Abstract
Replicative DNA polymerases require an RNA primer for leading and lagging strand DNA synthesis, and primase is responsible for the de novo synthesis of this RNA primer. However, the archaeal primase from Pyrococcus furiosus (Pfu) frequently incorporates mismatched nucleoside monophosphate, which stops RNA synthesis. Pfu DNA polymerase (PolB) cannot elongate the resulting 3′-mismatched RNA primer because it cannot remove the 3′-mismatched ribonucleotide. This study demonstrates the potential role of a RecJ-like protein from P. furiosus (PfRecJ) in proofreading 3′-mismatched ribonucleotides. PfRecJ hydrolyzes single-stranded RNA and the RNA strand of RNA/DNA hybrids in the 3′–5′ direction, and the kinetic parameters (Km and Kcat) of PfRecJ during RNA strand digestion are consistent with a role in proofreading 3′-mismatched RNA primers. Replication protein A, the single-stranded DNA–binding protein, stimulates the removal of 3′-mismatched ribonucleotides of the RNA strand in RNA/DNA hybrids, and Pfu DNA polymerase can extend the 3′-mismatched RNA primer after the 3′-mismatched ribonucleotide is removed by PfRecJ. Finally, we reconstituted the primer-proofreading reaction of a 3′-mismatched ribonucleotide RNA/DNA hybrid using PfRecJ, replication protein A, Proliferating cell nuclear antigen (PCNA) and PolB. Given that PfRecJ is associated with the GINS complex, a central nexus in archaeal DNA replication fork, we speculate that PfRecJ proofreads the RNA primer in vivo.
INTRODUCTION
DNA replication is a complex biochemical process that is catalyzed by numerous proteins; it is characterized by three stages: initiation, elongation and termination (1–5). After the replicative helicase melts the DNA duplex, the single-stranded (ss) DNA-binding protein, SSB or replication protein A (RPA), binds the ssDNA to prevent reannealing of the complementary strands (6–8). DNA primase can synthesize short oligoribonucleotides de novo using ssDNA as a template (9–11). The replicative DNA polymerase and other replisome subunits are recruited to these short RNA primers and start a highly processive DNA polymerization reaction (1,3–5). On the lagging strand of DNA synthesis, the DNA polymerase core complex disassociates from the replisome after Okazaki fragment synthesis. A new replisome is assembled for the next Okazaki fragment synthesis, which uses a new RNA primer (1–5).
DNA primase is a multifunctional enzyme that can polymerize diribonucleotides or di(deoxy)ribonucleotides on ssDNA, and in vitro, can elongate these di(deoxy)ribonucleotides into a long RNA or DNA primer (9–11,12). Aside from RNA and DNA polymerase activities, primase also has 3′-terminal nucleotidyl transferase activity and can add nucleotides to the 3′ terminus of a primer, independent of the template (12,13). The function of primase is highly conserved in three domains of life, but its subunit component differs in bacteria, archaea and eukaryotes. Bacterial primase is a single-subunit enzyme encoded by the dnaG gene (10). Eukaryotic primase consists of four subunits: a catalytic subunit, a regulative subunit, a B subunit and a polymerase α subunit (11). Archaeal primase is a two-subunit complex, consisting of homologs of the eukaryotic catalytic and regulative subunits (9,12,13). Several archaeal primases have been identified and biochemically characterized (9,12–15); the primase from Thermococcus kodakaraensis can form phosphodiester bonds between deoxyribonucleotide monophosphates and various hydroxyl acceptors (16).
The bacterial nuclease RecJ shows 5′–3′ exonuclease activity on ssDNA (17) and deoxyribose phosphatase activity (18). In bacteria, RecJ mainly participates in three DNA repair pathways: homologous recombination, mismatch repair and base excision repair (19–21). RecJ functions as a 5′–3′ ssDNA-specific exonuclease to generate a long 3′ ssDNA for strand exchange with homologous double-stranded (ds) DNA in recombination, or to generate a long ssDNA gap for DNA resynthesis by DNA polymerase in mismatch repair (19,20). In base excision repair, the 5′-deoxyribose phosphatase activity of RecJ removes deoxyribose phosphate from an abasic site, after cleavage of the DNA backbone by a class II apurinic/apyrimidinic (AP) endonuclease (18,21). RecJ belongs to an exonuclease superfamily with the conserved diagnostic motif of DHH (22,23); this family also includes RecJ-like protein and other nucleases (22–24,25). No RecJ homolog exists in eukaryotes, but the eukaryotic Cdc45 protein belongs to the DHH superfamily (25,26). Structurally, bacterial RecJ features an N-terminal catalytic core consisting of two domains and an oligonucleotide/oligosaccharide-binding (OB) domain located at the C terminus; the latter is conserved in numerous proteins with ssDNA/RNA-binding activity (27). Archaeal RecJ-like or its DHH superfamily member, eukaryotic Cdc45, interacts with several DNA replication proteins, including the GINS complex (a central nexus in the archaeal DNA replication fork) and the replicative minichromosome maintenance (MCM) helicase (23,28–31).
Although several DNA replication proteins have been identified and characterized in archaea, numerous aspects of DNA replication remain unclear. We have characterized the synthesis fidelity of RNA primers by Pyrococcus furiosus (Pfu) primase, as well as the effect of 3′-mismatched ribonucleotides on DNA elongation by DNA polymerase. P. furiosus primase was found to incorporate 3′-mismatched ribonucleotides during the extension of a short RNA primer. This 3′-mismatched RNA primer cannot be efficiently extended by Pfu DNA polymerase, a family B DNA polymerase (PolB). We have found that P. furiosus RecJ-like protein (PfRecJ) exhibits intrinsic 3′–5′ exonuclease activity on ssRNA and on the mismatched ribonucleotide of an RNA/DNA hybrid; the latter activity is stimulated by RPA. After this proofreading of the 3′-mismatched ribonucleotide by PfRecJ, Pfu DNA polymerase can efficiently extend the RNA primer to a full-length RNA–DNA chimeric strand. This study is the first to report proofreading of a 3′-mismatched RNA primer during DNA replication.
MATERIALS AND METHODS
Materials
The expression vector pDEST17 and expression bacterial host BL21 (DE3) pLysS and Rosetta were used throughout this study. Genomic DNA of P. furiosus was purchased from the American Type Culture Collection. KOD-plus DNA polymerase was purchased from Toyobo (Shanghai, China). Nickel–nitrilotriacetic acid resin was purchased from Bio-Rad. Oligodeoxyribonucleotides and oligoribonucleotides were synthesized by Invitrogen (Shanghai, China) and Takara (Dalian, China), respectively. All other chemicals and reagents were of analytical grade.
Preparation of recombinant P. furiosus proteins
Genes encoding the RecJ-like protein (PF2055), primase (PF0110 and PF0111), GINS (PF0483 and PF0982), Proliferating cell nuclear antigen (PCNA, PF0983), RPA (PF2018-2020) and PolB (PF0212) were amplified from P. furiosus genomic DNA by PCR using their respective primers (Supplementary Table S1) and then inserted into pDEST17 as described previously (32). Amino acid substitutions were introduced into RecJ and PolB with a QuikChange® Site-Directed Mutagenesis Kit using KOD-plus DNA polymerase and the appropriate primers (Supplementary Table S1). The nucleotide sequences were confirmed by DNA sequencing.
