Abstract
We investigated the taxonomic placement of phenazine-producing fluorescent Pseudomonas spp. in the Inland Pacific Northwest region of the United States. Five distinct species were identified, two of which were provisionally considered to be new. Agroclimatic zone and soil silt content affected the species diversity across the region.
TEXT
Phenazines are functionally diverse heterocyclic nitrogen-containing compounds produced by bacteria containing phz genes (1, 2). Indigenous phenazine-producing (Phz+) pseudomonads are abundant in certain agricultural soils (1, 3, 4), and such strains inhibit soilborne plant pathogens (3, 4). In the Inland Pacific Northwest region of the United States (IPNW), Phz+ pseudomonads are genetically and phenotypically diverse (5) and particularly abundant in the low-precipitation zone, where they occur at densities of 105 to 107 CFU g root−1 (4, 6). However, the taxonomy of these Phz+ populations and the extent to which Phz+ rhizobacterial community composition is impacted by abiotic factors remains unclear. Here, we addressed this knowledge gap by using multilocus sequence analysis (MLSA) (7) of Phz+ isolates and sequence analysis of phzF alleles cloned from IPNW commercial wheat fields. Our results provide new insights into the prevalence of Phz+ pseudomonads in the environment and serve as a foundation for characterizing such species globally.
During this study, we sampled several wheat-growing areas throughout the IPNW (Fig. 1; see also Fig. S1 in the supplemental material) and isolated indigenous Phz+ strains (see Table S1 and Table S2 in the supplemental material) and extracted DNA from the rhizosphere of field-grown wheat (Table 1). Indigenous populations of Phz+ Pseudomonas were quantified using the terminal endpoint dilution assay (1, 8). Phz+ strains were isolated from rhizosphere suspensions and subjected to BOX-PCR profiling (5, 9). The newly isolated Phz+ strains, along with those isolated in previous surveys (1, 5), were subjected to profiling by BOX-PCR (5, 10). Isolates of the same banding profile from each location were grouped, and one representative isolate was selected for multilocus sequence analysis (MLSA) (7). Phylogeny was defined with MLSA using partial DNA sequences for 16S rRNA (1,384 nucleotides [nt]), gyrB (794 nt), rpoB (903 nt), and rpoD (698 nt) that were amplified, respectively, with the primer sets 8F/1492R (11), Up-1G-/Up-2G- (1), rpoBup1/rpoBlow1 (1) and LAPS/LAPS27 (12), and PsEG30F and PsEG790R (13) (see Table S4 in the supplemental material). Genomic DNA was extracted from strains using a GenElute bacterial genomic DNA kit (Sigma-Aldrich Co., St. Louis, MO). PCR amplifications were performed using GoTaq DNA polymerase (Promega, Inc., Madison, WI) on a PTC-200 thermal cycler (Bio-Rad, Hercules, CA), and the amplicons were sequenced by Elim Biopharm (Hayward, CA). DNA sequences were processed using CLC bio Main Workbench v.6.6.2 (CLC Bio, Aarhus, Denmark) and aligned in MEGA5 (14) using MUSCLE (15). Phylogenetic distances were estimated using the Jukes-Cantor substitution model and bootstrapped with 1,000 replications. Simultaneous Alignment and Tree Estimation v.2.2.2 (SATé-II) (16) was used for maximum-likelihood MLSA phylogeny estimation with the suggested settings and GTR+G20 as the model. We also carried out detailed sequence analysis of phzF alleles amplified directly from rhizosphere DNA samples isolated from rhizosphere suspensions with a PowerSoil DNA isolation kit (Mo Bio, Carlsbad, CA). phzF alleles were amplified with the primers Ps_up1/Ps_low1 (1), and the amplicons were cloned into the pGEM-T Easy vector (Promega, Madison, WI). The resultant recombinant plasmids were purified using a MinElute 96 UF kit (Qiagen, Valencia, CA) and sequenced by Elim Biopharm (Hayward, CA). The nucleotide sequences were analyzed in SATé-II (16), and the resultant phylogeny was visualized in Dendroscope v.3.2.2 (17). The phzF-based taxonomy classification was performed in Mothur v.1.26.0 (18) using “classify.seqs” and a taxonomy database constructed from the reference alleles. An alignment without reference phzF alleles was used to calculate the inverse Simpson diversity (1/D) estimate (19). All calculations in Mothur were carried out using a phylogenetic distance cutoff of 0.04 nucleotide substitution per site.
