Abstract
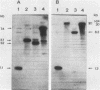
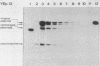
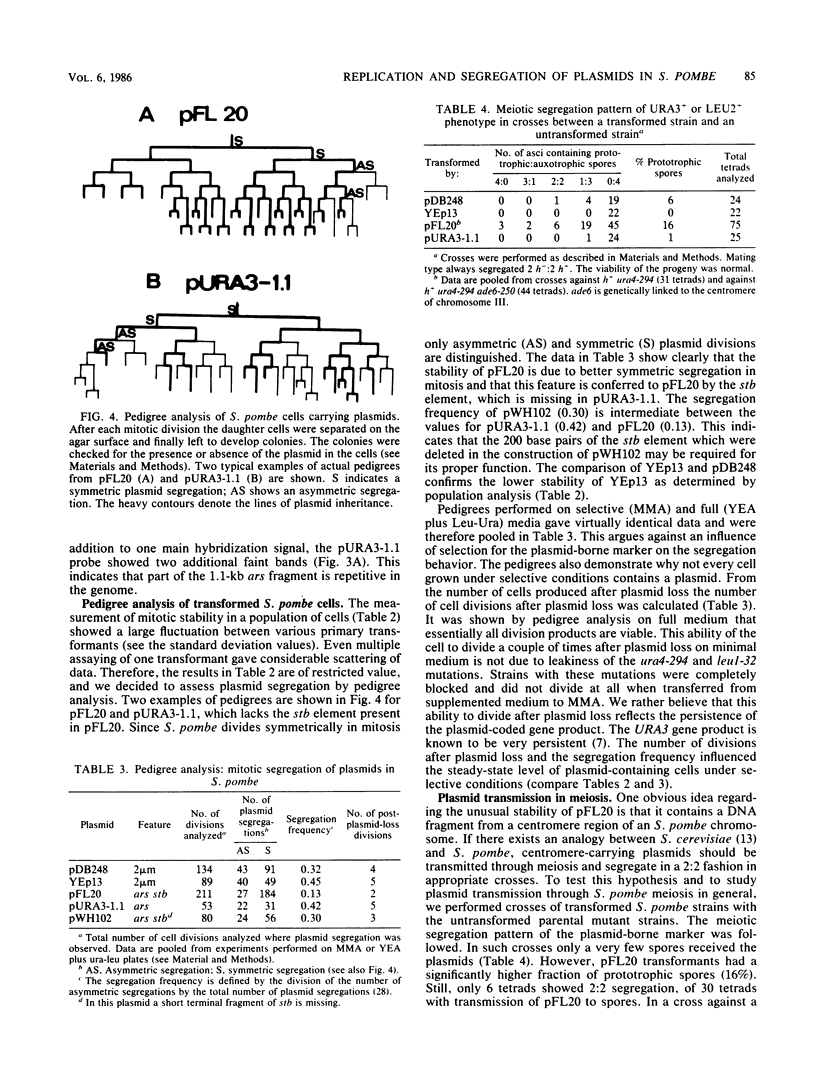
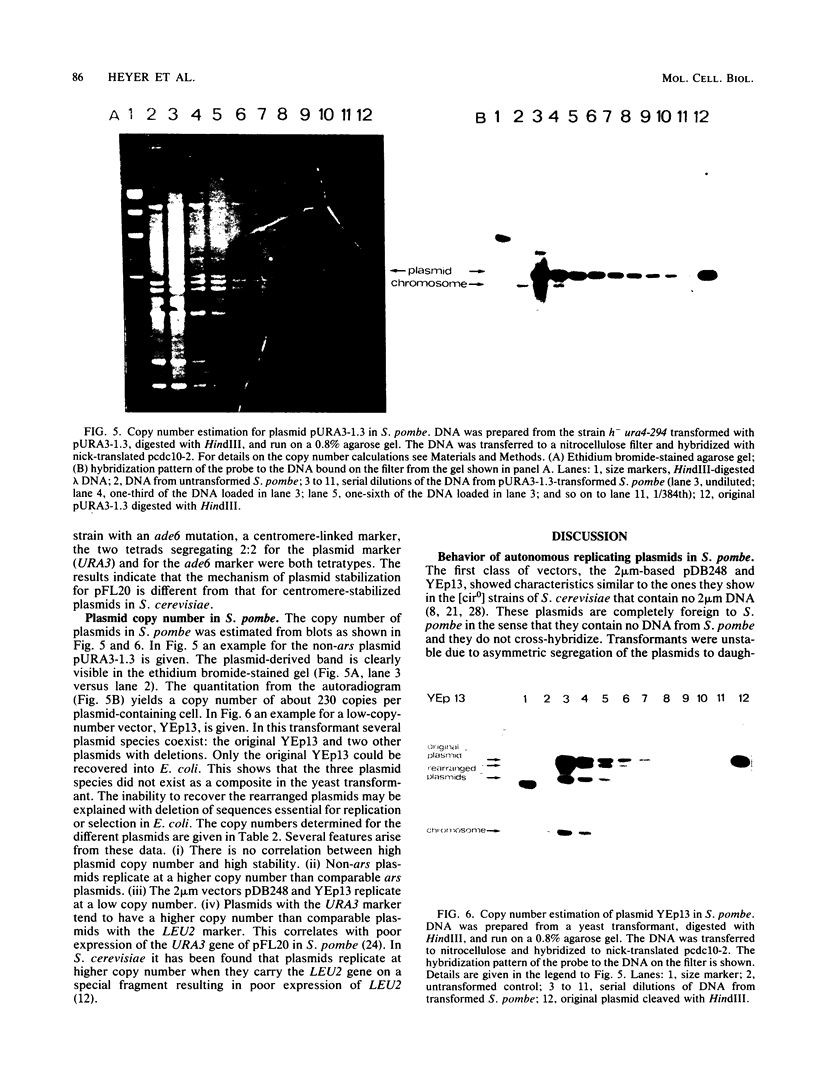
We characterized a number of widely used yeast-Escherichia coli shuttle vectors in the fission yeast Schizosaccharomyces pombe. The 2 micron vectors pDB248 and YEp13 showed high frequency of transformation, intermediate mitotic and low meiotic stability, and a low copy number in S. pombe, analogous to their behavior in [cir0] strains of Saccharomyces cerevisiae. The S. cerevisiae integration vectors pLEU2 and pURA3 transformed S. pombe at very low frequencies but, surprisingly, in a nonintegrative fashion. Instead, they replicated autonomously, and they showed very high copy numbers (up to 150 copies per plasmid-containing cell). This could reflect a lack of sequence specificity for replication of plasmid DNA in S. pombe. pFL20, an S. pombe ars vector, and a series of plasmids derived from it were studied to analyze the unusually high stability of this plasmid. Mitotic stability and partitioning of the plasmids was measured by pedigree analysis of transformed S. pombe cells. An S. pombe DNA fragment (stb) was identified that stabilizes pFL20 by improvement of plasmid partitioning in mitosis and meiosis.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amstutz H., Munz P., Heyer W. D., Leupoid U., Kohli J. Concerted evolution of tRNA genes: intergenic conversion among three unlinked serine tRNA genes in S. pombe. Cell. 1985 Apr;40(4):879–886. doi: 10.1016/0092-8674(85)90347-2. [DOI] [PubMed] [Google Scholar]
- Aves S. J., Durkacz B. W., Carr A., Nurse P. Cloning, sequencing and transcriptional control of the Schizosaccharomyces pombe cdc10 'start' gene. EMBO J. 1985 Feb;4(2):457–463. doi: 10.1002/j.1460-2075.1985.tb03651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach D., Nurse P., Egel R. Molecular rearrangement of mating-type genes in fission yeast. Nature. 1982 Apr 15;296(5858):682–683. doi: 10.1038/296682a0. [DOI] [PubMed] [Google Scholar]
- Beach D., Piper M., Nurse P. Construction of a Schizosaccharomyces pombe gene bank in a yeast bacterial shuttle vector and its use to isolate genes by complementation. Mol Gen Genet. 1982;187(2):326–329. doi: 10.1007/BF00331138. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Hicks J. B. Replication and recombination functions associated with the yeast plasmid, 2 mu circle. Cell. 1980 Sep;21(2):501–508. doi: 10.1016/0092-8674(80)90487-0. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Clancy S., Mann C., Davis R. W., Calos M. P. Deletion of plasmid sequences during Saccharomyces cerevisiae transformation. J Bacteriol. 