Abstract
Metabolomics is a rapidly growing field of research used in the identification and quantification of the small molecule metabolites within an organism, thereby providing insights into cell metabolism and bioenergetics as well as processes important in clinical medicine, such as disposition of pharmaceutical compounds. It offers comprehensive information on thousands of low molecular weight compounds (<1500 Da) that represent a wide range of pathways and intermediary metabolism. Due to its vast expansion in the last two decades mass spectrometry has become an indispensable tool in “omic” analyses. The use of different ionization techniques such as the more traditional electrospray (ESI) and matrix-assisted laser desorption (MALDI), as well as recently popular desorption electrospray ionization (DESI), has allowed the analysis of a wide range of biomolecules (e.g. peptides, proteins, lipids and sugars), and their imaging and analysis in the original sample environment in a workup free fashion. An overview of the current state of the methodology is given, as well as examples of application.
During the last decade metabolomics has become increasingly utilized as a tool in systems biology analyses and has been considered the latest of the “omics” technologies that could provide the most functional information in understanding biological systems. Comprehensive and quantitative study of small molecules (metabolites) as a read-out of biological processes is the focus of metabolomics. The metabolome can be thought of as encompassing the small molecular building blocks (e.g., nucleotides, sugars, amino acids), metabolic intermediates (e.g., fatty acids), and structural and signaling elements (e.g, lipids) which lie outside of cellular encoding mechanisms (genome, transcriptome, proteome). Unlike genomics, transcriptomics, and proteomics, where changes are not always associated with a different phenotype, metabolites are small molecules controlling cellular metabolism and represent functional entities that reveal physiological, pathological or developmental status of a biological system. More than two decades of experience with the lipidome, a subset of the metabolome which can be extracted from the organic layer in a physical separation, have been devoted to optimizing the resolution of specific classes of lipid molecules. It has become apparent that each class, due to its particular chemistry, requires a unique set of preparatory and chromatographic techniques. This is also the case for metabolite analyses as a whole, as we expect a similar set of chromatographic challenges based on their similar chemical nature. However, the number of analytes associated with cellular metabolism will be substantially greater than for lipids alone, so the field faces an even more daunting challenge.
Targeted and Untargeted Metabolomics
Unlike other “omics” technologies metabolomics is presented with the challenge of immense chemical diversity and number of small molecules in addition to presence of many experimental artifacts. Some have conjectured that up to 200,000 metabolites exist in plants and more than 100,000 small molecules exist in humans from the intake of foods and drugs1,2. In addition, the wide array of environmental chemicals, microbes, and viruses present in everyone can significantly influence the human metabolome and thus, considerably increase the total number of metabolites.
Liquid chromatography - electrospray mass spectrometry (LC-ESI-MS) has become a method of choice for metabolomics studies as it allows ionization of large number of metabolites with minimal fragmentation. The larger portion of scientific publications focuses on “targeted” metabolomics investigation as it involves analysis of metabolites for which there are synthetic standards. Intracellular metabolites are sometimes labile compounds and thus require specific quenching and extractions, which presents a significant challenge given the chemical diversity, instability and rapid turnover of the cellular metabolites. Metabolite analysis requires tailoring sample preparation, extractions and analysis methods to afford appropriate analysis in spite of diverse metabolite concentration range, polarities, and other matrix-dependent variabilities3–5.
Targeted metabolomics relies on a pre-established strategy used for metabolite identification [e.g., selected reaction monitoring (SRM) by tandem mass spectrometry] and is used for quantitative determination of a set number of metabolites with known and expected chemistry, thus providing information on a specific pathway. Examples of this for multiple classes of metabolites can be found in Table 1. An important step in sample preparation and analysis that has already been implemented in lipidomics analyses is instrument calibration24. It is achieved by use of internal (added before extraction) or external (added after extraction) standards which are used for minimizing the variability in different samples’ preparations and also matrix effects (e.g. tissue, biological fluids, cell preparation, etc.). An excellent review on application of targeted metabolomics for biomarker identification and qualification has been recently published25.
Table 1.
LC-MS and GC-MS protocols for analysis of various metabolite classes.
| Metabolite class | MS platform | References |
|---|---|---|
| glycerophospholipids | LC-MS | (6,7) |
| eicosanoids | LC-MS | (8–11) |
| fatty acids | GC-MS | (12) |
| sphingolipids | LC-MS | (13,14) |
| sterols | LC-MS | (15) |
| cholesterol esters | LC-MS | (16) |
| MAG/DAG/TAG | LC-MS | (17,18) |
| TCA cycle | LC-MS | (19) |
| glycolysis cycle | LC-MS | (19) |
| nucleotides | LC-MS | (19) |
| CoAs | LC-MS | (19) |
| sugars | GC-MS | (20) |
| pyrophosphates | LC-MS | (21–23) |
| amino acids | GC-MS | (20) |
According to Siuzdak, untargeted metabolomic methods “are global in scope and have the aim of simultaneously measuring as many metabolites as possible from biological samples without bias.”26. As opposed to targeted metabolomics which might only concentrate on one pathway with twenty or fewer analytes, untargeted metabolomics, or metabolite profiling, can involve spectra with thousands of peaks. Experiments of this nature are used to observe global differences between sample types which might lead to potential targets for a pathway of interest, disease states, or the identification of new classes of metabolites27–29. Targets of interest can be further analyzed following fraction collection and more extensive structural characterization using methods such as NMR and MSn.
In situ labeling of compounds using techniques such a bioorthogonal ligation can also be of utility in compound and pathway elucidation. Azide, alkyne, or biotin tags, to name a few can be used to identify trace levels species, determine spatial localization within a cell, and to follow compounds through their metabolic pathways30–41. In one such application, an alkyne tag has been used to conjugate and visualize cross-linked proteins42. Other researchers have used 13C labeling to better distinguish de novo fatty acid synthesis from elongation43 and to confirm a novel ethylmalonyl-CoA pathway for methanol assimilation in a bacterium by tracking the isotopically labeled intermediates44.
Sample Analysis
Optimized extraction for specific class
Analyses in metabolomics are presented with a number of challenges coming from the metabolites nature: diverse chemical properties, rapid turnover, and mixed abundance. These issues determine the necessity of special quenching and extraction methods. A valuable review of methods for quenching and extraction of certain metabolites from mammalian cell culture was recently published45. Based on evaluation of different extraction and quenching methods it was apparent that the extraction protocol should quantitatively extract as many metabolites as possible without causing a chemical of physical degradation. The polarity of the extraction solvents should be suitable to solubilize both polar and non-polar materials, and using highly polar, alkaline or acidic solvents should be avoided. In this study it was also determined that in order to obtain a true “snapshot” of the present metabolites a good quenching method is needed using a solvent that does not damage the cell membrane and cause leakage46.
Multiple species extractions
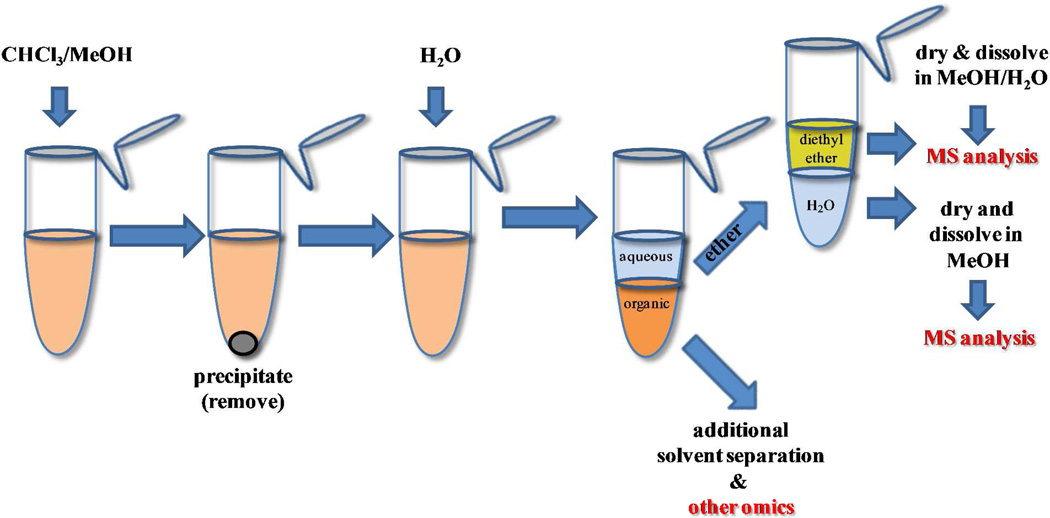
In order to extract metabolites with different polarity in a partially untargeted manner a three-phase solvent system can be utilized47. Solvents such as n-hexane, methyl acetate, acetonitrile, and water in a ratio (4:4:3:4) formed organic-aqueous solvent mixture with three mutually immiscible phases. It can be used as such for extraction of hydrophobic compounds (upper phase), moderately-polar compounds (middle phase) and polar compounds in the aqueous (lower phase) (Figure 1). This system can also be utilized in addition to the “traditional” aqueous-organic extraction when a more comprehensive analysis is needed (e.g. lipids, etc.). The lower layer of a traditional Bligh/Dyer extraction48 can be separated in parts for analysis for lipids and the rest can be subjected to a three-phase extraction for metabolite analysis.