Fig 1.
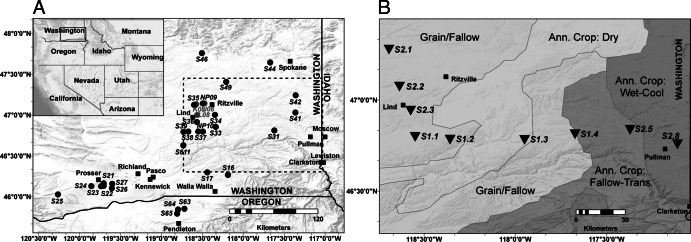
Distribution of sites sampled for Phz+ populations throughout the Inland Pacific Northwest. All sites labeled with an “S” in panel A were dryland winter wheat fields sampled for Phz+ Pseudomonas populations in a previous study (4) and used to isolate new Phz+ strains to establish MLSA-based phylogenetic placement. Sampled sites in panel B were used to isolate DNA from the rhizosphere of winter wheat for subsequent amplification of phzF for cloning and sequencing. The map in panel B also indicates agroclimatic zones (22). Three sites (K08, J08, and L08) sampled previously (5) were also included and are shaded in gray. Spatial data visualization was performed in ArcGIS v.9.3 (Esri, Inc.). The map was created with ArcGIS (base map from USGS, Esri, Tele Atlas, and Automotive Navigation Data [AND]).
Table 1.
Characteristics of sites sampled for total rhizosphere DNA
| Sample | Geographic coordinates | Total no. of phzF clones | Phz+ population density (log CFU g root−1 [fresh wt])a | Phz+ plant colonization frequency | Agroclimate zoneb | Mean annual precipitation (mm)c |
|---|---|---|---|---|---|---|
| S1.1 | 46°47′49″N, 118°32′57″W | 51 | 5.4 ± 0.6 | 1.0 | Grain/fallow | 259 |
| S1.2 | 46°46′53″N, 118°21′28″W | 54 | 5.6 ± 1.0 | 0.7 | Grain/fallow | 297 |
| S1.3 | 46°46′50″N, 117°56′58″W | 52 | 5.3 ± 0.8 | 0.8 | Annual crop; dry | 371 |
| S1.4 | 46°48′40″N, 117°40′22″W | 55 | 6.3 ± 0.6 | 0.6 | Grain/fallow | 430 |
| S2.1 | 47°16′33″N, 118°41′34″W | 52 | 5.4 ± 1.3 | 0.6 | Grain/fallow | 273 |
| S2.2 | 47°4′20″N, 118°38′7″W | 52 | 5.7 ± 0.9 | 0.6 | Grain/fallow | 265 |
| S2.3 | 46°56′26″N, 118°34′26″W | 46 | 5.3 ± 0.8 | 0.8 | Grain/fallow | 258 |
| S2.5 | 46°50′9″N, 117°22′18″W | 44 | 4.8 ± 0.5 | 0.2 | Annual crop; fallow-transition | 512 |
| S2.8 | 46°45′20″N, 117°6′41″W | 48 | 5.0 ± 0.6 | 0.3 | Annual crop; wet-cool | 581 |
At each location, 16 plants were randomly sampled and root suspensions were used to determine population density and plant colonization by the terminal endpoint dilution assay (1, 8). Values are means ± standard deviations.
Agroclimatic zones defined by Douglas et al. (22).
Mean annual precipitation data for 30 years (1971–2000) from the PRISM Climate Group (www.prism.oregonstate.edu/).