1984 Sep;159(3):1065–1067. doi: 10.1128/jb.159.3.1065-1067.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L., Carbon J. Functional expression of cloned yeast DNA in Escherichia coli: specific complementation of argininosuccinate lyase (argH) mutations. J Mol Biol. 1978 Apr 25;120(4):517–532. doi: 10.1016/0022-2836(78)90351-0. [DOI] [PubMed] [Google Scholar]
- Erhart E., Hollenberg C. P. The presence of a defective LEU2 gene on 2 mu DNA recombinant plasmids of Saccharomyces cerevisiae is responsible for curing and high copy number. J Bacteriol. 1983 Nov;156(2):625–635. doi: 10.1128/jb.156.2.625-635.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M., Clarke L., Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982 May;29(1):235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- Grant D. M., Lambowitz A. M., Rambosek J. A., Kinsey J. A. Transformation of Neurospora crassa with recombinant plasmids containing the cloned glutamate dehydrogenase (am) gene: evidence for autonomous replication of the transforming plasmid. Mol Cell Biol. 1984 Oct;4(10):2041–2051. doi: 10.1128/mcb.4.10.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks J. B., Hinnen A., Fink G. R. Properties of yeast transformation. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1305–1313. doi: 10.1101/sqb.1979.043.01.149. [DOI] [PubMed] [Google Scholar]
- Hyman B. C., Cramer J. H., Rownd R. H. Properties of a Saccharomyces cerevisiae mtDNA segment conferring high-frequency yeast transformation. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1578–1582. doi: 10.1073/pnas.79.5.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaram M., Li Y. Y., Broach J. R. The yeast plasmid 2mu circle encodes components required for its high copy propagation. Cell. 1983 Aug;34(1):95–104. doi: 10.1016/0092-8674(83)90139-3. [DOI] [PubMed] [Google Scholar]
- Kearsey S. Structural requirements for the function of a yeast chromosomal replicator. Cell. 1984 May;37(1):299–307. doi: 10.1016/0092-8674(84)90326-x. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y. Yeast plasmid requires a cis-acting locus and two plasmid proteins for its stable maintenance. Cell. 1983 Dec;35(2 Pt 1):487–493. doi: 10.1016/0092-8674(83)90182-4. [DOI] [PubMed] [Google Scholar]
- Losson R., Lacroute F. Plasmids carrying the yeast OMP decarboxylase structural and regulatory genes: transcription regulation in a foreign environment. Cell. 1983 Feb;32(2):371–377. doi: 10.1016/0092-8674(83)90456-7. [DOI] [PubMed] [Google Scholar]
