Abstract
Herpes simplex virus 1 (HSV-1), a ubiquitous human pathogen, expresses several viral microRNAs (miRNAs). These, along with the latency-associated transcript, represent the only viral RNAs detectable in latently infected neuronal cells. Here, for the first time, we analyze which HSV-1 miRNAs are loaded into the RNA-induced silencing complex (RISC), the key effector of miRNA function. Only 9 of the 17 reported HSV-1 miRNAs, i.e., miR-H1 to miR-H8 plus miR-H11, were found to actually load into the RISC. Surprisingly, this analysis also revealed that HSV-1 miRNAs loaded into the RISC with efficiencies that differed widely; <1% of the miR-H1-3p miRNA detectable in HSV-1-infected cells was loaded into the RISC. Analysis of HSV-1 mutants individually lacking the viral miR-H2, miR-H3, or miR-H4 miRNA revealed that loss of these miRNAs affected the rate of replication of HSV-1 in neuronal cells but not in fibroblasts. Analysis of mRNA and protein expression, as well as assays mapping viral miRNA binding sites in infected cells, showed that endogenous HSV-1 miR-H2 binds to viral ICP0 mRNA and inhibits its expression, while endogenous miR-H4 inhibits the expression of the viral ICP34.5 gene. In contrast, no viral mRNA target for miR-H3 could be detected, even though miR-H3, like miR-H4, is perfectly complementary to ICP34.5 mRNA. Together, these data demonstrate that endogenous HSV-1 miRNA expression can significantly alter viral replication in culture, and they also identify two viral mRNA targets for miR-H2 and miR-H4 that can partially explain this phenotype.
INTRODUCTION
Herpes simplex virus 1 (HSV-1) is the most common cause of oral herpes and the leading cause of infectious blindness in the United States. HSV-1 is typically acquired by infection of the oral mucosa, where it replicates locally, and it then enters the nervous system via sensory nerve termini. HSV-1 establishes a lifelong latent infection in sensory neurons and reactivates periodically in response to various stressors. The molecular processes that regulate the neurotropism of HSV-1 latency and the processes by which the lytic genes are silenced and subsequently reactivated are poorly understood (for a review, see reference 1). Only one transcript, the latency-associated transcript (LAT), is abundantly expressed during latency (2, 3). Recently this transcript has been shown to be the precursor of a number of HSV-1-encoded microRNAs (miRNAs), and these have been suggested to play a role in the regulation of various aspects of viral latency and/or reactivation (4, 5).
miRNAs are a family of short regulatory RNAs, ∼22 nucleotides (nt) long, that bind to complementary target sites on mRNAs and then inhibit their expression (6). miRNAs are derived from one arm of an imperfect stem-loop structure, with a characteristic stem length of ∼33 bp, that is located within a longer, largely unstructured transcript referred to as a primary miRNA (pri-miRNA) precursor (7–9). Recognition of this stem-loop structure by the nuclear microprocessor, consisting of the RNase III enzyme Drosha and the double-stranded RNA (dsRNA) binding protein DGCR8, results in cleavage of the stem ∼22 bp away from the terminal loop and generates an ∼2-nt 3′-terminal overhang (10–12). The resultant ∼60-nt pre-miRNA intermediate is then exported to the cytoplasm by Exportin 5 (13) and is there bound by a second processing complex, consisting of the RNase III enzyme Dicer and its cofactor TRBP. Dicer cleaves the pre-miRNA proximally to the stem-loop junction, leaving a second 2-nt 3′ overhang, to generate the miRNA duplex intermediate (14, 15). One strand of this duplex is then incorporated into the RNA-induced silencing complex (RISC), where it functions as a guide RNA to direct the RISC to fully or partially complementary mRNA targets (6, 16). The second strand of the duplex is degraded. The choice of which strand of the miRNA duplex intermediate is incorporated into the RISC, i.e., which strand functions as a miRNA, is thought to be regulated by the stability of the base pairing at the termini of the duplex: the strand with a 5′ end that is less tightly base paired is favored for incorporation into the RISC (17, 18). However, this discrimination is rarely complete, so that the less favored strand, referred to as the “star” or passenger strand, is often detectable at significant levels in the RISC. The two strands of the duplex are referred to as the “5p” and “3p” strands, to reflect their origins within the original pri-miRNA stem-loop.
While human cells are now known to encode >1,000 miRNAs, cellular miRNAs are not the only miRNAs that have been detected in human cells. Specifically, it is now well established that several human DNA viruses, including members of the herpesvirus, polyomavirus, and adenovirus families, express one or more miRNAs in infected cells (19). Herpesviruses, a number of which share the ability to establish long-term, latent infections in humans, are particularly adept at using miRNAs to control gene expression in infected cells and have been shown to express numerous distinct pre-miRNAs in latently and/or productively infected cells in culture and in vivo (19).
The first attempt to identify miRNAs expressed by HSV-1 used a combination of bioinformatic approaches and Northern blotting to identify a single HSV-1-encoded miRNA, miR-H1, that is expressed in productively infected cells (20). Subsequent analysis of small RNAs derived from mouse and human trigeminal ganglia latently infected with HSV-1, using a deep-sequencing approach, identified seven additional HSV-1 miRNAs, called miR-H2 to miR-H8 (4, 5). Of these eight HSV-1 miRNAs, miR-H1 is expressed largely or exclusively during productive infection, while miR-H6 is expressed at significant levels in both productively and latently infected cells. The remaining six HSV-1 miRNAs, all of which derive from the unstable, exonic regions of the HSV-1 LAT, are expressed at high levels in latently infected cells but only at modest levels during productive infection in culture, as is also seen for the LAT itself (4, 5, 21). In contrast, both miR-H1 and miR-H6 derive from currently undefined pri-miRNAs that are transcribed proximally to, but separately from, the LAT. Since the LAT has been implicated in reactivation (22, 23), the regulation of apoptosis (24, 25), and the control of latent transcription (26, 27), the identification of these miRNAs provided a potential regulatory mechanism for the multiple profound and varied phenotypes encoded within this region.
A more recent analysis of HSV-1 miRNA expression during productive infection in culture, again using deep sequencing, reported the existence of another eight HSV-1 miRNAs, miR-H11 to miR-H18, all of which were found to be expressed exclusively or predominantly in productively, rather than latently, infected cells (28). In agreement with this finding, these newer miRNAs are scattered over the entire genome of HSV-1, rather than being tightly clustered in or near the LAT gene, as reported for miR-H2 to miR-H8. Finally, another group recently reported another HSV-1 miRNA, miR-92944, which is, again, encoded distally to the LAT and is expressed during productive viral replication in culture (29).
In this report, we have sought to define carefully which HSV-1 miRNAs are functional by deep sequencing of small RNAs associated with the RISC in infected cells in culture, obtained by immunoprecipitation of the RISC using an antibody specific for mammalian Argonaute (Ago) proteins, including human Ago1, Ago2, and Ago3. Each of these Ago proteins is capable of being incorporated into the RISC and mediating RISC function (6, 30). Surprisingly, our results indicate that the profile of HSV-1-encoded miRNAs present in the RISC is substantially different from the expression pattern observed upon analysis of total small RNAs in HSV-1-infected cells. Moreover, these data strongly suggest that miR-H12 to miR-H18, as well as miR-92944, are not functional HSV-1 miRNAs.
To begin to define the functions of the RISC-associated HSV-1 miRNAs, we have generated mutant viruses individually lacking three of the most highly expressed HSV-1 miRNAs, i.e., miR-H2, miR-H3, and miR-H4. We demonstrate that endogenous miR-H2 is able to downregulate the expression of the HSV-1 regulatory protein ICP0, while miR-H4 downregulates the expression of the viral pathogenicity factor ICP34.5. Surprisingly, miR-H3, which, like miR-H4, derives from LAT sequences located antisense to the ICP34.5 mRNA, has no effect on ICP34.5 expression. In agreement with these data, miR-H2 and miR-H4 are shown to bind ICP0 and ICP34.5 mRNAs, respectively, in infected cells, while miR-H3 fails to bind to the latter, despite perfect complementarity. Analysis of the growth potential of these HSV-1 miRNA mutants shows, surprisingly, that both the miR-H3 and miR-H4 mutants replicate more rapidly than the wild type (WT) in neuronal cells in culture, while the miR-H2 mutant shows a modest reduction in replication potential.
In summary, the data presented here more fully define the miRNA expression profile of HSV-1 and provide the first demonstration of a phenotypic effect of these miRNAs in culture. These findings have begun to reveal an important role for viral miRNAs in HSV-1 replication and pathogenesis and should prompt additional mutational analyses targeting the other HSV-1 LAT-associated miRNAs shown here to be RISC associated.
MATERIALS AND METHODS
Cell culture and virus infection.
Rabbit skin (RS) cells, employed to prepare and titrate virus stocks and for transfection experiments, were propagated in minimum essential medium Eagle (MEME) supplemented with 5% calf serum and PSG (250 U of penicillin, 250 mg/ml of streptomycin, 2.5 mg/ml of amphotericin B, and 292 mg/ml of l-glutamine; Life Technologies, Inc.) (31). NIH 3T3 cells were propagated in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and PSG. Neuro2A cells were maintained in MEME with 10% FBS and 1% nonessential amino acids (NEAA). 293 cells were maintained in DMEM with 10% FBS. SY5Y cells were maintained in a 1:1 mixture of MEME and Ham's F-12 medium with 10% FBS and 1% NEAA. Cells were infected with HSV-1 strain 17syn+ (32) at the indicated multiplicity of infection (MOI) by incubation in a minimal volume of virus for 1 h at 37°C with occasional rocking. The virus was then removed, and cells were washed with phosphate-buffered saline (PBS) before the growth medium was replenished. Samples were harvested at the time points indicated in Tables 1 to 4 and Fig. 3 for RNA or protein analysis.
Table 1.
