Abstract
Monoterpenes can, upon hydrogenation, be used as light-fraction components of sustainable aviation fuels. Fermentative production of monoterpenes in engineered microorganisms, such as Saccharomyces cerevisiae, has gained attention as a potential route to deliver these next-generation fuels from renewable biomass. However, end product toxicity presents a formidable problem for microbial synthesis. Due to their hydrophobicity, monoterpene inhibition has long been attributed to membrane interference, but the molecular mechanism remains largely unsolved. In order to gain a better understanding of the mode of action, we analyzed the composition and structural integrity of the cell envelope as well as the transcriptional response of yeast cells treated with an inhibitory amount of d-limonene (107 mg/liter). We found no alterations in membrane fluidity, structural membrane integrity, or fatty acid composition after the solvent challenge. A 4-fold increase in the mean fluorescence intensity per cell (using calcofluor white stain) and increased sensitivity to cell wall-degrading enzymes demonstrated that limonene disrupts cell wall properties. Global transcript measurements confirmed the membrane integrity observations by showing no upregulation of ergosterol or fatty acid biosynthesis pathways, which are commonly overexpressed in yeast to reinforce membrane rigidity during ethanol exposure. Limonene shock did cause a compensatory response to cell wall damage through overexpression of several genes (ROM1, RLM1, PIR3, CTT1, YGP1, MLP1, PST1, and CWP1) involved with the cell wall integrity signaling pathway. This is the first report demonstrating that cell wall, rather than plasma membrane, deterioration is the main source of monoterpene inhibition. We show that limonene can alter the structure and function of the cell wall, which has a clear effect on cytokinesis.
INTRODUCTION
Monoterpenes are 10-carbon (C10) olefins composed of two 5-carbon (C5) isoprene units (1). Monoterpenes, such as limonene, pinene, and cymene, have traditionally been used as flavors and fragrances (2) but recently have gained attention as potential light-component precursors for drop-in jet fuels (3–5). In June 2012, a demonstration flight by Amyris Inc. proved that their biojet fuel, AMJ-700, successfully met engine performance requirements and required no changes to the aircraft (6). AMJ-700 contains 60% monoterpene-derived paraffins, which are critical for meeting strict fuel requirements (5). Although synthesis of monoterpene products in engineered biocatalysts, such as baker's yeast and Escherichia coli, is still in the developmental stages (only 30 μg/liter to 1 g/liter of monoterpene product has been achieved so far [7–10]), farnesene (a C15 sesquiterpene) uses the same precursors in the cell and is currently produced for diesel markets (11). However, the primary challenge facing microbial monoterpene synthesis is product toxicity, which adversely impacts production parameters such as titer, yield, and rate (12).
The mechanism behind monoterpene inhibition is poorly understood. While solvent stress in microorganisms has been studied extensively over the past 40 years, these reports have focused primarily on short-chain alcohols and organic acids (13), and although monoterpenes are well-known antifungal agents, only a few quantitative studies exist (3, 14–17). Due to their lipophilicity (log P = 4.5), monoterpene inhibition has been attributed to molecular toxicity, i.e., the direct interference of solvent molecules with membrane function (17–19). Consistent with this, increases in membrane fluidity (17) and structural membrane damage (20) were observed in whole yeast cells exposed to pinene.
Solvents partition between the membrane and the aqueous phase. For a sparingly water-soluble solvent in equilibrium, the membrane concentration will be proportional to the solvent's aqueous concentration, and molecular toxicity is typically observed to increase proportionally with the solvent's aqueous concentration (21–23). With the membrane and aqueous phases in equilibrium, molecular toxicity must peak when the aqueous concentration has reached its solubility limit, since both phases are saturated at this point.
Monoterpenes are essential oils; thus, they are sparingly soluble in water (e.g., Slimonene = 6 mg/liter) (24). In a recent study, we found that the inhibitory concentrations for five monoterpene products were at least an order of magnitude higher than their aqueous saturation limits (3). The cells were also able to grow at 97% of the maximum growth rate when their cellular compartments (e.g., plasma membrane) were saturated with monoterpene (3). Thus, monoterpene toxicity is per definition not molecular toxicity but rather phase toxicity (25). Phase toxicity has a phenomenological rather than a mechanistic definition: toxicity occurs in the presence of a distinct solvent phase (i.e., after water and membrane saturation) and is a nonequilibrium (kinetic) effect that increases with interfacial area (e.g., larger solvent phase volume and greater agitation).
Here we investigated the direct impact that limonene has on the yeast cell envelope. Many microorganisms, including yeast, Pseudomonas putida, and E. coli, respond to solvent stress by altering their lipid bilayers to counteract fluidization effects (13). To confirm whether or not molecular toxicity effects were indeed the source of limonene inhibition, membrane fluidity, structural membrane integrity, and membrane composition were measured after limonene exposure. Due to the lack of evidence for molecular toxicity, additional experiments such as cell wall integrity (CWI) staining and cell wall sensitivity assays were used to investigate potential sources of phase toxicity at the surface of the cell. Lastly, analysis of the global transcriptional response was used to support the physiological observations.
MATERIALS AND METHODS
Strain, chemicals, and growth conditions.
The Saccharomyces cerevisiae strain S288C (MATα SUC2 gal2 mal mel flo1 flo8-1 hap1) was provided by the Australian Wine Research Institute (AWRI), Adelaide, South Australia, Australia. Analytical-grade d-limonene (93%) was purchased from Sigma-Aldrich. The growth conditions and limonene challenge were as described previously (3) with the following modifications. Chemically defined medium (CBS) was used. CBS contained 20 g/liter sucrose, 5 g/liter (NH4)2SO4, 3 g/liter KH2PO4, 0.5 g/liter MgSO4 · 7H2O, and the exact components and concentration of vitamins and trace metals described by Brennan et al. (3). Overnight precultures (pH 5.0, 30°C) were used to inoculate 25 ml of fresh CBS to an optical density at 660 nm (OD660) of 0.5 against water, and then 2.8 μl of limonene (107 mg/liter) was added exogenously to the medium during mid-exponential growth phase (at ∼5 h; OD660 = 2.5). All of the measurements described below were taken 2 h after the limonene challenge for limonene-treated cultures and similarly for control cultures with no limonene present.
Fluorescence anisotropy.
The membrane fluidity of limonene-challenged and control cultures was determined using fluorescence anisotropy as described previously (26, 27) with the following modifications. After 2 h of limonene exposure, cells were harvested at 13,000 rpm for 1 min, washed twice with 15 mM Tris-HCl buffer, and resuspended to an OD660 of 0.2. A 1.0-μl volume of a 12 mM stock solution of diphenylhexatriene (DPH) in tetrahydrofuran was then added to the resuspension and incubated at 30°C for 30 min with continuous stirring (200 rpm) to allow the probe to intercalate into the plasma membrane (27). The anisotropy was measured with a Spectromax M5 (Sunnyvale, CA) at excitation and emission wavelengths of 358 and 428 nm, respectively, and recorded using Softmax Pro (v 5.3) software. All anisotropy measurements were analyzed in biological triplicate for limonene-treated and control cultures at 30°C. A positive control was used to test for increase fluidity by heat treating cells under the identical conditions described above at 50°C for 50 min. Heat-treated samples had a 16% increase in fluidity compared to the control (anisotropy value r = 0.138 ± 0.003 [n = 3]).
Confocal microscopy.
Two microliters of calcofluor white (CFW)-stained cells was loaded onto a glass slide. A coverslip was added on top, and a thin layer of nail polish was used to seal the sample between the glass slide and the coverslip. The images were taken throughout the coverslip with a Zeiss 710 confocal laser scanning microscope (CLSM) using a Plan-Apochromat 63×/1.40 oil DIC M27 objective. The 2,048- by 2,048-pixel map images were recorded using a 405-nm wavelength excitation laser, and the data were collected in the 409- to 517-nm window. All the images were analyzed using the ZEN 2011 software (Carl Zeiss Microscopy, Jena, Germany).
Cell wall isolation.
Cell walls were isolated and analyzed using the acid (H2SO4) treatment method (28). Cell wall extractions were performed in biological triplicate for both control and limonene-challenged cultures at 2 h after limonene addition. Cell wall monosaccharide concentrations were normalized using the total dry cell wall mass for individual samples.
Lipid extraction and methylation.
