Abstract
The nanopore sensor can detect cancer-derived nucleic acids biomarkers such as microRNAs (miRNAs), providing a non-invasive tool potentially useful in medical diagnostics. However, the nanopore-based detection of these biomarkers remains confounded by the presence of numerous of other nucleic acid species found in biofluid extracts. Their non-specific interactions with the nanopore inevitably contaminate the target signals, reducing the detection accuracy. Here we report a novel method that utilizes a polycationic peptide-PNA probe for selective miRNA detection in the nucleic acids mixture. The cationic probe hybridized with microRNA forms a dipole complex, which can be captured by the pore using a voltage polarity that is opposite the polarity used to capture negatively charged nucleic acids. As a result, non-target species are driven away from the pore opening, and the target miRNA can be detected accurately without interference. In addition, we demonstrate that the PNA probe enables accurate discrimination of miRNAs with single-nucleotide difference. This highly sensitive and selective nano-dielectrophoresis approach can be applied to the detection of clinically-relevant nucleic acid fragments in complex samples.
Keywords: nanopore, single molecule, biosensor, nucleic acids, microRNA (miRNA), HIV-1 TAT, peptide, PNA, probe, cancer, diagnostics
The nanopore provides a sensitive single-molecule platform for exploring a large variety of life sciences problems.1–14 Not only are nanopores are being widely developed for rapid and low cost gene sequencing,15–18 they also have been found to be able to analyse epigenetic changes such as DNA methylation19 and gene damage.20 In this rapidly evolved field, the nanopore sensor has recently been designed to electrically detect microRNAs (miRNAs),21,22 a class of tiny but extremely important regulatory RNA molecules.23–26 As miRNAs are potential cancer biomarkers,27–35 an accurate nanopore sensor for circulating miRNA detection would offer a potential non-invasive tool for screening and diagnostics of diseases.
However, translating the nanopore sensor into a clinically usable technology faces challenges due to the complexity of clinical samples. Generally, the clinical samples used to test for miRNA are RNA extractions from a patient's biofluids such as plasma. These extractions are a complex collection of various RNA species: miRNAs, mRNAs, tRNAs, etc. When the nanopore is used to detect the target miRNA, any free nucleic acids in the RNA mixture can also non-specifically interact with the pore. These interactions result in intensive “contaminative” signals that severely influence the target miRNA determination, and they should be eliminated.
We have devised a solution to this contamination problem (Figure 1): by using a polycationic probe, the nanopore can selectively capture and detect the target miRNA. The probe comprises a sequence of peptide nucleic acids (PNA) conjugated with a polycationic peptide lead. The PNA is designed to specifically capture the target miRNA. Upon hybridization, the positively charged peptide lead and the negatively charged miRNA together form a dipole. This structure can be driven into the nanopore by a large electric field gradient around the nanopore opening. At the same time, any free nucleic acids without probe hybridization would carry negative charge and migrate away from the pore opening. Consequently only the signatures for the miRNA•probe complex and probe alone in the nanopore will be identified, and any interference signal originating from free nucleic acids is completely eliminated.
Figure 1.
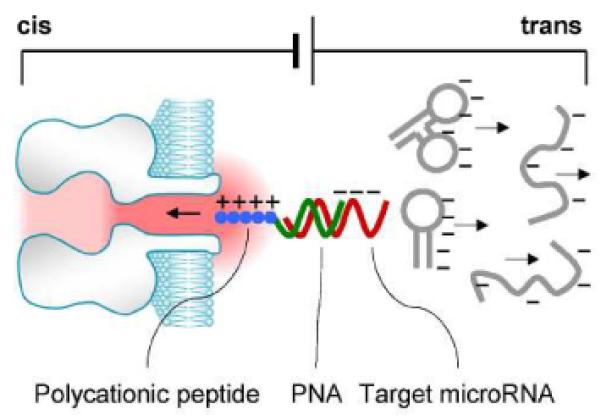
Cationic probe-enabled interference-free detection of miRNAs in the nanopore. The probe comprises a capture domain (PNA, green) attached with a polycationic polymer lead (peptide, blue). The capture domain hybridizes with the target miRNA (red). In an electrical field, the miRNA•probe complex is drawn into the nanopore, while any free nucleic acids without the probe binding carry negative charges (grey) and electrophoretically move away from the pore.
RESULTS AND DISCUSSION
Simultaneous enrichment and detection of miRNAs with a polycationic probe. We chose the Let-7 tumour-suppressing microRNA family31,36,37 as the target. Figure 2a and Table S1 shows the sequences of Let-7b and its probe P7b. P7b contained a 10 base PNA sequence designed to specifically hybridize with Let-7b. The PNA was extended at the N-terminal with an HIV-1 TAT polycationic peptide,38 which includes six arginines and two lysines, which are positively charged amino acids. All analytes, including the miRNA, the probe and their mixtures were presented on trans side of the nanopore. A positive voltage was applied from trans side to the grounded cis side. This voltage polarity can drive cationic molecules toward pore, while repelling anionic molecules away from the pore. We utilized the mutant α-hemolysin pore K131D as the sensor because the seven negatively charged Asp131 residues at trans opening of this mutant pore can enhance the attraction of cationic molecules.
Figure 2.
