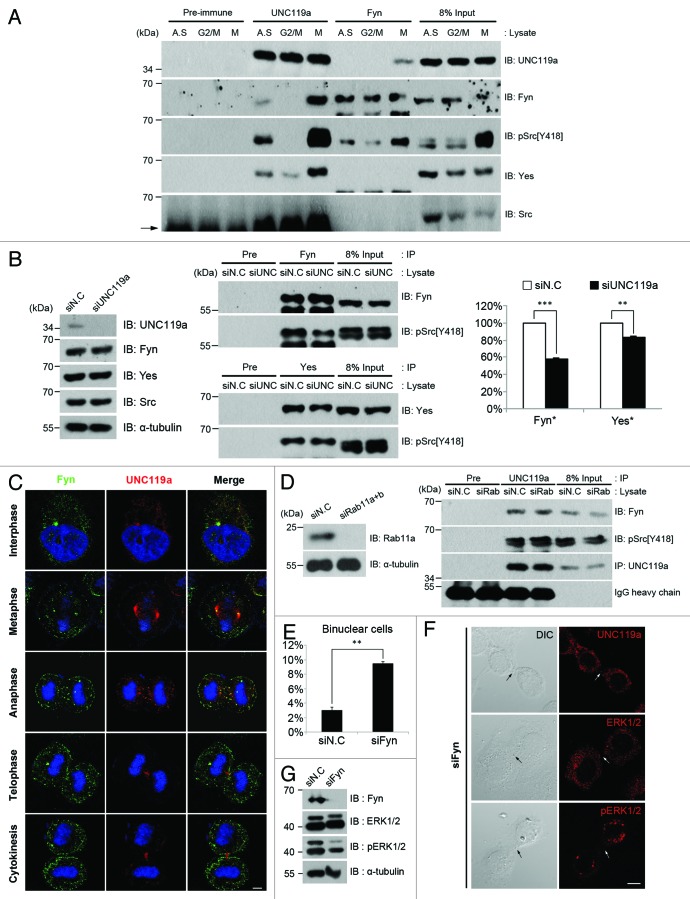
To identify the SFK that interacts with UNC119a and functions in cytokinesis, we performed immunoprecipitation experiments using UNC-Ab and HeLa cell extracts prepared from asynchronously growing cells, G2/M-arrested cells and mitotic cells (see “Materials and Methods”). Western blotting experiments showed that the kinases Fyn and Yes, but not Src, co-immunoprecipitated with UNC119a. This co-immunopreciptation of Fyn and Yes with UNC119 was observed strongly in mitotic cell extracts, suggesting that the interaction between these two SFKs and UNC119a is cell cycle-dependent (Fig. 5A). To confirm this interaction, immunoprecipitation experiments were performed using Fyn-Ab. Western blotting the precipitated proteins with UNC-Ab showed that UNC119a and Fyn indeed interacted in mitotic cell extracts (Fig. 5A). Yes was not detected in Fyn-Ab immunoprecipitated extracts, indicating that Fyn did not directly interact with Yes (Fig. 5A). Immunoprecipitation experiments using mitotic cell extracts and Yes-Ab and western blotting with UNC-Ab also confirmed that UNC119a and Yes interacted in the extracts (Fig. S4A).
Figure 5. UNC119a is required for Fyn signaling for the completion of cytokinesis. (A) Co-immunoprecipitation of Fyn with UNC119a. HeLa cell extracts were prepared as described in Figure 3B and immunoprecipitation was performed using UNC-Ab or Fyn-Ab. Western blots were performed with UNC-Ab, Fyn-Ab, phosphor-Src (pSrc)-Ab [pSrc(Y418)], Yes-Ab and Src-Ab. Pre-immune, immunoprecipitation with rabbit preimmune serum; UNC119a and Fyn, immunoprecipitation with respective Ab; Input, loading control as described in Figure 3B. Arrow indicates IgG heavy chain. (B) UNC119a activates Fyn. Left: UNC119a siRNA treatment depletes endogenous UNC119a but has no effect on the amount of SFKs. Western blotting analyses were performed with indicated antibodies. siN.C, extracts from control siRNA-treated cells; siUNC119, extracts from UNC119a siRNA-treated cells. Middle: UNC119a siRNA treatment inhibits the activation of Fyn and Yes. Upper panel: After immunoprecipitation with Fyn-Ab, western blotting was performed with Fyn-Ab and pSrc-Ab. Lower panel: After immunoprecipitation with Yes-Ab, western blotting was performed with Yes-Ab and pSrc-Ab. Pre, immunoprecipitation with rabbit preimmune serum; Input, loading control as above. Right: UNC119a-depletion has a more significant inhibitory effect on Fyn activation than on Yes activation. The images of three independent western blots were analyzed by LAS-4000 (Fuji Film) using Multi Gauge ver.3.1 to quantitate the degree of inhibition. ***p < 0.001; **p < 0.01. (C) Cell cycle-dependent co-localization of Fyn with UNC119a. Asynchronously growing HeLa cells were fixed with paraformaldehyde and double immunostained with monoclonal Fyn-Ab (green) and UNC-Ab (red). Bar, 10 μm. (D) The interaction of Fyn with UNC119a is independent of Rab11a. Left: Rab11 siRNA treatment depletes endogenous Rab11a. Western blots with the Rab11a-Ab and α-tubulin-Ab. siN.C; control siRNA-treated cell extracts. siRab11; Rab11 (a + b) siRNA-treated cells extracts. Right: After immunoprecipitation with UNC-Ab, western blotting was performed with Fyn-Ab, pSrc-Ab and UNC-Ab. Input, loading control as above. (E) Fyn siRNA treatment inhibits cytokinesis. HeLa cells were treated with Fyn siRNA for 72 h and fixed with paraformaldehyde. After permeabilization, the cells were immunostained with α-tubulin Ab and