Key Points
KIR gene copy number variation influences NK cell education at the repertoire level due to a linear effect on KIR expression.
No effect of KIR gene dose on NK cell education at the single cell level.
Abstract
Natural killer (NK) cells are functionally tuned by education via killer cell immunoglobulin receptors (KIRs) interacting with HLA class I molecules. We examined the effect of KIR gene copy number variation on the education of human NK cells. The frequency of NK cells expressing a given KIR correlated with the copy number of that gene. However, coexpression of multiple copies from a single locus, or duplicated loci, was infrequent, which is in line with independent transcriptional regulation of each allele or copy. Intriguingly, coexpression of 2 KIR alleles, resulting in higher surface expression, did not lead to enhanced functional responses in vitro or to selective advantages during in vivo responses to cytomegalovirus infection, suggesting that receptor density does not influence NK education at the single cell level. However, individuals with multiple KIR gene copies had higher frequencies of responding cells, consistent with heightened overall responsiveness.
Introduction
Killer cell immunoglobulin-like receptors (KIRs) interact with HLA molecules and shape the functionality of natural killer (NK) cells. At the genetic level, there is evidence that epistatic interactions between KIR and HLA influence outcomes in several clinical conditions, including infections, autoimmunity, and cancer (reviewed in Khakoo and Carrington1). More recently, the KIR gene dose was associated with simian immunodeficiency virus and HIV-1 viral load in rhesus macaque and humans, respectively, suggesting that KIR gene copy number variation (CNV) influences antiviral immunity.2,3 One suggested mechanism for how the KIR gene CNV affects the NK cell response to viral infection is by promoting education.3 During NK cell education, interactions between inhibitory KIRs and their cognate HLA class I ligands set the threshold for NK cell activation upon stimulation with target cells lacking the corresponding HLA class I ligands.4 Hence, the expression of multiple copies of an inhibitory KIR could potentially lead to enhanced NK cell education, thereby strengthening antiviral immunity.
KIR expression on NK cells is largely random and is determined by the KIR gene content, polymorphism, and stochastic epigenetic regulation at the promoter level.5,6 In addition, extensive CNV contributes to diversity of KIR genes.7 At the protein level, KIR2DL3 (2DL3) and 3DS1 expression frequencies were shown to be higher in individuals with 2 copies of the respective KIR gene compared with those with only 1 copy, suggesting a gene dose effect on KIR expression.3,5 However, the overarching impact of KIR CNV on human NK cell repertoires and functionality remains unknown.
Using a recently developed high throughput methodology for KIR gene typing, we investigated the effect of KIR CNV on the expression of 7 major inhibitory and activating KIRs and determined its influence on NK cell education at the single cell and repertoire level.
Study design
Healthy donors
This study was approved by the regional ethics committee in Stockholm, Sweden, with 204 healthy adults (mean age, 42.2 years; range, 18 to 69 years). This study was conducted in accordance with the Declaration of Helsinki.
Flow cytometry
Peripheral blood mononuclear cells were separated from buffy coats by density gravity centrifugation (Ficoll-Hypaque; GE Healthcare, Uppsala, Sweden). KIR repertoire staining and analysis were performed as described in detail elsewhere.8
Functional assays
Peripheral blood mononuclear cells (106 cells) were mixed with K562 cells at a ratio of 10:1 in U-bottomed 96-well plates, centrifuged at 300 rpm for 3 minutes, and incubated for 2 or 6 hours at 37°C and 5% CO2. CD107a APC-Cy7 or CD107a PE (H4A3) were added prior the start of the assay. After incubation, cells were harvested by centrifugation, surface stained, fixed, permeabilized (Fixation & Permeabilization Buffers; Ebioscience) for polyfunctional assays, stained with intracellular interferon (IFN)-γ-AF700 (B27) and tumor necrosis factor-α-eF450 (MAb11), and analyzed by flow cytometry.
KIR and HLA typing
Genomic DNAs were isolated from whole blood using a commercial kit (Qiagen) and stored at −20°C. KIR ligands were determined using the KIR HLA ligand kit (Olerup-SSP) for detecting the HLA-Bw4, HLA-C1, and HLA-C2 motifs. KIR genotyping was performed using a recently described high-throughput technology called qKAT as described in detail in Jiang et al.7
The 2DL3*05 genotype was determined using 2 polymerase chain reaction amplifications with the following pairs of sequence-specific primers (polymerase chain reaction-SSP)1: forward 5′-GTCCACAGAAAACCTTCCCTCAG-3′ and reverse 5′-GGTGCAAAGTGTCCTTAAACTTCCTT-3′ 2DL3*004/*005/*010-specific and2 forward 5′-GTGTCTCCTCTTCTTCCAGGTAATC-3′, reverse 5′-GCAGGCTCTTGGTCCATTACTA-3′ and product sequencing primer 5′-GGGACCATCCTGTCTGTGAG-3′ to distinguish 2DL3*005 from 2DL3*004 and 2DL3*010.