Recombinant plasmids were introduced into pLysS or Rosetta strains of Escherichia coli to express recombinant proteins. Isopropylthio-β-galactoside (0.5 mM final concentration) was added to a bacterial culture of OD600 = 0.5–0.6 to express the recombinant proteins. Recombinant proteins were purified via immobilized Ni2+ affinity chromatography as follows: the bacterial pellet was suspended in lysis buffer (20 mM Tris–HCl, pH 8.0, 0.3 M NaCl, 5 mM mercaptoethanol, 5 mM imidazole, 1 mM phenylmethylsulfonyl fluoride and 10% glycerol) and then disrupted by sonication. After incubation for 20 min at 75°C, the cell extract was clarified by centrifugation at 10 000 rpm for 30 min. After loading the supernatant onto a column pre-equilibrated with lysis buffer, the resin was washed with >25 column volumes of lysis buffer containing 20 mM imidazole. Finally, the bound protein was eluted from the column using elution buffer (20 mM Tris–HCl, pH 8.0, 0.3 M NaCl, 5 mM mercaptoethanol, 200 mM imidazole and 10% glycerol). After verifying the purity of the eluate using 15% sodium dodecyl sulfate–polyacrylamide gel electrophoresis, the preparations were dialyzed against a storage buffer (20 mM Tris–HCl, pH 8.0, 0.3 M NaCl and 50% glycerol) and then stored in small aliquots at −20°C.
Characterization of P. furiosus enzymes
P. furiosus primase was characterized in 40 mM HEPES (pH 6.4), 30 mM NaCl and 10 mM MnCl2. Pfu DNA polymerase (PolB) was characterized in 20 mM Tris–HCl (pH 8.8), 10 mM (NH4)2SO4, 10 mM KCl, 2 mM MgSO4, 0.1% Triton X-100 and 100 ng/μl bovine serum albumin (BSA) or the same buffer as primase. RecJ-like protein PF2055 was characterized in 20 mM Tris–HCl (pH 7.5), 30 mM NaCl, 10 mM KCl, 5 mM dithiothreitol (DTT), 0.25 mM MnCl2 and 100 ng/μl BSA. Nucleic acid binding experiments of RecJ were performed using the same buffer as its enzyme activity assay, but Mn2+ was omitted. The kinetic parameters (Km and Kcat) of P. furiosus RecJ, primase and PolB were calculated using double-reciprocal plotting. The oligoribonucleotides and oligodeoxyribonucleotides used in the activity assays of primase, PolB and RecJ are listed in Table 1. After incubation for a specified time at 50°C (Tm of the RNA/DNA hybrid is 53°C), an equal volume of a stopping buffer (90% formamide, 100 mM EDTA and 0.2% sodium dodecyl sulfate) was added to the reaction. Subsequently, the reactions were subjected to 15% 8 M urea–denatured polyacrylamide gel electrophoresis. After electrophoresis, images of the gels were quantitated using an FL5000 fluorescent scanner (FUJIFILM).
Table 1.
Oligonucleotides used in activity assays
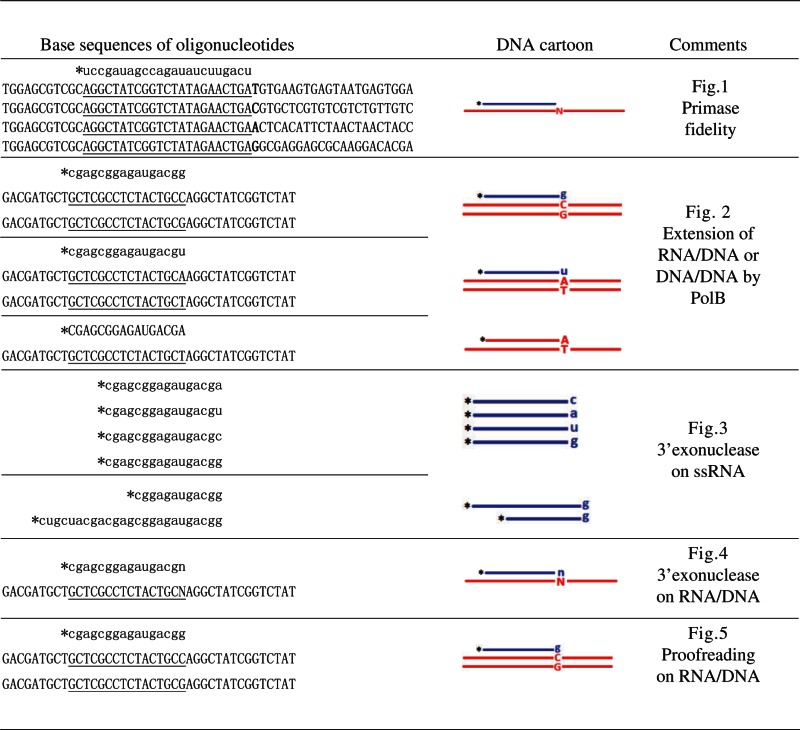 |
Asterisks denote the fluorescein (6-FAM) moiety at the 5′ end. The fluorescein-labeled strand and the complementary strand are shown in the 5′–3′ and 3′–5′ directions, respectively. Blue lines and lowercase letters represent RNA. Red lines and uppercase letters represent DNA. Letters n or N represents any of the four bases. The DNA bases complementary to labeled RNA are underlined. The complementary DNA strands used to prepare the RNA/DNA hybrids have four successive phosphorothioate modifications at the 5′ terminal to protect the 5′ digestion of ssDNA by PfRecJ.
Reconstitution of RNA primer–proofreading reaction
Proofreading of a 3′-mismatched ribonucleotide during DNA extension by PolB was reconstituted in the presence of PolB, RecJ, PCNA and RPA. A 3′-recessed RNA/DNA hybrid carrying a 3′-mismatched ribonucleotide was used as substrate in the proofreading reaction. Different enzyme combinations were added into the proofreading reaction to determine the function of each protein. The reactions were stopped and analyzed as described above.
Results
Primase incorporates mismatched nucleoside monophosphates
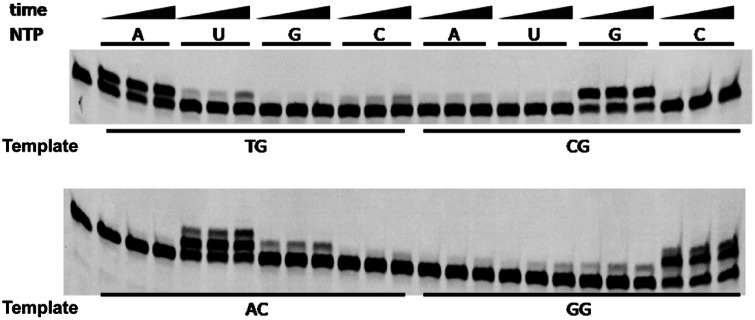
Previous studies have shown that primase can incorporate several mismatched nucleoside monophosphates (NMPs) in vitro (33,34). In the present study, we confirmed the identity of each misincorporated NMP using an artificially synthesized oligoribonucleotide as an RNA primer. During the extension of an RNA primer in the presence of a single NTP, several extended mismatches were generated (Figure 1). The mismatches detected were mainly u/T, c/T, a/C, u/C, g/A, u/G and g/G; their frequencies are listed in Table 2. Several mismatches (c/T, g/A, g/G and u/T) exceeded 10% of the total (perfectly matched) RNA/DNA hybrid. This result suggests that P. furiosus primase incorporates several mismatches, which will stop further extension of the mismatched RNA primer.
