Abstract
Solid-organ transplant recipients rely on pharmacological immunosuppression to prevent allograft rejection. The effect of such chronic immunosuppression on the microflora at mucosal surfaces is not known. We evaluated the salivary bacterial microbiome of 20 transplant recipients and 19 nonimmunosuppressed controls via 454 pyrosequencing of 16S rRNA gene amplicons. Alpha-diversity and global community structure did not differ between transplant and control subjects. However, principal coordinate analysis showed differences in community membership. Taxa more prevalent in transplant subjects included operational taxonomic units (OTUs) of potentially opportunistic Gammaproteobacteria such as Klebsiella pneumoniae, Pseudomonas fluorescens, Acinetobacter species, Vibrio species, Enterobacteriaceae species, and the genera Acinetobacter and Klebsiella. Transplant subjects also had increased proportions of Pseudomonas aeruginosa, Acinetobacter species, Enterobacteriaceae species, and Enterococcus faecalis, among other OTUs, while genera with increased proportions included Klebsiella, Acinetobacter, Staphylococcus, and Enterococcus. Furthermore, in transplant subjects, the dose of the immunosuppressant prednisone positively correlated with bacterial richness, while prednisone and mycophenolate mofetil doses positively correlated with the prevalence and proportions of transplant-associated taxa. Correlation network analysis of OTU relative abundance revealed a cluster containing potentially opportunistic pathogens as transplant associated. This cluster positively correlated with serum levels of C-reactive protein, suggesting a link between the resident flora at mucosal compartments and systemic inflammation. Network connectivity analysis revealed opportunistic pathogens as highly connected to each other and to common oral commensals, pointing to bacterial interactions that may influence colonization. This work demonstrates that immunosuppression aimed at limiting T-cell-mediated responses creates a more permissive oral environment for potentially opportunistic pathogens without affecting other members of the salivary bacteriome.
INTRODUCTION
Solid-organ transplantation in patients with end-stage organ failure requires lifelong immunosuppression to prevent transplant rejection. The goal of immunosuppressive therapies is to inhibit T-cell-mediated responses, since CD4+ and/or CD8+ lymphocytes have a requisite role in graft rejection (1). However, opportunistic infections, which could directly impact graft survival, are a common comorbidity of continued immunosuppression (2, 3). Common causes of infection, particularly in the late posttransplantation period, are Gram-negative rods such as Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa, in addition to Gram-positive cocci such as Enterococcus spp. and staphylococci (3, 4). Fungal infections, commonly caused by Candida spp., are also frequent in this patient population (3). While the source of infectious microorganisms could be exogenous, the endogenous flora may also serve as an important reservoir of such opportunistic pathogens.
The relationship between long-term, low-intensity immunosuppression aimed at limiting adaptive immune responses and the resident flora colonizing mucosal surfaces is unclear. Studies that characterized the microflora at mucosal surfaces in chronically immunosuppressed patients are scarce, with the available studies having used microbiological methodologies of limited scope (5, 6). Understanding long-term alterations in the mucosal resident microflora is important since pathogen-associated molecular patterns originating from commensal microorganisms may shape local and distal immune responses that could affect transplant survival (2). Moreover, disrupted homeostasis of the resident flora could promote colonization by nonresident microorganisms or increase carriage of opportunistic bacteria and fungi, augmenting the possibility of their translocation to distal sites. Thus, mucosal surfaces have the potential to become important infection portals or reservoirs. The oral cavity, in particular, harbors a diverse resident microbiome and represents a portal of entry for microorganisms into the host. Little is known, however, regarding the role of adaptive immunity and its disruption in shaping the oral commensal flora.
The advent of rRNA gene-based taxonomic identification combined with high-throughput sequencing technologies permits comprehensive characterization of the host microflora, providing a view of microbiome diversity not previously possible. This molecular approach allows evaluation of global microbial profiles, overcoming the limitations of cultivation, which reveals only 30 to 80% of the flora present at host sites (7, 8). High-throughput sequencing of rRNA gene libraries has never been used to evaluate the effect of long-term immunosuppression on the microbial communities that reside at mucosal surfaces. In a prior cultivation-based comparison of the frequency of oral carriage of Candida spp. in solid-organ transplant recipients and nonimmunosuppressed individuals, our group reported that the frequency of carriage of non-albicans Candida spp. is higher in transplanted subjects (9). In the present study, we evaluated the oral bacterial microbiome of a subgroup of these immunosuppressed individuals and compared their salivary bacteriomes to those of nonimmunosuppressed controls. Our objective was to define the alterations inflicted on the resident oral bacterial flora by chronic pharmacological immunosuppression.
MATERIALS AND METHODS
Studied populations, medical data collection, and sampling.
The population investigated was a subset from a larger study that recruited 90 renal and cardiac transplant recipients and 72 controls (9, 10). All study procedures were approved by the Institutional Review Boards from the University of Connecticut Health Center and Hartford Hospital. The current study included 20 subjects from the transplant group and 19 from the control group. Subjects were selected based on availability of saliva samples for microbial profiling. Transplant subjects met the following inclusion criteria: (i) at least 1 year posttransplant; (ii) clinically stable, as defined by serum creatinine levels (kidney only) and no signs of recurrent primary disease or acute rejection; and (iii) no history of antibiotic, antifungal, or antiviral usage during the previous 4 months. Control subjects had no immunological compromising condition and no history of antibiotic, antifungal, or antiviral usage during the previous 4 months.
Medical records of transplant subjects were reviewed, and all relevant information was collected using a standardized data extraction form, as previously described (10). A self-reported medical history was obtained from control subjects. Serum values of C-reactive protein (CRP) and interleukin-6 (IL-6) were determined in both groups via enzyme-linked immunosorbent assays. Descriptive statistics on the entire study population were previously reported (10).
Subjects also received a comprehensive oral examination, which included determination of the number of missing teeth (excluding third molars), plaque scores (% of surfaces positive for plaque), evaluation of periodontal health, and evaluation of mucosal disease. Unstimulated whole saliva and swabs from the oral mucosa were collected as previously described (9). Determinations of Candida load in saliva and oral mucosa and Candida species identification were performed via cultivation methods as previously reported (9).
DNA isolation from saliva, 16S rRNA gene library preparation, and sequencing.
DNA was isolated from whole unstimulated saliva according to previously described procedures using lysozyme and proteinase K treatments and a DNeasy blood and tissue kit (Qiagen) (11). Amplicon libraries of 16S rRNA gene V1-V2 hypervariable regions were generated in triplicate using fusion primers, which included universal primers 8F (5′AGAGTTTGATCMTGGCTCAG3′) and 361R (5′CYIACTGCTGCCTCCCGTAG3′) (12), Roche Life Sciences' 454 Lib-A adapters A and B, and a unique multiplex identifier. PCR and library preparation procedures have been described previously (11). Briefly, PCR mixtures contained 10 ng of purified DNA, 1 U platinum Taq polymerase (Invitrogen), 1.5 mM MgCl2, 200 μM deoxynucleoside triphosphates (dNTPs), Taq buffer (1×), 0.5 μM each forward and reverse primer, and molecular-grade water to a final volume of 25 μl. Thermal cycler conditions were initial denaturation at 95°C for 3 min; 25 cycles of denaturation at 95°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 1 min; and a final extension step at 72°C for 9 min. Negative controls included a DNA isolation control and a PCR control with no added template. Combined triplicate amplicon libraries were sequenced in the forward direction using 454 Titanium chemistry on the 454-GS-FLX platform (454 Life Sciences). Sequences are available at the Short Reads Archive (accession number SRA062231).
