Abstract
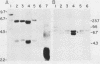
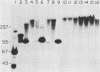
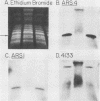
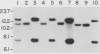
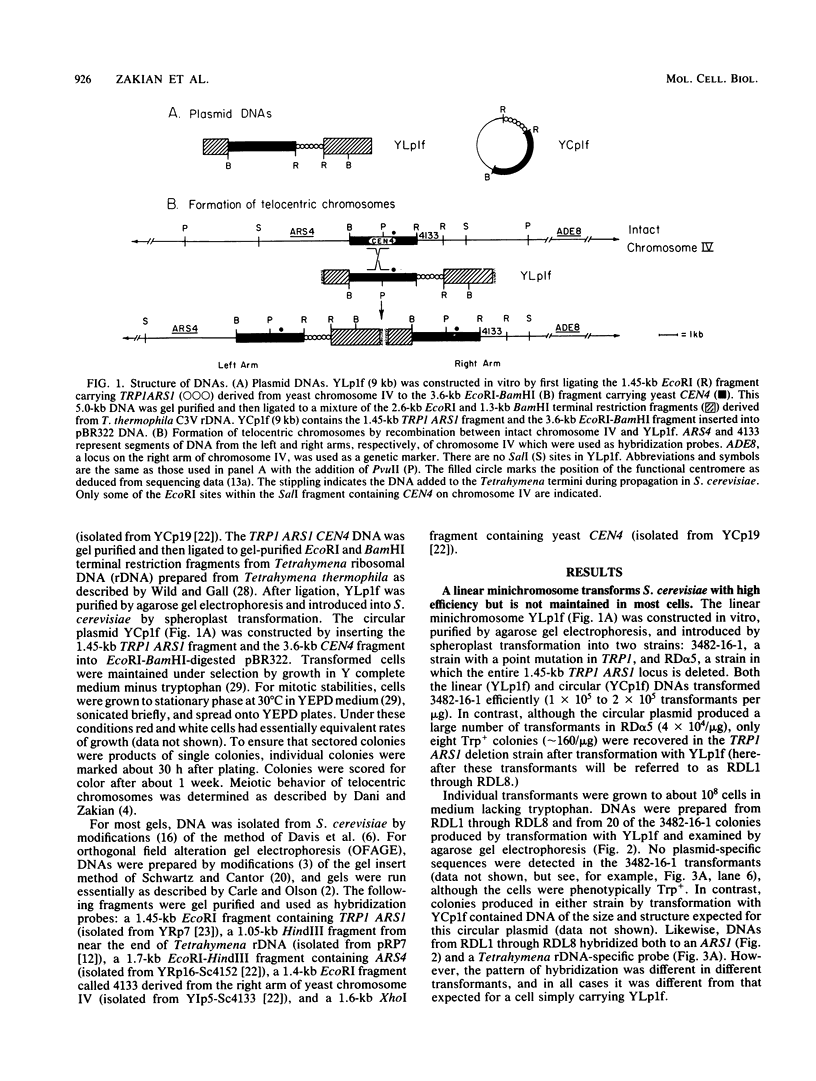
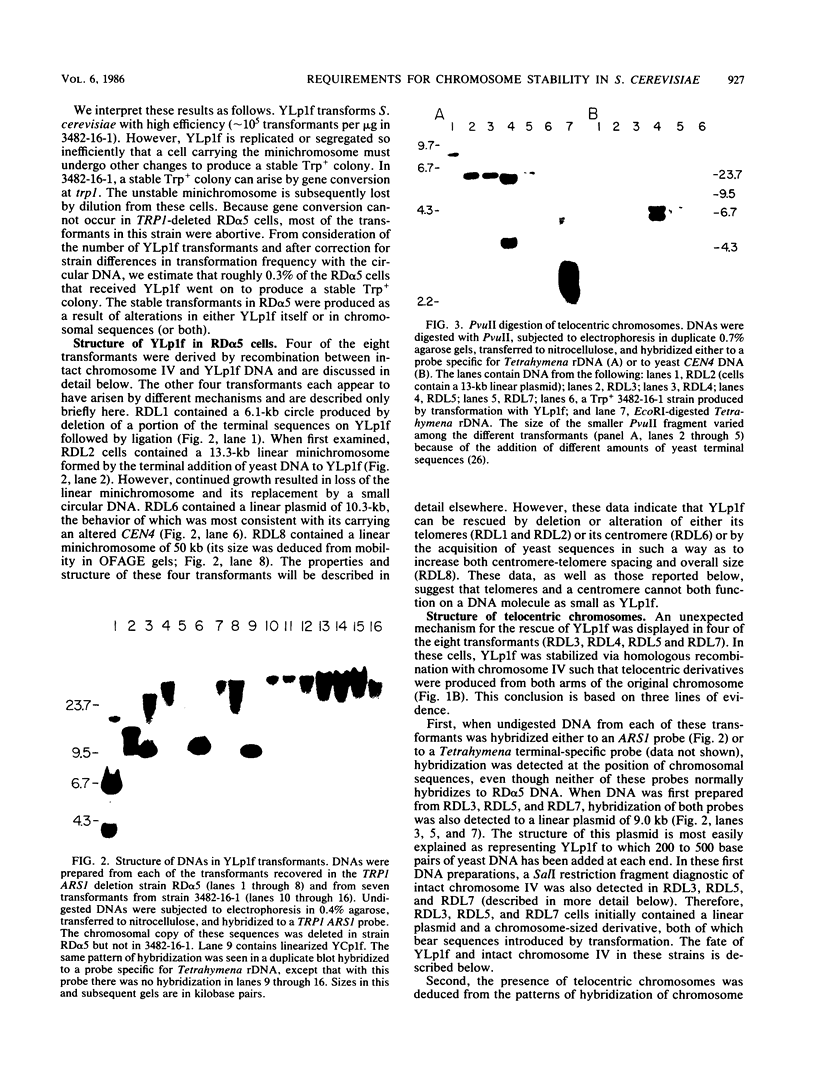
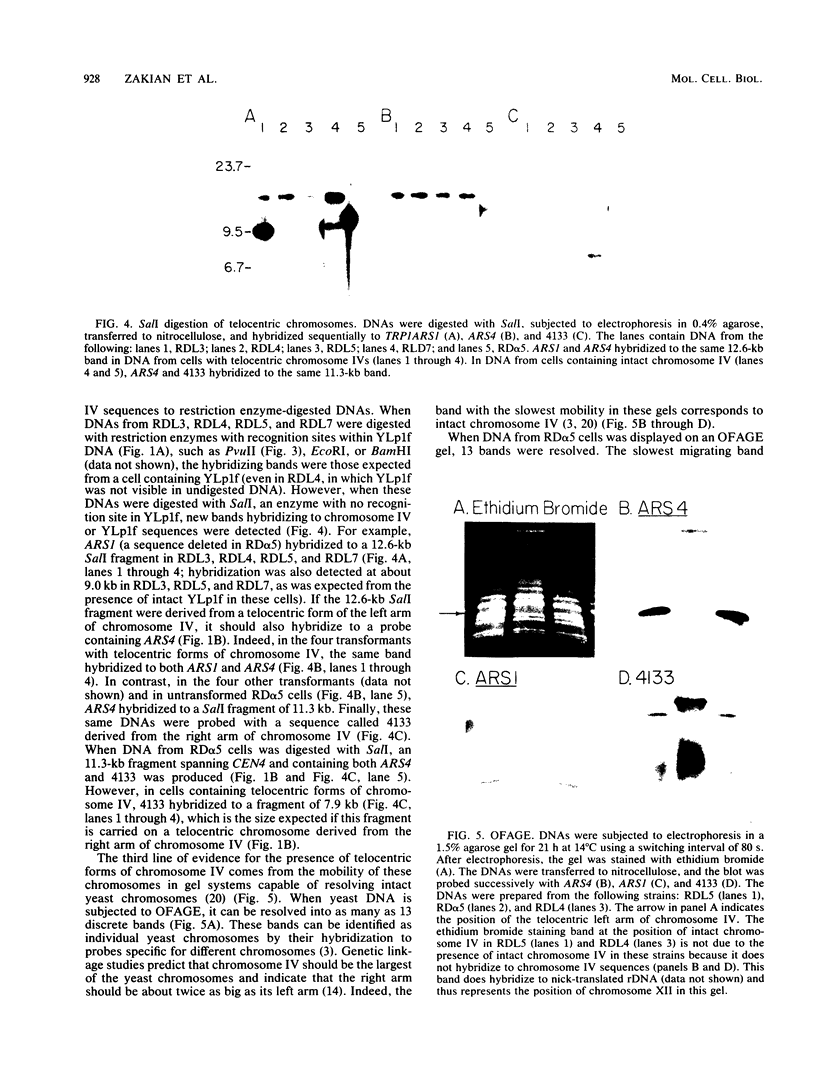
A 9-kilobase pair CEN4 linear minichromosome constructed in vitro transformed Saccharomyces cerevisiae with high frequency but duplicated or segregated inefficiently in most cells. Stable transformants were only produced by events which fundamentally altered the structure of the minichromosome: elimination of telomeres, alteration of the centromere, or an increase of fivefold or greater in its size. Half of the stable transformants arose via homologous recombination between an intact chromosome IV and the CEN4 minichromosome. This event generated a new chromosome from each arm of chromosome IV. The other "arm" of each new chromosome was identical to one "arm" of the unstable minichromosome. Unlike natural yeast chromosomes, these new chromosomes were telocentric: their centromeres were either 3.9 or 5.4 kilobases from one end of the chromosome. The mitotic stability of the telocentric chromosome derived from the right arm of chromosome IV was determined by a visual assay and found to be comparable to that of natural yeast chromosomes. Both new chromosomes duplicated, paired, and segregated properly in meiosis. Moreover, their structure, as deduced from mobilities in orthogonal field gels, did not change with continued mitotic growth or after passage through meiosis, indicating that they did not give rise to isochromosomes or suffer large deletions or additions. Thus, in S. cerevisiae the close spacing of centromeres and telomeres on a DNA molecule of chromosomal size does not markedly alter the efficiency with which it is maintained. Taken together these data suggest that there is a size threshold below which stable propagation of linear chromosomes is no longer possible.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Carle G. F., Olson M. V. An electrophoretic karyotype for yeast. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3756–3760. doi: 10.1073/pnas.82.11.