Abstract
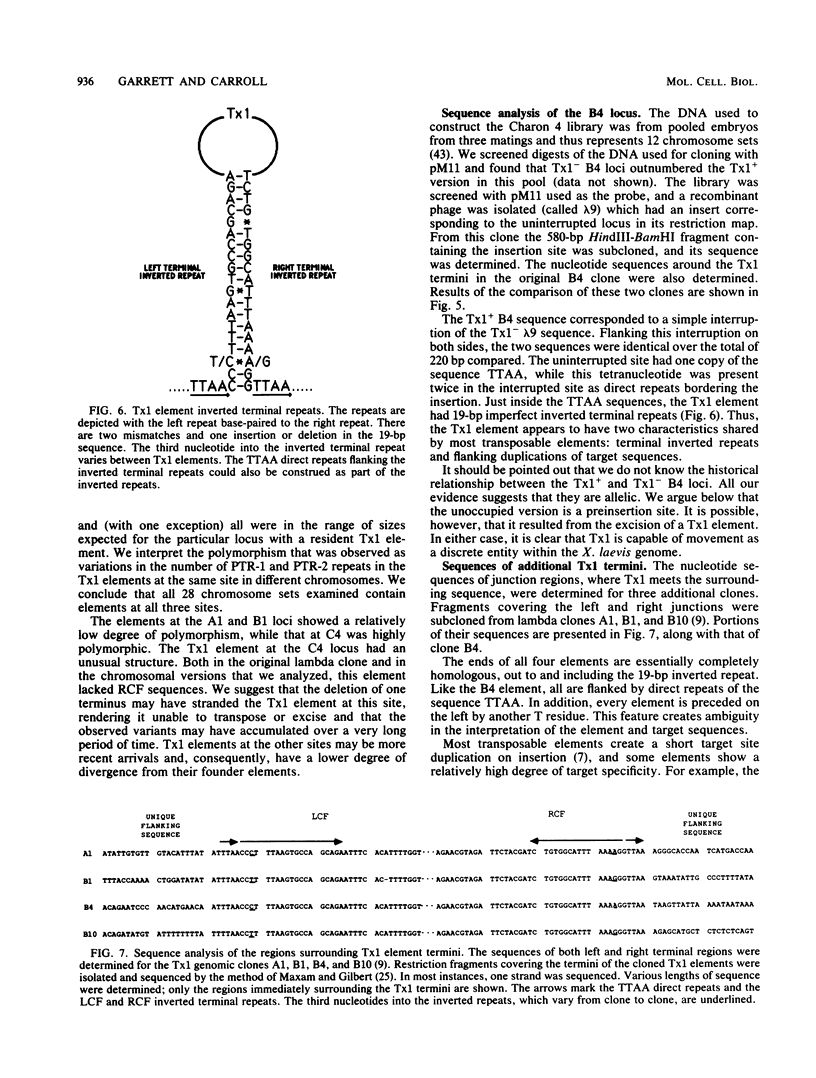
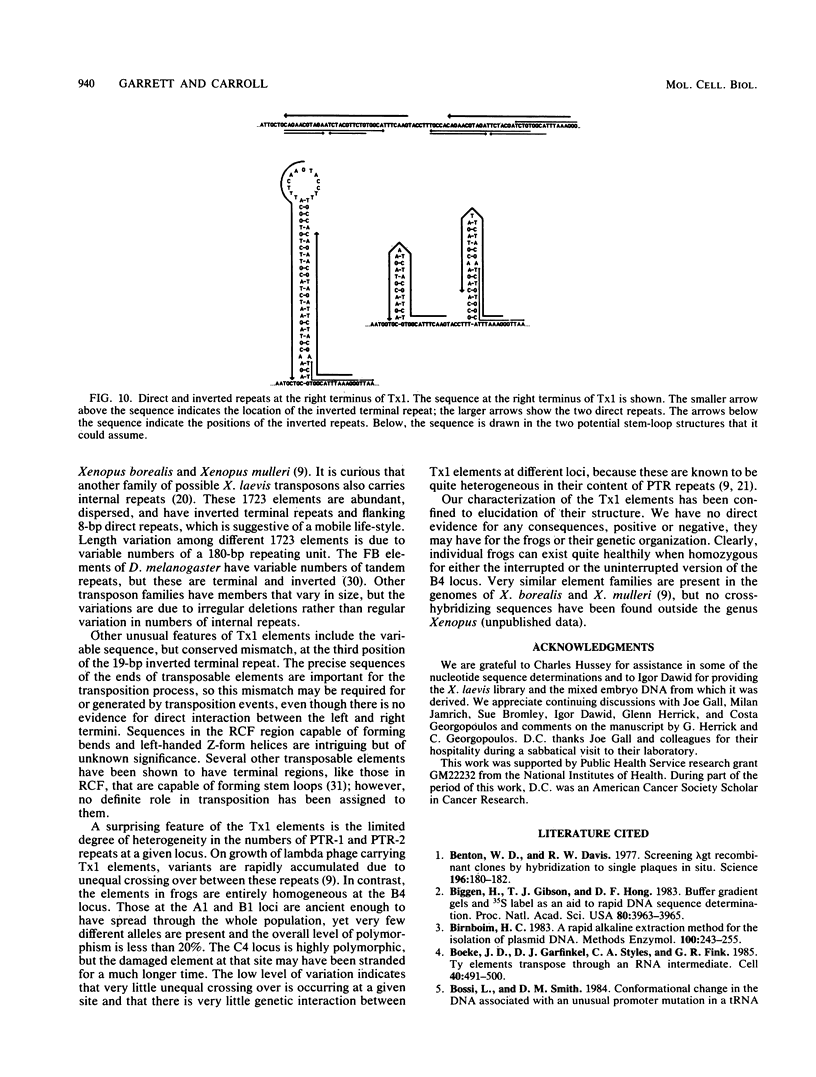
A family of transposable genetic elements in the genome of the frog, Xenopus laevis, is described. They are designated Tx1. Transposability of the elements was deduced by characterization of a chromosomal locus which is polymorphic for the presence or absence of a Tx1 element. Nucleotide sequence analysis suggested that Tx1 elements show target site specificity, as they are inserted at the pentanucleotide TTTAA in all four cases that were examined. The elements appear to have 19-base-pair (bp) inverted terminal repeats, and they are flanked by 4-bp target duplications (TTAA), although the possibility that they do not create target site duplications is discussed. Tx1 elements have several unusual characteristics: the central portion of each element is comprised of a variable number of two types of 393-bp repeating units; the rightmost 1,000 bp of the element contains separate regions potentially capable of forming bends, left-handed Z-form DNA, and alternative stem-loop structures. Comparisons among single frogs suggest that germ line transposition is relatively infrequent and that variations in numbers of internal repeats accumulate quite slowly at any locus.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C. A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol. 1983;100:243–255. doi: 10.1016/0076-6879(83)00059-2. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Garfinkel D. J., Styles C. A., Fink G. R. Ty elements transpose through an RNA intermediate. Cell. 1985 Mar;40(3):491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- Bossi L., Smith D. M. Conformational change in the DNA associated with an unusual promoter mutation in a tRNA operon of Salmonella. Cell. 1984 Dec;39(3 Pt 2):643–652. doi: 10.1016/0092-8674(84)90471-9. [DOI] [PubMed] [Google Scholar]
- Brown D. D., Wensink P. C., Jordan E. Purification and some characteristics of 5S DNA from Xenopus laevis. Proc Natl Acad Sci U S A. 1971 Dec;68(12):3175–3179. doi: 10.1073/pnas.68.12.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calos M. P., Miller J. H. Transposable elements. Cell. 1980 Jul;20(3):579–595. doi: 10.1016/0092-8674(80)90305-0. [DOI] [PubMed] [Google Scholar]
