Abstract
KIF7, the human ortholog of Drosophila Costal2, is a key component of the Hedgehog signaling pathway. Here we report mutations in KIF7 in individuals with hydrolethalus and acrocallosal syndromes, two multiple malformation disorders with overlapping features that include polydactyly, brain abnormalities and cleft palate. Consistent with a role of KIF7 in Hedgehog signaling, we show deregulation of most GLI transcription factor targets and impaired GLI3 processing in tissues from individuals with KIF7 mutations. KIF7 is also a likely contributor of alleles across the ciliopathy spectrum, as sequencing of a diverse cohort identified several missense mutations detrimental to protein function. In addition, in vivo genetic interaction studies indicated that knockdown of KIF7 could exacerbate the phenotype induced by knockdown of other ciliopathy transcripts. Our data show the role of KIF7 in human primary cilia, especially in the Hedgehog pathway through the regulation of GLI targets, and expand the clinical spectrum of ciliopathies.
Based on overlapping phenotypes, the rare hydrolethalus (HLS; MIM236680) and acrocallosal syndromes (ACLS; MIM200990), which share polydactyly, midline brain and facial abnormalities, have been predicted to be members of the ciliopathy group1. HLS is an autosomal recessive lethal syndrome characterized by the association of postaxial polydactyly of the hands, preaxial polydactyly of the feet, micrognathia and hydrocephaly or anencephaly with a key-hole defect of the occipital bone2. In 2005, a founder mutation in HYLS1 was identified in Finnish individuals with HLS3. To date, no other HYLS1 mutations have been found in individuals with HLS. Recently, the HYLS1 ortholog in Caenorhabditis elegans and Xenopus laevis was shown to be necessary for the apical anchoring of centrioles to the plasma membrane, a phenomenon required for ciliogenesis4. ACLS is an autosomal recessive syndrome with, typically, corpus callosum agenesis, occasional anencephaly and/or Dandy-Walker malformation, hypertelorism, postaxial polydactyly of the hands and preaxial polydactyly of the feet5. The molecular basis of ACLS is unknown, although researchers from a previous study proposed that ACLS and HLS are allelic conditions6.
We investigated the underlying causes of HLS and ACLS in three separate genetic studies. First, we studied an Algerian consanguineous family (family 1; Fig. 1) with four affected fetuses (Table 1) with hydrocephaly (two out of four fetuses), exencephaly (two out of four fetuses), postaxial polydactyly of the hands, hallux duplication and a cleft palate in two out of four cases. Neuropathological examination of fetus 3 showed widened ventricles and midbrain-hindbrain malformation evocative of the molar tooth sign (MTS). We considered this phenotype similar to HLS (Supplementary Table 1) and therefore sequenced HYLS1 but found no mutation. We then performed a genome-wide scan using an Affymetrix 250K SNP chip. We found a homozygous 14-Mb genomic region on chromosome 15q26 with a maximal log10 odds (LOD) score of 2.6 between rs62030374 and rs7497122 (Supplementary Fig. 1).
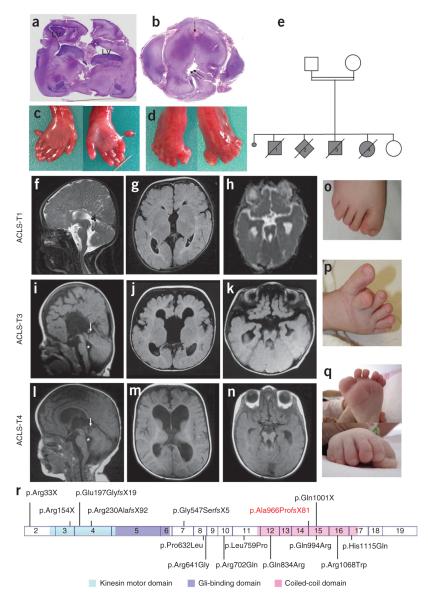
Figure 1.
Phenotypes of individuals with KIF7 mutations. (a) Coronal section of the cerebral hemispheres showing lateral ventricles (LV) dilatation in fetus 3 of family 1. (b) Transverse section of the brainstem at the midbrain-hindbrain junction showing deep interpeduncular fossa (arrow), malformed and stretched aqueduct of Sylvius (double arrow) in fetus 3 of family 1. (c,d) Postaxial polydactyly and hallux duplication in fetus 3. (e) Pedigree of family 1. (f,i,l) Sagittal brain MRI section in individuals ACLS-T1, ACLS-T3 and ACLS-T4 showing the corpus callosum agenesis (f,i) or the thin corpus callosum (l), dysplastic superior vermis (arrows) or dilated fourth ventricle (asterisks). (g,h,j,k,m,n) Axial view of the cerebral hemisphere showing corpus callosum agenesis (g,j) or hypoplasia (m). (h,k,n) Axial view of brainstem abnormalities with deep interpeduncular fossa and stretched cerebellar peduncles in three individuals with ACLS. (o) Hallux duplication in ACLS-T1. (p) Hallux duplication in ACLS-N2. (q) Hallux duplication and postaxial polydactyly in ACLS-T4. (r) Schematic representation of KIF7 complementary DNA and protein with the functional domains (kinesin motor, Gli-binding and coiled-coil domains26) and localization of KIF7 mutations identified in this study (ACLS and HLS mutations are shown above the scheme and heterozygous variations are shown below the scheme). The mutation found in HLS family 1 is shown in red.
