Abstract
It has been reported that the endocannabinoid anandamide (AEA) binds to a class of fatty acid-binding proteins and serum albumin which can serve as carrier proteins and potentiate the cellular uptake of AEA and its intracellular translocation. Here, we employed 19F nuclear magnetic resonance spectroscopy to study the interactions of serum albumin with two inhibitors of fatty acid amide hydrolase (FAAH), the enzyme involved in the deactivation of anandamide. We found that, for both inhibitors AM5206 and AM5207, the primary binding site on serum albumin is drug site 1 located at subdomain IIA. Neither inhibitor binds to drug site 2. While AM5207 binds exclusively to drug site 1, AM5206 also interacts with other fatty acid-binding sites on serum albumin. Additionally, AM5206 has an affinity for serum albumin approximately one order of magnitude higher than that of AM5207. The data suggest that interactions of FAAH inhibitors with albumin may provide added advantages for their ability to modulate endocannabinoid levels for a range of applications including analgesia, antiemesis, and neuroprotection.
Keywords: anandamide carrier proteins, FAAH, 19F-NMR, serum albumin
Introduction
Fatty acid amide hydrolase (FAAH) is a membrane-associated enzyme (1–3) that catalyzes the hydrolysis of the endocannabinoid anandamide (N-arachidonoylethanolamine; AEA) and other bioactive amides (1–6). The blockade of FAAH can lead to chronic elevation of endocannabinoid levels at the synapse and produce sustained analgesic responses that are devoid of adverse effects typically associated with classical cannabinergic agonists (7–10). Therefore, FAAH has been a target of intense research efforts aimed at developing potent and selective inhibitors. Recently, it was reported that the cellular uptake of AEA can also be significantly potentiated by a class of anandamide carrier proteins (11–13) such as serum albumin, heat shock proteins (Hsp70), and fatty acid-binding proteins. These findings provide an opportunity for a potential new therapeutic modality to the treatment of pain through dual inhibition of FAAH and anandamide carrier proteins.
In an effort to explore the role of carrier proteins in the transport of endocannabinoids, we studied the interaction of two selective FAAH inhibitors, AM5206 and AM5207 (Fig. 1), with a representative anandamide carrier protein serum albumin. These two trifluoromethyl ketone analogs were selected because they represent a new generation of reversible FAAH inhibitors that can cross the blood–brain barrier and protect against the neurodegenerative changes and cytoskeletal damages (14–17). Serum albumin, the major protein constituent of blood plasma, has long been regarded as a reservoir and a carrier protein for various lipophilic ligands to their specific targets (18–26). The crystal structure of serum albumin reveals that it is a globular “heart-shaped” protein consisting of three homologous subdomains (I, II, and III) (22,27–29). There are two major structurally selective drug-binding sites (drug site 1 and drug site 2) located within subdomains IIA and IIIA, respectively (19,27,30–32).
Fig. 1.
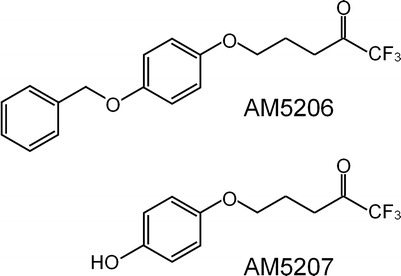
Structure of the two trifluoromethyl ketone FAAH inhibitors, AM5206 and AM5207
The absence of fluorine in proteins provides much simplified 19F nuclear magnetic resonance (19F-NMR) spectra when studying the interactions between 19F-labeled ligand and its target protein (33–38). Fluorine chemical shift anisotropy and exchange for screening has been commonly used as a rapid high-throughput screening NMR method to rank ligands for their binding affinity to a target protein (33,34). Here, our study was aimed at a more detailed characterization of the specific interactions between albumin and two of our lead FAAH inhibitors. We first employed fluorine NMR competition binding experiments to study the site-specific binding preference of these two FAAH inhibitors using site markers warfarin, l-tryptophan, and oleic acid (29,39–41). The binding affinity of each ligand was determined based on 19F pulsed-field gradient (PFG) NMR diffusion measurements (42).