Yeast lipids were extracted by using the chloroform-methanol protocol described previously (26) with the following modifications. At 2 h after limonene addition, the entire culture volume (22 to 25 ml) was weighed and harvested (4,025 × g) for 2 min at 25°C. The cell pellet was washed in phosphate-buffered saline (PBS) (Invitrogen, Life Technologies, Carlsbad, CA), resuspended, and added to a 1:1 solution of methanol and chloroform as described but with 0.27 μg of nonadecanoic acid (Sigma-Aldrich) to a final concentration of 0.018 μg/ml as an internal standard (ISTD). The lower chloroform phase was removed and dried down under nitrogen. Lipid extracts were saponified under reflux (2 h, 80°C) with 1 ml of 2 M NaOH and 2 ml of methanol. After acidification by the addition of 200 μl of 37.5% concentrated HCl, 4 ml (2 aliquots of 2 ml) of chloroform was added and vortexed. Upon phase separation, the chloroform phase (2 ml) was removed, and the second aliquot (2 ml) of chloroform was added and vortexed. Both chloroform aliquots were then recombined. To recover the lipids, the chloroform was evaporated under nitrogen. For methylation, 200 μl of 2% H2SO4 in methanol was added to the extracts and incubated for 2 h at 80°C. Once cooled, 200 μl of 0.9% NaCl was added and vortexed, and then the addition of 300 μl of hexane was used to recover the fatty acid methyl esters (FAME) for gas chromatography-mass spectrometry (GC-MS) analysis. Lipid extraction was carried out in biological triplicate for control and limonene-treated samples.
GC-MS.
For GC-MS analysis, 2 μl from the hexane-FAME sample was injected into the GC inlet in splitless mode at 350°C using helium as a carrier gas with a constant flow rate of 1 ml/min. Compounds were separated using a Varian factorFOUR capillary column (VF-5 ms; 0.25-mm inner diameter, 0.25-μm film, 30-m length with a 10-m fused guard column) on an Agilent 7890A gas chromatograph attached to an Agilent 5975C MSD. Initially the oven temperature was set to 70°C, and this was held for 5 min, followed by a temperature ramp of 9°C/min to 320°C and then 30°C/min to 350°C and holding for 6.3 min. Detection was achieved in scan mode at 9.26 scans/s from 30 to 800 amu. Thirty-two individual fatty acid standards (Sigma-Aldrich) were used to build a FAME library using their target ions and retention times. FAME peaks were detected in each sample by matching to compounds within the FAME library, and an alkane (C7-30) standard was used as a retention time index. The relative area for each fatty acid in a sample was calculated by dividing the FAME area by the ISTD area, multiplying by the ratio of actual ISTD to set ISTD mass, and then dividing by grams of dry cell weight (gDCW) of the sample. Biological triplicates were used for each condition.
Fluorochrome staining.
Aliquots from control and limonene-treated cultures were taken in mid-exponential phase 2 h after limonene addition. Propidium iodide (PI) was used for determination of cell permeability and membrane integrity (3). For cell wall staining, calcofluor white (CFW) (Sigma-Aldrich) was used as described previously (29) with the following modifications. One milliliter of culture was quickly harvested at 13,000 rpm for 1 min, washed with sterile Milli-Q water, and resuspended in 200 μl of water containing 100 μg/ml of CFW. The solution was then incubated at 30°C for 2 min, harvested, washed twice with Milli-Q water, and resuspended in 1 ml of water before flow cytometry analysis. All data were produced in biological triplicate.
Flow cytometry.
Cell viability was analyzed using PI exactly as described by Brennan et al. (3). For calcofluor white staining, cells were analyzed using a BD LSRII flow cytometer with a UV laser at 350 nm. Forward scatter (FSC), side scatter (SSC), and 450/50 band-pass (BP) filter excitation were acquired using BD FACSDiVa software (v 6.1.1). Postacquisition compensation analysis of total fluorescence intensity was carried out using Flow Jo software (v 10.0). All data were analyzed in biological triplicate.
Cell wall sensitivity assay.
The sensitivity of cell walls to enzymatic degradation was performed as described by Takahashi et al. (30). Briefly, 200 μl of culture was harvested at mid-exponential phase 2 h after limonene addition, washed with sterile water, and diluted to an OD660 of 2 in buffer containing NaH2PO4 (0.1 M, pH 7.5), 0.04% NaN3, and 40 μg/ml lyticase enzyme from Arthrobacter luteus (Sigma-Aldrich). The samples were placed in an Eppendorf rack at 30°C and 200 rpm, and the cell density was monitored hourly. An identical buffer lacking the enzyme was used as a negative control. The percentage of the original OD was reported for biological triplicates for both control and limonene-challenged cultures.
Transcriptomics. (i) RNA sampling and isolation.
RNA was isolated from cultures containing no limonene (control) at mid-exponential phase. For challenged cultures, limonene was added (107 mg/liter) at mid-exponential phase, and RNA was sampled 2 h later. All isolations were done with three biological replicates. For each set of replicates, 10 ml of culture was sampled and harvested for 1 min (4,025 × g), the supernatant was discarded, and the total RNA was immediately extracted using the RiboPure-Yeast kit (Ambion, Life Technologies, Carlsbad, CA) according to the manufacturer's protocol except that bead beating (Biospec Products, Bartlesville, OK) was used to fully lyse the cells (3 times for 20 s, followed by 1 min on ice). The total RNA was then digested (twice) using the Turbo DNA-free kit (Invitrogen, Life Technologies, Carlsbad, CA) and then cleaned and concentrated using the RNA Clean & Concentrator-25 kit (Zymo Research, Irvine, CA) according to both manufacturer's instructions. DNA contamination was assessed by quantitative reverse transcription-PCR (qRT-PCR). The total RNA sample quality was then measured using an Agilent 2100 Bioanalyzer and RNA 6000 Nano kit according to manufacturer's methods (Agilent, Santa Clara, CA). Sample labeling and hybridization to Affymetrix yeast genome 2.0 arrays were performed by the Ramaciotti Centre for Gene Function Analysis (University of New South Wales, Sydney, Australia).
(ii) Microarray data analysis.
Statistical analysis of the raw microarray data was carried out using GenePattern (31). The six Affymetrix CEL files were imported, normalized, and log transformed using the following software modules within GenePattern: GarvanCaArray2.3.0Importer, NormalizeAffymetrix3prime, and LogTransform. Detection of differentially expressed genes and statistical analysis of the samples were carried out using the limma package (32) within the LimmaGP module. The differentially expressed gene set was then filtered using a cutoff at a Bonferroni-corrected P value of ≤0.01. The filtered gene list was then used as input into g:Profiler to asses significantly changed pathways, reactions, and Gene Ontology (GO) terms using a hypergeometric test (33). The results were visualized using the MultiExperiment Viewer software (34). Using the unfiltered gene expression values (log2), the entire transcriptome data set was viewed using the Saccharomyces Genome Database (SGD) pathways tool (http://pathway.yeastgenome.org) for metabolic mapping and identification. The raw microarray data are publically available at http://pwbc.garvan.unsw.edu.au/caarray/project/details.action?project.id=607.
RESULTS
In order to study the response to limonene, the dose added must be sublethal. We have previously observed substantial cell death if inhibitory levels of limonene are added to the inoculum (3). When added in mid-exponential phase, however, cell growth is arrested (Fig. 1a), while viability remains greater than 98% after 2 h of limonene exposure (Fig. 1b). Viability is measured through PI stain exclusion and confirms structural membrane integrity. Further investigation of membrane properties revealed no significant changes in membrane fluidity as measured through fluorescence anisotropy (Fig. 1b), nor were there any significant changes to the fatty acid composition after limonene treatment (Fig. 2b).
Fig 1.
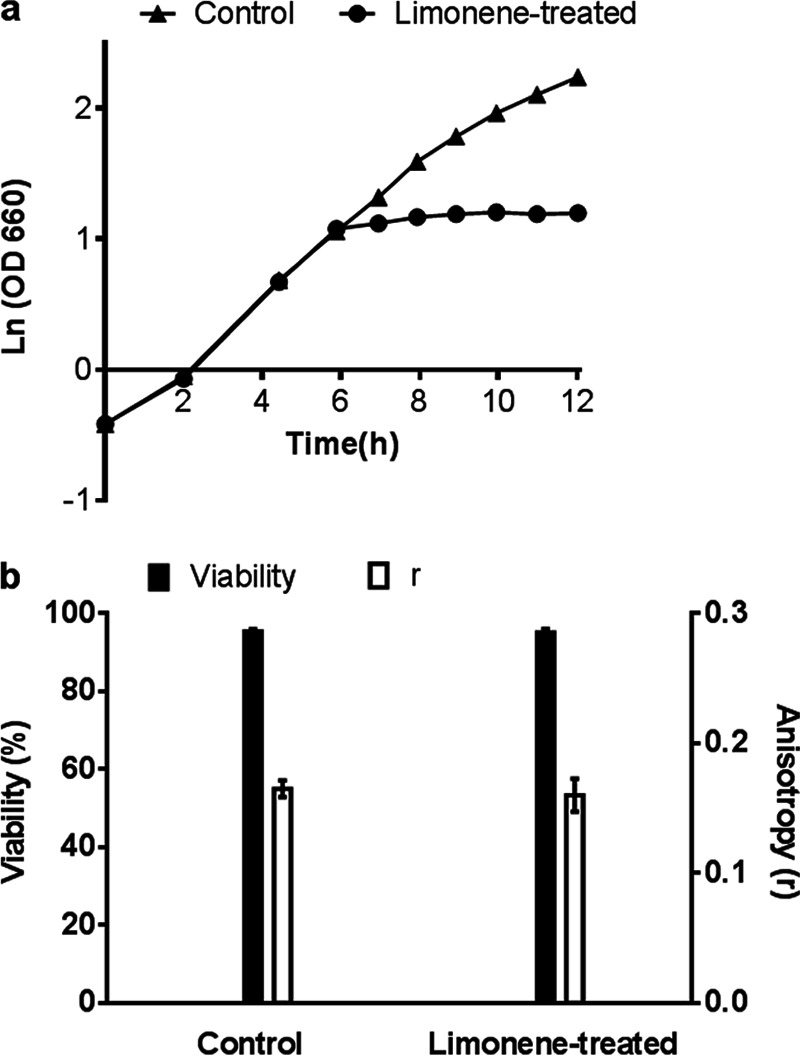
(a) Growth plot for control and limonene-treated (107 mg/liter) cultures. (b) Cell viability and membrane fluidity (anisotropy, r) measurements at 2 h after limonene addition. Error bars represent one standard deviation (SD) above and below the mean for biological replicates.