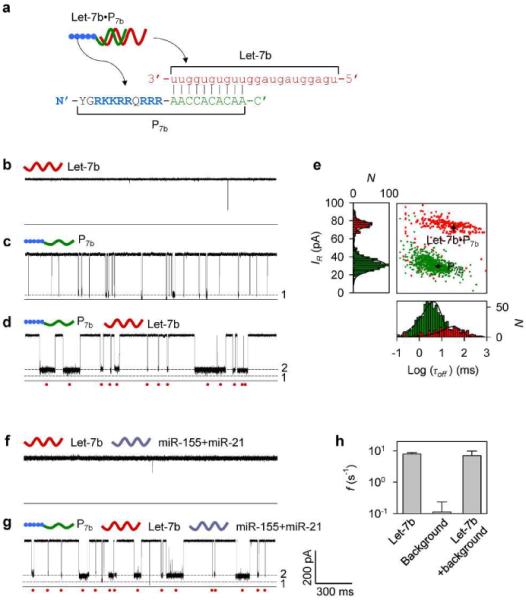
Detection of miRNA with a polycationic probe. a, Sequences of the target miRNA Let-7b and the polycationic peptide-PNA probe P7b. b–d, Current traces for the K131D pore in the presence of (b) Let-7b alone, (c) P7b alone, and (d) the Let-7b / P7b mixture, recorded at +180 mV in 1 M KCl solutions (pH7.2). Concentrations of Let-7b and P7b were 300 nM and 100 nM respectively. e, scattering plots and histograms showing the duration and residual currents of P7b and Let-7b•P7b. f–g, Current traces for (f) Let-7b, (g) the Let-7b / P7b mixture in the presence of background RNAs including miR-155 and miR-21, at +180 mV, in 1 M KCl solutions (pH7.2). Concentrations of Let-7b, miR-155 and miR-21 were 300 nM respectively, and P7b was 100 nM. h, Comparison of frequencies of Let-7b•P7b signatures in the absence of background, the background alone and Let-7b•P7b in the presence of background.
We found that the current trace for Let-7b alone in trans solution (Figure 2b, +180 mV) was similar to that in the absence of any nucleic acid analyte (Figure S1), indicating that Let-7b never blocked the pore itself without hybridization. This absence of Let-7b blocks is expected given that the positive voltage should prevent the negatively charged miRNA from interacting with the nanopore. In contrast to Let-7b, the current trace for P7b alone in trans solution shows a large number of Level 1 blocks (Figure 2c). The duration of these blocks τoff was 4.8±1.2 ms and their relative conductance IR/I was 8.2% (IR and I are currents of the block and the empty pore, Figure 2e). The presence of these of Level 1 blocks is expected given that the positive voltage should lead the positively charged probe toward the pore. When the Let-7b/P7b mixture was added in trans solution, the Level 1 blocks became rarely observed. Instead, we identified a large number of distinct Level 2 blocks (Figure 2d). Compared with the Level 1 blocks, the Level 2 blocks were 6-fold longer with duration of 28±4 ms and featured higher relative conductance at 26% (Figure 2e). These Level 2 blocks cannot be observed at negative voltage. Because the Level 2 blocks were only observed in the presence of both Let-7b and P7b, they are attributed to the formation of Let-7b•P7b hybrids that interact with the pore's trans opening. Therefore the Level 2 blocks serve as signatures for Let-7b identification.
To validate this finding in a more complex system, we mixed Let-7b with two background RNAs, miR-155 and miR-21, which have significantly different sequences from Let-7b. Indeed, no block was observed for Let-7b in the presence background RNAs (Figure 2f, +180 mV), suggesting that none of nucleic acids components can interact with the pore at this voltage. In the presence of P7b in the mixture, a large number of Level 2 blocks appeared (Figure 2g). As the background RNAs cannot hybridize with P7b, the Level 2 blocks should be attributed to the Let-7b•P7b hybrids. Their properties, including the current amplitude, the duration and occurring frequency (Figure 2h and Table S2), had no significant difference from that observed without background RNAs (Figure 2c), suggesting that the background RNAs do not affect the target miRNA detection. Therefore we conclude that a polycationic probe can electrically separate the target miRNA from free nucleic acids components. The probe-labeled miRNAs can be enriched around the nanopore and simultaneously detected. As free nucleic acids not bound to complementary probes cannot interact with the pore under these conditions, their affect on signature recognition is effectively eliminated.
Configuration of the miRNA•probe complex in the nanopore. Since the probe P7b (1.1 nm wide, Figure 3a left) is narrower than the α-hemolysin pore (1.5–2 nm),39 the entire polymer including the PNA domain can be trapped in the pore to generate the Level 1 block (Figure 3b). PNA itself can be only rarely trapped in the pore (Figure S1), so the trapping of P7b would likely be led by P7b's peptide domain. Interestingly, the Level 1 block reached the longest duration at +140 mV, and was shortened by either decreasing or increasing the voltage (Figure 3g). This “hill”-shaped voltage dependence suggests that P7b binds in the pore. Lower than the peak voltage, P7b tends to return to trans solution from the binding site, and higher than this voltage, P7b traverses the pore to cis solution. It is likely that the binding of P7b is contributed by PNA interaction with the pore, since peptide sequences alone simply translocate through the pore (Figure 3g).
Figure 3.