Sequencing data processing.
Sequence data were processed using a modification of the pipeline by Schloss et al. (13), as previously described (11), using mothur (14). For operational taxonomic unit (OTU)-based analysis, sequences were clustered using the average neighbor algorithm (15) and a 3% dissimilarity cutoff. Template taxonomies included the large ribosomal database project (RDP) reference data set and the Human Oral Microbiome Database (HOMD), a curated data set, including a large collection of species-level taxa from the oral cavity previously identified (16). OTUs were assigned a taxonomy based on the consensus taxonomic assignment for the majority of sequences within each OTU. If a consensus taxonomy was not possible at the species level (based on HOMD), the nearest taxonomical level at which a consensus was reached was reported. In such cases, the representative sequence from the OTU was also compared to the HOMD, and if results showed more than 97% similarity to an oral taxon (OT), the OT name of the top hit was reported in parentheses as part of the OTU taxonomy. Individually classified sequences were also grouped into phylotypes (from genus to phylum level) based on taxonomic identity.
Sequence libraries were subsampled to contain the same number of sequences for α-diversity comparisons. Richness was evaluated by the number of observed OTUs and number of estimated OTUs as calculated with CatchAll (17). Diversity was measured by the nonparametric Shannon index (18) and the inverse of the Simpson index (19). β-Diversity was measured with the Jaccard index for comparison of communities based on membership and the θYC distance (20) for comparison of communities according to their structure. Principal coordinate analysis (PCoA) of the distance among communities based on the Jaccard and θYC metrics was performed in mothur, and graphs were visualized using the rgl application within R (http://www.r-project.org/). Methods used for DNA isolation, amplicon library preparation, sequencing, and microbial profile analysis have been previously validated using a mock community of oral microorganisms (11).
Statistical analysis.
Clinical and demographic data were compared via t tests, chi-square tests, or Mann-Whitney tests, as appropriate. Differences in α-diversity were evaluated by t tests. Significant separation of clusters after PCoA was evaluated via analysis of molecular variance (AMOVA) (21), as implemented in mothur. Differences in relative abundances of individual OTUs and genera were determined via Metastats (22), while differences in taxon prevalence were tested via chi-square test. While all genera were included in these analyses, only OTUs with 10 or more sequences were considered for relative abundance and prevalence comparisons. Bivariate correlations between taxon-relative abundance, taxon prevalence, Candida-related variables, diversity measures, and dose of immunosuppressants were performed via Spearman rank order correlation tests. Prior to correlations, OTU and genus-relative abundances were transformed using the inverse hyperbolic sine method (23). The significance threshold for statistical tests was adjusted using the Benjamini-Hochberg false discovery rate method.
To facilitate interpretation of prevalence data, we used the Chernoff bound to calculate the minimal relative abundance for which we could have 95% certainty that we will observe at least one sequence at our sequencing effort (see Methods in the supplemental material).
OTU relative abundance data were used to perform correlation network analysis, which identifies clusters (modules) of highly correlated OTUs. Weighted correlation network analysis (WGCNA) (24) was performed as previously described (25). Nodes in undirected weighted networks represented OTUs, and edges represented their adjacency. We first identified outlier samples using absolute hierarchical cluster analysis and constructed a weighted network choosing a soft thresholding power β of 4 with an R2 value of 0.85. Network data were visualized in Cytoscape (26) using the weighted force-directed layout algorithm, with edge length representing the adjacency between nodes. Network centrality measures for the modules obtained were assessed using Cytoscape (26). To relate modules to clinical variables (traits), we also used the WGCNA package correlating the eigen gene for each module with the traits of interest and looking for significant associations based on their P values.
RESULTS
Demographic and clinical characteristics of subjects included in this study.
Table 1 shows the demographic and clinical information of transplant and control subjects. The two groups did not differ in terms of their age, gender, and body mass index (BMI). The transplant group, however, included more diabetics than the control group. In agreement with our previous findings pertaining to the entire study population (10), the transplant recipient subset included in this study showed significantly higher levels of serum CRP and IL-6 than the controls. Parameters related to oral health did not differ between the two groups.
Table 1.
Demographic and clinical characteristics of transplant and control subjectsa
| Characteristic | Value |
Statistic | |
|---|---|---|---|
| Transplant (n = 20) | Control (n = 19) | ||
| Age (yrs) | 51.5 ± 8.6 | 54.4 ± 10.4 | 0.356d |
| Gender (% female) | 30.0 | 36.8 | 0.455b |
| Diabetes (% yes) | 45.0 | 10.5 | 0.019b |
| Body mass index | 30.2 (27.6–35.0) | 27.5 (24.2–31.3) | 0.173c |
| No. of teeth | 26.0 (21.5–27.25) | 27.0 (25.0–28.0) | 0.138c |
| Plaque scores (% of surfaces with plaque) | 52.5 ± 25.5 | 51.7 ± 25.1 | 0.920d |
| Bleeding on probing (% of sites) | 18.3 ± 16.0 | 19.0 ± 13.0 | 0.882d |
| Mean probing depth (PD) (mm) | 2.7 ± 0.4 | 2.6 ± 0.4 | 0.454d |
| Mean clinical attachment level (mm) | 2.9 ± 0.5 | 2.9 ± 0.4 | 0.747d |
| % of sites with PD of ≥5 mm | 1.6 (0.4–8.4) | 1.8 (0.0–6.0) | 0.616c |
| % subjects with severe periodontitise | 15.0 | 26.3 | 0.317b |
| Presence of Candida albicans (%) | 35.0 | 47.4 | 0.372b |
| Presence of other Candida species (%) | 20.0 | 0.0 | 0.053b |
| Candida albicans load in salivaf | 0.0 (0.0–57.5) | 0.0 (0.0–72.5) | 0.729c |
| Candida spp. load in salivaf | 0.0 (0.0–6.0) | 0.0 (0.0–0.5) | 0.608c |
| Candida spp. load in mucosag | 0.0 (0.0–193.5) | 0.0 (0.0–72.5) | 0.783c |
| Serum IL-6 (pg ml−1) | 3.0 (2.2–5.0) | 2.1 (2.0–3.3) | 0.042c |
| Serum CRP (mg l−1) | 0.3 (0.1–0.6) | 0.1 (0.1–0.2) | 0.006c |
Data represent averages ± standard deviations for normally distributed variables, means (interquartile ranges) for nonnormally distributed variables, and frequencies (%) for dichotomous variables. Data in bold indicate a statistically significant difference between groups.