Materials and Methods
Materials
Fatty acid free bovine serum albumin (BSA), warfarin sodium, l-tryptophan, and oleic acid were purchased from Sigma-Aldrich (St. Louis, MO, USA). The two FAAH inhibitors AM5206 and AM5207 (Fig. 1) are trifluoromethyl ketone analogs synthesized in our laboratory, and the details of their synthesis will be published elsewhere. Deuterated dimethyl sulfoxide (DMSO) and deuterium oxide (D2O) were purchased from Cambridge Isotope Laboratories, Inc. (Cambridge, MA, USA).
Sample Preparation
BSA was dissolved in D2O, and the pH was 5.3 in concentrations ranging from 0.076 mM (0.5%) to 0.6 mM (4.0%). The two FAAH inhibitors AM5206 and AM5207 and the competing site markers warfarin, l-tryptophan, and oleic acid were separately prepared in concentrated stock solutions in DMSO-d6 or D2O. For the NMR diffusion experiments, each of the two FAAH inhibitors was added to BSA solutions and sonicated for 15 min in a water bath sonicator before experiments. The molar ratios of protein to each FAAH inhibitor are from 0.1 to 0.9. In the competitive binding experiments, each of the competing site markers was added from their respective stock solutions. The final concentrations of each competing site marker were 1, 3, 5, or 10 mM.
NMR Spectroscopy
All 19F-NMR experiments were carried out at 376.5 MHz using a Bruker Avance II 400 MHz NMR spectrometer. All spectra were acquired using a 9.5-μs 90°-pulse and a 2-s repetition time without proton decoupling. The 19F-NMR chemical shifts were referenced to an external standard (CF3COOH, −76.55 ppm). Diffusion measurements were performed using the stimulated echo (STE)-PFG pulse sequence (42–45), with a maximum gradient of 0.324 T/m. These STE experiments were designed to achieve an accurate diffusion measurement by minimizing the T2 relaxation effects (42).
Results and Discussion
Site-Selective Binding of AM5206 and AM5207
In aqueous solution, the trifluoromethyl group on both AM5206 and AM5207 shows a single sharp 19F-NMR resonance at −85.45 ppm (Figs. 2 and 3). Upon addition of BSA, the signal was significantly broadened with a downfield shift to −84.35 ppm for AM5206 and −84.83 ppm for AM5207. The significant line broadening suggests a much slower motion of both ligands after binding to the protein. The resonance at −85.45 ppm disappears completely, an indication that the exchange rate between the bound and unbound FAAH inhibitors in BSA solution was fast enough on the 19F-NMR timescale.
Fig. 2.
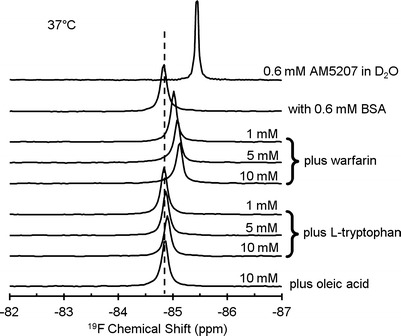
19F-NMR spectra of 0.6 mM AM5207, with 0.6 mM BSA, and in the presence of three different competing site markers: warfarin, l-tryptophan, and oleic acid at 37°C
Fig. 3.