Fig 2.
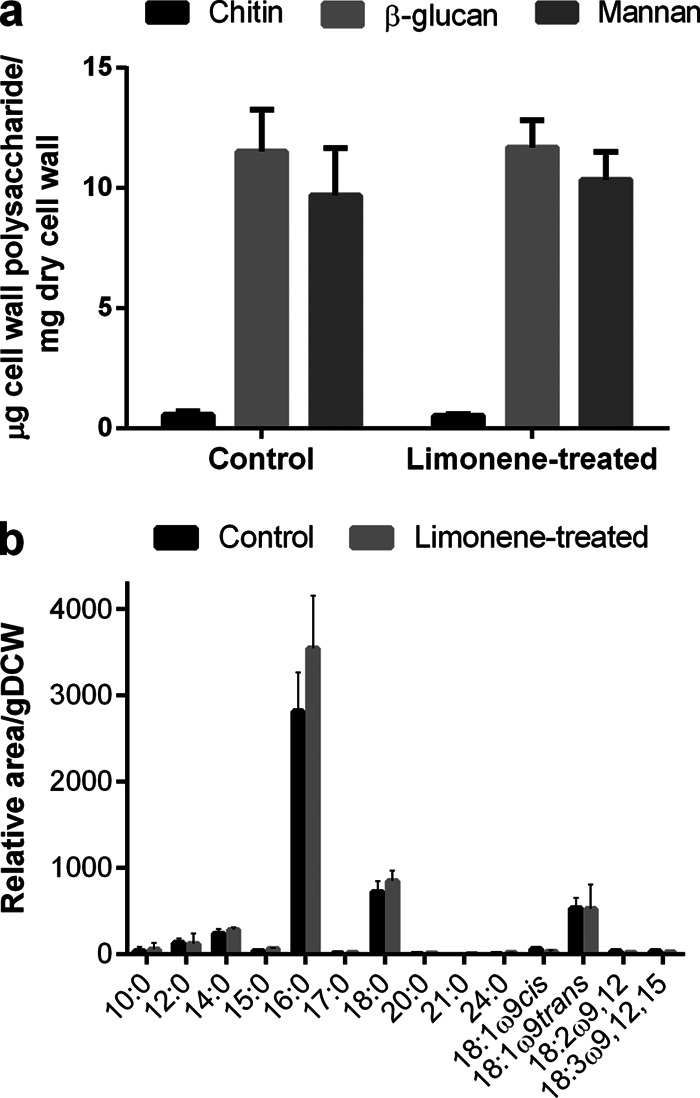
Cell envelope compositional changes due to limonene shock. (a) Cell wall polysaccharide compositions for control and limonene-treated cells. (b) Fatty acid composition at 2 h after limonene challenge. 10:0, decanoic acid; 12:0, dodecanoic acid; 14:0, myristic acid; 15:0, pentadecanoic acid; 16:0, palmitic acid; 17:0, heptadecanoic acid; 18:0, octadecanoic acid; 20:0, eicosanoic acid; 21:0, docosanoic acid; 24:0, tetracosanoic acid; 18:1ω9cis, oleic acid; 18:1ω9trans, elaidic acid; 18:2ω9,12, linoleic acid; 18:3ω9,12,15 linolenic acid. Error bars represent one SD above the mean (n = 3).
While 2 h of limonene treatment caused no significant changes in the cell wall polysaccharide composition (Fig. 2a), it greatly affected cell wall integrity. Limonene-treated cells were hypersensitive to the cell wall binding dye calcofluor white (CFW), exhibiting a 4-fold increase in mean fluorescence intensity (MFI) of 5,231 ± 471 compared to that of control cells (MFI = 1,270 ± 45) (Fig. 3a). In confocal microscopy (Fig. 3b and c), the hypersensitivity to CFW appears as enriched fluorescence in the bud neck region of the cells. Limonene-treated cells were also more susceptible to cell wall-degrading enzymes (Fig. 4). A 40% drop in cell density was observed after 1 h for limonene-challenged cells, compared to 20% found in the control.
Fig 3.
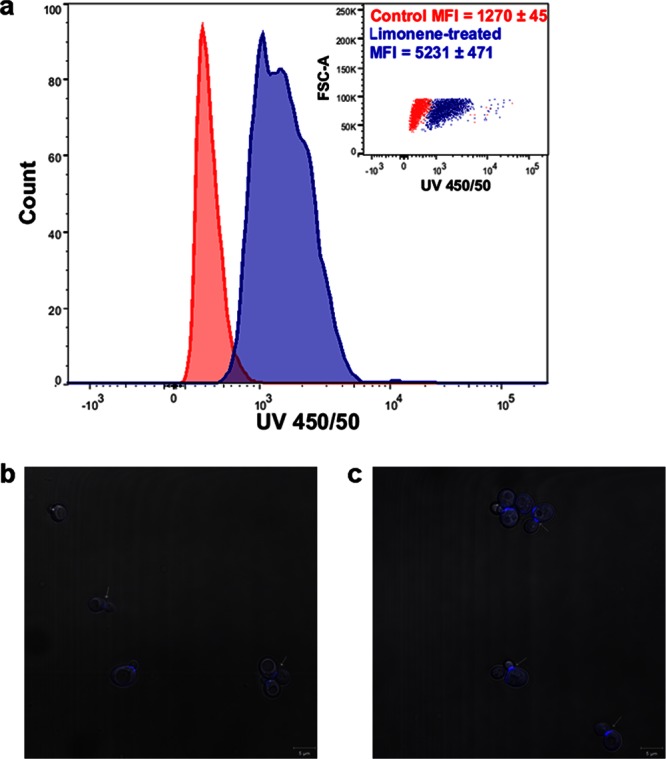
(a) Flow cytometry measurements of limonene-treated and control cells using the cell wall binding probe calcofluor white (CFW) at 2 h after limonene addition. Mean fluorescence intensity/cell (arbitrary units, n = 3) ± SD. Inset, forward side scatter versus CFW intensity. (b and c) Confocal microscopy images of control cells (b) and limonene-treated cells (c) stained with CFW. Arrows indicate the chitin-enriched bud neck.
Fig 4.
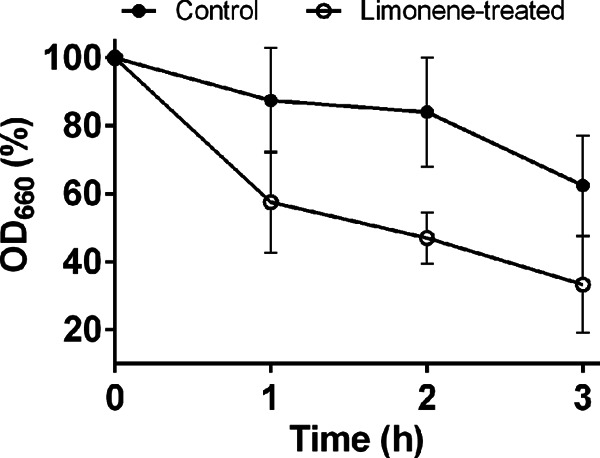
Cell wall sensitivity measurements using lyticase enzyme for limonene-treated cells. All data were carried out in biological triplicate and, error bars represent one SD above and below the mean.