DAPI. The number of bi-nucleated cells was counted. The graphs represent the mean ± SD of three independent experiments. siN.C: bi-nucleated cells, 2.84 ± 0.42%. n = 1,000: experiment 1, 500; experiment 2, 200; experiment 3, 300. siFyn: bi-nucleated cells, 9.44 ± 0.29%. n = 1,100: experiment 1, 400; experiment 2, 400; experiment 3, 300. siN.C; control siRNA-treated cells. siFyn; Fyn siRNA-treated cells. **p < 0.01. (F) Fyn siRNA treatment inhibits the midbody localization of UNC119a. Fyn siRNA-treated cells were fixed with paraformaldehyde and immunostained with UNC-Ab, ERK2-Ab or phosphor-ERK1/2-Ab. Dark arrows (DIC images) and white arrows (fluorescent images) indicate the midbody. UNC119a, ERK and pERK were not found at the midbody, which is indicated by white arrows. Bar, 20 μm. (G) Fyn siRNA treatment inhibits the phosphorylation of ERK. HeLa cells were treated with Fyn siRNA for 72 h and lysed with RIPA buffer. Western blotting experiments were performed with the indicated antibodies.
Using a phosphor-Src-Ab that recognizes the activated form of all three SFKs,10,47 we found that UNC119a co-immunoprecipitated with two separate proteins with relative molecular weights identical to Fyn and Yes. These results suggested that UNC119a associated with the activated forms of Fyn and Yes. To determine whether the interaction of UNC119a with Fyn and Yes was related to the activation of the respective kinases, we first examined the effect of UNC119a-depletion on the cellular content of Fyn, Yes and Src. Western blottings showed that the depletion of UNC119a had no effect on the cellular contents of the SFKs (Fig. 5B). Immunoprecipitation of the UNC119a-depleted cell extracts and western blottings showed that UNC119a depletion inhibited the activation of both Fyn and Yes; however, the inhibitory effect of UNC119a-depletion was more significant on the activation of Fyn than on the activation of Yes (Fig. 5B). In accordance, Gorska et al.30 reported that Fyn was specifically activated by the overexpression of UNC119a. Along with other reports indicating a possible function of Fyn in cytokinesis,7,9 collectively, these results strongly suggested that UNC119a might interact with Fyn kinase, which, in turn, functions in cytokinesis via Rab11a.
The possible cell cycle-dependent interaction between Fyn and UNC119a was further examined by double immunostaining HeLa cells with Fyn-Ab and UNC-Ab (Fig. 5C). As reported previously in T cells by Ley et al.,48 Fyn was observed at the centrosome in interphase cells and at the spindle poles in metaphase cells. Co-localization of Fyn with UNC119a was evident at both structures. Both Fyn and UNC119a were also present throughout the cytoplasm of these cells as distinct dots. However, co-localization of Fyn and UNC119a was not evident at these spots (Fig. 5C). In anaphase cells, the two proteins were present throughout the cytoplasm as distinct spots. Here, co-localization of the two proteins was observed mainly in spots located in the spindle midzone, suggesting that only a fraction of Fyn interacted with UNC119a. In telophase and cytokinetic cells, co-localization of Fyn with UNC119 was not evident, even though there were a few Fyn spots localized at the intercellular bridge and midbody (Fig. 5C). The possible co-localization of Rab11a with Fyn was also tested by immunostaining GFP-Rab11a transfected cells with Fyn-Ab. The co-localization of GFP-Rab11a and Fyn was apparent in anaphase cells and some telophase cells (Fig. S5A).