Figure 1.
Representation of a “three phase” extraction technique. Using methodology of this type, a full range of organic and aqueous partitioned metabolites can be recovered.
Another approach for more comprehensive extraction of metabolites that excludes components such as proteins or lipids that are not intended for analysis, is using a biphasic systems where solvent volumes, ratios, and aqueous solvent pH is carefully considered49. This method is based on a traditional organic/aqueous liquid/liquid extraction, but includes pH modifications to improve acidic and basic metabolite extractions. Addition of acid to the aqueous phase allows better solubility of the compounds containing basic groups while dilute base aids solubilization of acidic metabolites. Formic acid and ammonium hydroxide are often used for adjusting pH of the aqueous phase due to their volatility and compatibility with LC-MS techniques. By utilizing multiple extractions (of the same sample) at different pH of the aqueous component, improves recovery and allows for a more comprehensive analysis of metabolites. In conclusion, dependent on the type of analysis and the metabolites nature, a sample may have to be extracted by multiple methods in order to obtain a comprehensive representation of its metabolic composition.
Instrument Platforms
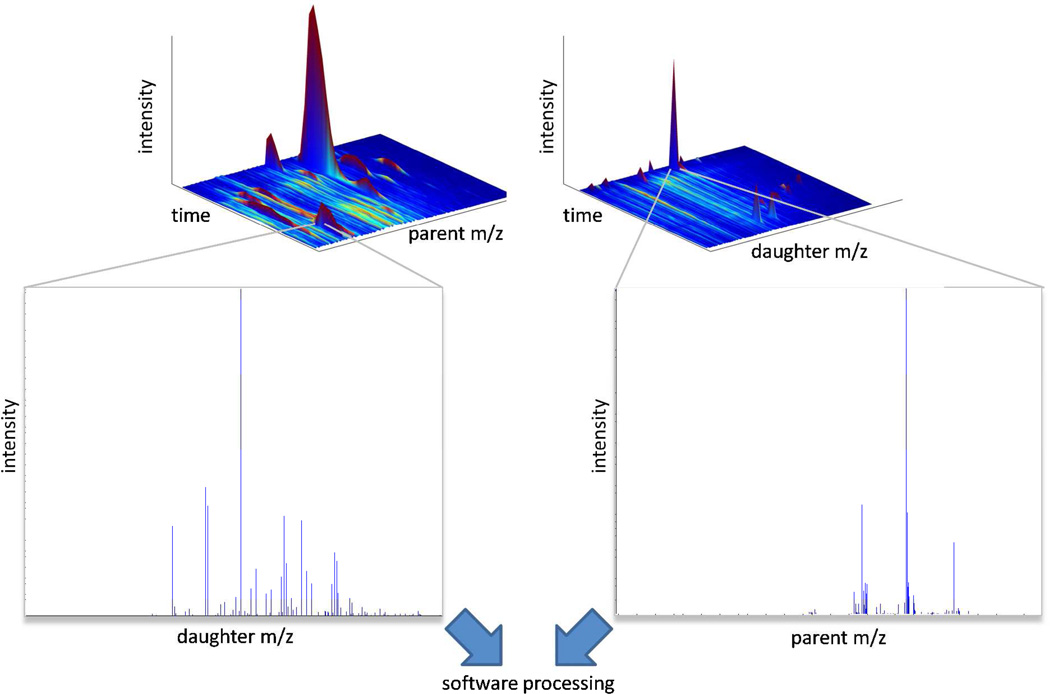
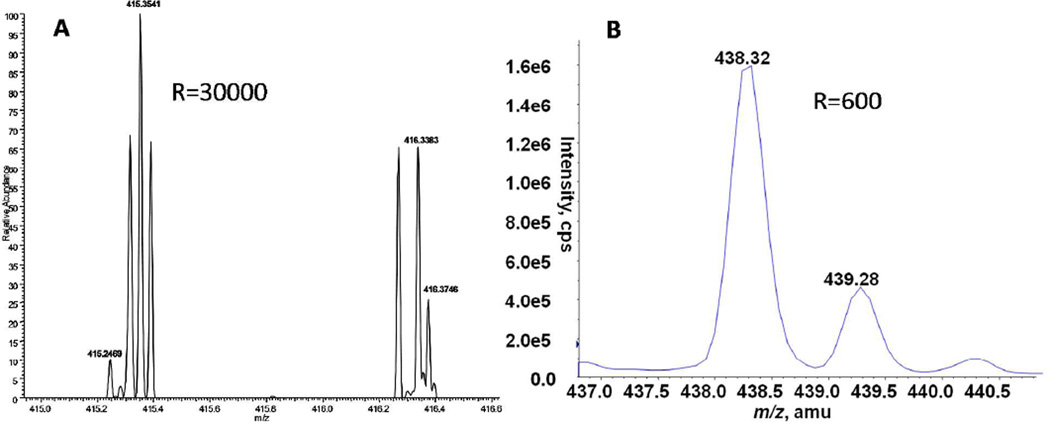
The majority of publications in the field of mass spectrometric metabolomics over the past decade have utilized HPLC-MS or UPLC-MS based systems due to their wide metabolome coverage using ionization methods such as ESI or MALDI. Recently, more groups have moved to UPLC-MS due to the shorter (90% less) run times and better peak separation inherent with these systems. Similarly, newer generation mass spectrometry instruments with very high scan rates can detect several fold more analytes per run compared to slower detectors present on older triple quadrupole and ion trap instruments. This advance in technology now makes possible highly data dependent processes such as SWATH “sequential window acquisition of all theoretical fragment-ion spectra”, GPS “global precursor ions scan mode”, or MSall 50–53. These techniques maximize metabolite detection by essentially collecting MS/MS data for every peak which can be used post-run to reconstruct MRM, neutral loss, and precursor ion scan like spectra (Figure 2). Exponentially more analytes can be detected and quantitated using a high resolution mass spectrometers compared to a triple quadrupole or ion trap instrument. As an example, Figure 3 illustrates this point with lysolipid spectra obtained on a low resolution AB Sciex 4000 Qtrap and a high resolution Thermo Fisher Orbitrap mass spectrometer. In this case, the Qtrap generated peak has a resolution of 600, whereas the Orbitrap peaks have a resolution of 30,000. Under these conditions, one could easily have 10 or more baseline separated peaks per m/z in an Orbitrap spectra whereas only 1 combined peak (containing multiple species) per m/z is obtainable on an ion trap. (FWHM 0.6 m/z compared to FWHM 0.015 m/z). The unambiguous identification of parent metabolites from their MS/MS spectra is greatly enhanced by the use of higher resolution instruments.
Figure 2.
“Fragment-everything” data mining approach. By acquiring MS/MS data for every parent and/or daughter ion, one can, post-analysis, reconstruct precursor ion scan, neutral loss scan and MRM type experimental data, which greatly enhances the ability of the analyst to identify metabolites within a sample. Rapid scanning technology in advanced commercial mass spectrometers is enabling analyses that can cycle intermittently over both (left) all possible parent ion m/z values to construct spectra of daughter ions and (right) over scans through all possible daughter m/z values to construct spectra of parent ions.
Figure 3.
Resolution comparisons between high and low resolution instruments. A) High resolution lysolipid spectra obtained on a Thermo Fisher Orbitrap with a resolution of 30,000. B) Low resolution lysolipid spectra obtained on an AB Sciex 4000 QTrap with a resolution of 600. Orbitrap spectra are courtesy of David Hachey and Vanderbilt MSRC Mass Spectrometry Core Lab.
GC-MS platforms
GC-MS based metabolomics, a complimentary method to LC-MS based metabolomics, has been pioneered by the Fiehn group who were among the first to use the term “metabolomics”54,55. The benefits of using a GC-based platform have been eloquently described by Hankemeier20: “As the full scan response in EI mode is approximately proportional to the amount of compound injected, i.e. more or less independently of the compound, all compounds suitable for GC analysis are detected non-discriminatively.” “Furthermore, problems with ion suppression of co-eluting compounds as observed in LC-MS are virtually absent in GC-EI-MS.”56. GC-MS and especially GC-GC-MS can detect a large number of species. One drawback to this platform is the need to derivatize samples, which can lead to less certainty in compound identification in GC/MS spectra (unknown number of methoximated and trimethylsilated positions) and can also lead to the loss of sample during derivatization. Since the detected GC metabolite set does not totally overlap with that observed by LC/MS, the use of both LC and GC-based methods, especially for untargeted metabolomics, would greatly increase the coverage of the metabolome.
