Background: Numerous proteins are exported across membranes by the translocon SecYEG, a highly conserved complex.
Results: Multiple structural conformations and oligomeric states of SecYEG observed in lipid bilayers.
Conclusion: Cytoplasmic membrane-external segments of SecYEG that orchestrate translocon function are highly dynamic.
Significance: Direct visualization of disordered, flexible structures and oligomeric states in lipid bilayers provides a near-native vista of the translocon.
Keywords: Atomic Force Microscopy, Membrane Biophysics, Membrane Proteins, Protein Conformation, Protein Dynamics, Protein Export, Secretion, Structural Biology
Abstract
Purified SecYEG was reconstituted into liposomes and studied in near-native conditions using atomic force microscopy. These SecYEG proteoliposomes were active in translocation assays. Changes in the structure of SecYEG as a function of time were directly visualized. The dynamics observed were significant in magnitude (∼1–10 Å) and were attributed to the two large loops of SecY linking transmembrane helices 6–7 and 8–9. In addition, we identified a distribution between monomers and dimers of SecYEG as well as a smaller population of higher order oligomers. This work provides a new vista of the flexible and dynamic structure of SecYEG, an intricate and vital membrane protein.
Introduction
Proteins are highly dynamic. This is especially true of the proteins that orchestrate translocation of polypeptides across membranes. More than 30% of proteins in any organism are transported from the site of synthesis into or through a membrane. In Escherichia coli as well as in all archaea and eubacteria, the general secretory or Sec system is the major route of export (1–3). The pathway through the membrane, the translocon, is provided by SecYEG, a protein complex that is highly conserved, with homologs across the kingdoms of life. In the E. coli SecYEG complex (∼75 kDa) the channel for passing proteins is found in SecY (4), the largest subunit, with 10 transmembrane (TM)2 helical spans. SecY also contains a lateral gate allowing protein insertion into the membrane. SecE and SecG are two smaller subunits located about the periphery of SecY. Proteins that are to be exported are synthesized as precursors. In a dynamic process, the mechanistic details of which are poorly understood, the motor protein SecA binds SecYEG and provides energy from a cycle of binding and hydrolysis of ATP to drive precursors through the channel. SecA interacts with SecY, making large surface area contact (∼6800 Å2) with cytoplasmic loops spanning TM helices 6–7 and 8–9 of SecY (5). Genetic analyses have determined that mutations in these loops deleteriously affect translocation activity (6, 7). As in many protein-protein interactions, SecA-SecYEG binding involves disordered loop regions. In a framework dubbed “fly casting” (8), disordered and flexible regions can increase the capture radius of a specific binding site and hence boost binding rates. Despite their functional significance, measurement of flexible and disordered protein regions remains a significant experimental challenge (9).
The stoichiometry of the active translocon has been a matter of contention (10–15). It has been reported that SecYEG exists in vivo as dimers and tetramers, and in vitro assays have shown that dimers are active. However, because SecYEG monomers contain a channel, it is not clear whether assembly into specific quaternary structures is necessary for activity. Better structural information, especially in a native lipid environment, may help to shed light on this interesting question.
Originally developed as a characterization tool for the surface science community (16), the atomic force microscope (AFM) has emerged as an important instrument for characterizing biological macromolecules in membranes (17–19). An AFM consists of a nanoscale force probe (i.e. a tip) interacting with a sample. The instrument, which operates in physiological solution, has been successfully used to map flexible and disordered regions of soluble proteins (20) and membrane proteins in two-dimensional crystalline arrays (21–23). Real-time probing of individual proteins distinguishes atomic force microscopy from traditional structural biology techniques, which typically require ensembles or cryogenically preserved samples. Furthermore, an AFM is capable of resolving absolute distances down to atomic length scales on unlabeled proteins in membrane and can provide a dynamic picture of macromolecules at work.
Here, we report the first AFM measurements of the translocon SecYEG and demonstrate the power of the technique in visualizing the structure of SecYEG and its dynamics in near-native conditions. Height analyses of the reconstituted SecYEG complex protruding from the bilayer allowed assignment of protein orientation. Diverse structural dynamics were observed. Height fluctuations of the cytoplasmic surface of SecYEG ranged from ∼1–10 Å and occurred over multiple timescales, the fastest of which was <100 ms. Direct visualization of individual proteins revealed the presence of monomers, dimers, and higher order oligomers. Taken all together, our work presents a view of the protein conducting channel in near-native, non-crystalline conditions and represents a novel characterization of the structure and time-varying conformations of SecYEG segments outside of the lipid bilayer.
EXPERIMENTAL PROCEDURES
Protein Purification
SecYEG was purified from strain C43(DE3) (24) harboring a plasmid encoding secE with a His tag at the N terminus, secYC329S, C385S, secG (25). Cells were broken by passage through a French pressure cell (8000 psi), and the membranes were isolated by centrifugation and solubilized in dodecyl-β-maltoside. SecYEG was purified by chromatography using a HisTrap column (GE Healthcare) and stored at −80 °C in 20 mm Tris-Cl, pH 8, 0.3 m NaCl, 10% glycerol, 0.6 mm dodecyl-β-maltoside, 2 mm DTT.
Proteoliposome Preparation
Lipids (E. coli polar lipid extract, Avanti) in chloroform were blown dry with N2 and placed in a vacuum chamber overnight. A dry mechanical vacuum pump (XDS5, Edwards) was used to prevent backstreaming of oil, a potential contaminant. Dried lipids were suspended in 10 mm HEPES, pH 7.6, 30 mm KAc, 1 mm Mg(Ac)2. Unilamellar liposomes were prepared by extrusion through membranes (∼100-nm pore diameter, Liposofast, Avestin). To form proteoliposomes the liposomes were swelled, but not disrupted, using a ratio of detergent to lipids of 4.65 mm dodecyl-β-maltoside to 5 mm lipids (26). After swelling for 3 h at room temperature, SecYEG was added at 5 μm. Incubation was continued for 1 h at room temperature followed by addition of BioBeads SM-2 (Bio-Rad) to remove the detergent. The proteoliposomes were isolated by centrifugation at 436,000 × g for 20 min, at 4 °C in a TL100.1 rotor (Beckman). The pellet was suspended in the same buffer and centrifuged again as described above. The final pellet was suspended to give a concentration of ∼8 mm lipid and 8 μm SecY. The suspension was stored at −80 °C.
