Abstract
Poly(ADP-ribose) polymerases (PARPs) are NAD+ dependent enzymes that were identified as DNA repair proteins, however, today it seems clear that PARPs are responsible for a plethora of biological functions. Sirtuins (SIRTs) are NAD+-dependent deacetylase enzymes involved in the same biological processes as PARPs raising the question whether PARP and SIRT enzymes may interact with each other in physiological and pathophysiological conditions. Hereby we review the current understanding of the SIRT-PARP interplay in regard to the biochemical nature of the interaction (competition for the common NAD+ substrate, mutual posttranslational modifications and direct transcriptional effects) and the physiological, or pathophysiological consequences of the interactions (metabolic events, oxidative stress response, genomic stability and ageing). Finally, we give an overview of the possibilities of pharmacological intervention to modulate PARP and SIRT enzymes either directly, or through modulating NAD+ homeostasis.
Keywords: poly(ADP-ribose) polymerase, sirtuins, NAD+, metabolism, oxidative stress, mitochondria
1. Introduction
Adaptative responses are the product of critical balances integrating a myriad of molecular changes. These molecular changes include covalent modifications of diverse protein and changes in enzymatic substrate bioavailability, amongst others.
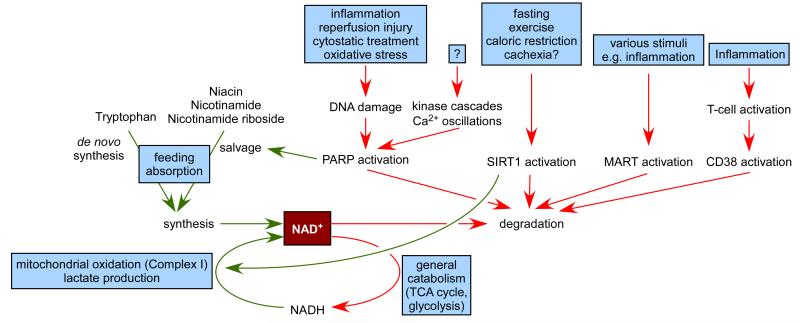
NAD+ (Figure 1) and its redox counterpart, NADH, are key metabolites influencing a large constellation of metabolic reactions. The most largely studied poly(ADP-ribose) polymerase (PARP) family members, PARP-1 and PARP-2, use NAD+ as a co-substrate in their catalytic activity. It has been observed that persistent PARP activation can deplete total intracellular levels by 80% and elevates nicotinamide (NAM), its reaction product. This depletion can have major metabolic impacts, due to the large spectrum of metabolic activities depending on NAD+ bioavailability (Bai and Canto 2012.).
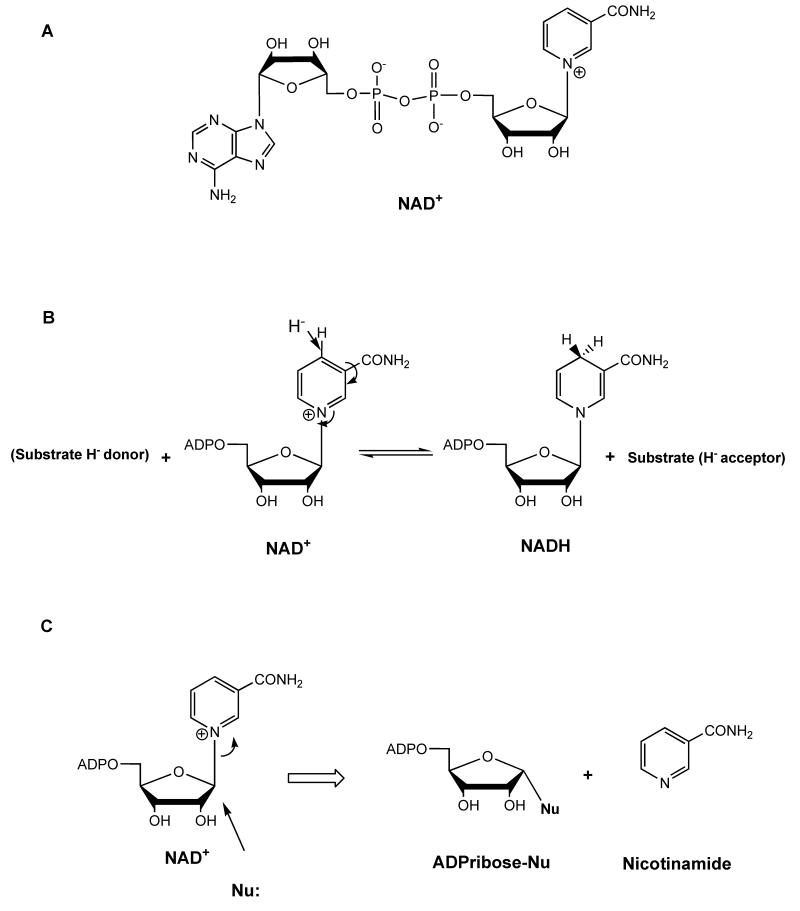
Figure 1. NAD and reactions of NAD+.
(A) Depiction of the chemical structure of NAD+. (B) Transfer of hydride to nicotinamide of NAD+ to form NADH. (C) ADP-ribosyltransfer reaction of NAD+ to a cellular nucleophile (acetyllysine, aspartate, glutamate, protein, etc).