Other technology
MALDI-TOF, which does not require extraction, can be used on intact biological samples. Dorrestein has demonstrated the utility of this technique in the monitoring of polymicrobial infections57. Using MALDI-TOF, he was able to map the metabolites and signaling molecules between or from different colonies. Additional examples of MALDI- TOF imaging experiments can be found in the recent reviews by Murphy and Wariishi58,59. DESI-MS is another useful technique which can be employed to monitor biological samples (such as bacterial colonies)60. One drawback of DESI is its spatial resolution of 200 µm compared to 20 µm for MALDI. Also, whereas abundant molecules in biological samples can easily be detected using DESI, lower abundance species and low molecular weight metabolites have not been widely reported using this method59. Recent advances in nanoDESI technology have led to substantial improvements in spatial resolution (less than 50 µm). Once this technology is readily available, this should greatly increase the capabilities of DESI imaging61.
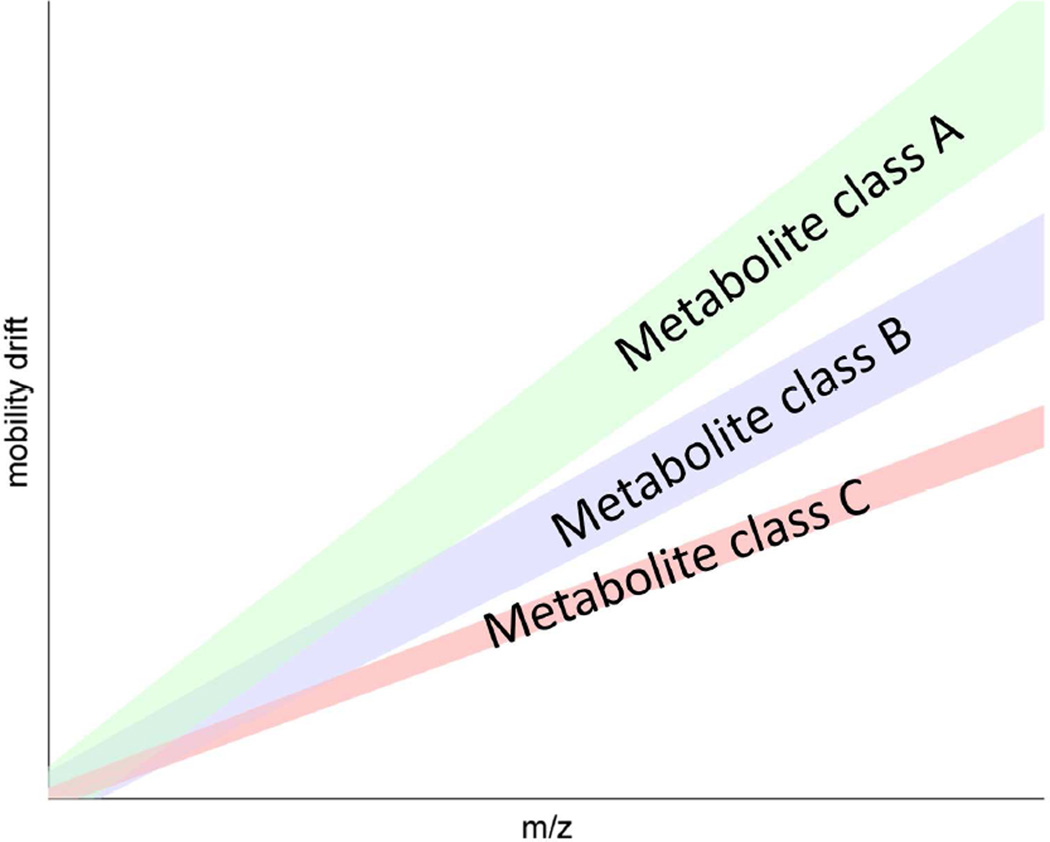
LC-MS platforms have difficulty measuring isobaric and structural isomers. Ion mobility coupled to MS (IM-MS) can routinely separate structurally similar molecules and at least double the peak capacity of a MS with MALDI as the ionization source62–64. Unlike traditional mass spectrometry, ions in IM-MS travel through a pressurized chamber containing a neutral gas. The mobility of ions through this chamber is governed by the size and structure of the compound. Subtle changes in structure within isobaric pairs, such as leucine and isoleucine, lead to different transition times through the ion mobility chamber. Using this technique, small molecules, peptides, and lipids can routinely be separated from the same sample (Figure 4). In addition, it has been shown that using ESI-IM-TOF-MS, peak capacity can be increased tenfold compared to MS alone62,63.
Figure 4.
Ion mobility mass spectral analysis of a complex mixture can be used to separate classes of metabolites (‘class A’, ‘class B’, ‘class C’) based on ion mobility drift time. Diagram patterned after results obtained in Kliman et al., (Figure 2)63, based on a methanol extract of whole rat blood, yielding a signal dominated by peptides, lipids, and other metabolites.
MS Issues (data analysis related)
From spectra to quantified result
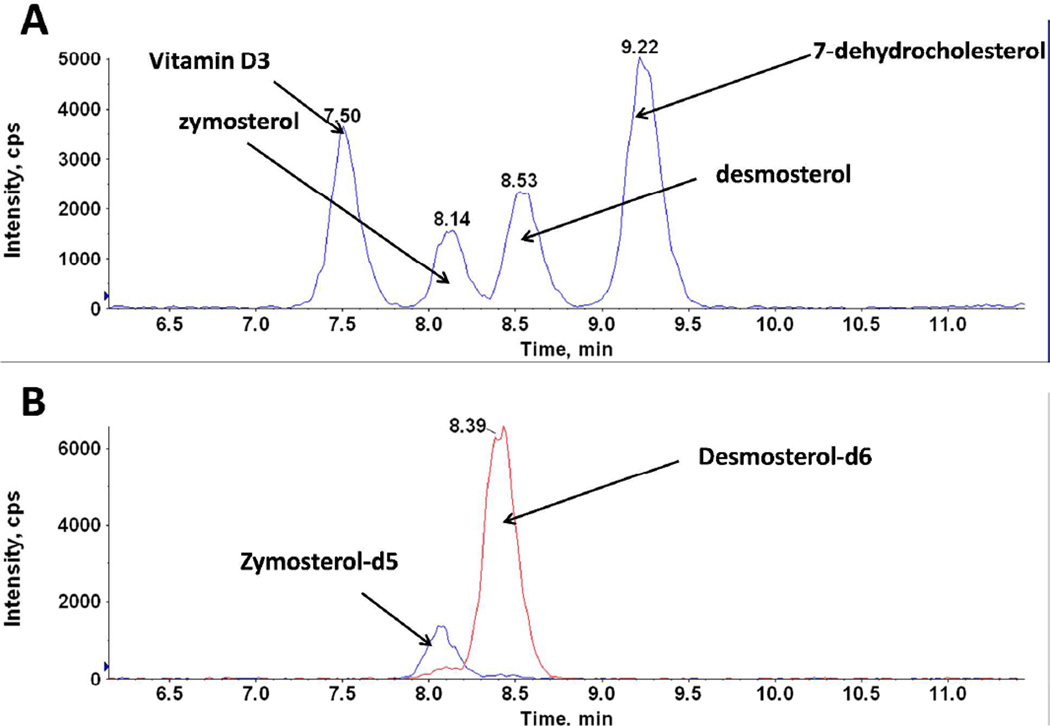
While there is a wide variety of unique molecules that are commonly encountered in metabolomic analysis, the number of non-natural internal standards available is far narrower. Perhaps the most significant challenge to quantitation of metabolomic data by mass spectral techniques stems from this disparity. In practice, once a particular subset of metabolites within a pathway or class is targeted for quantitative analysis, whether in the absolute or relative to defined benchmarks but without rigorous quantitation, suitable standards which elute and ionize relatively similarly to the molecules of interest must be used. The coverage that one particular standard affords across a swath of metabolomic analytes will necessarily vary. Larger biomolecules have a tendency for side chains to interfere with primary ionization sites for deprotonation, such as for lipids6,7,65,66, fatty acids, larger polypeptides, or multiply sugared conglomerates containing several backbones. In such cases, considerable care should be applied to determining whether coverage by the selected standards has truly been achieved. Classes that allow for each constituent to be monitored with an accompanying standard exist67,12,15, but are the exception and not the rule. Pathways with a limited number of common chemical fragments provide appealing targets for adequate coverage by a small number of standards68. An example of the problems that can occur when multiple analytes share common MRM pairs can be seen in the analysis of sterols (Figure 5). In this case, four sterols share common MRM pairs but are resolved by HPLC chromatography. Without deuterated standards for desmosterol and zymosterol, unambiguous lipid identification during a LC-MS run would be especially challenging for these nearly co-eluting isobaric compounds.