Chi-square test.
Mann-Whitney test.
t test.
Severe periodontitis defined according to Page and Eke (43).
Measured as CFU ml−1.
Measured as total CFU recovered from a single mucosal swab.
Assessment of oral colonization by Candida spp. in the transplant and control groups via culturing methods showed no differences in the presence and load of Candida albicans or in the total Candida species load. However, as previously reported by the parent study (9), there was a greater number of non-albicans species from the genus Candida present in saliva and oral mucosal of transplant subjects than in the controls. This difference, however, did not reach statistical significance in this cohort (Table 1).
In the transplant group, 19 subjects had received a kidney transplant, while 1 subject was a heart recipient. The average number of years posttransplant was 5.2 ± 4.2 (range of 2 to 16 years), with 45% of subjects having received a transplant from a living donor and 40% of subjects with a history of rejection. Subjects were taking multiple combinations of medications as part of their immunosuppressive regimens, with 90% of subjects on prednisone, 80% on mycophenolate mofetil, 65% on tacrolimus, 35% on cyclosporine, 15% on sirolimus, and 10% on azathioprine. In addition, 30% of subjects were taking calcium channel blockers.
Transplant status does not influence the diversity and global structure of salivary bacterial communities but affects community membership.
A total of 183,398 raw sequence reads were obtained, of which 152,301, with an average read length of 222 bp, were included in the final analysis. The range of sequence reads obtained per subject varied from 1,709 to 7,725; thus, for α- and β-diversity comparisons, all subject libraries were randomly subsampled to contain 1,709 reads.
A total of 642 OTUs were observed across all subjects after subsampling; however, the average number of observed OTUs per subject ranged from 59 to 178, indicating intersubject variability in the salivary microbiome at this sequencing effort. Calculations of the number of OTUs predicted to exist in each subject using CatchAll (17) revealed a range of estimated OTUs per subject of 70 to 262. Figure S1 in the supplemental material depicts comparisons of α-diversity between transplant and control subjects. Panel A shows that richness (both observed and estimated OTUs) did not differ between groups, while panel B demonstrates that diversity did not vary between transplant and control subjects.
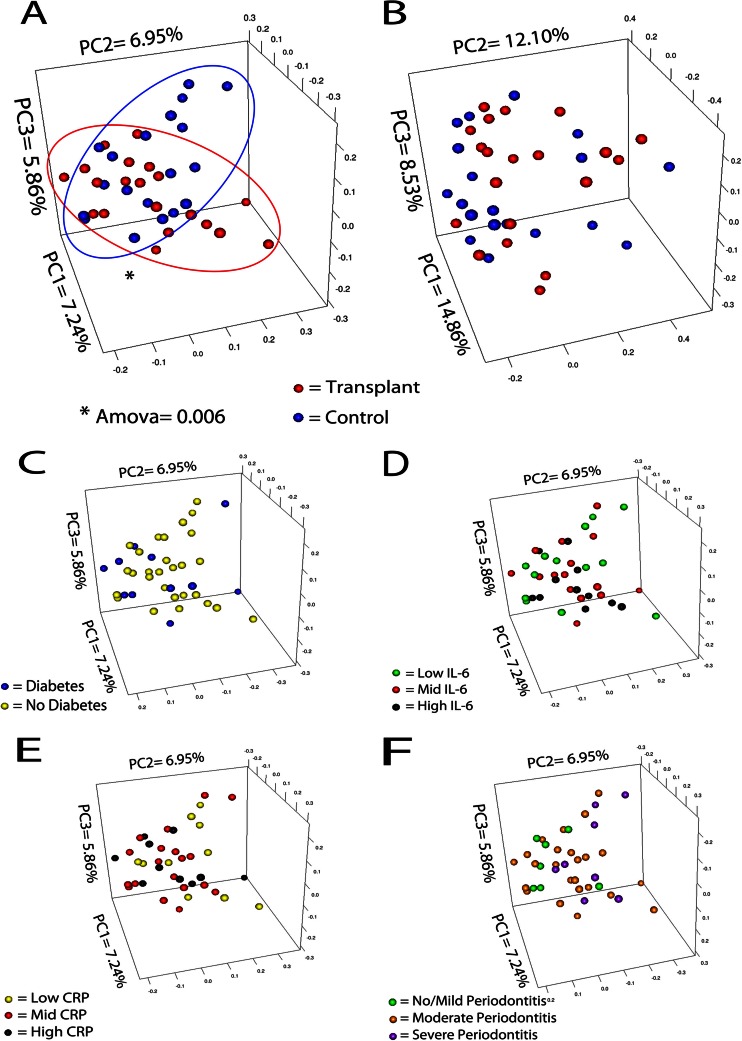
Next, we evaluated overall differences in community composition and in community structure between transplant and control groups. Figure 1 shows PCoA plots of distance among communities according to OTU-based Jaccard (Fig. 1A) and θYC (Fig. 1B) indices. Communities of transplant and control subjects did not form completely separate clusters in these plots. However, comparison of community membership (Fig. 1A) shows that despite some overlap, the two data clouds were statistically different from each other (AMOVA P = 0.006). These observations indicate that transplant status does partially influence community membership (presence/absence of species) but does not affect community structure (the distribution of species counts, i.e., relative abundance). Since transplant and control subjects differed in the number of diabetic subjects and in their serum CRP and IL-6 levels, we also evaluated if these variables were related to community membership and could have confounded the differences observed. As Fig. 1C, D, and E indicate, diabetes, CRP, and IL-6 were not drivers of community composition, further confirming that the observed differences were due to transplant status. Similarly, Fig. 1F shows that the presence of periodontitis did not explain the organization of data points in the Jaccard-based PCoA plot. Furthermore, BMI and other oral health parameters depicted in Table 1 were not found to be drivers of salivary community membership or structure (data not shown).
Fig 1.
β-Diversity comparisons between transplant and control groups. Principal coordinate plots were created based on the Jaccard distance (A) for comparison of communities of transplant and control subjects based on their composition, or from the θYC distance (B) for comparison of communities of transplant and control subjects according to their structure. Differences in the spatial separation of samples belonging to each group were tested using AMOVA, which was significant for the Jaccard distance. Panels C, D, E, and F show lack of clustering of samples (Jaccard) according to diabetes status (C), serum IL-6 levels (D), serum CRP levels (E), and periodontitis status (F). Data points were separated as high, medium, and low in panels D and E according to interquartile ranges. Periodontitis status in panel F was determined according to Page and Eke (43).