- Maundrell K., Wright A. P., Piper M., Shall S. Evaluation of heterologous ARS activity in S. cerevisiae using cloned DNA from S. pombe. Nucleic Acids Res. 1985 May 24;13(10):3711–3722. doi: 10.1093/nar/13.10.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montiel J. F., Norbury C. J., Tuite M. F., Dobson M. J., Mills J. S., Kingsman A. J., Kingsman S. M. Characterization of human chromosomal DNA sequences which replicate autonomously in Saccharomyces cerevisiae. Nucleic Acids Res. 1984 Jan 25;12(2):1049–1068. doi: 10.1093/nar/12.2.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A. W., Szostak J. W. Pedigree analysis of plasmid segregation in yeast. Cell. 1983 Oct;34(3):961–970. doi: 10.1016/0092-8674(83)90553-6. [DOI] [PubMed] [Google Scholar]
- Méchali M., Kearsey S. Lack of specific sequence requirement for DNA replication in Xenopus eggs compared with high sequence specificity in yeast. Cell. 1984 Aug;38(1):55–64. doi: 10.1016/0092-8674(84)90526-9. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 1983;101:228–245. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- Russell P. R., Hall B. D. The primary structure of the alcohol dehydrogenase gene from the fission yeast Schizosaccharomyces pombe. J Biol Chem. 1983 Jan 10;258(1):143–149. [PubMed] [Google Scholar]
- Sakaguchi J., Yamamoto M. Cloned ural locus of Schizosaccharomyces pombe propagates autonomously in this yeast assuming a polymeric form. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7819–7823. doi: 10.1073/pnas.79.24.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai K., Sakaguchi J., Yamamoto M. High-frequency cotransformation by copolymerization of plasmids in the fission yeast Schizosaccharomyces pombe. Mol Cell Biol. 1984 Apr;4(4):651–656. doi: 10.1128/mcb.4.4.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcomb D. T., Struhl K., Davis R. W. Isolation and characterisation of a yeast chromosomal replicator. Nature. 1979 Nov 1;282(5734):39–43. doi: 10.1038/282039a0. [DOI] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker W. T., Miller C. A., Cohen S. N. Structural and functional analysis of the par region of the pSC 10 1 plasmid. Cell. 1984 Aug;38(1):191–201. doi: 10.1016/0092-8674(84)90540-3. [DOI] [PubMed] [Google Scholar]
- Uemura T., Yanagida M. Isolation of type I and II DNA topoisomerase mutants from fission yeast: single and double mutants show different phenotypes in cell growth and chromatin organization. EMBO J. 1984 Aug;3(8):1737–1744. doi: 10.1002/j.1460-2075.1984.tb02040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakian V. A., Kupfer D. M. Replication and segregation of an unstable plasmid in yeast. Plasmid. 1982 Jul;8(1):15–28. doi: 10.1016/0147-619x(82)90037-3. [DOI] [PubMed] [Google Scholar]