Our results suggested that UNC119a binds to and activates Fyn kinase after that activated Fyn-UNC119a complex interacts with Rab11a. To test this possibility, we performed immunoprecipitation experiments using Rab11 (a + b) siRNA-treated cell extracts and examined whether Fyn co-immunoprecipitated with UNC119a (Fig. 5D). The results indicated that Fyn-UNC119a interaction was independent of Rab11. We next examined the effect of Fyn knockdown on the midbody localization of UNC119a and on the phosphorylation and midbody localization of ERK. Fyn siRNA treatment increased the percentage of bi-nucleated cells, suggesting that cytokinesis was inhibited (Fig. 5E). Interestingly, depletion of Fyn did not induce the accumulation of cytokinetic cells with long intercellular bridges, as was observed in UNC119a (or Rab11) siRNA-treated cells, but instead significantly decreased the number of cytokinetic cells. Whereas approximately 2% of the cell population was usually undergoing cytokinesis in control cells, only 72 out of more than 7,000 cells (~1%) were undergoing cytokinesis in the Fyn siRNA-treated cells. Localization of UNC119a at the midbody was not observed in the majority of these cells (75%; n = 72). Fyn siRNA treatment also inhibited the phosphorylation and midbody localization of ERK (Fig. 5F and G), though it did not have a noticeable effect on the interaction between UNC119a and Rab11a (data not shown). Fyn siRNA treatment had no effect on the concentration of UNC119a at centrosomes and spindle poles (data not shown). Together, these results suggested that the activity of Fyn, UNC119a and Rab11a are necessary for the phosphorylation and midbody localization of ERK, even though we did not measure Fyn kinase activity in this process. These results also suggested that UNC119a interacts with Rab11 in a manner independent of Fyn activity.
Discussion
Distinct regions of UNC119a are required for the cell cycle-dependent localization of UNC119a
Our data suggested that the C-terminal domain (amino acids 121–240) of UNC119a is essential for the localization of UNC119a at the centrosome, and that the centrosomal localization of UNC119a is not dependent on Rab11a (Fig. S3). At present, we do not understand how UNC119a localizes to centrosomes. Although our data suggest that UNC119a is not required either for the organization of cytoplasmic microtubules or the formation of spindle fibers, the concentration of the protein at centrosomes implies that UNC119a may function in other cellular events that are not examined in our current study. In this regard, it is interesting that Naegleria UNC119, which lacks the N-terminal 60 amino acid of human UNC119a but shows 46% amino acid sequence identity to the C-terminal 3/4 of human UNC119a (amino acids 60–240), is concentrated at the basal body,17 though is not involved in cytokinesis (Lee, unpublished observation).
The N-terminal domain of UNC119a (amino acids 1–59) was clearly involved in midbody localization. Gorska et al. (2004) previously showed that this domain is essential for the activation of Fyn by UNC119a, and our current results demonstrate that the activity of Fyn is necessary for the midbody localization of UNC119a. Collectively, this implies that the N-terminal domain plays a key role in transmitting Fyn signals to Rab11a and in the Rab11a-mediated midbody localization of UNC119a (see below). However, it has not yet been determined whether this domain alone can function in mediating the transmission of Fyn signals to Rab11a for the completion of cytokinesis.
The middle (M) domain of UNC119a (amino acids 60–120) was also capable of localizing to the midbody, suggesting this domain may interact with protein(s) other than Fyn and form a protein complex distinct from the Fyn-UNC119a complex. These different complexes may interact with either a common protein, Rab11a, or with distinct proteins for the midbody localization.
Simultaneous binding of two distinct proteins to the N- and M-domains of one UNC119a molecule might have a negative effect on the midbody localization of UNC119a, resulting in the less efficient midbody localization of GFP-NM (amino acids 1–120) than that of either GFP-N or GFP-M (Fig. S3). Wild type UNC119a fused to GFP (GFP-T) also showed weaker centrosomal localization (30%) and midbody localization (51.7%) compared with the centrosomal localization of GFP-C (73.3%) and to the midbody localization of GFP-N (82.4%). These data suggest that overexpression of UNC119a and, hence, unregulated and/or simultaneous binding of different proteins to the respective binding sites in UNC119a inhibits the regulated localization of UNC119a, and that UNC119a is a multi-functional protein that interacts with diverse proteins. Indeed, in addition to the interaction of UNC119a with different SFKs via the N-terminal SH3-binding motif,29 interactions of UNC119a with various other proteins have been reported. UNC119a interacts with Abl kinase34 and dynamin33 in NIH3T3 cells, RIBEYE in photoreceptor ribbon synapses23 and transducin α in C. elegans sensory neurons.36 Interaction of UNC119 with different myristoylated proteins has also been reported.37
Possible function of UNC119a in cytokinesis
Our results showed that UNC119a interacts sequentially with Fyn and Rab11a and is necessary for the completion of cytokinesis, suggesting that UNC119a plays important roles in mediating SFK signals for the completion of cytokinesis. However, there are several important points to be explored to fully understand the role of UNC119a in mediating SFK signals for the completion of cytokinesis.