Statistics
For comparisons of independent groups, Mann-Whitney U tests were performed. For comparisons of matched groups, paired Student t test or Wilcoxon matched test were performed. The above statistical analyses were performed using GraphPad software. Subsets with high KIR expression representing statistical outliers were identified using Chauvenet’s criterion (see Béziat et al8 for details).
Results and discussion
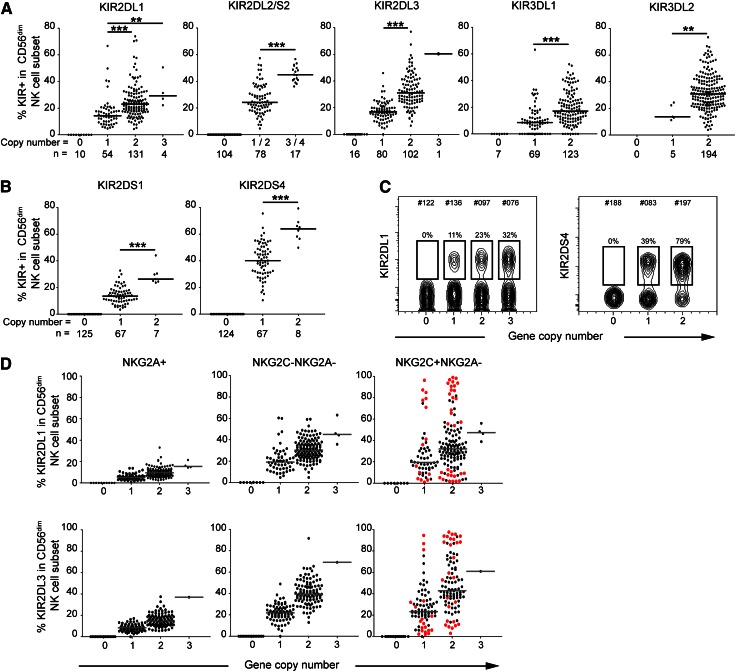
Previous studies have suggested a gene dose effect on KIR expression at the messenger RNA level for 2DL1, 2DL2/3, and 3DL1/S1,9 and at the protein level for 2DL2/3 and 3DS1.3,5,10 To determine whether such gene dose effects apply to a broader set of inhibitory and activating KIRs, we combined a recently developed methodology for assessing KIR gene CNV with a high-resolution phenotypic analysis of KIR repertoires. Strikingly, we observed a linear relationship between KIR gene copy number and the expression of all tested inhibitory and activating KIRs, including 2DL1, 2DL2/S2, 2DL3, 3DL1, 3DL2, 2DS1, and 2DS4 (Figure 1A-C). Hence, in line with the effect of multiple copies of 2DL3, previously reported by Li et al,5 the results suggest that independent regulation of distinct gene copies is common to all KIRs.
Figure 1.
The KIR expression frequency correlates linearly with KIR gene copy number. Frequency of CD56dim NK cells expressing the indicated inhibitory (A) and activating (B) KIR stratified based on KIR gene copy number. (C) Representative examples of expression of the indicated KIR in donors with distinct copy numbers. (D) Frequency of 2DL1+ (top) and 2DL3+ (bottom) NK cells within the indicated subsets and stratified based on KIR gene copy number. Red dots represent the donors with statistical outliers identified by the Chauvenet algorithm. **P < .01; ***P < .001.
Next we plotted KIR expression in distinct NK cell subsets, representing discrete stages of NK cell differentiation. This revealed a number of donors with very high frequencies of specific KIRs, particularly in the NKG2C+NKG2A− NK cell subset (Figure 1D). These outliers correspond to the recently described clonal-like expansions of educated NKG2C+ NK cells (expressing self-specific KIRs) in CMV seropositive individuals.8 Importantly, KIR gene copy number stratification revealed that such outliers were found independently of the KIR gene copy number (Figure 1D).