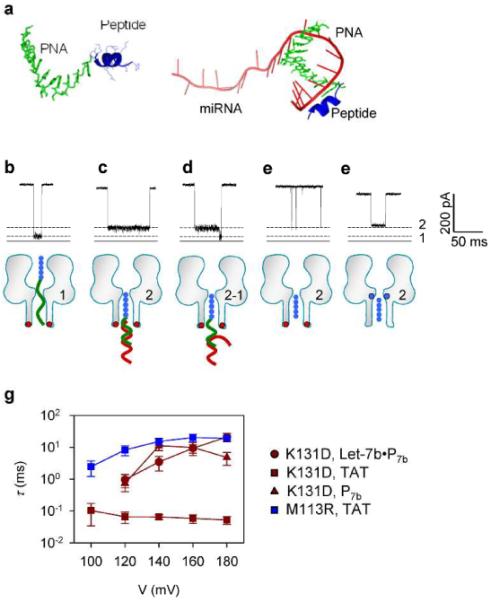
Molecular configurations for various polymers in nanopores. a, The most stable structures of P7b (left) and the Let-7b•P7b complex (right) in 1 M KCl at pH7.0. Simulation was detailed in Supplementary Information S1. b–d, typical signature blocks and molecular configurations for trapping of (b) P7b (peptide-PNA), (c) the Let-7b•P7b complex (back to trans solution) and (d) the Let-7b•P7b complex (unzipped upon trapped) in the K131D pore. e, Translocation of the HIV-TAT peptide through the K131D pore. f, Trapping of the HIV-TAT peptide in the M113R pore. Red dots in the K131D pore (b through e) represent the anionic D131 ring at trans opening, and blue dots in the M113R pore (f) mark the cationic R113 ring in the constrictive region. g, Voltage-dependent duration (τ) of blocks in panel b through f. Red circle: Let-7b•P7b complex in the K131D pore; Red square: TAT peptide in the K131D pore; Red triangle: P7b probe in the M113R pore; and blue square: TAT peptide in the M113R pore.
Structural comparison suggests that the folded peptide domain (~1.7 nm wide) of the Let-7b•P7b complex can enter the pore from the trans opening (~2 nm), but the Let-7b•PNA duplex (~2.3 nm wide) is too wide and cannot enter the pore. So the Level 2 block is likely generated by the unbound peptide domain occupying the β-barrel stem, which produces a partial block in the ionic current (Figure 3c). The trapped Let-7b•P7b complex is simply released back to trans solution, rather than translocate through the pore, because the Level 2 duration was consistently extended with increased voltage (Figure 3g). Other than Level 2 signatures, it was very rare to observe a multi-level block featuring a conductance transition from Level 2 to Level 1 (Figure 3d). This two-level block represents the unzipping of Let-7b•P7b: its peptide domain first enters the pore for Level 2; once unzipped, the dissociated P7b slides into the pore to produce a stepwise change in conductance to Level 1, while the dissociated Let-7c miRNA returns to trans solution. Additional support that the peptide domain is being trapped in the pore comes from observations made using the HIV-TAT peptide. This peptide is the same as our probe peptide. TAT translocation reduced the pore conductance to IR/I =27% (Figure 3e), very close to Level 2. We further encapsulated the TAT peptide in the mutant pore M113R to confirm this configuration (Figure 3f). In this case, the block duration was consistently extended as the voltage increased (Figure 3g), suggesting that the TAT peptide cannot pass through the M113R pore, but is encapsulated within the β-barrel between the arginine ring and trans entrance. Ultimately, the TAT peptide trapped in this position produced a similar block in conductance at 34%, which is similar to our system's Level 2 block in the K131D pore. Therefore, we conclude that the Level 2 block represents a β-barrel blockage by the single-stranded folded peptide domain of the probe.
Mechanism for probe-induced nucleic acids separation in electric field. The core finding in the above study is that the free nucleic acids are kept out of the pore while miRNA•probe complexes can enter the pore. This enables interference-free detection of target miRNA using the nanopore platform. However, the mechanism for miRNA•probe capture by the pore is under debate. The charges on the miRNA and the probe are −22e and +8e respectively. Although the effective charges can be greatly lowered in high salt concentration,40,41 the overall charge polarity retains a net negative charge without inversion.42 This is in agreement with the zeta potential (ζ) of the two polymers, ζ=−14.4±1.9 mV for Let-7b and +11.7±0.4 mV for P7b. Furthermore, we measured ζ potential of the Let-7b•P7b hybrid to be −10.0±3.6 mV, confirming that the miRNA•probe complex is negatively charged. Therefore, the negatively charged complex's movement is somehow in the opposite direction from the movement of the other negatively charged RNA species. This phenomenon is inconsistent with either electrophoretic12,17,41,43 or electroosmotic effect44–46 explanations for the capturing of this analyte in the nanopore.
We reason that it is the particular distribution of the electrostatic potential at the trans entrance of the nanopore that enables the capture of the miRNA•probe complex. Molecular dynamics simulations have shown that the major drop of the electrostatic potential in the α-hemolysin pore occurs near the nanocavity/β-barrel junction, and the electric field sharply decays within a very short distance (~1 nm) outside the nanopore.47,48 miRNA can diffuse toward the trans entrance up to this distance without feeling the effect of the transmembrane potential. However, once inside that distance, complexes with the positively charged peptide oriented closer to the pore may be captured and drawn into the nanopore. Once in the equilibrium configuration (Figure 1), the transmembrane potential acting on the peptide counterbalances repulsion between miRNA and the negatively charged trans end of the β-barrel. Overall, the highly asymmetrical distribution of charges within the complex coupled with the highly asymmetrical electric field along the pore's axis can allow for a net force pulling the complex into the pore even though the electrostatic simplification of this system would suggest otherwise.