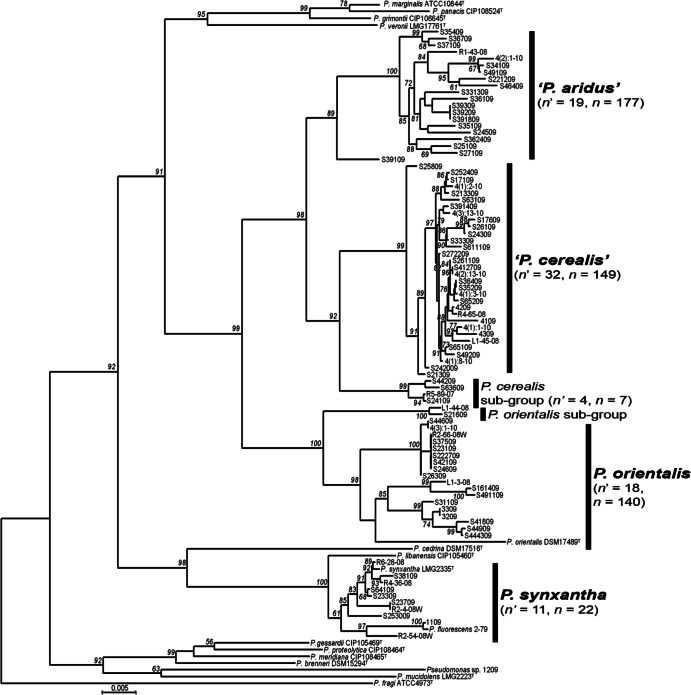
The largest clade found using BOX-PCR profiling and MLSA was provisionally named “Pseudomonas aridus” (n = 177) (Fig. 2), as it occurred only in fields that receive the least annual precipitation. The most closely related described species had between 4.1% and 4.5% sequence dissimilarity (Table 2). The second largest clade, provisionally named “Pseudomonas cerealis,” included a subgroup with ca. 2% MLSA dissimilarity (n = 156) (Fig. 2) and was more cosmopolitan throughout the IPNW. “Pseudomonas cerealis” was most closely related to strains of P. orientalis (ca. 3.7% dissimilarity) and “P. aridus.” “P. cerealis” and “P. aridus” were found at the majority of sites throughout the IPNW (Table 2) and comprised over 67% of our Phz+ isolates. The DNA sequence similarities of these groups were below 97%, the MLSA-defined cutoff for defining species within the Pseudomonas genus (7). Phenotypic differences in PCA and biosurfactant production in vitro also were observed between these two species (5), in addition to differential carbon substrate utilization profiles (5) (see Table S3 in the supplemental material).
Fig 2.
Phylogenetic placement of Phz+ Pseudomonas spp. isolates in the “P. fluorescens” and “P. gessardii” subgroups as defined by Mulet et al. (7). Eighty-eight representative Phz+ isolates (n′) were analyzed alongside 14 Pseudomonas type strains, including the outgroup P. fragi ATCC 4973T. The total number of strains defined by BOX-PCR in this study (n) is also indicated. The scale bar indicates the number of nucleotide substitutions per site.
Table 2.
MLSA-based sequence dissimilarity comparison of Phz+ strain groups to species of the P. fluorescens lineage
| Strain | Dissimilarity value (%)a |
||||
|---|---|---|---|---|---|
| P. aridus group | P. cerealis group | P. orientalis group | P. synxantha group | Pseudomonas sp. strain 1209 | |
| P. brenneri DSM15294T | 5.3 ± 0.3 | 5.4 ± 0.3 | 5.8 ± 0.4 | 5.0 ± 0.3 | 5.2 ± 0.3 |
| P. cedrina DSM17516T | 5.2 ± 0.3 | 5.0 ± 0.3 | 4.6 ± 0.3 | 4.5 ± 0.3 | 6.3 ± 0.4 |
| P. gessardi CIP105469T | 5.5 ± 0.3 | 5.4 ± 0.3 | 5.7 ± 0.4 | 5.0 ± 0.3 | 5.0 ± 0.3 |
| P. grimontii CIP106645T | 4.1 ± 0.3 | 4.2 ± 0.3 | 4.5 ± 0.3 | 4.6 ± 0.3 | 5.6 ± 0.4 |
| P. libanensis CIP105460T | 4.8 ± 0.3 | 4.9 ± 0.3 | 5.0 ± 0.3 | 1.5 ± 0.2 | 6.2 ± 0.4 |
| P. marginalis ATCC10844T | 4.3 ± 0.3 | 4.3 ± 0.3 | 4.6 ± 0.3 | 4.7 ± 0.3 | 5.7 ± 0.4 |
| P. meridiana CIP108465T | 4.8 ± 0.3 | 4.8 ± 0.3 | 5.2 ± 0.3 | 5.3 ± 0.3 | 4.8 ± 0.3 |
| P. mucidolens LMG2223T | 6.2 ± 0.4 | 6.0 ± 0.4 | 6.8 ± 0.4 | 5.8 ± 0.4 | 5.5 ± 0.3 |
| P. orientalis DSM17489T | 4.1 ± 0.3 | 3.7 ± 0.3 | 2.4 ± 0.2 | 4.8 ± 0.3 | 6.6 ± 0.3 |
| P. panacis CIP108524T | 4.5 ± 0.3 | 4.5 ± 0.3 | 4.5 ± 0.3 | 4.8 ± 0.3 | 5.6 ± 0.4 |
| P. proteolytica CIP108464T | 5.0 ± 0.3 | 5.0 ± 0.3 | 5.4 ± 0.3 | 4.9 ± 0.3 | 4.9 ± 0.3 |
| P. synxantha LMG2335T | 4.5 ± 0.3 | 4.5 ± 0.3 | 4.6 ± 0.3 | 0.6 ± 0.1 | 6.2 ± 0.4 |
| P. veronii LMG1776T | 4.5 ± 0.3 | 4.5 ± 0.3 | 4.5 ± 0.3 | 4.8 ± 0.4 | 5.2 ± 0.3 |