Translocation Assay
Translocation of 1 μm radioactive proOmpA (precursor of outer membrane protein OmpA), labeled with a 14C-labeled l-amino acid mixture, into proteoliposomes was carried out at 30 °C under conditions of limiting SecY (1 μm) with SecA (1.2 μm dimer), and SecB (1 μm tetramer), as described (27) with the following modifications. An ATP-regenerating system consisting of 7.5 mm phosphocreatine and 37 mg/ml of creatine phosphokinase was present in the reaction. Proteinase K (5 units/ml, 15 min on ice) was used for degradation of untranslocated proteins and digestion terminated by trichloroacetic acid precipitation. The washed precipitate was dissolved in gel sample buffer containing DTT (10 mm) for analysis by electrophoresis. The radioactivity in the protein bands in the gels of the translocation assays was measured using a Fujifilm FLA 3000 phosphorimager in the linear range of its response, and the molarity of the full-length protected precursors was estimated by comparison with samples taken from the same reaction mix, which had not been subjected to proteinase K digestion but had been applied to the same gel.
Atomic Force Microscope Imaging
Proteoliposome stock solution was diluted to 80 nm SecYEG, 80 μm lipid in imaging buffer (100 mm KAc, 5 mm MgAc2, 10 mm HEPES, pH 7.6), immediately deposited on a freshly cleaved muscovite mica surface (V-1 grade, Structure Probe, Inc.), and incubated for ∼20 min, over which time the proteoliposomes were allowed to deposit on the surface through vesicle fusion (28). The surface was then rinsed three times with ∼100 μl of imaging buffer. AFM images were acquired in imaging buffer in tapping mode using a commercial instrument (Cypher, Asylum Research). Biolever mini tips (BL-AC40TS, Olympus) with measured spring constants ∼0.06 n/m were used. Images were recorded at ∼30 °C with an estimated tip-sample force < 100 pN, deduced by comparing the free space tapping amplitude (∼5 nm) to the imaging set point amplitude. Under such conditions, minimal protein distortion is expected (29, 30).
Image Analysis
Prior to analysis, images were processed with standard AFM techniques: flattening to minimize background tilt and filtering to reduce the influence of noise. A flood mask was then applied to isolate protein protrusions with a manually set threshold (∼3 Å above the planar lipid surface). This introduced a small systemic height error that was subsequently corrected. Software (Igor Pro) was used to extract the volume and maximum height for each protein protrusion. Features exhibiting maximum heights < 5 Å were excluded from analysis. The base height used for the maximum feature height and volume calculations was taken to be the average height of the pixels at the perimeter of the feature. Finally, data were compiled into histograms for display.
Tip Deconvolution
AFM images convolve tip geometry with that of the sample. We implemented tip deconvolution software (31, 32) to more accurately deduce protein protrusion volumes. The program used blind tip estimation to determine the bluntest tip that could resolve the image. The generated tip geometry was then removed from the image, outputting a deconvolved image that more closely approximated the sample topography (supplemental Fig. 1).
RESULTS
Activity and Orientation of SecYEG
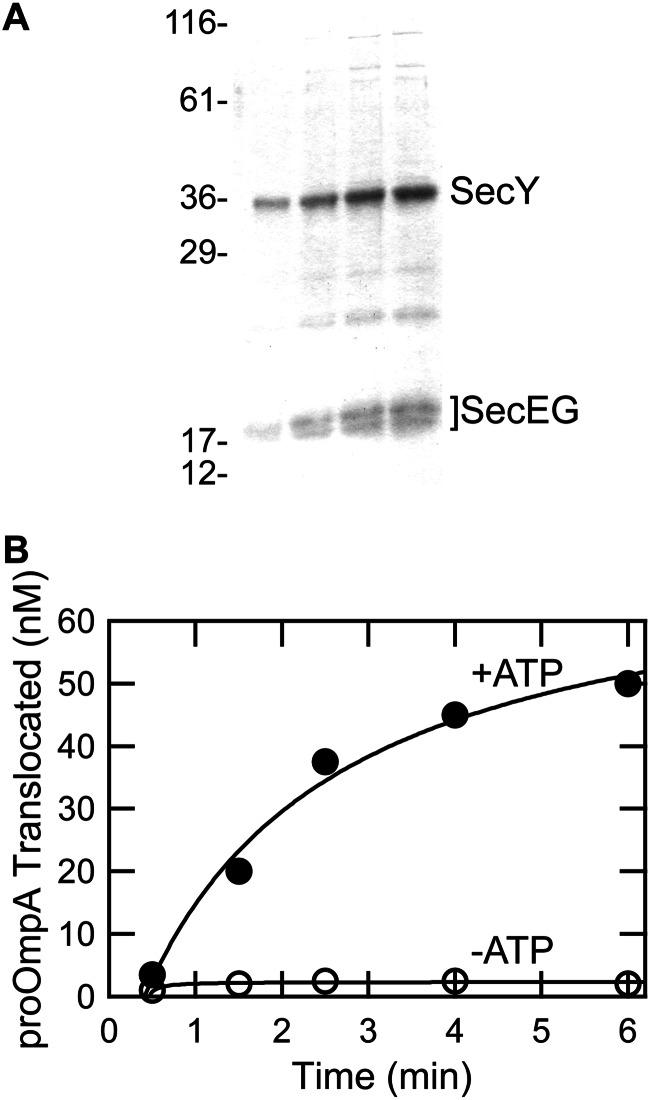
Purified SecYEG was reconstituted into liposomes and shown to be active for translocation. Fig. 1 shows an example of one preparation. Upon addition of SecA, SecB (a chaperone that keeps the precursor unfolded; Ref. 33), and proOmpA (precursor of outer membrane protein OmpA) in the presence of ATP, the precursor is translocated as assessed by protection from proteinase K. The protection is dependent on the presence of ATP (Fig. 1B, compare filled and unfilled circles), indicating that it represents active translocation.
