Abstract
RbpA is a small non–DNA-binding transcription factor that associates with RNA polymerase holoenzyme and stimulates transcription in actinobacteria, including Streptomyces coelicolor and Mycobacterium tuberculosis. RbpA seems to show specificity for the vegetative form of RNA polymerase as opposed to alternative forms of the enzyme. Here, we explain the basis of this specificity by showing that RbpA binds directly to the principal σ subunit in these organisms, but not to more diverged alternative σ factors. Nuclear magnetic resonance spectroscopy revealed that, although differing in their requirement for structural zinc, the RbpA orthologues from S. coelicolor and M. tuberculosis share a common structural core domain, with extensive, apparently disordered, N- and C-terminal regions. The RbpA–σ interaction is mediated by the C-terminal region of RbpA and σ domain 2, and S. coelicolor RbpA mutants that are defective in binding σ are unable to stimulate transcription in vitro and are inactive in vivo. Given that RbpA is essential in M. tuberculosis and critical for growth in S. coelicolor, these data support a model in which RbpA plays a key role in the σ cycle in actinobacteria.
INTRODUCTION
The high G + C, Gram-positive actinobacteria are of immense medical and industrial importance. For example, although the Streptomyces genus is a key source of bioactive compounds, including antibiotics, immunosuppressants and anti-cancer drugs, Mycobacterium tuberculosis has maintained its global notoriety, currently causing 9 million cases and 1.5 million tuberculosis deaths per year (1). As well as being a major target for anti-mycobacterial agents, such as rifampicin, RNA polymerase (RNAP) also has a major influence on the control of antibiotic biosynthesis in Streptomyces, and mutations in this enzyme complex can both positively and negatively impact production (2,3). Therefore, a deeper understanding of RNAP and its regulation in the Actinobacteria phylum can contribute to both the production and development of new medicines.
Transcription initiation in bacteria occurs in several steps. First, an RNAP holoenzyme binds to a promoter element to form an initial closed complex in which the DNA remains double-stranded. This triggers a process of isomerization in which the DNA unwinds, and localized DNA melting reveals the template strand to the active site of the enzyme, giving rise to an open complex. Subsequently, short abortive transcripts are produced in a cyclical manner before the RNAP escapes into the elongation phase (4). The RNAP holoenzyme can be divided into core (five subunits, 2α, β, β′ and ω), which is catalytically active for elongation, and a dissociable σ factor subunit that is required for promoter recognition. The σ subunit provides critical DNA-binding determinants in both closed and open complexes, but it stochastically dissociates from elongating RNAP soon after promoter escape (5,6). This scheme gives rise to a ‘σ cycle’ whereby the dissociated σ factor enters a pool of σ factors that compete for binding to core RNAP (7). Bacteria are, therefore, able to tune gene expression by controlling the cellular level and composition of σ factors (8). All bacteria seem to contain a single essential σ factor that is orthologous to σ70 of Escherichia coli and directs the transcription of most ‘housekeeping’ genes during exponential growth, such as those involved in ribosome production and central metabolism. Structural studies on the principal σ factors of E. coli and Thermus sp., either alone or in the context of holoenzyme, revealed four domains, σ1.1, σ2, σ3 and σ4, that are interconnected by flexible linkers and correspond to conserved regions 1.1, 1.2–2.4, 3.0–3.1 and 4.1–4.2, respectively (9–12). The σ2, σ3 and σ4 domains each comprise key contact points for both core enzyme and promoter DNA, whereas σ1.1 is thought to maintain free σ in a compact non–DNA-binding form through interaction with σ2 and σ4 (12).
In addition to a principal and essential σ factor, bacteria usually contain one or more alternative σ factors that can redirect the transcription machinery to specific regulons, allowing the cell to respond to a wide variety of environmental, physiological or developmental cues (8). Most of these are related to σ70 and have been classified into four main groups (Groups 1–4) based on phylogeny and biological function (13): Group 1 σ factors are the σ70 orthologues; Group 2 σ factors are closely related to Group 1 σ factors but have specialized non-essential functions; Group 3 σ factors are structurally and functionally diverse but include all three globular domains (σ2–σ4); Group 4 comprises the highly diverse extracytoplasmic function (ECF) σ factors that lack both Region 1 and the σ3 globular domain. The control of alternative σ factor activity is particularly well established and often occurs at the post-translational level through the action of anti-σ factors that bind to σ and prevent its association with core RNAP (14). The σ factor is released after the inactivation of the anti-σ factor through competition with an alternative binding partner (partner-switching), regulated proteolysis or an allosteric change in response to the direct sensing of a signal. Although the σ subunit can act as an essential contact point for DNA-binding activators, proteins that bind to and activate free σ factors are rare. One example, however, is the small protein Crl, which stimulates the activity of the stationary phase regulator σS in E. coli and Salmonella (15) through direct interaction, both with free σ and as part of the holoenzyme (16,17). The role of Crl seems to be to improve the association of σS with core RNAP through interaction with the σ2 globular domain (18–21).
RbpA is an RNAP-binding protein that is confined to, and widespread within, the actinobacteria, and evidence is accumulating that it plays a crucial role in transcription initiation. RbpA was discovered in the antibiotic-producing bacterium Streptomyces coelicolor where the 124 amino acid protein was shown to confer basal levels of resistance to rifampicin in vivo and stimulate transcription in vitro (22,23). The S. coelicolor RbpA (designated RbpASc for clarity) co-purifies as a major component of RNAP holoenzyme, suggesting a broad role in transcription, although stimulation of transcription was previously only demonstrated using the ribosomal RNA promoter rrnDp3 (22). The rrnDp3 promoter is dependent on the principal σ factor in S. coelicolor, σHrdB, and RbpASc failed to stimulate transcription from a promoter dependent on the alternative σ factor σR (22). Although S. coelicolor mutants that lack RbpASc grow slowly, they are viable. However, it was recently demonstrated that RbpA (Rv2050; designated RbpAMt for clarity) is essential in M. tuberculosis (24). Furthermore, RbpAMt was shown to stabilize the principal σA-RNAP holoenzyme and stimulate transcription from wide range of σA-dependent promoters in vitro, suggesting a general role in transcription initiation (25). Two biochemical studies have proposed that the primary contact point for RbpAMt is the β subunit. However, although one study proposed binding close to the active-site cleft near the rifampicin-binding site (26), the other study proposed that RbpAMt contacts the sandwich-barrel hybrid motif located on the surface of the β subunit (25). In either study, the apparent specificity of RbpAMt for the principal σ factor was not explained, although it was proposed that RbpAMt might allosterically modify core RNAP to improve its interaction with σ (25). Here, we show that RbpA binds specifically to the principal and certain related σ factors of S. coelicolor and M. tuberculosis, and that RbpASc is present at a σHrdB–RNAP transcription initiation complex in vivo. The orthologous RbpA proteins from these organisms comprise a core domain, for which we present the high-resolution solution structures, together with extensive N- and C-terminal regions that are disordered in the free proteins. Finally, we map the interaction between S. coelicolor σHrdB and RbpASc, and we show that binding occurs primarily between the σ2 domain of σHrdB and the C-terminal region of RbpASc. Our data support a model in which RbpA plays a key role in the σ cycle of the vegetative RNAP in actinobacteria, possibly acting as a chaperone-like protein to aid the formation of active holoenzyme, which might be particularly important during stress.
MATERIALS AND METHODS
Strains, plasmids and growth conditions
Strains and plasmids are listed in Supplementary Table S1, and oligonucleotides used for the construction of plasmids are listed in Supplementary Table S2. E. coli K-12 strain DH5α was used for general cloning and plasmid propagation, and E. coli ET12567 (pUZ8002) was used to conjugate plasmids into S. coelicolor (27). S. coelicolor A3(2) strains were cultivated on mannitol–soya (MS) agar or in a modified yeast extract–malt extract (YEME, 10% sucrose and 0.01% antifoam) broth as previously described (27). To construct S. coelicolor S129, a ΔrbpA::apr allele was introduced into the S. coelicolor J1981 (rpoC-His6) genome by double crossover recombination as described previously (22). To construct pSX190, the S. coelicolor rbpA gene and promoter region were amplified using oligonucleotide primers rbpA_H3_rev and DT1a (22), thereby introducing a HindIII site immediately upstream of the stop codon, then fused to a triple FLAG tag sequence in the integrative vector pSET152. Site-directed mutagenesis of the C-terminal region of rbpA was performed in pSX512, and the rbpA derivatives were cloned along with the native promoter into pSETΩ as a BglII fragment.
Protein overexpression and purification
Core RNAP was prepared from late-exponential YEME cultures of S. coelicolor S129. Harvested mycelium was re-suspended in binding buffer (20 mM Tris–HCl, pH 7.5, 500 mM NaCl, 20 mM imidazole, 3 mM 2-mercaptethanol, 5% glycerol and 25 μg/ml phenylmethanesulphonyl fluoride) plus complete ethylenediaminetetraacetic acid (EDTA)-free protease inhibitor (Roche). Mycelium was lysed by passing twice through a French press at 10 000 kPa before centrifugation to remove the cell debris. The lysate was immediately loaded on a Ni-IDA sepharose column and washed with 10 column volumes of binding buffer followed by 5 column volumes buffer A (20 mM Tris–HCl, pH 7.5, 3 mM 2-mercaptoethanol, 5% glycerol and 150 mM NaCl) containing 20 mM imidazole, then eluted with buffer A containing 250 mM imidazole. The eluate was immediately applied to a HiTrap Heparin HP column (GE Healthcare) equilibrated with buffer A, and protein was eluted from the column using a salt gradient (20 mM Tris–HCl, pH 7.5, 3 mM 2-mercaptoethanol, 5% glycerol and 1 M NaCl). RNAP fractions were dialysed against buffer C (20 mM Tris–HCl, pH 7.5, 3 mM 2-mercaptoethanol, 5% glycerol and 50 mM NaCl) and further purified by mono-Q (GE Healthcare) ion-exchange chromatography. Core-rich RNAP fractions were eluted in tail fractions from the mono-Q column. Native RbpASc, hexahistidine (His6)-tagged RbpASc and σHrdB were prepared as described previously (22). For in vitro transcription reactions, the His6-tag on RbpASc was removed by treatment with thrombin, and the protein was purified by gel filtration. For expression of truncated σHrdB domain fragments (σ2, σ2–σ4, σ3–σ4 and σ4) with N-terminal His6-tags, polymerase chain reaction (PCR) was used to amplify the corresponding hrdB regions with primers incorporating N-terminal NdeI and C-terminal XbaI sites and cloned into pET15b. Expression was achieved in E. coli BL21λDE3 (pLysS), and fragments were purified using Ni-NTA spin columns (Qiagen) as recommended by the manufacturer. For NMR analysis, the full-length M. tuberculosis rbpA gene (Rv2050) was amplified by PCR incorporating an N-terminal NdeI site and C-terminal BamHI site and cloned into pET20b to give pSX500. To solve the structure of the core domain of RbpASc, a truncated gene encoding the first 75 amino acids was chemically synthesized with codon optimization (Eurofins) for expression in E. coli and subsequently cloned into pET15b to give pSX505. E. coli BL21λDE3 (pLysS) containing pSX500 or pSX505 was grown in L-broth to OD600 of 3–5, then transferred to optimized high-cell density isopropyl-β-D-thiogalactopyranoside (IPTG)-induction minimal media (28,29) containing 0.1% 15NH4Cl and 1% 13C-glucose, grown for 2 h at 37°C, before induction with 1 mM IPTG at 30°C for 3 h. Oligonucleotides used to amplify each gene or domain are listed in Supplementary Table S2.