Figure 1.
Incorporation fidelity of P. furiosus primase. The extension fidelity of an RNA primer by P. furiosus primase was determined in a buffer consisting of 40 mM HEPES (pH 6.4), 30 mM NaCl, 10 mM MnCl2 and 4 U RNase inhibitor (Rnsin). P. furiosus primase (100 nM) was incubated with 100 nM recessed RNA primer–DNA template substrate at 50°C for 2, 4 and 8 min. Four types of NTP (50 μM) were added singly to each reaction to determine the fidelity of the first incorporated ribonucleotide. The corresponding template bases of each primer–template substrate are listed at the bottom of the image.
Table 2.
Frequency of mismatches incorporated by P. furiosus primase
| Template base | Incorporated NMP |
|||
|---|---|---|---|---|
| GMP | AMP | UMP | CMP | |
| T | 0.023 | – | 0.166 | 0.184 |
| C | – | 0.055 | 0.030 | 0.017 |
| G | 0.123 | 0.030 | 0.056 | – |
| A | 0.114 | 0.015 | – | 0.035 |
The frequency of mismatches incorporated by P. furiosus primase was calculated by dividing the mismatched product by the perfectly matched product.
PolB cannot extend a mismatched RNA primer
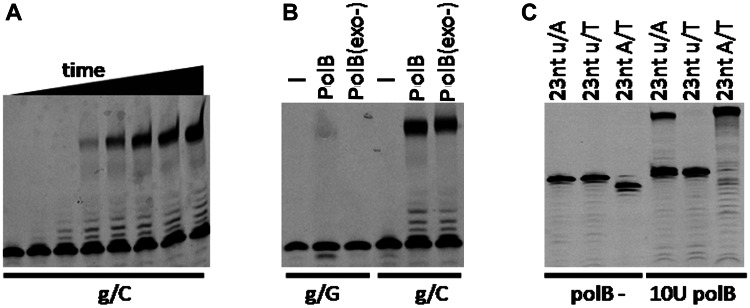
RNA oligoribonucleotides are used by various DNA polymerases as primers for extension (35–39). As a replicative DNA polymerase, PolB from P. furiosus efficiently elongates an RNA primer with a 3′-matched ribonucleotide in vitro into a full-length fragment (Figure 2A). The extension rate is relatively slow at the initial extension stage, as indicated by the accumulation of immediate bands elongated by 1–6 nt. When 3′-mismatched RNA was used as a substrate, 3′ exonuclease–deficient PolB did not generate any product (Figure 2B, lane 3), indicating that 3′-mismatched ribonucleotides completely block extension. However, wild-type (wt) PolB generated a small amount of extended product (Figure 2B, lane 2), indicating that the 3′ exonuclease activity of PolB on ssDNA can remove the 3′-mismatched RNA ribonucleotide but does so with low efficiency. To exclude the effect of sequence context, a different RNA primer was used as a substrate. The perfectly matched RNA primer was extended efficiently by wt PolB (Figure 2C, lane 4), whereas the 3′-mismatched primer was not (Figure 2C, lane 5). Moreover, a DNA primer was extended more efficiently than an RNA primer (Figure 2C, lanes 4 and 6).
Figure 2.
Extension of RNA primer by P. furiosus family B DNA polymerase. The extension of RNA primers annealed to complementary DNA templates was determined in a buffer consisting of 20 mM Tris–HCl, pH 8.8, 10 mM (NH4)2SO4, 10 mM KCl, 2 mM MgSO4, 0.1% Triton X-100, 100 ng/μl BSA, 100 μM dNTPs and 4 U Rnsin. Approximately 50 nM of RNA primer–DNA template substrates was incubated with 100 nM Pfu DNA polymerase at 50°C for 0, 1, 2, 4, 8, 15, 30 and 60 min (A) or 30 min (B and C). Different RNA primers and DNA templates were annealed to form the matched and mismatched RNA/DNA hybrids used in the extension reactions. Lowercase and uppercase denote RNA and DNA, respectively.
RecJ-like protein has intrinsic 3′ exonuclease activity on ssRNA
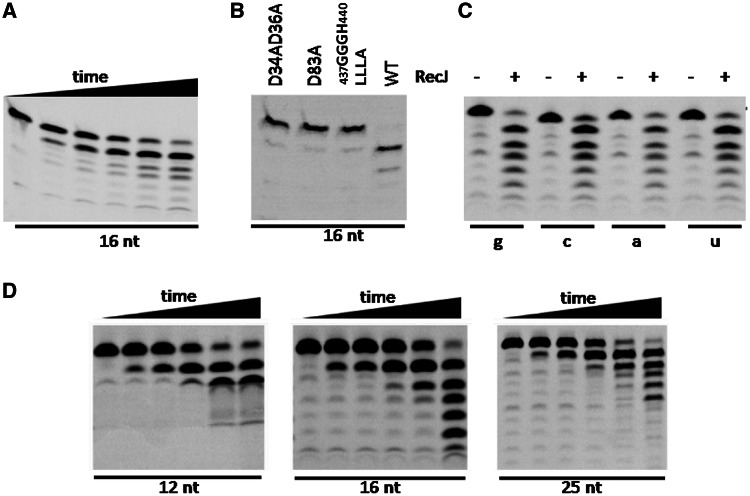
Primase is capable of synthesizing 3′-mismatched RNA primers (33,34). Thus, a proofreading activity should be present to remove 3′-mismatched ribonucleotides for efficient RNA primer extension by DNA polymerase. This proofreading protein might interact with a specific component of the replisome and should prefer ssRNA. Previous studies have shown that the archaeal RecJ-like protein specifically hydrolyzes ssDNA in the 5′–3′ direction and interacts with the GINS complex (22,23). Hence, we characterized the activity of PfRecJ on ssRNA and ssDNA. In contrast to its activity on ssDNA (Supplementary Figure S1A), PfRecJ hydrolyzed ssRNA in the 3′–5′ direction, leaving a hydroxyl group on the shortened 3′ end (Figure 3A). To investigate the possibility that this exonuclease activity was due to a contaminant protein that co-purifies with PfRecJ, we demonstrated that the 3′ exonuclease activity on ssRNA was intrinsic to PfRecJ. Several genes from P. furiosus (encoding PCNA, RPA, GINS and primase) were cloned in the pDEST17 expression vector, and recombinant proteins were induced and purified in an identical method to PfRecJ. None of these recombinant proteins, except RecJ, showed 3′ exonuclease activity on ssRNA (data not shown). RecJ protein has several conserved motifs, some of which are also conserved in the DHH superfamily (22,25,27). We mutated three conserved motifs to verify their effect on 3′ and 5′ exonuclease activities on ssRNA and ssDNA, respectively. Changing D34, D36 and D83 to alanine and replacing the 437GGGH440 motif with 437LLLA440 resulted in loss of both 3′ and 5′ exonuclease activities (Figure 3B and Supplementary Figure S1B), indicating that the three motifs are necessary for a potential proofreading function.