Primers used for the construction of recombinant viruses
| PCR | Forward/reversea | Sequence (5′–3′) |
|---|---|---|
| 17mH2 | ||
| In-Fusion | F | TCCCCCGGGCTGCAGGAATTCGCCTCCTGCTCGACAGAG |
| R | GATAAGCTTGATATCGAATTCCGCCCATGCCAGGCTC | |
| Mutagenesis | F | ATAGTAGTTTAGCGCAAGCCGCGAACCAAGAACAGAG |
| R | GCGCTAAACTACTATGGCTCTGGGGCACACGGCGCGCGTCCG | |
| 17mH3 | ||
| In-Fusion | F | GACTTTGAGCCGTGGGCACGGCGCCCGTGGGCCCGGGCGG |
| R | GATAAGCTTGATATCGAATTCCCCCCCGCGGCCGAGACTAG | |
| Mutagenesis | F | GGGCGGCGGGGGCCGCGATGGCGGC |
| R | GCCGCCATCGCGGCCCCCGCCGCCC | |
| 17dH4 | ||
| In-Fusion Left | F | TCCCCCGGGCTGCAGGAATTCCTGACCGCGGGTTCCGAGTTG |
| R | GGCCGAGCACTACTCGCCTCTGCACGCAC | |
| In-Fusion Right | F | GAGTAGTGCTCGGCCGCGGGGGGCCC |
| R | GATAAGCTTGATATCGAATTCGGCGGTCCGGGCGGCGTG | |
F, forward; R, reverse.
Table 4.
Overview of previously reported HSV-1 microRNAs identified by deep sequencing of RISC-associated small RNAs in HSV-1-infected Neuro2A, SY5Y, or 293 cells
| HSV-1 miRNAa | Total no. of reads in the following infected-cell type: |
||
|---|---|---|---|
| Neuro2A | SY5Y | 293 | |
| H1-5p | 115 | 3 | 34 |
| H1-3p | 230 | 72 | 83 |
| H2-5p | 14 | 3 | 7 |
| H2-3p | 485 | 50 | 76 |
| H3-5p | 18 | 6 | 95 |
| H3-3p | 1,072 | 18 | 544 |
| H4-5p | 64 | 6 | 39 |
| H4-3p | 180 | 39 | 1,240 |
| H5-5p | 13 | 2 | 10 |
| H5-3p | 1 | 0 | 1 |
| H6-5p | 9 | 1 | 12 |
| H6-3p | 1,245 | 291 | 976 |
| H7-5p | 3 | 0 | 2 |
| H7-3p | 6 | 2 | 7 |
| H8-5p | 3 | 1 | 3 |
| H8-3p | 0 | 0 | 0 |
| H11-3p | 100 | 3 | 118 |
| H13-5p | 4 | 0 | 3 |
The suffix “5p” or “3p” describes the arm of the miRNA precursor stem-loop from which the miRNA derives.
Fig 3.

Effect of miR-H2, miR-H3, or miR-H4 on the expression of HSV-1 ICP0 and ICP34.5. (A) 293 cells were infected with WT HSV-1 17syn+ in the presence or absence of miRNA mimics encoding miR-H2, miR-H3, or miR-H4, or with 17syn+ mutants lacking miR-H2 (mH2), miR-H3 (mH3), or miR-H4 (dH4). At 8 h postinfection, total RNA was harvested and was analyzed for viral ICP0 or ICP34.5 mRNA expression by qRT-PCR. The data obtained were normalized to those for actin mRNA and are averages for three experiments, with standard deviations indicated. (B) 293 cells were infected with HSV-1 and were harvested as described for panel A. Then the cells were subjected to Western blot analysis using antibodies specific for ICP0, ICP34.5, or cellular actin (as a loading control). The band intensities (given below each lane) were quantitated using a Syngene G:Box gel imaging system and were normalized to the actin signal. Uninfected 293 cells served as the negative control (CTRL).
Construction of viral recombinants.
HSV-1 recombinants with deletions/mutations of the viral miRNAs were constructed by cotransfecting a plasmid containing the mutated region/deletion and HSV-1 strain 17syn+ virion DNA into RS cells by calcium phosphate precipitation at a molecular ratio of 10:1, as described previously (33). Briefly, all recombination plasmids contained mutated or deleted target sites and ∼150 bp on either side of the sites to effect homologous recombination of the gene onto the viral genome. The sequences were constructed and subcloned into a pBluescript plasmid by using an In-Fusion Advantage PCR cloning kit (Clontech) according to the manufacturer's protocol. After subcloning, mutations of miR-H3 or miR-H2 were introduced using a QuikChange II site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. The primers used are listed in Table 1.
At 3 to 4 days posttransfection, the RS cells were harvested, and the cell lysate was frozen, thawed, and diluted to produce well-isolated plaques. RS monolayers were then infected with this dilution under agarose. Individual HSV-1 plaques were amplified on RS cells in a 96-well format and were then screened for recombinants by dot blot hybridization using oligonucleotide probes, as described previously (33). Five rounds of plaque purification were performed on three independent isolates (from separate transfections) for each HSV-1 miRNA mutant. The purity of the stocks was determined by PCR analysis, followed by sequencing of the PCR products. The integrity of each of the recombinants was further confirmed by Southern blotting.
DNA replication assay.
Quantities of viral DNA following infection of the recombinants in vitro were determined by quantitative real-time PCR under reaction conditions described previously (34). Infected cells (MOI, 0.01 or 3) at each point were harvested, resuspended in 200 μl of a lysis buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 0.001% Triton X-100, 0.001% SDS) containing 50 mg/ml of proteinase K, and incubated at 50°C overnight. Three replicates for each time point were performed in 24-well dishes, with each well containing 1.6 × 106 (Neuro2A) or 2.4 × 104 (NIH 3T3) cells. The quantitative real-time PCR assay was performed using primers and a probe within the viral polymerase gene (UL30) (35).
RNA recovery, sequencing, and bioinformatics.
RISC-bound miRNAs were isolated by immunoprecipitation using one of two different monoclonal antibodies specific for the human Ago proteins: ab57113 or 2A8 (Abcam) (36). Briefly, an ∼2-ml cell pellet (from 2 × 106 cells) was lysed in lysis buffer (50 mM HEPES [pH 7.5], 150 mM KCl, 2 mM EDTA, 1 mM NaF, 0.5% [vol/vol] NP-40, 0.5 mM dithiothreitol [DTT], protease inhibitors) and was then centrifuged to remove insoluble particulates. The lysate was then filtered through a 0.2-μm membrane prior to incubation on a rotator overnight at 4°C with protein G beads loaded with the anti-Ago antibody. The beads were then washed 10 times with NT2 buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM MgCl2, 0.05% NP-40). The washed beads were then incubated in NT2 buffer in the presence of proteinase K prior to extraction using acid-phenol. The aqueous phase was then used to isolate small RNAs (<200 nt) using a mirVana kit (Ambion). Typically, this resulted in the recovery of ∼250 ng of total small RNAs from each sample. The RNA immunoprecipitation and sequencing (RIP-Seq) cDNA library was constructed essentially as described previously (37) by using an Illumina TruSeq small-RNA kit prior to sequencing with an Illumina HiSeq 2000 system. The resulting reads were then processed using the FASTX-Toolkit and were aligned to the human genome (NCBI, Hg19), the mouse genome (NCBI, m37 Ensembl release 61), the HSV-1 genome (17syn+; NCBI, NC_001806), a human miRNA library (miRBase19), and an HSV-1 miRNA library (miRBase19) using Bowtie (38). Reads of at least 15 nt were retained if they mapped to the indicated library with a maximum of 1 mismatch.
PAR-CLIP library construction and bioinformatics.
The photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP) library was constructed as described previously (39–41) with minor modifications, i.e., an Illumina TruSeq small-RNA kit was used to prepare cDNA for sequencing, and an Illumina HiSeq 2000 system was used for sequencing. The resulting reads were then processed using the FASTX-Toolkit and were aligned to the HSV-1 genome using Bowtie (38). Reads of at least 15 nt were retained if they mapped to HSV-1 with ≤2 mismatches.
qRT-PCR.
ICP0 and ICP34.5 mRNA levels were analyzed by SYBR green quantitative reverse transcription-PCR (qRT-PCR). Briefly, RNA was isolated by TRIzol (Invitrogen) extraction according to the manufacturer's protocol. Reverse transcription was performed using a high-capacity cDNA reverse transcription kit (Applied Biosystems) with 500 ng of DNase-treated total RNA. Two microliters of cDNA was then used with Power SYBR green PCR master mix and gene-specific primers (Applied Biosystems) for qRT-PCR. Values were normalized to levels of β-actin mRNA, determined in parallel, and assays were performed in triplicate. Values are expressed as multiples of the signal detected using RNA from cells infected with WT HSV-1.
To confirm the loss of miRNA expression in the viral recombinants, RS cells (MOI, 5 or 1) were infected with each recombinant; the infected cells were harvested at 18 h postinfection; and total RNA was prepared using TRIzol. The relative abundance of the viral miRNAs was then determined using the TaqMan microRNA assay (Applied Biosystems). Assays were performed in triplicate, and miRNA values were normalized to the level of viral thymidine kinase (TK) gene (UL23) mRNA. The assay identification numbers (IDs) are 7322 for miR-H1-5p, 5632 for miR-H2-3p, 197242_mat for miR-H3-3p, 1979191_mat for miR-H4-3p, 197213_mat for miR-H5-3p, 197219_mat for miR-H6-3p, 462602_mat for miR-H7-5p, 241862_mat for miR-H8-5p, and 2542341 for the viral TK mRNA.
For analysis of viral miRNA expression levels in cells infected by WT HSV-1, 293 cells were infected (MOI, 2), and total RNA or RISC-associated RNA was isolated at different time points postinfection. miRNA levels were again determined by the TaqMan microRNA assay using 10 ng of total RNA per RT reaction. Assays were performed in triplicate, and miRNA values were normalized to the level of U6 RNA for total-RNA preparations or to the level of miR-16 for RISC-associated RNA preparations. The number of strands per cell was calculated using synthetic RNAs (Integrated DNA Technologies [IDT]) for miR-H1-5p, miR-H2-3p, miR-H3-3p, and miR-H6-3p by spiking known levels of each synthetic miRNA into noninfected 293 cell total RNA at decreasing concentrations, in order to establish a linear regression equation.
Transfection of small duplex RNAs.
