Abstract
Herpes simplex virus 1 (HSV-1), the prototypic member of herpesviruses, employs a virally encoded molecular machine called terminase to package the viral double-stranded DNA (dsDNA) genome into a preformed protein shell. The terminase contains a large subunit that is thought to cleave concatemeric viral DNA during the packaging initiation and completion of each packaging cycle and supply energy to the packaging process via ATP hydrolysis. We have determined the X-ray structure of the C-terminal domain of the terminase large-subunit pUL15 (pUL15C) from HSV-1. The structure shows a fold resembling those of bacteriophage terminases, RNase H, integrases, DNA polymerases, and topoisomerases, with an active site clustered with acidic residues. Docking analysis reveals a DNA-binding surface surrounded by flexible loops, indicating considerable conformational changes upon DNA binding. In vitro assay shows that pUL15C possesses non-sequence-specific, Mg2+-dependent nuclease activity. These results suggest that pUL15 uses an RNase H-like, metal ion-mediated catalysis mechanism for cleavage of viral concatemeric DNA. The structure reveals extra structural elements in addition to the RNase H-like fold core and variations in local architecture of the nuclease active site, which are conserved in herpesvirus terminases and bear great similarity to the phage T4 gp17 but are distinct from podovirus and siphovirus orthologs and cellular RNase H, delineating a new evolutionary lineage among a large family of eukaryotic viruses and simple and complex prokaryotic viruses.
INTRODUCTION
Herpes simplex virus 1 (HSV-1) is the prototypical member of the herpesvirus family and belongs to the alpha-herpesvirus subfamily (1). HSV-1 infects more than 60% of the United States population and is responsible for oral cold sores and rare but severe encephalitis (1). HSV-1 can establish latency in neurons and can replicate in brains. The virion comprises three major structural elements: a nucleocapsid containing the 152-kbp genome, an envelope consisting of a lipid bilayer with embedded glycoproteins, and a proteinaceous tegument in between (2). The capsid is an icosahedral shell of 1,250-Å diameter enclosing a double-stranded DNA (dsDNA) genome (3).
HSV-1 produces branched concatemeric viral DNA in the nuclei of infected cells. Individual genomes are generated by virally encoded machinery that recognizes packaging signals in concatemeric DNA and endonucleolytically cleaves the DNA within these signals, and the cleavage is tightly coupled to insertion of DNA into a preformed protein shell called procapsid (4–8). The first of these cleavages generates the long component terminus, while the second generates the terminus of the short component and frees the cleaved genome from the concatemer (9). The viral DNA is inserted into procapsid through a unique portal vertex, which is composed of a dodecameric ring of pUL6 protein with an internal diameter wide enough to accommodate passage of dsDNA (10–12). As the DNA is packaged into the capsid, it replaces the internal proteinaceous shell or scaffold, and the outer shell undergoes a dramatic and stabilizing conformational change (13, 14). Capsids containing DNA are termed nucleocapsids or type C capsids (15). In a default reaction that occurs in the absence of terminase components, immature capsids (termed procapsids or large-cored B capsids) undergo the conformational change and retain the internal shell (13, 14). These forms are believed to represent dead-end products and are termed B capsids or small-cored B capsids. In instances in which the scaffold is expelled but DNA is absent, type A capsids are produced. Such capsids are believed to result when the DNA-packaging process initiates but is subsequently aborted and the DNA slips out of the capsid.
DNA packaging of HSV-1 requires seven protein-encoding genes, namely, UL6, UL15, UL17, UL25, UL28, UL32, and UL33, as identified through genetic analysis (7). Mutations within these genes result in defects either in the cleavage of concatemeric DNA or DNA encapsidation (8, 16, 17). Like many tailed dsDNA bacteriophages, genome cleavage and packaging in HSV-1 are tightly coupled and require a terminase enzyme that harbors the DNA recognition, endonuclease, and ATPase activities (7, 18). Unlike bacteriophage terminases that consist of two subunits, evidence suggests that the herpes simplex virus terminase consists of three protein components encoded by the UL15, UL28, and UL33 genes (19, 20). It was reported that the HSV-1 terminase complex assembled in the cytoplasm, eventually interacting with the portal vertex of the procapsid in infected cell nuclei (21). Trafficking of HSV-1 terminase proteins into nuclei depends on a nuclear localization signal in the product of the UL15 gene (pUL15) (21). pUL28 has been shown to bind viral DNA-packaging sequences (22), while pUL33 associates with pUL28 and enhances the interaction between the pUL28-pUL33 complex and pUL15 (20).
The UL15 gene is composed of two exons separated by an intron of 3,587 bp, and it encodes a 735-residue protein (23, 24). The UL15 gene family of herpesviruses predicts a highly conserved ATPase motif that, at least in the herpes simplex virus, is required for viral DNA cleavage and packaging (25, 26). The structure of the C-terminal domain of the pUL15 ortholog (designated pUL89C) from human cytomegalovirus (HCMV), a beta-herpesvirus, exhibits an RNase H-like nuclease domain fold that is responsible for cleavage of concatemeric DNA into unit-length genomes (27, 28). Here, we report the 2.46-Å resolution crystal structure of the pUL15 C-terminal domain (pUL15C) arranged as a novel trimer. The structure shows a fold resembling those of RNase H, integrases, DNA polymerases, and topoisomerases, indicating that pUL15 utilizes a similar metal ion-mediated catalytic mechanism. Docking analysis enabled by better-defined surface loops shows a putative DNA-binding surface comprising numerous positively charged residues. Structural comparison with viral terminase nuclease domains and RNase H-like nucleases reveals conserved and variable features in the fold cores and the nuclease active sites, suggesting an evolutionary lineage among eukaryotic and prokaryotic viruses. As HSV-1 is the prototype of herpesviruses and much has been known regarding cis-acting elements on the viral genome required for DNA packaging and composition and cellular localization of the terminase components, the pUL15C structure can enable further studies to understand assembly of the HSV-1 terminase and the DNA-packaging mechanisms in herpesviruses.
MATERIALS AND METHODS
Protein expression and purification.
The pUL15C gene encoding residues 471 to 735 was cloned into the pET28b (Novagen, Madison, WI) expression vector between the NdeI and XhoI restriction sites, resulting in pUL15C with an N-terminal His tag. The protein was expressed in Escherichia coli B834(DE3) cells overnight at 15°C to an optical density at 600 nm (OD600) of 2.2. Protein expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1.0 mM at an OD600 of 0.59, and the cells were kept to grow overnight. Cells were harvested at 5,000 rpm, resuspended in a buffer containing 20 mM Tris-HCl (pH 8.5), 500 mM NaCl, and 10 mM 2-mercaptoethanol and sonicated. Insoluble materials were sedimented by centrifugation (15,000 rpm, 4°C, 60 min), and the supernatant was passed through a 0.45-μm-pore-size filter. Proteins were purified by Ni2+ affinity chromatography followed by size exclusion chromatography on a Sephacryl S-300 column (GE Healthcare) equilibrated in 20 mM Tris-HCl (pH 8.5), 150 mM NaCl, 1 mM EDTA, and 1 mM dithiothreitol (DTT).