Table 1.
Clinical features of individuals with KIF7 mutations
| Origin | C | Cases | Age | Vermis | Corpus callosum |
Other brain anomalies |
Upper limb polydactyly |
Lower limb polydactyly |
Cleft | Other | KIF7 homozygous mutations |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
Fam 1
Algeria |
+ | Fetus 1 | 15 gw | – | ? | Anencephaly | Postaxial R | Preaxial R+L | + | c.2896_2897del, p.Ala966ProfsX81 |
|
| Fetus 2 | 12 gw | ? | ? | Anencephaly | – | – | ? | ||||
| Fetus 3 | 15 gw | MTS | Absent | Hydrocephaly, arhinencephaly |
Postaxial R+L | Preaxial and postaxial R+L |
+ | Micrognathia | |||
| Fetus 4 | 11 gw | – | ? | Hydrocephaly | – | – | – | ||||
|
Fam 2
Turkey |
+ | ACLS-T1 | 4 y | MTS | Agenesis | – | – | Preaxial R | – | MR, facial dysmorphism | c.460C>T, p.Arg154X |
|
Fam 3
Turkey |
+ | ACLS-T2 | 2 m | MTS | Agenesis | Temporal pachygyria, malformed hippocampus |
Postaxial R+L | – | – | MR, macrocephaly, large fontanelle, depressed and wide nasal bridge, short philtrum, bilateral optic atrophy, ventricular septal defect |
c.1639_ 1640delinsT, p.Gly547SerfsX5 |
|
Fam 4
Turkey |
– | ACLS-T3 | 26 m | MTS | Agenesis | Wide ventricles | Postaxial R+L | Postaxial R+L | – | MR, hypotonia, macrocephaly, prominent forehead, hypertelorism, wide nasal bridge, short philtrum, high palate, nail dystrophy |
c.3001C>T, p.Gln1001X |
|
Fam 5
Turkey |
+ | ACLS-T4 | 33 m | MTS | Thin | Wide ventricles, hypomyelinization |
Cutaneous syndactyly |
Preaxial and postaxial R+L; cutaneous syndactyly |
– | MR, hypotonia, macrocephaly, prominent forehead, hypertelorism, depressed and wide nasal bridge, short philtrum, congenital hypothyroidism |
c.67C>T, p.Arg33X |
|
Fam 6
Pakistan |
+ | ACLS1 | 9 y | – | Agenesis | Wide ventricles, poor frontal cortical development |
Postaxial R+L | Preaxial and postaxial R+L |
– | MR, macrocephaly, frontal bossing, hypertelorism, broad nasal bridge, notched nasal tip, low-set ears |
c.687delG, p.Arg230AlafsX92 |
|
Fam 7
Finland |
? | ACLS2 | 12 y | – | Agenesis | – | Postaxial R+L | Postaxial R+L | – | MR, macrocephaly, broad forehead, hypertelorism, peri-orbital fullness, wide nasal bridge, high palate, micrognathia |
c.587dupT, p.Glu197GlyfsX19 |
|
Fam 8
Algeria |
+ | ACLS-N1 | 15 y | MTS | Thin ? | Para-callosal cyst | – | Preaxial | – | MR, hypertelorism, dental anomalies (teeth loss, implantation anomalies), retrognatism |
c.2896_2897del, p.Ala966ProfsX81 |
|
Fam 9
Egypt |
+ | ACLS-N2 | 12 m | MTS | Agenesis | – | Postaxial L | Preaxial | + | MR, macrocephaly, hypertelorism |
c.233_234del, p.Leu78ProfsX2 |
Age is given in gw (gestational weeks), m (months) or y (years). Fam, family; ACLS, acrocallosal syndrome; C, consanguinity; MR, mental retardation; MTS, molar tooth sign; L, left; R, right.
Parallel to, and independent from, this scan, we carried out genome-wide screens in two acrocallosal cohorts; analyses of individuals ACLS1-7 and ACLST1-T4 identified, respectively, an 8-Mb region (LOD score = 2.4 in ACLS1 and ACLS2) and a 1.82-Mb region of homozygosity between rs11637762 and rs2657947 on chromosome 15q26 (ACLS-T1-4). The smallest common interval contained 32 annotated genes. Among them, we selected KIF7, the vertebrate homolog of Drosophila costa (also called Costal2), which encodes a cilia-associated protein belonging to the kinesin family, as the best candidate because of its major role in Sonic hedgehog (Shh) transduction. Costal2 and its vertebrate homolog Kif7 have been identified as members of a protein complex that contains Sufu and kinases7 and plays a major role in activation of Hedgehog target genes through the regulation of Ci/Gli transcription factors8. Furthermore, loss of Kif7 in mice induces a Gli3-like phenotype that includes polydactyly and exencephaly9–11.