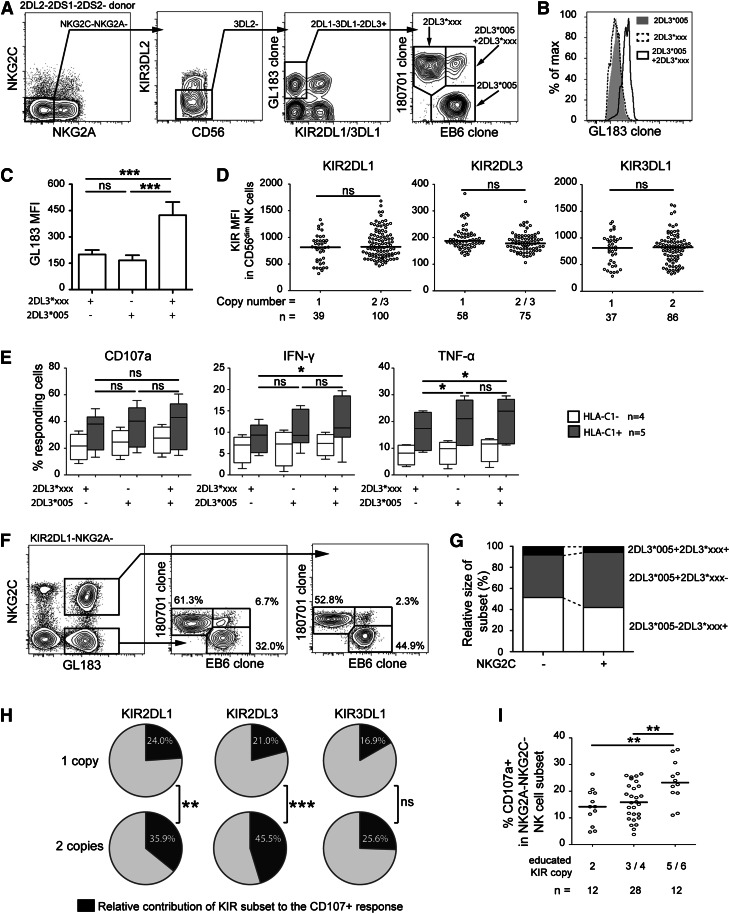
Because NK cell education is dependent on the strength of the interactions between inhibitory KIRs and their cognate ligands, we speculated that CNV could influence NK cell education at the single cell level. It is not possible to discriminate cells that express 1 or more copies of the same allele. Instead, to test this hypothesis, we took advantage of recently described cross-reactivity of the anti-2DL1/S1 mAb (EB6) with the 2DL3*005 allele.11 By combining distinct anti-2DL1/S1 and anti-2DL2/3/S2 clones, it is possible to discriminate 2DL3*005 single positive, 2DL3*xxx single positive, and 2DL3*005/2DL3*xxx double positive NK cells in individuals lacking 2DL2/S2 and 2DS1 (Figure 2A). We identified 9 donors with this particular configuration of KIR genes, out of which 5 had the educating ligand HLA-C1. In these sets of unique donors, a number of observations were made. First, NK cells expressing 2 alleles of 2DL3 had higher expression of total 2DL3 (Figure 2B-C). Second, the regulation of the 2 distinct alleles was independent and bi-allelic expression frequencies were near or slightly higher than those predicted by the product rule. Thus, despite an increase in overall frequencies of NK cells expressing a given KIR in individuals with multiple gene copies (Figure 1), most NK cells only transcribed from 1 of the available loci. Supporting this interpretation, the mean level of KIR expression on CD56dim NK cells did not differ in individuals with multiple KIR gene copies, as exemplified for 2DL1, 2DL3, and 3DL1 (Figure 2D). Third, simultaneous expression of 2 distinct alleles of 2DL3 did not influence the education at the single cell level as determined by polyfunctional responses against K562 targets (Figure 2E and supplementary Figure 1). Substantiating the latter finding, in vivo expansion of educated NK cells in CMV seropositive individuals occurred independently of the KIR gene copy number, because there was no preferential expansion of NK cells expressing 2 alleles among the expanded NKG2C+ NK cells (Figure 2F-G). The lack of effect of KIR density on NK cell education is in line with the observation in mice that the quality rather than the quantity of the receptor-ligand interactions determine NK cell education,12 as well as recent findings in humans suggesting that the dose of HLA-C does not influence education.13
Figure 2.