FIGURE 1.
Activity of SecYEG reconstituted into liposomes. A, SDS-PAGE of SecYEG proteoliposomes at 0.15, 0.3, 0.45, and 0.56 μg of SecY. B, translocation of radiolabeled precursor of outer membrane protein OmpA was assayed in vitro with (●) and without ATP (○) by protection from proteinase K. The lines are fits to the data.
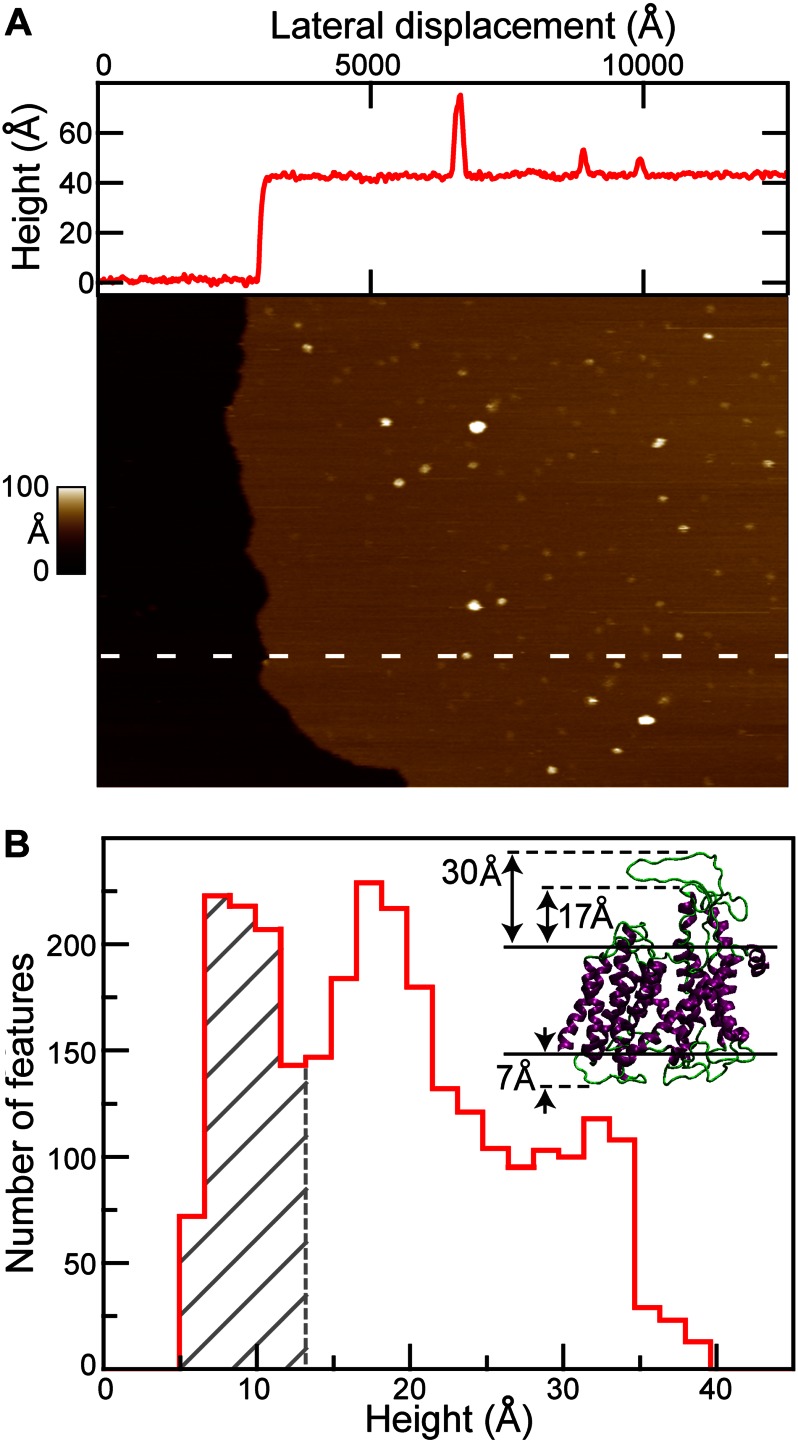
The SecYEG proteoliposomes were deposited onto mica surfaces for examination by AFM. The presence of a lipid bilayer was confirmed by observation of the characteristic ∼40-Å thickness (34, 35) of a bilayer at the edge of a membrane patch (Fig. 2A and line scan, red). Alternatively, pits with a depth of ∼40 Å were used. Proteoliposomes containing SecYEG displayed numerous punctuate features (Fig. 2A). The line scan profile through the image shown indicated the presence of three SecYEG protrusions above the lipid bilayer. In contrast, images of liposomes assembled without proteins were essentially featureless, exhibiting <1 protrusion per μm2 (supplemental Fig. 2).
FIGURE 2.