PNA-enabled specificity for single nucleotide discrimination. Both functional explorations and diagnostic applications require accurate discrimination between miRNAs that have similar sequences. Previous work has shown that miRNAs with a single-nucleotide difference can be statistically separated using a DNA probe, based on a 3–4 folds difference in block duration (de-hybridization time) between fully-matched and one-mismatched miRNA•probe complexes.21 However, this separation method is not practical in real-time detection, because theoretical analysis has indicated that there should be at least 8–10 folds duration difference in order to achieve 90% discrimination accuracy in the mixture.21 Therefore, we seek to overcome the limitations associated with DNA probes by using PNA. PNAs are a type of artificial nucleic acids which can complimentarily bind to nucleic acids.49,50 Due to their well-characterized binding strength and specificity, PNAs have been broadly applied in molecular biology, diagnostic assays and antisense therapies,50 as well as being used for gene detection in synthetic nanopores.51 Importantly, it has been demonstrated that a PNA•RNA complex shows higher hybridization strength compared with the same sequence of a DNA•RNA complex.52 This higher hybridization strength leads to greater specificity in binding to complementary DNAs or RNAs because a PNA/RNA base mismatch is therefore more destabilizing than a similar mismatch in a DNA/RNA duplex.52 Here we exploit this effect to demonstrate that the use of PNA in the probe can build single-base discrimination capability in the nanopore.
We used probe P7b to target miRNAs Let-7b and Let-7c. Because Let-7c and Let-7b has one nucleotide difference (Table S1), the Let-7c•P7b hybrid contains a single mismatched base pair. Figure 4a shows the current trace for the Let-7b / P7b mixture in trans side of the pore, monitored at +130 mV in 3 M/0.5 M cis/trans KCl (pH7.2). The duration of Level 2 signatures for fully-hybridized Let-7b•P7b was 2.3±0.5 s. When using P7b to detect Let-7c under the same conditions, the number of Level 2 signatures was reduced. Instead, the current trace in Figure 4b shows a distinct type of two-level block from Level 2 to Level 1. These blocks should be attributed to the unzipping of the Let-7b•P7b complex. Such two-level blocks were rarely observed in Let-7b•P7b, and their duration was 19±7 ms –about 120 times shorter compared with Level 2 blocks for Let-7b•P7b. All these findings suggests that the fully-matched miRNA•PNA duplex is sufficiently stable to resist de-hybridization, while a single mismatch introduced in the duplex of Let-7c•P7b significantly destabilizes its hybridization. This generates a 120-fold difference in block duration between two miRNAs, and the mismatch-induced unzipping of the complex ensures high-fidelity differentiation of the mismatch from a fully complementary complex. As a result, the current traces for Let-7b and Let-7c can easily be discriminated visually: the long single-step Level 2 block with a high occurrence represents Let-7b, and the short two-step block with greatly reduced occurrence represents Let-7c.
Figure 4.
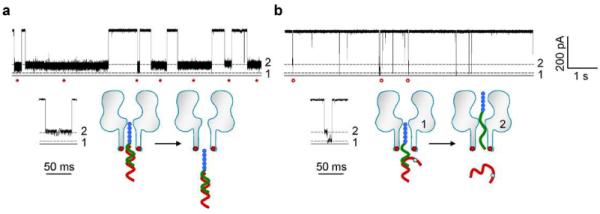
Discrimination of miRNAs with single-nucleotide difference. The probe P7b was used to detect Let-7b and Let-7c. The sequences of the two miRNAs have a single base difference. The nanopore was monitored at +130 mV in 3 M/0.5 M cis/trans KCl. a, Current trace showing the long Level 2 signatures produced by the fully-matched Let-7b•P7b complex. b, current traces showing short two-level signatures produced by Let-7c•P7b complex containing one mismatched base pair. The transition from Level 2 to Level 1 in these signatures suggests the unzipping of the complex in the electrical field.
This high-fidelity differentiation is also possible even at elevated voltages where de-hybridization occurs more frequently. At +180 mV, many of Let-7b•P7b blocks also featured a Level 2-Level 1 transition for unzipping. However, the duration of Let-7b•P7b blocks (1.7±0.6 s) was still over 150 times longer than Let-7c•P7b (11±3 ms) (Figure S4). Therefore both miRNAs can still be accurately discriminated based on the block duration even when unzipping does occur for the Let-7b•P7b blocks.
Enhancing sensitivity with optimized pore and probe. The integration of probe design and nanopore engineering enables the highly sensitive nucleic acid detection demonstrated above. This high sensitivity is driven in part by the judicious choice of pore mutants and probe designs.
In our K131D mutant pore, the anionic aspartic acids ring constructed at the trans opening of the pore play an important role in attracting cationic molecules, thus greatly increases the target capture rate kon. For example, the TAT peptide's kon at +180 mV was 4.1±0.9 μM−1·s−1 in the wild-type pore. The use of K131D vastly elevated the kno over 200-fold to 880±110 μM−1·s−1 (Figure 5a). Similarly, kon for P7b (peptide-PNA) was enhanced by 90-fold from 3.2±0.9 μM−1·s−1 in the wild-type pore to 280±70 μM−1·s−1 in the K131D pore (Figure 5a). Finally, the Let-7b•P7b complex was rarely trapped in the wild-type pore, while its kon was 80±9 μM−1·s−1 in the K131D pore. We have previously reported that kon for the miRNA in complex with a DNA probe was 1.4 μM−1·s−1.21 Therefore the combined used of the polycationic probe and the mutant pore enhanced kon over 50-fold. Figure 5b shows that the frequency of Let-7b•P7b signatures consistently increases with increasing Let-7b concentrations ranging from 50 pM to 5 nM at +180 mV. These data can be fitted to a straight line in the log-log scale, which shows us that target concentrations lower than 50 pM should be detectable. However, at concentrations that low, the frequency of miRNA signatures would be similar to the spontaneous gating events in the K131D pore53 (~6×10−3 s−1 above +140 mV). Fortunately, these gating events can be distinguished from miRNA signatures (Figure S5), and thus can be excluded from events used for miRNA quantification. Recent studies have demonstrated that further structure-directed protein engineering can prevent these intrinsic gate events from occurring,54,55 providing an approach for future improvement of this system's detection sensitivity.