The sequence dissimilarity values (means ± standard errors) were calculated as p distance in MEGA5 using 1,000 bootstrap replications.
The P. orientalis clade contained the type strain P. orientalis CFML-170 (20) and 140 IPNW Phz+ strains (Fig. 2) as well as a small subgroup at 2.8% dissimilarity (see Table S2 in the supplemental material). The P. synxantha clade included the type strain P. synxantha LMG 2335 and the model strain 2-79 (21) (n = 22) (Fig. 2) and clearly diverged from other species (Table 2). This report provides the first evidence for phenazine production by either P. orientalis or P. synxantha (20). Finally, Pseudomonas sp. strain 1209 was genetically distinct from other pseudomonads, as it was only 95.2% similar by MLSA to any previously described species (Fig. 2; also, see Table S2 in the supplemental material). We also found that three different dryland plant hosts (spring wheat, common yarrow, and needle-and-thread grass) supported largely different communities of Phz+ species, although spring wheat and common yarrow shared some closely related “P. cerealis” strains (Fig. 2; also, see Table S1 in the supplemental material).
Among 454 sequenced phzF amplicons from different locations, the majority of alleles belonged to one of the four dominant Phz+ species (Fig. 3). The largest percentage of these (45%) were classified as “P. cerealis.” Another 10% (45 of 454) were classified as Pseudomonas sp. 1209-like, and 44 of these originated from sample S2.8 in the wet-cool annual crop agroclimatic zone (22), suggesting that this group favors higher-precipitation areas. No phzF alleles were assigned to P. synxantha, and a small group of sequences from samples S1.3 and S2.8 did not match any of the known Phz+ species (Fig. 3). Rarefaction analysis of phzF revealed that the Phz+ communities were nearly completely sampled at seven of the eight sites and S2.8 had the greatest OTU richness (see Fig. S2 in the supplemental material). Taken together with population densities of 105 to 106 Phz+ CFU per gram of root, these results indicate very low overall diversity in the Phz+ community (Table 1). We also estimated the diversity of phzF by calculating the inverse Simpson's index (1/D) (19) and found that S2.5 had the highest diversity, at 2.47. Sites S2.8 and S1.4 had the next highest, at 2.01 and 1.58, respectively, whereas sites S1.1 and S2.1 had the lowest diversity, at 1.00. Mean annual precipitation had only a slight effect on diversity (t-test P value = 0.0560; analysis of variance [ANOVA] F-test P value = 0.0593); however, soil percent silt (t-test P value = 0.0220; ANOVA F-test P value = 0.0233) and agroclimatic zone (t-test P value = 0.0188; ANOVA F-test P value = 0.0217) showed a much greater effect where overall diversity was lowest under “grain/fallow” management (see Fig. S3 in the supplemental material). Future studies on the effect of soil type, crop management, and water availability on the Phz+ community composition under controlled conditions are needed to determine the specific influence of these factors on the ecology of Phz+ rhizobacteria.