Crystallization and data collection.
The proteins were concentrated using centrifugal filters with a molecular weight cutoff of 10 kDa to 10 mg/ml in 20 mM Tris-HCl (pH 8.5), 150 mM NaCl, 1 mM EDTA, and 1 mM DTT. Native crystals of pUL15C were obtained at 20°C by vapor diffusion in hanging drops containing equal volumes of the protein solution and a reservoir solution with 1 M ammonium citrate and 0.1 M sodium acetate at pH 4.6. The crystals were flash-cooled in 15% polyethylene glycol 400 as the cryoprotectant. The X-ray data were collected at 100 K at the Advanced Photon Source (APS) beamlines GM-CA/CAT 23ID-B and 23ID-D and at the Stanford Synchrotron Radiation Lightsource (SSRL). The data for final structure refinement were collected at SSRL beamline BL 11-1 and were processed with the HKL2000 (29). X-ray data processing statistics are summarized in Table 1.
Table 1.
Data collection and refinement statistics
| Statisticb | Valuea |
|---|---|
| Data collection | |
| Beamline | SSRL BL 11-1 |
| Wavelength (Å) | 0.97945 |
| Resolution (Å) | 50–2.46 (2.50–2.46) |
| No. of measurements | 421,384 |
| No. of unique reflections | 34,432 (1,635) |
| % completeness | 99.6 (95.2) |
| I/σ | 42.9 (2.6) |
| % Rmerge | 8.6 (48.9) |
| Space group | P43212 |
| Unit cell (Å) | a = 96.9, c = 194.0 |
| Refinement | |
| Resolution (Å) | 20–2.46 (2.53–2.46) |
| No. of reflections | 34,293 |
| % Rwork | 20.8 (26.4) |
| % Rfree | 24.5 (30.6) |
| RMS deviation | |
| Bond length (Å) | 0.01 |
| Bond angle (°) | 1.42 |
| Chiral vol (Å3) | 0.096 |
| Mean B values (Å2) | 55.7 |
| Ramachandran plot | |
| Most favorable regions (%) | 93.4 |
| Allowed region (%) | 5.8 |
| Outlier (%) | 0.8 |
Values in parentheses are for the outmost resolution shells.
Rmerge = ΣhklΣi|Ii(hkl) − <I(hkl)>|/ΣhklΣiIi(hkl), where Ii(hkl) is the observed intensity of reflection hkl and <I(hkl)> is the averaged intensity of symmetry-equivalent measurements; Rwork = Σ||Fobs| − |Fcal||/Σ|Fobs|; Rfree has the same formula as Rwork, except that calculation was made with the structure factors from the test set representing 5% of all data.
Structure determination.
The structure was determined by molecular replacement with the structure of the HCMV pUL89 C-terminal nuclease domain (47% sequence identity) as the initial search model using the program Phaser incorporated in PHENIX (30). The structure was first determined with X-ray data collected at APS and was later refined with higher-resolution data collected at SSRL. Crystal belonged to space group P43212, with three molecules in the asymmetric unit, and the cell dimensions are a = 96.9 and c = 194.0. The structure was built manually using the program COOT (31), and the refinement was performed with the program PHENIX using noncrystallographic symmetry restraints. The model was improved by alternating cycles of refinement. The final refinement cycles included TLS refinement. Refinement statistics are summarized in Table 1. In the final refined structure, residues 476 to 732 were modeled in A, B, and C molecules. Residues 512 to 519 and 544 to 547 were invisible in B and C molecules, and residues 604 to 613 where invisible in all the three chains. Residues 686 to 704 were invisible in A and B molecules, but partial density at a 0.70 sigma cutoff were found in C molecules, and alanine residues were added from 686 to 688 and 693 to 698. In the pUL15C DNA-binding model, a model for the disordered residues in loop L4 (residues 686 to 704) was built based on the C molecule using the COOT program.
Nuclease activity assay.
The purified pUL15C at various concentrations were incubated with ∼400 ng of supercoiled plasmid DNA (pET20b; 3,716 bp) in a reaction solution containing 20 mM Tris-HCl (pH 7.8), 10 mM NaCl, and 1.0 mM MgCl2 for 1 h at room temperature. The reactions were terminated by the addition of EDTA to a final concentration of 50 mM. To analyze the effects of EDTA and MgCl2 on pUL15C nuclease activity, different concentrations of pUL15C were incubated with the same plasmid DNA in the presence of 5 mM EDTA or 5 mM MgCl2 in a 10-μl reaction mixture containing 20 mM Tris-HCl (pH 7.8), 100 mM NaCl, and 1 mM DTT. The reactions were terminated as described above and analyzed by 1.0% (wt/vol) agarose gel electrophoresis followed by ethidium bromide staining.
Protein structure accession number.
The coordinates and reflection data have been deposited with RCSB Protein Data Bank with the accession code of 4IOX.
RESULTS AND DISCUSSION
Overall structure of pUL15C.