Figure 5.
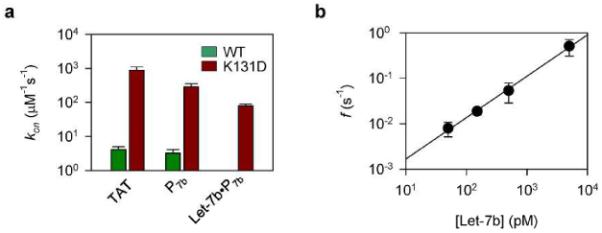
Quantification of miRNAs. a, Capture rates for the TAT peptide, the probe P7b and the Let-7b•P7b complex in the wild-type (WT) and K131D pores. b, Concentration-dependent frequency of Let-7b•P7b signatures in the K131D pore.
Concerning our polycationic probe, several key properties of the probe allow us to achieve these critical functions: separate the probe-bound miRNA from free nucleic acids, lead the miRNA•probe complex into the nanopore, and enhance the sensitivity by promoting the capture rate. First, the peptide's sequence and structure are programmable. Its properties can be tuned by adjusting the peptide length and, in particular, the number and position of charged amino acids. The peptide can also be functionalized at both the terminal end and at cysteines in any position of the sequence, making it possible to generate different signatures for multiplex detection. Finally, peptide-PNA probes have the advantage of being synthesized together as their units are all linked by the peptide bond, avoiding additional crosslinking. In future work, we plan to optimize the probe architecture, including net charge count and the charge distribution in order to enhanced the capture rate of the miRNA•probe dipole in the pore..
CONCLUSION AND PERSPECTIVE
A polycationic probe can be used for selective nucleic acid detection in the nanopore. While free nucleic acids are electrophoretically driven away from the mouth of the pore, the hybrid of the probe with the target nucleic acids forms a dipole that can be pulled into the asymmetrical electric field at the pore opening. This allows us to selectively detect only those nucleic acid sequences that hybridize with the probe, even when many other confounding species are present.
This result has the potential to be very useful for clinical detection; therefore the authors are motivated to perform future studies to elucidate the mechanism for the probe-induced nucleic acids separation in an asymmetric electric field, called nano-dielectrophoresis. Once the mechanism is determined, we will be able to further optimize and improve our approaches for highly selective and sensitive miRNA detection. If validated in clinical samples, for example the detection of target miRNA from RNA extractions derived from a patients' biofluids, this method would have applications in many areas such as: early disease diagnosis, cancer metastasis prediction, and the monitoring of a patient's response to therapy. In conclusion, this novel approach introduces a new method of detecting clinically relevant DNA or RNA fragments in a complex nucleic acids mixture.
MATERIALS AND METHODS
Chemicals and materials. Let-7b and Let-7c miRNA oligonucleotides were synthesized and electrophoresis-purified by Integrated DNA Technologies (Coralville, IA). The P7b peptide-PNA probe was synthesized and HPLC-purified by Bio-Synthesis Inc (Lewisville, TX) with a purity of 95%. All polymers were dissolved in RNAase-free water to 100 μM as stocks. Before the nanopore measurement, the miRNAs, probe and their mixtures at desired concentrations were heated to 90 °C for 5 minutes, then gradually cooled down to room temperature. Lipid 1,2-diphytanoyl-sn-glycerophosphatidylcholine (DPhPC) was purchased from Avanti Polar Lipids (Alabaster, AL). The 25-μm-thick Teflon film was obtained from Goodfellow Inc (Oakdale, PA). The mutant α-hemolysin proteins K131D and M113R were constructed according to a method similar to the previous report56. The wild-type and mutant proteins were synthesized using the in vitro Transcription and Translation kit provided by Promega Corporation (Madison, WI), and collected from the electrophoresis gel57.
Recording of single protein pores. Nanopore electrical recording was conducted according to previous reports21,58. The lipid bilayer membrane was formed over a 100–150 μm orifice in the center of the Teflon film that partitioned between cis and trans recording solutions. Both solutions contained KCl at a desired concentration and were buffered with 10 mM Tris (pH 7.2). The proteins were added in the cis solution, from which they were inserted into the bilayer to form a nanopore. miRNA and probe polymers were released to the trans solution. The voltage was applied from trans solution and cis solution was grounded, such that a positive voltage can pull the positively charged probe and miRNA•probe complex into the pore from trans entrance. The ionic flow through the pore were recorded with an Axopatch 200B amplifier (Molecular Device Inc., Sunnyvale, CA), filtered with a built-in 4-pole low-pass Bessel Filter at 5 kHz, and acquired with Clampex 9.0 software (Molecular Device Inc.) through a Digidata 1440 A/D converter (Molecular Device Inc.) at a sampling rate of 20 kHz. Single-channel event amplitude and duration were analyzed using Clampfit 9.0 (Molecular Device Inc.), Excel (Microsoft) and SigmaPlot (SPSS) software. The nanopore measurements were conducted at 22 ± 2 °C. Data was presented as mean±SD of at least three independent experiments.
Modeling of the miRNA•PNA duplex. The structure of the miRNA•PNA duplex was estimated using an experimentally determined template (PDB id: 176D); the peptide domain was constructed using the CABS and PULCHRA models59–61 and the miRNA segment using the Vfold model.62 The overall structure and charge were calculated from the ensemble average over 20 randomly selected flexible conformations and 100 miRNA conformations in total 2,000 conformations (Supplementary Information S1).