- Cappello J., Cohen S. M., Lodish H. F. Dictyostelium transposable element DIRS-1 preferentially inserts into DIRS-1 sequences. Mol Cell Biol. 1984 Oct;4(10):2207–2213. doi: 10.1128/mcb.4.10.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D., Garrett J. E., Lam B. S. Isolated clusters of paired tandemly repeated sequences in the Xenopus laevis genome. Mol Cell Biol. 1984 Feb;4(2):254–259. doi: 10.1128/mcb.4.2.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare J., Farabaugh P. Nucleotide sequence of a yeast Ty element: evidence for an unusual mechanism of gene expression. Proc Natl Acad Sci U S A. 1985 May;82(9):2829–2833. doi: 10.1073/pnas.82.9.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison R. A., Weiner A. M. Human U1 RNA pseudogenes may be generated by both DNA- and RNA-mediated mechanisms. Mol Cell Biol. 1982 Jul;2(7):815–828. doi: 10.1128/mcb.2.7.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dente L., Cesareni G., Cortese R. pEMBL: a new family of single stranded plasmids. Nucleic Acids Res. 1983 Mar 25;11(6):1645–1655. doi: 10.1093/nar/11.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring H. P., Starlinger P. Barbara McClintock's controlling elements: now at the DNA level. Cell. 1984 Dec;39(2 Pt 1):253–259. doi: 10.1016/0092-8674(84)90002-3. [DOI] [PubMed] [Google Scholar]
- Fedoroff N. V., Brown D. D. The nucleotide sequence of oocyte 5S DNA in Xenopus laevis. I. The AT-rich spacer. Cell. 1978 Apr;13(4):701–716. doi: 10.1016/0092-8674(78)90220-9. [DOI] [PubMed] [Google Scholar]
- Garoff H., Ansorge W. Improvements of DNA sequencing gels. Anal Biochem. 1981 Aug;115(2):450–457. doi: 10.1016/0003-2697(81)90031-2. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Evidence for the existence of stable curvature of DNA in solution. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4632–4636. doi: 10.1073/pnas.81.15.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman P. J. Evidence for the existence of stable curvature of DNA in solution. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4632–4636. doi: 10.1073/pnas.81.15.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Kay B. K., Dawid I. B. The 1723 element: a long, homogeneous, highly repeated DNA unit interspersed in the genome of Xenopus laevis. J Mol Biol. 1983 Nov 5;170(3):583–596. doi: 10.1016/s0022-2836(83)80122-3. [DOI] [PubMed] [Google Scholar]
- Lam B. S., Carroll D. Tandemly repeated DNA sequences from Xenopus laevis. II. Dispersed clusters of a 388 base-pair repeating unit. J Mol Biol. 1983 Apr 25;165(4):587–597. doi: 10.1016/s0022-2836(83)80268-x. [DOI] [PubMed] [Google Scholar]