Fig 3.
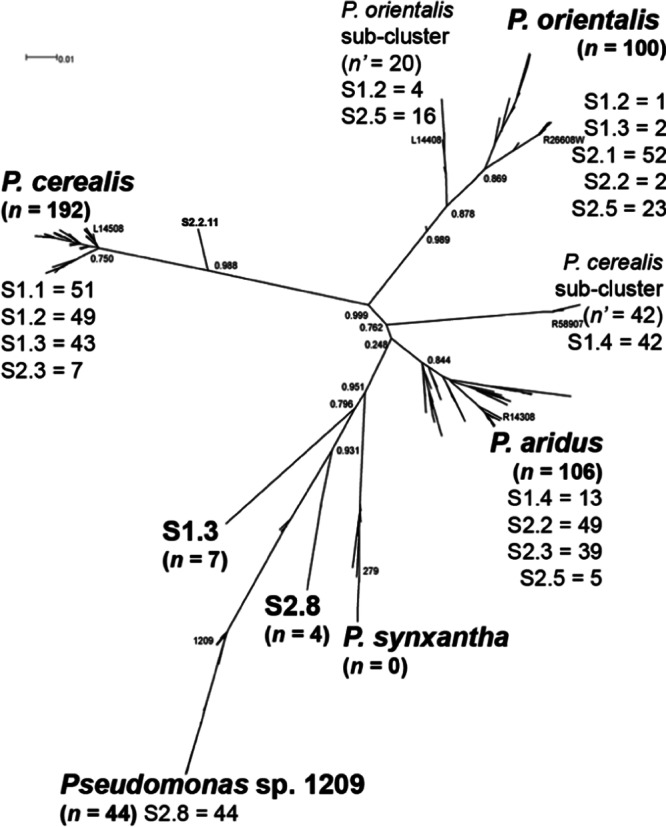
Unrooted maximum-likelihood phylogenetic tree of partial (389 nt) phzF DNA sequences obtained from representative strains featured in this (n = 76) and previous (n = 37) (1, 5) studies, as well as from samples of rhizosphere DNA (n = 454). The total number of clones within the cluster (n) or subcluster (n′) specific to each geographic site is indicated. A representative strain from the MLSA analysis is also shown for each cluster.
This study provides new insights into the evolutionary ecology and global distribution of beneficial plant-associated Phz+ bacteria. While it previously appeared that the capacity to produce phenazines was limited to just a few Pseudomonas species (1), we have now demonstrated the presence of an extensive complex of Phz+ species belonging to the P. fluorescens lineage in the IPNW. The provisional “P. aridus” and “P. cerealis” species are particularly abundant in this semiarid region and may have unique traits allowing them to thrive under dryland conditions. We also demonstrated that, in addition to mean annual precipitation, the agroclimatic zone and soil silt content significantly affect the diversity of indigenous Phz+ communities. These results contribute to the rational exploitation of beneficial microbial communities for the purpose of improving plant health and managing soilborne pathogens in agroecosystems.
Nucleotide sequence accession numbers.
MLSA and phzF DNA sequences were deposited in the NCBI GenBank database with accession numbers KC834074 to KC834377 and KC842530 to KC843059, respectively.
Supplementary Material
ACKNOWLEDGMENTS
J.A.P. was supported in part by award T32GM083864 from the National Institute of General Medical Sciences.
We are grateful to Bellevue College summer interns Jennifer Apple, Emilia Gan, and Chelsea Stone for their diligent and excellent help performing BOX-PCR and genomic DNA extractions. We also thank Irina Mavrodi for her help in purifying phzF amplicon DNA.
USDA is an equal opportunity provider and employer. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Footnotes
Published ahead of print 12 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03945-12.
