Abstract
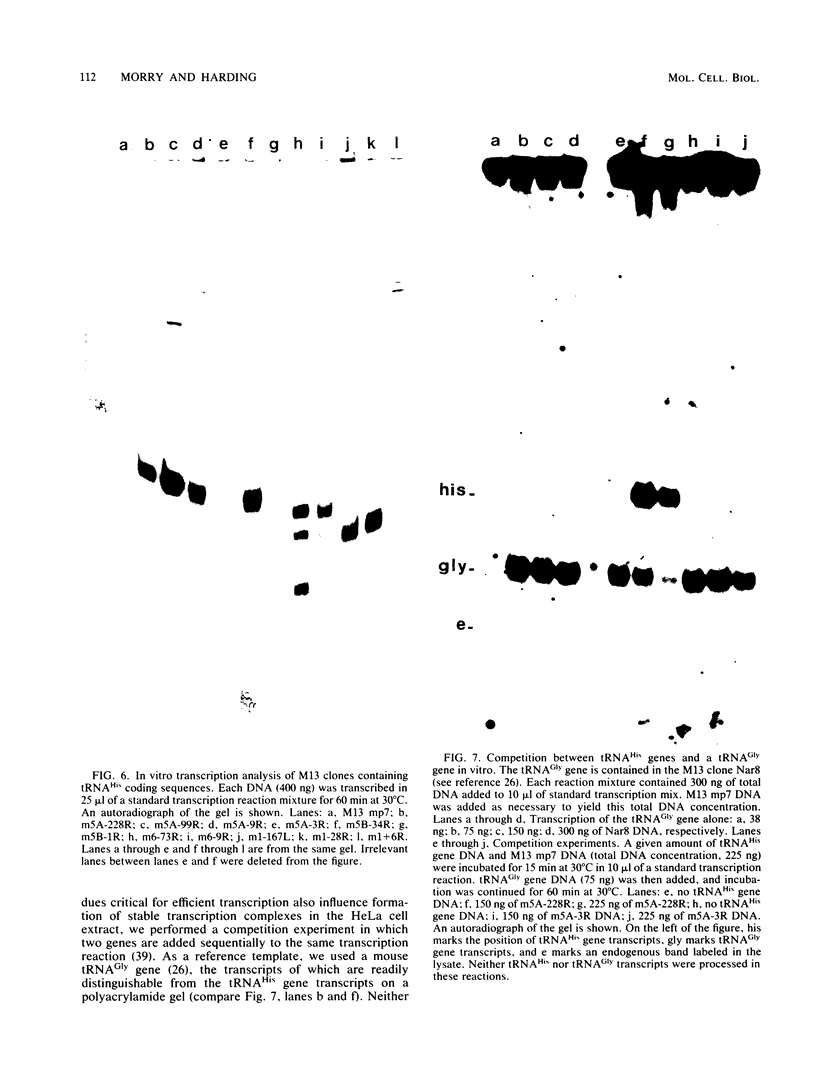
We determined the nucleotide sequences of three mouse tRNAHis genes and a tRNAGly gene present in two different lambda clones. One lambda clone contained two tRNAHis genes 600 base pairs (bp) apart in opposite orientations. The other clone contained a tRNAHis and a tRNAGly gene 569 bp apart in the same orientation. The coding regions of the three tRNAHis genes were identical to sequenced mammalian tRNAHis if posttranscriptional modifications are not considered. Notably, the three tRNAHis genes and a fourth gene previously sequenced by us contained within the flanking regions, various amounts of short, conserved 5' leader sequences and 3' trailer sequences directly abutting the coding regions. Otherwise the flanking regions were not homologous. Deletion mutants of one of the tRNAHis genes were constructed which contained 228, 99, 9, and 3 bp of the wild-type 5'-flanking region, respectively. Deletion of 5'-flanking sequences from positions -9 to -4 reduced transcriptional activity substantially (ca. fivefold) in a HeLa cell S-100 lysate. This effect was independent of the vector sequences in the deletion clone, implying that the region from -4 to -9 of the intact gene contains a positive modulatory element for transcription in vitro. The deletion mutant containing 3 bp of wild-type 5'-flanking sequence also had a greatly reduced ability to inhibit the transcription of a second tRNA gene in a competition assay. Thus, the normal 5'-flanking region influences the ability of the gene to form stable complexes with transcription factors. These data further indicate that a mammalian transcription extract is sensitive to 5'-flanking-region effects if a suitable tRNA gene is assayed.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Addison W. R., Astell C. R., Delaney A. D., Gillam I. C., Hayashi S., Miller R. C., Rajput B., Smith M., Taylor D. M., Tener G. M. The structures of genes hybridizing with tRNA4Val from Drosophila melanogaster. J Biol Chem. 1982 Jan 25;257(2):670–673. [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Burke D. J., Söll D. Functional analysis of fractionated Drosophila Kc cell tRNA gene transcription components. J Biol Chem. 1985 Jan 25;260(2):816–823. [PubMed] [Google Scholar]
- Ciliberto G., Castagnoli L., Melton D. A., Cortese R. Promoter of a eukaryotic tRNAPro gene is composed of three noncontiguous regions. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1195–1199. doi: 10.1073/pnas.79.4.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L., Carbon J. A colony bank containing synthetic Col El hybrid plasmids representative of the entire E. coli genome. Cell. 1976 Sep;9(1):91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- Cooley L., Appel B., Söll D. Post-transcriptional nucleotide addition is responsible for the formation of the 5' terminus of histidine tRNA. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6475–6479. doi: 10.1073/pnas.79.21.