In vitro transcription reactions
In vitro transcription assays were carried out using S. coelicolor S129 core RNAP and purified native σHrdB and RbpASc proteins, essentially as described previously (30). Templates containing promoters were amplified by PCR using primers listed in Supplementary Table S2. Assays (10 μl) were performed at 30°C, and they contained 5 nM DNA template in reaction buffer (50 mM Tris–HCl, pH 8, 10 mM MgCl2, 200 mM KCl, 0.1 mg/ml of bovine serum albumin and 10 mM β-mecaptoethanol). σHrdB, RbpASc and core RNAP were mixed in RNAP dilution buffer (10 mM Tris–HCl, pH 8, 10 mM KCl, 0.4 mg/ml of bovine serum albumin, 0.1 mM Na2EDTA, 0.1% Triton X-100 and 10 mM β-mecaptoethanol) (30) for 15 min on ice, and reactions were initiated by the addition of nucleotide mix (1 mM adenosine triphosphate, GTP, CTP and 50 μM UTP), including 5 μCi [α-32P] UTP (>800 Ci/mmol, GE Healthcare), and incubated for 15 min. Reactions were halted by addition of an equal volume of in vitro loading dye [80% (w/v) formamide, 0.01% (w/v) xylene cyanol and 0.01% (w/v) bromophenol blue], separated on denaturing 6% polyacrylamide gels and quantified by phosphorimaging.
Bacterial two-hybrid analysis
The complete rbpA genes from S. coelicolor and M. tuberculosis, as well as truncated derivatives as indicated in figure legends, were PCR amplified then cloned into pUT18, generating rbpA-T18 translational fusions to the T18 catalytic domain of Bordetella pertussis adenylate cyclase. The σ factor genes from S. coelicolor (hrdA, hrdB, hrdC, hrdD, sigB, sigE, sigR and whiG) and M. tuberculosis (sigA and sigB), as well as truncated derivatives of hrdB, were cloned into pKT25, generating T25-σ translational fusions to the T25 catalytic domain. The amino acids included in each truncated hybrid construct are indicated in the figure legends, and the oligonucleotides used for PCR amplification are indicated in Supplementary Table S2. Functional complementation between chimeric proteins was evaluated in E. coli BTH101 by performing β-galactosidase measurements on permeabilized overnight cultures grown in L-broth containing 0.5 mM IPTG. One unit of β-galactosidase activity corresponds to 1 nmol of o-nitrophenyl-β-D-galactoside (ONPG) hydrolysed per min at 28°C. Results were presented either as Miller units (31) or as % Miller units relative to a control.
In vitro pull-down assays
Experiments were performed using Dynabeads® His-Tag Isolation and Pulldown beads (Invitrogen) essentially as recommended by the manufacturer. Test proteins were mixed on ice in binding buffer (50 mM sodium phosphate, pH 8, 300 mM NaCl and 0.01% Tween®-20), then added to 20 μl magnetic Dynabeads and left to incubate for 30 min at ambient temperature with shaking. Samples were washed four times, and then eluted in 100 μl elution buffer (binding buffer plus 300 mM imidazole) for 15 min.
Chromatin immunoprecipitation–quantitativePCR
Strains were grown in modified YEME to mid-late exponential phase (OD450: 2.0), treated with rifampicin (140 µg/ml) for 20 min, then formaldehyde [1.1% (v/v); Sigma] for a further 20 min, with continued shaking at 200 rpm. After quenching with 0.5 M glycine for 5 min at 30°C, mycelium was collected by centrifugation, washed twice in phosphate-buffered saline and stored at −20°C. Chromatin extraction and immunoprecipitation protocols were essentially as described previously (32). Mycelial pellets were subjected to 3 × 90 s of cryogenic grinding (Retsch MM301, 30 Hz) then resuspended in 9 ml of IP buffer (32), and 3-ml aliquots were sonicated (35% amplitude) for 4, 8 and 12 cycles of 15 s ON and 45 s OFF. After clearing by centrifugation, 70 µl of samples were incubated with proteinase K (Roche), de-cross-linked and then analysed by gel electrophoresis (32). Samples comprising chromatin fragments of ∼0.5 kb were pre-cleared by incubation (rotation for 4 h at 4°C) with either protein A or protein G magnetic beads (NEB) before the addition of antibodies. Immunoprecipitations were conducted with M2 mouse monoclonal anti-FLAG antibody (Sigma), monoclonal 8RB13 anti-E. coli RNAP β subunit antibody (Abcam) or a custom rabbit polyclonal antiserum raised against σHrdB (Eurogentec). Corresponding mock immunoprecipitations (IPs) with no added antibody were performed as negative control. After incubation with antibodies (rotation for 12 h at 4°C), immunoprecipitations were performed using protein G magnetic beads (RbpASc and RNAP; NEB) or protein A magnetic beads (σHrdB; NEB). Subsequent steps to dissociate cross-links and purify DNA were as described previously (32), and DNA was resuspended in 25 µl of ultrapure water. Quantitative real-time PCR was performed using the standard curve absolute quantification approach using serial dilutions of sonicated S. coelicolor M145 genomic DNA to generate standard curves for each of the four primer pairs used (Supplementary Table S2). qPCR reactions were set-up in 0.1 ml of MicroAMP FAST optical 96-well reaction plates (Applied Biosystems) using GoTaq qPCR reaction mix (Promega), 10 ng of template DNA and each primer at 0.15 pmol/µl. The data were analysed using a modified method to that previously described (33). The mock immunoprecipitation (IP) signal was subtracted from the signal obtained using the corresponding immunoprecipitated sample DNA as input to correct for background signal. To allow visual comparison of the relative enrichments obtained using the three different antibodies, the background corrected signals obtained using a particular antibody were expressed relative to the highest corrected signal obtained using either of the four primer pairs.
Metal analysis
Native (un-tagged) RbpA proteins were produced in E. coli and purified by ion-exchange chromatography and gel filtration, before concentration as described previously (22). Approximately 0.5 mg of protein was vacuum-dried then re-suspended in 86 μl of 70% nitric acid (metal analysis grade) and heated for 10 min at 105°C. Water (metal analysis grade) was added to give a final concentration of 2% nitric acid, and inductively coupled plasma mass spectrometry was performed using an Agilent 7500 series instrument. A solution containing 1 ppm of each metal to be analysed (Mn, Fe, Ni, Cu and Zn) was used as standard, and values were normalized against protein storage buffer alone. Experiments were performed in duplicate.
NMR and solution structure determination
Samples were buffer-exchanged into 100 mM MES buffer, pH 6.0, with 150 mM KCl. NMR experiments were performed on >200 µM samples of uniformly 15N- and 13C-labelled protein.
The backbone resonance assignments of RbpASc and RbpAMt were completed by using the standard triple-resonance assignment approach (34), and data were analysed using in-house algorithms with the program NMRView (35). Hα and Hβ assignments were obtained using HBHA(CBCACO)NH. The side-chain assignments of the two proteins were extended using HCCH TOCSY and (H)CC(CO)NH TOCSY. The distance restraints were obtained from 1H-15N/13C nuclear Overhauser effect spectroscopy (NOESY)–heteronuclear single- or multiple-quantum coherence (HSQC/HMQC) experiments (mixing time, 100 ms at 800 MHz). Data were collected on Bruker AvanceIII (600 MHz) and AvanceII (800 MHz) spectrometers equipped with TCI and TXI cryoprobes, respectively. The ARIA protocol was used for completion of the NOE assignment and structure calculation (36). Dihedral angle restraints derived from TALOS were also incorporated in the calculation (37). The frequency window tolerances for assigning NOEs were ±0.05 ppm for direct proton dimensions and ±0.05 ppm for indirect proton dimensions, and ±0.5 ppm for nitrogen dimensions and ±1.3 ppm for carbon dimensions. One hundred structures were calculated in the final iteration, and these had no NOE violations >0.5 Å and no dihedral angle violations >5°. The 10 lowest energy structures were selected for further analysis and figure generation.