Sequencing of the 19 exons of KIF7 (Supplementary Table 2) in HLS family 1 identified a homozygous deletion of the first two base-pairs of exon 15 (c.2896_2897del, p.Ala966Profs*81) potentially affecting splicing. RT-PCR analysis on RNA extracted from lungs of affected fetuses showed no aberrant splicing or RNA decay. We next sequenced our ACLS families and found truncating mutations in all six ACLS pedigrees showing linkage to 15q26 and in two additional individuals with ACLS (Fig. 1r and Table 1). The majority of affected individuals had macrocephaly, mental retardation, abnormal facies and brain abnormalities (Table 1). Brain magnetic resonance imaging (MRI) (Fig. 1f–n) showed dilated ventricles, corpus callosum agenesis or hypoplasia and a superior vermis dysgenesis resulting in the MTS in four cases. Five affected individuals had postaxial polydactyly of the hands. In the feet, polydactyly was preaxial, hallux duplication was postaxial or preaxial and postaxial. We also observed other features (Table 1). We found a total of three nonsense and four frameshift KIF7 mutations in cases with ACLS spread throughout the gene (Fig. 1r). It is noteworthy that individual ACLS-N1, a 14 year boy, carries the same p.Ala966Profs*81 homozygous alteration found in the lethal HLS family. Therefore, no genotype-phenotype correlation can be made at this time.
Because of the similarities between HLS, ACLS and the Kif7 knockout mouse model, we reasoned that mutations in KIF7 might also cause or contribute to other predicted or known ciliopathies. We therefore sequenced 130 individuals with Bardet-Biedl (n = 96), hydrolethalus (n = 6), Pallister-Hall and Greig (n = 9), Oro-faciodigital type VI (n = 6) and Joubert and Meckel-Gruber (n = 11) syndromes. Overall, we found eight heterozygous missense changes (Fig. 1r and Supplementary Table 3). These alleles were predicted to be detrimental to protein function by PolyPhen-2 and/or SIFT (see URLs) and absent from ethnically matched controls, dbSNP or the 1000 Genomes Project database, except for p.Gln834Arg, which was present in 3 out of 384 control chromosomes. Importantly, we found no other non-synonymous changes in the controls we sequenced, indicating a significant enrichment for non-synonymous changes in cases (3 out of 384 chromosomes from controls compared to 11 out of 314 chromosomes from cases; P < 0.0039).
To investigate the pathogenic potential of the eight non-synonymous KIF7 changes identified, we used a previously characterized Shh somite defect as a phenotypic readout and an in vivo complementation approach to rescue morphant phenotypes with human mRNA12–14. Subsequent to injection of a previously validated15 kif7 translation-blocking morpholino (tb-MO) into wild-type embryos at the one-to-two cell stage, we were able to both recapitulate and quantify the aberrantly shaped somites (similar to multiple Shh component morphants, including shh, ptc and fu (refs. 16–18)) in embryos imaged at 30 h post fertilization (hpf) (Fig. 2a,b). Morphant embryos had a significantly wider mean somite angle in comparison to control embryos (99.1° versus 84.1°, respectively; P < 0.0001; n = 58–59 embryos per batch; masked scoring), which could be rescued with co-injection of full length human KIF7 mRNA (mean somite angle 88.1°; Fig. 2c and Supplementary Table 4). We then assayed each of the eight KIF7 missense changes and observed incomplete rescue that was statistically better than morpholino alone (P < 0.0001) but was significantly worse than wild-type rescue (P ≤ 0.0008), indicating that these changes are hypomorphic (Fig. 2c and Supplementary Table 4).
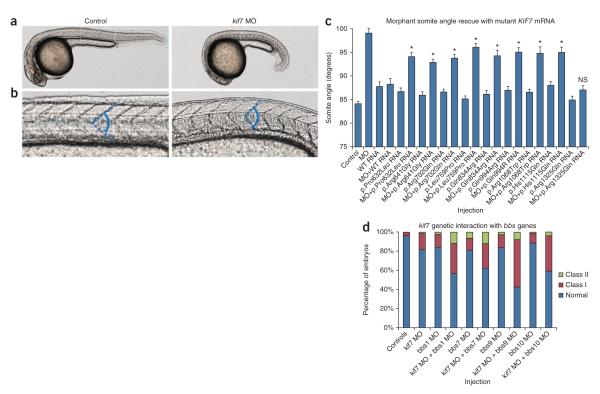
Figure 2.
Missense KIF7 variants are pathogenic and may interact genetically with other BBS loci. (a) Representative live lateral views of control (left) or kif7 morphants (right) imaged at 30 h post-fertilization. (b) Magnified view of a. Chevron-shaped somites in control (left) or abnormally shaped somites in kif7 morphant (right) embryos, attributable to ectopic Hedgehog signaling in the zebrafish myotome15. The dashed blue line denotes the somite angle measured in the in vivo complementation assay to test KIF7 allele pathogenicity. (c) All eight missense KIF7 alleles identified in cases with ciliopathy were hypomorphic as indicated by the partial ability of mutant human KIF7 mRNA to rescue the somite angle defect induced by kif7 morpholino (MO). The p.Arg1325Gln coding allele (rs73477443; present in the dbSNP database and not in cases of ciliopathy) was not significantly different (NS) from wild-type rescue, providing support for the specificity of the assay. We measured n = 43–71 embryos per injection. The asterisks indicate P < 0.001 for mutant versus wild-type rescue. Error bars, s.e.m. See Supplementary Table 4 for somite measurement data. (d) Co-injection of sub-effective doses of kif7 morpholino and four different bbs gene morpholinos resulted in gastrulation phenotypes in mid-somitic embryos that exceed the sum of affected embryos induced by either individual MO alone. For all co-injections, we observed a marked increase in the proportion of affected embryos with severe phenotypes (Class II; bbs1 and bbs7) or moderately affected embryos (Class I; bbs9 and bbs10), which exceeded the additive effects of kif7 translation-blocking morpholino or bbs morpholino alone. Objective phenotypic criteria have been described previously12. We scored n = 71–86 embryos per injection cocktail and repeated masked scoring twice.