Duplex HSV-1 miRNA mimics were synthesized by IDT. RNA was annealed by heating in 1 μM Tris (pH 8.0), 50 mM NaCl, 1 mM EDTA at 95°C for 5 min and then decreasing the temperature by 0.5°C every 30 s until a final temperature of 4°C was reached. Annealed duplex miRNA mimics were stored at −80°C until use. Transfections were performed by using RNAiMAX (Invitrogen) according to the manufacturer's protocol, with 2 × 106 cells. Cells were infected with HSV-1 40 h later and were then harvested for Western blotting and qRT-PCR analysis 8 h after infection.
Western blotting.
Western blotting was performed as described previously (4). ICP0 was detected using an anti-ICP0 mouse monoclonal antibody obtained from Virusys (HIA027) at a dilution of 1:8,000. β-Actin was detected using an anti-β-actin monoclonal antibody from Santa Cruz (catalog no. SC-47778) at a 1:5,000 dilution. A rabbit anti-ICP34.5 antiserum was generated by using a fusion protein, containing the N-terminal 69 amino acids of ICP34.5 fused to the C terminus of maltose binding protein (MBP), that was expressed in Escherichia coli BL21. The MBP fusion protein was purified from soluble bacterial lysates by binding to amylose resin and was then eluted with 20 mM maltose in PBS. The antigen was concentrated into PBS and was injected into rabbits at Pocono Rabbit Farm and Laboratory. Polyclonal sera were harvested from these rabbits and were used directly for Western blotting of ICP34.5 protein at a 1:3,000 dilution. Bound antibodies were detected using a horseradish peroxidase (HRP)-conjugated donkey anti-rabbit (catalog no. NA934V; GE Healthcare) or rabbit anti-mouse (catalog no. A9044; Sigma-Aldrich) antiserum and were visualized using an enhanced chemiluminescence (ECL) kit from GE (product code RPN2232) and a Syngene G:Box gel imaging system.
Luciferase assays.
Indicator vectors were generated by using the Renilla luciferase (RLuc) vector pNL-SIN-CMV-RLuc, as described previously (42). Briefly, two fully complementary synthetic binding sites for the miRNA of interest were inserted using ClaI and XbaI sites present in the 3′ untranslated region (3′ UTR) of RLuc. ICP34.5 RLuc was generated by PCR amplification of infected-cell genomic DNA to yield a 570-bp fragment derived from the HSV-1 ICP34.5 transcript, extending from position 125430 to 125999 in the viral genome, that fully encompasses both the predicted antisense miR-H3-3p and miR-H4-3p targets and flanking sequences. This DNA fragment was cloned into the ClaI and XbaI sites present in pNL-SIN-CMV-RLuc. RLuc expression constructs (500 ng) and small duplex RNAs (40 pmol) were cotransfected into a 24-well plate seeded with 293 cells by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Cells were harvested 24 h later, and RLuc levels were determined.
RESULTS
Identification of HSV-1 microRNAs loaded into the RISC.
Initial efforts to use a small-RNA deep-sequencing approach to identify HSV-1 miRNAs expressed in productively infected cells revealed large amounts of small, ∼21-nt RNA breakdown products, of both cellular and viral origin, in infected cells that made it difficult to distinguish miRNAs from small RNAs that are simply degradation intermediates (data not shown). These likely reflect the action of the HSV-1 virion host shutoff (Vhs) protein, which is known to induce the degradation of most cellular mRNAs, as well as a subset of viral mRNAs, in infected cells (43).
To select for authentic HSV-1 miRNAs, and also to define the small viral RNAs that are actually loaded into the RISC and therefore are likely biologically relevant, we decided to immunoprecipitate the RISC by using an antibody raised against amino acids 483 to 859 of the key RISC component Ago2. This region of Ago2 is well conserved across all four human and murine Ago proteins, and this antibody effectively recognized not only Ago2 but also the Ago1 and Ago3 proteins (data not shown) and is therefore able to immunoprecipitate essentially all RISC components, thus allowing us to selectively deep sequence RISC-bound small RNAs (by RIP-Seq). To select an optimal time point for this analysis, we infected 293 cells with HSV-1 strain 17syn+ and then harvested total RNA at various times postinfection. Analysis of these RNA samples by qRT-PCR, using TaqMan primers specific for the HSV-1 miRNAs miR-H1-5p, miR-H2-3p, miR-H3-3p, and miR-H6-3p, confirmed earlier work showing that miR-H1-5p and miR-H6-3p, which are encoded outside the LAT, are expressed at substantial levels during productive HSV-1 infection, while miR-H2-3p and miR-H3-3p, which are encoded within the second LAT exon, are expressed at more modest levels (Table 2) (4, 28, 29). On the basis of these data, we decided to use the time point of 8 h after infection to analyze viral miRNAs that are associated with the RISC in infected cells. We performed RIP-Seq on three different cell lines—human 293 cells, human SY5Y cells, and mouse Neuro2A cells—at 8 h after infection with the HSV-1 strain 17syn+ at a multiplicity of infection (MOI) of 2. As shown in Table 3, we obtained 9,495,950 to 14,905,951 total small-RNA reads from each cell type, of which 6,464,486 to 8,937,722 represent known cellular miRNAs. Total HSV-1-specific reads ranged from a low of 6,447 in SY5Y cells to a high of 40,704 in Neuro2A cells. Alignment of these reads to the HSV-1 genome showed that the large majority aligned with the viral LAT region, with very few reads aligning to the unique long or short region of the viral genome (Fig. 1A). Analysis of these viral small-RNA reads (Table 4; Fig. 1B) revealed that a substantial number derived from the known HSV-1 pre-miRNAs encoding miR-H1 to miR-H8, even though most of these miRNAs (the exceptions are miR-H1 and miR-H6) are derived from the exonic regions of the LAT and therefore are not expected to be expressed at high levels during lytic replication (4, 5). Of the more recently reported, lytic-cycle-specific miRNAs miR-H11 to miR-H18 and miR-92944 (28, 29), we recovered significant numbers of reads for miR-H11-3p but only a total of seven, from all three cell lines combined, for miR-H13-5p (Fig. 1A; Table 4). No reads specific for miR-92944, miR-H12, or miR-H14 through miR-H18 were detected in any of the three infected-cell lines analyzed.
Table 2.
qRT-PCR analysis of the levels of expression of HSV-1 microRNAs in infected 293 cellsa
| Time postinfection (h) | No. of microRNA strands per cell |
|||
|---|---|---|---|---|
| miR-H1-5p | miR-H2-3p | miR-H3-3p | miR-H6-3p | |
| 2 | 60 | 14 | 62 | 143 |
| 4 | 302 | 5 | 70 | 272 |
| 6 | 3,562 | 15 | 88 | 742 |
| 8 | 5,254 | 34 | 95 | 1,629 |
| 10 | 7,172 | 36 | 95 | 2,772 |
| 12 | 9,140 | 49 | 127 | 3,923 |
| 24 | 9,384 | 46 | 139 | 3,941 |
293 cells were infected with HSV-1 strain 17syn+, and samples were harvested at the indicated times postinfection. A total of 106 cells were used for RNA isolation, and the number of miRNA strands per cell was quantitated by qRT-PCR by comparison to standard curves obtained using synthetic versions of these HSV-1 miRNAs.
Table 3.
Overview of deep-sequencing data for RISC-associated small RNAs recovered from HSV-1 infected Neuro2A, 293, and SY5Y cellsa
| Infected-cell type | No. of deep-sequencing readsb |
||||
|---|---|---|---|---|---|
| Total | Aligned to the cellular genome | Aligned to known cellular pre-miRNAs | Aligned to the HSV-1 genome | Aligned to known HSV-1 pre-miRNAs | |
| Neuro2A | 14,905,951 | 13,028,073 | 8,683,033 | 40,704 | 3,560 |
| SY5Y | 9,495,950 | 9,100,690 | 8,937,722 | 6,447 | 499 |
| 293 | 9,557,538 | 8,404,460 | 6,464,486 | 14,195 | 3,250 |
Each cell line was infected with the HSV-1 WT isolate 17syn+ at an MOI of 2. RISC-associated small RNAs were isolated 8 h later.
Obtained for cDNAs generated from the 18- to 25-nt RNA fraction. Alignment to the cellular (human or mouse) and HSV-1 genomes is shown.
Fig 1.
Alignment of HSV-1 small-RNA sequences to the HSV-1 genome. HSV-1 derived small (18- to 24-nt) RNA sequence reads obtained from HSV-1-infected 293, SY5Y, and Neuro2A cells were aligned to the full-length HSV-1 genome sequence with the terminal repeats deleted (A) or to the HSV-1 LAT region (B). Known HSV-1 miRNAs are indicated.
An unexpected result is that miR-H1-5p, which was previously reported to be both the most highly expressed HSV-1 miRNA in lytically infected cells and the dominant arm of the miR-H1 miRNA (4, 28, 29), was recovered at levels lower than those for miR-H2-3p, miR-H3-3p, miR-H4-3p, miR-H6-3p, and even miR-H1-3p in all three cell lines analyzed (Table 4). To determine whether this resulted from the selective exclusion of miR-H1-5p during cDNA preparation, we performed qRT-PCR analysis of the levels of expression of miR-H1-5p, miR-H2-3p, miR-H3-3p, and miR-H6-3p in both total small RNA and RISC-associated small RNA prepared in parallel from HSV-1-infected Neuro2A cells at 8 h postinfection. As shown in Table 5, the levels of miR-H1-5p and, to a lesser extent, miR-H6-3p recovered were much lower in the RISC-associated small-RNA fraction, whether measured by qRT-PCR or by deep sequencing, than those in total small RNA (measured by qRT-PCR) and were also lower than previously published levels of HSV-1 miRNA expression during lytic replication, determined by deep-sequencing analyses (4). Similar results were obtained using a second monoclonal antibody, called 2A8, that is capable of immunoprecipitating all four human Ago proteins (36) (data not shown). Based on these data, it appears that miR-H1-5p is loaded into the RISC ∼200-fold less effectively than miR-H3-3p, while miR-H2-3p and miR-H6-3p are loaded into the RISC ∼2-fold and ∼10-fold less effectively, respectively, than miR-H1-3p. This surprising result is discussed in more detail below.
Table 5.