Figure 5.
The utility of using internal standards. Four isobaric sterol MRM peaks are shown in (A) which are baseline separated. Without appropriate internal standards (shown in B), it would be difficult to accurately identify zymosterol and desmosterol by this analysis.
Methods for handling peak integration depend on the mode of MS operation [e.g., feature extraction in untargeted69–72 vs. analysis of peak robustness in targeted applications73,74] and cannot be ignored. By nature of the types of questions posed, metabolomics is often concerned with quantitative analysis of low abundance molecules, and assessments of peak quality merits attention75. First, full scan information should be retained even for selective reaction monitoring (SRM) data collection, as the level of the background may be substantial for certain extraction protocols and can thereby be monitored more completely, and because the presence of nearby isotopic peaks to analytes of interest may not be as obvious in SRMs alone without examination of the full spectrum. Similarly, high-resolution MS76 provides the opportunity to retain peak shape information that could indicate interference from different metabolites with nearby/identical mass and/or retention, and thus capturing the spectra in this format is preferable to data that has already been centroided74.
Evolution of database approaches
For several years, considerable effort has gone toward cataloging information about metabolites in publicly accessible databases with approaches tailored to multiple goals. Searchable indices for unknowns within mass ranges such as HMDB, Metlin, and LipidMAPS2,77–81; provide one level of interface, while various pathway oriented solutions can enable researchers to leverage larger scale patterns among several analytes/peaks in their data simultaneously. Examples of such platforms include KEGG, BioSpider, and MetaCyc82–84. While most of the projects above have typically been scientifically organized as top-down initiatives with editorial control being somewhat centralized, a fully user-driven community library approach to warehousing relevant data and metadata (e.g. MS/MS spectra, pathway involvement, structure drawing, etc.) may be extremely useful.
Further development of tools that facilitate hypothesis driven research by bringing together the ability to track and quantify trends in both known (and/or quantified) and unknown analytes of interest will be needed to best take advantage of data being collected in the context of interrogating specific metabolic pathways. Algorithms that analyze similarities between unknown peaks in metabolite profiling spectra in order to assess a correlative70,85,86 or more heuristic quasi-match with a curated or community-based database of known MS/MS data87,88 are examples of ways in which the full extent of data sets can be used to place untargeted scanning within a more pathway- or network-oriented context.
Example of Applications to Pathways
Technological advances in LC-MS analysis have vastly improved upon the resolution of small molecule metabolites from complex biological mixtures. While these improvements have only recently allowed for the observation of metabolic flux within a system, the concept of metabolic phenotypes in disease is not new. Perhaps the most well-established metabolic phenotype is the increased ‘aerobic glycolysis’ observed in cancer cells, known widely as the Warburg Effect89. The increased resolution provided by ultra-sensitive mass spectrometers and the decreased run times afforded through the introduction of UPLC have made real-time analysis of these complex metabolic phenotypes possible. The information gathered with these technologies has made the deconvolution of complex and previously unknown metabolic phenotypes readily discernible. Ultimately, understanding these metabolic pathways is essential for the development of future disease treatments.
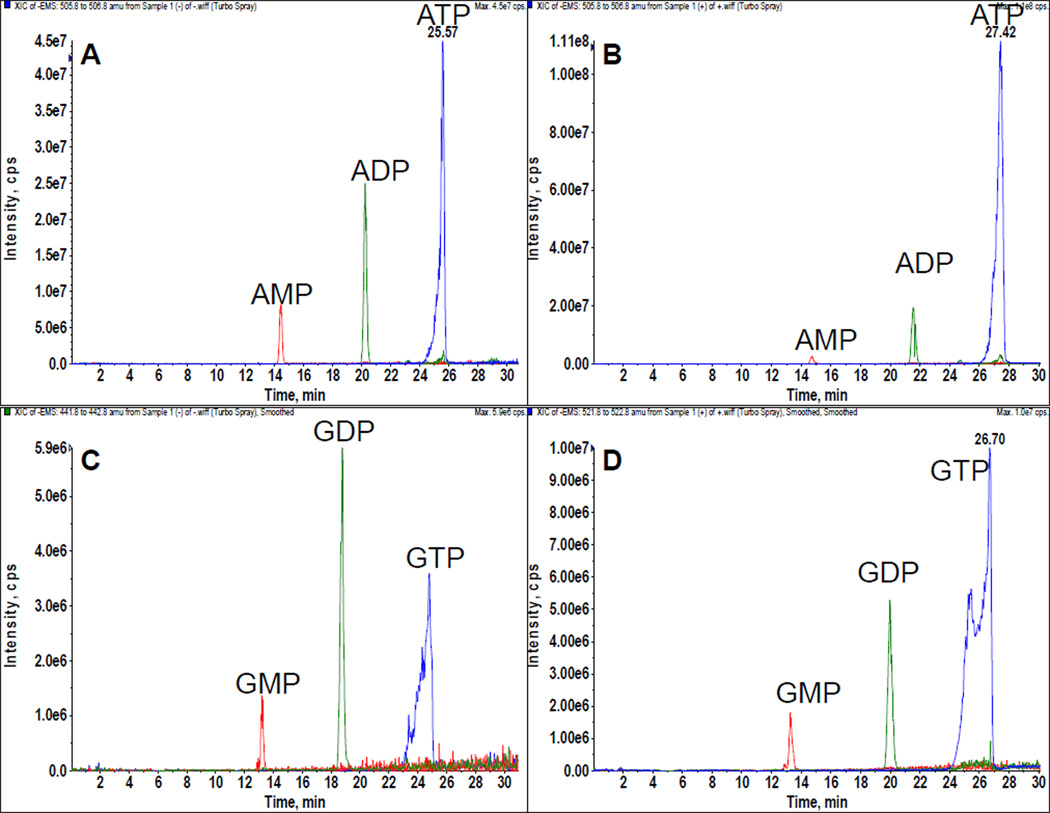
In particular, altered bioenergetic pathways serve as a potential target to slow or halt the growth of hyperproliferative diseases. Hyperproliferative cell types, such as cancer, rapidly consume ATP relative to the surrounding tissue to meet the energetic requirements of rapid cell division. This metabolic difference is regularly used in the clinic to image tumors with 18F positron imaging90. Increased metabolic demands render cells sensitive to slight alterations within a specific bioenergetic pathway and this has been put forward as an ‘Achilles heel’ of tumorigenesis91. HPLC-MS-based quantification of ATP/ADP/AMP ratios can be used to identify differences in cellular bioenergetics (Figure 6).
Figure 6.
LC-MS analysis of nucleotides. HEK cells treated with deoxyglucose (A) and (C) have a dramatically different nucleotide phosphate pattern compared to HEK cells treated with glucose (B) and (D).
To understand alterations in metabolic flux and energy production in hyperproliferative cell types, a recently published study92 melds the evolving metabolomics methodologies discussed in this review to functionally characterize a conserved bioenergetic mutation in glioblastomas. Isocitrate dehydrogenase (IDH) normally functions within the TCA cycle to convert isocitrate to α-ketoglutarate (αKG). This oxidation event yields one equivalent of NADH which proceeds to the electron transport chain for ATP production. In humans, IDH is expressed as 5 distinct isoforms93. In a genome-wide study of human gliomas, the IDH1 isoform was found to harbor a specific and conserved point mutation in up to 12% of the gliomas screened94. This mutation was tied to corresponding amino acid replacement (R132H) in the active site of the enzyme. Despite the striking conservation of this mutation, the functional implications are as yet still unknown.
Functional characterization of the R132H mutation observed in IDH1 required a multidisciplinary approach. Cells were first transfected with a dominant-negative IDH1 construct harboring R132H mutation92. An upfront non-targeted metabolomic analysis was performed on these cells using a stand-alone Orbitrap mass spectrometer. The increased mass sensitivity afforded by these instruments allows for the calculation of exact chemical masses of metabolites for identification – oftentimes out to the fourth of fifth decimal place. Coupling this technology, with a broad-spectrum extraction and reversed phase HPLC analysis recently reported by Rabinowitz and coworkers95, a peak was rapidly identified corresponding to 2-hydroxyglutarate (2HG). As a secondary confirmation step, the flux of 2HG within cells was verified through a targeted and more quantifiable LC-MS method. This analysis was performed on a triple quadrupole instrument using MRM transitions to verify structural components of the analyte of interest. An upfront HPLC method was developed specifically around 2HG to optimize its elution and concentration before reaching the ESI source. Thus, the exact degree to which 2HG was altered in cells overexpressing the R132H mutated IDH1 protein could be directly assessed.