REFERENCES
- 1. Mavrodi DV, Peever TL, Mavrodi OV, Parejko JA, Raaijmakers JM, Lemanceau P, Mazurier S, Heide L, Blankenfeldt W, Weller DM, Thomashow LS. 2010. Diversity and evolution of the phenazine biosynthesis pathway. Appl. Environ. Microbiol. 76:866–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mavrodi DV, Blankenfeldt W, Thomashow LS. 2006. Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu. Rev. Phytopathol. 44:417–445 [DOI] [PubMed] [Google Scholar]
- 3. Mazurier S, Corberand T, Lemanceau P, Raaijmakers JM. 2009. Phenazine antibiotics produced by fluorescent pseudomonads contribute to natural soil suppressiveness to Fusarium wilt. ISME J. 3:977–991 [DOI] [PubMed] [Google Scholar]
- 4. Mavrodi DV, Mavrodi OV, Parejko JA, Bonsall RF, Kwak YS, Paulitz TC, Thomashow LS, Weller DM. 2012. Accumulation of the antibiotic phenazine-1-carboxylic acid in the rhizosphere of dryland cereals. Appl. Environ. Microbiol. 78:804–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Parejko JA, Mavrodi DV, Mavrodi OV, Thomashow LS, Weller DM. 2012. Population structure and diversity of phenazine-1-carboxylic acid producing fluorescent Pseudomonas spp. from dryland cereal fields of central Washington State (U.S.). Microb. Ecol. 64:226–241 [DOI] [PubMed] [Google Scholar]
- 6. Mavrodi OV, Mavrodi DV, Parejko JA, Thomashow LS, Weller DM. 2012. Irrigation differentially impacts populations of indigenous antibiotic-producing Pseudomonas spp. in the rhizosphere of wheat. Appl. Environ. Microbiol. 78:3214–3220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mulet M, Lalucat J, García-Valdés E. 2010. DNA sequence-based analysis of the Pseudomonas species. Environ. Microbiol. 12:1513–1530 [DOI] [PubMed] [Google Scholar]
- 8. McSpadden Gardener BB, Mavrodi DV, Thomashow LS, Weller DM. 2001. A rapid polymerase chain reaction-based assay characterizing rhizosphere populations of 2,4-diacetylphloroglucinol-producing bacteria. Phytopathology 91:44–54 [DOI] [PubMed] [Google Scholar]
- 9. Versalovic J, Schneider M, De Bruijn F, Lupski J. 1994. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 5:25–40 [Google Scholar]
- 10. Rademaker JLW, De Bruijn FJ. 1997. Characterization and classification of microbes by rep-PCR genomic fingerprinting and computer assisted pattern analysis, p 151–171 In Caetano-Anollés G, Gresshoff PM. (ed), DNA markers: protocols, applications and overviews. Wiley, New York, NY [Google Scholar]
- 11. Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ait Tayeb L, Ageron E, Grimont F, Grimont PAD. 2005. Molecular phylogeny of the genus Pseudomonas based on rpoB sequences and application for the identification of isolates. Res. Microbiol. 156:763–773 [DOI] [PubMed] [Google Scholar]
- 13. Mulet M, Bennasar A, Lalucat J, García-Valdés E. 2009. An rpoD-based PCR procedure for the identification of Pseudomonas species and for their detection in environmental samples. Mol. Cell. Probes 23:140–147 [DOI] [PubMed] [Google Scholar]
- 14. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 10:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu K, Warnow TJ, Holder MT, Nelesen SM, Yu J, Stamatakis AP, Linder CR. 2012. SATe-II: very fast and accurate simultaneous estimation of multiple sequence alignments and phylogenetic trees. Syst. Biol. 61:90–106 [DOI] [PubMed] [Google Scholar]
- 17. Huson DH, Scornavacca C. 2012. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 61:1061–1067 [DOI] [PubMed] [Google Scholar]
- 18. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hill TCJ, Walsh KA, Harris JA, Moffett BF. 2003. Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 43:1–11 [DOI] [PubMed] [Google Scholar]
- 20. Dabboussi F, Hamze M, Elomari M, Verhille S, Baida N, Izard D, Leclerc H. 1999. Taxonomic study of bacteria isolated from Lebanese spring waters: proposal for Pseudomonas cedrella sp. nov. and P. orientalis sp. nov. Res. Microbiol. 150:303–316 [DOI] [PubMed] [Google Scholar]
- 21. Thomashow LS, Weller DM. 1988. Role of a phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var. tritici. J. Bacteriol. 170:3499–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Douglas CLJ, Rickman RW, Klepper BL, Zuzel JF. 1992. Agroclimatic zones for dryland winter wheat producing areas of Idaho, Washington, and Oregon. NW Sci. 66:26–34 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.