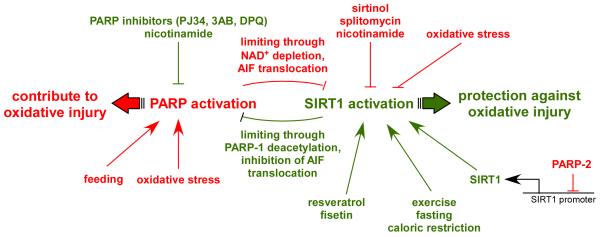
Sirtuins are a family of NAD+-dependent protein deacetylases with critical metabolic roles (Houtkooper, et al. 2012.). Early observations indicated that the decline of NAD+ and the rise of NAM promoted by enhanced PARP activity correlates with a downregulation of sirtuin activity (Bai, et al. 2011b, Pillai, et al. 2005.). Similarly, the activation of the most well-known mammalian sirtuin, SIRT1, led to reduced PARP activity (Kolthur-Seetharam, et al. 2006.). These observations supported a hypothesis raised a decade ago, where it was postulated that the activity of sirtuins and PARPs might compete for the availability of a common NAD+ pool (Zhang 2003.). In this review we will dissect the possible linkage of these two ancient pathways, PARPs and sirtuins, their possible competition for NAD+, and the physiological, or pathophysiological impact of these interactions. Furthermore, we will discuss how the vertexes of these interactions could be approached pharmacologically.
1.1. NAD+ metabolism
The metabolism of the pyridine dinucleotides is a long studied one with reports dating to the early parts of the twentieth century (Harden and Young 1906.) wherein Harden coined the term “co-zymase” to indicate NAD+. The redox activity of the dinucleotide compounds (NAD+ and NADP+) were first described to be a consequence of the pyridine moiety by Warburg in 1936 (Warburg and Christian 1936.). For most of the last century, the chemistry that converted the dinucleotides to their reduced counterparts (NADH and NADPH) constituted nearly the entire focus of interest on these important players in metabolism. In fact, cell metabolism has a plentitude of redox transformations that interconvert NAD+ and NADH (or NADP+ and NADPH), ranging from catabolism to biosynthesis (Pfleiderer 1970.).
In the latter part of the twentieth century, the non-redox reactivity of NAD+ was recognized as a second major function of NAD+, wherein ADP-ribose (ADPR) is transferred to cellular nucleophiles, such as proteins, in chemistry called ADP-ribosyl transfer (Honjo, et al. 1968, Nishizuka, et al. 1968.). This “newer” chemistry of NAD+ is diversified and has been expanded in mammalian organisms, where seven sirtuins (Sauve, et al. 2006.) and 17 PARP enzymes (Ame, et al. 2004.) harness this chemistry for signaling and cell adaptation.
The central role of NAD+ in metabolic transformations, as well as its incorporation into signaling pathways has made the study of NAD+ and how it is made in cells a rejuvenated topic of interest (Houtkooper, et al. 2010, Koch-Nolte, et al. 2009.). The manner in which NAD+ is made and utilized constitutes “NAD+ metabolism” and is a modern subject, with open ended questions on how it is biosynthesized, maintained in cells, incorporated into signaling, etc. This introduction surveys these topics in brief, but also provides an opportunity to highlight the variety of ways in which the study of NAD+ has blossomed over the years.
1. 1. 1. Redox properties of NAD+
The role of NAD+ as a direct player in catabolic metabolism is well known. NAD+ participates as a co-substrate in several steps of glycolysis, lactate pyruvate interconversion, pyruvate oxidation to acetyl-CoA catalyzed by pyruvate dehydrogenase complex, TCA cycle and is the donor of electron equivalents to Complex I in the electron transport chain (Ramakrishna, et al. 2001.). NAD+ is integrated centrally into energy metabolism. Consequently, NAD+ level is crucial for the proper maintenance of metabolic functions in cells. NAD+ level is normally maintained at a relative abundance to NADH level in cells (Williamson, et al. 1967.), and the NAD+/NADH ratio regulates numerous metabolic pathways, including glycolysis (Sun, et al. 2012.). NADH accumulation generally leads to feedback inhibition of metabolic processes upstream of the electron transport chain, except lactate production. Lactate production provides means to mitigate unbalanced NAD+/NADH ratio (Sun, et al. 2012.) and basis for the Cori cycle, in which lactate is released into the bloodstream and delivered to the liver for gluconeogenesis (Katz and Tayek 1998.). Not surprisingly, when excess NADH accumulates, lactate also accumulates and typically, high lactate is associated with hypoxia (Rimachi, et al. 2012.) or other mitochondrial deficiencies (Yamada, et al. 2012.). Lactate pyruvate ratio is a clinical surrogate for NAD+/NADH ratio in physiology.
The redox properties of NAD+ originate from the deficiency in electron density in the NAM ring. When conjugated to ribose, this electron deficiency becomes further accentuated by the quaternization of the pyridine nitrogen in the heterocycle (Figure. 1B). This quaternary pyridine group is made even more electron deficient by the carboxamide, which is a good electron withdrawing group. The positive charge on the nicotinamide group in NAD+ has been calculated to be 0.541 charge units (Cen and Sauve 2010.). The electron deficiency of the pyridine ring provides a driving force for acceptance of hydride ion at C4 (Figure 1B). Hydride ion acceptance breaks the aromaticity in the pyridine ring, but the energy expense is compensated by increased negative charge into the ring. The removal of the hydride ion in the reverse direction is driven by restoration of aromaticity. This redox chemistry reflects fine balancing of acceptance and removal of hydride, and evolution has centrally incorporated NAD+ into many metabolic processes requiring hydride ion transfer.
1. 1. 2. Non-redox properties of NAD+
The ribose ring is conjugated to nicotinamide in NAD+ via the anomeric carbon. The construction of this bond can occur via several pathways, as discussed in the next section; however, the decomposition of this bond is crucial to the action of ADPR transfer (ART) enzymes (Figure 1C). NAD+ is the electrophile, and the ADPR moiety is transferred to a variety of cellular nucleophiles, including proteins (Figure. 1C). This ADPR transfer chemistry is facilitated by the property of NAM as a good leaving group, with a pKa value of near 3.5 (Jackson, et al. 2003.). PARPs and sirtuins harness this general reactivity in addition to the NAD+ glycohydrolase/ADP-ribosyl cyclases CD38 (Sauve and Schramm 2004.) and CD157 (Ortolan, et al. 2002.). In addition there are a number of other putative ART enzymes encoded by the human genome with possible effects in modulating protein activities by ADP-ribosylation (Hottiger, et al. 2010.). The diverse ADP-ribosylation enzymes encoded by the mammalian genome suggest that ADP-ribosyl modifications are of fundamental importance for shaping mammalian physiology.