AFM imaging yields protein orientation. A, large-scale (1250 × 900 nm2) AFM image of SecYEG in lipid bilayer with a cross-section profile (top, red). The location of the profile is indicated in the image (white dashed line). Both cytoplasmic (bright) and periplasmic (less bright) features can be identified protruding the lipid bilayer surface (light brown) in the image. The vertical scale spans 0 to 100 Å. B, histogram of the maximum height of individual SecYEG features (n = 2766; bin size = 1.7 Å). Gray hatched region represents periplasmic SecYEG with one major peak at ∼7 Å. In contrast, the cytoplasmic side of SecYEG exhibited two peaks in its height distribution (∼17 and ∼32 Å), with a significant population of heights in between them. Inset, crystal structure of the SecYEG complex indicating asymmetry. The maximum height of the periplasmic protrusion is ∼7 Å. In contrast, it is ∼30 Å on the cytoplasmic side. The ends of the 8–9 helices extend ∼17 Å above the lipid bilayer; the 6–7 loop extends ∼13 Å above these helices. Membrane boundaries are indicated (solid black lines).
SecYEG displays topographic asymmetry between the maximum heights of the protrusion on the cytoplasmic face compared with the periplasmic face of the membrane. Analyses of several thousand protrusions (features) from different areas of lipids using software that determines the maximal height of each feature above the lipid bilayer are summarized in a histogram (Fig. 2B). Heights in excess of 40 Å were rare (<10% of total population) and were excluded from analysis. Comparison of the heights of the three peaks at 7, 17, and 32 Å with the crystal structure of SecYEG from Thermotoga maritima (Fig. 2B) allowed us to assign two of the three peaks. We chose T. maritima as a reference because it is homologous (>40% identity) to E. coli (for which there is no high resolution structure available). From this comparison, we assigned the region containing the 7 Å peak (Fig. 2B, gray hatched bars) to the periplasmic loops (7 Å maximum in the structure) with a cutoff for this region based on the clear minimum found at ∼13 Å in the height histogram.
The loops connecting TM 6–7 and 8–9 of T. maritima SecY are similar in length to those of E. coli. The close agreement between the 32-Å peak in the histogram with the 30-Å height of the loop connecting TM 6–7 in the crystal structure allows assignment of the peak to a similar loop configuration. It should be noted that the loop in the crystal structure was stabilized in an extended configuration by interaction with the co-crystallized SecA, which was removed from the figure for clarity.
The third peak observed at 17 Å is attributed to conformations of the cytoplasmic face of the translocon wherein the 6–7 and 8–9 loops are collapsed downwards, toward the membrane. Such a collapse would expose the AFM probe to the end of TM helices 8 and 9, which protrude ∼17 Å above the bilayer. The relative population of the cytoplasmic feature heights is biased toward the compact structure: 57% of cytoplasmic features exhibited a maximum height ∼17 Å; 43% achieve heights between 22 and 38 Å. This continuum of populated heights is likely to result from conformational dynamics.
Structural Dynamics
The unstructured loops at the cytoplasmic face of SecYEG are involved in critical interactions with SecA, which is the ATPase that enables transport. We addressed conformational dynamics of these loops on a time scale accessible by AFM scanning. Height analyses indicated that two conformations of the loops are preferred; however, a large population of molecules exhibited an intermediate range of heights. Data for the height histogram shown in Fig. 2 were extracted from a set of images acquired over multiple months. Thus, to probe dynamics, we acquired a series of images to track 128 SecYEG features over time. Four representative features from this data set are presented (Fig. 3, A–D) and enumerated by image frame number i–v, which is a proxy for time.
FIGURE 3.
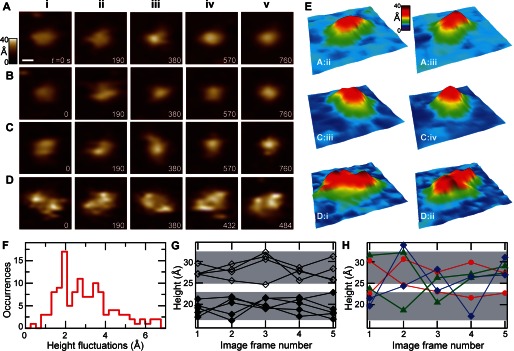
Dynamics of SecYEG. Successive AFM images of individual cytoplasmic SecYEG features are displayed. A, monomer with dynamic height (prominent in A, panel iii). B, monomer exhibiting a change in lateral structure (prominent in B, panel ii). C, monomer with significant conformational changes in both height and shape. D, dimer exhibiting changes in height, shape, and rotational orientation. The scale bar shown in A is 50 Å and applies to A–D. Elapsed time in seconds is indicated in each image frame. E, three-dimensional renderings are shown of selected images: A, panel ii and iii; C, panels iii and iv; and D, panels i and ii. The orientation of these renderings (fixed for each pair) was chosen to highlight prominent structural changes. F, histogram of the standard deviation in maximum height calculated from individual features (n = 128) in successive images. A population (37%) of features fluctuated with σ = 2 Å or less. Other peaks are observed with σ ∼ 2.7 and 3.5 Å, respectively, and the population extends out beyond σ ∼ 5 Å. G, height trajectories of individual SecYEG in five successive images are shown corresponding to σ = 2 Å features (n = 9, data are shown as black diamonds, lines between points guide the eye). Two clusters of trajectories (filled diamonds, low; unfilled diamonds, high) were delimitated (gray shaded regions). H, trajectories of representative features exhibiting height fluctuations σ > 2 Å, in particular: σ = 2.7 Å (red), σ = 3.5 Å (purple), and σ = 5 Å (blue).