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carle G. F., Olson M. V. Separation of chromosomal DNA molecules from yeast by orthogonal-field-alternation gel electrophoresis. Nucleic Acids Res. 1984 Jul 25;12(14):5647–5664. doi: 10.1093/nar/12.14.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani G. M., Zakian V. A. Mitotic and meiotic stability of linear plasmids in yeast. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3406–3410. doi: 10.1073/pnas.80.11.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. W., Thomas M., Cameron J., St John T. P., Scherer S., Padgett R. A. Rapid DNA isolations for enzymatic and hybridization analysis. Methods Enzymol. 1980;65(1):404–411. doi: 10.1016/s0076-6879(80)65051-4. [DOI] [PubMed] [Google Scholar]
- Dunn B., Szauter P., Pardue M. L., Szostak J. W. Transfer of yeast telomeres to linear plasmids by recombination. Cell. 1984 Nov;39(1):191–201. doi: 10.1016/0092-8674(84)90205-8. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H., Smith D. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics. 1985 Jul;110(3):381–395. doi: 10.1093/genetics/110.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieter P., Mann C., Snyder M., Davis R. W. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985 Feb;40(2):381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- Kiss G. B., Amin A. A., Pearlman R. E. Two separate regions of the extrachromosomal ribosomal deoxyribonucleic acid of Tetrahymena thermophila enable autonomous replication of plasmids in Saccharomyces cerevisiae. Mol Cell Biol. 1981 Jun;1(6):535–543. doi: 10.1128/mcb.1.6.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann C., Davis R. W. Structure and sequence of the centromeric DNA of chromosome 4 in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jan;6(1):241–245. doi: 10.1128/mcb.6.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer R. K., Schild D. Genetic map of Saccharomyces cerevisiae. Microbiol Rev. 1980 Dec;44(4):519–571. doi: 10.1128/mr.44.4.519-571.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A. W., Szostak J. W. Construction of artificial chromosomes in yeast. Nature. 1983 Sep 15;305(5931):189–193. doi: 10.1038/305189a0. [DOI] [PubMed] [Google Scholar]