If Fyn is the SFK that functions in regulating cytokinesis by interacting with UNC119a, it is expected that the depletion of Fyn and of UNC119a might have a similar inhibitory effect on the completion of cytokinesis. However, our results showed that depletion of UNC119a (Fig. 2D) is significantly more effective in inhibiting cytokinesis than depletion of Fyn (Fig. 5E). Our data also showed that the phenotype of UNC119a siRNA-treated cells (Fig. 2) (and cells treated with PP2 treatment; Fig. 4C) is different from that of Fyn siRNA-treated cells. These results suggest the presence of other SFKs that are involved in the regulation of cytokinesis by interacting with UNC119a. We found that both Fyn and Yes interact with UNC119a, and that the depletion of UNC119a inhibits the activation of both kinases. However, we focused our study on Fyn, because the depletion of UNC119a had a more significant inhibitory effect on the activation of Fyn than on the activation of Yes. Hence, future studies addressing the role of Yes and possibly other SKFs will be needed to fully elucidate the role of SFKs in the completion of cytokinesis.10 Localization of Yes at the midbody of cytokinetic cells has recently been reported.49
Similarly, the depletion of UNC119a was significantly more effective in inhibiting cytokinesis than the depletion of Rab11 (Figs. 2D and 3D). As discussed above, our data suggested that N-domain and M-domain of UNC119a can independently mediate the midbody localization of the protein. Our data also showed that the depletion of Rab11 was very effective but not complete in inhibiting the midbody localization of UNC119a. Taken together, our results suggest that, even though Rab11 might be the major protein that interacts with UNC119a to mediate SFK signals for the completion of cytokinesis, there are possibly other proteins that interact with UNC119a to mediate SFK signals for the completion of cytokinesis. Future studies on proteins that interact with UNC119a in cell cycle-dependent manner are necessary to fully understand the role of UNC119a in cytokinesis.
In addition, we demonstrated that knockdown of Rab11a alone or of both Rab11 isoforms in HeLa cells inhibited the completion of cytokinesis in the cells to a similar degree. One simple explanation for this finding is that Rab11a is important for the completion of cytokinesis, but Rab11b is not. Indeed, several reports have suggested that Rab11a and Rab11b play distinct roles.50-52 However, if Rab11a and Rab11b play different, but necessary, roles in a common pathway leading to the completion of cytokinesis, knockdown of either Rab11a alone or both Rab11 isoforms would have a similar inhibitory effect. Further studies are needed to clarify these two possibilities.
Human and mouse cells exhibit two UNC119 genes that encode two different proteins, UNC119a and UNC119b, which are approximately 60% identical in their amino acid sequences. However, a comparison of the amino acid sequences of the two proteins shows that the shared amino acid sequence identity between the two proteins mainly occurs in the C-terminal half of the proteins (amino acids 121–240), and that the SH3-binding motif, which is required for the interaction with SFKs,29 is not present in UNC119b. Recently, different localization patterns of the two UNC119 isoforms were also reported (UNC119a, centrosome; UNC119b, basal body and the primary cilium).37 These data suggest that UNC119a is the major protein functioning in SFK signal transduction leading to the completion of cytokinesis; however, this matter should be more definitively determined in future studies.
Materials and Methods
Production of the UNC119 antibody
We cloned the full-length mouse UNC119a cDNA, synthesized the protein in E. coli as an MBP-fusion protein, and raised a polyclonal antiserum by injecting the bacterially synthesized MBP-UNC119a into rabbits. The obtained polyclonal anti-UNC119a antibody (UNC-Ab) recognized both human and mouse UNC119a, which are 91% identical in their amino acid sequences. Western blotting experiments showed that the Ab recognized a human protein with a relative molecular mass of 36 kDa and a mouse protein with a relative molecular mass of 34 kDa, despite the fact that the calculated molecular weights of the two proteins are almost identical (26.99 kDa and 26.95 kDa, respectively) (Fig. S5B). The nature of this difference between the relative molecular masses of the two proteins was not clear. Pre-incubation of UNC-Ab with GST-tagged UNC119a completely inhibited the binding of the Ab to the presumed UNC119a proteins in each cell extract (Fig. S5B). Additionally, the Ab bound more intensely to the mouse protein than to the human protein in a reproducible manner, which could be due to mouse cells containing more UNC119a than the human cells or the Ab showing a stronger affinity for mouse UNC119a than for human UNC119a (Fig. S5B).
Cell culture and cell cycle synchronization
HeLa cells and U2OS cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin and 100 μg/ml streptomycin (Hyclone). For synchronization at G1/S, HeLa cells were cultured in the presence of 2 mM thymidine (Sigma) for 17 h, washed three times with PBS and cultured in fresh medium for 9 h. After a second thymidine treatment for 16 h, the cells were washed with PBS and released into fresh medium. To prepare G2/M cell extracts, HeLa cells were cultured in the presence of nocodazole (40 ng/ml) for 12 h and harvested. To prepare mitotic cell extracts, HeLa cells were cultured in the presence of nocodazole, as above, and round-shaped cells were collected using “the mitotic shake-off” procedure. The cells were then washed with PBS, incubated in fresh medium for 1 h 45 min and harvested. This harvest time was determined by immunostaining a small portion of the cells with an α-tubulin Ab at 15 min intervals during the incubation.