RESULTS
RbpASc activates a range of σHrdB-dependent promoters
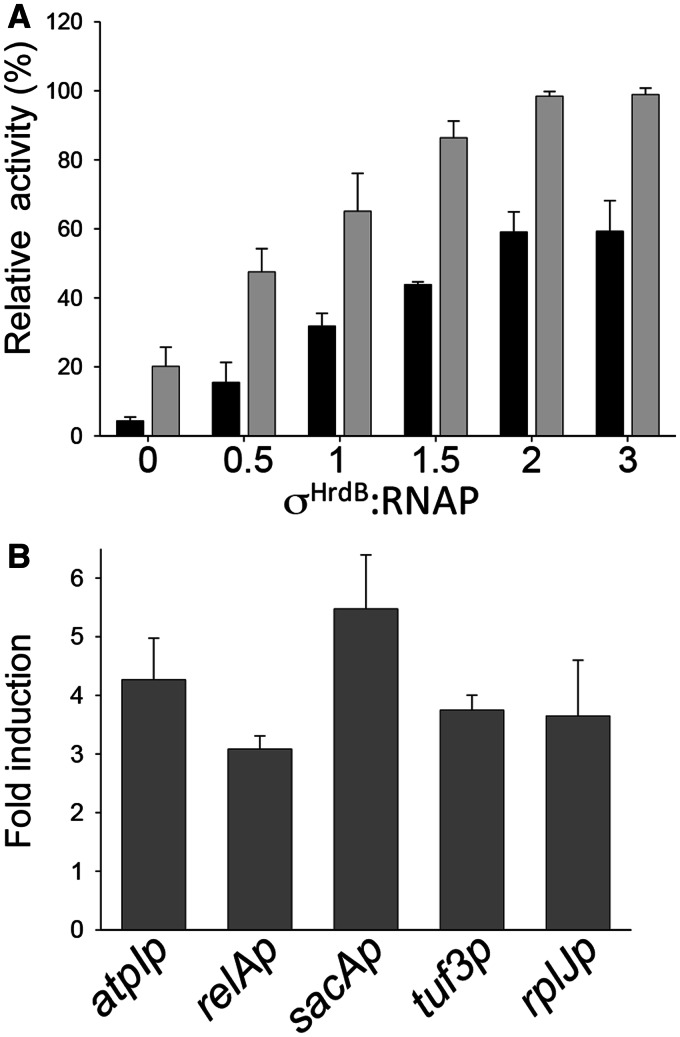
Previously, using total RNAP holoenzyme preparations, we showed that RbpASc stimulated transcription in vitro from the σHrdB-dependent promoter rrnDp3, but not from a σR-dependent promoter, rbpAp (22). Initially, we extended this work by showing that RbpA increased the maximal activity of σHrdB–RNAP at the rrnDp3 promoter by titrating increasing concentrations of σHrdB in the presence and absence of excess RbpASc (Figure 1A). We then tested whether the RbpASc activity was specific to rrnDp3 or whether it played a more general role in activating σHrdB-dependent promoters. Transcription run-off assays were performed with a selection of promoters that we or others have predicted to be dependent on σHrdB, based on the putative −10 promoter elements: sacAp (38), tuf3p (39), relAp2 (40), atpIp (unpublished observations) and rplJp (41). In each case, RbpASc stimulated transcription in assays consisting of core RNAP, σHrdB and linear DNA templates (Figure 1B). RbpASc also activated transcription initiation in single-round transcription assays, which suggests that the stimulatory effect is not a consequence of increased recycling or elongation (data not shown). Together with the previous demonstration that RbpAMt stimulates a range of σA-dependent promoters with no apparent specificity (25), these data support the idea that RbpA is a general activator of the principal RNAP holoenzyme in actinobacteria.
Figure 1.
RbpASc activates transcription from a range of σHrdB-dependent promoters in vitro. (A) Multi-round in vitro transcription reactions using the rrnDp3 promoter as template (22) and including core RNAP (75 nM) σHrdB at σHrdB: RNAP ratios of 0, 0.5, 1, 1.5 or 2, or 3, in the presence (grey bars) or absence (black bars) of excess RbpASc (40 μM). rrnDp3 transcription products are normalized relative to the highest signal. Note that these preparations of core RNAP have residual levels of σHrdB. (B) Multi-round in vitro transcription reactions contained core RNAP (75 nM) σHrdB (375 nM), RbpASc (750 nM) and DNA templates generated by PCR using primers listed in Supplementary Table S2. Data are presented as fold-difference relative to reactions lacking RbpA. Transcript levels were quantified by phosphorimaging from triplicate data, and standard deviation is indicated.
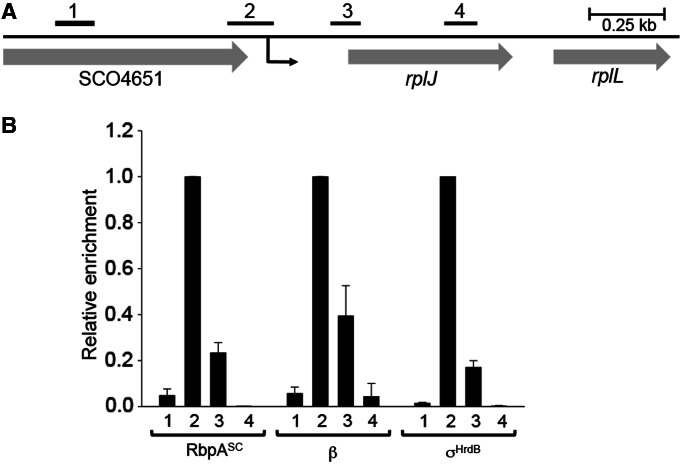
Although it is known that RbpASc is present in purified RNAP preparations, it has not yet been shown to be present in RNAP–promoter complexes in vivo. We, therefore, decided to test this using chromatin immunoprecipitation combined with quantitative PCR (ChIP–qPCR). The presence of RbpASc, σHrdB and the β subunit of RNAP was analysed at the rplJp promoter. Immunoprecipitation of β and σHrdB was performed using a commercially available monoclonal antibody (42) and a polyclonal antibody, respectively. To immunoprecipitate RbpASc, the rbpA gene was C-terminally tagged with tandem FLAG epitopes and integrated into the genome of the S. coelicolor rbpA mutant S129 at the ϕC31 attachment site using the vector pSET152. The resulting construct, pSX190, fully restored normal growth rate to S129, indicating that that the epitope tag did not impede normal RbpASc function (data not shown). S. coelicolor S129 (pSX190) was grown to mid-exponential phase before treatment with rifampicin to inhibit global RNA synthesis by trapping RNAP at promoters (43). As expected, the rplJ promoter region (qPCR product 2; Figure 2), centred −79 bp upstream from the transcription start point, was highly enriched for both σHrdB and the β subunit, with much lower enrichment seen for control regions centred −653 bp upstream, or +235 bp and +608 bp downstream, of the transcription initiation site, suggesting that σHrdB–RNAP was indeed trapped at the rplJ promoter (Figure 2). The enrichment patterns seen using the anti-FLAG antibody were similar to those obtained with the σHrdB and the β antibodies, indicating that RbpASc is present in these transcription initiation complexes, although it should be noted that differences in antibody–epitope affinities prevent the relative proportion of initiation complexes that contain RbpASc from being assessed. Control anti-FLAG immunoprecipitation experiments using an equivalent strain in which RbpASc was untagged showed no enrichment above that seen with the ‘no antibody’ control, confirming that this signal is specific to RbpASc (data not shown).
Figure 2.
Localization of RbpASc at a σHrdB-dependent promoter in vivo. (A) Schematic of the rplJ region. The transcription start point (+1) of rplJp is indicated by a black arrow located 240 bp upstream from the rplJ start codon. The bars above the genes show the relative positions of PCR products used for ChIP–qPCR that are centred with respect to +1 as follows: 1, −653 bp; 2, −79 bp; 3, +235 bp; 4, +235 bp. (B) Occupancy of RbpASc, RNAP β subunit, and σHrdB at the indicated regions in S. coelicolor S129 (pSX190), after treatment with rifampicin. Immunoprecipitations were performed using monoclonal anti-β, polyclonal anti-σHrdB and anti-FLAG antibody to detect RbpA–FLAG. To allow comparison of RbpASc, β and σHrdB localization, after absolute quantitation of co-immunoprecipitated DNA and background correction for each antibody, enrichment is presented relative to the highest corrected signal obtained using either of the four primer pairs. Standard deviations (calculated for two biological replicates) are indicated.