Relative recovery levels of selected HSV-1 microRNAs
| HSV-1 miRNA | Relative level of strandsa in: |
Ratio of relative recovery in total RNA to that in RISC-associated RNAb determined by: |
|||
|---|---|---|---|---|---|
| Total RNA (determined by qRT-PCR) | RISC-associated RNA |
qRT-PCR | Deep sequencing | ||
| Determined by qRT-PCR | Determined by deep sequencing | ||||
| miR-H1-5p | 87 | 0.44 | 0.11 | 198 | 791 |
| mir-H2-3p | 2.2 | 0.52 | 0.45 | 4.2 | 4.9 |
| miR-H3-3p | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| miR-H6-3p | 10.4 | 1.1 | 1.2 | 9.5 | 8.7 |
Recovered from Neuro2A cells infected (MOI, 2) with the HSV-1 isolate 17syn+. RNA was isolated at 8 h postinfection. The number of strands of each HSV-1 miRNA in a total small-RNA preparation or in an RNA sample isolated from a RISC immunoprecipitate from the same culture was normalized to the quantity of miR-H3-3p strands, which was set at 1.
The miR-H3-3p ratio was set at 1.0. The higher the value, the lower the level of recruitment of the indicated HSV-1 miRNA into the RISC.
The most critical region for mRNA targeting by miRNAs is the so-called seed region, extending from position 2 to 8 from the 5′ end of the miRNA, and a key characteristic of authentic miRNAs is therefore that the 5′ end of the miRNA is more invariant than the 3′ end, which plays a more limited role in mRNA targeting (6). As shown in Table S1 in the supplemental material, we in fact saw little sequence variation in the cloned HSV-1 miRNA 5′ ends; the exceptions were miR-H1-5p and -3p, miR-H7-5p (the star strand for miR-H7), and miR-H13-5p, each of which exists as variants differing by one or two 5′ nucleotides.
Generation of HSV-1 mutants lacking miR-H2, miR-H3, or miR-H4.
As reported previously (4, 5), the HSV-1 miRNAs miR-H2-3p and miR-H3-3p lie antisense to the coding regions of ICP0 and ICP34.5, respectively, while miR-H4-3p is located antisense to the 5′ UTR of ICP34.5 (Fig. 2A). These three miRNAs are expressed at readily detectable levels in productively HSV-1 infected cells (Table 4) and are highly expressed in latently infected primary neurons (4, 5). To begin to define the functions of these miRNAs during both productive and latent infection, we generated mutant HSV-1 variants, each lacking one of these viral miRNAs. For miR-H4-3p, which is located outside any gene coding region, the whole sequence was simply deleted (17dH4). However, for miR-H2-3p and miR-H3-3p, which are located antisense to viral coding regions, the pri-miRNA hairpins were mutated such that the miRNA seed region was mutated, and the predicted pri-miRNA stem-loop was extensively destabilized, without affecting the underlying coding sequence (17mH2 and 17mH3, respectively) (Fig. 2B). Several independent recombinants were constructed using techniques detailed previously (32, 33). We confirmed by HindIII digestion of the whole viral genome, followed by agarose gel electrophoresis, that no extra changes within the genomes of viral mutants were generated during recombination (data not shown). As expected, the expression of each mutated miRNA was eliminated in each recombinant HSV-1 clone (Fig. 2C), as determined by qRT-PCR. Finally, each of these three viral mutants was analyzed to determine whether the mutational inactivation of the miR-H2, miR-H3, or miR-H4 miRNA affected the expression of the other LAT region-encoded miRNAs. There was no observable impact of the mutations in 17mH3 on the expression of any of the other miRNAs (Fig. 2D). While the 17dH4 mutation had no effect on the expression of miR-H2, -H3, -H7, or -H8, the expression of miR-H1, miR-H5, and miR-H6 was higher than that in the wild type. Finally, although the mH2 mutation did induce modest decreases in the levels of miR-H1, miR-H3, miR-H4, miR-H5, miR-H6, and miR-H8 expression, relative to that in the wild type, these decreases were generally ≤2-fold.
Fig 2.
HSV-1 miRNAs and construction and characterization of HSV-1 recombinants lacking miR-H2, miR-H3, or miR-H4. (A) Schematic diagram of the HSV-1 genome and expanded view of the LAT locus. The diagram of the HSV-1 genome (∼150 kb) shows the unique long (UL) and short (US) regions, flanked by the terminal repeat long (TRL) and internal repeat long (IRL) and by the internal repeat short (IRS) and terminal repeat short (TRS), respectively. The location of the LAT within the RL is shown. The 8.5-kb LAT primary transcript is shown, with the exons and 2.0-kb stable intron indicated. Additionally, the lytic ICP0 and ICP34.5 transcripts are designated. Six miRNAs (miR-H2, -H3, -H4, -H5, -H7, and -H8) are encoded within the LAT exonic region, and two miRNAs (miR-H1 and -H6), which lie antisense to each other, are encoded within the LAT promoter (shaded arrowheads). (B) Inactivating mutations introduced into HSV-1 miR-H2 and miR-H3. Shown are the coding sequences of the HSV-1 ICP0 and ICP34.5 genes, located antisense to HSV-1 miR-H2-3p and miR-H3-3p miRNAs, respectively. In each case, the upper line gives the predicted amino acid sequence; the second line, the wild-type DNA sequence; and the third line, the mutant sequence. Changes are indicated by boldface, and the miRNA seed sequence targets are boxed. The mutations introduced do not affect the coding sequence but were predicted to disrupt the formation of the pri-miRNA stem-loop. (C) Characterization of HSV-1 mutants lacking miR-H2 (17mH2), miR-H3 (17mH3), or miR-H4 (17dH4). RS cells were infected with recombinant virus clones of 17mH2 (left), 17mH3 (center), 17dH4 (right), and 17syn+ (WT). The infected cells were harvested at 18 h postinfection, and miRNA abundance was quantified using qRT-PCR. The relative quantities were normalized to that of HSV-1 miR-H1-5p. The standard error of the mean was calculated from three independent experiments. (D) MicroRNA assay of HSV-1 mutants 17mH2, 17mH3, and 17dH4. Recombinant virus clones of 17mH2 (light green), 17mH3 (purple), 17dH4 (sky blue), and wild-type 17syn+ (blue) were used to infect RS cells. The infected cells were harvested at 18 h postinfection, and miRNA abundance was quantified using qRT-PCR. The relative quantities were normalized to the level of the viral thymidine kinase mRNA (UL23). Threshold cycle values indicated by asterisks were as high as those seen in mock infections (data not shown). The standard error of the mean was calculated from three independent experiments.
Regulation of ICP0 and ICP34.5 expression by HSV-1 miRNAs.
As noted above, miR-H2-3p is located antisense to an exonic region of ICP0 mRNA, while miR-H3 and miR-H4 are located antisense to the mature ICP34.5 mRNA (Fig. 2A) (4, 5). Previously, miR-H2-3p was shown to inhibit HSV-1 ICP0 expression in 293T cells cotransfected with an ICP0 and a miR-H2-3p expression vector (4). Whether HSV-1 miR-H3-3p or miR-H4-3p is able to inhibit HSV-1 ICP34.5 expression has not been examined previously. However, both miR-H3 and miR-H4 have positional homologs in HSV-2, although these miRNAs differ in their seed sequences, and both these HSV-2 miRNAs have been reported to inhibit HSV-2 ICP34.5 expression in cotransfected cells (44–46).
To test whether endogenous, virus-derived miR-H2, miR-H3, or miR-H4 is indeed able to inhibit endogenous ICP0 or ICP34.5 expression in infected cells, we used qRT-PCR to examine the level of ICP0 or ICP34.5 mRNA expression in 293 cells 8 h after infection with the WT 17syn+ strain of HSV-1 or with a 17syn+ mutant lacking miR-H2, miR-H3, or miR-H4, as described above. As shown in Fig. 3A, we observed a significant and specific increase in the level of ICP0 mRNA expression in cells infected with the mH2 mutant virus and an even more marked increase in ICP34.5 mRNA expression in cells infected with the dH4 mutant, relative to expression by either the WT virus or the mH3 mutant. Unexpectedly, however, the mH3 mutant virus did not differ from the WT in the level of ICP34.5 mRNA detected (Fig. 3A). To further extend these data, we also measured ICP0 and ICP34.5 mRNA expression in WT HSV-1-infected 293 cells transfected with miRNA mimics encoding miR-H2, miR-H3, or miR-H4. In agreement with the data generated using mutant HSV-1, we observed decreased levels of ICP0 mRNA in cells expressing ectopic miR-H2 and markedly decreased ICP34.5 mRNA levels in cells expressing ectopic miR-H4. Ectopically expressed miR-H3, again, had no significant effect (Fig. 3A). This does not reflect differences in miRNA function, since all three miRNA mimics effectively inhibited a cognate miRNA indicator vector (Fig. 4) and all three are effectively loaded into the RISC, as determined by deep sequencing of RISC-associated small RNAs from transfected cells (data not shown).
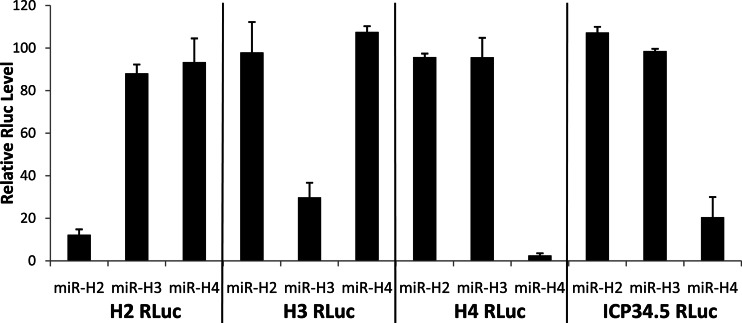
Fig 4.
HSV-1 miRNA mimics are biologically active. Indicator constructs were generated by the insertion of two perfect copies of the predicted full-length target site for miR-H2-3p (H2 RLuc), miR-H3-3p (H3 RLuc), or miR-H4-3p (H4 RLuc) into the 3′ UTR of the RLuc indicator gene. ICP34.5 RLuc is similar, except that it contains a 570-bp fragment of the HSV-1 ICP34.5 gene fully encompassing both the antisense miR-H3-3p and miR-H4-3p target sites. The indicator constructs were cotransfected into 293 cells along with synthetic duplex miRNA mimics for each of these HSV-1 miRNAs or with a control miRNA duplex. Cells were harvested 24 h later and RLuc activity determined. Data are expressed relative to the value for the control duplex, which was set at 100. Values are averages for three independent experiments, with standard deviations indicated.