Figure 3.
Biochemical characterization of P. furiosus RecJ on RNA substrates. The 3′–5′ exonuclease activity of PfRecJ on ssRNA was determined in a buffer consisting of 20 mM Tris–HCl (pH 7.5), 30 mM NaCl, 10 mM KCl, 5 mM DTT, 0.25 mM MnCl2, 100 ng/μl BSA and 4 U Rnsin. Substrates (50 nM) were incubated with 50 nM PfRecJ at 50°C for 0, 2, 5, 10, 20 and 30 min (A and D) or 30 min (B and C). Lowercase and uppercase denote RNA and DNA, respectively. (A) Time course of 16-nt ssRNA digestion by wt PfRecJ. (B) Identification of key residues. Three mutant PfRecJs were used to confirm the conserved amino acid residues essential for exonuclease activity. (C) Effect of 3′ ribonucleotide on ssRNA digestion by PfRecJ. A 16-nt ssRNA with four 3′ ribonucleotides (a, u, c and g) was digested by PfRecJ. (D) Effect of ssRNA length on hydrolysis efficiency. ssRNAs with different lengths (12, 16 and 25 nt) were digested by PfRecJ.
The 3′ exonuclease activity of PfRecJ was found to depend on the divalent metal ion Mn2+, with an optimal concentration range of 0.1–0.5 mM (Supplementary Figure S2A and B). Other divalent metal ions (Ca2+, Cu2+, Zn2+, Ni2+ and Co2+) inhibited the 3′ exonuclease activity even in the presence of Mn2+ (Supplementary Figure S2C). The biochemical properties of PfRecJ were then characterized using a 12-nt ssRNA (Supplementary Figure S3). PfRecJ exhibited highest activity in the following conditions: pH 7.0–7.5 (Supplementary Figure S3A), low ion strength (Supplementary Figure S3B) and 70°C reaction temperature (Supplementary Figure S3C). However, the reaction temperature used subsequently was 50°C because of the thermal instability of ssRNA at high temperatures (data not shown). The relative activities on ssDNA and ssRNA were comparable (Supplementary Figures S1A and S3D).
If PfRecJ has a proofreading function, then the four types of ribonucleotides should be removed with a comparable efficiency. Hence, four ssRNAs with various 3′ ribonucleotides were used to verify the selectivity of the 3′ exonuclease activity. Our result showed that the four ssRNAs were digested by PfRecJ in almost equal efficiency (Figure 3C), indicating that PfRecJ has no clear selectivity on the 3′ ribonucleotide. The RNA primer synthesized by primase is ∼12–20 nt long (11,40). Thus, we investigated the cleavage efficiency of PfRecJ on ssRNAs with different lengths. The results showed that the cleavage efficiencies on ssRNAs with different lengths were comparable, with some preference for 16-nt ssRNA (Figure 3D). The cleavage of the 12-nt ssRNA stopped at 10 nt, indicating that binding and cleavage of ssRNA by PfRecJ require a length of at least 10 nt.
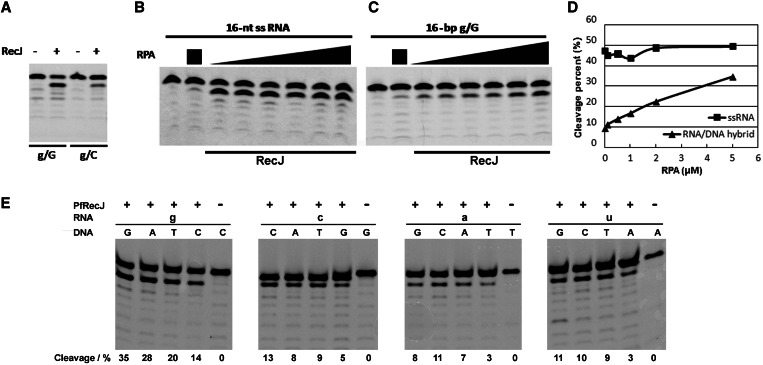
RecJ-like protein has preference for 3′-mismatched RNA/DNA hybrids
During DNA replication, an RNA primer forms a ds RNA/DNA hybrid with the DNA template (9–11). Therefore, we characterized the 3′ exonuclease activity of PfRecJ on a ds RNA/DNA hybrid with 3′-end recessed RNA. Results show that PfRecJ preferentially acts on the 3′-mismatched ribonucleotide in the ds RNA/DNA hybrid (Figure 4A); the removal of a g/G mismatch was approximately four times more efficient than that of a g/C match. This preference for the 3′ mismatch suggests that PfRecJ has a potential proofreading activity for mismatched RNA primers. Previous reports have shown that GINS, a core subcomplex in archaeal replisome, stimulates 5′ exonuclease activity and physically interacts with archaeal RecJ-like protein (23,31). However, GINS of P. furiosus did not stimulate the 3′ exonuclease activity of PfRecJ on ssRNAs of various lengths (Supplementary Figure S4A), and on the RNA/DNA hybrid (Supplementary Figure S4B); however, we did confirm the stimulation by GINS of 5′ exonuclease activity on ssDNA (Supplementary Figure S4C). Given that the ssDNA template is bound by RPA during DNA replication, the effects of RPA on the activity of PfRecJ were characterized. RPA exhibited differential effects on the 3′ exonuclease of RecJ on the ssRNA and the RNA/DNA hybrid. The activity on the ssRNA was not affected by RPA (Figure 4B), whereas the activity on the RNA/DNA hybrid was markedly stimulated by RPA (Figure 4C), which elevated cleavage of the RNA strand of the RNA/DNA hybrid by ∼3.5 times (Figure 4D). We then characterized 16 RNA/DNA hybrids with all possible base matches/mismatches in the presence of 1 μM RPA. All 3′-mismatched RNA/DNA hybrids were hydrolyzed with a higher efficiency than the 3′-matched hybrids (Figure 4E). These results indicate that PfRecJ can remove mismatched ribonucleotides incorporated by primase (Figure 1), and that RPA stimulates the ability of RecJ to proofread 3′-mismatched ribonucleotides by binding to the ssDNA template.
Figure 4.
Hydrolysis of the RNA strand of RNA/DNA hybrids by PfRecJ with preference for 3′-mismatches. The 3′–5′ exonuclease of PfRecJ on RNA/DNA hybrids (A, C–E) and ssRNA (B) was determined in a buffer consisting of 20 mM Tris–HCl (pH 7.5), 30 mM NaCl, 10 mM KCl, 5 mM DTT, 0.25 mM MnCl2, 100 ng/μl BSA and 4 U Rnsin. (A) Hydrolysis of the RNA/DNA hybrid. RNA/DNA hybrids (50 nM) with 3′-mismatched and 3′-matched ribonucleotides were incubated with 50 nM PfRecJ at 50°C for 30 min. The 3′–5′ exonuclease activity on ssRNA (B) and 3′-mismatched (g/G) RNA/DNA hybrid (C) was assayed in the presence of different RPA concentrations (0, 0.1, 0.5, 1, 2 and 5 μM). The substrates (50 nM) were incubated with 10 nM PfRecJ at 50°C for 30 min. Panel D is the quantitation of panels B and C. (E) Hydrolysis of the RNA/DNA hybrid with all possible base matches/mismatches. Sixteen RNA/DNA hybrids (50 nM) with all possible base matches/mismatches were incubated with 50 nM PfRecJ at 50°C for 10 min in the presence of 1 μM RPA. The cleavage percentages are listed at the bottom of panel E. Lowercase and uppercase denote RNA and DNA, respectively.