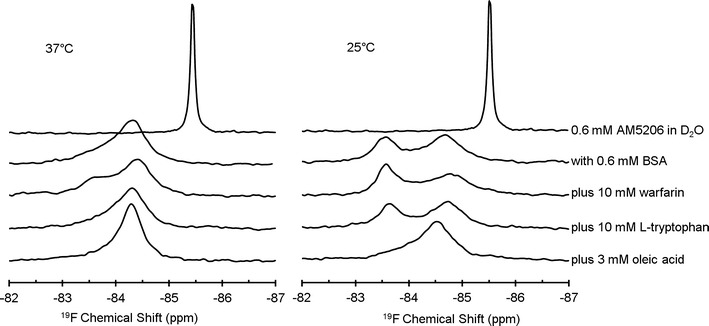
19F-NMR spectra of 0.6 mM AM5206, with 0.6 mM BSA, and in the presence of three different competing site markers: warfarin, l-tryptophan, and oleic acid at 37°C (left panel) or 25°C (right panel)
To further compare the site-selective binding of AM5206 and AM5207, we have employed a series of competition binding experiments using three well-characterized site markers: warfarin, l-tryptophan, and oleic acid. Warfarin is known to have high binding affinity to drug site 1 at subdomain IIA (39). l-Tryptophan is a typical ligand for drug site 2 at subdomain IIIA through hydrogen bonding and hydrophobic and electrostatic interactions (40,41). The long chain fatty acid, oleic acid, primarily binds to domain I and domain III on albumin (29). Figure 2 displays the 19F-NMR spectra of AM5207 in 0.6 mM BSA solution in the presence of warfarin, l-tryptophan, or oleic acid as a competing site marker. With increasing concentration of warfarin, the 19F-NMR signal shifts upfield toward −85.45 ppm. This clearly demonstrates that AM5207 can be displaced by warfarin, indicating that AM5207 binds to drug site 1 at subdomain IIA. On the other hand, there are no discernible changes in the 19F-NMR signal after adding l-tryptophan or oleic acid to the sample solution. We conclude that AM5207 primarily binds to drug site 1 at subdomain IIA, but not drug site 2 at subdomain IIIA or other domains on albumin.
Figure 3 shows the 19F-NMR spectra of a series of competition binding experiments of AM5206 in BSA solutions at two different temperatures. All spectra on the left panel were recorded at 37°C. The addition of warfarin produces a shoulder at −83.55 ppm in the 19F-NMR spectrum. However, no significant changes were observed with the addition of l-tryptophan. The 19F-NMR signal becomes significantly narrower after the addition of oleic acid to the solution. This observation indicates a more complicated binding scenario that AM5206 may have redistributed between different binding sites with the addition of a competing site marker. Indeed, after decreasing the temperature to 25°C, we can clearly detect two well-resolved resonances (−84.75 and −83.55 ppm) due to AM5206 in BSA solutions, indicating at least two different binding sites for AM5206 on BSA. This is understandable because the exchange rate between the bound states is significantly decreased from 37°C to 25°C. When using warfarin as a competing ligand, the signal at −84.75 ppm decreases, while the signal at −83.55 ppm increases. This demonstrates that a portion of AM5206 binds to drug site 1 at subdomain IIA and thus can be displaced by warfarin. On the other hand, the addition of l-tryptophan does not produce any changes in the spectrum, indicating that AM5206 does not bind to drug site 2 at subdomain IIIA. Upon the addition of 3 mM oleic acid, the signal at −83.55 ppm almost completely disappears, while the signal at −84.75 ppm increases correspondingly. This illustrates that the signal at −83.55 ppm corresponds to AM5206 at some of the fatty acid-binding sites and thus can be displaced by oleic acid.
The binding characteristics of AM5206 were further studied by exploiting the two well-resolved resonances at 25°C with a series of titration experiments. Figure 4 shows the 19F-NMR spectra with various BSA to AM5206 ratios. At a ratio of 1:0.5, the signal at −83.55 pm is much stronger than that at −84.75 ppm. With the addition of AM5206, the resonance at −84.75 ppm drastically increases. At a BSA/AM5206 ratio of 1:5, the signal at −84.75 ppm is ∼10 times stronger than that of −83.55 ppm. Based on earlier competition binding experiments, we have assigned the −84.75 ppm peak to AM5206 binding at drug site 1 (possibly undergoing fast exchange with non-specific binding sites) and the −83.55 ppm peak to AM5206 binding at fatty acid-binding sites. The titration experiments show that, while some of the fatty acid-binding sites can be easily accessible by AM5206 at low concentrations, drug site 1 at subdomain IIA is the primary binding site when there is an excess amount of ligand.