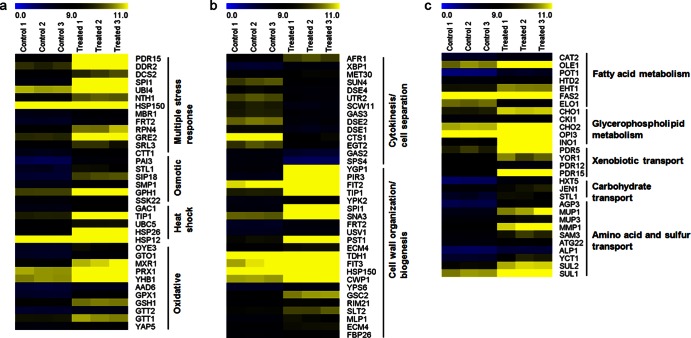
Global gene expression analysis identified 277 upregulated and 176 downregulated genes (Bonferroni-corrected P value of <0.01). Enrichment analysis identified the expected downregulation of biological processes linked to cell growth, e.g., cytokinesis, ribosome biogenesis, nucleotide biosynthesis, and amino acid biosynthesis (see the supplemental material). The only biological process identified as significantly enriched in the upregulated gene set was iron ion homeostasis. Though none of the stress-related biological processes were significantly enriched, several KEGG pathways linked to stress (glutathione metabolism, peroxisome, and autophagy) were identified (see the supplemental material).
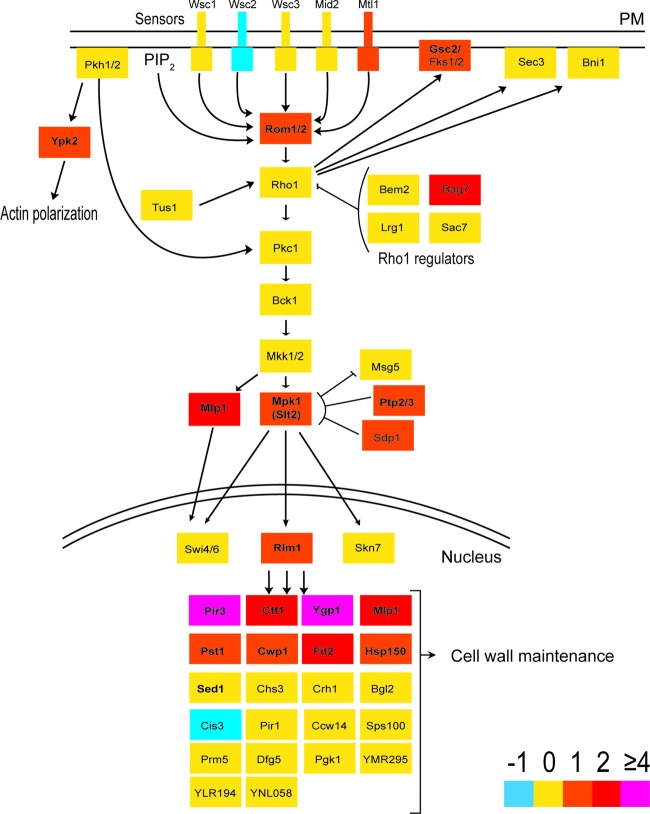
While global repression trends in amino acid synthesis, ribosome activity, and protein synthesis were to be expected for arrested cells, we were particularly interested in how the gene expression data related to the physiological observations of the cell envelope. Limonene-treated cells induced expression of seven fatty acid metabolism and six glycerophospholipid metabolism genes (Table 1 and Fig. 5c); however, global metabolic mapping showed no overexpression of either ergosterol or saturated and unsaturated fatty acid biosynthesis pathways (Table 1; see Table S1 in the supplemental material). Limonene treatment also caused the overexpression of carbohydrate, xenobiotic, amino acid, and sulfur transport activity (Table 1 and Fig. 5c), as well as the expression of several common stress-associated genes (Fig. 5a). Finally, limonene exposure induced a number of genes associated with cell wall organization and biogenesis (Table 1 and Fig. 5b). The observed impact on the structural integrity of the cell wall (Fig. 3 and 4) was further supported by the regulation of the cell wall integrity (CWI) signaling pathway. Several key signaling genes (MTL1, ROM1, MPK1, BAG7, and MLP1) and the major transcription factor gene RLM1 were upregulated (Fig. 6). Furthermore, 12 of RLM1's 25 gene targets were overexpressed, most of which encode proteins associated with cell wall organization and/or wall biogenesis (Fig. 6).
Table 1.
Differentially expressed genes associated with the plasma membrane and cell wall cellular compartments and functionsa
| Process (GO term, no. of genes,b P value) | Upregulated genes |
Downregulated genes |
||||
|---|---|---|---|---|---|---|
| Gene | Protein description | Fold changec | Gene | Protein description | Fold change | |
| Fatty acid metabolic process (0006631, 7, 3.58E−02) | CAT2 | Carnitine acetyl-CoA transferase present in both mitochondria and peroxisomes; transfers activated acetyl groups to carnitine to form acetylcarnitine, which can be shuttled across membranes | 1.65 | ELO1 | Elongase I, medium-chain acyl elongase, catalyzes carboxy-terminal elongation of unsaturated C12–C16 fatty acyl-CoAs to C16–C18 fatty acids | −2.21 |
| OLE1 | Delta(9) fatty acid desaturase, required for monounsaturated fatty acid synthesis and for normal distribution of mitochondria | 1.90 | ||||
| POT1 | 3-Ketoacyl-CoA thiolase with broad chain-length specificity; cleaves 3-ketoacyl-CoA into acyl-CoA and acetyl-CoA during beta-oxidation of fatty acids | 2.84 | ||||
| HTD2 | Mitochondrial 3-hydroxyacyl-thioester dehydratase involved in fatty acid biosynthesis; required for respiratory growth and for normal mitochondrial morphology | 0.98 | ||||
| EHT1 | Acyl-coenzyme A:ethanol O-acyltransferase that plays a minor role in medium-chain fatty acid ethyl ester biosynthesis; possesses short-chain esterase activity; localizes to lipid particles and the mitochondrial outer membrane | 0.92 | ||||
| FAS2 | Alpha subunit of fatty acid synthetase, which catalyzes the synthesis of long-chain saturated fatty acids; contains the acyl-carrier protein domain and beta-ketoacyl reductase, beta-ketoacyl synthase and self-pantetheinylation activities | 1.06 | ||||
| Glycerophospholipid metabolism (0564 [KEGG], 6, 1.42E−02 | INO1 | Inositol 1-phosphate synthase, involved in synthesis of inositol phosphates and inositol-containing phospholipids; transcription is coregulated with other phospholipid biosynthetic genes by Ino2p and Ino4p, which bind the UASINO DNA element | 4.10 | |||
| CLD1 | Mitochondrial cardiolipin-specific phospholipase; functions upstream of Taz1p to generate monolyso-cardiolipin; transcription increases upon genotoxic stress; involved in restricting Ty1 transposition; has homology to mammalian CGI-58 | 1.89 | ||||
| CHO1 | Phosphatidylserine synthase; functions in phospholipid biosynthesis; catalyzes the reaction CDP-diacylglycerol + l-serine = CMP + l-1-phosphatidylserine; transcriptionally repressed by myo-inositol and choline | 1.15 | ||||
| CKI1 | Choline kinase, catalyzing the first step in phosphatidylcholine synthesis via the CDP-choline (Kennedy pathway); exhibits some ethanolamine kinase activity contributing to phosphatidylethanolamine synthesis via the CDP-ethanolamine pathway | 1.28 | ||||
| CHO2 | PEMT, catalyzes the first step in the conversion of phosphatidylethanolamine to phosphatidylcholine during the methylation pathway of phosphatidylcholine biosynthesis | 1.13 | ||||
| OPI3 | Phospholipid methyltransferase (methylene-fatty-acyl-phospholipid synthase); catalyzes the last two steps in phosphatidylcholine biosynthesis | 0.95 | ||||
| Xenobiotic transporter activity (0042910, 3, 2.18E−02) | PDR5 | Plasma membrane ABC transporter; multidrug transporter actively regulated by Pdr1p; also involved in steroid transport, cation resistance, and cellular detoxification during exponential growth | 2.00 | PDR12 | Plasma membrane ABC transporter; weak-acid-inducible multidrug transporter required for weak organic acid resistance; induced by sorbate and benzoate and regulated by War1p; mutants exhibit sorbate hypersensitivity | −1.54 |
| YOR1 | Plasma membrane ABC transporter; multidrug transporter mediates export of many different organic anions, including oligomycin; similar to human CFTR | 1.65 | ||||
| PDR15 | Plasma membrane ABC transporter, multidrug transporter, and general stress response factor implicated in cellular detoxification; regulated by Pdr1p, Pdr3p, and Pdr8p; promoter contains a PDR-responsive element | 3.19 | ||||
| Amino acid and sulfur transmembrane transport (0003333 and 0072348, 9, 0.00136–0.01) | AGP3 | Low-affinity amino acid permease; may act to supply the cell with amino acids as nitrogen source under nitrogen-poor conditions; transcription is induced under conditions of sulfur limitation; plays a role in regulating Ty1 transposition | 5.09 | |||
| MUP1 | High-affinity methionine permease; integral membrane protein with 13 putative membrane-spanning regions; also involved in cysteine uptake | 4.63 | ||||
| MUP3 | Low-affinity methionine permease similar to Mup1p | 1.87 | ||||
| MMP1 | High-affinity S-methylmethionine permease; required for utilization of S-methylmethionine as a sulfur source; has similarity to S-adenosylmethionine permease Sam3p | 2.28 | ||||
| SAM3 | High-affinity S-adenosylmethionine permease; required for utilization of S-adenosylmethionine as a sulfur source; has similarity to S-methylmethionine permease Mmp1p | 2.23 | ||||
| ALP1 | Arginine transporter; expression is normally very low, and it is unclear what conditions would induce significant expression | 1.22 | ||||
| YCT1 | High-affinity cysteine-specific transporter with similarity to the Dal5p family of transporters; GFP fusion protein localizes to the endoplasmic reticulum; YCT1 is not an essential gene | 4.33 | ||||
| SUL2 | High-affinity sulfate permease; sulfate uptake is mediated by specific sulfate transporters Sul1p and Sul2p, which control the concn of endogenous activated sulfate intermediates | 1.47 | ||||
| SUL1 | High-affinity sulfate permease of the SulP anion transporter family; sulfate uptake is mediated by specific sulfate transporters Sul1p and Sul2p, which control the concn of endogenous activated sulfate intermediates | 1.80 | ||||