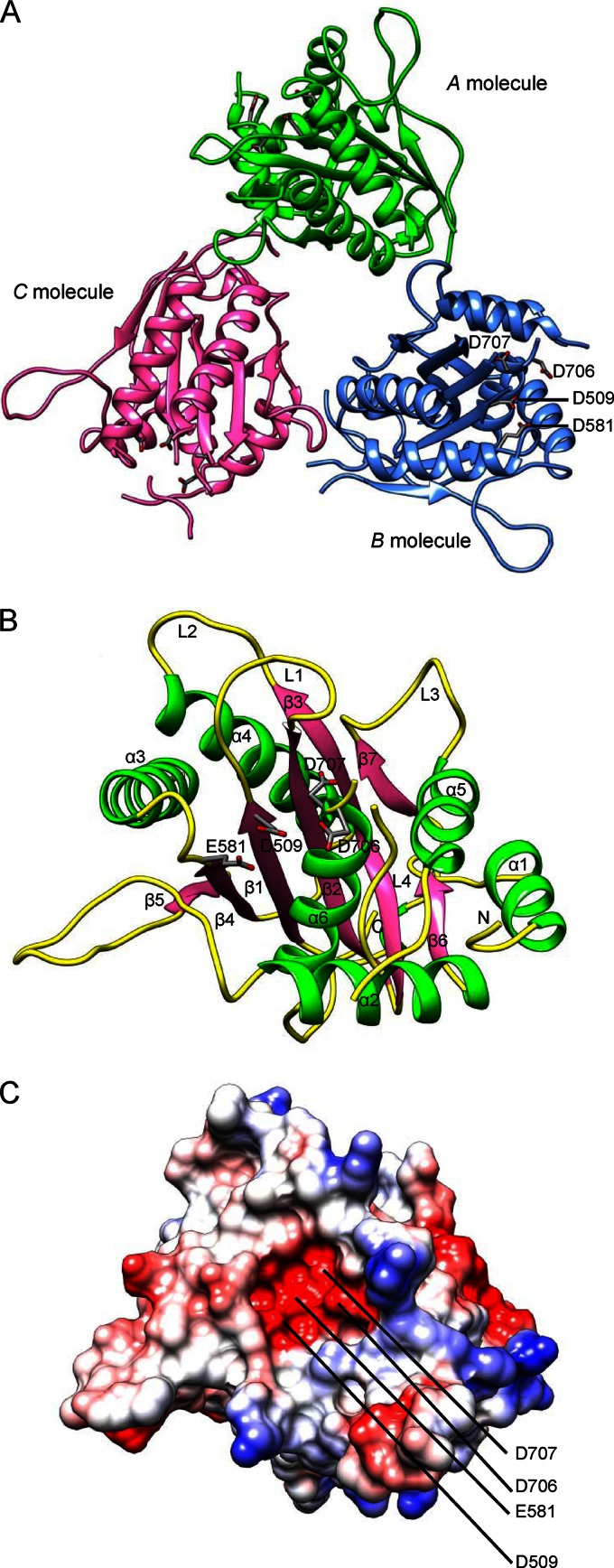
The pUL15C crystallizes with three molecules per crystallographic asymmetric unit (Fig. 1A; Table 1). The root mean square (RMS) deviations after pairwise superposition are 0.45 Å, 0.50 Å, and 0.61 Å for 211, 212, and 214 equivalent C-α, respectively, indicating that they are essentially identical. The A molecule is more complete compared to the other two molecules. The Phe512-Gly520 loop (L1) and Lys543-Gly548 loop (L2) are well defined in the electron density map for the A molecule but are missing in B and C molecules (Fig. 1B). The Lys604-Ser613 loop is invisible in the electron density map for all three molecules and thus is disordered. The loop Glu686-Ala704 (L4) is disordered in A and B molecules but displays weak main-chain electron density for C molecule and thus is modeled as poly-alanine between 686 to 688 and 693 to 698 in the C molecule. These loops are located in close proximity to the putative nuclease active site and presumably make contact with the DNA substrate, indicating that these loops are flexible and may become ordered upon DNA binding (Fig. 1B). The three molecules are arranged about a noncrystallographic, approximate 3-fold rotational symmetry axis, and the pairwise rotations between molecules are 119.50 Å, 120.02 Å, and 120.51 Å, respectively. The buried solvent accessible surface areas are 639.3, 871.9, and 974.5 A2, consistent with slight discrepancy from a proper 3-fold symmetry. Intermolecular interactions involve the N-terminal (α1), C-terminal, α2-β4, and β6-α5 regions, in which loop 3 (residues 665 to 672) plays a major role. There is a channel at the center of the crystallographic trimer that is of ∼10 Å in diameter and is unlikely for passage of DNA.
Fig 1.
The overall structure of pUL15C. (A) Ribbon representation of the pUL15C trimer viewed down the noncrystallographic 3-fold axis. A, B, and C molecules are shown in green, blue, and pink, respectively. Conserved acidic residues in the active site, D509, E581, D706, and D707, are labeled. (B) Overall structure of pUL15C in a ribbon representation. The α-helices, β-strands, and loops are in green, pink, and yellow, respectively. The loops L1, L2, L3, and L4 and the N terminus are indicated. (C) The electrostatic potential surface of pUL15C in the same view as shown in panel B. The positive potential is shown in blue, whereas the negative potential is in red.
The pUL15C exists mainly as a monomer in solution, although a very small fraction that corresponds to a potential trimer is observed (data not shown). The biological implication of the pUL15C crystallographic trimer is unclear. Terminase large subunits of phages T4 and phi29 assemble into ringlike pentamers upon binding to the procapsid (32, 33), and gpA of phage lambda forms a tetramer in complex with the terminase small-subunit gpNu1 (34). The pUL89 nuclease domain structure displays a dimer of dimers, and the intermolecular interactions involve regions different from those in pUL15C and is mediated through the central β-sheet (27). Nevertheless, it is interesting to note that the active sites in the crystallographic trimer of pUL15C all face outside (Fig. 1A), making them accessible for potential DNA binding and cleavage.
The pUL15C belongs to the RNase H-like endonucleases and polynucleotidyl transferases.
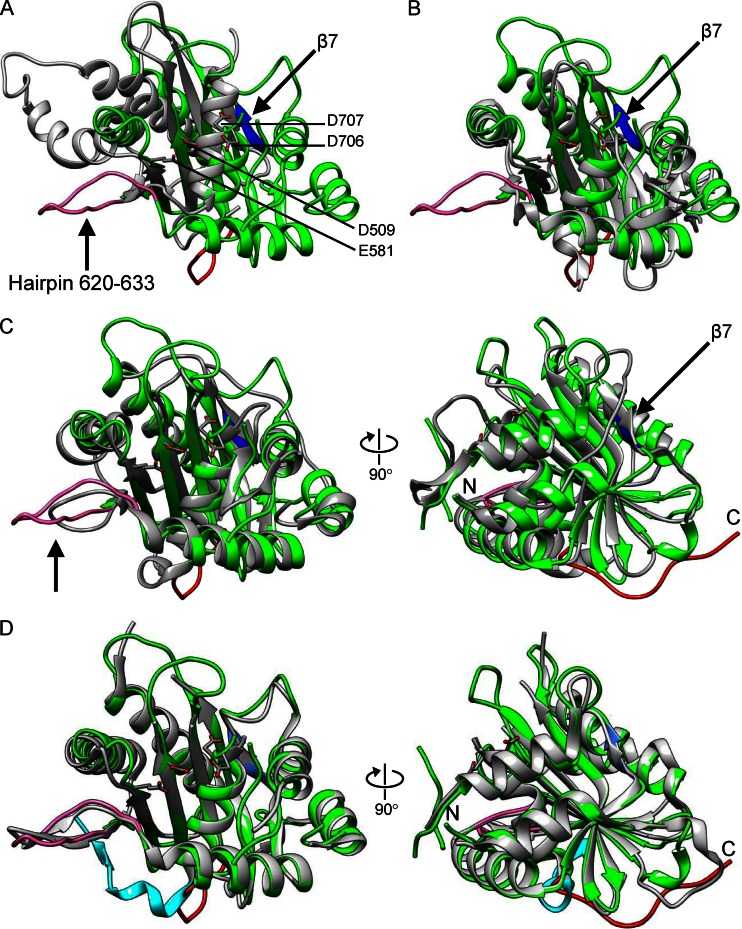
The pUL15C molecule measures about 48 by 47 by 42 Å3 and consists of a seven-stranded β-sheet, with parallel and antiparallel strands sandwiched between six α-helices (Fig. 1B). The central β-sheet (β1 to β5) curves around the helix α6, which is situated on the concave face of the central β-sheet. Negatively and positively charged areas disperse along the surface of pUL15C (Fig. 1C). The active-site groove is largely negatively charged, consistent with recruiting of metal ions for DNA binding and cleavage. The core of pUL15C, that is, the central β-sheet (β1 to β5) surrounded by α-helices, exhibits a characteristic fold similar to those of the RNase H-like superfamily of nucleases and polynucleotidyl transferases (35), despite the lack of an apparent amino acid sequence identity (Fig. 2). Structural superposition shows RMS deviations of 2.8 Å for 80 equivalent C-α with 9% identity with RNase H (36), 3.0 Å for 104 equivalent C-α with 7% identity with the Holliday junction resolvase RuvC (37), 3.0 Å for 79 equivalent C-α with 5% identity with RNase H1 (38) (Fig. 2A), and 3.0 Å for 90 equivalent C-α with 4.4% identity with the Tn5 transposase (39) (Table 2). The core folds in these proteins display a conserved 5-stranded mixed β-sheet arranged as β5-β4-β1-β2-β3, with four parallel strands and an antiparallel β2.