Supplementary Material
Acknowledgements
We appreciate Dr. ZhiQiang Hu's laboratory for the help with zeta potential measurement. This investigation was partially supported by grants from the National Science Foundation 0546165 (L.Q.G), the National Institutes of Health GM079613 (L.Q.G.), Coulter Translational Partner Program at the University of Missouri (L.Q.G.), and was conducted in a facility that was constructed with support from the Research Facilities Improvement Program Grant Number C06-RR-016489-01 from the National Centre for Research Resources, National Institutes of Health.
Footnotes
Supporting Information is available free of charge via the Internet at http://pubs.acs.org. Competing Financial Interests. There is no competing financial interest
Reference List
- 1.Bayley H, Jayasinghe L. Functional Engineered Channels and Pores - (Review) Molecular Membrane Biology. 2004;21:209–220. doi: 10.1080/09687680410001716853. [DOI] [PubMed] [Google Scholar]
- 2.Bayley H, Cronin B, Heron A, Holden MA, Hwang WL, Syeda R, Thompson J, Wallace M. Droplet Interface Bilayers. Mol.Biosyst. 2008;4:1191–1208. doi: 10.1039/b808893d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu LQ, Shim JW. Single Molecule Sensing by Nanopores and Nanopore Devices. Analyst. 2010;135:441–451. doi: 10.1039/b907735a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall AR, Scott A, Rotem D, Mehta KK, Bayley H, Dekker C. Hybrid Pore Formation by Directed Insertion of Alpha-Haemolysin into Solid-State Nanopores. Nat Nanotechnol. 2010;5:874–877. doi: 10.1038/nnano.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hornblower B, Coombs A, Whitaker RD, Kolomeisky A, Picone SJ, Meller A, Akeson M. Single-Molecule Analysis of DNA-Protein Complexes Using Nanopores. Nat Methods. 2007;4:315–317. doi: 10.1038/nmeth1021. [DOI] [PubMed] [Google Scholar]
- 6.Howorka S, Siwy Z. Nanopore Analytics: Sensing of Single Molecules. Chemical Society Reviews. 2009;38:2360–2384. doi: 10.1039/b813796j. [DOI] [PubMed] [Google Scholar]
- 7.Ma L, Cockroft SL. Biological Nanopores for Single-Molecule Biophysics. Chembiochem. 2010;11:25–34. doi: 10.1002/cbic.200900526. [DOI] [PubMed] [Google Scholar]
- 8.Majd S, Yusko EC, Billeh YN, Macrae MX, Yang J, Mayer M. Applications of Biological Pores in Nanomedicine, Sensing, and Nanoelectronics. Current Opinion in Biotechnology. 2010;21:439–476. doi: 10.1016/j.copbio.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Movileanu L. Interrogating Single Proteins Through Nanopores: Challenges and Opportunities. Trends Biotechnol. 2009;27:333–341. doi: 10.1016/j.tibtech.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 10.Olasagasti F, Lieberman KR, Benner S, Cherf GM, Dahl JM, Deamer DW, Akeson M. Replication of Individual DNA Molecules Under Electronic Control Using a Protein Nanopore. Nat.Nanotechnol. 2010;5:798–806. doi: 10.1038/nnano.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venkatesan BM, Bashir R. Nanopore Sensors for Nucleic Acid Analysis. Nat Nanotechnol. 2011;6:615–624. doi: 10.1038/nnano.2011.129. [DOI] [PubMed] [Google Scholar]
- 12.Wanunu M, Morrison W, Rabin Y, Grosberg AY, Meller A. Electrostatic Focusing of Unlabelled DNA into Nanoscale Pores Using a Salt Gradient. Nat.Nanotechnol. 2010;5:160–165. doi: 10.1038/nnano.2009.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wendell D, Jing P, Geng J, Subramaniam V, Lee TJ, Montemagno C, Guo P. Translocation of Double-Stranded DNA Through Membrane-Adapted Phi29 Motor Protein Nanopores. Nat Nanotechnol. 2009;4:765–772. doi: 10.1038/nnano.2009.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langecker M, Arnaut V, Martin TG, List J, Renner S, Mayer M, Dietz H, Simmel FC. Synthetic Lipid Membrane Channels Formed by Designed DNA Nanostructures. Science. 2012;338:932–936. doi: 10.1126/science.1225624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X, Jovanovich SB, Krstic PS, Lindsay S, Ling XS, Mastrangelo CH, Meller A, Oliver JS, Pershin YV, Ramsey JM, Riehn R, Soni GV, Tabard-Cossa V, Wanunu M, Wiggin M, Schloss JA. The Potential and Challenges of Nanopore Sequencing. Nature Biotechnology. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cherf GM, Lieberman KR, Rashid H, Lam CE, Karplus K, Akeson M. Automated Forward and Reverse Ratcheting of DNA in a Nanopore at 5-A Precision. Nat Biotechnol. 2012;30:344–348. doi: 10.1038/nbt.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of Individual Polynucleotide Molecules Using a Membrane Channel. Proc.Natl.Acad.Sci.U.S.A. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manrao EA, Derrington IM, Laszlo AH, Langford KW, Hopper MK, Gillgren N, Pavlenok M, Niederweis M, Gundlach JH. Reading DNA at Single-Nucleotide Resolution With a Mutant MspA Nanopore and Phi29 DNA Polymerase. Nat Biotechnol. 2012;30:349–353. doi: 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wallace EV, Stoddart D, Heron AJ, Mikhailova E, Maglia G, Donohoe TJ, Bayley H. Identification of Epigenetic DNA Modifications With a Protein Nanopore. Chem.Commun.