| Cell wall organization (0005576, 22, 7.86E−02) | YGP1 | Cell wall-related secretory glycoprotein; induced by nutrient deprivation-associated growth arrest and upon entry into stationary phase; may be involved in adaptation prior to stationary-phase entry; has similarity to Sps100p | 4.03 | HPF1 | Haze-protective mannoprotein that reduces the particle size of aggregated proteins in white wines | −2.52 |
| PIR3 | O-glycosylated covalently bound cell wall protein required for cell wall stability; expression is cell cycle regulated, peaking in M/G1, and also subject to regulation by the cell integrity pathway | 4.99 | GAS3 | Low-abundance, possibly inactive member of the GAS family of GPI-containing proteins; putative 1,3-beta-glucanosyltransferase with similarity to other GAS family members; localizes to the cell wall; mRNA induced during sporulation | −2.27 | |
| FIT2 | Mannoprotein that is incorporated into the cell wall via a GPI anchor; involved in the retention of siderophore iron in the cell wall | 2.01 | UTR2 | Chitin transglycosylase that functions in the transfer of chitin to beta(1-6) and beta(1-3) glucans in the cell wall; similar to and functionally redundant with Crh1; GPI-anchored protein localized to bud neck | −2.56 | |
| SUC2 | Invertase, sucrose-hydrolyzing enzyme; a secreted, glycosylated form is regulated by glucose repression, and an intracellular, nonglycosylated enzyme is produced constitutively | 2.63 | SUN4 | Cell wall protein related to glucanases; possibly involved in cell wall septation; member of the SUN family | −2.21 | |
| TIP1 | Major cell wall mannoprotein with possible lipase activity; transcription is induced by heat and cold shock; member of the Srp1p/Tip1p family of serine-alanine-rich proteins | 2.06 | DSE4 | Daughter cell-specific secreted protein with similarity to glucanases; degrades cell wall from the daughter side, causing daughter to separate from mother | −1.89 | |
| SPI1 | GPI-anchored cell wall protein involved in weak acid resistance; basal expression requires Msn2p/Msn4p; expression is induced under conditions of stress and during the diauxic shift; similar to Sed1p | 4.74 | SCW11 | Cell wall protein with similarity to glucanases; may play a role in conjugation during mating based on its regulation by Ste12p | −2.99 | |
| PST1 | Cell wall protein that contains a putative GPI attachment site; secreted by regenerating protoplasts; upregulated by activation of the cell integrity pathway, as mediated by Rlm1p; upregulated by cell wall damage via disruption of FKS1 | 1.88 | DSE2 | Daughter cell-specific secreted protein with similarity to glucanases; degrades cell wall from the daughter side, causing daughter to separate from mother; expression is repressed by cAMP | −3.16 | |
| FIT3 | Mannoprotein that is incorporated into the cell wall via a GPI anchor; involved in the retention of siderophore iron in the cell wall | 1.73 | CTS1 | Endochitinase; required for cell separation after mitosis; transcriptional activation during the G1 phase of the cell cycle is mediated by transcription factor Ace2p | −2.36 | |
| HSP150 | O-mannosylated heat shock protein that is secreted and covalently attached to the cell wall via beta-1,3-glucan and disulfide bridges; required for cell wall stability; induced by heat shock, oxidative stress, and nitrogen limitation | 0.97 | EGT2 | GPI-anchored cell wall endoglucanase required for proper cell separation after cytokinesis; expression is activated by Swi5p and tightly regulated in a cell cycle-dependent manner | −2.09 | |
| CWP1 | Cell wall mannoprotein that localizes specifically to birth scars of daughter cells, linked to a beta-1,3- and beta-1,6-glucan heteropolymer through a phosphodiester bond; required for propionic acid resistance | 1.07 | ||||
| PRB1 | Vacuolar proteinase B (yscB), a serine protease of the subtilisin family; involved in protein degradation in the vacuole and required for full protein degradation during sporulation; activity inhibited by Pbi2p | 1.30 | ||||
| DIA3 | Protein of unknown function involved in invasive and pseudohyphal growth | 1.35 | ||||
| TDH1 | Glyceraldehyde-3-phosphate dehydrogenase, isozyme 1, involved in glycolysis and gluconeogenesis; tetramer that catalyzes the reaction of glyceraldehyde-3-phosphate to 1,3-bis-phosphoglycerate; detected in the cytoplasm and cell wall | 1.15 | ||||
Abbreviations: CoA, coenzyme A; PEMT, phosphatidylethanolamine methyltransferase; ABC, ATP binding cassette; CFTR, cystic fibrosis transmembrane receptor; GFP, green fluorescent protein; GPI, glycosylphosphatidylinositol; cAMP, cyclic AMP.
Number of genes associated with the reported Gene Ontology accession number. The total number of differentially expressed genes was 453.
Log2 ratio of expression in treated cells to that in control cells.
Fig 5.
Gene expression values (log2) for each biological replicate (control versus limonene-treated cells) for genes involved in stress responses (a), cell wall organization and cytokinesis (b), and plasma membrane transport and fatty acid metabolism (c).
Fig 6.
Cell wall integrity (CWI) signaling pathway with transcriptomic expression changes during limonene stress. Colors indicate the fold change (log2) cutoffs. Genes in bold represent genes that were differentially expressed (Bonferroni-corrected P value of <0.01). PM, plasma membrane; PIP2, phosphatidylinositol-4,5-bisphosphate. (Adapted from reference 70 with permission.)
DISCUSSION
Monoterpene toxicity is generally assumed to be due to membrane interference, as observed for ethanol (17–19). Yeast cells respond to ethanol exposure by increasing their ergosterol and unsaturated fatty acid composition to increase membrane rigidity (35–37). An early microarray study of α-terpinene toxicity observed global upregulation of genes involved in ergosterol synthesis and sterol uptake (15). In the current study, however, we observed no evidence of membrane interference by limonene. Membrane integrity, fluidity (Fig. 1), and fatty acid composition (Fig. 2b) did not change significantly in response to limonene. Furthermore, no increase in the expression of genes in the ergosterol and fatty acid biosynthesis pathways was observed (Table 1; see Fig. S1 in the supplemental material).
We would have expected results similar to those observed for α-terpinene (15). Like limonene, terpinenes are cyclic monoterpenes. Terpinene was used at a higher concentration (170 mg/liter rather than 107 mg/liter) but is also slightly less toxic; the terpinene concentration was chosen as the 50% inhibitory concentration (IC50) in the previous study (15), while our study used nearly two times the IC50 for limonene (3). The monoterpenes were added to cells in mid-exponential phase, i.e., 5 h in culture in our study and overnight in the terpinene study. In both studies, the response was measured at 2 h after monoterpene addition. One significant difference was that complex (YPD) medium was used in the terpinene study, compared to minimal medium in our study. Another major difference was that a number of genes involved in redox activity and glutathione metabolism (see the supplemental material) were found for limonene but not for terpinene exposure. Dioxygenase and oxidoreductase genes JLP1 and OYE3, for example, were highly upregulated (6.7- and 3.6-fold changes, respectively [see the supplemental material]) during limonene exposure. Glutathione biosynthesis (GSH1, 1.3-fold induced) and peroxide protection CTT1 and GPX1 (2.2- and 3-fold induced, respectively) genes (see the supplemental material) were also found in this work but not in the terpinene study. Differences between the two transcriptional responses may stem from culture conditions or monoterpene load, as well as differences in chemical properties between the two compounds and their potential to form epoxides.