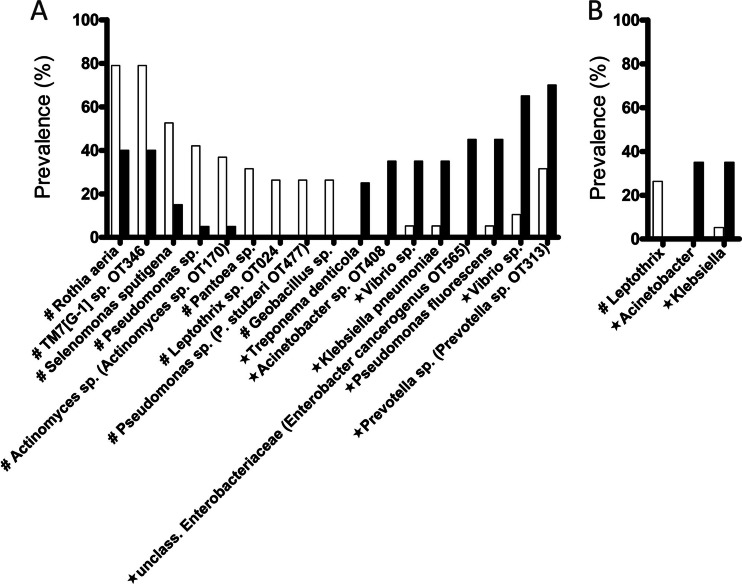
Differences in prevalence of individual taxa between transplant and control groups were then evaluated. We found 17 OTUs and 3 genera with significantly different prevalence between groups (Fig. 2). None of these taxa, however, were among the most frequently detected taxa in salivary communities (depicted in Fig. S2A and S3A in the supplemental material). Among the microorganisms with increased prevalence in transplant subjects (Fig. 2) were Gammaproteobacteria such as Klebsiella pneumoniae, Acinetobacter sp. OT408, which is a close phylogenetic relative of Acinetobacter baumannii, and Pseudomonas fluorescens, all of which are frequently associated with extraoral infections in immunocompromised patients (4, 27, 28). The periodontal pathogen Treponema denticola was also more frequently found in the transplant population, although both groups did not differ in terms of their periodontal health (Table 1). Interestingly, control subjects showed increased prevalence of an OTU of Pseudomonas sp. closely related to Pseudomonas stutzeri, a Pseudomonas sp. very rarely seen as a human pathogen (29).
Fig 2.
OTUs (A) and genera (B) with different prevalence in control (white) and transplant (black) subjects. Taxa with increased prevalence in control subjects are marked with #, while taxa with increased prevalence in transplant subjects are marked with a star.
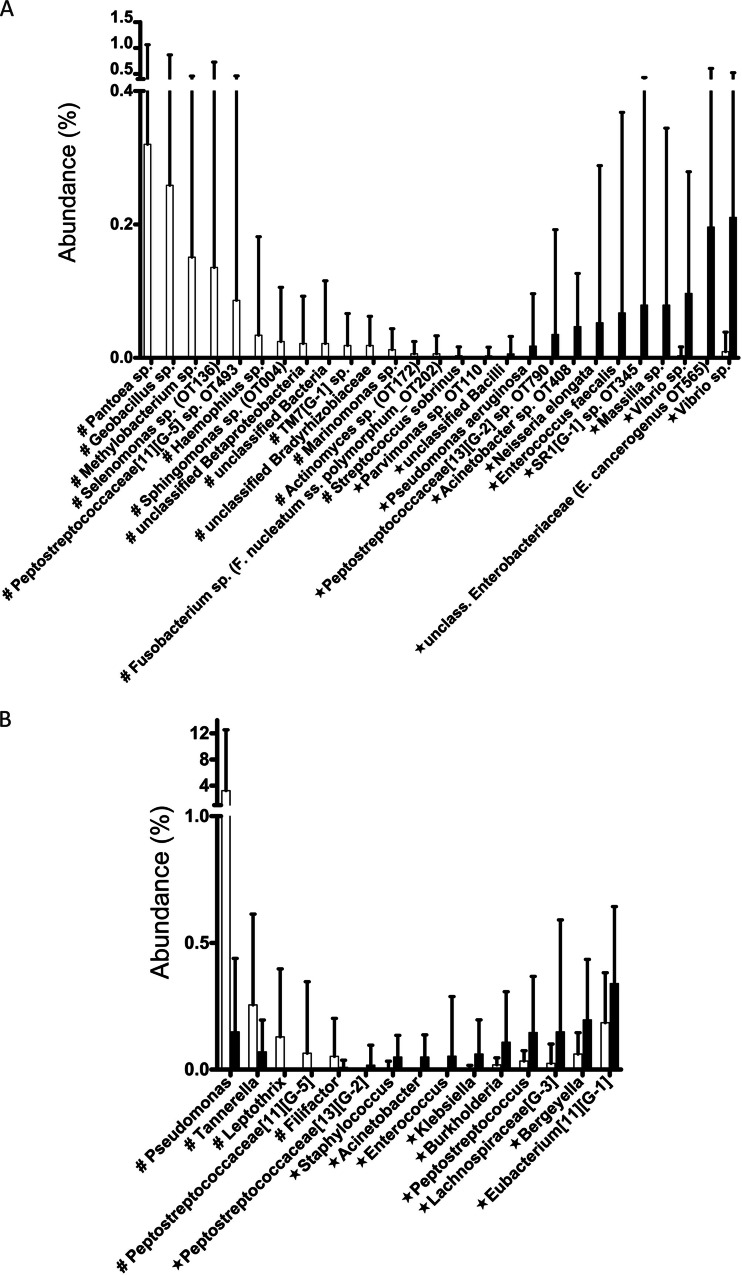
Differences in relative abundance of individual taxa were also evaluated. The B panels in Fig. S2 and S3 in the supplemental material show the most abundant OTUs and genera in transplant and control subjects. Salivary communities were dominated by Streptococcus spp. comprising about 25% of sequence reads, followed by Prevotella spp., Veillonella spp., uncultured TM7 G-1 and Fusobacterium spp., all found at 5 to 10% abundance. Figure 3A shows OTUs differentially represented in terms of their relative abundance in each group. In agreement with prevalence data, we found Gammaproteobacteria such as Pseudomonas aeruginosa, an Acinetobacter sp., and an Enterobacteriaceae sp. closely related to Enterobacter cancerogenus increased in transplant subjects. In addition, Enterococcus faecalis, an organism associated with infections in immunocompromised hosts (30), was increased in the transplant group. In Fig. 3B, genera with different relative abundance between groups are depicted. Among these, we found of interest that the genus Staphylococcus, commonly associated with extraoral infections in immunosuppressed populations (3), was increased in the transplant group.
Fig 3.
OTUs (A) and genera (B) with different relative abundance in control (white) and transplant (black) subjects. Taxa with increased relative abundance in control subjects are marked with #, while taxa with increased relative abundance in transplant subjects are marked with a star.
Relationship between dose of immunosuppressants and the prevalence and relative abundance of individual oral taxa.