1. 1. 2. 1. NAD+ biosynthesis
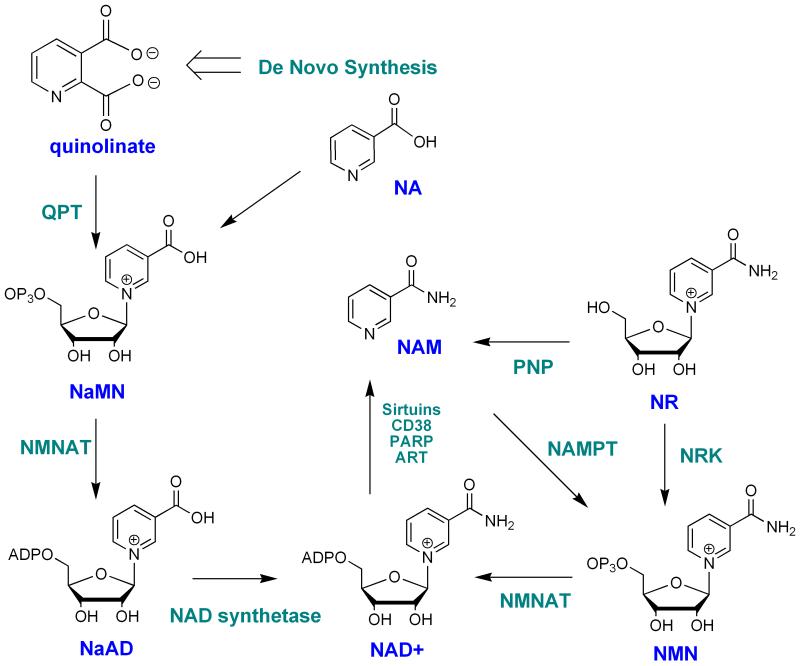
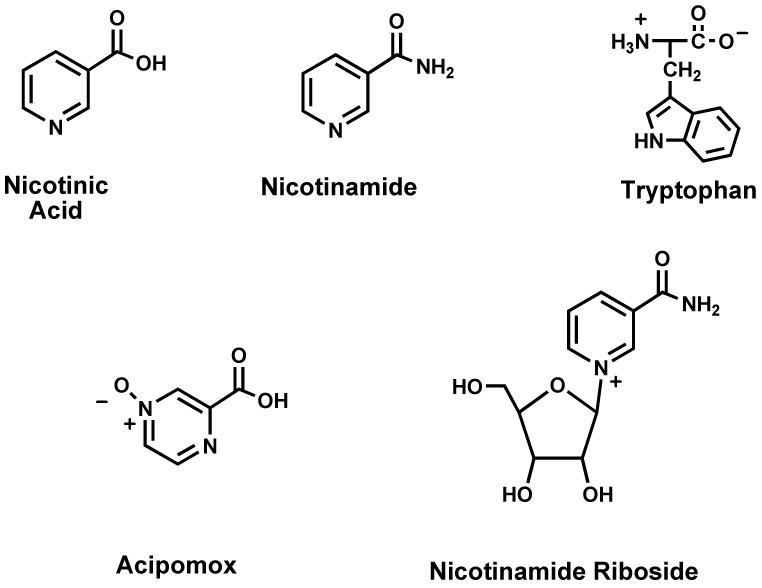
NAD+ is biosynthesized by a number of different pathways in humans (Bogan and Brenner 2008, Sauve 2008, Xu and Sauve 2010.). In broader terms, these can be broken down into de novo and salvage pathways. The de novo pathway in humans derives from the essential amino acid tryptophan, which is catabolized through the kynurenic pathway to quinolinic acid (Figure. 2) (Satyanarayana and Rao 1980.). Quinolinic acid is the universal metabolite in biology that generates the aromatic pyridine ring of NAD+ (Colabroy and Begley 2005, Kurnasov, et al. 2003.). This metabolite is coupled to the activated sugar metabolite 5-phospho-ribosyl-1-pyrophosphate (PRPP) to produce nicotinic acid mononucleotide (NaMN) with decarboxylation (Gholson, et al. 1964.). NaMN intersects the salvage pathway of nicotinic acid (niacin, NA), which was first characterized by Preiss and Handler in human erythrocytes (Preiss and Handler 1958a, Preiss and Handler 1958b.). NA is coupled to PRPP via a separate enzyme nicotinic acid phosphoribosyltransferase which has interesting biochemical properties, in that it appears to couple NA and PRPP in a coupled reaction with ATP hydrolysis (Galassi, et al. 2012, Vinitsky and Grubmeyer 1993.). This ATPase activity assists forward progress of the reaction via energy coupling (Vinitsky and Grubmeyer 1993.). NaMN is subsequently adenylated to nicotinic acid adenine dinucleotide by one of three mammalian adenylyltransferases (Lau, et al. 2009, Schweiger, et al. 2001.) (NMNAT1, NMNAT2 or NMNAT3) and then the acid is converted to an amide via a glutamine dependent NAD+ synthetase (Bembenek, et al. 2005, Bieganowski and Brenner 2003.). These reactions complete the biosynthetic process that culminates in NAD+ synthesis from de novo NA synthesis and NA salvage.
Figure 2. NAD+ biosynthetic pathways in mammals.
Naming derives from mammalian abbreviations. NA: nicotinic acid; NR: NAM riboside; NAM: nicotinamide; NAM NMN: NAM mononucleotide; NaMN: nicotinic acid mononucleotide; NaAD: nicotinic acid adenine dinucleotide; QPT: nicotinic acid phosphoribosyl-transferase; NRK: NAM riboside kinase; ART: ADP-ribosyl transferase; PARP: poly-ADP-polymerase; Nampt: NAM phosphoribosyltransferase; PNP: purine nucleoside phosphorylase; NMNAT: NMN/NaMN adenylyltransferase.