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley L., Schaack J., Burke D. J., Thomas B., Söll D. Transcription factor binding is limited by the 5'-flanking regions of a Drosophila tRNAHis gene and a tRNAHis pseudogene. Mol Cell Biol. 1984 Dec;4(12):2714–2722. doi: 10.1128/mcb.4.12.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco D., Schmidt O., Söll D. Two control regions for eukaryotic tRNA gene transcription. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3365–3368. doi: 10.1073/pnas.77.6.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco D., Sharp S., Söll D. Identification of regulatory sequences contained in the 5'-flanking region of Drosophila lysine tRNA2 genes. J Biol Chem. 1981 Dec 10;256(23):12424–12429. [PubMed] [Google Scholar]
- Dingermann T., Burke D. J., Sharp S., Schaack J., Söll D. The 5- flanking sequences of Drosophila tRNAArg genes control their in vitro transcription in a Drosophila cell extract. J Biol Chem. 1982 Dec 25;257(24):14738–14744. [PubMed] [Google Scholar]
- Dingermann T., Sharp S., Schaack J., Söll D. Stable transcription complex formation of eukaryotic tRNA genes is dependent on a limited separation of the two intragenic control regions. J Biol Chem. 1983 Sep 10;258(17):10395–10402. [PubMed] [Google Scholar]
- Fuhrman S. A., Engelke D. R., Geiduschek E. P. HeLa cell RNA polymerase III transcription factors. Functional characterization of a fraction identified by its activity in a second template rescue assay. J Biol Chem. 1984 Feb 10;259(3):1934–1943. [PubMed] [Google Scholar]
- Galli G., Hofstetter H., Birnstiel M. L. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature. 1981 Dec 17;294(5842):626–631. doi: 10.1038/294626a0. [DOI] [PubMed] [Google Scholar]
- Han J. H., Harding J. D. Isolation and nucleotide sequence of a mouse histidine tRNA gene. Nucleic Acids Res. 1982 Aug 25;10(16):4891–4900. doi: 10.1093/nar/10.16.4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipskind R. A., Clarkson S. G. 5'-flanking sequences that inhibit in vitro transcription of a xenopus laevis tRNA gene. Cell. 1983 Oct;34(3):881–890. doi: 10.1016/0092-8674(83)90545-7. [DOI] [PubMed] [Google Scholar]
- Hofstetter H., Kressman A., Birnstiel M. L. A split promoter for a eucaryotic tRNA gene. Cell. 1981 May;24(2):573–585. doi: 10.1016/0092-8674(81)90348-2. [DOI] [PubMed] [Google Scholar]
- Hosbach H. A., Silberklang M., McCarthy B. J. Evolution of a D. melanogaster glutamate tRNA gene cluster. Cell. 1980 Aug;21(1):169–178. doi: 10.1016/0092-8674(80)90124-5. [DOI] [PubMed] [Google Scholar]
- Hu J. C., Cote B. D., Lund E., Dahlberg J. E. Isolation and characterization of genomic mouse DNA clones containing sequences homologous to tRNAs and 5S rRNA. Nucleic Acids Res. 1983 Jul 25;11(14):4809–4821. doi: 10.1093/nar/11.14.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. D., Raymond G. J. Three regions of a yeast tRNALeu3 gene promote RNA polymerase III transcription. J Biol Chem. 1984 May 10;259(9):5990–5994. [PubMed] [Google Scholar]