Because miRNAs have been reported previously to occasionally block translation without causing mRNA degradation (6), we extended these mRNA analyses by performing Western blotting on analogous samples to detect ICP0 or ICP34.5 protein expression, with cellular actin functioning as a loading control (Fig. 3B). As may be observed, these protein data closely paralleled the RNA data presented in Fig. 3A. Specifically, loss of miR-H2, in the mH2 HSV-1 mutant, resulted in enhanced ICP0 expression, while ectopic miR-H2 reduced ICP0 expression in WT HSV-1-infected cells (Fig. 3B). Similarly, but more strikingly, loss of miR-H4 expression, in the dH4 mutant, strongly enhanced ICP34.5 expression, while ectopic miR-H4 inhibited ICP34.5 expression in WT HSV-1-infected cells by ∼8-fold (Fig. 3B). Again, both loss of miR-H3 expression, in the mH3 mutant, and overexpression of ectopic miR-H3 had no significant effect. Therefore, the lack of effect of miR-H3 on ICP34.5 expression is consistent, regardless of whether mRNA or protein levels are examined.
One possible explanation for the lack of effect of miR-H3-3p on ICP34.5 mRNA and protein expression (Fig. 3) despite the presence of a perfect target site in ICP34.5 is that this mRNA target is somehow occluded. To further address this possibility, we cloned a segment of the ICP34.5 gene encompassing both the miR-H3-3p and miR-H4-3p target sites, as well as flanking viral sequences, into the 3′ UTR of RLuc. As shown in Fig. 4, this indicator construct was strongly inhibited by a cotransfected miR-H4 miRNA duplex but was unaffected by an analogous miR-H3 miRNA duplex.
It could be argued that the enhanced expression of ICP34.5 protein seen in the HSV-1 17dH4 mutant (Fig. 3B) had nothing to do with the loss of the perfect target site for miR-H4 present in the 5′ UTR of the ICP34.5 mRNA but rather reflected the intrinsically more efficient translation of mutant ICP34.5 mRNAs bearing a deletion of the 5′ UTR located antisense to the pri-miR-H4 stem-loop. To control for this possibility, we generated ICP34.5 expression vectors bearing the full-length WT 5′ UTR or bearing the 5′ UTR present in the dH4 virus. After transfection into 293T cells, Western blot analysis revealed no difference in ICP34.5 expression between the WT and dH4-derived ICP34.5 vectors (data not shown), thus arguing that the increased ICP34.5 expression seen in Fig. 3B indeed reflects the loss of miR-H4 targeting of this mRNA.
Analysis of viral mRNA target site occupancy by miR-H2, miR-H3, and miR-H4.
The data presented in Fig. 3 demonstrate that both miR-H2, which is transcribed antisense to ICP0 mRNAs, and miR-H4, which is transcribed antisense to ICP34.5 mRNAs, are capable of inhibiting the expression of these mRNAs in productively HSV-1 infected cells. In contrast, miR-H3 is incapable of inhibiting ICP34.5 mRNA and protein expression, even though it is also transcribed antisense to the mature ICP34.5 mRNAs and is therefore fully complementary to these mRNAs. This lack of inhibition could result either from an inability of RISCs guided by miR-H3 to bind to ICP34.5 mRNAs or from an inability of bound RISCs to inhibit ICP34.5 mRNA function. Clearly, RISCs loaded with ectopic miR-H3, as tested in the experiments for which results are shown in Fig. 3, are indeed functional, since they inhibit luciferase reporter constructs bearing an artificial, perfect target site in their 3′ UTR (i.e., the same site that is present in ICP34.5) essentially as effectively as miR-H2 or miR-H4 inhibit their cognate indicator plasmids (Fig. 4).
To address whether miR-H2, miR-H3, and miR-H4 are indeed bound to their antisense mRNA target sites in infected cells, we utilized the photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP) technique, as described previously (39, 40, 41). In PAR-CLIP, cells are pulsed with 4-thiouridine (4SU), a nontoxic uridine analog that is incorporated into newly synthesized mRNAs and is efficiently cross-linked to mRNA-bound proteins after brief exposure to UV light at 365 nm. RISC-mRNA complexes are recovered by immunoprecipitation of the RISC and are then treated with RNase T1 to remove nonbound mRNA sequences. The residual ∼20- to 40-nt mRNA fragments are then converted to cDNAs and are sequenced using an Illumina HiSeq system. This technique captures essentially all mRNA sites bound by miRNAs in the cells being analyzed.
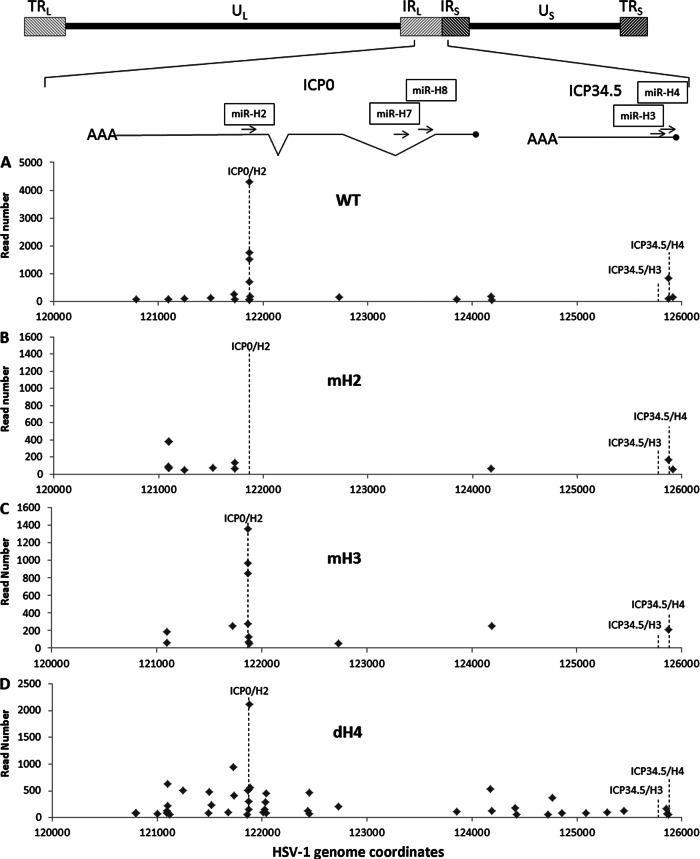
To perform this analysis, Neuro2A cells were pulsed with 4SU for 16 h, as described previously (40, 41), and were then immediately infected with HSV-1 at an MOI of 2. After infection, the cells were maintained in a 4SU-containing medium and were then cross-linked and harvested at 4 h postinfection. The reads obtained were then aligned to the region of the HSV-1 genome lying antisense to the second exon of the LAT, where miR-H2, miR-H3, and miR-H4 are encoded. As shown in Fig. 5A, PAR-CLIP of cells infected with WT HSV-1 revealed an intense cluster of reads at the site in the ICP0 mRNA located precisely antisense to miR-H2-3p, while a second, more modest cluster of reads was located at the site in ICP34.5 mRNA located antisense to miR-H4-3p. In fact, in the latter case, we recovered PAR-CLIP reads induced not only by the dominant miR-H4-3p strand but also by the miR-H4-5p star strand (Fig. 5A and Table 4; also data not shown). No reads were recovered from the ICP34.5 mRNA sequences located antisense to miR-H3-3p (Fig. 5A). Analysis of Neuro2A cells infected with the miR-H2 mutant reveals the complete loss of the miR-H2-specific cluster in ICP0, as expected, but no effect on the binding of miR-H4 to ICP34.5 mRNA (Fig. 5B). The pattern of RISC binding seen in cells infected with the HSV-1 mH3 mutant was comparable to that seen in WT-infected cells, again arguing that miR-H3 is not recruited to this region of the HSV-1 genome (Fig. 5C). Finally, in cells infected with the dH4 mutant, the miR-H2 cluster in ICP0 remained detectable but the miR-H4 cluster in ICP34.5 was lost (Fig. 5D). A small number of reads detected 5′ to the miR-H4 cluster are actually located ∼30 nt away from the predicted miR-H4 binding site on the ICP34.5 mRNA, as is more readily visible if the scale of the x axis is changed to allow small differences in read location to become visible (data not shown). We also detected a variable number of other, fairly weak signals in this region that do not have seed homology to either strand of miR-H2, miR-H3, or miR-H4 and therefore likely reflect weak and/or variable binding of RISCs bearing cellular miRNAs to viral transcripts. As expected, we did not see any clusters of reads located antisense to HSV-1 miR-H7 and miR-H8, both of which are expressed at low levels in productively infected cells (Table 4) and are located antisense to an intronic region of the primary ICP0 transcript (Fig. 5) (5). Together, these data demonstrate that miR-H2 and miR-H4 are indeed bound to the antisense target sites found in the HSV-1 ICP0 and ICP34.5 transcripts, respectively. In contrast, miR-H3 not only fails to inhibit ICP34.5 mRNA function (Fig. 3); it also fails to bind to the perfect target site located on this mRNA (Fig. 5).
Fig 5.
PAR-CLIP analysis of RISC binding to transcripts encoded within the LAT region of HSV-1. PAR-CLIP reads mapping to the HSV-1 LAT genomic region, extending from genome coordinates 120000 to 126000, are shown. A schematic of the HSV-1 mRNAs and miRNAs known to be encoded in this region is shown at the top, with the ICP0 introns indicated. (A) Reads mapping to this region in Neuro2A cells infected with WT HSV-1 17syn+, aligned by the most 5′ nucleotide of each read. The expected locations of the antisense mRNA targets for miR-H2, miR-H3, and miR-H4 are indicated. (B) PAR-CLIP reads obtained using the mH2 virus mutant. Note the loss of reads located antisense to miR-H2. (C) PAR-CLIP reads obtained using the mH3 virus mutant. (D) PAR-CLIP reads obtained using the dH4 virus mutant. Reads located antisense to miR-H4 were absent in this analysis.
Effects of miR-H2, miR-H3, and miR-H4 on HSV-1 replication in culture.