A separate salvage pathway is known, although only recently characterized, wherein NAM is coupled to PRPP to form NMN, via an enzyme called nicotinamide phosphoribosyltransferase (Revollo, et al. 2004, Rongvaux, et al. 2002.) (Nampt). This latter enzyme has a weakly coupled ATPase activity, thereby having some similarity to the corresponding nicotinate coupling enzyme (Burgos and Schramm 2008.). This enzyme has a very low Km for NAM, ranging from 1 μM to 5 nM (Burgos and Schramm 2008.) and is key to setting NAD+ levels in cells (Revollo, et al. 2004, Yang, et al. 2007a.). NAM recycling is of singular importance, because of the abundance of ART activities in cells, which generate continuous flux of NAM, which sustain in vivo tissue concentrations of NAM well above 20 μM (Qin, et al. 2006.). The importance of NAM recycling activity in regulating NAD+ biosynthesis is discussed in the following section. Importantly, the Nampt activity is not found in lower metazoans, suggesting that this enzyme is a mammalian adaptation (although the last common ancestor is unidentified) (Yang, et al. 2007a.).
In addition there has been an identification of two kinases encoded in mammalian cells called nicotinamide riboside kinase 1 and 2 (Nrk1 and 2) (Bieganowski and Brenner 2004, Tempel, et al. 2007.). These enzymes catalyze the efficient phosphorylation of nicotinamide riboside (NR) and nicotinic acid riboside (NaR) in vitro (Tempel, et al. 2007.). The Km and kcat parameters are as follows: human Nrk1 NR: kcat=0.6 s-1, Km =88 μM, NaR: kcat=0.21 s-1, Km=51 μM; human Nrk2 NR kcat=0.34 s-1 Km =190 μM; NaR: kcat=0.34 s-1, Km =63 μM (Tempel, et al. 2007.). A structural study of the human Nrk1 enzyme complexed with NR and a non-hydrolyzable ATP analogue has confirmed that the enzyme accommodates NR into a geometry that places the 5-OH of the ribose into close proximity to the terminal phosphate position of the ATP for efficient phosphorylation (Tempel, et al. 2007.). Studies of the Nrk1 and Nrk2 roles in mammalian NAD+ biosynthesis are very limited, although studies of the human enzymes in yeast establish that they can complement loss of the corresponding Nrk1 in yeast (Bieganowski and Brenner 2004.). Moreover, yeast can grow on NR if NAD+ synthetase is deleted (Δqns1), indicating that NR is metabolized differently from nicotinamide or nicotinic acid (Bieganowski and Brenner 2004.). The putative role of NR as a mammalian metabolite is supported by detection of NR in milk (Bieganowski and Brenner 2004.), although quantitative information on its abundance in milk is currently unavailable. Detection of NR in liver tissues has been reported by the Imai laboratory although quantitation was not provided (Yoshino, et al. 2011.).
The relative contributions of the different pathways of NAD+ synthesis in mammals is only generally understood, and is subject to many factors including diet (Rodgers and Puigserver 2007, Yang, et al. 2007a.). Humans do not encode efficient pathways for nicotinic acid synthesis, suggesting that nicotinic acid is not an abundant cellular metabolite. On the other hand, plant and fermented food sources are likely fortified with NA, since plants, yeast and bacteria encode nicotinamidases (French, et al. 2010.). Meats are enriched in NAM and have less NA.
1. 1. 2. 2. Regulation of NAD+ biosynthesis
The ability of cells to regulate NAD+ synthesis was only recently appreciated. Consistent with the centrality of NAM recycling as the ultimate regulator of NAD+ levels in cells and tissues, it has been determined that the enzyme Nampt is subject to dynamic regulation (Fulco, et al. 2008, Yang, et al. 2007a.), and that it is also subject to circadian influences (Nakahata, et al. 2009, Ramsey, et al. 2009.). The NAM salvage pathway is thought to be central to mammalian NAD+ homeostasis, since NAD+ has a limited lifetime in tissues. For example NAD+ has a reported half-life of 5-10 hours in liver (Ijichi, et al. 1966.). Sauve laboratory experiments in cell culture measure NAD+ half-lives in the timeframe of 3-5 hours in unstressed cells. Several laboratories have examined the responsiveness of NAD+ metabolism to level of Nampt expression, and have determined that Nampt level determines cellular NAD+ level (Yang, et al. 2007a.). Nampt appears to be induced by different stresses, such as reduced nutrient availability and exercise (Costford, et al. 2010.). Fulco et al. established a link with Nampt transcription linked to AMP-activated protein kinase (AMPK) activation (Fulco, et al. 2008.). Recent work by the Chang laboratory has also established that cAMP production can activate NAD+ biosynthesis, presumably also through AMPK activation (Park, et al. 2012.). Some other enzymes that may be dynamically regulated include NMNAT-2. This adenylyltransferase is limiting in injured axons, and its targeted degradation may lead to rapid NAD+ depletion and may stimulate axon degeneration (Gilley and Coleman 2010.).
Key questions of interest include why NAD+ metabolism should be regulated in the first place? One possible explanation is that NAD+ levels are important for optimizing metabolic performance during different nutritional situations; in light of the key involvement of NAD+ in key metabolic pathways (glycolysis, fermentation, pyruvate dehydrogenase, TCA cycle and oxidative phosphorylation). In fact, dynamic regulation of NAD+ metabolism by nutritional stress, while not preserved in specific details, is phylogenetically conserved from yeast to humans. The downstream coupling of powerful signaling enzymes called sirtuins, which are sensitive to NAD+ concentrations, establishes a second set of effectors that are cued by these NAD+ biosynthetic changes.
1.2. Sirtuins as NAD+ consuming enzymes
Sirtuins have emerged in the last decade as an essential family of enzymes in the regulation of eukaryotic metabolism. In mammalians, sirtuins control whole body metabolic homeostasis and are postulated as promising targets for multiple pathophysiological states, including insulin resistance, cardiovascular disease, neurodegeneration and cancer (Houtkooper, et al. 2012, Nakagawa and Guarente 2011.).