(Camb.) 2010;46:8195–8197. doi: 10.1039/c0cc02864a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.An N, Fleming AM, White HS, Burrows CJ. Crown Ether-Electrolyte Interactions Permit Nanopore Detection of Individual DNA Abasic Sites in Single Molecules. Proc Natl Acad Sci U S A. 2012;109:11504–11509. doi: 10.1073/pnas.1201669109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Zheng D, Tan Q, Wang MX, Gu LQ. Nanopore-Based Detection of Circulating MicroRNAs in Lung Cancer Patients. Nat.Nanotechnol. 2011;6:668–674. doi: 10.1038/nnano.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wanunu M, Dadosh T, Ray V, Jin J, McReynolds L, Drndi-ç M. Rapid Electronic Detection of Probe-Specific MicroRNAs Using Thin Nanopore Sensors. Nat.Nanotechnol. 2010;5:807–814. doi: 10.1038/nnano.2010.202. [DOI] [PubMed] [Google Scholar]
- 23.Carthew RW, Sontheimer EJ. Origins and Mechanisms of MiRNAs and SiRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee RC, Feinbaum RL, Ambros V. The C. Elegans Heterochronic Gene Lin-4 Encodes Small RNAs With Antisense Complementarity to Lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 25.Kim VN, Han J, Siomi MC. Biogenesis of Small RNAs in Animals. Nat.Rev.Mol.Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 26.Wightman B, Ha I, Ruvkun G. Posttranscriptional Regulation of the Heterochronic Gene Lin-14 by Lin-4 Mediates Temporal Pattern Formation in C. Elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 27.Boeri M, Verri C, Conte D, Roza L, Modena P, Facchinetti F, Calabr+╎ E, Croce CM, Pastorino U, Sozzi G. MicroRNA Signatures in Tissues and Plasma Predict Development and Prognosis of Computed Tomography Detected Lung Cancer. Proc.Natl.Acad.Sci.U.S.A. 2011;108:3713–3718. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu Y, Chen Y, Xu L, Zen K, Zhang C, Shen H. Serum MicroRNA Signatures Identified in a Genome-Wide Serum MicroRNA Expression Profiling Predict Survival of Non-Small-Cell Lung Cancer. J Clin.Oncol. 2010;28:1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 29.Hunt EA, Goulding AM, Deo SK. Direct Detection and Quantification of MicroRNAs. Anal.Biochem. 2009;387:1–12. doi: 10.1016/j.ab.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iorio MV, Croce CM. MicroRNAs in Cancer: Small Molecules With a Huge Impact. J.Clin.Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landi MT, Zhao Y, Rotunno M, Koshiol J, Liu H, Bergen AW, Rubagotti M, Goldstein AM, Linnoila I, Marincola FM, Tucker MA, Bertazzi PA, Pesatori AC, Caporaso NE, McShane LM, Wang E. MicroRNA Expression Differentiates Histology and Predicts Survival of Lung Cancer. Clin.Cancer Res. 2010;16:430–441. doi: 10.1158/1078-0432.CCR-09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating MicroRNAs As Stable Blood-Based Markers for Cancer Detection. Proc.Natl.Acad.Sci.U.S.A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen J, Todd NW, Zhang H, Yu L, Lingxiao X, Mei Y, Guarnera M, Liao J, Chou A, Lu CL, Jiang Z, Fang H, Katz RL, Jiang F. Plasma MicroRNAs As Potential Biomarkers for Non-Small-Cell Lung Cancer. Lab Invest. 2011;91:579–587. doi: 10.1038/labinvest.2010.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sozzi G, Roz L, Conte D, Mariani L, Andriani F, Lo VS, Verri C, Pastorino U. Plasma DNA Quantification in Lung Cancer Computed Tomography Screening: Five-Year Results of a Prospective Study. Am.J Respir.Crit Care Med. 2009;179:69–74. doi: 10.1164/rccm.200807-1068OC. [DOI] [PubMed] [Google Scholar]
- 35.Zheng D, Haddadin S, Wang Y, Gu LQ, Perry MC, Freter CE, Wang MX. Plasma Micrornas As Novel Biomarkers for Early Detection of Lung Cancer. Int.J.Clin.Exp.Pathol. 2011;4:575–586. [PMC free article] [PubMed] [Google Scholar]
- 36.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annual Review of Medicine. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 37.Ortholan C, Puissegur MP, Ilie M, Barbry P, Mari B, Hofman P. MicroRNAs and Lung Cancer: New Oncogenes and Tumor Suppressors, New Prognostic Factors and Potential Therapeutic Targets. Curr.Med.Chem. 2009;16:1047–1061. doi: 10.2174/092986709787581833. [DOI] [PubMed] [Google Scholar]
- 38.Takeshima K, Chikushi A, Lee KK, Yonehara S, Matsuzaki K. Translocation of Analogues of the Antimicrobial Peptides Magainin and Buforin Across Human Cell Membranes. Journal of Biological Chemistry. 2003;278:1310–1315. doi: 10.1074/jbc.M208762200. [DOI] [PubMed] [Google Scholar]
- 39.Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Structure of Staphylococcal Alpha-Hemolysin, a Heptameric Transmembrane Pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 40.Nakane J, Wiggin M, Marziali A. A Nanosensor for Transmembrane Capture and Identification of Single Nucleic Acid Molecules. Biophysical Journal. 2004;87:615–621. doi: 10.1529/biophysj.104.040212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Dorp S, Keyser UF, Dekker NH, Dekker C, Lemay SG. Origin of the Electrophoretic Force on DNA in Solid-State Nanopores. Nature Physics. 2009;5:347–351. [Google Scholar]