Fig 2.
Structural comparison of pUL15C with RNase H family nucleases and viral terminase nuclease domains. The pUL15C (green) is superposed with RNase H1 (A; PDB code 2QKB), phage SPP1 (B; PDB code 2WC9), phage T4 gp17 (C; PDB code 3CPE), and pUL89C (D; PDB code 3N4P). In all panels, the other protein is in gray. In panels C and D, the right views are 90° from the left ones. The pUL15C active-site residues are shown as stick models for side chains and are labeled in panel A. The β7 of pUL15C is shown in blue and the hairpin 620 to 633 is shown in pink, both of which are indicated with an arrow. In panel D, the C-terminal region encompassing residues 720 to 732 of pUL15C and the corresponding region of pUL89C are shown in red and cyan, respectively. The amino and carboxyl termini are indicated with N and C, respectively, in panels C and D.
Table 2.
Structural superposition of pUL15C with pUL89C, RNase H family nucleases, and bacteriophage terminases
| Protein family | PDB ID | No. of amino acid | No. of equivalent C-α atoms | RMS deviation (Å) |
|---|---|---|---|---|
| Herpesvirus | ||||
| pUL89C | 3N4P | 216 | 202 | 1.3 |
| RNase H | ||||
| RNase H | 1ZBI | 135 | 80 | 2.8 |
| RNase H1 | 2QKB | 143 | 79 | 3.0 |
| RuvC resolvase | 1HJR | 131 | 104 | 3.0 |
| Tn5 transposase | 1MM8 | 164 | 90 | 3.0 |
| Bacteriophage | ||||
| T4 gp17 | 3CPE | 201 | 157 | 2.7 |
| RB49 gp17 | 3C6H | 198 | 162 | 2.8 |
| SPP1 G2P | 2WBN | 178 | 149 | 2.9 |
| P22 gp2 | 4DKW | 194 | 145 | 2.4 |
The pUL15C structure is readily superimposable with those of phage terminase large-subunit nuclease domains despite <15% sequence identity, which shows RMS deviations of 2.8 Å for 162 equivalent C-α with 12% identity for RB49 gp17, 2.7 Å for 157 equivalent C-α with 11% identity for T4 gp17, 2.9 Å for 149 equivalent C-α with 2% identity for SPP1 G2P, and 2.4 Å for 145 equivalent C-α with 9% identity for P22 gp2 (Table 2). The pUL15C structure superimposes well onto the HCMV pUL89C, with RMS deviation of 1.3 Å for C-α atoms (Fig. 2D). The surface loops L1 and L2 are observed in pUL15C, which were invisible in HCMV pUL89C. These loops are located next to the nuclease active site, indicating that they may undergo conformational changes upon DNA binding. A major difference between the pUL15C and HCMV pUL89C structures is that the C-terminal region encompassing residues 720 to 732 of pUL15C adopts a remarkably different conformation, resulting in a 31-Å distance between C-α atoms of visible C-terminal residues of the two proteins (Fig. 2D). In pUL15C, the C-terminal region is involved in intermolecular interactions in the crystallographic trimer (Fig. 1A). The C-terminal portions of terminase large subunits of phages T4, T3, and lambda were suggested to interact with the portal proteins (40–43), although the C-terminal nuclease domain of phage T4 gp17 was positioned distal to the portal in an assembly model based on low-resolution electron cryo-microscopic studies (32). The C-terminal portions in phage terminase large subunits were disordered and invisible in X-ray structures (32, 44, 45). These observations in herpesviruses and phages suggest that the C-terminal regions of terminase large subunits are mobile and may adopt defined conformations upon interactions with other components of the viral DNA-packaging machinery, such as the portal proteins.
Structural elements in pUL15C exhibit variations to phage orthologs.
Comparative structural analysis reveals two structural elements in pUL15C that exhibit variations compared to phage orthologs and cellular RNase H1 (arrows in Fig. 2). The first structural element is an extended surface hairpin structure formed by residues 620 to 633, which follows β5 in the RNase H-like fold core. This hairpin is absent in RNase H1 and SPP1 G2P (45) (Fig. 2B) and P22 gp2 (44), but a similar hairpin structure, albeit shorter, is observed in phage T4 gp17 (Fig. 2C). This hairpin is in proximity to DNA in our DNA-binding model (see below) and thus presumably helps properly position the DNA for cleavage. The second structural element is the segment consisting of residues 661 to 665, which forms a β-strand (β7), adding an extra parallel strand to β3 of the RNase H-like fold core. Like the hairpin 620 to 633 in pUL15C, this region is absent in RNase H1, SPP1 G2P, and P22 gp2. However, a similar β-strand is present in phage T4 gp17 but adopts an opposite orientation (Fig. 2C). Interestingly, segment β7 is located between α4 and α5 in pUL15C, but the corresponding region in phage T4 gp17 is located immediately downstream to the α-helix at the N-terminal proximity of the nuclease domain (corresponding to helix α1 in pUL15C). In the T4 gp17 nuclease domain, the N-terminal α-helix is followed by that β segment, which then extends to β1 of the RNase H-like fold core. In comparison, in pUL15C, the N-terminal helix α1 is followed by a loop that extends to β1 of the RNase H-like fold core, and that β-strand is located between α4 and α5. This can be viewed as if this β7 segment jumped from between the N-terminal helix α1 and β1 in the case of T4 gp17 to a position between helices α4 and α5 in the case of pUL15C, which might result from transferring of the genetic fragments that encode those polypeptides. These results indicate that the terminase large-subunit nuclease domains of herpesviruses and myoviruses are closely related but are only remotely related to cellular RNase H family nucleases, while siphovirus and podovirus orthologs are closer relatives of RNase H family nucleases.