Our data argue that endogenous miR-H2 inhibits ICP0 expression, while miR-H4 inhibits ICP34.5 expression, in cells infected with HSV-1 in culture. Since both ICP0 and ICP34.5 have been shown to function as inhibitors of innate immune responses to HSV-1 (47–50), and the immediate early ICP0 protein also functions to facilitate HSV-1 early gene expression (47, 48), one might predict that loss of miR-H2 and/or miR-H4 would enhance HSV-1 replication, while overexpression would inhibit HSV-1 replication. Of course, it remains possible that the most critical targets for miR-H2 and miR-H4, especially in latently infected neurons, are actually cellular mRNAs. In the case of miR-H3, this would seem likely, since miR-H3 does not target ICP34.5 mRNA or any other viral mRNA identified thus far.
To address this question, we therefore analyzed the replication potential of the mH2, mH3, and dH4 HSV-1 mutants in Neuro2A cells, which are of neuronal origin, and NIH 3T3 cells, which are of fibroblastic origin, infected at a low or high MOI (0.01 or 3, respectively). For these analyses, three independent clones of each miRNA recombinant were analyzed, and replicative yields were assessed by quantitative PCR (Q-PCR) for viral DNA. As shown in Fig. 6, mutational inactivation of either miR-H3 or miR-H4 resulted in significant enhancement, as high as 10-fold (P, <0.01 by Student's t test), of viral replication at 24 h in Neuro2A cells infected at a low MOI (Fig. 6A). At a high MOI, only the mH3 recombinants replicated significantly better than the WT in Neuro2A cells (Fig. 6B). In contrast, in NIH 3T3 cells, the viral mH3 and dH4 mutants replicated at levels comparable to that of WT HSV-1 (Fig. 6C and D). Mutational inactivation of miR-H2, in the three mH2 recombinants, resulted in a significantly lower level (P, <0.01 by Student's t test) of viral replication at 24 and 72 h in Neuro2A cells infected at a low MOI (Fig. 6A). However, in Neuro2A cells infected at a high MOI, or in NIH 3T3 cells infected at either a high or a low MOI, the levels of replication of the WT and mH2 viruses were comparable (Fig. 6B, C, and D). We therefore conclude that the miR-H2, miR-H3, and miR-H4 mutant viruses can indeed influence HSV-1 replication in culture but that, interestingly, the effects exerted by these miRNAs differ depending upon the cell lines and MOIs used, and a significant effect on replication in all cases is observed only in the neuronal-cell-derived cell line Neuro2A.
Fig 6.
Analysis of the replication capacities of HSV-1 mutants lacking miR-H2, miR-H3, or miR-H4. Each recombinant virus and wild-type HSV-1 (strain 17syn+) were used to infect two different types of cultured cells, Neuro2A cells (A and B) or NIH 3T3 cells (C and D), at a low MOI of 0.01 (A and C) or a high MOI of 3 (B and D). In each case, results are shown for the 17mH2 (green) (top), 17mH3 (purple) (center), and 17dH4 (light blue) (bottom) recombinant virus clones compared with WT 17syn+ (blue). Infected cells were harvested at the indicated hours postinfection. Levels of viral DNA were determined by quantitative real-time PCR using primers and a probe within the viral polymerase gene (UL30) and were plotted as ratios to the corresponding values for uninfected cells (0 h postinfection). The standard errors of the means were from three independent experiments.
DISCUSSION
An initial goal of this study was to determine which of the 17 HSV-1 miRNAs that have been reported in the literature (4, 5, 28, 29) are functionally relevant by determining which of these miRNAs, several of which have been observed only by a single group and/or exclusively in productively HSV-1 infected cells, are actually loaded into the RISC. While our data do effectively address this question (see below), the most surprising result to arise from this analysis is that deep sequencing of total small RNAs, at least in HSV-1-infected cells, does not give an accurate picture of which miRNAs are actually loaded into the RISC and, hence, are likely to be biologically relevant. Specifically, while both deep sequencing (28, 29) and qRT-PCR analysis (Table 2) (4) of total small-RNA preparations indicate that HSV-1 miR-H1-5p and miR-H6-3p are the dominant HSV-1 miRNAs expressed in productively HSV-1 infected cells, deep sequencing and qRT-PCR analysis of HSV-1 miRNAs actually present in the RISC indicate that this is not, in fact, the case (Tables 4 and 5). Remarkably, our data indicate that miR-H1-5p is loaded into the RISC ∼200-fold less effectively than miR-H3-3p, while miR-H6-3p is loaded ∼10-fold less effectively than miR-H3-3p. This is true not only of virally encoded miRNAs; our data demonstrate that a subset of cellular miRNAs also show a clear discrepancy when total small-RNA reads are compared to RISC-associated miRNA reads (data not shown). While historically it was thought that miRNAs would be highly labile when not present in the RISC, and that total small-RNA reads therefore give an accurate picture of RISC-associated miRNAs, recent data indicate that only ∼8% of miRNAs are actually in the RISC at any given time (51). This suggests that variable RISC loading has the potential to regulate miRNA function and that the ability to be stably recruited into a limited amount of the RISC may represent a key rate-limiting step in determining miRNA function. At present, we have no idea where the ∼99% of miR-H1-5p detected by deep sequencing but not present in the RISC is located, but it has been suggested that miRNAs not bound to the RISC may nevertheless bind to complementary sites on mRNAs, although it is not clear whether this binding would have any functional consequences (51). Future work will address whether this phenomenon is restricted to HSV-1-infected cells, whether miRNAs that are loaded into the RISC inefficiently share any sequence characteristics, and whether inefficient loading into the RISC predicts a reduced ability to inhibit target mRNA expression.
Several proposed HSV-1 miRNAs are not loaded into the RISC.
While the differential loading of HSV-1 miRNAs was unexpected, our data did allow us to address which of the 17 proposed HSV-1 miRNAs are loaded into the RISC in productively HSV-1 infected cells. As noted above, previous data (4, 5, 28, 29) have identified six HSV-1 mRNAs that are encoded within the LAT and hence are expressed predominantly during latent infection (miR-H2 to miR-H5, miR-H7, and miR-H8), one HSV-1 miRNA expressed both during latency and during lytic replication (miR-H6), and nine that have been observed only during productive HSV-1 replication (miR-H11 to miR-H18 and miR-92944). Our cloning data, using a RISC-associated small-RNA preparation, recovered reads specific for miR-H1 through miR-H8, although, as noted above, these were recovered at levels distinct from those observed when total small RNA was analyzed. With regard to the more recently reported HSV-1 miRNAs (miR-H11 to miR-H18 and miR-92944), we failed to recover any reads for miR-H12, miR-H14 to miR-H18, and miR-92944 in any of the three HSV-1-infected cell lines analyzed. Since we recovered 6,467,736 to 8,686,593 miRNA reads by deep sequencing, and an average cell is thought to express ≤200,000 miRNAs (51), we can roughly calculate that these miRNAs, if they exist at all, are present in the RISC at <0.03 copy per infected cell. We therefore do not believe that these proposed HSV-1 miRNAs are functionally relevant.
Two other recently proposed lytic-cycle-specific miRNAs, i.e., miR-H11 and miR-H13, were recovered at ≤118 total reads for miR-H11 and at ≤4 total reads for miR-H13. In the case of miR-H13, we again do not believe that this is a physiologically relevant HSV-1 miRNA. Not only does the low number of reads obtained for miR-H13 indicate that this “miRNA” is expressed at <0.1 copy/cell, but also the small number of reads we did obtain show extensive 5′ sequence heterogeneity (see Table S1 in the supplemental material), and therefore, the few copies of miR-H13 that are expressed do not share a conserved seed sequence. In contrast, the miRNA strands derived from miR-H2 to miR-H8 all have tightly conserved 5′ ends, and hence seed sequences, as is also true of most cellular miRNAs (6). Finally, while miR-H11-3p is certainly RISC associated in HSV-1-infected cells (Table 4), it remains unclear whether this is indeed an authentic miRNA. One issue relates to the structure of the proposed pri-miRNA stem-loop from which miR-H11 is excised. Drosha-dependent miRNAs are located in the upper part of an ∼33-bp imperfect stem that is flanked by unstructured RNA sequences and that ends in a large (≥10-nt) terminal loop (7, 8). This is indeed the structure adopted by all eight of the validated HSV-1 miRNAs miR-H1 to miR-H8, as shown in Fig. 7 for miR-H3. However, the stem that encompasses miR-H11 is instead a perfect 130-bp inverted repeat, with a very small terminal loop, derived from the HSV-1 origin of replication OriL (28) (Fig. 7). Moreover, more than half the reads obtained for miR-H11 bear a 5′ end distinct from the major 5′-end sequence shown in Table S1 in the supplemental material, and finally, no reads derived from the predicted miR-H11 star strand were obtained either in our analysis or previously (28, 29). Therefore, if miR-H11 is indeed a functional miRNA, it is processed via a mechanism very different from the canonical, Drosha-dependent processing pathway used not only by HSV-1 miR-H1 to miR-H8 but also by the large majority of cellular miRNAs. Whether miR-H11 is a functional miRNA is currently unknown.
Fig 7.
Predicted structure of the pri-miRNA stem-loops for miR-H3 and miR-H11, as well as a possible stem-loop located antisense to miR-H3. Functional pri-miRNA stem-loops generally contain an ∼33-bp imperfect stem and a large terminal loop and are flanked by unstructured RNA sequences (7, 8). This is the case for the eight HSV-1 pri-miRNAs encoding miR-H1 to miR-H8, including the miR-H3 pri-miRNA stem-loop shown here as an example. In contrast, miR-H11 derives from the HSV-1 origin of replication OriL, which consists of a 130-bp perfect inverted repeat, as shown. The principal mature miRNA sequences and, for miR-H3, the passenger strand sequences, are shown in boldface. This figure also presents a potential RNA structure adopted by the ICP34.5 coding sequences located directly antisense to the miR-H3 pri-miRNA stem-loop. See the text for a discussion.
HSV-1 ICP34.5 transcripts are targeted by miR-H4 but not by miR-H3.