RbpA binds specifically to principal and related σ factors
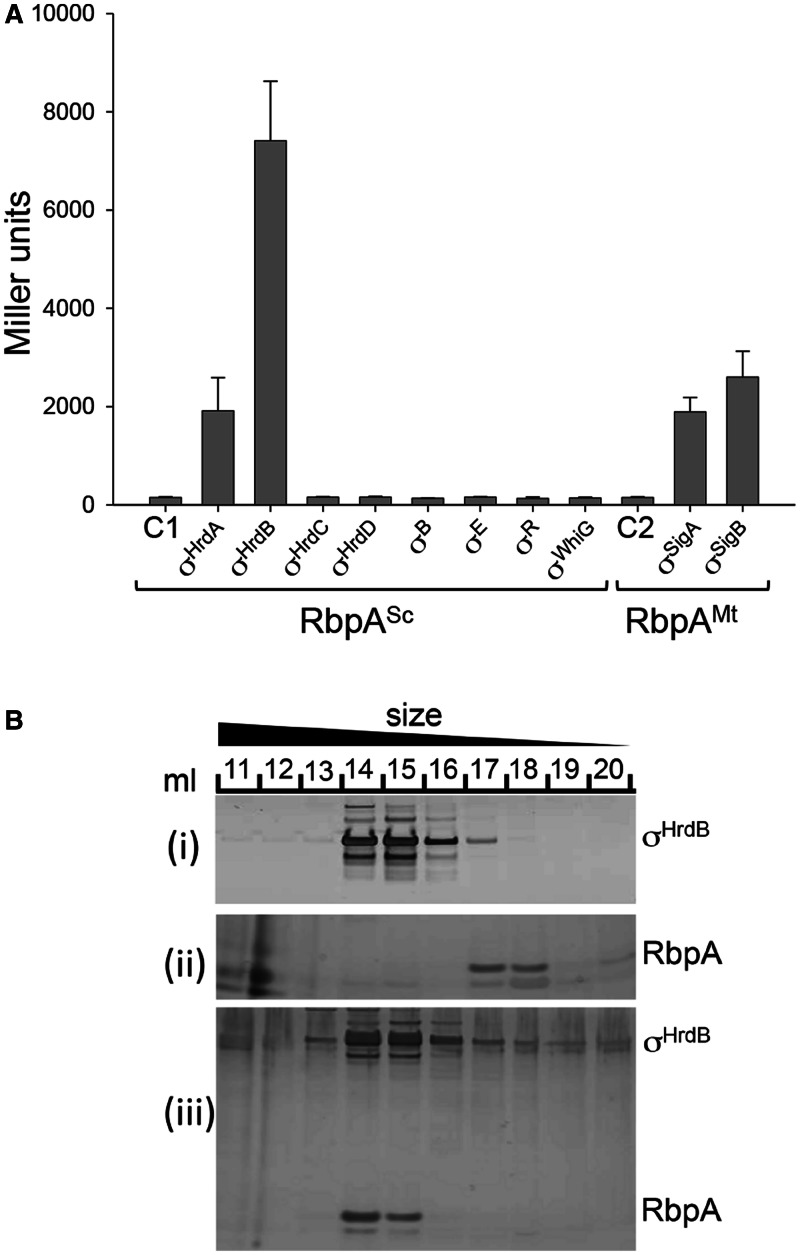
We considered the possibility that the apparent specificity of RbpA activity towards promoters recognized by the principal σ factors might be determined by a direct interaction with the σ factor itself. To test this idea, we applied a bacterial two-hybrid (BACTH) assay based on the B. pertussis adenylate cyclase (44). The S. coelicolor rbpA gene was fused to the gene that encodes the T18 domain of adenylate cyclase such that RbpASc was at the N-terminus of the fusion protein. A range of σ factor genes were fused to the gene that encodes the T25 domain, such that the σ factors were located at the C-terminus of the fusion proteins. Initially we tested for interaction between RbpASc and the following S. coelicolor σ factors: σHrdB (Group 1); σHrdA, σHrdC and σHrdD (Group 2); σWhiG and σB (Group 3); and σE and σR (Group 4) (Figure 3A). The Group 1 and Group 2 σ factors were N-terminally truncated such that they included only conserved regions 1.2–4.2, which comprise structural domains σ2, σ3 and σ4. This was necessary because full-length T25–hrdB fusions seemed to be toxic to E. coli. A positive result was detected for σHrdB and σHrdA, whereas none of the remaining σ factors bound to RbpASc. This suggests that RbpASc interacts only with principal and closely related σ factors. To broaden this idea, we tested for interaction between RbpAMt and M. tuberculosis σ factors σA (Group 1) and σB (Group 2). Binding was detected in both cases (Figure 3A), which suggests that the interaction with principal and related σ factors is a general feature of RbpA proteins in actinobacteria. In principal, the interactions observed during in vivo two-hybrid analysis could involve E. coli RNAP in hybrid higher order complexes. To confirm that the interaction between RbpASc and σHrdB does not involve any other factors, the two proteins were analysed, separately or mixed, by gel filtration chromatography (Figure 3B). σHrdB (55.9 kDa) and RbpASc (14.1 kDa) eluted at different volumes with apparent molecular masses of 183.2 and 29.9 kDa, respectively, suggesting trimeric and dimeric complexes. When mixed together, a complex of the two proteins eluted close to the σHrdB peak with an apparent mass of 211.4 kDa. Although it is clear that the two proteins interact, the difference in size between σHrdB and RbpASc, and our suspicion that σHrdB might run anomalously during gel filtration, makes it difficult to predict precise stoichiometry at this point.
Figure 3.
RbpA binds to the principal and closely related σ factors. (A) BACTH analysis of RbpA from S. coelicolor (RbpASc) and M. tuberculosis (RbpAMt) interactions with σ factors from S. coelicolor (Group 1, σHrdB; Group 2, σHrdA, σHrdC and σHrdD; Group 3, σHrdB and σWhiG; Group 4, σE and σR) and M. tuberculosis (Group 1, σA; Group 2, σB). The rbpA genes were fused to the T18 subunit of B. pertussis adenylate cyclase, whereas the σ factor genes were fused to the T25 subunit. Groups 1 and 2 σ factor hybrid fusions included only σ domains σ2, σ3 and σ4, whereas the remaining σ factor fusions were full length. β-galactosidase assays were performed in triplicate, and standard deviations are indicated. Control strains contained C1, pUT18-rbpASc, pKT25; C2, pUT18-rbpAMt, pKT25. (B) Co-elution of RbpASc and σHrdB during size-exclusion chromatography. (i) σHrdB (5 nmol), (ii) RbpASc (5 nmol) or a mixture of the two (iii) were passed through a Superose 6 10/300 GL size-exclusion column, and 1 ml of fractions (indicated by numbers) were separated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) and silver stained.
RbpA orthologues differ in their zinc content
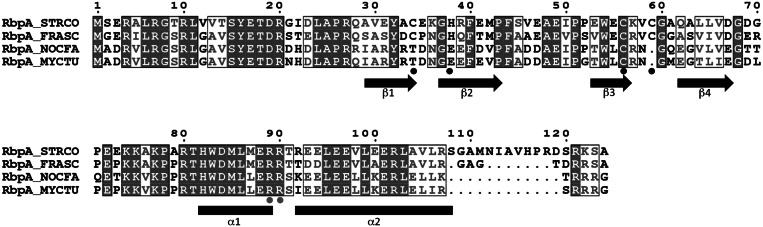
Protein sequence alignments suggested that the RbpA orthologues from S. coelicolor and M. tuberculosis might differ in their metal content (Figure 4). S. coelicolor RbpASc contains three cysteine residues, Cys35, Cys56 and Cys59, and a histidine His38, each of which are widely conserved (e.g. in Bifidobacterium sp. and Frankia sp.) and arranged as a putative C(H/C)CC zinc-ribbon motif. Surprisingly, RbpAMt, along with RbpA orthologues from Corynebacterium sp. and Nocardia sp., lacks all of these residues apart from the Cys56-equivalent cysteine. To investigate metal content, we isolated RbpASc and RbpAMt from overproducing E. coli strains in their native (untagged) forms. Inductively coupled plasma mass spectrometry (ICP-MS), set to detect Mn, Fe, Ni, Cu and Zn, revealed zinc as the only metal present at significant quantities in RbpASc, at a stoichiometry of 0. 83 (±0.04) mol zinc mol−1 protein. Conversely, no metal was detected at significant levels in RbpAMt. To test the importance of the presumed cysteine zinc ligands in RbpASc, each was mutated to alanine, and the resultant genes were introduced into S. coelicolor S101 (ΔrbpA::hyg) using the integrative expression vector pIJ6902 to test for complementation. In each case, the mutant genes failed to restore normal growth rate (Supplementary Figure S1), suggesting that each residue is crucial for the function or stability of the protein. Each of the three histidine residues in RbpASc was also changed to alanine, although in each case the mutant protein complemented the S101 strain (Supplementary Figure S1), suggesting that the protein retains function even when the functional requirement of zinc is conferred solely by the cysteine ligands.
Figure 4.
Alignment of the amino acid sequences of selected RbpA orthologues. STRCO, S. coelicolor, NP_625703; FRASC, Frankia sp. CcI3, YP_481367; NOCFA, Nocardia farcinica, YP_119325; M. tuberculosis H37Rv, NP_216566. Residues that coordinate a zinc atom in RbpASc, or form a stabilizing hydrogen bonding network in RbpAMt, are indicated with black dots. Arginine residues that when mutated prevent σHrdB interaction are indicated by grey dots (Figure 6). The β-strand secondary structure elements in the RCD domain are indicated, as are predicted α-helices in the C-terminal region.