The nuclease active site.
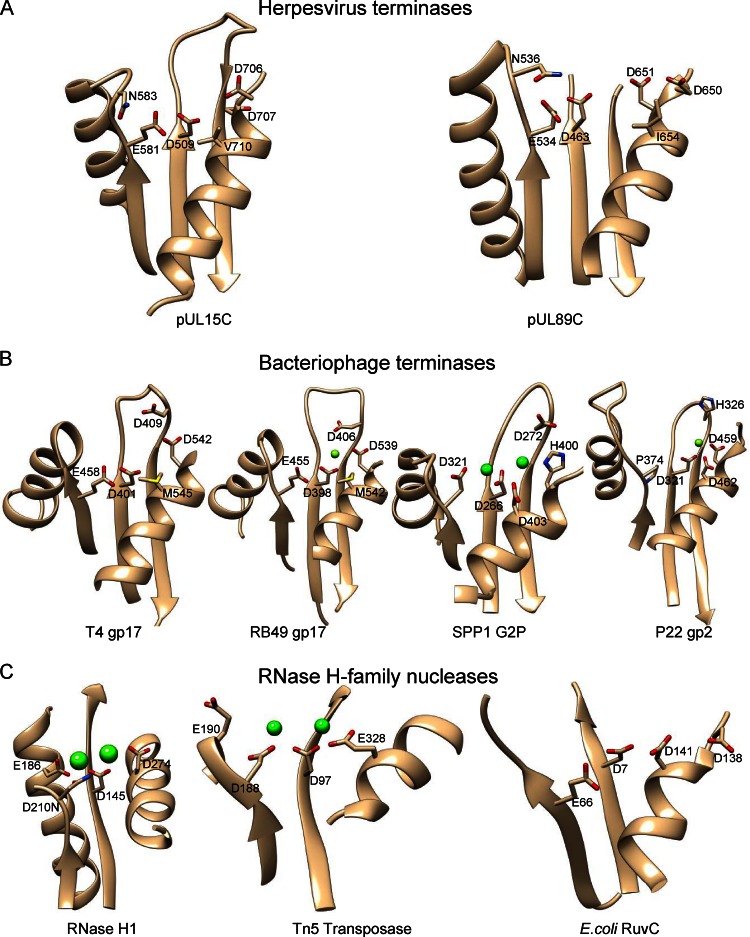
The active site is located at one side of the central five-stranded β-sheet, forming a cleft region clustered with acidic residues (Fig. 1B; Fig. 3). The local structure of the pUL15 nuclease active site resembles those in HCMV pUL89, phage terminase large subunits, and the RNase H family nucleases, while variations are clearly observed (Fig. 3). Three acidic residues, Asp509, Glu581, and Asp707, in pUL15C form the conserved triad in the active site, corresponding to residues Asp463, Glu534, and Asp651 in HCMV pUL89C, respectively. Metal ions, usually Mg2+, are required for the functioning of RNase H family nucleases, and a two-metal-ion catalysis mechanism has been proposed (35, 46, 47). Two Mn2+ ions were observed in the HCMV pUL89C structure, the first coordinated by Asp463 and Glu534 and the second by Asp463 and Asp651 (27). No Mg2+ ion density was observed in the nuclease active center of pUL15C, but water molecules were observed. These acidic residues are strictly conserved among human herpesvirus pUL15 orthologs (data not shown), suggesting that they are essential for nucleolytic catalysis. These acidic residues generate a strongly electronegative environment in the active site, consistent with the metal ion-mediated mechanism for DNA binding and cleavage.
Fig 3.
Comparison of the pUL15C active-site region with those of HCMV pUL89C (A), phage terminases (B), and RNase H family nucleases (C) showing conserved features and structural variations. Conserved residues in the active sites are labeled with side chains shown as stick models. Metal ions in RNase H family nucleases are shown as green spheres. All structures are in the same view after superimposition.
In RNase H1, a conserved acidic residue triad (D210-D145-D274) plus a fourth acidic residue (E186) located on the fold core is responsible for metal ion-mediated nucleolytic catalysis (38) (Fig. 3C). In particular, the D210-D145-D274 triad helps coordinate an Mg2+ ion, which is presumed to form the nucleophile, while D210, D145, and E186 help coordinate a second Mg2+ ion, which is thought to stabilize the transition state (35, 47). Structural comparison of pUL15C, phage terminases, and RNase H family nucleases shows that the active-site residues are readily superimposable, indicating conserved catalytic mechanisms. The central residue of the triad (D509 in pUL15C, D401 in T4 gp17, D266 in SPP1 G2P, and D145 in RNase H1) is invariably an aspartic acid, indicating a central role of this residue in catalysis and strict conservation during evolution. However, interesting structural variations are observed in other parts of the active site. In Siphovirus SPP1 G2P, a catalytic triad comprised of residues D321, D266, and D403 superimposes well with that of RNase H1, while the fourth acidic residue corresponding to E186 in RNase H1 is missing and, instead, residue H400 on the opposite side of the catalytic triad participates in coordination of a second metal ion (45). In phage T4 gp17, further variations are observed: (i) the residue corresponding to the first one of the RNase H1 catalytic triad is a glutamic acid (E458) instead of an aspartic acid; (ii) the residue corresponding to the third aspartic acid of the RNase H1 catalytic triad is a methionine (M545) that is conserved in a related phage RB49; and (iii) an additional acidic residue D542 was proved to be essential for catalysis, may participate in coordination of a metal ion (32), and occupies a position similar to that of residue H400 in SPP1 G2P. The local architecture of the pUL15C active site closely resembles that of T4 gp17, in which a valine (V710) occupies the same position as residue M545 in T4 gp17. Position 710 is invariably a hydrophobic residue in human herpesviruses (data not shown). Based on these analyses, it becomes clear that there is a trajectory of accumulative structural variations in the nuclease active sites, which changes from (E186)-D210-D145-D274 in RNase H1 to D321-D266-D403-(H400) in SPP1 G2P, then to E458-D401-(M545)-D542 in T4 gp17, and finally to E581-D509-(V710)-D707 in HSV-1 pUL15. HSV-1 pUL15 and T4 gp17 are closely related to each other but are remotely related to SPP1 G2P and RNase H1.
The pUL15c shows metal ion-dependent, non-sequence-specific nuclease activity.