- Larson D., Bradford-Wilcox J., Young L. S., Sprague K. U. A short 5' flanking region containing conserved sequences is required for silkworm alanine tRNA gene activity. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3416–3420. doi: 10.1073/pnas.80.11.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassar A. B., Martin P. L., Roeder R. G. Transcription of class III genes: formation of preinitiation complexes. Science. 1983 Nov 18;222(4625):740–748. doi: 10.1126/science.6356356. [DOI] [PubMed] [Google Scholar]
- Lin V. K., Farkas W. R., Agris P. F. Specific changes in Q-ribonucleoside containing transfer RNA species during Friend leukemia cell erythroid differentiation. Nucleic Acids Res. 1980 Aug 11;8(15):3481–3489. doi: 10.1093/nar/8.15.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looney J. E., Harding J. D. Structure and evolution of a mouse tRNA gene cluster encoding tRNAAsp, tRNAGly and tRNAGlu and an unlinked, solitary gene encoding tRNAAsp. Nucleic Acids Res. 1983 Dec 20;11(24):8761–8775. doi: 10.1093/nar/11.24.8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews M. B., Bernstein R. M. Myositis autoantibody inhibits histidyl-tRNA synthetase: a model for autoimmunity. Nature. 1983 Jul 14;304(5922):177–179. doi: 10.1038/304177a0. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munz P., Amstutz H., Kohli J., Leupold U. Recombination between dispersed serine tRNA genes in Schizosaccharomyces pombe. Nature. 1982 Nov 18;300(5889):225–231. doi: 10.1038/300225a0. [DOI] [PubMed] [Google Scholar]
- Page G. S., Hall B. D. Characterization of the yeast tRNA Ser genomic organization and DNA sequence. Nucleic Acids Res. 1981 Feb 25;9(4):921–934. doi: 10.1093/nar/9.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond G. J., Johnson J. D. The role of non-coding DNA sequences in transcription and processing of a yeast tRNA. Nucleic Acids Res. 1983 Sep 10;11(17):5969–5988. doi: 10.1093/nar/11.17.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson R. R., Davidson N. Analysis of a drosophila tRNA gene cluster: two tRNALeu genes contain intervening sequences. Cell. 1981 Jan;23(1):251–259. doi: 10.1016/0092-8674(81)90289-0. [DOI] [PubMed] [Google Scholar]
- Rosa M. D., Hendrick J. P., Jr, Lerner M. R., Steitz J. A., Reichlin M. A mammalian tRNAHis-containing antigen is recognized by the polymyositis-specific antibody anti-Jo-1. Nucleic Acids Res. 1983 Feb 11;11(3):853–870. doi: 10.1093/nar/11.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Santos T., Zasloff M. Comparative analysis of human chromosomal segments bearing nonallelic dispersed tRNAimet genes. Cell. 1981 Mar;23(3):699–709. doi: 10.1016/0092-8674(81)90433-5. [DOI] [PubMed] [Google Scholar]
- Schaack J., Sharp S., Dingermann T., Burke D. J., Cooley L., Söll D. The extent of a eukaryotic tRNA gene. 5'- and 3'-flanking sequence dependence for transcription and stable complex formation. J Biol Chem. 1984 Feb 10;259(3):1461–1467. [PubMed] [Google Scholar]