Accumulating evidence suggests that primary ciliopathy loci can be modulated by pathogenic lesions in other ciliary genes to either exacerbate overall severity or induce specific endophenotypes12,19,20. Notably, four unrelated individuals with Bardet-Biedl syndrome carrying a KIF7 missense change also had mutations in BBS1, BBS7, BBS9 and BBS10, respectively, suggesting genetic interaction between KIF7 and these loci. To test this possibility, we compared embryos injected with sub-effective doses of kif7 tb-MO and bbs morpholino together or alone (n = 71–86 embryos per injection; repeated twice). For each of the co-injections, we observed a marked increase in the proportion of affected embryos (Fig. 2d). Taken together, these results suggest that the heterozygous hypomorphic KIF7 alleles identified in our cohort likely interact in trans with lesions at primary BBS loci, although the small number of KIF7 and BBS gene mutational events precludes us from making specific statements about the modulation of penetrance or expressivity.
Depletion of proteins involved in the Shh pathway has been shown to result in ciliogenesis defects21. To analyze ciliogenesis in individuals with KIF7 mutations, we stained cultured fibroblasts derived from individuals ACLS-T1 and ACLS-N1 (carrying the p.Arg154X and p.Ala966Profs*81 alterations, respectively) and from an unaffected control with antibodies against pericentrin and acetylated α-tubulin. We found that primary cilia were present and protruded normally from the surface of each fibroblast cell after two days of serum starvation (Supplementary Fig. 2a–c). However, cilia were significantly longer (P < 0.05) in cells from ACLS-N1 compared with control fibroblasts (Supplementary Fig. 2b,d), although ultrastructural analysis (using scanning electron microscopy) showed normal cilia components (Supplementary Fig. 2e–g). These findings suggest that, whereas KIF7 is not necessary for cilia formation, it might be involved in regulating cilia length.
To gain insight into the pathophysiological mechanism underlying the clinical features of individuals with KIF7 mutations, we undertook a transcriptome-wide analysis using total RNA extracted from lung tissue from three affected fetuses from family 1 and three age-matched controls. We subjected the data to Ingenuity Pathway Analysis (see URLs) to model protein networks with relevant biological dysregulation. This analysis showed that most known direct and/or secondary targets of GLI1, GLI2 and/or GLI3 were significantly (P < 0.05) upregulated in cases compared to age-matched controls (fold change >1.2; Fig. 3a and Supplementary Table 5), whereas KIF7 and components of the Shh pathway acting upstream of KIF7 such as SMO or proteins that interact with KIF7 (SUFU, GLI1, GLI2 and GLI3) are all transcribed similarly in cases and controls. We confirmed these results by real-time RT-PCR for several genes, including PTCH1, on RNA extracted from fibroblasts from ACLS-T1 and ACLS-N1 (Fig. 3b and Supplementary Table 5). These data are in agreement with previous studies in mice and zebrafish lacking Kif7 activity, which showed an increase in Shh-mediated target gene expression9–11,15.
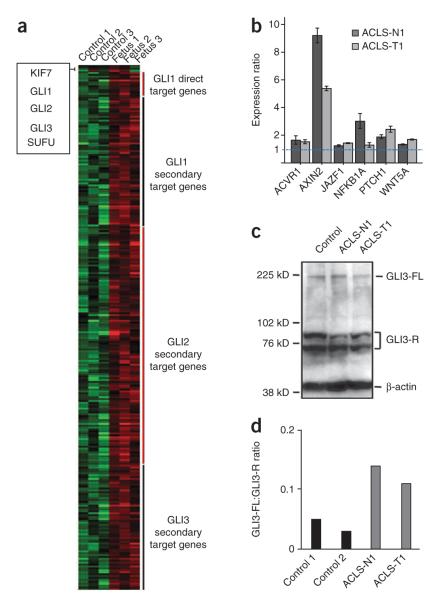
Figure 3.
Hedgehog signaling in individuals with KIF7 mutations. (a) Heat map representation of microarray expression analysis in the three KIF7 mutated fetuses from family 1 compared to three aged-matched controls (Supplementary Table 4). The heat map shows the upregulation of many of the GLI1, GLI2 and GLI3 direct and/or secondary target genes (upregulated genes in red boxes), confirmed by semiquantitative RT-PCR in fibroblasts for several selected genes relative to GAPDH (b). Error bars, s.d. (c,d) Protein blot analysis for GLI3 on proteins extracted from control and KIF7 mutated fibroblasts. (c) Immunoblot shows the amounts of GLI3-A (GLI3-FL) and GLI3-R (bands are in accordance with a previous study27 and included a Gli3−/− mouse extract as a control) in control and case fibroblasts and revealed reduced GLI3-R in KIF7 mutated fibroblasts. (d) Graphical evaluation of the GLI3-FL:GLI3-R ratio in control and case fibroblasts. We used β-actin as the loading control.