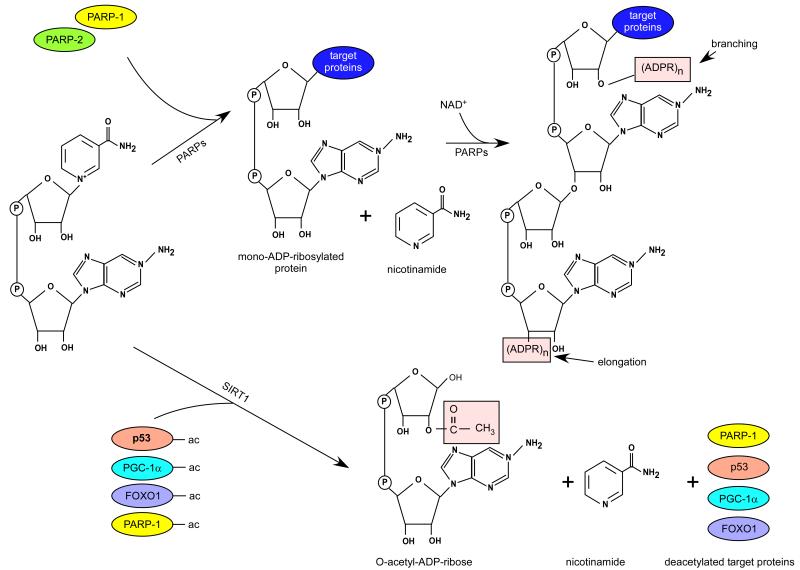
The first sirtuin, Sir2 (silent information regulator 2) was identified almost three decades ago as a protein contributing to gene silencing (Ivy, et al. 1986, Shore, et al. 1984.). However, Sir2 remained as a largely overseen protein until Kaeberlein and collaborators demonstrated in 1999 how Sir2 could influence yeast replicative lifespan (Kaeberlein, et al. 1999.). Additional copies of Sir2 increased yeast replicative lifespan by 30%, while ablation of the Sir2 gene had the opposite effects, reducing life span by 50% (Kaeberlein, et al. 1999.). A critical breakthrough in the sirtuin world came immediately after, when Sir2 was demonstrated to be an NAD+-dependent deacetylase enzyme (Imai, et al. 2000.). Unlike all previously described deacetylases (HDACs Type I and II), Sir2 coupled the removal of acetyl modifications on lysine residues to the consumption of NAD+, providing NAM and O-acetyl-ADP ribose as side products (Figure 3). The coupling of the deacetylase reaction to NAD+ at a Km around the intracellular concentration of NAD+ immediately suggested a potential link between Sir2 activity and the metabolic/redox status of the cell (Guarente 2000, Imai, et al. 2000.). This notion was further supported by a possible implication of Sir2 and its invertebrate orthologs as effectors of the metabolic adaptations triggered by caloric restriction (see (Canto and Auwerx 2009.) for review). However, the consistency and amplitude of the effects of Sir2 orthologs in organismal lifespan and their role as key mediators by which calorie restriction enhances lifespan in lower eukaryotes are still a matter of debate (Burnett, et al. 2011, Kaeberlein and Powers 2007, Lombard, et al. 2011, Viswanathan and Guarente 2011.). While there are also some caveats on the mammalian translation of the link between Sir2 and lifespan, it is nevertheless true that the mammalian sirtuins have key role in metabolic regulation, as will be discussed below.
Figure 3. An overview of the deacetylation reactions catalyzed by SIRT1 and mono/poly(ADP-ribosyl)ation reactions catalyzed by PARPs.
Sir2 unfolded into 7 mammalian homolog family members (SIRT1-7). The 7 mammalian sirtuins share a conserved catalytic domain of 275 aminoacids and their expression is quite ubiquitous (Michan and Sinclair 2007.). The different members of the sirtuin family, however, show distinct features that might endow them with specific functions. An initial difference can be found in their subcellular localization: SIRT1 can shuttle between the nucleus and the cytosol, and its predominant localization varies depending on the cell type and environmental cues (Michishita, et al. 2005, Tanno, et al. 2007.). SIRT2 is predominantly cytosolic (Michishita, et al. 2005.). In contrast, SIRT3, SIRT4 and SIRT5 are considered mitochondrial proteins (Hallows, et al. 2008, Michishita, et al. 2005.), whereas SIRT6 and SIRT7 are nuclear. However, while SIRT6 is located in the heterochromatin, SIRT7 is mostly found in the nucleolus (Michishita, et al. 2005.).
A second key difference between sirtuins can be found at the level of their catalytic activity. Originally, Sir2 was characterized as a deacetylase enzyme (Imai, et al. 2000.). However, its spectrum of functions has largely expanded in mammals. SIRT1, SIRT2 and SIRT3 maintain a strong (North, et al. 2003, Schwer, et al. 2002, Vaziri, et al. 2001.), while, SIRT4-6 display weak deacetylase activity. Instead, SIRT4 and SIRT6 might rather act as NAD+-dependent mono-ADP-ribosyltransferases (Haigis, et al. 2006, Liszt, et al. 2005.). SIRT5 has recently been reported to amplify the spectrum or sirtuin functions, being able to act as a demalonylase and desuccinylase enzyme (Du, et al. 2011.). In this sense, it wouldn’t be surprising if new de-acylation activites are identified within the sirtuin family in the near future. SIRT7 seems to predominantly act as a deacetylase, but only a few substrates have been identified, such as p53 (Vakhrusheva, et al. 2008b.) and H3K18 (Barber, et al. 2012.).