A key goal of the studies reported here was to generate mutant HSV-1 variants lacking key viral miRNAs with a view to identifying target mRNA species and observing the consequences for viral replication. As shown in Table 4, the most prominent HSV-1 miRNAs that are recovered from a RISC-associated RNA preparation are miR-H1, miR-H2, miR-H3, miR-H4, and miR-H6, and since it has been reported recently that an HSV-1 mutant lacking both miR-H1 and miR-H6, which lie precisely antisense to one another in the HSV-1 genome (Fig. 2A), grows normally in culture (52), we decided to focus our attention on miR-H2, miR-H3, and miR-H4. After the derivation of a set of recombinant viruses each lacking one of these HSV-1 miRNAs (Fig. 2), we first asked where we could detect any effect on the expression of the HSV-1 ICP0 mRNA, which lies antisense to miR-H2, or the ICP34.5 mRNA, which lies antisense to both miR-H3 and miR-H4. As shown in Fig. 3, loss of endogenous miR-H2 indeed enhances the expression of endogenous ICP0 mRNA and protein (Fig. 3), and miR-H2 binding to ICP0 transcripts could be readily detected by PAR-CLIP using WT HSV-1, but not the mH2 mutant viruses (Fig. 5). Similarly, loss of endogenous miR-H4 enhances ICP34.5 mRNA and protein expression (Fig. 3), and both miR-H4-3p and miR-H4-5p were readily detected bound to the predicted antisense mRNA target site in cells infected with WT HSV-1 but not in those infected with dH4 (Fig. 5A). Surprisingly, however, loss of miR-H3 did not affect ICP34.5 expression measured at either the mRNA or the protein level (Fig. 3), and miR-H3 was not found to bind ICP34.5 transcripts detectably (Fig. 5). Moreover, while a synthetic miR-H3-3p duplex RNA readily inhibited the expression of RLuc when the RLuc open reading frame was linked to a perfect, 22-bp 3′ UTR target site, the same site presented in the context of a 570-bp fragment of the ICP34.5 gene encompassing both the miR-H3-3p and miR-H4-3p target sites responded to the latter but not to the former in cotransfected cells (Fig. 4). ICP34.5 mRNAs therefore are clearly not a target for repression by miR-H3. Since the same target is active in the absence of flanking sequences but is not effectively targeted when these flanking sequences are present, we hypothesize that the antisense target for miR-H3-3p is occluded by an RNA secondary structure formed by these flanking sequences. One possibility is that the sequences located antisense to the miR-H3 pri-miRNA stem-loop are also able to form an RNA secondary structure, as shown in Fig. 7, that blocks miR-H3-3p access. The formation of “mirror image” stem-loops in pri-miRNAs and in transcripts antisense to pri-miRNAs is actually fairly common and can sometimes give rise to miRNAs that are derived from both strands of the DNA being transcribed, as is seen with miR-H1 and miR-H6 in HSV-1 (Fig. 2A) (4). Whether this is indeed the case will require further investigation.
Impact of miR-H2, miR-H3, and miR-H4 on HSV-1 replication in culture.
Analysis of the HSV-1 recombinants containing mutations/deletions of the viral miRNA miR-H2, miR-H3, or miR-H4 revealed that the mH3 and dH4 recombinants replicated 4- to 10-fold better than the WT in murine Neuro2A cells, while no difference in replication was seen in murine NIH 3T3 cells (Fig. 6). This effect on replication is consistent with the ability of miR-H4 to target ICP34.5 RNA (Fig. 3) and the fact that ICP34.5 is known to confer a significant advantage on HSV-1 replication within neurons (49, 50, 53, 54). The fact that a significant effect on HSV-1 replication was also exhibited by the mH3 recombinant in the absence of a detectable effect on ICP34.5 expression (Fig. 3) suggests that this miRNA acts on an mRNA target(s) other than ICP34.5 and that this activity (like that of miR-H4) results in a decrease in HSV-1 replication within neurons. While at first glance it might seem counterintuitive that a virus would evolve to express miRNAs that reduce replicative fitness, this actually does make sense when one considers the biphasic life cycle of HSV-1. The ability of HSV-1 to persist within the nervous system depends on its ability to establish a latent infection in neurons without killing them and then to reactivate periodically from these cells over time. It has long been known that HSV-1 replicative yields in neurons are lower than those in other cell types, and this has been assumed to be due to the low abundance of nucleotide precursors and replicative infrastructure in these terminally differentiated cells. While this may indeed be the case, it seems plausible that these viral miRNAs also help restrict replication and perhaps help shift the balance toward viral latency in these cells. While Neuro2A cells have provided a good surrogate cell line in the past, mirroring many aspects of HSV-1 transcription and replication seen in primary neurons, further studies are under way in actual urine sensory neurons (both in vivo and in vitro). It is possible that we will see an even greater effect of these miRNAs on HSV-1 replication in these nondividing cells.
In contrast to the results with miR-H3 and miR-H4, loss of miR-H2 expression actually reduced viral replication, but only in Neuro2A cells infected at a low multiplicity (Fig. 6). Since the only currently known target for miR-H2 is the viral ICP0 mRNA (Fig. 3), it is possible that miR-H2 acts to reduce ICP0 expression to a more favorable level. ICP0 is a protein encoded by HSV-1 that is not essential for in vitro replication but is required for efficient lytic infection both in vitro and in vivo and for efficient reactivation from latency (for a review, see reference 55). ICP0 mutants display reduced viral yields in most cell types at low MOIs but replicate to near-wild-type levels at high multiplicities and are more restricted in some cell types than in others (56, 57). Based on the phenotype of the ICP0 mutants, it might at first seem counterintuitive that HSV-1 would need a means to downregulate ICP0 expression via miRNAs. However, in addition to being a promiscuous transactivator of both viral and cellular genes, ICP0 is an E3 ubiquitin ligase that is well known to play a role in degrading cellular proteins involved in a number of critical pathways, including DNA damage response, innate signaling, and chromatin modification. It is therefore possible that regulating the overall amount of ICP0, or modulating ICP0 accumulation temporally, could be advantageous for the virus. Of course, it remains possible that the key targets for miR-H2 are, in fact, cellular mRNAs that remain to be identified. We also note that mutational inactivation of miR-H2 did appear to globally reduce the expression of the other HSV-1 miRNAs by ∼2-fold (Fig. 2D), despite the subtle nature of the mutation introduced (Fig. 2B), and this could also contribute to the reduction in HSV-1 replication that we observed in Neuro2A cells in culture (Fig. 6). It should also be noted that the mutation of miR-H4 also affected the expression of several of the other miRNAs in culture as well. This highlights the importance of examining how the loss of these three viral miRNAs affects the establishment of latency, viral loads, viral pathogenesis, and viral reactivation in experimentally HSV-1 infected animals.
In conclusion, we have derived HSV-1 mutants individually lacking three of the most highly expressed viral miRNAs, and we show that each of these three mutations can significantly affect viral replication in culture (Fig. 6). To our knowledge, this is the first demonstration that virally encoded miRNAs can affect the productive replication of the cognate virus in culture, although several reports have argued that viral miRNAs affect the establishment or maintenance of viral latency (58–61). While our efforts to identify the mRNA targets for miR-H2, miR-H3, and miR-H4 remain at an early stage, we were able to demonstrate unequivocally that endogenous miR-H2 binds to and inhibits endogenous ICP0 mRNAs, while endogenous miR-H4 binds to and inhibits endogenous ICP34.5 mRNAs (Fig. 3 and 5), and we believe that this may partially (miR-H2) or entirely (miR-H4) explain the mutant phenotypes observed (Fig. 6). Surprisingly, however, miR-H3, which, like miR-H4, lies antisense to ICP34.5 transcripts, does not bind to or inhibit ICP34.5 expression (Fig. 3 and 5). Nevertheless, ablation of miR-H3 miRNA does affect viral replication (Fig. 6), although the underlying mechanism is not apparent at this time.
The most unexpected and exciting result reported here is that the efficiency of RISC loading of miRNAs in HSV-1-infected cells differs widely; HSV-1 miR-H1-5p, for example, is loaded into the RISC >100-fold less efficiently than miR-H3-3p (Table 5; also data not shown). It could be argued that this result reflects the differential recovery of miRNAs from the RISC immunoprecipitate, and indeed, one group has reported previously that miRNAs with “low G-C content” can be selectively lost during extraction from a small number of cells (62). However, this seems very unlikely to explain the unequal levels of recovery of miR-H1-3p from total RNA and from RISC immunoprecipitate RNA reported here. First, miR-H1-3p does not have a low G-C content; 12 of the 22 nt in miR-H1-5p are G or C (see Table S1 in the supplemental material). Second, Kim et al. (62) reported that unequal miRNA recovery was seen with TRIzol extraction but not after small-RNA recovery using the Ambion mirVana kit. All our experiments used small RNAs recovered using the mirVana kit, again arguing against the hypothesis that these data reflect unequal miRNA recovery during isolation. Therefore, while the basis for this phenomenon is currently unclear, it does clearly imply that deep sequencing of total small RNAs can give a highly misleading picture of which miRNAs are actually recruited into the RISC effector and hence are likely to be of biological importance.
Supplementary Material
ACKNOWLEDGMENTS
We thank Chris Pierick for assistance with bioinformatic analysis and figure construction, Christine Krupa for assistance with manuscript preparation, and Levi Watson and Nat Johns for comments on the manuscript. We also thank Zissimos Mourelatos for the gift of the 2A8 antibody.
This research was supported by an NIH grant (R01-AI097376). O.F. was supported by a postdoctoral fellowship from Hoffman-LaRoche, Inc., while S.N. was supported in part by a Japan Herpesvirus Infections Forum Scholarship Award in Herpesvirus Infection Research.
Footnotes
Published ahead of print 27 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00504-13.