Because redox and glutathione metabolism activity was observed, we wanted to exclude oxidative stress as possible source of molecular toxicity. Expression of GSH1, CTT1, and GPX1 is a well-characterized response to oxidative stress in S. cerevisiae (38). Signature genes involved in antioxidant defense systems, however, were not overexpressed. Genes for key transcriptional regulators (YAP1 and SKN7), thioredoxins (TRX1, TRX2, TRR1, and TRR2), glutaredoxins (GLX1, -2, 3, -4, and -5), and superoxide dismutases (SOD1 and -2) as well as for two key stress response element (STRE) regulators (MSN2 and MSN4) were not induced during limonene shock (38, 39). A potential cause of increased redox activity may come from oxygenated limonene compounds such as limonene epoxides, which can form when limonene is exposed to air for long periods of time (40). Epoxide compounds have been reported to cause oxidative damage in yeast (41), and limonene-1,2-epoxide, for example, is 23 times more soluble in water than limonene (137 mg/liter) (42). There may be differences between the toxic effects of monoterpene hydrocarbons and oxy-functionalized monoterpene compounds. For example, 1,8-cineole (an epoxy-monoterpene) was endogenously produced in yeast with no report of toxicity limitations up to 1 g/liter (10), while a separate study reported that limonene stops growth at 60 mg/liter (3). While limonene itself is toxic at the phase level, limonene epoxides may have caused redox imbalances to cells on the molecular level. In order to rule this out, the experiments were repeated under anaerobic conditions. We found no limonene epoxide formation in GC-MS but observed the same adverse effects on growth (see Fig. S2 in the supplemental material). This means that although limonene epoxides can form in aerobic cultures (see Fig. S3 in the supplemental material), limonene itself remains the primary source of toxicity.
Membrane-bound efflux pumps play a critical role in solvent-tolerance in P. putida (43). Expression of the AcrAB-TolC efflux proteins in E. coli led to higher tolerance of cyclohexane (44) and increased tolerance and production of pinene and limonene (8). Similar to the bacterial pumping system, cellular detoxification in yeast is driven primarily by pleiotropic drug resistance (PDR) pumps, which are a subfamily of yeast's ABC proteins (45). We found three ABC transporter genes induced under limonene stress (YOR1, PDR15, and PDR5) (Fig. 5c and Table 1). The same three transporters were identified in a recent study, but overexpression failed to improve tolerance for limonene (46), and the pumps may lack affinity to limonene.
Absence of a membrane effect is consistent with limonene toxicity being phase toxicity rather than molecular toxicity. There is no evidence of molecular toxicity, as the system is saturated at a water concentration of 6 mg/liter and the IC50 is 10 times greater than the solubility. The solvent phase contacts the cell wall rather than the cell membrane. Having primary roles in protection from turgor pressure and cell division, the cell wall is essential for survival (47). Its latticework is tightly held together by strong hydrogen bonding networks between cellulose and chitin chains as well as covalent glycosidic linkages between all three wall components (glucan, mannoprotein, and chitin) (48). Calcofluor white is a fluorochrome that binds to the cell wall by hydrogen bonding with chitin and β-linked polysaccharides (49–51), but in fungi, CFW preferentially binds to the chitin, which is localized in the bud neck (29). In Fig. 3a, a 4-fold increase in the MFI demonstrates that more fluorochrome is bound per cell after limonene treatment. Increased sensitivity to CFW is indicative of cell wall damage in S. cerevisiae, Candida albicans (52), Aspergillus niger (53), and a number of other fungi (54). Disruption of the crystalline lattice of chitin polymers has been shown to weaken the cell wall, causing cell arrest and accumulation of chitin deposits in S. cerevisiae (55). Although we did not find higher levels of chitin (Fig. 2a), this mechanism is in concert with our CFW hypersensitivity and growth inhibition results. A decrystallizing effect is also seen in Fig. 4, where the cell walls were found to be more susceptible to glucan-specific degradation with lyticase enzymes after having been treated with limonene. Because we found no changes in the total wall composition after limonene exposure (Fig. 2a), the data suggest that limonene can alter the properties of the cell wall. Given its pivotal role in the budding process, particularly in septum formation (47, 56), our results indicate that by disrupting the cell wall structure, limonene has a profound effect on cell growth.
The cellular response to limonene further demonstrates cell wall stress. Twelve differentially expressed genes that were found in this study (CWP1, PIR3, SED1, PST1, SLT2, MLP1, ECM4, HSP12, DDR2, SLR3, FBP26, and AFR1) belong to a cluster of 20 well-characterized genes that represent the main transcriptional fingerprint of cell wall stress (57, 58). Furthermore, several genes directly involved in the cell wall integrity (CWI) signaling pathway, the sole purpose of which is to respond to cell wall stress (59), were upregulated in response to limonene exposure (Fig. 6). The guanosine nucleotide exchange factor gene ROM1, the cell surface sensor gene MLT1, and the genes SLT2 and MLP1, coding for the mitogen-activated protein kinase (MAPK) proteins cascade, were induced (Fig. 6). RLM1 codes for the transcription factor responsible for the majority of the transcriptional output of CWI (59). RLM1 was slightly overexpressed (1.2-fold [log2]), while five of its targets (PST1, CWP1, PIR3, CTT1, and YGP1 in Fig. 6) were highly upregulated. In particular, PIR3 and YGP1, which are required for cell wall organization and stability, were two of the most highly induced genes found, having fold changes of 5 and 4, respectively (Fig. 6 and Table 1). Lastly, the cellular response to limonene caused decreased transcript levels for transcription, ribosomal activity, and purine nucleotide metabolism. These repression trends were also found in two independent studies profiling the global transcriptional response to cell wall damage (57, 58).
The physical interaction between an insoluble limonene phase and a cell is still unclear. We have shown here that limonene dispersions can alter surface properties of yeast cells by disrupting the normal structure of the cell wall, but the relationship between limonene's rheological behavior in an aqueous culture and its apparent toxicity to a microorganism is yet to be determined. In a recent study (3), limonene toxicity was dramatically reduced in the presence of an inert extractant. The extractive solvents dibutyl phthalate (DBP) and isopropyl myristate (IPM), for example, are harmless to yeast cells and are at least an order of magnitude more viscous than limonene (μlimonene = 0.008 P [60], μDBP = 0.166 P [61], and μIPM = 0.043 P [62]). The viscosity of a fluid has a strong influence on droplet size, which affects the dispersion's overall surface area (63). Future work is required to characterize how fluid properties (e.g., viscosity, droplet size, and interfacial tension) affect the interfacial contact between cells and monoterpene dispersions. This information could render more insight into the phase mechanisms responsible for inhibition in biphasic systems.
The impact on cell physiology can change if surfactants are used to solvate monoterpene compounds before they are added. Due to their low aqueous solubility, monoterpenes are commonly administered in biological cultures via emulsifying agents, such as Tween 80 and dimethylformamide (DMF) (8, 17, 64). Because of their amphiphilic nature, surfactants and cosolvents change the interfacial properties at the oil-water interface (65, 66). The result is the formation of micelles, with monoterpene-rich interiors and watery exteriors (65–67). In a recent study, a limonene-Tween 80 solvent mixture caused membrane deterioration and upregulation of ergosterol biosynthesis in yeast (64). These authors found that exogenous ergosterol addition enhanced tolerance (64). In our study, no cosolvent was used; limonene exposure caused no upregulation of ergosterol biosynthesis (see Fig. S4 in the supplemental material) or damage to the membrane (Fig. 1). Furthermore, growth did not improve with ergosterol supplementation in this work (see Fig. S5 in the supplemental material), but we did observe changes in the dose response when Tween 80 was used as a cosolvent compared to limonene alone. At a constant limonene concentration (107 mg/liter), the limonene-Tween 80 mixture caused no growth disturbance, while limonene without Tween 80 caused growth to cease (see Fig. S6 in the supplemental material). While Tween 80 is nontoxic to yeast cells (68), there are clearly multifunctional effects at play when surfactants and toxic solvents are used simultaneously. A detailed mechanism of cosolvent effects on microorganisms is unknown. However, the introduction of cosolvents with hydrophobic compounds, such as monoterpenes, may facilitate more favorable interactions or access of micelle fluid structures with biological membranes, which has been reported for some common detergents (69). Variations in experimental conditions, such as the type of surfactant used and the surfactant concentration, may explain the differences in inhibitory concentrations and mechanistic conclusions found in most monoterpene toxicity studies. Nevertheless, the ultimate goal is to develop yeast strains that produce monoterpenes endogenously in the absence of specialized surfactants. Therefore, understanding the physiological impact of limonene alone was of particular interest in this work.