The activity of sirtuins is characterized by its NAD+ dependence. Kinetic studies have determined that the Km of most sirtuins for NAD+ are in the range of 100-300 μM (see (Houtkooper, et al. 2010.) for review). Intracellular concentrations of bioavailable NAD+ are still to this date not easy to determine. While most papers report fluctuations of NAD+ concentrations between 200 and 500 μM (Houtkooper, et al. 2010.), these estimations do not take into account cellular compartmentalization or whether the measured NAD+ is freely available or protein-bound. Considering that freely available NAD+ is only a fraction of the total NAD+ content of the cell, it is likely that the activity of sirtuins could truly be rate-limited by NAD+ in certain scenarios.
A number of interventions aimed to increase NAD+ bioavailability have been shown to impact on sirtuin activity. For example, dietary supplementation with NAD+ precursors, such as NMN or NR enhances sirtuin, at least SIRT1 and SIRT3, activation in rodent tissues (Canto, et al. 2012, Yoshino, et al. 2011.). Physiologically, NAD+ levels generally fluctuate within a 2-fold range (Chen, et al. 2008, Houtkooper, et al. 2010, Rodgers, et al. 2005.), which is a fine range to affect sirtuin activity. In general, it has been observed that sirtuins are activated in situations of energy stress, including exercise (Canto, et al. 2009, Canto, et al. 2010.), and nutrient deprivation (fasting or caloric restriction) (Canto, et al. 2010, Rodgers, et al. 2005.). All these situations are also characterized by increases in NAD+ levels (Canto, et al. 2009, Canto, et al. 2010, Chen, et al. 2008, Costford, et al. 2010, Rodgers, et al. 2005.). In addition, NAD+ fluctuates in a circadian fashion according to feeding/fasting cycles (Nakahata, et al. 2009, Ramsey, et al. 2009.). While a causal link has not been demonstrated to date, the fact that SIRT1 activity also changes in a circadian fashion (Asher, et al. 2008, Nakahata, et al. 2008.) strongly suggest that NAD+ levels could act as a determinant for these shifts.
It is important to note that sirtuins are also tightly regulated by NAM, a product of their catalytic activity. Actually, NAM is also a reaction product of other NAD+ consuming enzymes, such as PARPs or cADP-ribose synthases (CD38 and CD157) (Houtkooper, et al. 2010.). This way, enhanced activity of non-sirtuin NAD+ consuming enzymes might not only influence sirtuin activity by reducing the availability for NAD+, but also by increasing NAM levels. In this sense, it is important to note that NAD+ can be generated from NAM via salvage pathways, initiated and rate-limited in mammals by Nampt (Revollo, et al. 2004.). The overexpression of Nampt favours NAD+ synthesis while lowering NAM levels in virtually any mammalian cell tested (Fulco, et al. 2008, Pittelli, et al. 2010, Rongvaux, et al. 2008, van der Veer, et al. 2005.). Consequently, overexpression, or knock-down of Nampt were associated with increases or reductions, respectively, of, at least, SIRT1 activity (Fulco, et al. 2008, Revollo, et al. 2007, van der Veer, et al. 2007, van der Veer, et al. 2005.).
Amongst the whole family of mammalian sirtuins, SIRT1 is the one more deeply studied. SIRT1 might play a crucial role in metabolic homeostasis by regulating the activity of a number of transcriptional regulators (Canto and Auwerx 2012.). The deacetylation by SIRT1 can lead to direct activation or inhibition of the target transcriptional regulator, as well as the modification of their interaction profiles. The spectrum of transcriptional targets for SIRT1 includes key controllers of mitochondrial biogenesis (peroxisome proliferator activated receptor γ coactivator (PGC)-1α), lipid and carbohydrate metabolism (peroxisome proliferator activated receptors (PPARs), sterol regulatory element binding protein (SREBP)-1, liver X receptor (LRX), FOXOs, cAMP response element binding protein (CREB), CREB regulated transcription coactivator 2 (CRTC2), etc.) and cellular proliferation (p53). Given the dual localization of SIRT1 in both the cytoplasmatic and nuclear compartment, it is not surprising that SIRT1 also deacetylates a constellation of cytosolic proteins, including acetyl-coA synthase 1, endothelial nitrogen monoxide synthase (eNOS) and components of the authophagy machinery, including the Atg family of proteins. For an extensive overview on SIRT1 targets, we refer the reader to other recent reviews (Canto and Auwerx 2012.). Broadly, the activation of SIRT1 leads to changes in the acetylation status of these targets, which co-ordinately orchestrate cellular and whole-body metabolism to extract energy from non-carbohydrate sources and using respiration based-routes. This perfectly matches the fact that SIRT1 is activated in situations of nutrient scarcity. Further pinpointing the interaction between SIRT1 and the metabolic status, SIRT1 expression is triggered by nutrient scarcity and other energy stresses, while blocked by nutrient abundance. A number of transcription factors can regulate the expression of SIRT1 under fasting conditions, such as CREB, PPARs, FOXOs or p53 (see Canto and Auwerx 2012). Conversely, transcription factors activated by high glucose availability, such as ChREBP, downregulate SIRT1 levels (Noriega, et al. 2011).
SIRT2 is the only sirtuin residing primarily in the cytoplasm (Michishita, et al. 2005.). An initial functional clue was provided by the finding that SIRT2 acts as a tubulin deacetylase (North, et al. 2003.). At the same time, SIRT2 was demonstrated to be downregulated in human gliomas (Hiratsuka, et al. 2003.), the most frequent malignant brain tumors, which suggested a tumor suppression role. The interest on SIRT2 has re-emerged recently as the identification of SIRT2 targets unfolds. SIRT2 has been shown to target also key metabolic regulators, such as FOXOs (Jing, et al. 2007.), the p65 subunit of NF-kB (Rothgiesser, et al. 2010.) and phosphoenolpyruvate carboxykinase (PEPCK) (Jiang, et al. 2011.), suggesting a role in the regulation of inflammation, gluconeogenesis and the responses to caloric restriction. In addition, SIRT2 has been linked to Hungtinton disease (HD), by acting as a key regulator of sterol biosynthesis (Luthi-Carter, et al. 2010.). Surprisingly, experiments in SIRT2 knock-out mice do not support a major role of SIRT2 in tubulin acetylation, cholesterol biosynthesis or the progression of HD (Bobrowska, et al. 2012.), indicating that either it is dispensable or that compensatory activities might exist. In all, the role of SIRT2 in mammalian biology is still far from established. Transgenic models currently arising will help uncovering the roles of SIRT2.