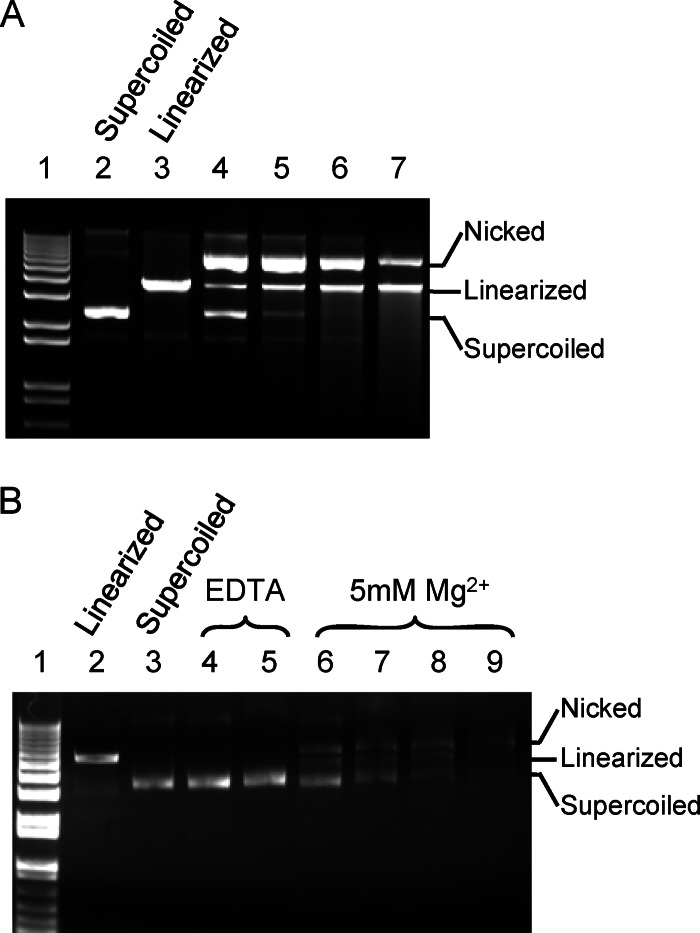
In vitro assay shows that pUL15C possesses non-sequence-specific nuclease activity (Fig. 4A). Both nicked and linearized dsDNA are observed as reaction products. At lower protein concentrations, there is an accumulation of nicked DNA, while the fraction of linearized DNA increases when the protein concentration is higher (Fig. 4A). At higher protein concentrations, smeared DNA appears, representing cleavage of DNA into variable, smaller-size fragments. Similar behavior was observed for HCMV pUL89C (27). These data are implicative of a stepwise course of the nucleolytic catalysis, that is, pUL15C may bind to DNA and make a cut on one strand of the dsDNA before cutting the other strand, thus distinguishing pUL15C from dimeric nucleases, such as restriction endonucleases that cooperatively cut the two strands of dsDNA. In vivo cleavage of HSV-1 genomic DNA by pUL15 occurs within the DR1 motif, generating a 3′ single-base overhang (48). Cleavage of the two DNA strands likely requires two copies of the pUL15 molecules. As pUL15C exists as a monomer in solution, molecular determinants for oligomerization of pUL15C may be located elsewhere in the HSV-1 DNA-packaging machinery or in the N-terminal domain of pUL15. EDTA eliminates the nuclease activity of pUL15C, whereas Mg2+ shows stimulatory effect (Fig. 4B). These results indicate that the pUL15C nuclease activity is dependent on metal ions, consistent with structural homology to RNase H family nucleases.
Fig 4.
In vitro nuclease activity assay. (A) Non-sequence-specific nuclease activity of pUL15C. Lane 1, 1 kb plus DNA ladder (Invitrogen); lane 2, supercoiled plasmid DNA; lane 3, plasmid DNA linearized with restriction enzyme; lanes 4 to 7, pUL15C at a concentration of 17.5 μM, 35 μM, 70 μM, and 140 μM, respectively, was incubated with supercoiled plasmid DNA at room temperature for 1 h. Positions for the nicked, linearized, and supercoiled DNA are indicated. (B) Effects of EDTA or MgCl2 on pUL15C nuclease activity. Lane 1, 1 kb plus DNA ladder (Invitrogen); lane 2, plasmid DNA linearized with restriction enzyme; lane 3, supercoiled plasmid DNA; lanes 4 and 5, pUL15C at 35 μM and 70 μM, respectively, was incubated with supercoiled plasmid DNA in the presence of 5 mM EDTA; lanes 6 to 9, pUL15C at 17.5 μM, 35 μM, 70 μM, and 140 μM, respectively, was incubated with supercoiled plasmid DNA in the presence of 5 mM MgCl2.
The DNA binding mode of pUL15.
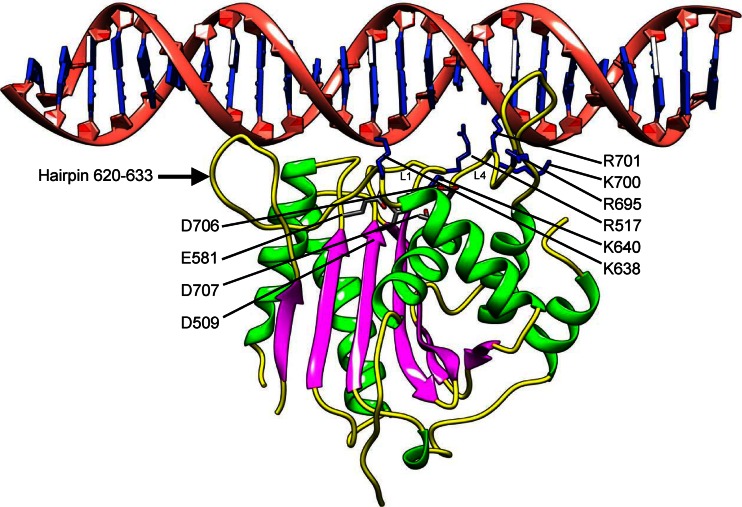
To understand how DNA binds to pUL15C for cleavage, we modeled dsDNA on the pUL15C nuclease active site by superimposing the pUL15C structure onto that of the RNase H1 in complex with a DNA-RNA hybrid, which was then replaced with dsDNA (38) (Fig. 5). In this model, the dsDNA spans the nuclease active site, approaching the catalytic active residues. Loops L1 and L4 make contact with the DNA, in which positively charged residues R517, R695, K700, and R701 may interact with the DNA phosphate backbones. Additional positively charged residues K638 and K640 may also contribute to DNA binding. The hairpin (residues 620 to 633) also appears to interact with DNA, presumably helping hold the DNA in an appropriate position and orientation with respect to the nuclease active site. The loop L4 is located at a position similar to that of a β-hairpin structure in the SPP1 G2P nuclease domain (45) and in T4 gp17 (32). In the SPP1 G2P structure, it was proposed that the conformational flexibility of this β-hairpin might mediate DNA binding and thus the nuclease activity. In a T4-related phage, RB49 gp17, this β-hairpin is disordered (32). While it is ordered in the T4 gp17 structure (32), this β-hairpin also showed flexibility and was proposed to interact with DNA in a later molecular dynamics study of T4 gp17 (49). In the pUL15C structure, the loop L4 is disordered in the A and B molecules but is partially ordered in the C molecule, indicative of conformational flexibility. In our DNA binding model, the loop L4 is adjacent to DNA and the positively charged residues R695, K700, and R701 are in close proximity from DNA to make direct interactions. Additionally, the loop L1 is well defined in the electron density map for the A molecule but is disordered in B and C molecules, suggesting flexibility of this loop and induced fit upon DNA binding.