A bivariate correlation analysis was performed to evaluate whether exposure to immunosuppressants was associated in a dose-dependent manner with the prevalence or relative abundance of transplant-associated OTUs and genera. The relationship between fungal colonization as determined by culturing methods (9) and immunosuppressant doses was also evaluated. From all the individual immunosuppressant drugs subjects were receiving, only prednisone and mycophenolate mofetil were significantly associated with microbial colonization of the oral cavity (Table 2). Prednisone dose showed a positive correlation with both the relative abundance and prevalence of 2 OTUs, identified as an unclassified Enterobacteriaceae species and a Vibrio species, and with the genera Klebsiella and Acinetobacter. The dose of mycophenolate mofetil also correlated with some of the bacterial variables evaluated, showing a positive correlation with the relative abundance of the genera Staphylococcus, Acinetobacter, and Klebsiella and with the prevalence of Klebsiella. Interestingly, the dose of prednisone and not that of mycophenolate mofetil showed a positive correlation with OTU richness, an indication that prednisone-related systemic immunosuppression is associated with a more permissive oral environment, conducive to increased bacterial diversity. In contrast, mycophenolate mofetil, and not prednisone, showed a positive correlation with the load and number of Candida spp. in the oral mucosa, which suggests that mycophenolate mofetil-related immunosuppression exerts a stronger influence on fungal colonization than prednisone.
Table 2.
Correlation between dose of immunosuppressants and bacterial and fungal oral colonizationa
| Organism | Value (mg kg−1) |
|
|---|---|---|
| Prednisone | Mycophenolate mofetil | |
| Relative abundance of bacterial taxab | ||
| OTU 14 Vibrio sp. | 0.595 (0.006) | 0.400(0.080) |
| OTU 99 Unclassified Enterobacteriaceae (E. cancerogenus OT 565) | 0.635 (0.003) | 0.452 (0.045) |
| OTU 17 Vibrio sp. | 0.460 (0.041) | 0.313(0.180) |
| Klebsiella | 0.677 (0.001) | 0.695 (0.001) |
| Acinetobacter | 0.532 (0.016) | 0.446 (0.049) |
| Staphylococcus | 0.600 (0.005) | 0.638 (0.002) |
| Prevalence of bacterial taxab | ||
| OTU 49 Treponema denticola | 0.551 (0.012) | 0.302(0.196) |
| OTU 99 unclassified Enterobacteriaceae (E. cancerogenus OT 565) | 0.602 (0.005) | 0.385(0.094) |
| OTU 17 Vibrio sp. | 0.473 (0.035) | 0.201(0.396) |
| Klebsiella | 0.664 (0.001) | 0.703 (0.001) |
| Acinetobacter | 0.555 (0.011) | 0.420(0.065) |
| Bacterial diversity metricsc | ||
| Observed OTUs | 0.479 (0.033) | 0.129(0.587) |
| Estimated OTUs | 0.527 (0.017) | 0.281(0.230) |
| Candida colonization | ||
| Candida species load in mucosad | 0.322(0.166) | 0.541 (0.014) |
| No. of Candida spp. detected in mucosa | 0.217(0.294) | 0.496 (0.026) |
Data in this table correspond to Spearman rank order correlation coefficients (rs), with respective P values in parenthesis. Data in bold indicate significant correlations.
Taxonomic levels evaluated included OTU and genus.
Data correspond to OTU richness measures per sample. Total richness (estimated OTUs) was determined with CatchAll (17).
Candida spp. colonization was measured as total CFUs recovered from a single mucosal swab.
We also tested if transplant-associated OTUs and genera (Fig. 2 and 3) correlated with indicators of transplant function (history of rejection, fibrosis, inflammatory changes) or other transplant-related variables (posttransplant years, live or cadaveric donor), but no significant correlations were found (data not shown).
To test if local factors related to oral health could be associated with increased colonization by opportunistic pathogens, a correlation analysis of oral health variables and prevalence or relative abundance of transplant-associated OTUs and genera was performed. No correlations were found, however, between oral health indicators and carriage of these bacterial taxa. Interestingly, in this analysis, salivary carriage of taxa classically associated with periodontitis, such as T. denticola, did not show a positive correlation with specific periodontal health-related parameters. Since we had reported a positive correlation between the prevalence of T. denticola and prednisone dose (Table 2), we also tested if prednisone dose correlated with oral health parameters but found no correlation. With respect to fungal colonization, we found that the number of Candida spp. in mucosa positively correlated with plaque scores (rs = 0.536, P = 0.015), and therefore oral hygiene status is a potential confounder of the relationship between mycophenolate mofetil and Candida diversity in mucosa reported in Table 2.
Correlations among salivary bacterial OTUs and the relationship of these networks to transplant status.
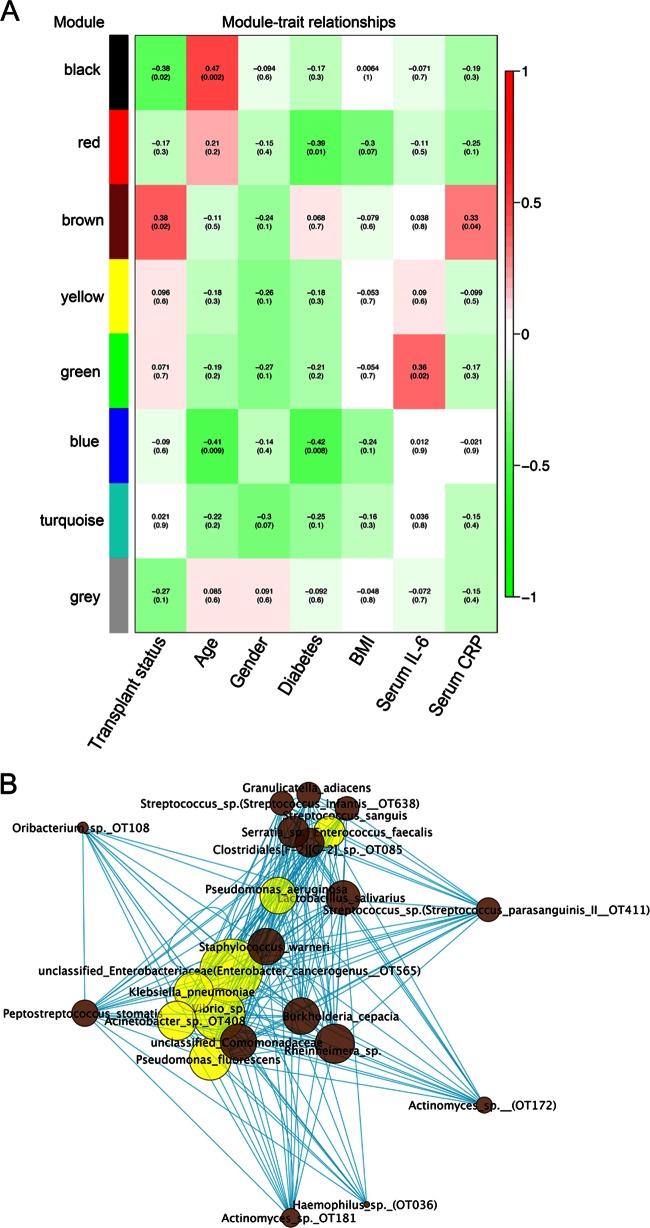
Next, we used WGNCA to find modules of salivary OTUs highly correlated to each other in terms of their frequency distributions. Figure S4 in the supplemental material shows the initial hierarchical clustering of samples. Eight modules of closely correlated OTUs were found (see Fig. S5 in the supplemental material), which were arbitrarily assigned to a color for their identification. Figure 4 shows the correlation of each module with subject demographic characteristics, transplant status, and levels of serum inflammatory markers. As seen in Fig. 4A, the brown module was the only one showing a positive correlation with transplant status. Interestingly, this module also showed a positive correlation with serum levels of CRP, a marker of systemic inflammation. Figure 4B shows the OTUs that form the brown module and their connections. In accordance with analysis of individual taxa, the brown module contained OTUs shown to be increased in transplant subjects or OTUs from genera individually associated with the transplant group (as per Fig. 2 and 3) and representing potentially opportunistic pathogens. The brown module also contained OTUs highly prevalent and abundant in salivary bacterial communities (as per Fig. S2 in the supplemental material) and considered to be prime oral commensals, such as Streptococcus sp. (S. parasanguinis II OT411), Streptococcus sp. (S. infantis OT638), Granulicatella adiacens, Oribacterium sp. OT108, and Actinomyces sp. (OT172). An uncultured Clostridiales sp., very abundant in saliva, was also part of this network. The relationship of OTUs within this network was further explored by measuring degree centrality (number of connections a node has). It was found that transplant-associated opportunistic pathogens displayed the highest degree centrality. These results are displayed in Fig. 4B and indicate that relative abundances among opportunistic species appeared to be highly correlated, while these microorganisms also interact closely with all commensal nonhub microorganisms in the network.