In conclusion, microbial synthesis of monoterpene products will not be viable unless the toxicity issue is solved. In order to engineer tolerant strains successfully, a greater understanding of the mechanism causing inhibition is first required. While monoterpene inhibition has long been attributed to the disruption of membrane properties (17–19), this is the first study in S. cerevisiae demonstrating that monoterpene toxicity is not due to membrane deterioration. Increasing membrane rigidity through changes in fatty acid content or by actively pumping monoterpene compounds from the cell (46) was not found to increase tolerance. Our results underscore the position that monoterpene inhibition is not at the molecular level (e.g., membrane interference effects) (3) and that the mechanism of action must stem from the physical interaction between an insoluble monoterpene phase and the surface of a cell. To this end, we have demonstrated here that limonene can alter the properties of yeast cell walls while triggering a compensatory transcriptional response to cell wall damage. Our data indicate chitin, a critical cell wall component, to be a primary target for limonene action, but the exact mechanism remains unclear. This study reveals that the development of monoterpene-resistant yeast strains will most likely not require alterations to the plasma membrane. Instead, the presented work suggests that strategies that focus on maintaining cell wall integrity and cell surface properties are likely to be more useful targets for strain improvement in the future.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Queensland government (National and International Research Alliances Program) for financial support.
We thank the Australian Wine Research Institute (Adelaide, South Australia, Australia) for providing us with the yeast strain. We thank Chris Paddon (Amyris Inc., Emeryville, CA) and Andreas Schmid (Technical University Dortmund, Germany) for useful discussions and Colin Archer (University of Queensland) for technical assistance with the RNA isolation.
We declare no conflicting interests.
Footnotes
Published ahead of print 29 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00463-13.
REFERENCES
- 1. Keasling JD. 2010. Microbial production of isoprenoids, p 2951–2966 In Timmis KN. (ed), Handbook of hydrocarbon and lipid microbiology. Springer, Berlin, Germany [Google Scholar]
- 2. van der Werf M, de Bont J, Leak D. 1997. Opportunities in microbial biotransformation of monoterpenes, p 147–177 In Berger R, Babel W, Blanch H, Cooney C, Enfors S, Eriksson K, Fiechter A, Klibanov A, Mattiasson B, Primrose S, Rehm H, Rogers P, Sahm H, Scḧugerl K, Tsao G, Venkat K, Villadsen J, von Stockar U, Wandrey C. (ed), Biotechnology of aroma compounds, vol 55 Springer, Berlin, Germany [Google Scholar]
- 3. Brennan TCR, Turner CD, Krömer JO, Nielsen LK. 2012. Alleviating monoterpene toxicity using a two-phase extractive fermentation for the bioproduction of jet fuel mixtures in Saccharomyces cerevisiae. Biotechnol. Bioeng. 109:2513–2522 [DOI] [PubMed] [Google Scholar]
- 4. Harvey BG, Wright ME, Quintana RL. 2009. High-density renewable fuels based on the selective dimerization of pinenes. Energy Fuels 24:267–273 [Google Scholar]
- 5. Ryder JA. February 2009. Fuel composition, useful to power any equipment such as an emergency generator or internal combustion engine, which requires a fuel such as jet fuels or missile fuels, comprises limonene and farnesane. U.S. patent 7589243-B1
- 6. Amyris 2012. Azul Brazilian airlines makes successful demonstration flight with Amyris renewable jet fuel produced from sugarcane. http://www.amyris.com/NEWS/112/Azul-Brazilian-Airlines-makes-Successful-Demonstration-Flight-with-Amyris-Renewable-Jet-Fuel-Produced-from-Sugarcane
- 7. Carrau FM, Medina K, Boido E, Farina L, Gaggero C, Dellacassa E, Versini G, Henschke PA. 2005. De novo synthesis of monoterpenes by Saccharomyces cerevisiae wine yeasts. FEMS Microbiol. Lett. 243:107–115 [DOI] [PubMed] [Google Scholar]
- 8. Dunlop MJ, Dossani ZY, Szmidt HL, Chu HC, Lee TS, Keasling JD, Hadi MZ, Mukhopadhyay A. 2011. Engineering microbial biofuel tolerance and export using efflux pumps. Mol. Syst. Biol. 7:487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fischer MJC, Meyer S, Claudel P, Bergdoll M, Karst F. 2011. Metabolic engineering of monoterpene synthesis in yeast. Biotechnol. Bioeng. 108:1883–1892 [DOI] [PubMed] [Google Scholar]
- 10. Ignea C, Cvetkovic I, Loupassaki S, Kefalas P, Johnson C, Kampranis S, Makris A. 2011. Improving yeast strains using recyclable integration cassettes, for the production of plant terpenoids. Microb. Cell Fact. 10:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Renninger N, McPhee D. April 2008. Fuel compositions including farnesane and farnesene derivatives and methods of making and using same. U.S. patent WO2008045555
- 12. Hill J, Nelson E, Tilman D, Polasky S, Tiffany D. 2006. Environmental, economic, and energetic costs and benefits of biodiesel and ethanol biofuels. Proc. Natl. Acad. Sci. U. S. A. 103:11206–11210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nicolaou SA, Gaida SM, Papoutsakis ET. 2010. A comparative view of metabolite and substrate stress and tolerance in microbial bioprocessing: from biofuels and chemicals, to biocatalysis and bioremediation. Metab. Eng. 12:307–331 [DOI] [PubMed] [Google Scholar]
- 14. Cristani M, D'Arrigo M, Mandalari G, Castelli F, Sarpietro MG, Micieli D, Venuti V, Bisignano G, Saija A, Trombetta D. 2007. Interaction of four monoterpenes contained in essential oils with model membranes: implications for their antibacterial activity. J. Agric. Food Chem. 55:6300–6308 [DOI] [PubMed] [Google Scholar]
- 15. Parveen M, Hasan MK, Takahashi J, Murata Y, Kitagawa E, Kodama O, Iwahashi H. 2004. Response of Saccharomyces cerevisiae to a monoterpene: evaluation of antifungal potential by DNA microarray analysis. J. Antimicrob. Chemother. 54:46–55 [DOI] [PubMed] [Google Scholar]
- 16. Uribe S, Pena A. 1990. Toxicity of allelopathic monoterpene suspensions on yeast dependence on droplet size. J. Chem. Ecol. 16:1399–1408 [DOI] [PubMed] [Google Scholar]
- 17. Uribe S, Ramirez J, Pena A. 1985. Effects of beta-pinene on yeast membrane functions. J. Bacteriol. 161:1195–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schrader J. 2007. Microbial flavour production, p 507–574 In Berger RG. (ed), Flavours and fragrances. Springer, Berlin, Germany [Google Scholar]
- 19. Sikkema J, de Bont J, Poolman B. 1995. Mechanisms of membrane toxicity of hydrocarbons. Microbiol. Rev. 59:201–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Andrews RE, Parks LW, Spence KD. 1980. Some effects of douglas-fir terpenes on certain microorganisms. Appl. Environ. Microbiol. 40:301–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. León R, Fernandes P, Pinheiro HM, Cabral JMS. 1998. Whole-cell biocatalysis in organic media. Enzyme Microb. Technol. 23:483–500 [Google Scholar]
- 22. Osborne SJ, Leaver J, Turner MK, Dunnill P. 1990. Correlation of biocatalytic activity in an organic aqueous 2-liquid phase seystem with solvent concentration in the cell-membrane. Enzyme Microb. Technol. 12:281–291 [DOI] [PubMed] [Google Scholar]
- 23. Sikkema J, de Bont JA, Poolman B. 1994. Interactions of cyclic hydrocarbons with biological membranes. J. Biol. Chem. 269:8022–8028 [PubMed] [Google Scholar]
- 24. Schmid C, Steinbrecher R, Ziegler H. 1992. Partition-coefficients of plant cuticles for monoterpenes. Trees Struct. Funct. 6:32–36 [Google Scholar]
- 25. Bar R. 1987. Phase toxicity in a water-solvent two-liquid phase microbial system, p 147–154 In Laane C, Tramper J, Lilly MD. (ed)., Studies in organic chemistry, vol 29 Biocatalysis in organic media. Elsevier Science Publishers B.V., Amsterdam, Netherlands [Google Scholar]
- 26. Huffer S, Clark ME, Ning JC, Blanch HW, Clark DS. 2011. The role of alcohols in growth, lipid composition, and membrane fluidity of yeasts, bacteria, and archaea. Appl. Environ. Microbiol. 77:6400–6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shinitzky M, Yuli I. 1982. Lipid fluidity at the submacroscopic level: determination by fluorescence polarization. Chem. Phys. Lipids 30:261–282 [Google Scholar]