This approach aptly combines two distinct metabolomic approaches to functionally characterize R132H IDH1 mutations. Unrestricted, exact mass analysis performed with a high-resolution Orbitrap instrument rapidly identified 2HG as the altered metabolite in this system. Once identified, a broad-spectrum mass analysis was insufficient for exact quantification of flux. Using a more traditional, targeted LC-MS approach, 2HG was reliably quantified with a triple quadrupole mass spectrometer. Each approach, targeted and untargeted, present an inverse set of advantages and disadvantages. The marriage of these techniques in this study illustrates the power of an integrated metabolomics platform.
It should be noted that this systematic approach to associate genetic and metabolic phenotypes was confirmed by a concurrent metabolomic study which identified 2HG fluctuations in acute myeloid leukemia. This altered metabolic phenotype was tied to a mutation in the second IDH isoform, IDH296. Since this time, studies have focused on the determination of the molecular mechanisms through which 2HG production leads to oncogenic transformation97 or decreased proliferation of human gliomas98. In light of the potential importance of this metabolite, a recent paper describes mice expressing IDH1 mutations known to increase 2HG production exhibit an early leukemic histone methylation phenotype99.
While the IDH1 report did not identify altered bioenergetics in gliomas per se, it does show the emerging science of metabolomics in a new light: as a platform to discover novel routes of flux within a system. As we learn more about complex disease states, such as cancer, metabolomics offers a powerful discovery platform. For example, alterations in TCA cycle metabolic flux have recently been demonstrated in Rho GTPase-transformed fibroblasts which are sensitized to the glutaminase inhibitor 968 that was developed in the Cerione laboratory100. In differentiated tissues, metabolites enter the TCA cycle through glycolysis, yet this process is often disregulated in hyperproliferative diseases due to functional alterations in glycolytic enzymes101. In keeping with this phenotype, Rho GTPase-transformed cells appear to obtain carbon atoms for TCA cycle oxidation from the amino acid glutamine. Glutaminase performs the rate-limiting hydrolysis reaction necessary for αKG production and entrance into the TCA cycle102. Thus treatment with 968 effectively starves these cells of fuel for TCA-cycle oxidation, decreasing cell viability and proliferation (Figure 7).
Figure 7.
Studying metabolic flux by use of labeled inputs. Glycolytic and TCA cycle analytes are particularly amenable to labeling of initial metabolite pools using stable isotopes (e.g., 13C glucose or acetate) and subsequent monitoring of intermediates and bottlenecks in those subsystems. Other similar opportunities exist as well; fatty acid incorporation into lipids, 15N labeling of nucleotide precursors of deoxynucleotides, and bioorthogonal approaches are all examples. Cells with increased glutaminase activity can be identified by supplementation of isotopically labeled glutamine during introduction into the TCA cycle. Dang et al.92 monitored TCA cycle metabolites to functionally characterize R132H mutation to isocitrate dehydrogenase in gliomas. The product, D-2-hydroxyglutarate, has been implicated as affecting oncogenic transformation although its precise role is as yet undefined97,98.
The commercial availability of isotopically-labeled metabolites has had a positive impact on the field of metabolomics as they are ideal tools to document how small molecule inhibitors such as 968 alter metabolic flux within a system. Approaches using isotopically-labeled metabolites have proven themselves as excellent tools to investigate TCA cycle metabolic flux. While these techniques have been used previously for 13C-NMR detection103, their application to a mass spectrometry-based metabolomics study can be equally informative. In particular, this approach has served as a useful paradigm for the study of deoxynucleotides in antiretroviral systems68. Figure 7 outlines several labeling opportunities across a number of integrated bioenergetic pathways including glycolysis, Krebs cycle and other pathways (Figure 7). Cellular media supplemented with isotopically-labeled glutamine could serve as an ideal assay condition for cellular glutaminase activity to test the activity of 968. Conversion of glutamine to αKG can be tracked by following isotopically-labeled metabolites through the TCA cycle. Since isotopically-labeled reagents are available commercially, most metabolic pathways of interest can be examined in this fashion.
Energetically, the TCA cycle is the most efficient mode for hyper-proliferative cell types to generate the massive amounts of ATP necessary for growth and replication. By inhibiting conversion of glutamine to glutamate and ultimately αKG, the small-molecule glutaminase inhibitor 968 slows the proliferation of malignantly transformed cells. In recent years, we have begun to recognize the importance of cellular energetics in hyper-proliferative disease. With the advent of small-molecules capable of attenuating metabolic flux, such as 968, the rapid diagnosis of a disregulated bioenergetic phenotype would have a profound clinical impact. Given that increased metabolic flux often corresponds directly to increased enzymatic activity in diseased cell types, these phenotypes may not be readily identifiable through a genetic screen. Rather, these specific phenotypes require the clinical application of a metabolomics platform. Employing commercially available UPLC systems dramatically decreases run times, allowing one to quickly identify aberrant metabolic phenotypes from primary tissues.
Concluding Remarks
Over the last ten years we have learned a number of valuable lessons in the emerging fields of lipidomics and metabolomics that have advanced biomedical research. First, direct infusion or shotgun approaches can be informative with respect to initial identification of changes in cellular analytes. Some caution must be observed that these changes are not the result of ion suppression and other instrumental artifacts24,104, but the combination of profiling analysis using shotgun approaches with subsequent quantitative analysis using appropriate chemically defined internal standards is a powerful advantage. Defects in metabolic pathways are detected by comparisons of analyte profiles, then subsequently quantitated. The effects of remediation can then be interrogated using molecular genetics and small molecule approaches.
In the next ten years we predict an increasing integration of the multiomic (e.g., genomic, proteomic, and metabolomic) approaches that will transform the way in which we look at biochemical pathways and think about cellular networks. The interconnectedness of pathways that at present seem distinct will increasingly be shown to have feedback loops that allow rapid responses to external stimuli and cellular injury. This kind of approach is represented in a recent, elegant report by Burant and colleagues105, where they show a plethora of changes in intracellular responses that arise during glucose stimulated insulin secretion. This work provides new insights into chronic insulin resistance associated with obesity and potentially new targets for treatment of diabetes mellitus. A better understanding of the downstream consequences of various genetic mutations on cell metabolism will advance the ways in which small molecule therapeutics are developed by providing more direct measurement of cellular defects present in a disease at a network level. Metabolomics applications in drug discovery research hold a great potential for identification of new drug targets, evaluation of lead compounds and toxicity screening, and tracking drug metabolites, thus contributing to the development of safer and more efficacious drugs. Increasingly metabolomic analysis will allow monitoring of the downstream consequences of essential cellular metabolites and the consequences of complex feedback loops. This facilitates the ability to better predict where combination therapies might be effectively used to address the multiple cellular defects associated with cellular transformation and tumor growth. This approach will also continue to drive the field of personalized medicine where the responses to specific therapeutic agents can be used to subdivide patients into differential responders based on categories of metabolomic profiles. Mass spectrometry and systems biology analysis are transforming the ways in which the molecular basis of diseases is understood and the approaches used to discover new therapeutics. Over the next decade metabolomics is likely to evolve into an invaluable screening and diagnostic tool to determine whether patients respond in a beneficial way to specific pharmacological therapies. The prospect of future diagnostics will rely on the ability of researchers to hone the selectivity and specificity of their analyses. By fine tuning the identification of biomarkers and pathway responses that are predictors of efficacy, this will make both drug discovery and therapeutics more cost efficient and effective.
Acknowledgments
Funding Sources
This work was partially supported by the NIH Grants (PO1-ES013125 and U54 069338), McDonnell Foundation for Brain Cancer Research and T32 MH093366 (Post Doctoral Training in CNS Drug Discovery Research support for TPM).
ABBREVIATIONS
- ESI-MS
electrospray ionization mass spectrometry
- GC-MS
gas chromatography mass spectrometry
- MALDI-MS
matrix-assisted laser desorption/ionization mass spectrometry
- MALDI-TOF
matrix-assisted laser desorption/ionization mass spectrometry-time of flight
- LC
liquid chromatography
- HPLC
high performance liquid chromatography
- UPLC
ultra performance liquid chromatography
- DESI-MS
desorption electrospray ionization mass spectrometry
- EI
electron ionization
- MRM
multiple reaction monitoring
- FWHM
full width at half maximum
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
NOTES
The authors declare no competing financial interest.