- Lemischka I., Sharp P. A. The sequences of an expressed rat alpha-tubulin gene and a pseudogene with an inserted repetitive element. Nature. 1982 Nov 25;300(5890):330–335. doi: 10.1038/300330a0. [DOI] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miesfeld R., Krystal M., Arnheim N. A member of a new repeated sequence family which is conserved throughout eucaryotic evolution is found between the human delta and beta globin genes. Nucleic Acids Res. 1981 Nov 25;9(22):5931–5947. doi: 10.1093/nar/9.22.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E., Löfdahl S. Transposition of Tn554 does not generate a target duplication. Nature. 1984 Jan 19;307(5948):292–294. doi: 10.1038/307292a0. [DOI] [PubMed] [Google Scholar]
- Nishioka Y., Leder A., Leder P. Unusual alpha-globin-like gene that has cleanly lost both globin intervening sequences. Proc Natl Acad Sci U S A. 1980 May;77(5):2806–2809. doi: 10.1073/pnas.77.5.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hare K., Rubin G. M. Structures of P transposable elements and their sites of insertion and excision in the Drosophila melanogaster genome. Cell. 1983 Aug;34(1):25–35. doi: 10.1016/0092-8674(83)90133-2. [DOI] [PubMed] [Google Scholar]
- Potter S. S. DNA sequence of a foldback transposable element in Drosophila. Nature. 1982 May 20;297(5863):201–204. doi: 10.1038/297201a0. [DOI] [PubMed] [Google Scholar]
- Rhodes P. R., Vodkin L. O. Highly structured sequence homology between an insertion element and the gene in which it resides. Proc Natl Acad Sci U S A. 1985 Jan;82(2):493–497. doi: 10.1073/pnas.82.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich A., Nordheim A., Wang A. H. The chemistry and biology of left-handed Z-DNA. Annu Rev Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- Rogers J. A straight LINE story. Nature. 1983 Nov 10;306(5939):113–114. doi: 10.1038/306113a0. [DOI] [PubMed] [Google Scholar]
- Rosenzweig B., Liao L. W., Hirsh D. Target sequences for the C. elegans transposable element Tc1. Nucleic Acids Res. 1983 Oct 25;11(20):7137–7140. doi: 10.1093/nar/11.20.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmus H. E. Reverse transcriptase rides again. Nature. 1985 Apr 18;314(6012):583–584. doi: 10.1038/314583a0. [DOI] [PubMed] [Google Scholar]
- Vodkin L. O., Rhodes P. R., Goldberg R. B. cA lectin gene insertion has the structural features of a transposable element. Cell. 1983 Oct;34(3):1023–1031. doi: 10.1016/0092-8674(83)90560-3. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahli W., Dawid I. B. Isolation of two closely related vitellogenin genes, including their flanking regions, from a Xenopus laevis gene library. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1437–1441. doi: 10.1073/pnas.77.3.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981 Feb 26;289(5800):751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]