Rich and varied dynamic structural behavior was observed. Examples of this behavior included a SecYEG monomer with significant changes in maximum height h (Fig. 3A, Δh = 7 Å, prominent in panel iii); a monomer with a significant lateral conformational change (Fig. 3B, prominent in panel ii); a monomer with changes in both height and shape (Fig. 3C, Δh = 6 Å); and finally, a SecYEG dimer with significant changes in conformation and rotational orientation in all images (Fig. 3D) from panels i to v. All data presented here have the same lateral (250 × 250 Å2) and vertical (0–40 Å) scales. For the purpose of visualization, pseudo-three-dimensional renderings of selected images are presented to highlight the dynamics (Fig. 3E, identified as row:column).
Temporal dynamics were further quantified by deducing the magnitude of the observed height fluctuations of individual features. We calculated the S.D. σ of the maximum height over time from individual cytoplasmic SecYEG features (n = 128, total temporal duration varied from 260 to 950 s). The histogram of height fluctuations (Fig. 3F) shows a peak at σ = 2 Å. Roughly one-third of the features were relatively quiescent, fluctuating at or below σ = 2 Å. However, the majority of the population (63%) exhibited height fluctuations exceeding σ = 2 Å. A broad distribution of fluctuations existed near the peaks at σ = 2.7 and 3.5 Å, and a small population (∼10%) extended out beyond σ = 4 Å.
Height trajectories, plots of feature height versus image frame number (i.e. time), were organized using the height fluctuation histogram as a guide. The elapsed time between frames varied from 52 to 190 s, which is very slow compared with the timescale of molecular fluctuations. Fig. 3G represents a subset of minority SecYEG trajectories that were relatively stable, exhibiting small amplitude height fluctuations (σ = 2 Å, n = 9). The data are plotted as points (black diamonds); the lines between them guide the eye. We delimitated two clusters of trajectories from this subset (gray shaded regions). The lower region (filled diamonds) is centered at a height of ∼18 Å, and the upper region (unfilled diamonds) is centered at ∼28 Å; the two regions encompass the first and second cytoplasmic peaks in the height histogram (Fig. 2B), respectively. Although the trajectories plotted here exhibit height fluctuations at or below 2 Å, it is possible that significantly greater excursions in height could be absent from this data due to the limited time resolution.
In contrast to the minimally fluctuating population, the majority of SecYEG features exhibited significantly greater (σ > 2 Å) height fluctuations. Representative σ > 2 Å height trajectories are presented (Fig. 3H) and color-coded by the magnitude of their S.D.: σ = 2.7 Å (red), σ = 3.5 Å (green), and σ = 5 Å (blue). Gray demarcated regions are carried over from Fig. 3G. All of these trajectories cross between gray regions; some cross once (red), some cross twice (green), and some cross three times (blue). This observation implies that collapsed loops can extend and vice versa, in a reversible fashion. We note that individual features within the same image sequence exhibited both positive and negative changes in heights, suggesting that the AFM imaging force was minimal and did not bias the observed dynamics.
In addition to height fluctuations, structural changes parallel to the membrane surface also occur. For example, the full width half-maximum of certain monomeric features (Fig. 3, B and C) changed by ∼10 Å from one image to the next, indicating a significant overall shape change of the protein.
We sought to place firmer bounds upon the time scale of the observed dynamics. As is typical in AFM, we recorded pairs of images, with the tip traveling left to right (trace) and right to left (retrace) over the same region of the membrane surface (neglecting protein diffusion and instrumental drift). Thus, trace and retrace line scan profiles from the same features, which are separated by ∼100 ms, could be compared, thereby increasing the temporal resolution >500-fold.
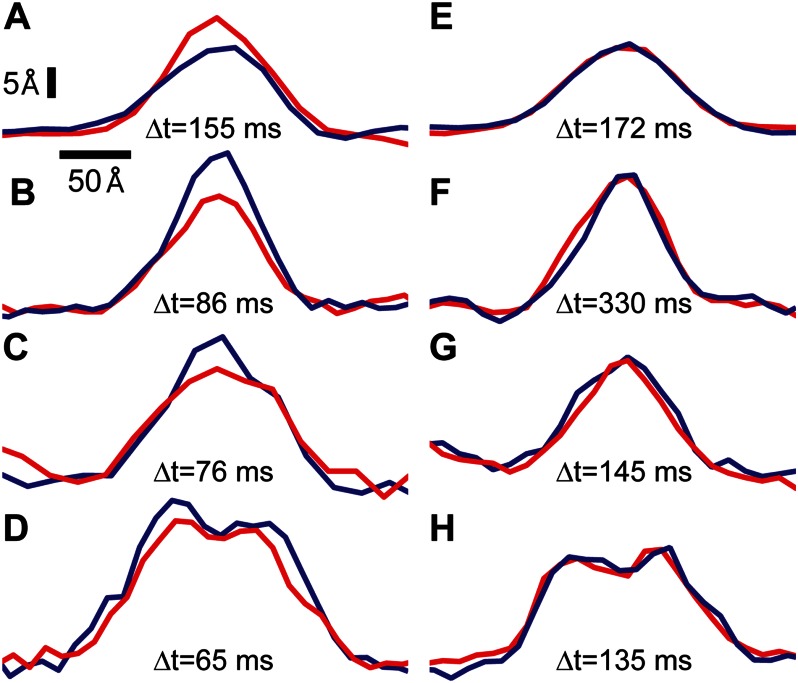
Pairs of line scan profiles of representative cytoplasmic loops are displayed in Fig. 4 (red, trace; blue, retrace). Features in the left hand column (Fig. 4, A–D) exhibited significant height differences in the time elapsed between the trace and retrace line scans. These structural changes occurred on time scales of ∼100 ms, the shortest of which was 65 ms. As a control, we also display representative features that were imaged under identical conditions and were quiescent on the ∼100-ms timescale (Fig. 4, E–H).
FIGURE 4.