Plasmids, siRNAs and transient transfection
Full-length and truncation mutants of human UNC119 cDNAs were subcloned into the pEGFP-C1 vector (Clontech). U2OS cells were transfected with plasmid DNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. siRNA oligonucleotides that were designed and targeted to human UNC119a mRNA were purchased from GenePharma. The dsRNA oligonucleotides were as follows: Oligo-1, (sense) 5′-CAGAACGGUUGCCCAUCAATT-3′, (antisense) 5′-UUGAUGGGCAACCGUUCUGAG-3′; Oligo-2, (sense) 5′-GGUUUAAGAUUCGGGACAUTT-3′, (anti-sense) 5′-AUGUCCCGAAUCUUAAACCTG-3′; Oligo-3 (sense) 5′-CGUAUGAGACCCAGUCUGATT-3′, (antisense) 5′-UCAGACUGGGUCUCAUACGGG-3′. Negative control dsRNA oligonucleotide: (sense) 5′-UUCUCCGAACGUGUCACGUTT-3′, (antisense) 5′-ACGUGACACGUUCGGAGAATT-3′. siRNA oligonucleotides targeting human Rab11 were designed as described by Wilson et al.11 and purchased from GenePharma. Fyn siRNA was purchased from Santa Cruz Biotechnology (sc-29321). HeLa cells were transfected with siRNA duplexes (30 nM) using Lipofectamine 2000 for 48~72 h according to the manufacturer’s instructions.
Chemical
PP2 was purchased from Calbiochem (529573). HeLa cells were treated with 10 μM PP2 in DMSO or DMSO alone (control) for 24 h. The cells were then fixed and immunostained with UNC-Ab.
Antibodies
An anti-UNC119 antibody (UNC-Ab) was generated as described above. The antibodies used in this study and the suppliers are listed below. Monoclonal anti-α-tubulin-Ab (Santa Cruz Biotechnology, sc-23948), monoclonal anti-Rab11a-Ab (Abcam, ab78337), monoclonal anti-γ-tubulin-Ab (Sigma, T6557), polyclonal anti-γ-tubulin-Ab (Sigma, T3559), monoclonal anti-p230 TGN-Ab (BD Transduction, 611281), polyclonal anti-ERK2-Ab (C-14, Santa Cruz Biotechnology, sc-154), polyclonal anti-phosphor-p44/42 MAPK (ERK1/2) (Thr202/Tyr204)-Ab (Cell Signaling, #9101), polyclonal anti-phosphor-histone H3 (Ser 10)-Ab (Millipore, #06-570), monoclonal anti-Fyn-Ab (FYN-01, Santa Cruz, sc-51598), monoclonal anti-Src [pY418]-Ab (phosphor-Src: Invitrogen, AHO0051), polyclonal anti-Src-Ab (N-16, Santa Cruz, sc-19), monoclonal c-Yes-Ab (BD Transduction, 610376), polyclonal anti-Rab4-Ab (Cell Signaling, #2167), Alexa 488-conjugated goat anti-mouse IgG (Invitrogen, A11001) and Alexa 546-conjugated goat anti-rabbit IgG (Invitrogen, A11010).
Cell fixation, immunocytochemistry and confocal microscopy
To perform cold methanol fixation, cells were incubated in −20°C methanol for 5 min and washed with PBS. For paraformaldehyde fixation, cells were fixed with 4% paraformaldehyde in PBS for 20 min and permeabilized with 0.2% Triton X-100 in PBS for 10 min at room temperature, then washed with PBS. For immunocytochemistry, cells were incubated in blocking solution (5% normal goat serum in PBS) for 1 h and then incubated with the primary antibody in PBS for 1 h 30 min at room temperature. After washing with PBS-T (PBS containing 0.1% Tween-20), the cells were incubated with a second antibody conjugated with a fluorescent dye for 1 h at room temperature. The cells were mounted on slides with Vectashield mounting medium (Vector Laboratories), and the samples were observed under an LSM 510 META confocal laser scanning microscope equipped with epifluorescence and a digital image analyzer (Carl Zeiss). Z-stacked images were acquired at 400× magnification.
Immunoprecipitation
Cells were collected and lysed in IP lysis buffer (50 mM TRIS-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100) containing protease inhibitors (5 μg/ml aprotinin, 5 μg/ml leupeptin, 5 μg/ml pepstatin A) and phosphatase inhibitors (1 mM sodium orthovanadate and 5 mM sodium fluoride). The cell lysates were precleared with equilibrated protein A Sepharose beads (Amersham) for 1 h. After centrifugation, the supernatant was incubated with 1~2 μg of the appropriate antibody or preimmune serum overnight at 4°C and then incubated with equilibrated protein A sepharose beads for 1 h at 4°C. The beads were collected via centrifugation, washed three times with IP washing buffer (50 mM TRIS-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.5% Triton X-100) and resuspended in 1× SDS protein sample buffer. Proteins were separated using SDS-PAGE for western blotting analysis.
Western blotting
Cells were lysed in RIPA buffer (50 mM TRIS-HCl pH 7.4, 1% NP-40, 0.25% Na-deoxycholate, 150 mM NaCl, 1 mM EDTA, 0.1% SDS) containing protease inhibitors. The protein concentration of each cell lysate was measured via the Bradford assay, and an equal amount of protein from each sample extract was separated using SDS-PAGE and transferred to a nitrocellulose membrane.