Kif7 has been shown to have both negative and positive roles in Shh signal transduction. In the absence of ligand, Kif7 localizes to the base of the cilium11 where it forms a complex with Gli proteins and other pathway components and promotes processing of GliRs. Upon ligand stimulation, Kif7 translocates to the cilium tip10, promoting Gli accumulation at that location10,11, where Kif7 may also block the function of Sufu, resulting in activation of the Gli proteins. To study the role of KIF7 in GLI3 processing in humans, we performed protein blot analysis using proteins extracted from control versus case fibroblasts. We found an increase in the GLI3-FL:GLI3-R ratio in fibroblasts from both cases (Fig. 3c,d), indicating a similar biological function of KIF7 in human and mouse to ensure normal levels of Gli3-R production. This result is concordant with the clinical features observed in humans and mice lacking KIF7 function. Indeed, in the limb, the preaxial polydactyly is consistent with ectopic de-repression of GLI3 target genes in anterior regions where Shh does not normally act but could also be ascribed in the central nervous system to an upregulation of GLI2-A9,11.
In this study, we have identified truncating mutations in KIF7, encoding an evolutionarily conserved component of the Shh signaling pathway, in individuals with two overlapping but distinct disorders. These findings highlight that extreme lethal phenotypes may represent the far end of the spectrum of less severe human disorders, as shown previously for two other ciliopathies—Meckel and Joubert syndromes22,23. Taken together, our results are of potential importance in several areas. First, they implicate this locus as the first bona fide gene for ACLS and the second locus for HLS, affording us the opportunity to improve our understanding of the pathomechanism of these disorders. Second, they expand the ciliopathy spectrum by providing experimental evidence for the prior prediction that each of these phenotypes is a ciliopathy1, while suggesting a potential mechanistic convergence in ciliary disorders of the developing cerebral cortex and cerebellum. Third, hypomorphic KIF7 alleles found in other ciliopathies and our in vivo genetic interaction studies indicate that KIF7 could exacerbate the phenotype induced by the loss of other cilia components. Fourth, our data complete a missing link of human Hedgehogopathies24 by showing that mutations in core Hedgehog signaling components cause ciliopathies and by showing the major role of KIF7 in the human Hedgehog pathway through the regulation of GLI transcription factor targets. The fact that the reported loss-of-function alleles are concomitant with GLI target dysregulation indicates that the known mechanics of the pathway, as elucidated through the previous studies of both invertebrate and vertebrate model organisms, are both functionally relevant and of clinical importance in humans. Given the possibility that the Hedgehog pathway can be manipulated pharmacologically, these findings might be of therapeutic value. Finally, our data suggest a potential new role for KIF7 in regulating cilia length as previously shown for several proteins25. The observed increase in cilia length might reflect a homeostatic response in which cells try to compensate for lack of Shh signaling, as suggested for other Shh signaling mutants (rpgrip1l)21. Further studies are necessary to elucidate the role of KIF7 in the regulation of ciliogenesis.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturegenetics/.
ONLINE METHODS
Research subjects
We used standard methods to isolate genomic DNA from peripheral blood of the affected children and family members or from frozen fetal tissue or amniocytes. Chromosome analysis was performed for at least one affected individual of each family. Two fetuses from family 1 had a complete autopsy and karyotyping after genetic counseling and had parental consent in conformity with French law. Informed consent was obtained from all participating families, and the study was approved by the Ethics Boards of Hôpital Necker-Enfants Malades and Great Ormond Street Hospital for Children.
SNPs analysis and homozygosity mapping
Genomic DNA was isolated by phenol/chloroform extraction and purified using Microcon YM-30 filters (Millipore). Genotyping was performed on the 250K NspI array of the Affymetrix 250K system, which consists of 262,000 SNPs (Affymetrix), according to the manufacturer’s instructions. Chips and data were processed on the Affymetrix platform with the Command Console Software. SNP genotypes were called using the Affymetrix BRLMM algorithm in the Genotyping Console 3.0.2 Software (Affymetrix). We performed multipoint linkage analysis using MERLIN software (see URLs) assuming a fully penetrant recessive model and a disease allele frequency of 0.001 and allowed for heterogeneity between families. Areas of homozygosity on chromosome 15 were confirmed through high-resolution haplotype analysis in HLS family 1.
Mutational screening
Mutational screening of KIF7 was performed by direct sequencing of PCR products of the 19 coding exons and the adjacent intronic junctions in families with HLS and ACLS showing potential linkage to the locus and in various individuals with ciliopathies. Segregation of the identified mutations was investigated in all available family members. The entire coding sequence of KIF7 was sequenced in 384 control chromosomes. PCR primers (Supplementary Table 2) were selected with the Primer3 program (see URLs) according to reference sequence NM_198525. PCR products were purified with the Exo-SAP cleanup kit (USB) and sequenced with BigDye chemistry and the ABI 3100 (Applied Biosystems) automated sequencer. Sequences were analyzed with SeqScape software (Applied Biosystems).