SecYEG dynamics on ∼100-ms time scales. Individual line scans are displayed (red indicates trace; blue indicates retrace). The time difference between trace and retrace is displayed for each pair of profiles. The first column (A–D) displays features that underwent significant conformational changes between trace and retrace. Specifically, the height differences were: 5, 8, 6, and 4 Å for A–D, respectively. In contrast, features in the second column (E–H) were quite stable. Vertical and horizontal scale bars are indicated.
Oligomeric States
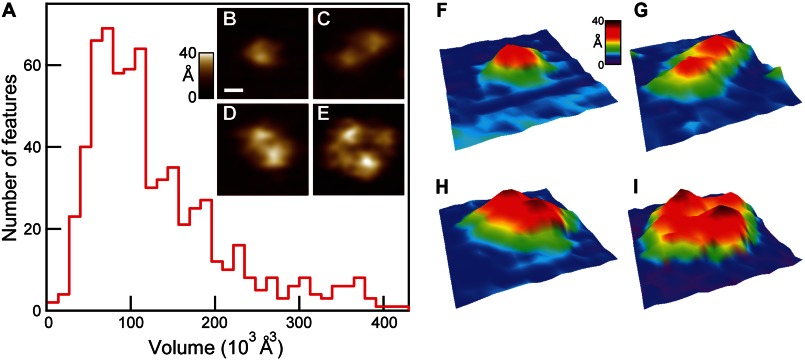
It has been debated whether SecYEG populates an equilibrium between monomer, dimer, and higher order structures. We determined oligomeric state via direct inspection and by calculating the volumes of SecYEG cytoplasmic protrusions. Fig. 5A presents a volume histogram of features on the cytoplasmic face (n = 676) after applying the tip deconvolution algorithm. Peaks in the volume distribution were found at 73, 112, 150, and 190 × 103 Å3, with lower populations of larger features also observed. Inspection of individual SecYEG complexes allowed assignment to one of three general classes: monomers, dimers, and tetramers. Features with characteristics of monomers (i.e. single Gaussian-like protrusions, Fig. 5B) occupied a volume range roughly between ∼40 and 120 × 103 Å3 (Fig. 5A). Within this range of volumes, two distinct peaks (73 and 112 × 103 Å3) were observed. Dimeric features (i.e. those exhibiting two Gaussian-like protrusions, Fig. 5, C and D) occurred between ∼80 and 240 × 103 Å3, and as was the case for monomers, two principle peaks (∼150 and 190 × 103 Å3) were detected. Conformational changes of the same quaternary structures can lead to differing volumes (see “Discussion” and supplemental Fig. 3). A low number of tetramers were also observed (Fig. 5E) with protrusion volumes >200 × 103 Å3. Our experiments, carried out with ∼80 μm lipid, ∼80 nm SecYEG, revealed that ∼40% of total SecYEG (expressed as monomers) existed as dimers with an additional ∼15% as tetramers. Pseudo-three-dimensional renderings of the four images are also shown (Fig. 5, F–I). The image size was 250 × 250 Å2; the vertical scale spanned 0–40 Å.
FIGURE 5.
Oligomeric state analysis. A, volume histogram of individual cytoplasmic SecYEG features (n = 676, bin size = 13 × 103 Å3). The first two peaks fall within the monomer volume range: 40 to 120 × 103 Å3. Dimers were found primarily in the subsequent two peaks: ∼150 and 190 × 103 Å3. Representative images were as follows: monomer (B), dimers (C and D), and tetramer (E). A scale bar is shown (B, white, 50 Å), and it applies to B–E. Three-dimensional renderings of the same four images are shown (F–I).
DISCUSSION
Proteins are dynamic and many protein-protein interactions involve highly dynamic disordered regions. Such regions can enhance capture radii and hence also increase the probability of binding. In the general secretory system, loop motion of SecY is likely to enhance recruitment of SecA to SecYEG and may play a key role in translocation through direct contact with the helix scaffold domain of SecA (5).
This report is the first observation of dynamics in the membrane external segments of the translocon. We observed two prominent peaks in the height distribution on the cytoplasmic face of SecYEG with a continuum of populated heights between them. We attribute these observations to a dynamic and reversible conversion between extended and collapsed configurations of the two major loops revealed in the crystal structure of SecY.
We followed the structural dynamics by observing individual cytoplasmic SecYEG features over time. The majority of proteins studied (63%) exhibited structural dynamics characterized by large deviations σ > 2 Å in height over time. These height fluctuations occurred spontaneously and in a temporally punctuated manner, sometimes with height differences exceeding 10 Å for an individual protein in successive images (e.g. Fig. 3H). Although large, these structural deviations are consistent with the 6–7 and 8–9 flexible loops, both of which exceed 35 amino acid residues in length. Assuming a rise per residue of 3.5 Å for an extended polypeptide chain and a tight (four residues) turn, the maximum extension of these loops is >50 Å.
We also observed conformational changes parallel to the membrane surface on the order of ∼10 Å (Fig. 3, B and C). Opening of the lateral gate of the translocon involves lateral displacements > 5 Å, which have been shown previously via simulations and cross-linking experiments (36–38). In particular, molecular dynamics simulations have shown large-scale conformational changes of the 6–7 loop (∼10 Å vertical, ∼30 Å lateral) and analogous results for the 8–9 loop (39). Thus, our results, based on direct single molecule observations, are in rough agreement with previous reports.
By analysis of trace and retrace profiles, we conclude that these conformational dynamics occur on time scales <100 ms. The position of a typical protein undergoing tethered Brownian motion (40) becomes uncorrelated on time scales ≫100 ns. Therefore, if the SecYEG loop dynamics were random and rapid with respect to the time scale of the experiment, one would expect the structural dynamics to average out (i.e. the trace and the retrace data would agree). In contrast, our data indicate rich and complex dynamics are occurring over multiple timescales, the shortest of which is <100 ms.