Fig. 4.
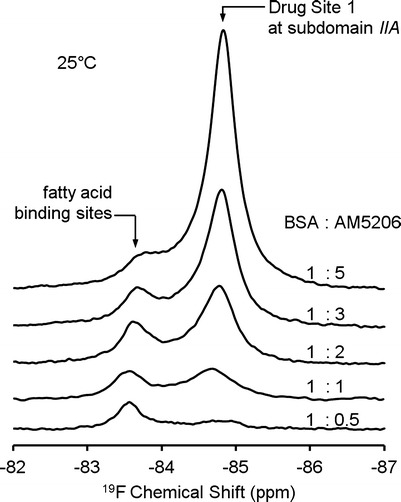
19F-NMR spectra of AM5206 in 0.6 mM BSA solutions with various molar ratios of BSA and AM5206
The Binding Affinity of AM5206 and AM5207 to Serum Albumin
NMR diffusion measurements can provide quantitative information on the ligand–protein interactions (42). Using 19F STE-PFG NMR (42–45), the diffusion coefficients of FAAH inhibitors AM5206 and AM5207 were determined in the presence of BSA at different concentrations. This method involves acquiring a series of spectra with increasing gradient strengths. The signal intensity I can be described by (44):
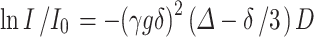 |
1 |
where I/I0 is the spin-echo attenuation ratio of the signals in the presence and absence of gradient pulses, γ is the gyromagnetic ratio, g is the gradient strength, δ is the duration of the gradient pulses, ∆ is the diffusion time, and D is the apparent self-diffusion coefficient. The top panels in Fig. 5 display the dependence of the apparent diffusion coefficient of AM5206 and AM5207 at different protein to ligand ratios ([P0]/[L0]). For both ligands, the apparent diffusion coefficient decreases with increasing protein to ligand ratio.
Fig. 5.
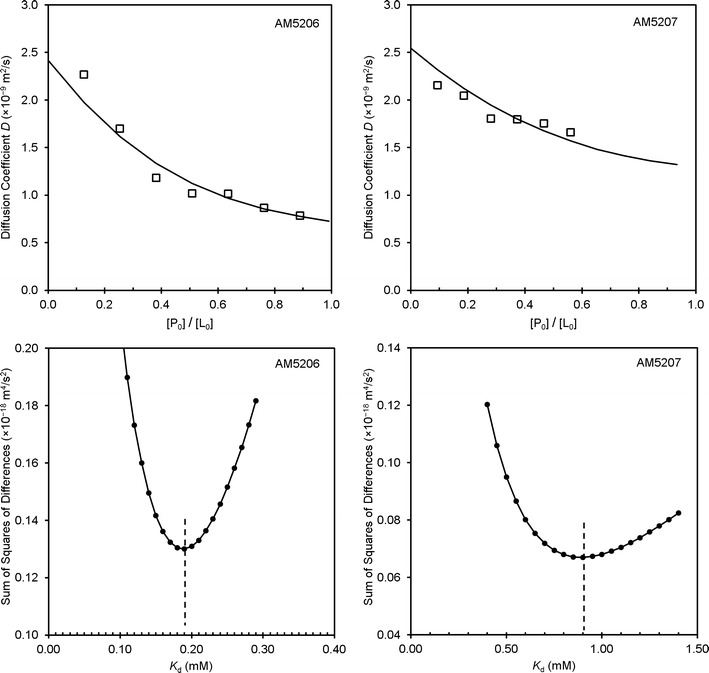
Experimental and calculated diffusion coefficients as a function of protein to ligand ratio (top panels) and the determination of K d values using least square curve fittings (bottom panels)
This set of diffusion coefficients allows us to determine the binding affinity (Kd) of each ligand to BSA. Under fast exchange conditions, the apparent diffusion coefficient (D) is the weighted average of two diffusion coefficients from ligand in its free and bound forms, which can be described as:
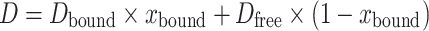 |
2 |
where xbound is the fraction of the bound ligand and Dbound and Dfree represent the diffusion coefficients of bound and free ligand, respectively. The bound fraction xbound can be expressed in terms of the dissociation constant Kd, the number of binding sites n, and the total ligand and protein concentrations [L0] and [P0] (42):
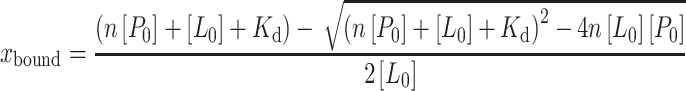 |
3 |