Influence of KIR gene copy number on NK cell education. (A) Gating strategy to identify single and bi-allelic expression of 2DL3 in donors lacking 2DS1, 2DS2, and 2DL2 genes. First, gates were set on NKG2C−NKG2A− NK cells to avoid differences in functionality due to NKG2C expression and/or education by NKG2A. Thereafter, 2DL3+2DL1− cells were gated out for further stratification into 2DL3*005 single positive cells (180701−EB6+), KIR2DL3*xxx single positive cells (180701+EB6−) and 2DL3*005/2DL3*xxx double positive cells (180701+EB6+). KIR2DL3*xxx represents all 2DL3 alleles recognized by mAb 180701. All donors were confirmed to be 2DL3*005 at the genetic level (B) Representative histogram and (C) bar chart (n = 9), showing the mean fluorescence intensity (MFI) (+/− standard deviation) of single and bi-allelic expression of 2DL3 (GL183+). (D) Expression levels (MFI) of 2DL1, 2DL3, and 3DL1 as a function of CNV. (E) Function (CD107a, IFN-γ and tumor necrosis factor-α) of single KIR-positive NK cells in donors stratified based on KIR gene CNV and the presence or absence of the corresponding KIR ligand. (F) One representative example and (G) recapitulative bar chart (mean of 3 donors) showing the allelic distribution in the expanded (NKG2C+) and the nonexpanded (NKG2C-) subset after gating on GL183+2DL1−NKG2A− CD56dim NK cells. All donors had the 2DL3*005/2DL3*xxx genotype. (H) Average contribution of the indicated KIR expressing subset to the CD107a response in donors with 1 or 2 copies of the KIR gene and with the cognate HLA-class I ligand. (I) Overall effect of KIR gene CNV on degranulation in the CD56dim NKG2A−NKG2C− NK cell compartment after exclusion of 2DL2/S2+ donors. *P < .05; **P < .01; ***P < .001. ns, not significant.
Together, the analyses made possible in these unique donors revealed that NK cells rarely express more than 1 copy, and when they do, it does not influence their overall functional responsiveness. Corroborating these 2 observations, an aggregated analysis of NK cells in the whole cohort revealed that education of single KIR+ NK cells, harboring 1 or more gene copies, was independent of the KIR gene CNV (supplementary Figure 2).
However, it is important to consider that differences in thresholds between distinct readouts may influence the interpretation of the results.4 Thus, in mice, the gene dose of H-2Dd did not influence the percentage of degranulating NK cells expressing the cognate Ly49A receptor but was associated with higher mean fluorescence intensity of CD107a, stronger IFN-γ responses, and more efficient in vivo rejection of major histocompatibility complex class I negative targets.14 Furthermore, education by NKG2A and KIRs seem to provide differential effects on IFN-γ vs degranulation responses by NK cells after allogeneic stem cell transplantation.15 Despite this potential limitation of current assays to monitor NK cell education, the CD107a assay is robust enough to consistently detect subtle differences in NK cell functionality caused by education via different KIR alleles.6 Indeed, in the present study the effects of combinations of 2DL3*xxx and 2DL3*005 alleles on NK education were consistent, irrespective of the readout used (Figure 2E and supplementary Figure 1).
Finally, even if CNV had no impact on education at the single cell level, it could still influence NK cell education at the level of NK cell repertoires. Indeed, high copy number of an educating KIR (ligand present) led to a relatively larger contribution of NK cells expressing this particular KIR to the responding (CD107a+) population (Figure 2H). To further isolate the effect of KIR gene copy number, the analysis was restricted to donors with a fixed number of KIR ligands (ie, HLA-C1, HLA-C2, and HLA-Bw4). Strikingly, donors with 5 or more copies of 2DL1, 2DL3, and 3DL1 responded better to stimulation with K562 cells than those with fewer copies (Figure 2I).
In summary, our results reveal a linear effect of KIR gene CNV on the frequency of cells expressing both activating and inhibitory KIRs and on the overall functional responsiveness of the NK cell repertoire. The results may have implications for NK cell-mediated alloreactivity in settings of allogeneic stem cell transplantation, where education in the donor has a major influence on outcome.16
Supplementary Material
Acknowledgments
This work was supported by grants from the Swedish Research Council, the Swedish Children’s Cancer Society, the Swedish Cancer Society, the Royal Swedish Academy of Sciences, the Tobias Foundation, the Karolinska Institutet, the Wenner-Gren Foundation, Oslo University Hospital, the MRC and Wellcome Trust with partial funding from the Cambridge BRC-NIHR.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: V.B., L.L.L., J.A.T., J.T., and K.-J.M. designed research; S.L. organized sampling of healthy blood donors; V.B., J.J., L.L.L., M.E., and J.A.T. performed research and analyzed data; and V.B., J.A.T., J.T., and K.-J.M. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Vivien Béziat, Karolinska Institutet, 14186, Stockholm, Sweden; e-mail: vivien.beziat@ki.se; and Karl-Johan Malmberg, Center for Infectious Medicine, F59, Karolinska University Hospital Huddinge, Stockholm, 14186 Sweden; e-mail: kalle.malmberg@ki.se or k.j.malmberg@medisin.uio.no.