KIF7 RT-PCR
RNA from lung tissues of the three affected fetuses and three age-matched controls and from fibroblasts of the case from family 2 and one age-matched control were extracted with the QIAGEN RNeasy Kit, including on-column DNase digestion. Complementary DNA (cDNA) synthesis from total RNA was conducted using the GeneAmp RNA PCR Core Kit (Applied Biosystems) with random hexamer primers. Primers were selected in exon 13 (forward), 16 (reverse), 1 (forward) and 4 (reverse), preventing potential contaminating genomic DNA amplification (Supplementary Table 2). Quantitative real time PCR for 14 genes of interest and 1 endogenous control gene was performed in triplicate using TaqMan Gene Expression Assays on an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). Gene expression was calculated using the relative quantification. The TaqMan Assays of selected GLI targets used are listed in Supplementary Table 5.
DNA constructs
We amplified the full-length human KIF7 open reading frame (ORF) from a human primary skin fibroblast cDNA library, cloned it into a Gateway entry vector (pCR8/GW/TOPO TA Cloning Kit; Invitrogen) and confirmed the ORF by direct sequencing. The insert was then transferred into the pCS2+ Gateway destination expression vector using LR clonase II (Invitrogen)– mediated recombination. Missense variants were introduced into the KIF7 ORF using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer’s instructions and were confirmed by direct sequencing.
Zebrafish embryo manipulation and morpholino injection
The kif7 translation-blocking morpholino (tb-MO; GeneTools) was characterized previously15,17 (kif7-tb: 5-GCCGACTCCTTTTGGAGACATAGCT-3) and diluted to the appropriate concentrations in deionized sterile water (8 ng/nl for rescue experiments and 4 ng/nl for genetic interaction studies). We injected 1 nl into wild-type zebrafish embryos at the 1–2 cell stage and reared the embryos at 28.5 °C according to standard procedures. To rescue morphant somite phenotypes, we transcribed mRNA from a linearized pCS2+-KIF7 vector with the SP6 mMessage mMachine kit (Ambion) (KIF7 human versus zebrafish: 57% identity and 70% similarity). Morpholinos targeting BBS genes have been characterized previously (bbs1, bbs7, bbs9 and bbs10)14. For somite angle rescue assays, embryos were phenotyped at 30 hpf using live imaging (lateral views, taken at 6× magnification) with a Nikon AZ100 stereoscope followed by measurement of one somite angle per embryo at the midpoint between the proximal hindgut and the anus. Images were acquired and analyzed with Nikon NIS Elements software. For the kif7 genetic interaction studies, embryos were scored for gastrulation defects at the 9-somite stage according to previously established objective phenotypic criteria12.
Immunocytochemistry
After two days of serum starvation, confluent fibroblasts were fixed in 4% paraformaldehyde, treated in 50 mM NH4Cl, 0.3% Triton X-100 followed by 1 h in BSA blocking solution. Cells were then incubated with mouse monoclonal acetylated α-tubulin (Sigma) and rabbit polyclonal pericentrin (AbCam) antibodies for 1 h and with alexa Fluor 488 goat anti-rabbit IgG and alexa Fluor 555 donkey anti-mouse IgG (Invitrogen). To measure cilium length, confocal images were taken using Leica SP5. We performed three-dimensional reconstruction of cilia using Imaris software, which allowed for length assessment irrespective of angle of orientation.
Expression analysis on human exon arrays
Total RNA from lung tissues of the three affected fetuses and three age-matched controls were isolated using an RNeasy Kit (QIAGEN). RNA quality was assessed by using the Agilent Model 2100 Bioanalyzer (Agilent Technologies). An expression analysis was performed using a GeneChip Human Exon 1.0 ST Array (Affymetrix). Two kits (NuGEN Technologies) were used for the amplification and post-amplification treatments of the samples preceding the hybridization. Total RNA (100 ng) were amplified by Ribo-SPIA RNA Amplification (SPIA, for Single Primer Isothermal Amplification), fragmented and labeled with biotin using the Applause WT-Amp Plus ST System and the Encore Biotin Module as recommended by the manufacturer. The arrays were scanned using the Affymetrix Model 3000 scanner and the Command Console software for piloting the GeneChip systems. Fluorescence data were imported into Affymetrix Expression Console and R Bioconductor analysis software. Gene expression levels were calculated using the RMA algorithm in Expression Console, and flags were computed using a custom algorithm within R (see URLs). To limit potentially biased measurement (background or saturating), all probes whose normalized intensity measures were outside of a confidence interval were flagged as 0. The confidence interval was 2× the s.d. from the mean intensity of each chip. The group comparisons were done using a Student’s t test. To estimate the false discovery rate, we filtered the resulting P values at 5% and used the Benjamini and Hochberg (BH) method, the Bonferroni method (B) or no correction (SC). Data were subsequently submitted to Ingenuity Pathway Analysis (IPA; see URLs) to model networks among genes and proteins and to construct putative pathways and relevant biological processes.
Protein blot analysis
For whole cell, lysates were prepared from dissected lung or fibroblasts using ice-cold RIPA buffer plus protease inhibitor cocktail (Roche). Nuclear and cytoplasmic protein extraction were performed using a NE-PER Kit (Thermoscientific). We loaded 40 μg of proteins onto 7% SDS-PAGE gels. Immunoblotting were performed using rabbit GLI-3 H280 (Santa Cruz Biotechnology) followed by HRP-conjugated antibody and ECL-Plus detection (Amersham). An antibody against β-actin (Santa Cruz Biotechnology; sc-81178) was used as a loading control. Densitometry was used to compare protein levels between the full length activator and short repressor forms of GLI3 (see URLs).