Fig 5.
A DNA-binding model for pUL15C. A model of 25-bp dsDNA docked onto the pUL15C structure. The docking was performed by superimposing the pUL15C structure onto that (PDB code 2QKB) of the RNase H1 in complex with a DNA-RNA hybrid, which was replaced with dsDNA. The loops L1 and L4 and the hairpin 620 to 633 are indicated. The active-site residues are shown as stick models for side chains and are labeled with D509, E581, D706, and D707. The positively charged residues that potentially interact with DNA are shown as blue stick models for side chains and are labeled.
An evolutionary lineage among the herpesviruses and bacteriophages.
Our comparative structural analysis reveals two structural elements in pUL15C that are in addition to the RNase H-like fold core, that is, the hairpin formed by residues 620 to 633 and β7 (Fig. 2). Both structural elements show close similarity between pUL15 and phage T4 gp17 but are absent in terminase large subunits of simple phages such as SPP1 and P22 and RNase H family nucleases. Likewise, a trajectory of accumulative structural variations in the nuclease active sites is observed from the (E186)-D210-D145-D274 motif in RNase H1 to D321-D266-D403-(H400) in SPP1 G2P to E458-D401-(M545)-D542 in T4 gp17 to E581-D509-(V710)-D707 in HSV-1 pUL15. These structural variations of the protein folds and the local architectures of the nuclease active sites consistently indicate a relationship among these proteins, that is, the terminase large-subunit nuclease domains of herpesviruses and myoviruses are closely related to each other but are only remotely related to cellular RNase H family nucleases, while siphovirus and podovirus orthologs are closer relatives of RNase H family nucleases. Thus, an evolutionary lineage can be delineated based on the structures of the viral DNA-packaging nuclease modules, which might originate from cellular RNase H family nucleases by, for example, gene acquisition, and first evolve to simple phages such as podoviruses and siphoviruses and then to complex phages such as myoviruses and to herpesviruses. Evolutionary relationships between herpesviruses and tailed dsDNA bacteriophages have been proposed based on similarity of the virus assembly pathways, ringlike dodecameric portal proteins, and folds of major capsid proteins (50, 51). Genome packaging is among the most fundamental processes in virus life cycles and may represent an evolutionarily ancient event. Thus, the relationship observed in our comparative structural analysis of pUL15 and phage terminases and the lineage revealed thereby provide new, strong evidence for an evolutionary path from simple phages such as podoviruses and siphoviruses to complex phages such as myoviruses and then to eukaryotic herpesviruses. It is worth pointing out that amino acid sequence identity of proteins between herpesviruses and tailed dsDNA phages is usually too low to detect apparent homologies, and comparative analysis based on three-dimensional structures can provide exceptionally valuable information for understanding evolutionary relationships among viruses.
ACKNOWLEDGMENTS
We thank the staff at Stanford Synchrotron Radiation Lightsource and the Advanced Photon Source beamlines GM-CA/CAT 23ID-B and 23ID-D for assistance in X-ray data collection.
This work was supported by the NIH grant R01GM090010 to L.T.
Footnotes
Published ahead of print 17 April 2013
REFERENCES
- 1. Roizman B, Knipe DM, Whitley RJ. 2007. Herpes simplex virus, p 2501–2601 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5 ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 2. Roizman B, Furlong D. 1974. Comprehensive virology, p 229–403 In Fraenkel-Conrat H, Wagner R. (ed). Plenum, New York, NY [Google Scholar]
- 3. Steven AC, Spear PG. 1997. Structural biology of viruses, p 312–351 In Chiu W, Burnett RM, Garcea RL. (ed). Oxford University Press, New York, NY [Google Scholar]
- 4. Deiss LP, Chou J, Frenkel N. 1986. Functional domains within the a sequence involved in the cleavage-packaging of herpes simplex virus DNA. J. Virol. 59:605–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nasseri M, Mocarski ES. 1988. The cleavage recognition signal is contained within sequences surrounding an a-a junction in herpes simplex virus DNA. Virology 167:25–30 [DOI] [PubMed] [Google Scholar]
- 6. Varmuza SL, Smiley JR. 1985. Signals for site-specific cleavage of HSV DNA: maturation involves two separate cleavage events at sites distal to the recognition sequences. Cell 41:793–802 [DOI] [PubMed] [Google Scholar]
- 7. Conway JF, Homa FL. 2011. Nucleocapsid structure, assembly and DNA-packaging of herpes simplex virus, p 175–193 In Weller SK. (ed), Alphaherpesviruses: molecular virology. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 8. Homa FL, Brown JC. 1997. Capsid assembly and DNA-packaging in herpes simplex virus. Rev. Med. Virol. 7:107–122 [DOI] [PubMed] [Google Scholar]
- 9. Stow ND. 2001. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J. Virol. 75:10755–10765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cardone G, Winkler DC, Trus BL, Cheng N, Heuser JE, Newcomb WW, Brown JC, Steven AC. 2007. Visualization of the herpes simplex virus portal in situ by cryo-electron tomography. Virology 361:426–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Newcomb WW, Juhas RM, Thomsen DR, Homa FL, Burch AD, Weller SK, Brown JC. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923–10932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Trus BL, Cheng N, Newcomb WW, Homa FL, Brown JC, Steven AC. 2004. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J. Virol. 78:12668–12671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Newcomb WW, Homa FL, Thomsen DR, Booy FP, Trus BL, Steven AC, Spencer JV, Brown JC. 1996. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J. Mol. Biol. 263:432–446 [DOI] [PubMed] [Google Scholar]
- 14. Trus BL, Booy FP, Newcomb WW, Brown JC, Homa FL, Thomsen DR, Steven AC. 1996. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J. Mol. Biol. 263:447–462 [DOI] [PubMed] [Google Scholar]
- 15. Gibson W, Roizman B. 1972. Proteins specified by herpes simplex virus. 8. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J. Virol. 10:1044–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Salmon B, Baines JD. 1998. Herpes simplex virus DNA cleavage and packaging: association of multiple forms of U(L)15-encoded proteins with B capsids requires at least the U(L)6, U(L)17, and U(L)28 genes. J. Virol. 72:3045–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lamberti C, Weller SK. 1998. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J. Virol. 72:2463–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Black LW. 1989. DNA-packaging in dsDNA bacteriophages. Annu. Rev. Microbiol. 43:267–292 [DOI] [PubMed] [Google Scholar]