References
- 1.Khakoo SI, Carrington M. KIR and disease: a model system or system of models? Immunol Rev. 2006;214:186–201. doi: 10.1111/j.1600-065X.2006.00459.x. [DOI] [PubMed] [Google Scholar]
- 2.Hellmann I, Lim SY, Gelman RS, Letvin NL. Association of activating KIR copy number variation of NK cells with containment of SIV replication in rhesus monkeys. PLoS Pathog. 2011;7(12):e1002436. doi: 10.1371/journal.ppat.1002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pelak K, Need AC, Fellay J, et al. NIAID Center for HIV/AIDS Vaccine Immunology. Copy number variation of KIR genes influences HIV-1 control. PLoS Biol. 2011;9(11):e1001208. doi: 10.1371/journal.pbio.1001208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Höglund P, Brodin P. Current perspectives of natural killer cell education by MHC class I molecules. Nat Rev Immunol. 2010;10(10):724–734. doi: 10.1038/nri2835. [DOI] [PubMed] [Google Scholar]
- 5.Li H, Pascal V, Martin MP, Carrington M, Anderson SK. Genetic control of variegated KIR gene expression: polymorphisms of the bi-directional KIR3DL1 promoter are associated with distinct frequencies of gene expression. PLoS Genet. 2008;4(11):e1000254. doi: 10.1371/journal.pgen.1000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yawata M, Yawata N, Draghi M, Partheniou F, Little AM, Parham P. MHC class I-specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood. 2008;112(6):2369–2380. doi: 10.1182/blood-2008-03-143727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang W, Johnson C, Jayaraman J, et al. Copy number variation leads to considerable diversity for B but not A haplotypes of the human KIR genes encoding NK cell receptors. Genome Res. 2012;22(10):1845–1854. doi: 10.1101/gr.137976.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Béziat V, Liu LL, Malmberg JA, et al. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood. 2013;121(14):2678–2688. doi: 10.1182/blood-2012-10-459545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McErlean C, Gonzalez AA, Cunningham R, Meenagh A, Shovlin T, Middleton D. Differential RNA expression of KIR alleles. Immunogenetics. 2010;62(7):431–440. doi: 10.1007/s00251-010-0449-9. [DOI] [PubMed] [Google Scholar]
- 10.Pascal V, Yamada E, Martin MP, et al. Detection of KIR3DS1 on the cell surface of peripheral blood NK cells facilitates identification of a novel null allele and assessment of KIR3DS1 expression during HIV-1 infection. J Immunol. 2007;179(3):1625–1633. doi: 10.4049/jimmunol.179.3.1625. [DOI] [PubMed] [Google Scholar]
- 11.Falco M, Romeo E, Marcenaro S, et al. Combined genotypic and phenotypic killer cell Ig-like receptor analyses reveal KIR2DL3 alleles displaying unexpected monoclonal antibody reactivity: identification of the amino acid residues critical for staining. J Immunol. 2010;185(1):433–441. doi: 10.4049/jimmunol.0903632. [DOI] [PubMed] [Google Scholar]
- 12.Brodin P, Lakshmikanth T, Mehr R, et al. Natural killer cell tolerance persists despite significant reduction of self MHC class I on normal target cells in mice. PLoS ONE. 2010;5(10) doi: 10.1371/journal.pone.0013174. pii:e13174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charoudeh HN, Schmied L, Gonzalez A, et al. Quantity of HLA-C surface expression and licensing of KIR2DL+ natural killer cells. Immunogenetics. 2012;64(10):739–745. doi: 10.1007/s00251-012-0633-1. [DOI] [PubMed] [Google Scholar]
- 14.Brodin P, Lakshmikanth T, Kärre K, Höglund P. Skewing of the NK cell repertoire by MHC class I via quantitatively controlled enrichment and contraction of specific Ly49 subsets. J Immunol. 2012;188(5):2218–2226. doi: 10.4049/jimmunol.1102801. [DOI] [PubMed] [Google Scholar]
- 15.Foley B, Cooley S, Verneris MR, et al. NK cell education after allogeneic transplantation: dissociation between recovery of cytokine-producing and cytotoxic functions. Blood. 2011;118(10):2784–2792. doi: 10.1182/blood-2011-04-347070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venstrom JM, Pittari G, Gooley TA, et al. HLA-C-dependent prevention of leukemia relapse by donor activating KIR2DS1. N Engl J Med. 2012;367(9):805–816. doi: 10.1056/NEJMoa1200503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.