- Schaack J., Sharp S., Dingermann T., Söll D. Transcription of eukaryotic tRNA genes in vitro. II. Formation of stable complexes. J Biol Chem. 1983 Feb 25;258(4):2447–2453. [PubMed] [Google Scholar]
- Schmidt O., Mao J., Ogden R., Beckmann J., Sakano H., Abelson J., Söll D. Dimeric tRNA precursors in yeast. Nature. 1980 Oct 23;287(5784):750–752. doi: 10.1038/287750a0. [DOI] [PubMed] [Google Scholar]
- Sekiya T., Nishizawa R., Matsuda K., Taya Y., Nishimura S. A rat tRNA gene cluster containing the genes for tRNAPro and tRNALys. Analysis of nucleotide sequences of the genes and the surrounding regions. Nucleic Acids Res. 1982 Oct 25;10(20):6411–6419. doi: 10.1093/nar/10.20.6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp S., DeFranco D., Dingermann T., Farrell P., Söll D. Internal control regions for transcription of eukaryotic tRNA genes. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6657–6661. doi: 10.1073/pnas.78.11.6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp S., Dingermann T., Schaack J., DeFranco D., Söll D. Transcription of eukaryotic tRNA genes in vitro. I. Analysis of control regions using a competition assay. J Biol Chem. 1983 Feb 25;258(4):2440–2446. [PubMed] [Google Scholar]
- Shaw K. J., Olson M. V. Effects of altered 5'-flanking sequences on the in vivo expression of a Saccharomyces cerevisiae tRNATyr gene. Mol Cell Biol. 1984 Apr;4(4):657–665. doi: 10.1128/mcb.4.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya K., Noguchi S., Nishimura S., Sekiya T. Characterization of a rat tRNA gene cluster containing the genes for tRNAAsp, tRNAGly and tRNAGlu, and pseudogenes. Nucleic Acids Res. 1982 Jul 24;10(14):4441–4448. doi: 10.1093/nar/10.14.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Gauss D. H. Compilation of tRNA sequences. Nucleic Acids Res. 1984;12 (Suppl):r1–57. [PMC free article] [PubMed] [Google Scholar]
- Stillman D. J., Geiduschek E. P. Differential binding of a S. cerevisiae RNA polymerase III transcription factor to two promoter segments of a tRNA gene. EMBO J. 1984 Apr;3(4):847–853. doi: 10.1002/j.1460-2075.1984.tb01895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela P., Venegas A., Weinberg F., Bishop R., Rutter W. J. Structure of yeast phenylalanine-tRNA genes: an intervening DNA segment within the region coding for the tRNA. Proc Natl Acad Sci U S A. 1978 Jan;75(1):190–194. doi: 10.1073/pnas.75.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil P. A., Segall J., Harris B., Ng S. Y., Roeder R. G. Faithful transcription of eukaryotic genes by RNA polymerase III in systems reconstituted with purified DNA templates. J Biol Chem. 1979 Jul 10;254(13):6163–6173. [PubMed] [Google Scholar]
- Yen P. H., Davidson N. The gross anatomy of a tRNA gene cluster at region 42A of the D. melanogaster chromosome. Cell. 1980 Nov;22(1 Pt 1):137–148. doi: 10.1016/0092-8674(80)90162-2. [DOI] [PubMed] [Google Scholar]