Statistical analyses of data
Data are presented as mean ± standard deviation (S.D.) from at least three independent experiments. Significant differences between groups were determined by Student’s t-tests. Values of ***p < 0.001, **p < 0.01 and *p < 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
Authors thank to Dr Jon Soderholm for proofreading this manuscript. We also thank to Waterborne Virus Bank (The Catholic University) for generous gift of materials. This work was supported by a grant from the National Research Foundation (NRF 2009-0079383, Mid-Career Researcher Program) funded by Ministry of Education, Science, and Technology, Republic of Korea.
Glossary
Abbreviations:
- UNC119
uncoordinated 119
- SFKs
Src family protein tyrosine kinases
- ERK
extracellular signal-regulated kinase
- Ab
antibody
- GFP
green fluorescent protein
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/24404
References
- 1.Barr FA, Gruneberg U. Cytokinesis: placing and making the final cut. Cell. 2007;131:847–60. doi: 10.1016/j.cell.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 2.Fededa JP, Gerlich DW. Molecular control of animal cell cytokinesis. Nat Cell Biol. 2012;14:440–7. doi: 10.1038/ncb2482. [DOI] [PubMed] [Google Scholar]
- 3.Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307:1735–9. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- 4.Normand G, King RW. Understanding cytokinesis failure. Adv Exp Med Biol. 2010;676:27–55. doi: 10.1007/978-1-4419-6199-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guarino M. Src signaling in cancer invasion. J Cell Physiol. 2010;223:14–26. doi: 10.1002/jcp.22011. [DOI] [PubMed] [Google Scholar]
- 6.Thomas SM, Brugge JS. Cellular functions regulated by Src family kinases. Annu Rev Cell Dev Biol. 1997;13:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- 7.Campbell KS, Cooper S, Dessing M, Yates S, Buder A. Interaction of p59fyn kinase with the dynein light chain, Tctex-1, and colocalization during cytokinesis. J Immunol. 1998;161:1728–37. [PubMed] [Google Scholar]
- 8.Tominaga T, Sahai E, Chardin P, McCormick F, Courtneidge SA, Alberts AS. Diaphanous-related formins bridge Rho GTPase and Src tyrosine kinase signaling. Mol Cell. 2000;5:13–25. doi: 10.1016/S1097-2765(00)80399-8. [DOI] [PubMed] [Google Scholar]
- 9.Yasunaga M, Yagi T, Hanzawa N, Yasuda M, Yamanashi Y, Yamamoto T, et al. Involvement of Fyn tyrosine kinase in progression of cytokinesis of B lymphocyte progenitor. J Cell Biol. 1996;132:91–9. doi: 10.1083/jcb.132.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasahara K, Nakayama Y, Nakazato Y, Ikeda K, Kuga T, Yamaguchi N. Src signaling regulates completion of abscission in cytokinesis through ERK/MAPK activation at the midbody. J Biol Chem. 2007;282:5327–39. doi: 10.1074/jbc.M608396200. [DOI] [PubMed] [Google Scholar]
- 11.Wilson GM, Fielding AB, Simon GC, Yu X, Andrews PD, Hames RS, et al. The FIP3-Rab11 protein complex regulates recycling endosome targeting to the cleavage furrow during late cytokinesis. Mol Biol Cell. 2005;16:849–60. doi: 10.1091/mbc.E04-10-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pelissier A, Chauvin JP, Lecuit T. Trafficking through Rab11 endosomes is required for cellularization during Drosophila embryogenesis. Curr Biol. 2003;13:1848–57. doi: 10.1016/j.cub.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 13.Yu X, Prekeris R, Gould GW. Role of endosomal Rab GTPases in cytokinesis. Eur J Cell Biol. 2007;86:25–35. doi: 10.1016/j.ejcb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Riggs B, Rothwell W, Mische S, Hickson GR, Matheson J, Hays TS, et al. Actin cytoskeleton remodeling during early Drosophila furrow formation requires recycling endosomal components Nuclear-fallout and Rab11. J Cell Biol. 2003;163:143–54. doi: 10.1083/jcb.200305115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maduro M, Pilgrim D. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics. 1995;141:977–88. doi: 10.1093/genetics/141.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higashide T, Murakami A, McLaren MJ, Inana G. Cloning of the cDNA for a novel photoreceptor protein. J Biol Chem. 1996;271:1797–804. doi: 10.1074/jbc.271.3.1797. [DOI] [PubMed] [Google Scholar]