Probably SIRT3 is the sirtuin that has attracted most attention in the last few years. SIRT3, together with SIRT4 and SIRT5, was identified as a mitochondrial sirtuin. Interestingly, only the deletion of SIRT3, but not other mitochondrial sirtuins, led to mitochondrial protein hyperacetylation (Lombard, et al. 2007.). The target proteins of SIRT3 include mitochondrial respiratory complexes, TCA cycle proteins and enzymes related to lipid metabolism and reactive oxygen intermediates (ROI) detoxification (for review, see (Giralt and Villarroya 2012.)). While no robust phenotype is found on SIRT3−/− mice in normal conditions, they show many layers of defects when nutritionally challenged. For example, fasted SIRT3−/− mice show defects in fatty acid oxidation (Hirschey, et al. 2010.) and ketogenesis (Shimazu, et al. 2010.). Upon caloric restriction, SIRT3 also determines isocitrate dehydrogenase (IDH)2 (Someya, et al. 2010.) and superoxide dysmutase (SOD)2 (Qiu, et al. 2010.) acetylation, which act as key controllers of ROI levels. The impact of SIRT3 in the function of these proteins also provides a possible explanation on why SIRT3 seems protective against cancer development (Bell, et al. 2011, Kim, et al. 2010a.). In general, the activation of SIRT3 procures optimal mitochondrial function and energy synthesis. In agreement with this notion, SIRT3 is positively regulated at the transcriptional level by PGC-1α, a master orchestrator of mitochondrial biogenesis, and in response to fasting and other energy stresses (Hirschey, et al. 2010; Kong, et al. 2010; Palacios, et al. 2009). Fully confirming the critical role of SIRT3 in energy homeostasis, SIRT3−/− mice were more prone to obesity and metabolic disease upon a fat regime (Hirschey, et al. 2011.). Of note, the defects of the SIRT3 null mice do not seem to be explained by a single tissue deficiency (i.e: liver-specific or muscle-specific defects) (Fernandez-Marcos, et al. 2012.), suggesting that the coordinated defect of SIRT3 in multiple tissues might be required to prompt these metabolic phenotypes.
The role of another mitochondrial sirtuin, SIRT4, is far less known. Initial studies identified SIRT4 as a mono-ADP-ribosylase for the glutamate dehydrogenase (GDH) enzyme. Mono-ADP ribosylation by SIRT4 impaired GDH activity, compromising amino-acid induced insulin secretion (Haigis, et al. 2006.). SIRT4 deficient mice display no gross phenotyping abnormalities, but have increased plasma insulin levels in fed, fasted and aminoacid-stimulated situations (Haigis, et al. 2006.). Recently, SIRT4 has also been shown to act as a modulator of fat metabolism in hepatocytes and myocytes. In an opposed fashion to SIRT3, the downregulation of SIRT4 potentiates fatty acid oxidation (Nasrin, et al. 2010.). Given that SIRT4 promotes opposite effects to those of SIRT1 on insulin secretion (Bordone, et al. 2006, Moynihan, et al. 2005.), or SIRT3 on fat oxidation (Hirschey, et al. 2010.), it will be crucial to understand how the activation of these enzymes is regulated and physiologically integrated. In addition, it suggests that mitochondrial sirtuin activation might not just depend on NAD+ availability, and that multiple other regulatory layers might exist.
As SIRT4, SIRT5 is a mitochondrial sirtuin with weak deacetylase activity (Du, et al. 2011.). Still, SIRT5 has been shown to regulate the activity of the carbamoyl phosphate synthase 1 (CPS-1) enzyme through direct deacetylation (Nakagawa, et al. 2009.). CPS-1 plays a crucial role in ammonia detoxification, as it is a critical step in the urea cycle. The deacetylation of CPS-1 by SIRT5 enhances CPS-1 catalytic activity in situations of fasting, allowing to handle ammonia detoxification during this higher amino acid catabolism state (Nakagawa, et al. 2009.). A major breakthrough in the sirtuin field came recently with the finding that the primary function of SIRT5 might not be to act as a deacetylase, but rather as a demalonylase and desuccinylase (Du, et al. 2011.). The relevance of malonylation and succinylation events in the mitochondria will be fertile ground for research in the upcoming years.
SIRT6 is another sirtuin that is gaining a lot of attention recently, due to its crucial roles in genomic DNA stability, metabolism and ageing. Initially, SIRT6 was described as a mono-ADP-ribosylation enzyme (Liszt, et al. 2005.). Later studies, however, indicated that SIRT6 had also critical actions as a histone deacetylase (Michishita, et al. 2008.). SIRT6 null mice die prematurely, displaying severe defects, such as lymphopenia, loss of subcutaneous fat, decreased bone mineral density, hypoglycemia and reduced levels of insulin-like growth factor (IGF)-1 (Mostoslavsky, et al. 2006.). At least some of these effects might be explained by the overactivation of the hypoxia-inducible factor 1α (HIF-1α), which leads to abnormally high glycolytic rates (Zhong, et al. 2010.). In this scenario, SIRT6 was found to act as a co-repressor of HIF-1α function (Zhong, et al. 2010.). In line with the above results, liver-specific deletion of SIRT6 led to increased glycolysis, triglyceride synthesis, reduced beta oxidation, and fatty liver formation (Kim, et al. 2010b.). Strikingly, mice with a neuron-specific defect of SIRT6 are also smaller at birth, but recover normal body weight later and even develop obesity in late life stages (Schwer, et al. 2010.). The mechanisms regulating these phenotypes are not clear yet. Additional knowledge on SIRT6 has been provided by gain-of-function strategies. Overexpression of SIRT6 renders protection against high-fat diet obesity (Kanfi, et al. 2010.) and has been recently shown to increase lifespan in mice (Kanfi, et al. 2012.). SIRT6, therefore, becomes the first sirtuin with genetic evidence for a direct effect on mammalian lifespan.