The solution structure of the core domain of RbpA from M. tuberculosis and S. coelicolor
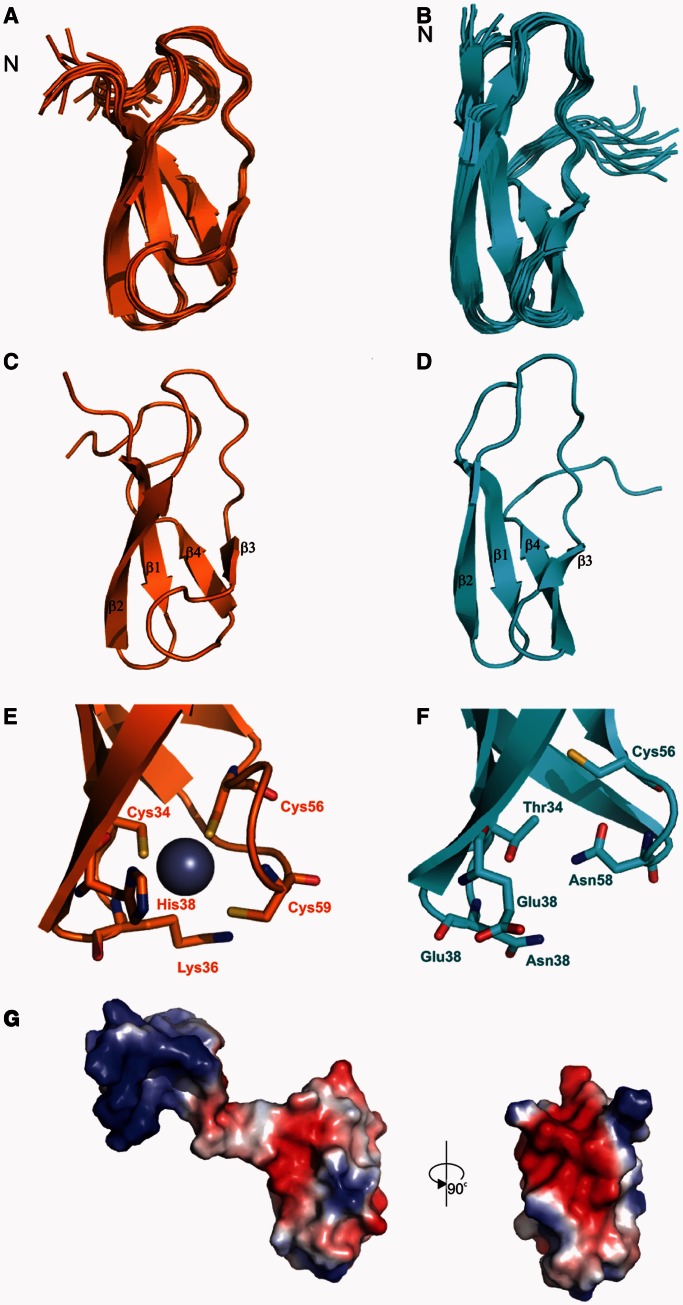
To investigate why RbpASc and RbpAMt differ in their zinc content and to dissect the structure–function relationship of RbpA, we used NMR spectroscopy to determine the solution structure of each protein. The dispersion of resonances in proton NMR spectra recorded on both RbpA proteins clearly showed that the proteins were at least partially structured (data not shown), although the M. tuberculosis protein gave better spectra and was, therefore, our initial focus. To determine the structured portions of the protein, attempts were made to assign backbone amides. Despite efforts to prevent proteolysis, the samples degraded rapidly, and complete assignments were obtained only for the core domain. Resonance line widths and chemical shift dispersion indicate that regions outside these domains are highly disordered. The solution structure of the core domain was then determined using standard multidimensional NMR spectroscopy methods. After this, we determined the structure of the RbpASc core domain using a truncated clone that included RbpASc residues 1–75. The structures of the RbpA orthologues (Figure 5; see Supplementary Table S3 for NMR and refinement statistics) reveal a compact core domain (residues 28–72; designated the RbpA core domain, RCD) comprising four β strands that form a partially open β barrel. Residues 29–34, 37–43, 53–56 and 62–67 make up strands β1, β2, β3 and β4, respectively. In RbpASc, the three Cys residues cluster together with His38 to form the likely metal-binding site, which is, together with the presence of Zn, consistent with a CHCC Zn-ribbon fold, a major subclass of the zinc-finger family, particularly common in proteins involved in translation and transcription (45). In RbpAMt, in place of the key residues (Cys34, His38, Cys56 and Cys59) are the following conserved hydrophilic residues: Thr34, Glu38, Cys56 and Asn58, which form a stabilizing hydrogen bond network (Figure 5E and F). Interestingly, although the RCD domain of RbpASc is largely negatively charged, the flexible N-terminal tail (1–26) is positively charged (Figure 5G), which may implicate this region in an interaction with DNA in transcription initiation complexes. Although the classical zinc-finger structure in the RbpASc RCD may suggest that it is specialized for DNA binding, the electrostatic potential and the absence of the Zn binding in other orthologues suggest that it is primarily a structural scaffold. The β-barrel structure is seen in other proteins (Supplementary Figure S2), including domains from mini-chromosome maintenance complexes (46), ribosomal protein L27 (47) and polypeptide chain release factors (48). Although the precise role of this domain in these proteins is unclear, they, along with adjacent sequences, seem to contribute to stabilizing either protein–protein or protein–RNA interactions. Interestingly, for example, the ribosomal protein L27 has a disordered N-terminal region (residues 1–19) in the oligonucleotide-free form, but in complex it forms an extended structure and interacts with the acceptor stem of the P-site tRNA and contributes to an increase in catalysis (49).
Figure 5.
Solution structures of RbpA from S. coelicolor (RbpASc) and M. tuberculosis (RbpAMt). NMR ensemble of the structure calculation for RbpASc (orange, A) and RbpAMt (cyan, B). Ribbon representation of the structure of RbpASc (C) and RbpAMt (D). Comparison of the Zn-binding site of RbpASc (E) with the equivalent residues in RbpAMt (F). Electrostatic surface potential for RbpASc (G) shown in two orientations. The flexible N-terminus in RbpASc is included in the orientation shown on the left and removed for clarity on the right.
The C-terminal region of RbpASc is required for interaction with σHrdB
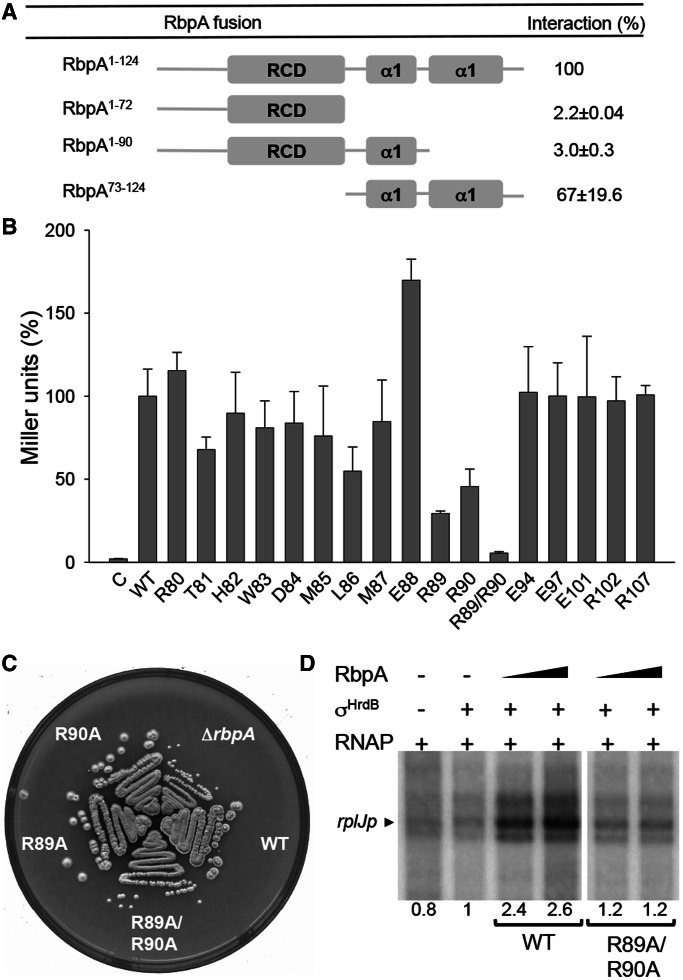
Although the NMR structures of the RbpA core domain did not include the C-terminal region, and indeed suggested that this region was disordered, sequence-based structural predictions using the Phyre 2 package (50) suggested the presence of two α-helices (designated α1 and α2), the second of which would be particularly amphipathic with a negatively charged face (Figure 4). To identify the region of RbpASc that interacts with σHrdB, we used BACTH to test three truncated S. coelicolor rbpA-T18 fusions for interaction with T25-σHrdB (Figure 6): RbpASc lacking both predicted C-terminal helices (residues 1–72, RbpA1-72); RbpASc lacking only α2 (RbpA1-90); and RbpASc lacking the N-terminal region and RCD domain but including α1 and α2 (RbpA73−124). In contrast to RbpA1−72 and RbpA1−90 for which an interaction with σHrdB was not detected, RbpA73−124 did interact, indicating that the C-terminal helical region of RbpASc is necessary and sufficient for binding. To identify critical residues involved in this interaction, we performed alanine-scanning mutagenesis, focusing on highly conserved residues in the C-terminal region. Apparent binding effects were observed for the highly conserved ERR motif (amino acid residues 88–90). Although binding seemed to be improved for the E88A mutation, binding was significantly reduced for R89A and R90A mutants, and a double R89A/R90A mutant was completely defective in σHrdB binding. To test the importance of this interaction in the function of RbpASc, we cloned the mutant genes into the integrative plasmid pSETΩ and tested for complementation of S. coelicolor S129 (ΔrbpA::apr). Although the introduction of wild-type rbpA and the E88A mutant (data not shown) completely restored normal growth to S129, the rbpA R89A/R90A mutant did not. The individual R89A and R90A mutants also failed to completely restore normal growth, each exhibiting a slight, but reproducible, reduction in colony size and a delay in sporulation timing. As the inability of the R89A/R90A double mutant to complement S. coelicolor S129 might be due to protein instability in vivo, we overexpressed the protein in E. coli and tested whether the pure protein would stimulate transcription in vitro. Using the rplJp promoter as template, the RbpASc R89A/R90A mutant failed to stimulate transcription, which suggests that a direct interaction with σHrdB is required for the action of RbpASc.
Figure 6.
The C-terminal region of RbpASc is necessary and sufficient for σHrdB interaction. (A) A schematic diagram indicating the regions of RbpASc tested for interaction with σHrdB using BACTH analysis. The RbpASc fragments and σHrdB σ2–σ4 were fused to the T18 and T25 domains of B. pertussis adenylate cyclase, respectively. (B) BACTH analysis of alanine point mutations in conserved residues in the C-terminal region of RbpASc. C, control strain containing pKT25–hrdB and pUT18. For (A) and (B), experiments were performed in triplicate (standard deviations indicated), and data are presented as % Miller units relative to results obtained with the full-length wild-type RbpASc. Control strains with pKT25–hrdB and pUT18 exhibited <1% the activity of the full-length RbpASc interaction. (C) Growth of S129 (ΔrbpA), S129 containing pSETΩ::rbpA (WT), or equivalent constructs with the rbpA mutations as indicated. Strains were streaked to MS agar and photographed after 4 days incubation at 30°C. (D) In vitro transcription from the rplJ promoter in the presence of RbpASc or RbpASc R89A/R90A mutant proteins. Multi-round in vitro transcription reactions contained core RNAP (50 nM) σHrdB (250 nM), RbpASc (250 nM or 1.25 µM) and a DNA template generated by PCR using primers listed in Supplementary Table S2. Transcript levels were quantified by phosphorimaging, and data are presented relative to reactions performed in the presence of σHrdB but the absence of RbpA.