- 17.Chung S, Kang S, Paik S, Lee J. NgUNC-119, Naegleria homologue of UNC-119, localizes to the flagellar rootlet. Gene. 2007;389:45–51. doi: 10.1016/j.gene.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 18.Ohshima S, Ohashi-Suzuki M, Miura Y, Yabu Y, Okada N, Ohta N, et al. TbUNC119 and its binding protein complex are essential for propagation, motility, and morphogenesis of Trypanosoma brucei procyclic form cells. PLoS ONE. 2010;5:e15577. doi: 10.1371/journal.pone.0015577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maduro MF, Gordon M, Jacobs R, Pilgrim DB. The UNC-119 family of neural proteins is functionally conserved between humans, Drosophila and C. elegans. J Neurogenet. 2000;13:191–212. doi: 10.3109/01677060009084494. [DOI] [PubMed] [Google Scholar]
- 20.Manning AG, Crawford BD, Waskiewicz AJ, Pilgrim DB. unc-119 homolog required for normal development of the zebrafish nervous system. Genesis. 2004;40:223–30. doi: 10.1002/gene.20089. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi A, Higashide T, Hamasaki D, Kubota S, Sakuma H, An W, et al. HRG4 (UNC119) mutation found in cone-rod dystrophy causes retinal degeneration in a transgenic model. Invest Ophthalmol Vis Sci. 2000;41:3268–77. [PubMed] [Google Scholar]
- 22.Higashide T, McLaren MJ, Inana G. Localization of HRG4, a photoreceptor protein homologous to Unc-119, in ribbon synapse. Invest Ophthalmol Vis Sci. 1998;39:690–8. [PubMed] [Google Scholar]
- 23.Alpadi K, Magupalli VG, Käppel S, Köblitz L, Schwarz K, Seigel GM, et al. RIBEYE recruits Munc119, a mammalian ortholog of the Caenorhabditis elegans protein unc119, to synaptic ribbons of photoreceptor synapses. J Biol Chem. 2008;283:26461–7. doi: 10.1074/jbc.M801625200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haeseleer F. Interaction and colocalization of CaBP4 and Unc119 (MRG4) in photoreceptors. Invest Ophthalmol Vis Sci. 2008;49:2366–75. doi: 10.1167/iovs.07-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishiba Y, Higashide T, Mori N, Kobayashi A, Kubota S, McLaren MJ, et al. Targeted inactivation of synaptic HRG4 (UNC119) causes dysfunction in the distal photoreceptor and slow retinal degeneration, revealing a new function. Exp Eye Res. 2007;84:473–85. doi: 10.1016/j.exer.2006.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kubota S, Kobayashi A, Mori N, Higashide T, McLaren MJ, Inana G. Changes in retinal synaptic proteins in the transgenic model expressing a mutant HRG4 (UNC119) Invest Ophthalmol Vis Sci. 2002;43:308–13. [PubMed] [Google Scholar]
- 27.Mori N, Ishiba Y, Kubota S, Kobayashi A, Higashide T, McLaren MJ, et al. Truncation mutation in HRG4 (UNC119) leads to mitochondrial ANT-1-mediated photoreceptor synaptic and retinal degeneration by apoptosis. Invest Ophthalmol Vis Sci. 2006;47:1281–92. doi: 10.1167/iovs.05-0493. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi A, Kubota S, Mori N, McLaren MJ, Inana G. Photoreceptor synaptic protein HRG4 (UNC119) interacts with ARL2 via a putative conserved domain. FEBS Lett. 2003;534:26–32. doi: 10.1016/S0014-5793(02)03766-3. [DOI] [PubMed] [Google Scholar]
- 29.Cen O, Gorska MM, Stafford SJ, Sur S, Alam R. Identification of UNC119 as a novel activator of SRC-type tyrosine kinases. J Biol Chem. 2003;278:8837–45. doi: 10.1074/jbc.M208261200. [DOI] [PubMed] [Google Scholar]
- 30.Gorska MM, Stafford SJ, Cen O, Sur S, Alam R. Unc119, a novel activator of Lck/Fyn, is essential for T cell activation. J Exp Med. 2004;199:369–79. doi: 10.1084/jem.20030589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vepachedu R, Gorska MM, Singhania N, Cosgrove GP, Brown KK, Alam R. Unc119 regulates myofibroblast differentiation through the activation of Fyn and the p38 MAPK pathway. J Immunol. 2007;179:682–90. doi: 10.4049/jimmunol.179.1.682. [DOI] [PubMed] [Google Scholar]
- 32.Gorska MM, Liang Q, Karim Z, Alam R. Uncoordinated 119 protein controls trafficking of Lck via the Rab11 endosome and is critical for immunological synapse formation. J Immunol. 2009;183:1675–84. doi: 10.4049/jimmunol.0900792. [DOI] [PubMed] [Google Scholar]
- 33.Karim Z, Vepachedu R, Gorska M, Alam R. UNC119 inhibits dynamin and dynamin-dependent endocytic processes. Cell Signal. 2010;22:128–37. doi: 10.1016/j.cellsig.2009.09.022. [DOI] [PubMed] [Google Scholar]