REFERENCES
- 1.Fiehn O. Metabolomics – the link between genotypes and phenotypes. Plant Mol. Biol. 2002;48:155–171. [PubMed] [Google Scholar]
- 2.Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, et al. HMBD: the Human Metabolome Database. Nucleic Acids Res. 2007;35:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leclercq L, Cuyckens F, Mannens GSJ, de Vries R, Timmerman P, Evans DC. Which human metabolites have we MIST? Retrospective analysis, practical aspects, and perspectives for metabolite identification and quantification in pharmaceutical development. Chem. Res. Toxicol. 2009;22:280–293. doi: 10.1021/tx800432c. [DOI] [PubMed] [Google Scholar]
- 4.Lu W, Bennett BD, Rabinowitz JD. Analytical strategies for LC-MS-based targeted metabolomics. J. Chromatogr. B. 2008;871:236–242. doi: 10.1016/j.jchromb.2008.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Urban M, Enot DP, Dallmann G, Körner L, Forcher V, Enoh P, Koal T, Keller M, Deigner H-P. Complexity and pitfalls of mass spectrometry-based targeted metabolomics in brain research. Anal. Biochem. 2010;406:124–131. doi: 10.1016/j.ab.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Myers DS, Ivanova PT, Milne SB, Brown HA. Quantitative analysis of glycerophospholipids by LC-MS: acquisition, data handling, and interpretation. Biochim. Biophys. Acta. 2011;1811:748–757. doi: 10.1016/j.bbalip.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivanova PT, Milne SB, Byrne MO, Xiang Y, Brown HA. Glycerophospholipid identification and quantitation by electrospray ionization mass spectrometry. Methods Enzymol. 2007;432:21–57. doi: 10.1016/S0076-6879(07)32002-8. [DOI] [PubMed] [Google Scholar]
- 8.Yang J, Eiserich JP, Cross CE, Morrissey BM, Hammock BD. Metabolomic profiling of regulatory lipid mediators in sputum from adult cystic fibrosis patients. Free Radic. Biol. Med. 2012;53:160–171. doi: 10.1016/j.freeradbiomed.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J, Schmelzer K, Georgi K, Hammock BD. Quantitative Profiling Method for Oxylipin Metabolome by Liquid Chromatography Electrospray Ionization Tandem Mass Spectrometry. Anal. Chem. 2009;81:8085–8093. doi: 10.1021/ac901282n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneider C, Yu Z, Boeglin WE, Zheng Y, Brash AR. Enantiomeric Separation of Hydroxy and Hydroperoxy Eicosanoids by Chiral Column Chromatography. Methods Enzymol. 2007;433:145–157. doi: 10.1016/S0076-6879(07)33008-5. [DOI] [PubMed] [Google Scholar]
- 11.Kingsley PJ, Marnett LJ. LC-MS-MS Analysis of Neutral Eicosanoids. Methods Enzymol. 2007;433:91–112. doi: 10.1016/S0076-6879(07)33005-X. [DOI] [PubMed] [Google Scholar]
- 12.Quehenberger O, Armando AM, Dennis EA. High sensitivity quantitative lipidomics analysis of fatty acids in biological samples by gas chromatography-mass spectrometry. Biochim. Biophys. Acta. 2011;1811:648–656. doi: 10.1016/j.bbalip.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sullards MC, Allegood JC, Kelly S, Wang E, Haynes CA, Park H, Chen Y, Merrill AH., Jr Structure-Specific, Quantitative Methods for Analysis of Sphingolipids by Liquid Chromatography-Tandem Mass Spectrometry: “Inside-Out” Sphingolipidomics. Methods Enzymol. 2007;432:83–115. doi: 10.1016/S0076-6879(07)32004-1. [DOI] [PubMed] [Google Scholar]
- 14.Signorelli P, Hannun YA. Analysis and quantitation of ceramide. Methods Enzymol. 2002;345:275–294. doi: 10.1016/s0076-6879(02)45023-9. [DOI] [PubMed] [Google Scholar]
- 15.McDonald JG, Smith DD, Stiles AR, Russell DW. A comprehensive method for extraction and quantitative analysis of sterols and secosteroids from human plasma. J. Lipid Res. 2012;53:1399–1409. doi: 10.1194/jlr.D022285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy RC, Leiker TJ, Barkley RM. Glycerolipid and cholesterol ester analyses in biological samples by mass spectrometry. Biochim. Biophys. Acta. 2011;1811:776–783. doi: 10.1016/j.bbalip.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krank J, Murphy RC, Barkley RM, Duchoslav E, McAnoy A. Qualitative Analysis and Quantitative Assessment of Changes in Neutral Glycerol Lipid Molecular Species Within Cells. Methods Enzymol. 2007;432:1–20. doi: 10.1016/S0076-6879(07)32001-6. [DOI] [PubMed] [Google Scholar]
- 18.Leiker TJ, Barkley RM, Murphy RC. Analysis of diacylglycerol molecular species in cellular lipid extracts by normal-phase LC-electrospray mass spectrometry. Int. J. Mass Spectrom. 2011;305:103–108. doi: 10.1016/j.ijms.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coulier L, van Kampen JJ, de Groot R, Gerritsen HW, Bas RC, van Dongen WD, Brüll LP, Luider TM. Simultaneous determination of endogenous deoxynucleotides and phosphorylated nucleoside reverse transcriptase inhibitors in peripheral blood mononuclear cells using ion-pair liquid chromatography coupled to mass spectrometry. Proteomics Clin. Appl. 2008;2:1557–1562. doi: 10.1002/prca.200800002. [DOI] [PubMed] [Google Scholar]
- 20.Koek MM, Muilwijk B, van der Werf MJ, Hankemeier T. Microbial metabolomics with gas chromatography/mass spectrometry. Anal. Chem. 2006;78:1272–1281. doi: 10.1021/ac051683+. [DOI] [PubMed] [Google Scholar]
- 21.Hooff GP, Patel N, Wood WG, Müller WE, Eckert GP, Volmer DA. A rapid and sensitive assay for determining human brain levels of farnesyl-(FPP) and geranylgeranylpyrophosphate (GGPP) and transferase activities using UHPLC-MS/MS. Anal. Bioanal. Chem. 2010;398:1801–1808. doi: 10.1007/s00216-010-4088-7. [DOI] [PubMed] [Google Scholar]
- 22.Räikkönen J, Mönkkönen H, Auriola S, Mönkkönen J. Mevalonate pathway intermediates downregulate zoledronic acid-induced isopentenyl pyrophosphate and ATP analog formation in human breast cancer cells. Biochem. Pharmacol. 2010;79:777–783. doi: 10.1016/j.bcp.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Lee SH, Raboune S, Walker JM, Bradshaw HB. Distribution of endogenous farnesyl pyrophosphate and four species of lysophosphatidic acid in rodent brain. Int. J. Mol. Sci. 2010;11:3965–3976. doi: 10.3390/ijms11103965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown HA, Murphy RC. Working towards the exegesis for lipids in biology. Nat. Chem. Biol. 2009;5(9):602–606. doi: 10.1038/nchembio0909-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffiths WJ, Koal T, Wang Y, Kohl M, Enot D, Deigner H-P. Targeted metabolomics for biomarker discovery. Angew. Chem. Int. Ed. 2010;49:5426–5445. doi: 10.1002/anie.200905579. [DOI] [PubMed] [Google Scholar]
- 26.Patti GJ, Yanes O, Siuzdak G. Innovation: Metabolomics: the apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012;13:263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saghatelian A, Trauger SA, Want EJ, Hawkins EG, Siuzdak G, Cravatt BF. Assignment of endogenous substrates to enzymes by global metabolite profiling. Biochemistry. 2004;43:14332–14339. doi: 10.1021/bi0480335. [DOI] [PubMed] [Google Scholar]
- 28.Saghatelian A, Cravatt BF. Global strategies to integrate the proteome and metabolome. Curr. Opin. Chem. Biol. 2005;9:62–68. doi: 10.1016/j.cbpa.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 29.Saghatelian A, Cravatt BF. Discovery metabolite profiling--forging functional connections between the proteome and metabolome. Life Sci. 2005;77:1759–1766. doi: 10.1016/j.lfs.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 30.Willems LI, Li N, Florea BI, Ruben M, van der Marel GA, Overkleeft HS. Triple bioorthogonal ligation strategy for simultaneous labeling of multiple enzymatic activities. Angew. Chem. Int. Ed. Engl. 2012;51:4431–4434. doi: 10.1002/anie.201200923. [DOI] [PubMed] [Google Scholar]
- 31.Milne SB, Tallman KA, Serwa R, Rouzer CA, Armstrong MD, Marnett LJ, Lukehart CM, Porter NA, Brown HA. Capture and release of alkynederivatized glycerophospholipids using cobalt chemistry. Nat. Chem. Biol. 2010;6:205–207. doi: 10.1038/nchembio.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saxon E, Bertozzi CR. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 33.Agard NJ, Prescher JA, Bertozzi CR. A strain-promoted [3 + 2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J. Am. Chem. Soc. 2004;126:15046–15047. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- 34.Wang Q, Chan TR, Hilgraf R, Fokin VV, Sharpless KB, Finn MG. Bioconjugation by copper(I)-catalyzed azide-alkyne [3 + 2] cycloaddition. J. Am. Chem. Soc. 2003;125:3192–3193. doi: 10.1021/ja021381e. [DOI] [PubMed] [Google Scholar]