- Pluta A. F., Dani G. M., Spear B. B., Zakian V. A. Elaboration of telomeres in yeast: recognition and modification of termini from Oxytricha macronuclear DNA. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1475–1479. doi: 10.1073/pnas.81.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROMAN H. Studies of gene mutation in Saccharomyces. Cold Spring Harb Symp Quant Biol. 1956;21:175–185. doi: 10.1101/sqb.1956.021.01.015. [DOI] [PubMed] [Google Scholar]
- Rhoades M M. A Cytogenetic Study of a Chromosome Fragment in Maize. Genetics. 1936 Sep;21(5):491–502. doi: 10.1093/genetics/21.5.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M M. Studies of a Telocentric Chromosome in Maize with Reference to the Stability of Its Centromere. Genetics. 1940 Sep;25(5):483–520. doi: 10.1093/genetics/25.5.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Sora S., Lucchini G., Magni G. E. Meiotic Diploid Progeny and Meiotic Nondisjunction in SACCHAROMYCES CEREVISIAE. Genetics. 1982 May;101(1):17–33. doi: 10.1093/genetics/101.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcomb D. T., Mann C., Davis R. W. Centromeric DNA from Saccharomyces cerevisiae. J Mol Biol. 1982 Jun 25;158(2):157–190. doi: 10.1016/0022-2836(82)90427-2. [DOI] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surosky R. T., Tye B. K. Construction of telocentric chromosomes in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Apr;82(7):2106–2110. doi: 10.1073/pnas.82.7.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W., Blackburn E. H. Cloning yeast telomeres on linear plasmid vectors. Cell. 1982 May;29(1):245–255. doi: 10.1016/0092-8674(82)90109-x. [DOI] [PubMed] [Google Scholar]
- Walmsley R. M., Szostak J. W., Petes T. D. Is there left-handed DNA at the ends of yeast chromosomes? Nature. 1983 Mar 3;302(5903):84–86. doi: 10.1038/302084a0. [DOI] [PubMed] [Google Scholar]
- Wild M. A., Gall J. G. An intervening sequence in the gene coding for 25S ribosomal RNA of Tetrahymena pigmentosa. Cell. 1979 Mar;16(3):565–573. doi: 10.1016/0092-8674(79)90030-8. [DOI] [PubMed] [Google Scholar]
- Zakian V. A., Scott J. F. Construction, replication, and chromatin structure of TRP1 RI circle, a multiple-copy synthetic plasmid derived from Saccharomyces cerevisiae chromosomal DNA. Mol Cell Biol. 1982 Mar;2(3):221–232. doi: 10.1128/mcb.2.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]