Dbound and Dfree were obtained using similar 1H or 19F STE-PFG NMR measurements from pure protein and pure ligand preparations, respectively. For the number of binding sites n, it is reasonable to use n = 2 for AM5206 in our calculations because it primarily binds to drug site 1, while a small fraction binds to the fatty acid-binding sites. On the other hand, we have used n = 1 for AM5207 because it only binds to drug site 1. We can then obtain the dissociation constant (Kd) of AM5206 and AM5207 by fitting our experimental diffusion measurements using the least square method. The bottom panels in Fig. 5 are plots of the sum of squared differences between the experimentally determined diffusion coefficients (D) and the calculated D using a range of Kd values. For AM5206, the minimum squared error corresponds to the best fit for a two-site binding with a mean Kd value of 190 μM. Conversely, the best fit for AM5207 provides a Kd value of 900 μM with a single binding site on albumin.
Conclusions
The interactions of two trifluoromethyl ketone FAAH inhibitors with serum albumin were characterized by a series of competitive binding experiments and self-diffusion measurements using 19F-NMR. We found that the primary binding site for both AM5206 and AM5207 is drug site 1 located at subdomain IIA. Neither of these two FAAH inhibitors binds to drug site 2 on albumin. While AM5207 binds exclusively to drug site 1, AM5206 also interacts with other fatty acid-binding sites on albumin. Interestingly, AM5206 tends to first bind to these fatty acid-binding sites especially at lower ligand concentrations, suggesting a much lower energy barrier for the ligand to access these sites. Furthermore, AM5206 has an affinity for serum albumin approximately one order of magnitude higher than that of AM5207. Arguably FAAH inhibitors such as AM5206 may compete with anandamide for binding with albumin and reduce the uptake of anandamide into cells. Such FAAH inhibitors can, thus, act by elevating the levels of available anandamide at the synapse through two converging mechanisms: the inactivation of FAAH and the inhibition of carrier protein-mediated transport of anandamide.
Acknowledgments
This work was supported by NIH grants DA003801 (A.M.), DA007215 (A.M.), DA007312 (A.M.), and DA032020 (J.G.) from the National Institute on Drug Abuse.
Abbreviations
- AEA
N-arachidonoylethanolamine or anandamide
- BSA
bovine serum albumin
- FAAH
fatty acid amide hydrolase
Contributor Information
Jianxin Guo, Phone: +1-617-3734219, Email: j.guo@neu.edu.
Alexandros Makriyannis, Phone: +1-617-3734200, Email: a.makriyannis@neu.edu.