RbpA binds to the σ2 domain of the principal sigma factor
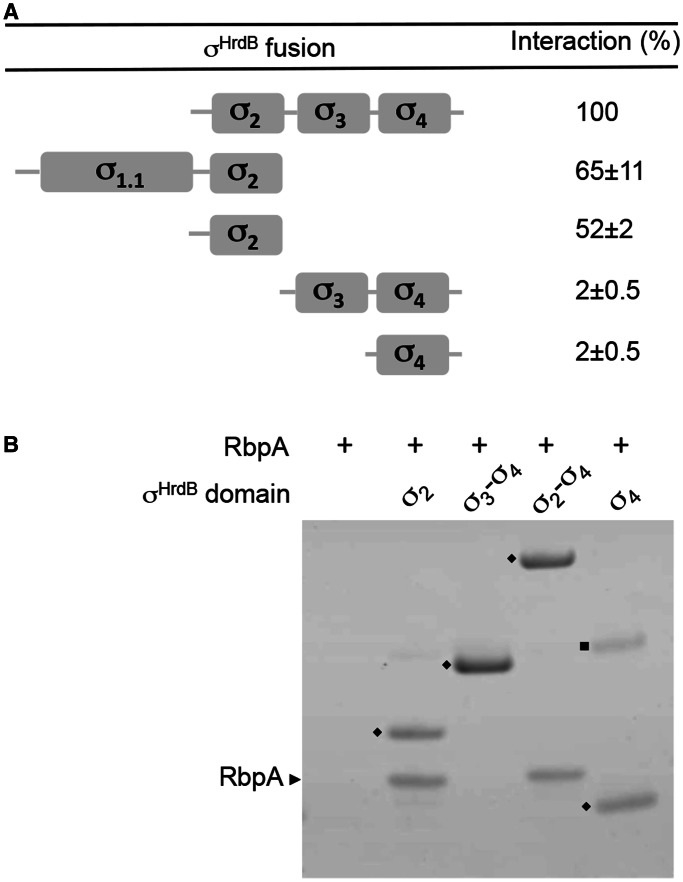
The different domains of σ have distinct functions; therefore, to better understand the role of RbpA in transcription initiation, we sought to localize the interaction to individual domain(s) of σHrdB. The σ2, σ3 and σ4 structural domains of σHrdB were predicted based on the structures of σA from Thermus sp. (11,51), and five overlapping T25–hrdB fusions were constructed and tested for interaction with the rbpA–T18 fusion in BACTH analysis (Figure 7). No interaction was detected for T25–hrdB (σ3–σ4) or T25–hrdB (σ4). However, all fragments that included σ2 interacted, including T25–hrdB (σ2) (amino acid residues 211–347), which comprises conserved regions 1.2–2.4. To confirm these data, each of the hrdB constructs used in the BACTH analysis was overexpressed with an N-terminal His6-tag, and the corresponding proteins purified, then tested for interaction with RbpASc using an in vitro pull-down assay (apart from σ1.1–σ2, which was insoluble). The pure σHrdB fragments (domains σ2, σ2–σ4, σ3–σ4 and σ4) were mixed with native RbpASc for 15 min and then isolated using magnetic Ni-affinity beads. RbpASc was co-isolated with the σ2 and σ2–σ4 fragments of σHrdB but not with σ3–σ4 and σ4, confirming a direct and specific interaction with the σ2 domain. Using both BACTH and in vitro pull downs, we also detected equivalent interactions between the σ2 domain of σA and the RbpAMt (data not shown). Furthermore, when equivalent σ2 and RbpA proteins from S. coelicolor and M. tuberculosis are co-expressed in E. coli, with one partner His6-tagged, they co-elute as a complex during Ni-affinity chromatography (data not shown). Taken together, these data suggest that the σ2–RbpA interaction involving the principal σ factor is conserved across the actinobacteria.
Figure 7.
RbpASc binds to the σ2 domain of σHrdB. (A) A schematic diagram indicating the four conserved regions/globular domains of σHrdB together with BACTH interaction data between truncated T25–σHrdB fusions and RbpASc–T18. The T25–σHrdB fusions included the following amino acids: σ1.1–σ2, 1–347; σ2, 211–347; σ3–σ4, 348–511; σ2–σ4, 211–511; σ4, 435–511. Control strains with pKT25 and pUT18–rbpA exhibited <1% the activity of the σ2–σ4 interaction. Experiments were performed in triplicate, and standard deviations are indicated. (B) Interaction between His6-tagged σHrdB fragments and RbpASc as judged using in vitro pull-down experiments. Purified His6-tagged σHrdB fragments (closed diamond) were mixed with RbpASc (∼0.5–1 μM each) before purification using Ni-affinity magnetic beads. Eluted proteins were separated by 4–12% Bis–Tris SDS–PAGE and stained using Coomassie brilliant blue. RbpASc is indicated with a black arrowhead. Closed square, unknown contaminating protein.
DISCUSSION
Here, we provide an explanation for why the RNA polymerase-binding protein RbpA stimulates transcription from promoters that are dependent on the principal σ factor in S. coelicolor and M. tuberculosis (σHrdB and σA), but not from promoters dependent on alternative σ factors. Although RbpASc was originally discovered as a component of RNAP holoenzyme, and it is demonstrated here to be present at transcription initiation complexes in S. coelicolor, we found that RbpASc can also form a complex with the free principal σ factor. We failed to detect RbpASc interactions with selected Group 3 (σB and σWhiG) and Group 4 (σE and σR) σ factors in S. coelicolor, although we found that RbpASc and RbpAMt can bind to the Group 2 σ factors, σHrdA in S. coelicolor and σB in M. tuberculosis, respectively. In S. coelicolor, the three Group 2 σ factors (σHrdA, σHrdC and σHrdD) are collectively non-essential (52), as is σB in M. tuberculosis (53), which implies that the interaction with, and activation of, the essential primary σ factor underlies the critical importance of RbpA for growth. Consistent with this, an RbpASc mutant that failed to bind σHrdB (RbpAR89A/R90A) did not stimulate transcription in vitro and could not complement an S. coelicolor rbpA null mutant. Given that σHrdB–RbpASc complexes are stably maintained during gel filtration and Ni-affinity chromatography, we speculate that the two proteins are able to form an initial binary complex before the formation of holoenzyme. One role of RbpA might, therefore, be to act in a chaperone-like fashion to remodel σ for optimal binding to core RNAP, and thereby be an integral component of the ‘σ cycle’ in actinobacteria. This is consistent with the observation that RbpAMt stabilizes M. tuberculosis σA–RNAP holoenzyme, but it argues for a more direct role rather than a proposed allosteric effect through changing the conformation of the β lobes (25). Such a role might account for the induction of rbpA during stress, as also proposed by others (25). Although rbpA is continually expressed in unstressed S. coelicolor cultures, consistent with its general importance for growth (23), transcription is also induced in response to disulphide stress, directed by the alternative ECF σ factor σR (23). In M. tuberculosis, the rbpA gene is also induced by stress through the σR orthologue, σH (54). The induction of rbpA might, therefore, ensure that the increased cellular levels of free alternative σ factors during stress do not adversely affect σHrdB/σA-dependent ‘house keeping’ gene expression, and it may also help the cell to reset the normal state once the stress has been resolved. Interestingly, it was recently discovered that the gene that encodes σHrdB is also controlled by two ECF σ factors: σShbA during normal growth and σR during disulphide stress, which would also help to maintain σHrdB activity when stress-induced alternative σ factors accumulate in the cell (55,56). Furthermore, the Group 2 σ factors σHrdD in S. coelicolor and σB in M. tuberculosis are induced by stress (23,57), which might also contribute to maintaining the ‘house keeping’ duties of RNAP during stress.
Although RbpA might play an important role in promoting the formation of fully active σHrdB/σA holoenzyme, we show that RbpA is also poised to play additional roles in the subsequent stages of transcription initiation, such as promoter binding, and the formation and stability of open complexes. Our ChIP–qPCR data suggest that RbpASc is present at RNAP–promoter complexes in vivo that have been trapped using rifampicin. As rifampicin does not seem to influence promoter binding or open complex formation, but rather blocks transcription when the nascent transcript is ∼3 nt in length (58), RbpA might play a role in any of the preceding stages of initiation. We mapped the primary binding site of RbpASc to the σ2 domain of σHrdB (conserved regions 1.2 and 2.1–2.4), although we cannot rule out additional weaker or transient contacts elsewhere on the protein. The σ2 domain has several key roles in transcription initiation. As well as forming the largest interface between σ and RNAP (10), it confers promoter specificity for the −10 element and provides aromatic side chains that nucleate and maintain the strand-separated state during the formation of the open complex (6,59). Furthermore, variation in the sequence of the discriminator element that lies between the −10 element and the +1 transcription start point can influence stabilizing interactions with σ region 1.2, which is exploited by factors such as ppGpp that further destabilize open complexes (60). An understanding of the mechanism of action of RbpA is an important future goal.
In a similar manner to RbpA, the Crl protein of enteric bacteria interacts with the σ2 domain of the stationary phase σ factor σS and stimulates holoenzyme formation, thereby contributing to the ability of σS to compete with σ70 during stationary phase or under stress, despite its lower abundance (19,21). Crl seems to be specific for σS, and although crl mutants were originally discovered by their defect in curli fimbriae formation, they are in fact defective in the expression of a large subset of σS-dependent genes. Nonetheless, it was found that Crl also improved the activity of σ70–RNAP when mixed with σ70 before formation of holoenzyme (18), suggesting that Crl might also play a non-essential role in formation of the vegetative holoenzyme. Although Crl and RbpA are not obviously related in protein sequence, the two proteins might play an analogous role in transcription initiation, for example, through modulating the interaction of σ factors with RNAP and promoting the formation and activation of certain holoenzyme complexes.
How do we reconcile our observations that RbpA interacts with the σ subunit with previous proposals that RbpAMt principally interacts with the β-subunit of RNAP (25,26)? First, it should be noted that previous studies involved attaching chemical probes to the single cysteine residue in RbpAMt located in the RCD, which does not interact with σ. Moreover, the cysteine is likely to be important in proper folding of the RCD (Figure 5), which might have caused anomalous results, although evidence was provided that the labelled proteins were functional. Alternatively, and perhaps more likely, RbpA might interact with both σ and β, through distinct regions of RbpA. For example, although the σ2 and the β sandwich-barrel hybrid motif are distantly located in the holoenzyme structure, the disordered regions of RbpA would have significant conformational freedom to bridge between these regions. In the structurally related ribosomal protein L27, the N-terminal 1–19 residues are unstructured in its free form, but extend to a distance of >35 Å from the core domain when assembled with the ribonucleoprotein complex.