Electron microscopy
After two days of serum starvation, confluent fibroblasts were fixed with glutaraldehyde 3% (TAAB) in 0.1 M Sorensen buffer, pH 7.4 at room temperature (20–22 °C) for 1 h. After washing with Sorensen buffer, cells were postfixed with osmic acid (OsO4) 1% for 30 min at 4 °C. Cells were dehydrated in increasing concentrations of ethanol and embedded in epon. After polymerization at 60 °C for 28 h, the coverslips were removed from the solidified blocks containing the cells. Ultrathin sections (70 ± 10 nm) were prepared, counterstained with uranyl acetate and lead citrate and then observed with a transmission electron microscope (JEOL 1011). Data acquisition was performed with GATAN CCD cameras. Image acquisition and image analysis were performed on the Cochin Imaging Facility.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to families and to the French Society of Fetal Pathology (SOFFOET) for participating in the study, to C. Dubourg, P. Wieacker, B. Leroy, N. Laurent, V. Fermeaux, S. Odent for fetuses’ referral and to A. Schinzel and A. David for ACLS samples. We thank M. Zarhrate, A. Aguilar, N. Spasky and L. Besse for technical help. We thank N. Boddaert for helpful discussion.
This work was supported by grants from ANR (FETALCILIOPATHIES number 07-MRAR-010-02 and FOETOCILPATH number BLAN-1122-01), E-RARE (Cranirare number 07-ERare-001-01) the Scientific and Technological Research Council of Turkey (TUBITAK, grant number 108S420 to N.A.A.), the US National Institutes of Health grant R01HD04260 from the National Institute of Child Health and Development (N.K.), R01DK072301 from the National Institute of Diabetes, Digestive and Kidney Disorders (N.K.) and the European Union (EU-SYSCILIA; E.E.D., N.K. and P.L.B.). A.P. was granted a fellowship from the Académie de Médecine. S.T. is supported by NIH ‘Hereditary Basis of Neural Tube Defects’ N° NS039818-07 to M. Speer. K.L.M.C. is supported by the Huygens Scholarship Programme and the Netherlands Organization for Scientific Research (NWO Toptalent-021.001.014). M.W. was a fellow of the Guggenheim Foundation and was supported by the March of Dimes Foundation (1-FY07-422). P.L.B. is a Wellcome Trust Senior Research Fellow. N.K. is a Distinguished George W. Brumley Professor.
Footnotes
URLs. Primer3, http://frodo.wi.mit.edu/primer3/input.htm; R, http://www.r-project.org/; Ingenuity Pathway Analysis, http://www.ingenuity.com; ImageJ software, http://rsb.info.nih.gov/ij/index.html; PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/; SIFT, http://sift.jcvi.org/; MERLIN Software, http://www.sph.umich.edu/csg/abecasis/Merlin/index.html.
Accession numbers. Affymetrix expression data are available from the MIAMExpress web-based tool under accession number E-MEXP-2707. The human KIF7 complementary DNA sequence is available from GenBank under accession number NM_198525.
Note: Supplementary information is available on the Nature Genetics website.
AUTHOR CONTRIBUTIONS S. Patrier, M.-H.S.F., F.E.-R. and J.-P.S. performed the fetal examinations. N.J. did the fetal karyotyping. L.R. and M.G. referred the HLS family. D.B. did the ultrasound diagnosis. G.O., Y.A., G.E.U., K.B., R.H., P.L.B., C.T., V.C.-D. and E.C. referred subjects. C.L.B., M.W., K.C., N.A.A., C.B.-F., E.U. and P.N. performed the homozygosity mapping and LOD score calculation. C.G., N.E., L.F., K.L.M.C., M.W. and C.B.-F. performed linkage and sequence analyses. S. Pruvost and E.U. did the chip experiments. S.T. and N.C. performed the expression analysis. A.S. performed the electron microscopy. N.G. and A.P. performed the confocal microscopy and measurement of cilia length. E.E.D. and N.K. did the zebrafish experiments. A.M., S.L. and M.V. advised the work. T.A.-B. designed the study and wrote the manuscript.