- 42.Luan B, Aksimentiev A. Electric and Electrophoretic Inversion of the DNA Charge in Multivalent Electrolytes. Soft.Matter. 2010;6:243–246. doi: 10.1039/b917973a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meller A, Nivon L, Branton D. Voltage-Driven DNA Translocations Through a Nanopore. Phys Rev Lett. 2001;86:3435–3438. doi: 10.1103/PhysRevLett.86.3435. [DOI] [PubMed] [Google Scholar]
- 44.Firnkes M, Pedone D, Knezevic J, Doblinger M, Rant U. Electrically Facilitated Translocations of Proteins Through Silicon Nitride Nanopores: Conjoint and Competitive Action of Diffusion, Electrophoresis, and Electroosmosis. Nano.Lett. 2010;10:2162–2167. doi: 10.1021/nl100861c. [DOI] [PubMed] [Google Scholar]
- 45.Maglia G, Restrepo MR, Mikhailova E, Bayley H. Enhanced Translocation of Single DNA Molecules Through α-Hemolysin Nanopores by Manipulation of Internal Charge. Proc.Natl.Acad.Sci.U.S.A. 2008;105:19720–19725. doi: 10.1073/pnas.0808296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gu LQ, Cheley S, Bayley H. Electroosmotic Enhancement of the Binding of a Neutral Molecule to a Transmembrane Pore. Proc.Natl.Acad.Sci.U.S.A. 2003;100:15498–15503. doi: 10.1073/pnas.2531778100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aksimentiev A, Schulten K. Imaging Alpha-Hemolysin With Molecular Dynamics: Ionic Conductance, Osmotic Permeability, and the Electrostatic Potential Map. Biophys.J. 2005;88:3745–3761. doi: 10.1529/biophysj.104.058727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bhattacharya S, Muzard L, Payet L, Mathe J, Bockelmann U, Aksimentiev A, Viasnoff V. Rectification of the Current in Alpha-Hemolysin Pore Depends on the Cation Type: the Alkali Series Probed by MD Simulations and Experiments. J.Phys.Chem.C.Nanomater.Interfaces. 2011;115:4255–4264. doi: 10.1021/jp111441p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nielsen PE, Egholm M. An Introduction to Peptide Nucleic Acid. Curr.Issues Mol.Biol. 1999;1:89–104. [PubMed] [Google Scholar]
- 50.Nielsen PE, Shiraishi T. Peptide Nucleic Acid (PNA) Cell Penetrating Peptide (CPP) Conjugates As Carriers for Cellular Delivery of Antisense Oligomers. Artif.DNA PNA.XNA. 2011;2:90–99. doi: 10.4161/adna.18739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singer A, Wanunu M, Morrison W, Kuhn H, Frank-Kamenetskii M, Meller A. Nanopore Based Sequence Specific Detection of Duplex DNA for Genomic Profiling. Nano Lett. 2010;10:738–742. doi: 10.1021/nl100058y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Natsume T, Ishikawa Y, Dedachi K, Tsukamoto T, Kurita N. Hybridization Energies of Double Strands Composed of DNA, RNA, PNA and LNA. Chem Phys Lett. 2007;434:133–138. doi: 10.1016/j.cplett.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mohammad MM, Movileanu L. Impact of Distant Charge Reversals Within a Robust Beta-Barrel Protein Pore. J.Phys.Chem.B. 2010;114:8750–8759. doi: 10.1021/jp101311s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mohammad MM, Iyer R, Howard KR, McPike MP, Borer PN, Movileanu L. Engineering a Rigid Protein Tunnel for Biomolecular Detection. J.Am.Chem.Soc. 2012;134:9521–9531. doi: 10.1021/ja3043646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen M, Khalid S, Sansom MS, Bayley H. Outer Membrane Protein G: Engineering a Quiet Pore for Biosensing. Proc.Natl.Acad.Sci.U.S.A. 2008;105:6272–6277. doi: 10.1073/pnas.0711561105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Howorka S, Bayley H. Improved Protocol for High-Throughput Cysteine Scanning Mutagenesis. BioTechniques. 1998;25:764–6. 768–770. doi: 10.2144/98255bm03. [DOI] [PubMed] [Google Scholar]
- 57.Shim JW, Yang M, Gu LQ. In Vitro Synthesis, Tetramerization and Single Channel Characterization of Virus-Encoded Potassium Channel Kcv. FEBS Lett. 2007;581:1027–1034. doi: 10.1016/j.febslet.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 58.Shim JW, Tan Q, Gu LQ. Single-Molecule Detection of Folding and Unfolding of the G-Quadruplex Aptamer in a Nanopore Nanocavity. Nucleic Acids Res. 2009;37:972–982. doi: 10.1093/nar/gkn968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kolinski A. Protein Modeling and Structure Prediction With a Reduced Representation. Acta Biochim.Pol. 2004;51:349–371. [PubMed] [Google Scholar]
- 60.Kolinski A, Skolnick J. Reduced Models of Proteins and Their Applications. Polymer. 2004;45:511–524. [Google Scholar]
- 61.Rotkiewicz P, Skolnick J. Fast Procedure for Reconstruction of Full-Atom Protein Models From Reduced Representations. J Comput.Chem. 2008;29:1460–1465. doi: 10.1002/jcc.20906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cao S, Chen SJ. Predicting RNA Folding Thermodynamics With a Reduced Chain Representation Model. RNA. 2005;11:1884–1897. doi: 10.1261/rna.2109105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