- 35.Hang HC, Wilson JP, Charron G. Bioorthogonal chemical reporters for analyzing protein lipidation and lipid trafficking. Acc. Chem. Res. 2011;44:699–708. doi: 10.1021/ar200063v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hang HC, Linder ME. Exploring protein lipidation with chemical biology. Chem Rev. 2011;111:6341–6358. doi: 10.1021/cr2001977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Devaraj NK, Weissleder R, Hilderbrand SA. Tetrazine-based cycloadditions: application to pretargeted live cell imaging. Bioconjug. Chem. 2008;19:2297–2299. doi: 10.1021/bc8004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bleijerveld OB, Houweling M, Thomas MJ, Cui Z. Metabolipidomics: profiling metabolism of glycerophospholipid species by stable isotopic precursors and tandem mass spectrometry. Anal. Biochem. 2006;352:1–14. doi: 10.1016/j.ab.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 39.Best MD, Rowland MM, Bostic HE. Exploiting bioorthogonal chemistry to elucidate protein-lipid binding interactions and other biological roles of phospholipids. Acc. Chem. Res. 2011;44:686–698. doi: 10.1021/ar200060y. [DOI] [PubMed] [Google Scholar]
- 40.Rowland MM, Bostic HE, Gong D, Speers AE, Lucas N, Cho W, Cravatt BF, Best MD. Phosphatidylinositol 3,4,5-trisphosphate activity probes for the labeling and proteomic characterization of protein binding partners. Biochemistry. 2011;50:11143–11161. doi: 10.1021/bi201636s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tallman K, Armstrong MD, Milne SB, Marnett LJ, Brown HA, Porter NA. Cobalt carbonyl complexes as probes for alkyne-tagged lipids. J. Lipid Res. 2013;54:859–868. doi: 10.1194/jlr.D033332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cisar JS, Cravatt BF. Fully functionalized small-molecule probes for integrated phenotypic screening and target identification. J. Am. Chem. Soc. 2012;134:10385–10388. doi: 10.1021/ja304213w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamphorst JJ, Fan J, Lu W, White E, Rabinowitz JD. Liquid Chromatography-High Resolution Mass Spectrometry Analysis of Fatty Acid Metabolism. Anal. Chem. 2011;83:9114–9122. doi: 10.1021/ac202220b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reaves ML, Rabinowitz JD. Metabolomics in systems microbiology. Curr. Opin. Biotechnol. 2011;22:17–25. doi: 10.1016/j.copbio.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dietmair S, Timmins NE, Gray PP, Nielsen LK, Krömer JO. Towards quantitative metabolomics of mammalian cells: Development of a metabolite extraction protocol. Anal. Biochem. 2010;404:155–164. doi: 10.1016/j.ab.2010.04.031. [DOI] [PubMed] [Google Scholar]
- 46.Yanes O, Tautenhahn R, Patti GJ, Siuzdak G. Expanding coverage of the metabolome for global metabolite profiling. Anal. Chem. 2011;83:2152–2161. doi: 10.1021/ac102981k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shibusawa Y, Yamakawa Y, Noji R, Yanagida A, Shindo H, Ito Y. Three-phase solvent system for comprehensive separation of a wide variety of compounds by high-speed counter-current chromatography. J. Chromatogr. A. 2006;1133:119–125. doi: 10.1016/j.chroma.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 48.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 49.Sana T, Fischer S. Maximizing metabolite extraction for comprehensive metabolomics studies of erythrocytes. 2007 http://www.chem.agilent.com/Library/applications/5989-7407EN.pdf. [Google Scholar]
- 50.Plumb RS, Johnson KA, Rainville P, Smith BW, Wilson ID, Castro-Perez JM, Nicholson JK. UPLC/MS(E); a new approach for generating molecular fragment information for biomarker structure elucidation. Rapid Commun. Mass Spectrom. 2006;20:1989–1994. doi: 10.1002/rcm.2550. [DOI] [PubMed] [Google Scholar]
- 51.Cho R, Huang Y, Schwartz JC, Chen Y, Carlson TJ, Ma J. MS(M), an efficient workflow for metabolite identification using hybrid linear ion trap Orbitrap mass spectrometer. J. Am. Soc. Mass Spectrom. 2012;23:880–888. doi: 10.1007/s13361-012-0351-9. [DOI] [PubMed] [Google Scholar]
- 52.Hopfgartner G, Tonoli D, Varesio E. High-resolution mass spectrometry for integrated qualitative and quantitative analysis of pharmaceuticals in biological matrices. Anal. Bioanal. Chem. 2012;402:2587–2596. doi: 10.1007/s00216-011-5641-8. [DOI] [PubMed] [Google Scholar]
- 53.Gillet LC, Navarro P, Tate S, Röst H, Selevsek N, Reiter L, Bonner R, Aebersold R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics. 2012;1 doi: 10.1074/mcp.O111.016717. O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fiehn O. Combining genomics, metabolome analysis, and biochemical modeling to understand metabolic networks. Comp. Funct. Genomics. 2001;2:155–168. doi: 10.1002/cfg.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ryan D, Robards K. Metabolomics: The greatest omics of them all? Anal. Chem. 2006;78:7954–7958. doi: 10.1021/ac0614341. [DOI] [PubMed] [Google Scholar]
- 56.Koek MM, Jellema RH, van der Greef J, Tas AC, Hankemeier T. Quantitative metabolomics based on gas chromatography mass spectrometry: status and perspectives. Metabolomics. 2011;7:307–328. doi: 10.1007/s11306-010-0254-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moree WJ, Phelan VV, Wu CH, Bandeira N, Cornett DS, Duggan BM, Dorrestein PC. Interkingdom metabolic transformations captured by microbial imaging mass spectrometry. Proc. Natl. Acad. Sci. USA. 2012;109:13811–13816. doi: 10.1073/pnas.1206855109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berry KA, Hankin JA, Barkley RM, Spraggins JM, Caprioli RM, Murphy RC. MALDI imaging of lipid biochemistry in tissues by mass spectrometry. Chem. Rev. 2011;111:6491–6512. doi: 10.1021/cr200280p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miuraa D, Fujimuraa Y, Wariishi H. In situ metabolomic mass spectrometry imaging: Recent advances and difficulties. J. Proteomics. 2012;75:5052–5060. doi: 10.1016/j.jprot.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 60.Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD, van der Voort M, Pogliano K, Gross H, Raaijmakers JM, Moore BS, Laskin J, Bandeira N, Dorrestein PC. Mass spectral molecular networking of living microbial colonies. Proc. Natl. Acad. Sci. U S A. 2012;109:E1743–E1752. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lanekoff I, Geydebrekht O, Pinchuk GE, Konopka AE, Laskin J. Spatially resolved analysis of glycolipids and metabolites in living Synechococcus sp. PCC 7002 using nanospray desorption electrospray ionization. Analyst. 2013 doi: 10.1039/c3an36716a. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 62.Dwivedi P, Puzon G, Tam M, Langlais D, Jackson S, Kaplan K, Siems WF, Schultz AJ, Xun L, Woods A, Hill HH., Jr Metabolic profiling of Escherichia coli by ion mobility-mass spectrometry with MALDI ion source. J. Mass. Spectrom. 2010;45:1383–1393. doi: 10.1002/jms.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kliman M, May JC, McLean JA. Lipid analysis and lipidomics by structurally selective ion mobility-mass spectrometry. Biochim. Biophys. Acta. 2011;1811:935–945. doi: 10.1016/j.bbalip.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.May JC, Goodwin CR, McLean JA. Gas-phase ion mobility-mass spectrometry and tandem IM-MS strategies for metabolism studies and metabolomics. In: Alexander Lyubimov., editor. Encyclopedia of Drug Metabolism & Drug Interactions. New Jersey: John Wiley & Sons, Hoboken; 2012. (in press). [Google Scholar]
- 65.Koivusalo M, Haimi P, Heikinheimo L, Kostiainen R, Somerharju P. Quantitative determination of phospholipid compositions by ESI-MS: effects of acyl chain length, unsaturation, and lipid concentration on instrument response, J. Lipid Res. 2001;42:663–672. [PubMed] [Google Scholar]
- 66.Brügger B, Erben G, Sandhoff R, Wieland FT, Lehmann WD. Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc. Natl. Acad. Sci. USA. 1997;94:2339–2344. doi: 10.1073/pnas.94.6.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo K, Ji C, Li L. Stable-isotope dimethylation labeling combined with LC-ESI MS for quantification of amine-containing metabolites in biological samples. Anal. Chem. 2007;79:8631–8638. doi: 10.1021/ac0704356. [DOI] [PubMed] [Google Scholar]