The RCD domains of S. coelicolor and M. tuberculosis have virtually identical structures, but differ in their ability to bind zinc. This correlates with the replacement of three of the likely zinc ligands in RbpASc with hydrophilic residues in RbpAMt that stabilize the domain through a hydrogen bond network. As the zinc-coordinating ligands are only absent from the Corynebacterium–Mycobacterium–Nocardia (CMN) group of actinobacteria, but are present in certain deeply rooted actinobacteria, such as Bifidobacterium sp. (61), it is likely that zinc-free RbpA evolved soon after the emergence of the CMN group. The RbpA proteins, therefore, present an example of how the evolution of a structural domain can contribute to a reduction in zinc requirements, which has also been described for alternative zinc-free ribosomal proteins (62,63). However, it is unlikely that this adaptation is linked to pathogenesis because the majority of CMN group comprises free-living non-pathogenic strains. Although mutations of the three zinc-coordinating cysteine residues seemed to inactivate or destabilize RbpASc, the RCD domain does not seem to be required for σ interaction. BACTH analysis and alanine scanning mutagenesis of RbpASc revealed that the C-terminal predicted helical region was required for this interaction, and that the highly conserved amino acid residues R89 and R90 play a critical role, although it is not yet clear whether they contact σ directly, or whether their mutation disrupts the overall conformation of this region. Although the C-terminal region is predicted to be helical, NMR analysis of the full-length RbpA proteins from both S. coelicolor and M. tuberculosis suggested that this region in each protein is unstructured, and despite extensive attempts to prevent it, it is readily proteolysed to the RCD during storage. The binding to σ, therefore, likely promotes folding of the RbpA C-terminal region, thereby stabilizing the complex. Clearly, future structural characterization of RbpA either in a binary complex with σ or in higher order complexes with RNAP will be highly valuable in understanding further mechanistic aspects of the function of this protein. This is likely to be of particular importance in the development of new approaches to target the process of transcription in M. tuberculosis and other pathogenic actinobacteria.
ACCESSION NUMBERS
Atomic coordinates for RbpASc and RbpAMt have been deposited with the Protein Data Bank under accession codes 2m6o and 2m6p, respectively. NMR chemical shifts for RbpASc and RbpAMt have been deposited with the Biological Magnetic Resonance DataBank under accession codes 19148 and 19149, respectively.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1 and 2 and Supplementary References [64–73].
ACKNOWLEDGEMENTS
The authors are grateful to Chris Dadswell, Sophie Buitenhuis and Yichao Zhou for contributions to experiments.
FUNDING
A.T.-S. B.L. and R.J.L. were funded by BBSRC grant [BB/I003045 to M.S.P. and S.J.M.]; P.D. was funded by a BBSRC grant [BB/D018293/1 to M.S.P.]; A.T.-S. was funded by the University of Sussex and the Overseas Research Students Awards Scheme. Funding for open access charge: BBSRC grant [BB/1003045].
Conflict of interest statement. None declared.
REFERENCES
- 1.WHO. Global Tuberculosis Report. Geneva: WHO Press; 2012. [Google Scholar]
- 2.Hu H, Zhang Q, Ochi K. Activation of antibiotic biosynthesis by specified mutations in the rpoB gene (encoding the RNA polymerase beta subunit) of Streptomyces lividans. J. Bacteriol. 2002;184:3984–3991. doi: 10.1128/JB.184.14.3984-3991.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhuo Y, Zhang W, Chen D, Gao H, Tao J, Liu M, Gou Z, Zhou X, Ye BC, Zhang Q, et al. Reverse biological engineering of hrdB to enhance the production of avermectins in an industrial strain of Streptomyces avermitilis. Proc. Natl Acad. Sci. USA. 2010;107:11250–11254. doi: 10.1073/pnas.1006085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saecker RM, Record MT, Dehaseth PL. Mechanism of bacterial transcription initiation: RNA polymerase—promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J. Mol. Biol. 2011;412:754–771. doi: 10.1016/j.jmb.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raffaelle M, Kanin EI, Vogt J, Burgess RR, Ansari AZ. Holoenzyme switching and stochastic release of sigma factors from RNA polymerase in vivo. Mol. Cell. 2005;20:357–366. doi: 10.1016/j.molcel.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 6.Feklistov A, Darst SA. Structural basis for promoter-10 element recognition by the bacterial RNA polymerase σ subunit. Cell. 2011;147:1257–1269. doi: 10.1016/j.cell.2011.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burgess RR, Travers AA, Dunn JJ, Bautz EK. Factor stimulating transcription by RNA polymerase. Nature. 1969;221:43–46. doi: 10.1038/221043a0. [DOI] [PubMed] [Google Scholar]
- 8.Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- 9.Young BA, Gruber TM, Gross CA. Views of transcription initiation. Cell. 2002;109:417–420. doi: 10.1016/s0092-8674(02)00752-3. [DOI] [PubMed] [Google Scholar]
- 10.Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 11.Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 12.Schwartz EC, Shekhtman A, Dutta K, Pratt MR, Cowburn D, Darst S, Muir TW. A full-length group 1 bacterial sigma factor adopts a compact structure incompatible with DNA binding. Chem. Biol. 2008;15:1091–1103. doi: 10.1016/j.chembiol.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paget MS, Helmann JD. The σ70 family of sigma factors. Genome Biol. 2003;4:203. doi: 10.1186/gb-2003-4-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Österberg S, del Peso-Santos T, Shingler V. Regulation of alternative sigma factor use. Annu. Rev. Microbiol. 2011;65:37–55. doi: 10.1146/annurev.micro.112408.134219. [DOI] [PubMed] [Google Scholar]
- 15.Pratt LA, Silhavy TJ. Crl stimulates RpoS activity during stationary phase. Mol. Microbiol. 1998;29:1225–1236. doi: 10.1046/j.1365-2958.1998.01007.x. [DOI] [PubMed] [Google Scholar]
- 16.Bougdour A, Lelong C, Geiselmann J. Crl, a low temperature-induced protein in Escherichia coli that binds directly to the stationary phase sigma subunit of RNA polymerase. J. Biol. Chem. 2004;279:19540–19550. doi: 10.1074/jbc.M314145200. [DOI] [PubMed] [Google Scholar]
- 17.England P, Westblade LF, Karimova G, Robbe-Saule V, Norel F, Kolb A. Binding of the unorthodox transcription activator, Crl, to the components of the transcription machinery. J. Biol. Chem. 2008;283:33455–33464. doi: 10.1074/jbc.M807380200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaal T, Mandel MJ, Silhavy TJ, Gourse RL. Crl facilitates RNA polymerase holoenzyme formation. J. Bacteriol. 2006;188:7966–7970. doi: 10.1128/JB.01266-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monteil V, Kolb A, Mayer C, Hoos S, England P, Norel F. Crl binds to domain 2 of σS and confers a competitive advantage on a natural rpoS mutant of Salmonella enterica serovar Typhi. J. Bacteriol. 2010;192:6401–6410. doi: 10.1128/JB.00801-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monteil V, Kolb A, D’Alayer J, Beguin P, Norel F. Identification of conserved amino acid residues of the Salmonella σS chaperone Crl involved in Crl-σS interactions. J. Bacteriol. 2010;192:1075–1087. doi: 10.1128/JB.01197-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Typas A, Barembruch C, Possling A, Hengge R. Stationary phase reorganisation of the Escherichia coli transcription machinery by Crl protein, a fine-tuner of sigmas activity and levels. EMBO J. 2007;26:1569–1578. doi: 10.1038/sj.emboj.7601629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newell KV, Thomas DP, Brekasis D, Paget MS. The RNA polymerase-binding protein RbpA confers basal levels of rifampicin resistance on Streptomyces coelicolor. Mol. Microbiol. 2006;60:687–696. doi: 10.1111/j.1365-2958.2006.05116.x. [DOI] [PubMed] [Google Scholar]
- 23.Paget MS, Molle V, Cohen G, Aharonowitz Y, Buttner MJ. Defining the disulphide stress response in Streptomyces coelicolor A3(2): identification of the σR regulon. Mol. Microbiol. 2001;42:1007–1020. doi: 10.1046/j.1365-2958.2001.02675.x. [DOI] [PubMed] [Google Scholar]
- 24.Forti F, Mauri V, Dehò G, Ghisotti D. Isolation of conditional expression mutants in Mycobacterium tuberculosis by transposon mutagenesis. Tuberculosis (Edinb.) 2011;91:569–578. doi: 10.1016/j.tube.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Hu Y, Morichaud Z, Chen S, Leonetti JP, Brodolin K. Mycobacterium tuberculosis RbpA protein is a new type of transcriptional activator that stabilizes the σA-containing RNA polymerase holoenzyme. Nucleic Acids Res. 2012;40:6547–6557. doi: 10.1093/nar/gks346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dey A, Verma AK, Chatterji D. Role of an RNA polymerase interacting protein, MsRbpA, from Mycobacterium smegmatis in phenotypic tolerance to rifampicin. Microbiology. 2010;156:873–883. doi: 10.1099/mic.0.033670-0. [DOI] [PubMed] [Google Scholar]
- 27.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich: The John Innes Foundation; 2000. [Google Scholar]
- 28.Studier FW. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 29.Sivashanmugam A, Murray V, Cui C, Zhang Y, Wang J, Li Q. Practical protocols for production of very high yields of recombinant proteins using Escherichia coli. Protein Sci. 2009;18:936–948. doi: 10.1002/pro.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu LM. Monitoring abortive initiation. Methods. 2009;47:25–36. doi: 10.1016/j.ymeth.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X, Bremer H. Control of the Escherichia coli rrnB P1 promoter strength by ppGpp. J. Biol. Chem. 1995;270:11181–11189. doi: 10.1074/jbc.270.19.11181. [DOI] [PubMed] [Google Scholar]