Kinetic parameters of PfRecJ support proofreading of 3′-mismatched RNA primers
Considering that PfRecJ hydrolyzed RNA at relative low rate, we confirmed its potential to proofread the 3′-mismatched RNA primer by comparing its kinetic parameters with those of primase and PolB. The kinetics of P. furiosus primase, PolB and RecJ were calculated by double-reciprocal plotting (Table 3). The Kcat/Km of the mismatched RNA/DNA hybrid (in the presence/absence of RPA) was ∼3-fold higher than that of the matched hybrid. However, the lower Kcat/Km of the matched RNA/DNA hybrid was comparable with that of primase and PolB. For primase and PolB, the Kcat/Km of the mismatched RNA/DNA hybrid cannot be determined accurately because of the very low reaction rate (Supplementary Figure S5). These results indicate that PfRecJ can efficiently remove the 3′-mismatched ribonucleotide, but would not block the incorporation of a matched NMP and dNMP by primase and PolB, respectively. Moreover, the presence of a 3′-mismatched ribonucleotide completely blocks the incorporation of the next nucleoside phosphate by PolB (Supplementary Figure S5), indicating that the removal of the 3′-mismatched ribonucleotide is essential for efficient DNA synthesis. Our findings that Thermus thermophilus RecJ preferably binds ssDNA (Supplementary Figure S6A), and that PfRecJ preferentially binds ssRNA compared with RNA/DNA hybrids (Supplementary Figure S6B and C), are consistent with the higher 3′ exonuclease activity of PfRecJ on ssRNA.
Table 3.
Comparison of Km and Kcat of P. furiosus RecJ, primase and PolB
| Enzymes | Substrates | Kinetic parameter |
||
|---|---|---|---|---|
| Km (μM) | Kcat (min−1) | Kcat/Km (min−1·μM−1) | ||
| Pol B | 3′-matched RNA/DNAa | 0.0080 | 0.00034 | 0.042 |
| 3′-mismatched RNA/DNA | NDb | ND | ND | |
| Primase | 3′-matched RNA/DNA | 0.14 | 0.017 | 0.12 |
| 3′-mismatched RNA/DNA | ND | ND | ND | |
| RecJc | ssRNA | 0.48 | 0.24 | 0.50 |
| 3′-mismatched RNA/DNA | 0.62 | 0.040 | 0.065 | |
| 3′-matched RNA/DNA | 0.70 | 0.015 | 0.022 | |
| RecJd | ssRNA | 0.73 | 0.34 | 0.47 |
| 3′-mismatched RNA/DNA | 0.29 | 0.12 | 0.42 | |
| 3′-matched RNA/DNA | 0.27 | 0.037 | 0.14 | |
Km and Kcat were calculated by double-reciprocal plotting using the initial reaction rates of ssRNA as well as matched (g/C) and mismatched (g/G) RNA/DNA hybrids at various substrate concentrations (0.1, 0.2, 0.5, 1, 2.5 and 10 μM).
aTo compare the catalytic efficiency of each enzyme for different substrates, various substrates (including ssRNA, 3′-matched RNA/DNA hybrids, or mismatched RNA/DNA hybrids) were used to calculate the kinetic parameter of each enzyme.
bThe kinetic parameters cannot be calculated accurately because the extension efficiency is too low to produce a clear product (Supplementary Figure S5).
cThe kinetic parameters were determined in the absence of RPA.
dThe kinetic parameters were determined in the presence of 1 μM RPA. ND, not determined.
RecJ proofreads 3′-mismatched ribonucleotides in the DNA elongation reaction of PolB
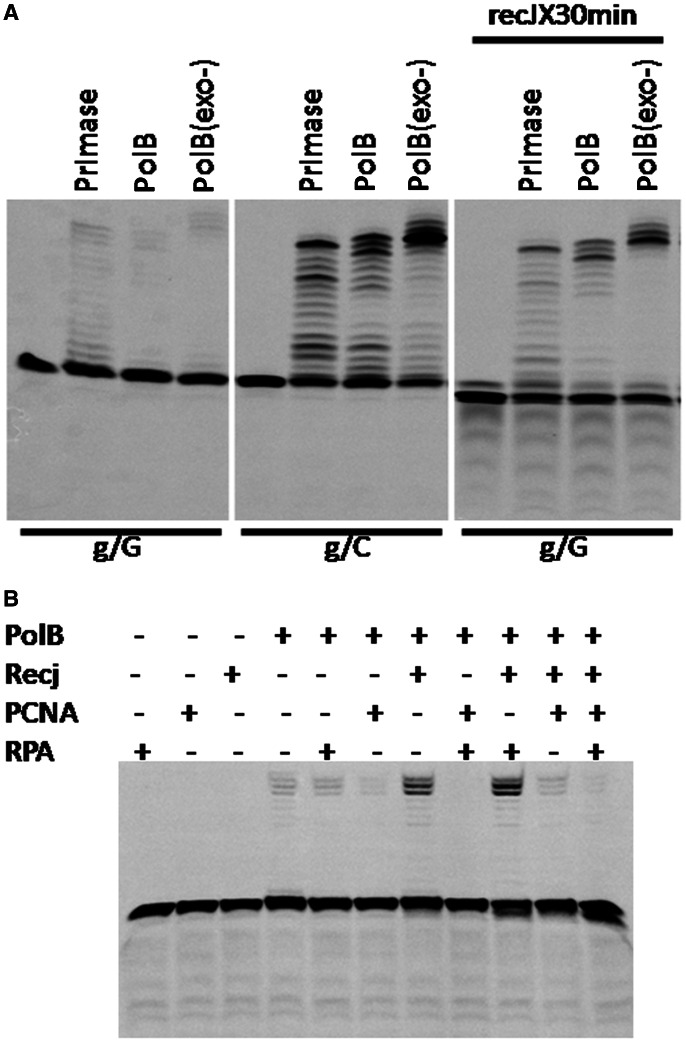
Neither wt nor 3′-exonuclease–deficient PolB can efficiently extend an RNA primer if the 3′ ribonucleotide is mismatched (Figure 5A, lanes 2 and 3). On incubation with PfRecJ, the two forms of PolB can extend the 3′-mismatched RNA primer into long fragments (Figure 5A, lanes 11 and 12). The wt PolB also has 3′–5′ exonuclease activity on ssRNA (<5% of the activity of RecJ, Supplementary Figure S7). However, it could not efficiently proofread the 3′-mismatched RNA primer (Figure 5A, lane 2). After the 3′-mismatched RNA/DNA hybrid was incubated with PfRecJ, the yield of fragments extended by primase also increased (Figure 5A, lanes 2 and 10). Incubation with RecJ decreased the extension yield of the 3′-mismatched RNA primer compared with the matched primer (Figure 5A, lanes 7–9 and 10–12). This result indicates that more than one ribonucleotide of 3′-mismatched RNA primer was removed, and that the shortened RNA primer possibly separated from the DNA template, and was therefore not extended by PolB and primase.