Fig 4.
Network correlation analysis identified a cluster (brown) associated with transplant status and containing OTUs from potentially opportunistic pathogens. Panel A shows correlations between demographic and clinical subject characteristics and the 8 different salivary OTU clusters identified in all subjects (transplant and control). Correlations are color coded, with red indicating the highest positive correlation and green the lowest negative correlation. Coefficients and P values appear in parenthesis. Panel B shows the OTUs that comprise the brown cluster and their connections. Each node in the network represents an OTU. Edge length represents adjacency between pairs of nodes. OTUs that were identified as transplant associated in individual taxa analysis are shown in yellow. Size of each node represents OTU connectivity, measured as degree centrality (number of connections with other nodes), with the bigger nodes representing those highly connected.
Figure S6 in the supplemental material shows the black module, which was found to be negatively associated with transplant status. This network contained taxa seen in individual analysis as associated with control subjects. Interestingly, Pseudomonas spp. with low pathogenic potential in humans were part of this cluster.
DISCUSSION
We have conducted the first comprehensive evaluation of the effect of long-term transplantation-related immunosuppression on the oral bacterial microbiome. Previous studies that evaluated the oral microflora in organ recipients suffered from the limitations of using a cultivation approach, which allowed identification of only a small number of species, and were confounded by the effects of antibiotics on the microflora of the studied subjects (31, 32). Using high-throughput sequencing of 16S rRNA gene libraries, we found that pharmacological immunosuppression aimed at increasing allograft tolerance does not affect the most common and abundant bacterial species in saliva but increases the frequency of detection of microorganisms known to be the cause of extraoral infections in transplant recipients (4, 27, 30). Oral colonization by Gram-negative rods such as Klebsiella pneumoniae, Pseudomonas aeruginosa, Pseudomonas fluorescens, and Acinetobacter baumannii and by Gram-positive cocci such as Staphylococcus aureus and Enterococcus faecalis has been previously noted in other medically compromised subjects, particularly in bed-ridden elderly individuals (33). Although it could be suspected that these pathogens colonize hospitalized elderly populations more frequently because of their compromised immunological responses, most studies attributed their increased frequency of detection to local factors, such as deficient salivary flow, lack of adequate oral hygiene, or antibiotic use, which are thought to disrupt the resident commensal flora (33). Subjects in our study, however, had not taken any type of antimicrobial for 4 months prior to sampling and did not have oral health or hygiene (plaque score) different from that of control individuals. Moreover, we found no correlation between the prevalence and relative abundance of transplant-associated OTUs and genera from potentially opportunistic pathogens and oral health parameters. Thus, the differences found can be directly attributed to the effect of immunosuppressive regimens on oral colonization.
Traditionally, Klebsiella spp., Acinetobacter spp., Pseudomonas spp., and staphylococci are not considered to form part of the regular oral commensal flora during health (33), although several studies using cultivation methods have revealed their presence in the oropharynx of a minority of subjects (34, 35). Our study agrees with the latter studies in finding these opportunistic pathogens in a small number of samples in the control group. These results are also in agreement with a recent analysis of data from the Human Microbiome Project, which found S. aureus, K. pneumoniae, and E. faecalis at very low prevalence and in low proportions (∼0.01% to 0.1% of the total flora) in saliva samples of healthy individuals (36). However, when interpreting prevalence data, it should be considered that lack of detection of a taxon in a given sample depends on its relative abundance and on sampling effort. Richness coverage in our study ranged from 54% to 87% of the OTUs predicted to exist in individual samples by the CatchAll estimator, indicating that a portion of the flora was not observed. By using the Chernoff bound, we calculated that at a sequencing effort of 1,709 sequences, 0.0052% is the lowest relative abundance at which a species could exist in a sample and be detected with 95% confidence. Therefore, opportunistic pathogens could have been part of the regular oral commensal flora of a greater number of individuals from the control group but present in very low abundance (less than 0.0052%) and therefore not detectable by our depth of sequencing. Thus, the increased frequency of detection of opportunistic pathogens in transplant subjects could reflect higher relative abundance than in controls, and it is not necessarily a consequence of differences in carriage. It should also be taken into account that the lysis method used for this study has been validated only for oral taxa (11). Some of the opportunistic species detected require special lysis protocols, and they may be underrepresented in our samples. It is then quite possible that opportunistic pathogens are common low-abundance inhabitants of the oral cavity in health. This question can be answered in future studies by utilizing a deeper sequencing approach and targeted lysis methods. The increased detection and higher proportions of opportunistic pathogens in transplant recipients is, however, an indication that systemic immune defenses serve as gatekeepers for these species, limiting their success in the oral cavity. The specificity of immunological protection against mucosal colonization by opportunistic pathogens is highlighted by the finding that common commensals were not affected by the altered immune responses associated with transplant-related immunosuppression.