- 32.Bucca G, Laing E, Mersinias V, Allenby N, Hurd D, Holdstock J, Brenner V, Harrison M, Smith CP. Development and application of versatile high density microarrays for genome-wide analysis of Streptomyces coelicolor: characterization of the HspR regulon. Genome Biol. 2009;10:R5. doi: 10.1186/gb-2009-10-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McClellan MJ, Khasnis S, Wood CD, Palermo RD, Schlick SN, Kanhere AS, Jenner RG, West MJ. Downregulation of integrin receptor-signaling genes by Epstein-Barr virus EBNA 3C via promoter-proximal and -distal binding elements. J. Virol. 2012;86:5165–5178. doi: 10.1128/JVI.07161-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nucl. Magn. Reson. Spectrosc. 1999;34:93–158. [Google Scholar]
- 35.Marchant J, Sawmynaden K, Saouros S, Simpson P, Matthews S. Complete resonance assignment of the first and second apple domains of MIC4 from Toxoplasma gondii, using a new NMRView-based assignment aid. Biomol. NMR Assign. 2008;2:119–121. doi: 10.1007/s12104-008-9100-1. [DOI] [PubMed] [Google Scholar]
- 36.Rieping W, Habeck M, Bardiaux B, Bernard A, Malliavin T, Nilges M. ARIA2: automated NOE assignment and data integration in NMR structure calculation. Bioinformatics. 2007;23:381–382. doi: 10.1093/bioinformatics/btl589. [DOI] [PubMed] [Google Scholar]
- 37.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 38.Viollier PH, Nguyen KT, Minas W, Folcher M, Dale GE, Thompson CJ. Roles of aconitase in growth, metabolism, and morphological differentiation of Streptomyces coelicolor. J. Bacteriol. 2001;183:3193–3203. doi: 10.1128/JB.183.10.3193-3203.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Wezel GP, Takano E, Vijgenboom E, Bosch L, Bibb MJ. The tuf3 gene of Streptomyces coelicolor A3(2) encodes an inessential elongation factor Tu that is apparently subject to positive stringent control. Microbiology. 1995;141:2519–2528. doi: 10.1099/13500872-141-10-2519. [DOI] [PubMed] [Google Scholar]
- 40.Chakraburtty R, White J, Takano E, Bibb M. Cloning, characterization and disruption of a (p)ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) Mol. Microbiol. 1996;19:357–368. doi: 10.1046/j.1365-2958.1996.390919.x. [DOI] [PubMed] [Google Scholar]
- 41.Vockenhuber MP, Sharma CM, Statt MG, Schmidt D, Xu Z, Dietrich S, Liesegang H, Mathews DH, Suess B. Deep sequencing-based identification of small non-coding RNAs in Streptomyces coelicolor. RNA Biol. 2011;8:468–477. doi: 10.4161/rna.8.3.14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bergendahl V, Thompson NE, Foley KM, Olson BM, Burgess RR. A cross-reactive polyol-responsive monoclonal antibody useful for isolation of core RNA polymerase from many bacterial species. Protein Expr. Purif. 2003;31:155–160. doi: 10.1016/s1046-5928(03)00145-1. [DOI] [PubMed] [Google Scholar]
- 43.Herring CD, Raffaelle M, Allen TE, Kanin EI, Landick R, Ansari AZ, Palsson B. Immobilization of Escherichia coli RNA polymerase and location of binding sites by use of chromatin immunoprecipitation and microarrays. J. Bacteriol. 2005;187:6166–6174. doi: 10.1128/JB.187.17.6166-6174.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl Acad. Sci. USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krishna SS, Majumdar I, Grishin NV. Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 2003;31:532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fletcher RJ, Bishop BE, Leon RP, Sclafani RA, Ogata CM, Chen XS. The structure and function of MCM from archaeal M. Thermoautotrophicum. Nat. Struct. Biol. 2003;10:160–167. doi: 10.1038/nsb893. [DOI] [PubMed] [Google Scholar]
- 47.Wang H, Takemoto CH, Murayama K, Sakai H, Tatsuguchi A, Terada T, Shirouzu M, Kuramitsu S, Yokoyama S. Crystal structure of ribosomal protein L27 from Thermus thermophilus HB8. Protein Sci. 2004;13:2806–2810. doi: 10.1110/ps.04864904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mantsyzov AB, Ivanova EV, Birdsall B, Alkalaeva EZ, Kryuchkova PN, Kelly G, Frolova LY, Polshakov VI. NMR solution structure and function of the C-terminal domain of eukaryotic class 1 polypeptide chain release factor. FEBS J. 2010;277:2611–2627. doi: 10.1111/j.1742-4658.2010.07672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 50.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 51.Campbell EA, Muzzin O, Chlenov M, Sun JL, Olson CA, Weinman O, Trester-Zedlitz ML, Darst SA. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol. Cell. 2002;9:527–539. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- 52.Buttner MJ, Chater KF, Bibb MJ. Cloning, disruption, and transcriptional analysis of three RNA polymerase sigma factor genes of Streptomyces coelicolor A3(2) J. Bacteriol. 1990;172:3367–3378. doi: 10.1128/jb.172.6.3367-3378.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fontán PA, Voskuil MI, Gomez M, Tan D, Pardini M, Manganelli R, Fattorini L, Schoolnik GK, Smith I. The Mycobacterium tuberculosis sigma factor σB is required for full response to cell envelope stress and hypoxia in vitro, but it is dispensable for in vivo growth. J. Bacteriol. 2009;191:5628–5633. doi: 10.1128/JB.00510-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manganelli R, Voskuil MI, Schoolnik GK, Dubnau E, Gomez M, Smith I. Role of the extracytoplasmic–function sigma factor σH in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 2002;45:365–374. doi: 10.1046/j.1365-2958.2002.03005.x. [DOI] [PubMed] [Google Scholar]
- 55.Otani H, Higo A, Nanamiya H, Horinouchi S, Ohnishi Y. An alternative sigma factor governs the principal sigma factor in Streptomyces griseus. Mol. Microbiol. 2013;87:1223–1236. doi: 10.1111/mmi.12160. [DOI] [PubMed] [Google Scholar]
- 56.Kim MS, Dufour YS, Yoo JS, Cho YB, Park JH, Nam GB, Kim HM, Lee KL, Donohue TJ, Roe JH. Conservation of thiol-oxidative stress responses regulated by SigR orthologues in actinomycetes. Mol. Microbiol. 2012;85:326–344. doi: 10.1111/j.1365-2958.2012.08115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Manganelli R, Voskuil MI, Schoolnik GK, Smith I. The Mycobacterium tuberculosis ECF sigma factor σE: role in global gene expression and survival in macrophages. Mol. Microbiol. 2001;41:423–437. doi: 10.1046/j.1365-2958.2001.02525.x. [DOI] [PubMed] [Google Scholar]
- 58.Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst SA. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell. 2001;104:901–912. doi: 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y, Feng Y, Chatterjee S, Tuske S, Ho MX, Arnold E, Ebright RH. Structural basis of transcription initiation. Science. 2012;338:1076–1080. doi: 10.1126/science.1227786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haugen SP, Berkmen MB, Ross W, Gaal T, Ward C, Gourse RL. rRNA promoter regulation by nonoptimal binding of sigma region 1.2: an additional recognition element for RNA polymerase. Cell. 2006;125:1069–1082. doi: 10.1016/j.cell.2006.04.034. [DOI] [PubMed] [Google Scholar]
- 61.Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, van Sinderen D. Genomics of actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 2007;71:495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gabriel SE, Helmann JD. Contributions of Zur-controlled ribosomal proteins to growth under zinc starvation conditions. J. Bacteriol. 2009;191:6116–6122. doi: 10.1128/JB.00802-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Panina EM, Mironov AA, Gelfand MS. Comparative genomics of bacterial zinc regulons: enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc. Natl Acad. Sci. USA. 2003;100:9912–9917. doi: 10.1073/pnas.1733691100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kelemen GH, Plaskitt KA, Lewis CG, Findlay KC, Buttner MJ. Deletion of DNA lying close to the glkA locus induces ectopic sporulation in Streptomyces coelicolor A3(2) Mol. Microbiol. 1995;17:221–230. doi: 10.1111/j.1365-2958.1995.mmi_17020221.x. [DOI] [PubMed] [Google Scholar]
- 65.Babcock MJ, Buttner MJ, Keler CH, Clarke BR, Morris RA, Lewis CG, Brawner ME. Characterization of the rpoC gene of Streptomyces coelicolor A3(2) and its use to develop a simple and rapid method for the purification of RNA polymerase. Gene. 1997;196:31–42. doi: 10.1016/s0378-1119(97)00179-0. [DOI] [PubMed] [Google Scholar]
- 66.Paget MS, Chamberlin L, Atrih A, Foster SJ, Buttner MJ. Evidence that the extracytoplasmic function sigma factor sigmaE is required for normal cell wall structure in Streptomyces coelicolor A3(2) J. Bacteriol. 1999;181:204–211. doi: 10.1128/jb.181.1.204-211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 68.Paget MS, Hintermann G, Smith CP. Construction and application of streptomycete promoter probe vectors which employ the Streptomyces glaucescens tyrosinase-encoding gene as reporter. Gene. 1994;146:105–110. doi: 10.1016/0378-1119(94)90842-7. [DOI] [PubMed] [Google Scholar]
- 69.Alting-Mees MA, Short JM. pBluescript II: gene mapping vectors. Nucleic Acids Res. 1989;17:9494. doi: 10.1093/nar/17.22.9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Karimova G, Ullmann A, Ladant D. Protein-protein interaction between Bacillus stearothermophilus tyrosyl-tRNA synthetase subdomains revealed by a bacterial two-hybrid system. J. Mol. Microbiol. Biotechnol. 2001;3:73–82. [PubMed] [Google Scholar]
- 71.Bierman M, Logan R, O’Brien K, Seno ET, Rao RN, Schoner BE. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 72.O’Connor TJ, Kanellis P, Nodwell JR. The ramC gene is required for morphogenesis in Streptomyces coelicolor and expressed in a cell type-specific manner under the direct control of RamR. Mol. Microbiol. 2002;45:45–57. doi: 10.1046/j.1365-2958.2002.03004.x. [DOI] [PubMed] [Google Scholar]
- 73.Huang J, Shi J, Molle V, Sohlberg B, Weaver D, Bibb MJ, Karoonuthaisiri N, Lih CJ, Kao CM, Buttner MJ, et al. Crossregulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor. Mol. Microbiol. 2005;58:1276–1287. doi: 10.1111/j.1365-2958.2005.04879.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.