Our data show that SecYEG exists in multiple oligomeric states and conformations. We observed monomers, dimers (40% of total monomer), and tetramers (15%). This distribution of quaternary structures is consistent with previous work based on electron microscopy, which involved freeze fracturing (41). Our conclusions are based on direct inspection and volume analyses of individual SecYEG in lipid bilayers at ∼30 °C.
The AFM data are in general agreement with the volumes determined from the crystal structure. By inscribing a cylinder (radius ∼ 27 Å; height ∼ 30 Å) over the cytoplasmic protrusion of the crystallographic structure, we coarsely estimate the monomeric SecYEG protrusion volume to be 69 × 103 Å3, which is within 6% of the measured volume peak. We note that two stable monomeric conformations were observed. In general, macromolecular volumes measured via AFM are not conserved under conformational changes (supplemental Fig. 3). Even if the tip deconvolution were perfect, structural changes resulting in differing voids within the protein structure are not always accessible to the probe, which traces only the surface of the protein.
Two modes of dimerization of SecYEG have been proposed: namely, front to front and back to back (2, 13, 42). The two distinct dimer peaks we observed suggest that two modes are likely to coexist; however, assignment of the peaks to either back to back or front to front will require further experimentation. We also note that different monomeric conformations could give rise to multiple dimeric volume peaks independent of dimeric orientation. The conformational changes introduced an overlap between the monomer and dimer regions in the volume histogram (supplemental Fig. 4). Thus, direct inspection was required to unambiguously distinguish monomers from dimers. These results provide a foundation for future work addressing the oligomeric state of the translocon in association with SecA during protein transport, a disputed topic.
AFM imaging allowed single molecule study of the integral membrane translocon SecYEG embedded in a native lipid environment at physiologically relevant temperature and ionic strength. Such studies are in contrast to many techniques that require solubilization in detergent, which often results in decreased activity. Our results shed new light on the intricate basal dynamics of SecYEG. The inherent flexibility and dynamics of the loops of SecY may play important roles in orchestrating protein secretion. Taken together, direct observation of quaternary structures and structural dynamics in near-native conditions could lead to a better understanding of the fully active complex. We thus expect that AFM has a bright future in addressing central questions in protein export.
Acknowledgments
We are grateful to Tom Rapoport and Julie Chen for advice regarding placement of membrane boundaries on crystal structure data. We also thank Krishna Sigdel, Allison Churnside, and Thomas Perkins for fruitful discussions.
This work was supported by National Science Foundation CAREER Award 1054832 (to G. M. K.), the Burroughs Wellcome Fund Career Award at the Scientific Interface (to G. M. K.), National Institutes of Health Grant GM29798 (to L. L. R.), and an endowment from the Hugo Wurdack Trust at the University of Missouri.

This article contains supplemental Figs. 1–4.
- TM
- transmembrane
- AFM
- atomic force microscope
- proOmpA
- precursor of outer membrane protein OmpA.
REFERENCES
- 1. Park E., Rapoport T. A. (2012) Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu. Rev. Biophys. 41, 21–40 [DOI] [PubMed] [Google Scholar]
- 2. Papanikou E., Karamanou S., Economou A. (2007) Bacterial protein secretion through the translocase nanomachine. Nat. Rev. Microbiol. 5, 839–851 [DOI] [PubMed] [Google Scholar]
- 3. Lycklama A., Nijeholt J. A., Driessen A. J. (2012) The bacterial Sec-translocase: structure and mechanism. Phil. Trans. R. Soc. B 367, 1016–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Van den Berg B., Clemons W. M., Jr., Collinson I., Modis Y., Hartmann E., Harrison S. C., Rapoport T. A.(2004) X-ray structure of a protein-conducting channel. Nature 427, 36–44 [DOI] [PubMed] [Google Scholar]
- 5. Zimmer J., Nam Y., Rapoport T. A. (2008) Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature 455, 936–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mori H., Ito K. (2001) An essential amino acid residue in the protein translocation channel revealed by targeted random mutagenesis of SecY. Proc. Natl. Acad. Sci. U.S.A. 98, 5128–5133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shiba K., Ito K., Yura T., Cerretti D. P. (1984) A defined mutation in the protein export gene within the spc ribosomal protein operon of Escherichia coli: isolation and characterization of new temperature-sensitive SecY mutant. EMBO J. 3, 631–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shoemaker B. A., Portman J. J., Wolynes P. G. (2000) Speeding molecular recognition by using the folding funnel: The fly-casting mechanism. Proc. Natl. Acad. Sci. U.S.A. 97, 8868–8873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dyson H. J., Wright P. E. (2005) Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 197–208 [DOI] [PubMed] [Google Scholar]
- 10. Manting E. H., van Der Does C., Remigy H., Engel A., Driessen A. J. (2000) SecYEG assembles into a tetramer to form the active protein translocation channel. EMBO J. 19, 852–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bessonneau P., Besson V., Collinson I., Duong F. (2002) The SecYEG preprotein translocation channel is a conformationally dynamic and dimeric structure. EMBO J. 21, 995–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Duong F. (2003) Binding, activation and dissociation of the dimeric SecA ATPase at the dimeric SecYEG translocase. EMBO J. 22, 4375–4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dalal K., Chan C.S., Sligar S.G., Duong F. (2012) Two copies of the SecY channel and acidic lipids are necessary to activate the SecA translocation ATPase. Proc. Natl. Acad. Sci. U.S.A. 109, 4104–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deville K., Gold V. A., Robson A., Whitehouse S., Sessions R. B., Baldwin S. A., Radford S. E., Collinson I. (2011) The oligomeric state and arrangement of the active bacterial translocon. J. Biol. Chem. 286, 4659–4669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kedrov A., Kusters I., Krasnikov V. V., Driessen A. J. (2011) A single copy of SecYEG is sufficient for preprotein translocation. EMBO J. 30, 4387–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Binnig G., Quate C. F., Gerber C. (1986) Atomic force microscope. Phys. Rev. Lett. 56, 930–933 [DOI] [PubMed] [Google Scholar]