- 19. Beard PM, Taus NS, Baines JD. 2002. DNA cleavage and packaging proteins encoded by genes U(L)28, U(L)15, and U(L)33 of herpes simplex virus type 1 form a complex in infected cells. J. Virol. 76:4785–4791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang K, Baines JD. 2006. The putative terminase subunit of herpes simplex virus 1 encoded by UL28 is necessary and sufficient to mediate interaction between pUL15 and pUL33. J. Virol. 80:5733–5739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang K, Homa F, Baines JD. 2007. Putative terminase subunits of herpes simplex virus 1 form a complex in the cytoplasm and interact with portal protein in the nucleus. J. Virol. 81:6419–6433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Adelman K, Salmon B, Baines JD. 2001. Herpes simplex virus DNA-packaging sequences adopt novel structures that are specifically recognized by a component of the cleavage and packaging machinery. Proc. Natl. Acad. Sci. U. S. A. 98:3086–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Costa RH, Draper KG, Kelly TJ, Wagner EK. 1985. An unusual spliced herpes simplex virus type 1 transcript with sequence homology to Epstein-Barr virus DNA. J. Virol. 54:317–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dolan A, Arbuckle M, McGeoch DJ. 1991. Sequence analysis of the splice junction in the transcript of herpes simplex virus type 1 gene UL15. Virus Res. 20:97–104 [DOI] [PubMed] [Google Scholar]
- 25. Davison AJ. 1992. Channel catfish virus: a new type of herpesvirus. Virology 186:9–14 [DOI] [PubMed] [Google Scholar]
- 26. Yu D, Weller SK. 1998. Genetic analysis of the UL 15 gene locus for the putative terminase of herpes simplex virus type 1. Virology 243:32–44 [DOI] [PubMed] [Google Scholar]
- 27. Nadal M, Mas PJ, Blanco AG, Arnan C, Sola M, Hart DJ, Coll M. 2010. Structure and inhibition of herpesvirus DNA-packaging terminase nuclease domain. Proc. Natl. Acad. Sci. U. S. A. 107:16078–16083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Scheffczik H, Savva CG, Holzenburg A, Kolesnikova L, Bogner E. 2002. The terminase subunits pUL56 and pUL89 of human cytomegalovirus are DNA-metabolizing proteins with toroidal structure. Nucleic Acids Res. 30:1695–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Otwinowski ZMW. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 30. Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1948–1954 [DOI] [PubMed] [Google Scholar]
- 31. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 32. Sun S, Kondabagil K, Draper B, Alam TI, Bowman VD, Zhang Z, Hegde S, Fokine A, Rossmann MG, Rao VB. 2008. The structure of the phage T4 DNA-packaging motor suggests a mechanism dependent on electrostatic forces. Cell 135:1251–1262 [DOI] [PubMed] [Google Scholar]
- 33. Morais MC, Koti JS, Bowman VD, Reyes-Aldrete E, Anderson DL, Rossmann MG. 2008. Defining molecular and domain boundaries in the bacteriophage phi29 DNA-packaging motor. Structure 16:1267–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maluf NK, Gaussier H, Bogner E, Feiss M, Catalano CE. 2006. Assembly of bacteriophage lambda terminase into a viral DNA maturation and packaging machine. Biochemistry 45:15259–15268 [DOI] [PubMed] [Google Scholar]
- 35. Yang W. 2011. Nucleases: diversity of structure, function and mechanism. Q. Rev. Biophys. 44:1–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. 2005. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell 121:1005–1016 [DOI] [PubMed] [Google Scholar]
- 37. Ariyoshi M, Vassylyev DG, Iwasaki H, Nakamura H, Shinagawa H, Morikawa K. 1994. Atomic structure of the RuvC resolvase: a holliday junction-specific endonuclease from E. coli. Cell 78:1063–1072 [DOI] [PubMed] [Google Scholar]
- 38. Nowotny M, Gaidamakov SA, Ghirlando R, Cerritelli SM, Crouch RJ, Yang W. 2007. Structure of human RNase H1 complexed with an RNA/DNA hybrid: insight into HIV reverse transcription. Mol. Cell 28:264–276 [DOI] [PubMed] [Google Scholar]
- 39. Steiniger-White M, Bhasin A, Lovell S, Rayment I, Reznikoff WS. 2002. Evidence for “unseen” transposase-DNA contacts. J. Mol. Biol. 322:971–982 [DOI] [PubMed] [Google Scholar]
- 40. Lin H, Rao VB, Black LW. 1999. Analysis of capsid portal protein and terminase functional domains: interaction sites required for DNA-packaging in bacteriophage T4. J. Mol. Biol. 289:249–260 [DOI] [PubMed] [Google Scholar]
- 41. Yeo A, Feiss M. 1995. Specific interaction of terminase, the DNA-packaging enzyme of bacteriophage lambda, with the portal protein of the prohead. J. Mol. Biol. 245:141–150 [DOI] [PubMed] [Google Scholar]
- 42. Morita M, Tasaka M, Fujisawa H. 1995. Structural and functional domains of the large subunit of the bacteriophage T3 DNA-packaging enzyme: importance of the C-terminal region in prohead binding. J. Mol. Biol. 245:635–644 [DOI] [PubMed] [Google Scholar]
- 43. Kanamaru S, Kondabagil K, Rossmann MG, Rao VB. 2004. The functional domains of bacteriophage t4 terminase. J. Biol. Chem. 279:40795–40801 [DOI] [PubMed] [Google Scholar]
- 44. Roy A, Cingolani G. 2012. Structure of p22 headful packaging nuclease. J. Biol. Chem. 287:28196–28205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Smits C, Chechik M, Kovalevskiy OV, Shevtsov MB, Foster AW, Alonso JC, Antson AA. 2009. Structural basis for the nuclease activity of a bacteriophage large terminase. EMBO Rep. 10:592–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nowotny M, Yang W. 2006. Stepwise analyses of metal ions in RNase H catalysis from substrate destabilization to product release. EMBO J. 25:1924–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang W, Lee JY, Nowotny M. 2006. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell 22:5–13 [DOI] [PubMed] [Google Scholar]
- 48. Mocarski ES, Roizman B. 1982. Structure and role of the herpes simplex virus DNA termini in inversion, circularization and generation of virion DNA. Cell 31:89–97 [DOI] [PubMed] [Google Scholar]
- 49. Ghosh-Kumar M, Alam TI, Draper B, Stack JD, Rao VB. 2011. Regulation by interdomain communication of a headful packaging nuclease from bacteriophage T4. Nucleic Acids Res. 39:2742–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. McGeoch DJ, Rixon FJ, Davison AJ. 2006. Topics in herpesvirus genomics and evolution. Virus Res. 117:90–104 [DOI] [PubMed] [Google Scholar]
- 51. Krupovic M, Bamford DH. 2011. Double-stranded DNA viruses: 20 families and only five different architectural principles for virion assembly. Curr. Opin. Virol. 1:118–124 [DOI] [PubMed] [Google Scholar]