References
- 1.Cravatt BF, Giang DK, Mayfield SP, Boger DL, Lerner RA, Gilula NB. Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature. 1996;384(6604):83–87. doi: 10.1038/384083a0. [DOI] [PubMed] [Google Scholar]
- 2.Giang DK, Cravatt BF. Molecular characterization of human and mouse fatty acid amide hydrolases. Proc Natl Acad Sci U S A. 1997;94(6):2238–2242. doi: 10.1073/pnas.94.6.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKinney MK, Cravatt BF. Structure and function of fatty acid amide hydrolase. Annu Rev Biochem. 2005;74:411–432. doi: 10.1146/annurev.biochem.74.082803.133450. [DOI] [PubMed] [Google Scholar]
- 4.Ahn K, Johnson DS, Fitzgerald LR, Liimatta M, Arendse A, Stevenson T, et al. Novel mechanistic class of fatty acid amide hydrolase inhibitors with remarkable selectivity. Biochemistry. 2007;46(45):13019–13030. doi: 10.1021/bi701378g. [DOI] [PubMed] [Google Scholar]
- 5.Piomelli D, Tarzia G, Duranti A, Tontini A, Mor M, Compton TR, et al. Pharmacological profile of the selective FAAH inhibitor KDS-4103 (URB597) CNS Drug Rev. 2006;12(1):21–38. doi: 10.1111/j.1527-3458.2006.00021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scherma M, Medalie J, Fratta W, Vadivel SK, Makriyannis A, Piomelli D, et al. The endogenous cannabinoid anandamide has effects on motivation and anxiety that are revealed by fatty acid amide hydrolase (FAAH) inhibition. Neuropharmacology. 2008;54(1):129–140. doi: 10.1016/j.neuropharm.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang L, Luo L, Palmer JA, Sutton S, Wilson SJ, Barbier AJ, et al. Inhibition of fatty acid amide hydrolase produces analgesia by multiple mechanisms. Br J Pharmacol. 2006;148(1):102–113. doi: 10.1038/sj.bjp.0706699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cravatt BF, Demarest K, Patricelli MP, Bracey MH, Giang DK, Martin BR, et al. Supersensitivity to anandamide and enhanced endogenous cannabinoid signaling in mice lacking fatty acid amide hydrolase. Proc Natl Acad Sci U S A. 2001;98(16):9371–9376. doi: 10.1073/pnas.161191698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jayamanne A, Greenwood R, Mitchell VA, Aslan S, Piomelli D, Vaughan CW. Actions of the FAAH inhibitor URB597 in neuropathic and inflammatory chronic pain models. Br J Pharmacol. 2006;147(3):281–288. doi: 10.1038/sj.bjp.0706510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Solinas M, Tanda G, Justinova Z, Wertheim CE, Yasar S, Piomelli D, et al. The endogenous cannabinoid anandamide produces delta-9-tetrahydrocannabinol-like discriminative and neurochemical effects that are enhanced by inhibition of fatty acid amide hydrolase but not by inhibition of anandamide transport. J Pharmacol Exp Ther. 2007;321(1):370–380. doi: 10.1124/jpet.106.114124. [DOI] [PubMed] [Google Scholar]
- 11.Fu J, Bottegoni G, Sasso O, Bertorelli R, Rocchia W, Masetti M, et al. A catalytically silent FAAH-1 variant drives anandamide transport in neurons. Nat Neurosci. 2012;15(1):64–69. doi: 10.1038/nn.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaczocha M, Glaser ST, Deutsch DG. Identification of intracellular carriers for the endocannabinoid anandamide. Proc Natl Acad Sci U S A. 2009;106(15):6375–6380. doi: 10.1073/pnas.0901515106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oddi S, Fezza F, Pasquariello N, D’Agostino A, Catanzaro G, De Simone C, et al. Molecular identification of albumin and Hsp70 as cytosolic anandamide-binding proteins. Chem Biol. 2009;16(6):624–632. doi: 10.1016/j.chembiol.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 14.Karanian DA, Brown QB, Makriyannis A, Kosten TA, Bahr BA. Dual modulation of endocannabinoid transport and fatty acid amide hydrolase protects against excitotoxicity. J Neurosci. 2005;25(34):7813–7820. doi: 10.1523/JNEUROSCI.2347-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karanian DA, Nikas SP, Zhao J, Wood JT, Williams JS, Makriyannis A, et al. Enhancement of endogenous cannabinoid responses through FAAH inhibition provides cellular and functional protection against excitotoxic brain damage. FASEB J. 2007;21:883.5. [Google Scholar]
- 16.Makriyannis A, Nikas SP, Alapafuja SO, Shukla VG (inventors); University of Connecticut (assignee). Fatty acid amide hydrolase inhibitors. Patent WO/2008/013963.