- 34.Vepachedu R, Karim Z, Patel O, Goplen N, Alam R. Unc119 protects from Shigella infection by inhibiting the Abl family kinases. PLoS ONE. 2009;4:e5211. doi: 10.1371/journal.pone.0005211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gopalakrishna KN, Doddapuneni K, Boyd KK, Masuho I, Martemyanov KA, Artemyev NO. Interaction of transducin with uncoordinated 119 protein (UNC119): implications for the model of transducin trafficking in rod photoreceptors. J Biol Chem. 2011;286:28954–62. doi: 10.1074/jbc.M111.268821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang H, Constantine R, Vorobiev S, Chen Y, Seetharaman J, Huang YJ, et al. UNC119 is required for G protein trafficking in sensory neurons. Nat Neurosci. 2011;14:874–80. doi: 10.1038/nn.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wright KJ, Baye LM, Olivier-Mason A, Mukhopadhyay S, Sang L, Kwong M, et al. An ARL3-UNC119-RP2 GTPase cycle targets myristoylated NPHP3 to the primary cilium. Genes Dev. 2011;25:2347–60. doi: 10.1101/gad.173443.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gleeson PA, Anderson TJ, Stow JL, Griffiths G, Toh BH, Matheson F. p230 is associated with vesicles budding from the trans-Golgi network. J Cell Sci. 1996;109:2811–21. doi: 10.1242/jcs.109.12.2811. [DOI] [PubMed] [Google Scholar]
- 39.Pohl C, Jentsch S. Final stages of cytokinesis and midbody ring formation are controlled by BRUCE. Cell. 2008;132:832–45. doi: 10.1016/j.cell.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 40.Buck RC, Tisdale JM. The fine structure of the mid-body of the rat erythroblast. J Cell Biol. 1962;13:109–15. doi: 10.1083/jcb.13.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mullins JM, Biesele JJ. Terminal phase of cytokinesis in D-98s cells. J Cell Biol. 1977;73:672–84. doi: 10.1083/jcb.73.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mullins JM, McIntosh JR. Isolation and initial characterization of the mammalian midbody. J Cell Biol. 1982;94:654–61. doi: 10.1083/jcb.94.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fabbro M, Zhou BB, Takahashi M, Sarcevic B, Lal P, Graham ME, et al. Cdk1/Erk2- and Plk1-dependent phosphorylation of a centrosome protein, Cep55, is required for its recruitment to midbody and cytokinesis. Dev Cell. 2005;9:477–88. doi: 10.1016/j.devcel.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 44.Gromley A, Jurczyk A, Sillibourne J, Halilovic E, Mogensen M, Groisman I, et al. A novel human protein of the maternal centriole is required for the final stages of cytokinesis and entry into S phase. J Cell Biol. 2003;161:535–45. doi: 10.1083/jcb.200301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chircop M, Oakes V, Graham ME, Ma MP, Smith CM, Robinson PJ, et al. The actin-binding and bundling protein, EPLIN, is required for cytokinesis. Cell Cycle. 2009;8:757–64. doi: 10.4161/cc.8.5.7878. [DOI] [PubMed] [Google Scholar]
- 46.van der Horst A, Simmons J, Khanna KK. Cep55 stabilization is required for normal execution of cytokinesis. Cell Cycle. 2009;8:3742–9. doi: 10.4161/cc.8.22.10047. [DOI] [PubMed] [Google Scholar]
- 47.Kuga T, Nakayama Y, Hoshino M, Higashiyama Y, Obata Y, Matsuda D, et al. Differential mitotic activation of endogenous c-Src, c-Yes, and Lyn in HeLa cells. Arch Biochem Biophys. 2007;466:116–24. doi: 10.1016/j.abb.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 48.Ley SC, Marsh M, Bebbington CR, Proudfoot K, Jordan P. Distinct intracellular localization of Lck and Fyn protein tyrosine kinases in human T lymphocytes. J Cell Biol. 1994;125:639–49. doi: 10.1083/jcb.125.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jung J, Lee MK, Jin Y, Fu SB, Rosales JL, Lee KY. Clues for c-Yes involvement in the cell cycle and cytokinesis. Cell Cycle. 2011;10:1502–3. doi: 10.4161/cc.10.9.15495. [DOI] [PubMed] [Google Scholar]
- 50.Scapin SM, Carneiro FR, Alves AC, Medrano FJ, Guimarães BG, Zanchin NI. The crystal structure of the small GTPase Rab11b reveals critical differences relative to the Rab11a isoform. J Struct Biol. 2006;154:260–8. doi: 10.1016/j.jsb.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 51.Lapierre LA, Dorn MC, Zimmerman CF, Navarre J, Burnette JO, Goldenring JR. Rab11b resides in a vesicular compartment distinct from Rab11a in parietal cells and other epithelial cells. Exp Cell Res. 2003;290:322–31. doi: 10.1016/S0014-4827(03)00340-9. [DOI] [PubMed] [Google Scholar]
- 52.Khvotchev MV, Ren M, Takamori S, Jahn R, Südhof TC. Divergent functions of neuronal Rab11b in Ca2+-regulated versus constitutive exocytosis. J Neurosci. 2003;23:10531–9. doi: 10.1523/JNEUROSCI.23-33-10531.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.