REFERENCES
- 1. Bloom DC, Giordani NV, Kwiatkowski DL. 2010. Epigenetic regulation of latent HSV-1 gene expression. Biochim. Biophys. Acta 1799:246–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stevens JG, Wagner EK, Devi-Rao GB, Cook ML, Feldman LT. 1987. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science 235:1056–1059 [DOI] [PubMed] [Google Scholar]
- 3. Spivack JG, Fraser NW. 1987. Detection of herpes simplex virus type 1 transcripts during latent infection in mice. J. Virol. 61:3841–3847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. 2008. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454:780–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Umbach JL, Nagel MA, Cohrs RJ, Gilden DH, Cullen BR. 2009. Analysis of human alphaherpesvirus microRNA expression in latently infected human trigeminal ganglia. J. Virol. 83:10677–10683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136:215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zeng Y, Yi R, Cullen BR. 2005. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 24:138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. 2006. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 125:887–901 [DOI] [PubMed] [Google Scholar]
- 9. Lee Y, Jeon K, Lee JT, Kim S, Kim VN. 2002. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 21:4663–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415–419 [DOI] [PubMed] [Google Scholar]
- 11. Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. 2004. Processing of primary microRNAs by the Microprocessor complex. Nature 432:231–235 [DOI] [PubMed] [Google Scholar]
- 12. Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature 432:235–240 [DOI] [PubMed] [Google Scholar]
- 13. Yi R, Qin Y, Macara IG, Cullen BR. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17:3011–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293:834–838 [DOI] [PubMed] [Google Scholar]
- 15. Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436:740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hammond SM, Bernstein E, Beach D, Hannon GJ. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293–296 [DOI] [PubMed] [Google Scholar]
- 17. Khvorova A, Reynolds A, Jayasena SD. 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell 115:209–216 [DOI] [PubMed] [Google Scholar]
- 18. Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115:199–208 [DOI] [PubMed] [Google Scholar]
- 19. Cullen BR. 2011. Viruses and microRNAs: RISCy interactions with serious consequences. Genes Dev. 25:1881–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cui C, Griffiths A, Li G, Silva LM, Kramer MF, Gaasterland T, Wang XJ, Coen DM. 2006. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J. Virol. 80:5499–5508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wagner EK, Guzowski JF, Singh J. 1995. Transcription of the herpes simplex virus genome during productive and latent infection. Prog. Nucleic Acid Res. Mol. Biol. 51:123–165 [DOI] [PubMed] [Google Scholar]
- 22. Hill JM, Sedarati F, Javier RT, Wagner EK, Stevens JG. 1990. Herpes simplex virus latent phase transcription facilitates in vivo reactivation. Virology 174:117–125 [DOI] [PubMed] [Google Scholar]
- 23. Leib DA, Bogard CL, Kosz-Vnenchak M, Hicks KA, Coen DM, Knipe DM, Schaffer PA. 1989. A deletion mutant of the latency-associated transcript of herpes simplex virus type 1 reactivates from the latent state with reduced frequency. J. Virol. 63:2893–2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Perng GC, Jones C, Ciacci-Zanella J, Stone M, Henderson G, Yukht A, Slanina SM, Hofman FM, Ghiasi H, Nesburn AB, Wechsler SL. 2000. Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript. Science 287:1500–1503 [DOI] [PubMed] [Google Scholar]
- 25. Thompson RL, Sawtell NM. 2001. Herpes simplex virus type 1 latency-associated transcript gene promotes neuronal survival. J. Virol. 75:6660–6675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kwiatkowski DL, Thompson HW, Bloom DC. 2009. The polycomb group protein Bmi1 binds to the herpes simplex virus 1 latent genome and maintains repressive histone marks during latency. J. Virol. 83:8173–8181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cliffe AR, Garber DA, Knipe DM. 2009. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J. Virol. 83:8182–8190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jurak I, Kramer MF, Mellor JC, van Lint AL, Roth FP, Knipe DM, Coen DM. 2010. Numerous conserved and divergent microRNAs expressed by herpes simplex viruses 1 and 2. J. Virol. 84:4659–4672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Munson DJ, Burch AD. 2012. A novel miRNA produced during lytic HSV-1 infection is important for efficient replication in tissue culture. Arch. Virol. 157:1677–1688 [DOI] [PubMed] [Google Scholar]
- 30. Wang D, Zhang Z, O'Loughlin E, Lee T, Houel S, O'Carroll D, Tarakhovsky A, Ahn NG, Yi R. 2012. Quantitative functions of Argonaute proteins in mammalian development. Genes Dev. 26:693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thompson RL, Stevens JG. 1983. Biological characterization of a herpes simplex virus intertypic recombinant which is completely and specifically non-neurovirulent. Virology 131:171–179 [DOI] [PubMed] [Google Scholar]
- 32. Bloom DC, Jarman RG. 1998. Generation and use of recombinant reporter viruses for study of herpes simplex virus infections in vivo. Methods 16:117–125 [DOI] [PubMed] [Google Scholar]
- 33. Bloom DC. 1998. HSV vectors for gene therapy. Methods Mol. Med. 10:369–386 [DOI] [PubMed] [Google Scholar]
- 34. Isomura H, Stinski MF, Kudoh A, Nakayama S, Murata T, Sato Y, Iwahori S, Tsurumi T. 2008. A cis element between the TATA box and the transcription start site of the major immediate-early promoter of human cytomegalovirus determines efficiency of viral replication. J. Virol. 82:849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Giordani NV, Neumann DM, Kwiatkowski DL, Bhattacharjee PS, McAnany PK, Hill JM, Bloom DC. 2008. During herpes simplex virus type 1 infection of rabbits, the ability to express the latency-associated transcript increases latent-phase transcription of lytic genes. J. Virol. 82:6056–6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nelson PT, De Planell-Saguer M, Lamprinaki S, Kiriakidou M, Zhang P, O'Doherty U, Mourelatos Z. 2007. A novel monoclonal antibody against human Argonaute proteins reveals unexpected characteristics of miRNAs in human blood cells. RNA 13:1787–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Keene JD, Komisarow JM, Friedersdorf MB. 2006. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 1:302–307 [DOI] [PubMed] [Google Scholar]
- 38. Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141:129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gottwein E, Corcoran DL, Mukherjee N, Skalsky RL, Hafner M, Nusbaum JD, Shamulailatpam P, Love CL, Dave SS, Tuschl T, Ohler U, Cullen BR. 2011. Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines. Cell Host Microbe 10:515–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Skalsky RL, Corcoran DL, Gottwein E, Frank CL, Kang D, Hafner M, Nusbaum JD, Feederle R, Delecluse HJ, Luftig MA, Tuschl T, Ohler U, Cullen BR. 2012. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog. 8:e1002484 doi: 10.1371/journal.ppat.1002484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gottwein E, Cai X, Cullen BR. 2006. A novel assay for viral microRNA function identifies a single nucleotide polymorphism that affects Drosha processing. J. Virol. 80:5321–5326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kwong AD, Frenkel N. 1989. The herpes simplex virus virion host shutoff function. J. Virol. 63:4834–4839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tang S, Bertke AS, Patel A, Wang K, Cohen JI, Krause PR. 2008. An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor. Proc. Natl. Acad. Sci. U. S. A. 105:10931–10936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tang S, Patel A, Krause PR. 2009. Novel less-abundant viral microRNAs encoded by herpes simplex virus 2 latency-associated transcript and their roles in regulating ICP34.5 and ICP0 mRNAs. J. Virol. 83:1433–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Umbach JL, Wang K, Tang S, Krause PR, Mont EK, Cohen JI, Cullen BR. 2010. Identification of viral microRNAs expressed in human sacral ganglia latently infected with herpes simplex virus 2. J. Virol. 84:1189–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hagglund R, Roizman B. 2004. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J. Virol. 78:2169–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Everett RD, Parsy ML, Orr A. 2009. Analysis of the functions of herpes simplex virus type 1 regulatory protein ICP0 that are critical for lytic infection and derepression of quiescent viral genomes. J. Virol. 83:4963–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. He B, Gross M, Roizman B. 1997. The γ134.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1α to dephosphorylate the α subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. U. S. A. 94:843–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Orvedahl A, Alexander D, Talloczy Z, Sun Q, Wei Y, Zhang W, Burns D, Leib DA, Levine B. 2007. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe 1:23–35 [DOI] [PubMed] [Google Scholar]
- 51. Janas MM, Wang B, Harris AS, Aguiar M, Shaffer JM, Subrahmanyam YV, Behlke MA, Wucherpfennig KW, Gygi SP, Gagnon E, Novina CD. 2012. Alternative RISC assembly: binding and repression of microRNA-mRNA duplexes by human Ago proteins. RNA 18:2041–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jurak I, Silverstein LB, Sharma M, Coen DM. 2012. Herpes simplex virus is equipped with RNA- and protein-based mechanisms to repress expression of ATRX, an effector of intrinsic immunity. J. Virol. 86:10093–10102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Leib DA, Machalek MA, Williams BR, Silverman RH, Virgin HW. 2000. Specific phenotypic restoration of an attenuated virus by knockout of a host resistance gene. Proc. Natl. Acad. Sci. U. S. A. 97:6097–6101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yordy B, Iijima N, Huttner A, Leib D, Iwasaki A. 2012. A neuron-specific role for autophagy in antiviral defense against herpes simplex virus. Cell Host Microbe 12:334–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Boutell C, Everett RD. 2013. Regulation of alphaherpesvirus infections by the ICP0 family of proteins. J. Gen. Virol. 94:465–481 [DOI] [PubMed] [Google Scholar]
- 56. Everett RD, Boutell C, Orr A. 2004. Phenotype of a herpes simplex virus type 1 mutant that fails to express immediate-early regulatory protein ICP0. J. Virol. 78:1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yao F, Schaffer PA. 1995. An activity specified by the osteosarcoma line U2OS can substitute functionally for ICP0, a major regulatory protein of herpes simplex virus type 1. J. Virol. 69:6249–6258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Feederle R, Linnstaedt SD, Bannert H, Lips H, Bencun M, Cullen BR, Delecluse HJ. 2011. A viral microRNA cluster strongly potentiates the transforming properties of a human herpesvirus. PLoS Pathog. 7:e1001294 doi: 10.1371/journal.ppat.1001294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lu F, Stedman W, Yousef M, Renne R, Lieberman PM. 2010. Epigenetic regulation of Kaposi's sarcoma-associated herpesvirus latency by virus-encoded microRNAs that target Rta and the cellular Rbl2-DNMT pathway. J. Virol. 84:2697–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bellare P, Ganem D. 2009. Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe 6:570–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lin X, Liang D, He Z, Deng Q, Robertson ES, Lan K. 2011. miR-K12-7-5p encoded by Kaposi's sarcoma-associated herpesvirus stabilizes the latent state by targeting viral ORF50/RTA. PLoS One 6:e16224 doi: 10.1371/journal.pone.0016224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kim YK, Yeo J, Kim B, Ha M, Kim VN. 2012. Short structured RNAs with low GC content are selectively lost during extraction from a small number of cells. Mol. Cell 46:893–895 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.