- 28. Francois JM. 2006. A simple method for quantitative determination of polysaccharides in fungal cell walls. Nat. Protoc. 1:2995–3000 [DOI] [PubMed] [Google Scholar]
- 29. Pringle JR. 1991. Staining of bud scars and other cell wall chitin with Calcofluor. Methods Enzymol. 194:732–735 [DOI] [PubMed] [Google Scholar]
- 30. Takahashi TT, Shimoi HS, Ito KI. 2001. Identification of genes required for growth under ethanol stress using transposon mutagenesis in Saccharomyces cerevisiae. Mol. Genet. Genomics 265:1112–1119 [DOI] [PubMed] [Google Scholar]
- 31. Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. 2006. GenePattern 2.0. Nat. Genet. 38:500–501 [DOI] [PubMed] [Google Scholar]
- 32. Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:Article3 [DOI] [PubMed] [Google Scholar]
- 33. Reimand J, Kull M, Peterson H, Hansen J, Vilo J. 2007. g:Profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 35:W193–W200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374–378 [DOI] [PubMed] [Google Scholar]
- 35. Ding J, Huang X, Zhang L, Zhao N, Yang D, Zhang K. 2009. Tolerance and stress response to ethanol in the yeast Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 85:253–263 [DOI] [PubMed] [Google Scholar]
- 36. Swan TM, Watson K. 1998. Stress tolerance in a yeast sterol auxotroph: role of ergosterol, heat shock proteins and trehalose. FEMS Microbiol. Lett. 169:191–197 [DOI] [PubMed] [Google Scholar]
- 37. You KM, Rosenfield C-L, Knipple DC. 2003. Ethanol tolerance in the yeast Saccharomyces cerevisiae is dependent on cellular oleic acid content. Appl. Environ. Microbiol. 69:1499–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Perrone GG, Tan SX, Dawes IW. 2008. Reactive oxygen species and yeast apoptosis. Biochim. Biophys. Acta Mol. Cell Res. 1783:1354–1368 [DOI] [PubMed] [Google Scholar]
- 39. Estruch F. 2000. Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol. Rev. 24:469–486 [DOI] [PubMed] [Google Scholar]
- 40. Nilsson U, Bergh M, Shao LP, Karlberg AT. 1996. Analysis of contact allergenic compounds in oxidized d-limonene. Chromatographia 42:199–205 [Google Scholar]
- 41. Hayes JD, McLellan LI. 1999. Glutathione and glutathione-dependent enzymes represent a co-ordinately regulated defence against oxidative stress. Free Radical Res. 31:273–300 [DOI] [PubMed] [Google Scholar]
- 42. SRC 12 October 2012, posting date Physical property database. http://www.syrres.com/what-we-do/databaseforms.aspx?id=386
- 43. Kieboom J, Dennis JJ, de Bont JAM, Zylstra GJ. 1998. Identification and molecular characterization of an efflux pump involved in Pseudomonas putida S12 solvent tolerance. J. Biol. Chem. 273:85–91 [DOI] [PubMed] [Google Scholar]
- 44. Aono R, Tsukagoshi N, Yamamoto M. 1998. Involvement of outer membrane protein TolC, a possible member of the mar-sox regulon, in maintenance and improvement of organic solvent tolerance of Escherichia coli K-12. J. Bacteriol. 180:938–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bauer BE, Wolfger H, Kuchler K. 1999. Inventory and function of yeast ABC proteins: about sex, stress, pleiotropic drug and heavy metal resistance. Biochim. Biophys. Acta Biomembr. 1461:217–236 [DOI] [PubMed] [Google Scholar]
- 46. Hu F, Liu J, Du G, Hua Z, Zhou J, Chen J. 2012. Key cytomembrane ABC transporters of Saccharomyces cerevisiae fail to improve the tolerance to d-limonene. Biotechnol. Lett. 34:1505–1509 [DOI] [PubMed] [Google Scholar]
- 47. Cabib E, Roh D-H, Schmidt M, Crotti LB, Varma A. 2001. The yeast cell wall and septum as paradigms of cell growth and morphogenesis. J. Biol. Chem. 276:19679–19682 [DOI] [PubMed] [Google Scholar]
- 48. Lipke PN, Ovalle R. 1998. Cell wall architecture in yeast: new structure and new challenges. J. Bacteriol. 180:3735–3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Haigler C, Brown R, Benziman M. 1980. Calcofluor white ST alters the in vivo assembly of cellulose microfibrils. Science 210:903–906 [DOI] [PubMed] [Google Scholar]
- 50. Klis FM, Boorsma A, De Groot PWJ. 2006. Cell wall construction in Saccharomyces cerevisiae. Yeast 23:185–202 [DOI] [PubMed] [Google Scholar]
- 51. Wood PJ. 1980. Specificity in the interaction of direct dyes with polysaccharides. Carbohydr. Res. 85:271–287 [Google Scholar]
- 52. Popolo L, Vai M. 1998. Defects in assembly of the extracellular matrix are responsible for altered morphogenesis of a Candida albicans phr1 mutant. J. Bacteriol. 180:163–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Damveld RA, vanKuyk PA, Arentshorst M, Klis FM, van den Hondel CAMJJ, Ram AFJ. 2005. Expression of agsA, one of five 1,3-α-d-glucan synthase-encoding genes in Aspergillus niger, is induced in response to cell wall stress. Fungal Genet. Biol. 42:165–177 [DOI] [PubMed] [Google Scholar]
- 54. Ram AFJ, Klis FM. 2006. Identification of fungal cell wall mutants using susceptibility assays based on Calcofluor white and Congo red. Nat. Protoc. 1:2253–2256 [DOI] [PubMed] [Google Scholar]
- 55. Elorza MV, Rico H, Sentandreu R. 1983. Calcofluor white alters the assembly of chitin fibrils in Saccharomyces cerevisiae and Candida albicans cells. J. Gen. Microbiol. 129:1577–1582 [DOI] [PubMed] [Google Scholar]
- 56. Cabib E. 2004. The septation apparatus, a chitin-requiring machine in budding yeast. Arch. Biochem. Biophys. 426:201–207 [DOI] [PubMed] [Google Scholar]
- 57. Boorsma A, Hd. Nobel Bt. Riet Bargmann B, Brul S, Hellingwerf KJ, Klis FM. 2004. Characterization of the transcriptional response to cell wall stress in Saccharomyces cerevisiae. Yeast 21:413–427 [DOI] [PubMed] [Google Scholar]
- 58. García R, Bermejo C, Grau C, Pérez R, Rodríguez-Peña JM, Francois J, Nombela C, Arroyo J. 2004. The global transcriptional response to transient cell wall damage in Saccharomyces cerevisiae and its regulation by the cell integrity signaling pathway. J. Biol. Chem. 279:15183–15195 [DOI] [PubMed] [Google Scholar]
- 59. Levin DE. 2011. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics 189:1145–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Francesconi R, Castellari C, Comelli F. 2001. Densities, viscosities, refractive indices, and excess molar enthalpies of methyl tert-butyl ether + components of pine resins and essential oils at 298.15 K. J. Chem. Eng. Data 46:1520–1525 [Google Scholar]
- 61. French CM, Singer N. 1956. The conductivity of solutions in which the solvent molecule is “large.” I. Solutions of tetraethylammonium picrate in some phthalate esters. J. Chem. Soc. 1956:1424–1429 [Google Scholar]
- 62. Zhang X, Chen Y, Liu J, Zhao C, Zhang H. 2012. Investigation on the structure of water/AOT/IPM/alcohols reverse micelles by conductivity, dynamic light scattering, and small angle X-ray scattering. J. Phys. Chem. B 116:3723–3734 [DOI] [PubMed] [Google Scholar]
- 63. Stone HA. 1994. Dynamics of drop deformation and breakup in viscous fluids. Annu. Rev. Fluid Mech. 26:65–102 [Google Scholar]
- 64. Liu J, Zhu Y, Du G, Zhou J, Chen J. 2013. Exogenous ergosterol protects Saccharomyces cerevisiae from d-limonene stress. J. Appl. Microbiol. 114:482–491 [DOI] [PubMed] [Google Scholar]
- 65. Chandler D. 2005. Interfaces and the driving force of hydrophobic assembly. Nature 437:640–647 [DOI] [PubMed] [Google Scholar]
- 66. Narang AS, Delmarre D, Gao D. 2007. Stable drug encapsulation in micelles and microemulsions. Int. J. Pharm. 345:9–25 [DOI] [PubMed] [Google Scholar]
- 67. Garti N, Yaghmur A, Leser ME, Clement V, Watzke HJ. 2001. Improved oil solubilization in oil/water food grade microemulsions in the presence of polyols and ethanol. J. Agric. Food Chem. 49:2552–2562 [DOI] [PubMed] [Google Scholar]
- 68. Wei G, Li Y, Du G, Chen J. 2003. Effect of surfactants on extracellular accumulation of glutathione by Saccharomyces cerevisiae. Process Biochem. 38:1133–1138 [Google Scholar]
- 69. Schreier S, Malheiros SVP, de Paula E. 2000. Surface active drugs: self-association and interaction with membranes and surfactants. Physicochemical and biological aspects. Biochim. Biophys. Acta Biomembr. 1508:210–234 [DOI] [PubMed] [Google Scholar]
- 70. Levin DE. 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69:262–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.