An intriguing possibility that arises from our results is whether the oral cavity could serve as a reservoir of microorganisms for opportunistic infections at distal sites in immunocompromised hosts. Short-duration bacteremias are documented to occur following daily oral hygiene practices such as tooth brushing (37). The higher proportions in which opportunistic pathogens are present in the oral cavity of transplant subjects increase their chances of forming part of such bacteremic challenges. It is also possible that immunosuppression favors colonization of these species at other mucosal sites, which could in turn serve as infection reservoirs. A recent report on the vaginal bacterial colonization in kidney transplant recipients showed no differences in lactobacilli isolated from transplant subjects and control women (6). This study, however, did not conduct a comprehensive microflora characterization, and no study to date has evaluated the mucosal microbiome at different body sites in immunocompromised individuals by using a global molecular approach. Therefore, epidemiological studies using high-throughput sequencing of 16S rRNA genes to characterize microbial communities at different mucosal surfaces in immunocompromised hosts are necessary. Furthermore, the use of metagenomic approaches in prospective trials could clarify whether infections in immunosuppressed subjects are caused by opportunistic pathogens that are not only phylogenetically related but also genetically similar to those present at mucosal surfaces. In addition, the finding that an oral salivary network containing opportunistic pathogens is associated with CRP levels, a systemic inflammatory marker, requires further investigation. It is possible that low-grade systemic inflammation triggered by the allograft plays a role in shaping the mucosal microbial ecology. Conversely, opportunistic pathogens at mucosal surfaces could be contributing to the systemic inflammatory burden. This question is particularly important in transplant recipients, as increased systemic inflammation triggered by nonsymptomatic colonization of mucosal surfaces by infectious agents could have grave consequences for graft survival.
Our results also show that there is an association between the dose of certain immunosuppressants taken by transplant subjects and oral colonization by opportunistic pathogens. It should be noted, however, that immunosuppressant regimes consist of a combination of pharmacological agents. Thus, we not only tested the association between single immunosuppressant doses and selected taxa, but we also included drug combinations in our correlation analysis (data not shown). However, our results revealed that the individual doses of two agents, prednisone and mycophenolate mofetil, showed the strongest correlations with prevalence and abundance of individual taxa. These results are possibly due to an independent effect of each agent on the oral microbiome. We found, for example, that although the dose of both agents correlated with some of the transplant-associated bacterial taxa, prednisone showed a higher number of significant correlations than mycophenolate mofetil. Moreover, prednisone, and not mycophenolate mofetil, correlated with bacterial richness, a suggestion that prednisone has a more profound effect on oral bacterial colonization. It is difficult to pinpoint, however, the mechanisms behind the apparently more permissive oral environment for bacteria created by prednisone, as this drug has a wide range of effects on immune responses (38). In contrast, we found that the dose of mycophenolate mofetil (and not that of prednisone) correlated with Candida species load and diversity in the oral mucosa. It is worth noting that no correlation was found when C. albicans loads alone were considered, a result that indicates that mycophenolate mofetil may create a more permissive environment only for non-albicans Candida spp. Mycophenolate mofetil has a somewhat narrow effect on host cells, specifically targeting activated T and B lymphocytes (39). It is then possible that non-albicans Candida spp. are more deeply affected by these altered acquired immune responses than the bacterial flora.
Network correlation analysis also confirmed the association between opportunistic pathogens and transplant status. This analysis revealed that opportunistic pathogens tend to cooccur in the same subjects. Not all transplant subjects, however, were colonized by opportunistic pathogens, and therefore the correlation of the brown module and transplant status was only weakly significant. It is also worth noting that due to the limited sample size, the relationships uncovered are preliminary. It was interesting to observe, however, that potential opportunistic pathogens are not only associated with each other but also connected to common and abundant oral commensals. These findings suggest that the ability of potential opportunistic pathogens to colonize oral surfaces not only depends on decreased immune surveillance, but it may also be facilitated by certain members of the commensal flora.
Although not the main purpose of this study, our evaluation of the salivary microbiome in this cohort of 39 subjects provides further confirmation of the microorganisms that reside in human saliva. Our results mostly agree with those from published 16S rRNA gene-based characterizations of the salivary bacterial flora (40–42), showing that despite the inclusion of different populations and utilization of various sampling methods, primers, and sequencing strategies, a concordant picture is emerging on the salivary bacteriome. For example, Segata et al. (40) reported that Streptococcus, Veillonella, and Prevotella are the most abundant genera in saliva, in agreement with our results. Our analysis, however, utilizes the HOMD (16), a curated data set for oral taxa, which allows identification of most OTUs at a species level and permits to assign traceable sequence identities to uncultured taxa. For instance, in the case of uncultured TM7, our analysis allowed to ascertain the identity of the most abundant TM7 in saliva as TM7 genospecies 1 (according to the HOMD phylogenetic classification).
In conclusion, this study evaluated the salivary microbiome of solid-organ transplant recipients, finding that immunosuppression does not exert a significant effect on the most abundant bacterial taxa but increases the frequency of detection and relative abundance of taxa documented to behave as opportunistic pathogens in immunocompromised hosts. Among the immunosuppressant agents subjects received, prednisone had the most significant effect on bacterial diversity and on the colonization of potentially opportunistic pathogens, while mycophenolate mofetil had a more limited effect on the bacterial flora but was associated with increased colonization by non-albicans Candida spp. Further studies should clarify the specific mechanisms mediating the effects of these pharmacological agents at oral mucosal surfaces and the association between an increase in the nonsymptomatic carriage of opportunistic pathogens at mucosal sites and risk for distal infections in transplant recipients.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grants R21DE016466 and R01DE021578 from the National Institutes of Health (NIH).
Footnotes
Published ahead of print 24 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/CVI.00734-12.