Figure 5.
Proofreading of PfRecJ on 3′-mismatched ribonucleotides during RNA primer extension by Pfu DNA polymerase. (A) Proofreading of 3′-mismatched ribonucleotides by PfRecJ during primer extension catalyzed by primase and Pfu DNA polymerase. A pair of recessed RNA/DNA hybrids (3′-matched/mismatched recess) was used to confirm the proofreading function of PfRecJ in a polymerization reaction catalyzed by P. furiosus PolB and primase. The substrates (50 nM) were incubated with 50 nM of PolB and primase in the presence/absence of PfRecJ (400 nM) at 50°C for 30 min in a buffer consisting of 20 mM HEPES (pH 7.5), 30 mM NaCl, 10 mM KCl, 5 mM MnCl2, 100 μM dNTPs, 4 U Rnsin and 100 ng/μl BSA. (B) Effect of RPA and PCNA on the proofreading of 3′-mismatched ribonucleotides by PfRecJ. Proofreading of the 3′-mismatched ribonucleotide during extension by PolB was performed in the presence of RPA and PCNA using a RNA/DNA hybrid carrying a 3′-mismatched ribonucleotide as a substrate. The RNA/DNA hybrid (50 nM) was incubated with PfRecJ (100 nM), PolB (50 nM), RPA (1 μM), PCNA (1 μM) or enzyme mixtures for 30 min at 50°C in a buffer consisting of 20 mM HEPES (pH 7.5), 30 mM NaCl, 10 mM KCl, 2 mM MnCl2, 100 ng/μl BSA, 4 U Rnsin and 100 μM dNTPs. Lowercase and uppercase denote RNA and DNA, respectively.
The primer extension reaction by the multiprotein system was also characterized using a 3′-mismatched RNA/DNA hybrid as a substrate. PolB alone extended the 3′-mismatched RNA primer by only a small degree (Figure 5B, lane 4), whereas the collaboration of RecJ and PolB increased the extension yield significantly (Figure 5B, lane 7). After the inclusion of RPA in the reaction, the yield of extended fragments increased further. This finding indicates that RPA promotes the removal of the 3′-mismatched ribonucleotide (Figure 4C). However, RPA did not promote the proofreading activity of PolB on the 3′-mismatched ribonucleotide (data not shown). PCNA strongly inhibited the extension of a 16-bp RNA/DNA hybrid by PolB (Figure 5B, lanes 6, 8, 10 and 11), and this inhibition was not relieved by the presence of RecJ and RPA (Figure 5B, lanes 8, 10 and 11). The RNA primer used may be too short to be bound by both PCNA and PolB, and this spatial hindrance might result in the inhibition by PCNA of RNA primer extension by PolB. Therefore, the effect of PCNA on the extension of a longer RNA primer by PolB was characterized. When a 25-nt RNA primer was used as substrate, the inhibition by PCNA disappeared (Supplementary Figure S8).
Discussion
As an essential enzyme for the initiation of DNA replication, primase has the capability to synthesize short RNA primers de novo. However, its fidelity is not 100%. When a mismatched NMP is incorporated into the RNA by primase, the 3′-mismatched ribonucleotide generally blocks further extension of RNA primer and fixes the mismatch on the 3′ terminus of the RNA primer (Figure 5A, lane 1). For efficient extension of the 3′-mismatched RNA primer by DNA polymerase PolB, the 3′-mismatched NMP must be removed by specific protein(s) with ribonuclease activity. Although archaeal replicative DNA polymerases have a strong 3′ exonuclease activity on 3′-mismatched deoxyribonucleotides (35–39), the 3′-mismatched ribonucleotide of RNA primer is not an efficient substrate for PolB (Supplementary Figure S7).
The PfRecJ protein has an intrinsic 3′–5′ exonuclease activity that is specific to ssRNA and 3′-mismatched RNA/DNA hybrids. We speculate that the RNA-specific 3′–5′ exonuclease of PfRecJ is responsible for removing the 3′-mismatched ribonucleotides from RNA primers in chromosomal DNA replication. Based on our results and a previous report on interactions between archaeal GINS and MCM helicase, primase and RecJ-like protein (23,29–31), we propose a model for the proofreading function of PfRecJ protein on 3′-mismatched RNA primers. RecJ, GINS and PolB function as a three-part complex in DNA replication. When PolB encounters a 3′-mismatched RNA primer, the complex arrests on the RNA primer. Then, the RecJ subunit removes the mismatched NMP and eliminates the hindrance to strand extension by PolB. Finally, PolB resumes DNA synthesis by extending the proofread RNA primer. Whether other DNA polymerases, such as DNA polymerase D (36–38), require archaeal RecJ-like protein to remove 3′-mismatched ribonucleotides needs to be experimentally verified. RNA primer proofreading is only necessary when the majority of RNA primers are mismatched. If both matched and mismatched RNA primers exist in a ssDNA template, the replicative DNA polymerases can use the matched primers to assemble the replisome. Moreover, considering that PfRecJ exhibits higher specific activity on ssRNA than RNA/DNA hybrids in vitro, this enzyme might be involved in the degradation of diverse ssRNAs (such as mRNA), as observed with other members of the DHH phosphoesterase superfamily (24).
The pfRecJ and its archaeal homologs are defined as RecJ-like proteins based on sequence similarity to bacterial RecJ (22,23). The T.thermophilus RecJ structure can be divided into four domains (27). Domains I (residues 47–291) and II (residues 323–425) are interconnected by a long helix (residues 292–322), forming an active center. Domain III comprises the N-terminal region (residues 1–46) and the internal region of ∼110 residues (residues 426–535). Domain IV comprises the C-terminal region of ∼120 residues (residues 536–658). The majority of bacterial RecJs, such as those of E.coli and Chlamydophila pneumoniae, only feature domains I, II and III (Supplementary Figure S9A). A sequence alignment (Supplementary Figure S9B) shows that archaeal RecJ-like proteins only have the two domains corresponding to the bacterial catalytic core domain, which includes residues 40–425 of the T.thermophilus RecJ (27,41). Hence, we propose that archaeal RecJ-like proteins, such as PfRecJ, TkoRecJ (23) and MjRecJ (22), should be classified into a domain-truncated RecJ subfamily (OB-fold domain-deleted RecJ subfamily). Moreover, archaeal RecJ-like proteins are longer by ∼100 amino acid residues than the bacterial RecJ core domain (owing to a longer domain I, Supplementary Figure S9C and D). The full-length protein and N-terminal catalytic core domain of T.thermophilus RecJ have 5′–3′ exonuclease activity on ssDNA, which is dependent on Mn2+ and Mg2+ (27). The Kcat value of the catalytic core domain is approximately the same as that of full-length RecJ, whereas the Km value of the catalytic core domain is ∼500 times higher than that of full-length RecJ (27). Hence, the OB-fold domain mainly functions in improving the ssDNA-binding capability of bacterial RecJ (Supplementary Figure S6A). Several members of the DHH phosphoesterase superfamily efficiently digest ssDNA and ssRNA shorter than 5 nt in the 5′–3′ direction (24). This activity of DHH phosphoesterase is proposed to play a role in RNA and DNA recycling. The differences in conserved residues and motifs between the DHH phosphoesterase superfamily and PfRecJ possibly result in the differing hydrolysis polarities on ssRNA.