- 17. Scheuring S., Sturgis J. N. (2005) Chromatic adaptation of photosynthetic membranes. Science 309, 484–487 [DOI] [PubMed] [Google Scholar]
- 18. Shibata M., Yamashita H., Uchihashi T., Kandori H., Ando T. (2010) High-speed atomic force microscopy shows dynamic molecular processes in photoactivated bacteriorhodopsin. Nat. Nanotechnol. 5, 208–212 [DOI] [PubMed] [Google Scholar]
- 19. Bippes C., Muller D. (2011) High-resolution atomic force microscopy and spectroscopy of native membrane proteins. Rep. Prog. Phys. 74, 086601 [DOI] [PubMed] [Google Scholar]
- 20. Miyagi A., Tsunaka Y., Uchihashi T., Mayanagi K., Hirose S., Morikawa K., Ando T. (2008) Visualization of intrinsically disordered regions of proteins by high-speed atomic force microscopy. Chemphyschem 9, 1859–1866 [DOI] [PubMed] [Google Scholar]
- 21. Müller D. J., Fotiadis D., Engel A. (1998) Mapping flexible protein domains at subnanometer resolution with the atomic force microscope. FEBS Lett. 430, 105–111 [DOI] [PubMed] [Google Scholar]
- 22. Rico F., Su C., Scheuring S. (2011) Mechanical mapping of single membrane proteins at submolecular resolution. Nano Lett. 11, 3983–3986 [DOI] [PubMed] [Google Scholar]
- 23. Scheuring S., Ringler P., Borgnia M., Stahlberg H., Müller D. J., Agre P., Engel A. (1999) High resolution AFM topographs of the Escherichia coli water channel aquaporin Z. EMBO J. 18, 4981–4987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Miroux B., Walker J. E. (1996) Overproduction of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260, 289–298 [DOI] [PubMed] [Google Scholar]
- 25. Cannon K. S., Or E., Clemons W. M., Jr., Shibata Y., Rapoport T. A. (2005) Disulfide bridge formation between SecY and a translocating polypeptide localizes the translocation pore to the center of SecY. J. Cell Biol. 169, 219–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rigaud J. L., Lévy D. (2003) Reconstitution of membrane proteins into liposomes. Methods Enzymol. 372, 65–86 [DOI] [PubMed] [Google Scholar]
- 27. Mao C., Hardy S. J., Randall L. L. (2009) Maximal efficiency of coupling between ATP hydrolysis and translocation of polypeptides mediated by SecB requires two protomers of SecA. J. Bacteriol. 191, 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Richter R. P., Bérat R., Brisson A. R. (2006) Formation of solid-supported lipid bilayers: an integrated view. Langmuir 22, 3497–3505 [DOI] [PubMed] [Google Scholar]
- 29. Müller D. J., Fotiadis D., Scheuring S., Müller S. A., Engel A. (1999) Electrostatically balanced subnanometer imaging of biological specimens by atomic force microscope. Biophys. J. 76, 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schaap I. A., Carrasco C., de Pablo P. J., MacKintosh F. C., Schmidt C. F. (2006) Elastic response, buckling, and instability of microtubules under radial indentation. Biophys. J. 91, 1521–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Villarubia J. S. (1997) Algorithms for scanned probe microscope image simulation, surface reconstruction and tip estimation. J. Res. Natl. Inst. Stand. Technol. 102, 425–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Todd B. A., Eppell S. J. (2001) A method to improve the quantitative analysis of SFM images at the nanoscale. Surface Science 491, 473–483 [Google Scholar]
- 33. Randall L. L., Hardy S. J. (2002) SecB, one small chaperone in the complex milieu of the cell. Cell Mol. Life Sci. 59, 1617–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Luckey M. (2008) Membrane Structural Biology, Cambridge University Press, Cambridge, UK [Google Scholar]
- 35. Müller D. J., Engel A. (1997) The height of biomolecules measured with the atomic force microscope depends on electrostatic interactions. Biophys. J. 73, 1633–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tsukazaki T., Mori H., Fukai S., Ishitani R., Mori T., Dohmae N., Perederina A., Sugita Y., Vassylyev D. G., Ito K., Nureki O. (2008) Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature 455, 988–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gumbart J., Schulten K. (2007) Structural determinants of lateral gate opening in the protein translocon. Biochemistry 46, 11147–11157 [DOI] [PubMed] [Google Scholar]
- 38. du Plessis D. J., Berrelkamp G., Nouwen N., Driessen A. J. (2009) The lateral gate of SecYEG opens during protein translocation. J. Biol. Chem. 284, 15805–15814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Frauenfeld J., Gumbart J., Sluis E. O., Funes S., Gartmann M., Beatrix B., Mielke T., Berninghausen O., Becker T., Schulten K., Beckmann R. (2011) Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat. Struct. Mol. Biol. 18, 614–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Howard J. (2001) Mechanics of Motor Proteins and the Cytoskeleton, Sinauer Associates, Inc., Sunderland, MA [Google Scholar]
- 41. Scheuring J., Braun N., Nothdurft L., Stumpf M., Veenendaal A. K., Kol S., van der Does C., Driessen A. J., Weinkauf S. (2005) The oligomeric distribution of SecYEG is altered by SecA and translocation ligands. J. Mol. Biol. 354, 258–271 [DOI] [PubMed] [Google Scholar]
- 42. Mitra K., Schaffitzel C., Shaikh T., Tama F., Jenni S., Brooks C. L., 3rd, Ban N., Frank J. (2005) Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature 438, 318–324 [DOI] [PMC free article] [PubMed] [Google Scholar]