REFERENCES
- 1. Lechler RI, Sykes M, Thomson AW, Turka LA. 2005. Organ transplantation—how much of the promise has been realized? Nat. Med. 11:605–613 [DOI] [PubMed] [Google Scholar]
- 2. Chong AS, Alegre ML. 2012. The impact of infection and tissue damage in solid-organ transplantation. Nat. Rev. Immunol. 12:459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hlava N, Niemann CU, Gropper MA, Melcher ML. 2009. Postoperative infectious complications of abdominal solid organ transplantation. J. Intensive Care Med. 24:3–17 [DOI] [PubMed] [Google Scholar]
- 4. Winters HA, Parbhoo RK, Schafer JJ, Goff DA. 2011. Extended-spectrum beta-lactamase-producing bacterial infections in adult solid organ transplant recipients. Ann. Pharmacother. 45:309–316 [DOI] [PubMed] [Google Scholar]
- 5. Brown LR, Mackler BF, Levy BM, Wright TE, Handler SF, Moylan JS, Perkins DH, Keene HJ. 1979. Comparison of the plaque microflora in immunodeficient and immunocompetent dental patients. J. Dent. Res. 58:2344–2352 [DOI] [PubMed] [Google Scholar]
- 6. Martirosian G, Radosz-Komoniewska H, Pietrzak B, Ekiel A, Kaminski P, Aptekorz M, Dolezych H, Samulska E, Jozwiak J. 2012. Characterization of vaginal lactobacilli in women after kidney transplantation. Anaerobe 18:209–213 [DOI] [PubMed] [Google Scholar]
- 7. Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, Podar M, Leys EJ. 2011. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 6:1176–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fraher MH, O'Toole PW, Quigley EM. 2012. Techniques used to characterize the gut microbiota: a guide for the clinician. Nat. Rev. Gastroenterol. Hepatol. 9:312–322 [DOI] [PubMed] [Google Scholar]
- 9. Dongari-Bagtzoglou A, Dwivedi P, Ioannidou E, Shaqman M, Hull D, Burleson J. 2009. Oral Candida infection and colonization in solid organ transplant recipients. Oral Microbiol. Immunol. 24:249–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shaqman M, Ioannidou E, Burleson J, Hull D, Dongari-Bagtzoglou A. 2010. Periodontitis and inflammatory markers in transplant recipients. J. Periodontol. 81:666–672 [DOI] [PubMed] [Google Scholar]
- 11. Diaz PI, Dupuy AK, Abusleme L, Reese B, Obergfell C, Choquette L, Dongari-Bagtzoglou A, Peterson DE, Terzi E, Strausbaugh LD. 2012. Using high throughput sequencing to explore the biodiversity in oral bacterial communities. Mol. Oral Microbiol. 27:182–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sundquist A, Bigdeli S, Jalili R, Druzin ML, Waller S, Pullen KM, El-Sayed YY, Taslimi MM, Batzoglou S, Ronaghi M. 2007. Bacterial flora-typing with targeted, chip-based pyrosequencing. BMC Microbiol. 7:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schloss PD, Gevers D, Westcott SL. 2011. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One 6:e27310 doi: 10.1371/journal.pone.0027310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schloss PD, Westcott SL. 2011. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl. Environ. Microbiol. 77:3219–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. 2010. The human oral microbiome. J. Bacteriol. 192:5002–5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bunge J. 2011. Estimating the number of species with CatchAll. Pac. Symp. Biocomput. 2011:121–130 [DOI] [PubMed] [Google Scholar]
- 18. Chao A, Shen TJ. 2003. Nonparametric estimation of Shannon's index of diversity when there are unseen species in sample. Environ. Ecol. Stat. 10:429–443 [Google Scholar]
- 19. Simpson EH. 1949. Measurement of diversity. Nature 163:688 [Google Scholar]
- 20. Yue JC, Clayton MK. 2005. A similarity measure based on species proportions. Comm. Stat. Theor. Meth. 34:2123–2131 [Google Scholar]
- 21. Excoffier L, Smouse PE, Quattro JM. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. White JR, Nagarajan N, Pop M. 2009. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 5(4):e1000352 doi: 10.1371/journal.pcbi.1000352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Burbidge JB, Lonnie M, Robb AL. 1988. Alternative transformations to handle extreme values of the dependent variable. J. Am. Stat. Assoc. 83:123–127 [Google Scholar]
- 24. Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Duran-Pinedo AE, Paster B, Teles R, Frias-Lopez J. 2011. Correlation network analysis applied to complex biofilm communities. PLoS One 6(12):e28438 doi: 10.1371/journal.pone.0028438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. 2011. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27:431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim YJ, Yoon JH, Kim SI, Hong KW, Kim JI, Choi JY, Yoon SK, You YK, Lee MD, Moon IS, Kim DG, Kang MW. 2011. High mortality associated with Acinetobacter species infection in liver transplant patients. Transplant. Proc. 43:2397–2399 [DOI] [PubMed] [Google Scholar]
- 28. Hsueh PR, Teng LJ, Pan HJ, Chen YC, Sun CC, Ho SW, Luh KT. 1998. Outbreak of Pseudomonas fluorescens bacteremia among oncology patients. J. Clin. Microbiol. 36:2914–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Noble RC, Overman SB. 1994. Pseudomonas stutzeri infection. A review of hospital isolates and a review of the literature. Diagn. Microbiol. Infect. Dis. 19:51–56 [DOI] [PubMed] [Google Scholar]
- 30. Lee SO, Kang SH, Abdel-Massih RC, Brown RA, Razonable RR. 2011. Spectrum of early-onset and late-onset bacteremias after liver transplantation: implications for management. Liver Transpl. 17:733–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Saraiva L, Lotufo RF, Pustiglioni AN, Silva HT, Jr, Imbronito AV. 2006. Evaluation of subgingival bacterial plaque changes and effects on periodontal tissues in patients with renal transplants under immunosuppressive therapy. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 101:457–462 [DOI] [PubMed] [Google Scholar]
- 32. Ahmadieh A, Baharvand M, Fallah F, Djaladat H, Eslani M. 2010. Oral microflora in patients on hemodialysis and kidney transplant recipients. Iran J. Kidney Dis. 4:227–231 [PubMed] [Google Scholar]
- 33. Tada A, Hanada N. 2010. Opportunistic respiratory pathogens in the oral cavity of the elderly. FEMS Immunol. Med. Microbiol. 60:1–17 [DOI] [PubMed] [Google Scholar]
- 34. Rosenthal S, Tager IB. 1975. Prevalence of Gram-negative rods in the normal pharyngeal flora. Ann. Intern. Med. 83:355–357 [DOI] [PubMed] [Google Scholar]
- 35. Conti S, dos Santos SS, Koga-Ito CY, Jorge AO. 2009. Enterobacteriaceae and Pseudomonadaceae on the dorsum of the human tongue. J. Appl. Oral Sci. 17:375–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Conlan S, Kong HH, Segre JA. 2012. Species-level analysis of DNA sequence data from the NIH Human Microbiome Project. PLoS One 7(10):e47075 doi: 10.1371/journal.pone.0047075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lockhart PB, Brennan MT, Sasser HC, Fox PC, Paster BJ, Bahrani-Mougeot FK. 2008. Bacteremia associated with toothbrushing and dental extraction. Circulation 117:3118–3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zen M, Canova M, Campana C, Bettio S, Nalotto L, Rampudda M, Ramonda R, Iaccarino L, Doria A. 2011. The kaleidoscope of glucorticoid effects on immune system. Autoimmun. Rev. 10:305–310 [DOI] [PubMed] [Google Scholar]
- 39. Ritter ML, Pirofski L. 2009. Mycophenolate mofetil: effects on cellular immune subsets, infectious complications, and antimicrobial activity. Transpl. Infect. Dis. 11:290–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, Huttenhower C, Izard J. 2012. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 13:R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lazarevic V, Whiteson K, Hernandez D, Francois P, Schrenzel J. 2010. Study of inter- and intra-individual variations in the salivary microbiota. BMC Genomics 11:523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zaura E, Keijser BJ, Huse SM, Crielaard W. 2009. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 9:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Page RC, Eke PI. 2007. Case definitions for use in population-based surveillance of periodontitis. J. Periodontol. 78(Suppl 7):1387–1399 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.