- 68.Fromentin E, Gavegnano C, Obikhod A, Schinazi RF. Simultaneous quantification of intracellular natural and antiretroviral nucleosides and nucleotides by liquid chromatography-tandem mass spectrometry. Anal. Chem. 2010;82:1982–1989. doi: 10.1021/ac902737j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Patti GJ, Tautenhahn R, Siuzdak G. Meta-analysis of untargeted metabolomic data from multiple profiling experiments. Nat. Protoc. 2012;7:508–516. doi: 10.1038/nprot.2011.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tautenhahn R, Patti GJ, Rinehart D, Siuzdak G. XCMS Online: a web-based platform to process untargeted metabolomic data. Anal. Chem. 2012;84:5035–5039. doi: 10.1021/ac300698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takahashi H, Morimoto T, Ogasawara N, Kanaya S. AMDORAP: non-targeted metabolic profiling based on high-resolution LC-MS. BMC Bioinformatics. 2011;12:259. doi: 10.1186/1471-2105-12-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 73.Melamud E, Vastag L, Rabinowitz JD. Metabolomics analysis and visualization engine for LC-MS data. Anal. Chem. 2010;82:9818–9826. doi: 10.1021/ac1021166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Crutchfield CA, Lu W, Melamud E, Rabinowitz JD. Mass spectrometry-based metabolomics of yeast. Methods Enzymol. 2010;470:393–426. doi: 10.1016/S0076-6879(10)70016-1. [DOI] [PubMed] [Google Scholar]
- 75.Matsuda F, Shinbo Y, Oikawa A, Hirai MY, Fiehn O, Kanaya S, Saito K. Assessment of metabolome annotation quality: a method for evaluating the false discovery rate of elemental composition searches. PLoS One. 2009;4:e7490. doi: 10.1371/journal.pone.0007490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wei X, Sun W, Shi X, Koo I, Wang B, Zhang J, Yin X, Tang Y, Bogdanov B, Kim S, Zhou Z, McClain C, Zhang X. MetSign: a computational platform for high-resolution mass spectrometry-based metabolomics. Anal. Chem. 2011;83:7668–7675. doi: 10.1021/ac2017025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bowen BP, Northen TR. Dealing with the unknown: metabolomics and metabolite atlases. J. Am. Soc. Mass Spectrom. 2010;21:1471–1476. doi: 10.1016/j.jasms.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 78.Tautenhahn R, Cho K, Uritboonthai W, Zhu Z, Patti GJ, Siuzdak G. An accelerated workflow for untargeted metabolomics using the METLIN database. Nat. Biotechnol. 2012;30:826–828. doi: 10.1038/nbt.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Orešic M. Bioinformatics and computational approaches applicable to lipidomics. Eur. J. Lipid. Sci. Technol. 2009;111:99–106. [Google Scholar]
- 80.Fahy E, Sud M, Cotter D, Maer A, Zhou Y, Byrnes R, Subramaniam S. Bioinformatics for lipidomics. Meth. Enzymol. 2007;432:245–271. doi: 10.1016/S0076-6879(07)32011-9. [DOI] [PubMed] [Google Scholar]
- 81.Sud M, Fahy E, Cotter D, Brown A, Dennis E, Glass C, Merrill A, Murphy R, Raetz C, Russell D, Subramaniam S. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007;35:D527–D532. doi: 10.1093/nar/gkl838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Arak iM, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Knox C, Shrivastava S, Stothard P, Eisner R, Wishart DS. BIOSPIDER: A Web Server For Automating Metabolome Annotations. Pacific Symposium on Biocomputing. 2007;12:145–156. [PubMed] [Google Scholar]
- 84.Caspi R, Foerster H, Fulcher CA, Kaipa P, Krummenacker M, Latendresse M, Paley S, Rhee SY, Shearer AG, Tissier C, Walk TC, Zhang P, Karp PD. The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2008;36:D623–D631. doi: 10.1093/nar/gkm900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xiao JF, Zhou B, Ressom HW. Metabolite identification and quantitation in LC-MS/MS-based metabolomics. Trends Anal. Chem. 2012;32:1–14. doi: 10.1016/j.trac.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Peironcely JE, Reijmers T, Coulier L, Bender A, Hankemeier T. Understanding and classifying metabolite space and metabolite-likeness. PLoS One. 2011;6:e28966. doi: 10.1371/journal.pone.0028966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rojas-Cherto M, Peironcely JE, Kasper PT, van der Hooft JJ, de Vos RC, Vreeken R, Hankemeier T, Reijmers T. Metabolite identification using automated comparison of high-resolution multistage mass spectral trees. Anal. Chem. 2012;84:5524–5534. doi: 10.1021/ac2034216. [DOI] [PubMed] [Google Scholar]
- 88.Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD, van der Voort M, Pogliano K, Gross H, Raaijmakers JM, Moore BS, Laskin J, Bandeira N, Dorrestein PC. Mass spectral molecular networking of living microbial colonies. Proc. Natl. Acad. Sci. U S A. 2012;109:E1743–E1752. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat. Rev. Cancer. 2011;11:325–327. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 90.Gambhir SS. Molecular imaging of cancer with positron emission tomography. Nat. Rev. Cancer. 2002;2:683–693. doi: 10.1038/nrc882. [DOI] [PubMed] [Google Scholar]
- 91.Jones NP, Schulze A. Targeting cancer metabolism – aiming at a tumor’s sweet-spot. Drug Discov. Today. 2012;17:232–241. doi: 10.1016/j.drudis.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 92.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2010;465:739–743. doi: 10.1038/nature09132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Geisbrecht BV, Gould SJ. The human PICD gene encodes a cytoplasmic and peroxisomal NADP(+)-dependent isocitrate dehydrogenase. J. Biol. Chem. 1999;274:30527–30533. doi: 10.1074/jbc.274.43.30527. [DOI] [PubMed] [Google Scholar]
- 94.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lu W, Clasquin MF, Melamud E, Amador-Noguez D, Caudy AA, Rabinowitz JD. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone Orbitrap mass spectrometer. Anal. Chem. 2010;82:3212–3221. doi: 10.1021/ac902837x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, Rabinowitz JD, Carroll M, Su SM, Sharp KA, Levine RL, Thompson CB. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen J, McKay R, Parada L. Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell. 2012;149:36–47. doi: 10.1016/j.cell.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bralten L, Kloosterhof N, Balvers R, Sacchetti A, Lapre L, Lamfers M, Sieger, Leenstra S, de Jonge H, Kros J, Jansen E, Struys E, Jakobs C, Salomons G, Diks S, Peppelenbosch M, Kremer A, Hoogenraad C, Smitt P, French P. IDH1 R132H Decreases Proliferation of Glioma Cell Lines In Vitro and In Vivo. Annals of Neurology. 69:455–463. doi: 10.1002/ana.22390. [DOI] [PubMed] [Google Scholar]
- 99.Sasaki M, Knobbe CB, Munger JC, Lind EF, Brenner D, Brüstle A, Harris IS, Holmes R, Wakeham A, Haight J, You-Ten A, Li WY, Schalm S, Su SM, Virtanen C, Reifenberger G, Ohashi PS, Barber DL, Figueroa ME, Melnick A, Zúñiga-Pflücker JC, Mak TW. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488:656–659. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang JB, Erickson JW, Fuji R, Ramachandran S, Gao P, Dinavahi R, Wilson KF, Ambrosio AL, Dias SM, Dang CV, Cerione RA. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell. 2010;18:207–219. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Anastasiou D, Yu Y, Israelsen WJ, Jiang JK, Boxer MB, Hong BS, Tempel W, Dimov S, Shen M, Jha A, Yang H, Mattaini KR, Metallo CM, Fiske BP, Courtney KD, Malstrom S, Khan TM, Kung C, Skoumbourdis AP, Veith H, Southall N, Walsh MJ, Brimacombe KR, Leister W, Lunt SY, Johnson ZR, Yen KE, Kunii K, Davidson SM, Christofk HR, Austin CP, Inglese J, Harris MH, Asara JM, Stephanopoulos G, Salituro FG, Jin S, Dang L, Auld DS, Park HW, Cantley LC, Thomas CJ, Vander Heiden MG. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat. Chem. Biol. 2012;6:839–847. doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Erickson JW, Cerione RA. Glutaminase: a hot spot for regulation of cancer cell metabolism? Oncotarget. 2010;1:734–740. doi: 10.18632/oncotarget.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cheng T, Sudderth L, Yang C, Mullen AR, Jin ES, Matés JM, DeBerardinis RJ. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc. Natl. Acad. Sci. USA. 2011;108:8674–8679. doi: 10.1073/pnas.1016627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brown HA. Lipidomics: When apocrypha becomes canonical. Curr. Opin. Chem. Biol. 2012;16:221–226. doi: 10.1016/j.cbpa.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lorenz MA, El Azzouny MA, Kennedy RT, Burant CF. Metabolome response to glycose in the β-cell line INS-1 832/13. J. Biol. Chem. 2013 Feb. doi: 10.1074/jbc.M112.414961. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]