- 17.Naidoo V, Nikas S, Karanian D, Hwang J, Zhao J, Wood J, et al. A new generation fatty acid amide hydrolase inhibitor protects against kainate-induced excitotoxicity. J Mol Neurosci. 2011;43(3):493–502. doi: 10.1007/s12031-010-9472-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bojesen IN, Hansen HS. Binding of anandamide to bovine serum albumin. J Lipid Res. 2003;44(9):1790–1794. doi: 10.1194/jlr.M300170-JLR200. [DOI] [PubMed] [Google Scholar]
- 19.Jisha VS, Arun KT, Hariharan M, Ramaiah D. Site-selective binding and dual mode recognition of serum albumin by a squaraine dye. J Am Chem Soc. 2006;128(18):6024–6025. doi: 10.1021/ja061301x. [DOI] [PubMed] [Google Scholar]
- 20.Kumar CV, Buranaprapuk A. Site-specific photocleavage of proteins. Angew Chem Int Ed. 1997;36(19):2085–2087. doi: 10.1002/anie.199720851. [DOI] [Google Scholar]
- 21.Kumar CV, Buranaprapuk A, Opiteck GJ, Moyer MB, Jockusch S, Turro NJ. Photochemical protease: site-specific photocleavage of hen egg lysozyme and bovine serum albumin. Proc Natl Acad Sci U S A. 1998;95(18):10361–10366. doi: 10.1073/pnas.95.18.10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mao H, Gunasekera AH, Fesik SW. Expression, refolding, and isotopic labeling of human serum albumin domains for NMR spectroscopy. Protein Expr Purif. 2000;20(3):492–499. doi: 10.1006/prep.2000.1330. [DOI] [PubMed] [Google Scholar]
- 23.Sandberg A, Fowler CJ. Measurement of saturable and non-saturable components of anandamide uptake into P19 embryonic carcinoma cells in the presence of fatty acid-free bovine serum albumin. Chem Phys Lipids. 2005;134(2):131–139. doi: 10.1016/j.chemphyslip.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 24.Simard JR, Zunszain PA, Hamilton JA, Curry S. Location of high and low affinity fatty acid binding sites on human serum albumin revealed by NMR drug-competition analysis. J Mol Biol. 2006;361(2):336–351. doi: 10.1016/j.jmb.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 25.Sułkowska A, Bojko B, Równicka J, Rezner P, Sułkowski WW. The competition of drugs to serum albumin in combination chemotherapy: NMR study. J Mol Struct. 2005;744–747:781–787. doi: 10.1016/j.molstruc.2004.11.050. [DOI] [Google Scholar]
- 26.Zucker SD, Goessling W, Gollan JL. Kinetics of bilirubin transfer between serum albumin and membrane vesicles. Insight into the mechanism of organic anion delivery to the hepatocyte plasma membrane. J Biol Chem. 1995;270(3):1074–1081. doi: 10.1074/jbc.270.3.1074. [DOI] [PubMed] [Google Scholar]
- 27.Carter DC, Ho JX. Structure of serum albumin. Adv Protein Chem. 1994;45:153–203. doi: 10.1016/S0065-3233(08)60640-3. [DOI] [PubMed] [Google Scholar]
- 28.He XM, Carter DC. Atomic structure and chemistry of human serum albumin. Nature. 1992;358(6383):209–215. doi: 10.1038/358209a0. [DOI] [PubMed] [Google Scholar]
- 29.Petitpas I, Grune T, Bhattacharya AA, Curry S. Crystal structures of human serum albumin complexed with monounsaturated and polyunsaturated fatty acids. J Mol Biol. 2001;314(5):955–960. doi: 10.1006/jmbi.2000.5208. [DOI] [PubMed] [Google Scholar]
- 30.Dockal M, Carter DC, Ruker F. The three recombinant domains of human serum albumin. Structural characterization and ligand binding properties. J Biol Chem. 1999;274(41):29303–29310. doi: 10.1074/jbc.274.41.29303. [DOI] [PubMed] [Google Scholar]