The crystal structure of full-length T.thermophilus RecJ shows that the OB-fold domain functions mainly in binding to ssDNA (27,41). Archaeal RecJ cleaves both ssDNA and ssRNA, but their digestion polarities are different. The only difference between RNA and DNA is the 2′-OH group; therefore, it would appear that the 2′ OH directs binding and hydrolysis of the phosphodiester bond in the 3′–5′ direction. Given that PfRecJ preferentially hydrolyzes ssDNA/ssRNA hybrids, the DNA-binding region must be relatively narrow and select for ss nucleic acids over ds nucleic acids. The specific removal of 3′-mismatched ribonucleotides from RNA/DNA hybrids (Figure 4A and E) also supports the binding specificity of the enzyme to ssRNA (Supplementary Figure S6C). The RecJs from E.coli and T.thermophilus did not exhibit 3′–5′ exonuclease activity on ssRNA (data not shown). The sequence differences in the RecJ core domain (the longer domain I for archaeal RecJ-like protein) and the addition of OB-fold domain into bacterial RecJ could result in their different substrate specificities.
Despite the high sequence conservation of RecJ-like protein in bacteria and archaea, no homolog of RecJ exists in eukaryotes. Bioinformatic analysis has shown that Cdc45, an essential replication initiation protein, has significant sequence similarity to the N-terminal–conserved DHH domain of RecJ family proteins (25,26). In addition, biochemical results have shown that Cdc45 and RecJ specifically bind ssDNA and ssRNA, but the exonuclease activity of Cdc45 has not been confirmed (26). Cdc45 stably interacts with MCM2–7 and GINS to form a complex of Cdc45/Mcm2–7/GINS (CMG) that is believed to act as the DNA helicase at the replication fork (28,42,43). Aside from forming a complex with MCM2–7 and GINS, Cdc45 also interacts with other replication factors, including MCM10, RPA and DNA polymerases (44). Cdc45 lacks most of the conserved motifs that are essential for bacterial and archaeal enzyme activities (26), and is therefore unlikely to have a similar exonuclease activity. Perhaps the loss of exonuclease activity has allowed Cdc45 to evolve into a protein with specialized functions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–9.
FUNDING
The National Basic Research Program of China [2009CB118906]; the National Science Foundation of China [30700131, 31070090 and 21135004]; the National Natural Science Foundation of Shanghai City, China [12ZR1413700]. Funding for open access charge: the National Science Foundation of China [21135004].
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Hamdan SM, Richardson CC. Motors, switches, and contacts in the replisome. Annu. Rev. Biochem. 2009;78:205–243. doi: 10.1146/annurev.biochem.78.072407.103248. [DOI] [PubMed] [Google Scholar]
- 2.Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M. Eukaryotic chromosome DNA replication: where, when, and how? Annu. Rev. Biochem. 2010;79:89–130. doi: 10.1146/annurev.biochem.052308.103205. [DOI] [PubMed] [Google Scholar]
- 3.Grabowski B, Kelman Z. Archeal DNA replication: eukaryal proteins in a bacterial context. Annu. Rev. Microbiol. 2003;57:487–516. doi: 10.1146/annurev.micro.57.030502.090709. [DOI] [PubMed] [Google Scholar]
- 4.McHenry CS. DNA replicases from a bacterial perspective. Annu. Rev. Biochem. 2011;80:403–436. doi: 10.1146/annurev-biochem-061208-091655. [DOI] [PubMed] [Google Scholar]
- 5.Hubscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu. Rev. Biochem. 2002;71:133–163. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- 6.Costes A, Lecointe F, McGovern S, Quevillon-Cheruel S, Polard P. The C-terminal domain of the bacterial SSB protein acts as a DNA maintenance hub at active chromosome replication forks. PLoS Genet. 2010;6:e1001238. doi: 10.1371/journal.pgen.1001238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bochkarev A, Bochkareva E, Frappier L, Edwards AM. The crystal structure of the complex of replication protein A subunits RPA32 and RPA14 reveals a mechanism for single-stranded DNA binding. EMBO J. 1999;18:4498–4504. doi: 10.1093/emboj/18.16.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robbins JB, Murphy MC, White BA, Mackie RI, Ha T, Cann IK. Functional analysis of multiple single-stranded DNA-binding proteins from Methanosarcina acetivorans and their effects on DNA synthesis by DNA polymerase BI. J. Biol. Chem. 2004;279:6315–6326. doi: 10.1074/jbc.M304491200. [DOI] [PubMed] [Google Scholar]
- 9.Liu L, Komori K, Ishino S, Bocquier AA, Cann IK, Kohda D, Ishino Y. The archaeal DNA primase: biochemical characterization of the p41-p46 complex from Pyrococcus furiosus. J. Biol. Chem. 2001;276:45484–45490. doi: 10.1074/jbc.M106391200. [DOI] [PubMed] [Google Scholar]
- 10.Bailey S, Eliason WK, Steitz TA. Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science. 2007;318:459–463. doi: 10.1126/science.1147353. [DOI] [PubMed] [Google Scholar]
- 11.Núñez-Ramírez R, Klinge S, Sauguet L, Melero R, Recuero-Checa MA, Kilkenny M, Perera RL, García-Alvarez B, Hall RJ, Nogales E, et al. Flexible tethering of primase and DNA Pol α in the eukaryotic primosome. Nucleic Acids Res. 2011;39:8187–8199. doi: 10.1093/nar/gkr534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lao-Sirieix SH, Bell SD. The heterodimeric primase of the hyperthermophilic archaeon Sulfolobus solfataricus possesses DNA and RNA primase, polymerase and 3′-terminal nucleotidyl transferase activities. J. Mol. Biol. 2004;344:1251–1263. doi: 10.1016/j.jmb.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 13.De Falco M, Fusco A, De Felice M, Rossi M, Pisani FM. The DNA primase of Sulfolobus solfataricus is activated by substrates containing a thymine-rich bubble and has a 3′-terminal nucleotidyl-transferase activity. Nucleic Acids Res. 2004;32:5223–5230. doi: 10.1093/nar/gkh865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desogus G, Onesti S, Brick P, Rossi M, Pisani FM. Identification and characterization of a DNA primase from the hyperthermophilic archaeon Methanococcus jannaschii. Nucleic Acids Res. 1999;27:4444–4450. doi: 10.1093/nar/27.22.4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chemnitz GW, Pan M, Kelman Z, Hurwitz J. Characterization of DNA primase complex isolated from the archaeon, Thermococcus kodakaraensis. J. Biol. Chem. 2012;287:16209–16219. doi: 10.1074/jbc.M111.338145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chemnitz GW, Pan M, Giulian G, Yuan W, Li S, Edwards JL, Marino JP, Kelman Z, Hurwitz J. Formation of dAMP-glycerol and dAMP-Tris derivatives by Thermococcus kodakaraensis DNA primase. J. Biol. Chem. 2012;287:16220–16229. doi: 10.1074/jbc.M111.338160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han ES, Cooper DL, Persky NS, Sutera VA, Jr, Whitaker RD, Montello ML, Lovett ST. RecJ exonuclease: substrates, products and interaction with SSB. Nucleic Acids Res. 2006;34:1084–1091. doi: 10.1093/nar/gkj503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dianov G, Sedgwick B, Daly G, Olsson M, Lovett S, Lindahl T. Release of 5′-terminal deoxyribose-phosphate residues from incised abasic sites in DNA by the Escherichia coli RecJ protein. Nucleic Acids Res. 1994;22:993–998. doi: 10.1093/nar/22.6.