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Baker K, Beales PL. Making sense of cilia in disease: the human ciliopathies. Am. J. Med. Genet. C. Semin. Med. Genet. 2009;151C:281–295. doi: 10.1002/ajmg.c.30231. [DOI] [PubMed] [Google Scholar]
- 2.Salonen R, Herva R, Norio R. The hydrolethalus syndrome: delineation of a “new,” lethal malformation syndrome based on 28 patients. Clin. Genet. 1981;19:321–330. doi: 10.1111/j.1399-0004.1981.tb00718.x. [DOI] [PubMed] [Google Scholar]
- 3.Mee L, et al. Hydrolethalus syndrome is caused by a missense mutation in a novel gene HYLS1. Hum. Mol. Genet. 2005;14:1475–1488. doi: 10.1093/hmg/ddi157. [DOI] [PubMed] [Google Scholar]
- 4.Dammermann A, et al. The hydrolethalus syndrome protein HYLS-1 links core centriole structure to cilia formation. Genes Dev. 2009;23:2046–2059. doi: 10.1101/gad.1810409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schinzel A, Schmid W. Hallux duplication, postaxial polydactyly, absence of the corpus callosum, severe mental retardation, and additional anomalies in two unrelated patients: a new syndrome. Am. J. Med. Genet. 1980;6:241–249. doi: 10.1002/ajmg.1320060308. [DOI] [PubMed] [Google Scholar]
- 6.Schinzel A, Kaufmann U. The acrocallosal syndrome in sisters. Clin. Genet. 1986;30:399–405. doi: 10.1111/j.1399-0004.1986.tb01897.x. [DOI] [PubMed] [Google Scholar]
- 7.Robbins DJ, et al. Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein costal2. Cell. 1997;90:225–234. doi: 10.1016/s0092-8674(00)80331-1. [DOI] [PubMed] [Google Scholar]
- 8.Kalderon D. Hedgehog signaling: Costal-2 bridges the transduction gap. Curr. Biol. 2004;14:R67–R69. doi: 10.1016/j.cub.2003.12.047. [DOI] [PubMed] [Google Scholar]
- 9.Cheung HO, et al. The kinesin protein Kif7 is a critical regulator of Gli transcription factors in mammalian hedgehog signaling. Sci. Signal. 2009;2:ra29. doi: 10.1126/scisignal.2000405. [DOI] [PubMed] [Google Scholar]
- 10.Endoh-Yamagami S, et al. The mammalian Cos2 homolog Kif7 plays an essential role in modulating Hh signal transduction during development. Curr. Biol. 2009;19:1320–1326. doi: 10.1016/j.cub.2009.06.046. [DOI] [PubMed] [Google Scholar]
- 11.Liem KF, Jr., He M, Ocbina PJ, Anderson KV. Mouse Kif7/Costal2 is a cilia-associated protein that regulates Sonic hedgehog signaling. Proc. Natl. Acad. Sci. USA. 2009;106:13377–13382. doi: 10.1073/pnas.0906944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khanna H, et al. A common allele in RPGRIP1L is a modifier of retinal degeneration in ciliopathies. Nat. Genet. 2009;41:739–745. doi: 10.1038/ng.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leitch CC, et al. Hypomorphic mutations in syndromic encephalocele genes are associated with Bardet-Biedl syndrome. Nat. Genet. 2008;40:443–448. doi: 10.1038/ng.97. [DOI] [PubMed] [Google Scholar]
- 14.Zaghloul NA, et al. Functional analyses of variants reveal a significant role for dominant negative and common alleles in oligogenic Bardet-Biedl syndrome. Proc. Natl. Acad. Sci. USA. 2010;107:10602–10607. doi: 10.1073/pnas.1000219107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tay SY, Ingham PW, Roy S. A homologue of the Drosophila kinesin-like protein Costal2 regulates Hedgehog signal transduction in the vertebrate embryo. Development. 2005;132:625–634. doi: 10.1242/dev.01606. [DOI] [PubMed] [Google Scholar]
- 16.Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat. Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 17.Wilson CW, et al. Fused has evolved divergent roles in vertebrate Hedgehog signalling and motile ciliogenesis. Nature. 2009;459:98–102. doi: 10.1038/nature07883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolff C, Roy S, Ingham PW. Multiple muscle cell identities induced by distinct levels and timing of hedgehog activity in the zebrafish embryo. Curr. Biol. 2003;13:1169–1181. doi: 10.1016/s0960-9822(03)00461-5. [DOI] [PubMed] [Google Scholar]
- 19.Badano JL, et al. Dissection of epistasis in oligogenic Bardet-Biedl syndrome. Nature. 2006;439:326–330. doi: 10.1038/nature04370. [DOI] [PubMed] [Google Scholar]
- 20.Davis EE, et al. TTC21B contributes both causal and modifying alleles across the ciliopathy spectrum. Nat. Genet. 2011;43:189–196. doi: 10.1038/ng.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patzke S, et al. CSPP is a ciliary protein interacting with Nephrocystin 8 and required for cilia formation. Mol. Biol. Cell. 2010;21:2555–2567. doi: 10.1091/mbc.E09-06-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delous M, et al. The ciliary gene RPGRIP1L is mutated in cerebello-oculo-renal syndrome (Joubert syndrome type B) and Meckel syndrome. Nat. Genet. 2007;39:875–881. doi: 10.1038/ng2039. [DOI] [PubMed] [Google Scholar]
- 23.Valente EM, et al. Mutations in TMEM216 perturb ciliogenesis and cause Joubert, Meckel and related syndromes. Nat. Genet. 2010;42:619–625. doi: 10.1038/ng.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat. Rev. Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J, et al. Functional genomic screen for modulators of ciliogenesis and cilium length. Nature. 2010;464:1048–1051. doi: 10.1038/nature08895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monnier V, Ho KS, Sanial M, Scott MP, Plessis A. Hedgehog signal transduction proteins: contacts of the Fused kinase and Ci transcription factor with the kinesin-related protein Costal2. BMC Dev. Biol. 2002;2:4. doi: 10.1186/1471-213X-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinelli DC, Fan CM. Gas1 extends the range of Hedgehog action by facilitating its signaling. Genes Dev. 2007;21:1231–1243. doi: 10.1101/gad.1546307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.