- 31.Sudlow G, Birkett DJ, Wade DN. Further characterization of specific drug binding sites on human serum albumin. Mol Pharmacol. 1976;12(6):1052–1061. [PubMed] [Google Scholar]
- 32.Sulkowska A, Bojko B, Rownicka J, Sulkowski W. Competition of drugs to serum albumin in combination therapy. Biopolymers. 2004;74(3):256–262. doi: 10.1002/bip.20031. [DOI] [PubMed] [Google Scholar]
- 33.Dalvit C, Fagerness PE, Hadden DT, Sarver RW, Stockman BJ. Fluorine-NMR experiments for high-throughput screening: theoretical aspects, practical considerations, and range of applicability. J Am Chem Soc. 2003;125(25):7696–7703. doi: 10.1021/ja034646d. [DOI] [PubMed] [Google Scholar]
- 34.Dalvit C, Vulpetti A. Technical and practical aspects of 19F NMR-based screening: toward sensitive high-throughput screening with rapid deconvolution. Magn Reson Chem MRC. 2012;50(9):592–597. doi: 10.1002/mrc.3842. [DOI] [PubMed] [Google Scholar]
- 35.Jenkins BG, Lauffer RB. Detection of site-specific binding and co-binding of ligands to human serum albumin using 19F NMR. Mol Pharmacol. 1990;37(1):111–118. [PubMed] [Google Scholar]
- 36.Kitamura K, Kume M, Yamamoto M, Takegami S, Kitade T. 19F NMR spectroscopic study on the binding of triflupromazine to bovine and human serum albumins. J Pharm Biomed Anal. 2004;36(2):411–414. doi: 10.1016/j.jpba.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 37.Shikii K, Sakurai S, Utsumi H, Seki H, Tashiro M. Application of the 19F NMR technique to observe binding of the general anesthetic halothane to human serum albumin. Anal Sci. 2004;20(10):1475–1477. doi: 10.2116/analsci.20.1475. [DOI] [PubMed] [Google Scholar]
- 38.Xu Y, Tang P, Firestone L, Zhang TT. 19F nuclear magnetic resonance investigation of stereoselective binding of isoflurane to bovine serum albumin. Biophys J. 1996;70(1):532–538. doi: 10.1016/S0006-3495(96)79599-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghuman J, Zunszain PA, Petitpas I, Bhattacharya AA, Otagiri M, Curry S. Structural basis of the drug-binding specificity of human serum albumin. J Mol Biol. 2005;353(1):38–52. doi: 10.1016/j.jmb.2005.07.075. [DOI] [PubMed] [Google Scholar]
- 40.McMenamy RH, Oncley JL. The specific binding of l-tryptophan to serum albumin. J Biol Chem. 1958;233:1436–1447. [PubMed] [Google Scholar]
- 41.Pardridge WM, Fierer G. Transport of tryptophan into brain from the circulating, albumin-bound pool in rats and in rabbits. J Neurochem. 1990;54(3):971–976. doi: 10.1111/j.1471-4159.1990.tb02345.x. [DOI] [PubMed] [Google Scholar]
- 42.Price WS. Recent advances in NMR diffusion techniques for studying drug binding. Aust J Chem. 2003;56(9):855–860. doi: 10.1071/CH03128. [DOI] [Google Scholar]
- 43.Cotts RM, Hoch MJR, Sun T, Markert JT. Pulsed field gradient stimulated echo methods for improved NMR diffusion measurements in heterogeneous systems. J Magn Reson. 1989;83(2):252–266. [Google Scholar]
- 44.Stilbs P. Fourier transform pulsed-gradient spin-echo studies of molecular diffusion. Prog Nucl Magn Reson Spectrosc. 1987;19(1):1–45. doi: 10.1016/0079-6565(87)80007-9. [DOI] [Google Scholar]
- 45.Tanner JE. Use of the stimulated echo in NMR diffusion studies. J Chem Phys. 1970;52(5):2523–2526. doi: 10.1063/1.1673336. [DOI] [Google Scholar]