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thoms B, Borchers I, Wackernagel W. Effects of single-strand DNases ExoI, RecJ, ExoVII, and SbcCD on homologous recombination of recBCD+ strains of Escherichia coli and roles of SbcB15 and XonA2 ExoI mutant enzymes. J. Bacteriol. 2008;190:179–192. doi: 10.1128/JB.01052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burdett V, Baitinger C, Viswanathan M, Lovett ST, Modrich P. In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl-directed mismatch repair. Proc. Natl Acad. Sci. USA. 2001;98:6765–6770. doi: 10.1073/pnas.121183298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dianov G, Lindahl T. Reconstitution of the DNA base excision-repair pathway. Curr. Biol. 1994;4:1069–1076. doi: 10.1016/s0960-9822(00)00245-1. [DOI] [PubMed] [Google Scholar]
- 22.Rajman LA, Lovett ST. A thermostable single-strand DNase from Methanococcus jannaschii related to the RecJ recombination and repair exonuclease from Escherichia coli. J. Bacteriol. 2000;182:607–612. doi: 10.1128/jb.182.3.607-612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Z, Pan M, Santangelo TJ, Chemnitz W, Yuan W, Edwards JL, Hurwitz J, Reeve JN, Kelman Z. A novel DNA nuclease is stimulated by association with the GINS complex. Nucleic Acids Res. 2011;39:6114–6123. doi: 10.1093/nar/gkr181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wakamatsu T, Kim K, Uemura Y, Nakagawa N, Kuramitsu S, Masui R. Role of RecJ-like protein with 5′-3′ exonuclease activity in oligo(deoxy)nucleotide degradation. J. Biol. Chem. 2011;286:2807–2816. doi: 10.1074/jbc.M110.161596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanchez-Pulido L, Ponting CP. Cdc45: the missing RecJ ortholog in eukaryotes? Bioinformatics. 2011;27:1885–1888. doi: 10.1093/bioinformatics/btr332. [DOI] [PubMed] [Google Scholar]
- 26.Krastanova I, Sannino V, Amenitsch H, Gileadi O, Pisani FM, Onesti S. Structural and functional insights into the DNA replication factor Cdc45 reveal an evolutionary relationship to the DHH family of phosphoesterases. J. Biol. Chem. 2012;287:4121–4128. doi: 10.1074/jbc.M111.285395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wakamatsu T, Kitamura Y, Kotera Y, Nakagawa N, Kuramitsu S, Masui R. Structure of RecJ exonuclease defines its specificity for single-stranded DNA. J. Biol. Chem. 2010;285:9762–9769. doi: 10.1074/jbc.M109.096487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2–7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl Acad. Sci. USA. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshimochi T, Fujikane R, Kawanami M, Matsunaga F, Ishino Y. The GINS complex from Pyrococcus furiosus stimulates the MCM helicase activity. J. Biol. Chem. 2008;283:1601–1609. doi: 10.1074/jbc.M707654200. [DOI] [PubMed] [Google Scholar]
- 30.Marinsek N, Barry ER, Makarova KS, Dionne I, Koonin EV, Bell SD. GINS, a central nexus in the archaeal DNA replication fork. EMBO Rep. 2006;7:539–545. doi: 10.1038/sj.embor.7400649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z, Santangelo TJ, Cuboňová L, Reeve JN, Kelman Z. Affinity purification of an archaeal DNA replication protein network. MBio. 2010;1:e00221–10. doi: 10.1128/mBio.00221-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu XP, Liu JH. The terminal 5′ phosphate and proximate phosphorothioate promote ligation-independent cloning. Protein Sci. 2010;19:967–973. doi: 10.1002/pro.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urban M, Joubert N, Hocek M, Alexander RE, Kuchta RD. Herpes simplex virus-1 DNA primase: a remarkably inaccurate yet selective polymerase. Biochemistry. 2009;48:10866–10881. doi: 10.1021/bi901476k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramirez-Aguilar KA, Kuchta RD. Herpes simplex virus-1 primase: a polymerase with extraordinarily low fidelity. Biochemistry. 2004;43:9084–9091. doi: 10.1021/bi049335+. [DOI] [PubMed] [Google Scholar]
- 35.Choi JY, Eoff RL, Pence MG, Wang J, Martin MV, Kim EJ, Folkmann LM, Guengerich FP. Roles of the four DNA polymerases of the crenarchaeon Sulfolobus solfataricus and accessory proteins in DNA replication. J. Biol. Chem. 2011;286:31180–31193. doi: 10.1074/jbc.M111.258038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cann IK, Komori K, Toh H, Kanai S, Ishino Y. A heterodimeric DNA polymerase: evidence that members of Euryarchaeota possess a distinct DNA polymerase. Proc. Natl Acad. Sci. USA. 1998;95:14250–14255. doi: 10.1073/pnas.95.24.14250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jokela M, Eskelinen A, Pospiech H, Rouvinen J, Syväoja JE. Characterization of the 3′ exonuclease subunit DP1 of Methanococcus jannaschii replicative DNA polymerase D. Nucleic Acids Res. 2004;32:2430–2440. doi: 10.1093/nar/gkh558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henneke G, Flament D, Hübscher U, Querellou J, Raffin JP. The hyperthermoPhilic euryarchaeota Pyrococcus abyssi likely requires the two DNA polymerases D and B for DNA replication. J. Mol. Biol. 2005;350:53–64. doi: 10.1016/j.jmb.2005.04.042. [DOI] [PubMed] [Google Scholar]
- 39.Rouillon C, Henneke G, Flament D, Querellou J, Raffin JP. DNA polymerase switching on homotrimeric PCNA at the replication fork of the euryarchaea Pyrococcus abyssi. J. Mol. Biol. 2007;369:343–355. doi: 10.1016/j.jmb.2007.03.054. [DOI] [PubMed] [Google Scholar]
- 40.Frick DN, Richardson CC. DNA primase. Annu. Rev. Biochem. 2001;70:39–80. doi: 10.1146/annurev.biochem.70.1.39. [DOI] [PubMed] [Google Scholar]
- 41.Yamagata A, Kakuta Y, Masui R, Fukuyama K. The crystal structure of exonuclease RecJ bound to Mn2+ ion suggests how its characteristic motifs are involved in exonuclease activity. Proc. Natl Acad. Sci. USA. 2002;99:5908–5912. doi: 10.1073/pnas.092547099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 43.Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell. 2006;21:581–587. doi: 10.1016/j.molcel.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 44.Pospiech H, Grosse F, Pisani FM. The initiation step of eukaryotic DNA replication. Subcell. Biochem. 2010;50:79–104. doi: 10.1007/978-90-481-3471-7_5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.