Finally, SIRT7 might still be the less known sirtuin. SIRT7 is localized in the nucleolus and was described as a component o the RNA polymerase I (Pol I) transcriptional machinery (Ford, et al. 2006.). However, the specific enzymatic activity of SIRT7 and its targets in these complexes remain unclear. Initial hints of a likely deacetylase activity of SIRT7 were confirmed when SIRT7 was reported to be a p53 deacetylase in cardiomyocytes (Vakhrusheva, et al. 2008b.). This way, mice lacking SIRT7 display cardiac hypertrophy, linked to p53 hyperacetylaion. The defects in cardiac morphology dampen the mean and maximum lifespan of SIRT7 null mice (Vakhrusheva, et al. 2008b.). In addition, a role for SIRT7 in cancer, while hypothesized a few years ago (Vakhrusheva, et al. 2008a.), has been recently confirmed by elegant studies showing how the deacetylation of H3K18Ac by SIRT7 is necessary for maintaining essential features of human cancer cells (Barber, et al. 2012.). The possible roles of SIRT7 in chromatin regulation, cellular transformation programs and tumour formation in vivo warrants future research and might also unveil further links between metabolic sensing and tumor development.
When viewed as a whole, it is clear that sirtuins play a key role in metabolic adaptation and in all the processes in the cell that are governed or require changes in energy substrate utilization: from caloric restriction to cell growth and proliferation control. Still, the many ways by which sirtuins might be regulated are still unclear. Their catalytic reaction is NAD+-dependent, but to this date it is still difficult to unequivocally demonstrate that sirtuin activity is determined by physiological fluctuations in NAD+. This does not rule out, however, than in extreme toxicity situations, where NAD+ levels sharply drop by 50-70% (Goodwin, et al. 1978, Pillai, et al. 2005, Skidmore, et al. 1979.), NAD+ might truly become rate-limiting for the sirtuin reaction. The Km of most sirtuins for NAD+ is still not well-determined (Houtkooper, et al. 2010.), however seems different for the members of the sirtuin family suggesting that all sirtuins are not activated at the same time that seems logical given the often opposing biological effect of these proteins. Compartmentalization of NAD+ bioavailability may also refine sirtuin activation, as it might allow sirtuin activation in a compartment specific fashion. Additionally, there are a few examples of proteins whose deacetylation in vivo is primed or impeded by other post-translational marks (for examples, see (Canto, et al. 2009, Murray-Zmijewski, et al. 2008.)) which could help refining subsets of targets to be deacetylated. A canonical example of how sirtuin activity specification must be required is the one constituted by SIRT3 and SIRT4, both of which share cellular compartment and NAD+-dependence, but drive apparently opposite metabolic adaptations. Altogether, logic dictates that sirtuins activity might be influenced by NAD+, but that many additional regulatory layers must exist in order to achieve specific substrate deacetylation and fine-tune their activity to the cellular metabolic needs.
1.3. Enzymology, function and biological significance of poly(ADP-ribose) polymerases
Poly(ADP-ribosyl)ation (PARylation) was identified by Pierre Chambon and colleagues (Chambon, et al. 1963.) initiating a half century long quest of understanding PARP enzyme action. PARP-1, the main enzyme responsible for that biochemical activity, was recognized in 1967 (Shimizu, et al. 1967.). Recently several other PARP enzymes, possessing a catalytic domain similar to that of PARP-1, were identified (PARP-1 to -17 in humans, PARP-1 to -16 in mice) (Ame, et al. 2004.). Besides the PARP domain, responsible for catalytic activity, PARPs are equipped with numerous other domains enabling the execution of a plethora of molecular functions (reviewed in (Ame, et al. 2004, Hottiger, et al. 2010.)). Among others, there are domains for DNA binding (e.g the zinc fingers in PARP-1 (Langelier, et al. 2008, Mazen, et al. 1989, Menissier-de Murcia, et al. 1989.), or SAP domain in PARP-2 (Huber, et al. 2004.)), protein-protein interaction (e.g. BRCT domain in PARP-1 (de Murcia, et al. 1994.), or ankyrin repeats in tankyrases (Smith, et al. 1998.)), or the macro domain in the macro-PARPs for PAR binding (Karras, et al. 2005.). In certain PARP enzymes nuclear, or nucleolar localization signals guide protein transport between organelles (Meder, et al. 2005, Schreiber, et al. 1992.).
Poly(ADP-ribosyl)ation (PARylation) is considered to be an ancient and evolutionarily conserved biochemical reaction. In line with that PARP catalytic domain is highly conserved throughout evolution as shown in sequence analysis studies (Otto, et al. 2005.) and by the discovery of PARP enzymes in plants (Doucet-Chabeaud, et al. 2001, Lepiniec, et al. 1995.), in lower animals (Tewari, et al. 1995.), or certain eubacteria, arhaebacteria and double-stranded DNA viruses (Hassa, et al. 2006, Otto, et al. 2005.). The catalytic domain of the chicken PARP-1 enzyme had been crystallized first (Ruf, et al. 1996.) giving insight into PARP action. The structure of the known catalytic domains of other members of the PARP superfamily displayed high sequence and structural homology with each other (Hottiger, et al. 2010.). Moreover, despite the poor sequence homology, considerable structural homology was observed with the catalytic domain of bacterial ARTs (Hottiger, et al. 2010, Ruf, et al. 1996.).
PARP-1, considered as the prototypical PARP enzyme, cleaves NAD+ and forms large, negatively charged poly(ADP-ribose) (PAR) polymers on a large set of target proteins. The poly(ADP-ribosyl)ation reaction (PARylation) can be divided into three steps: initiation, elongation and branching (Figure 3) (Alvarez-Gonzalez and Mendoza-Alvarez 1995.). In the initiation phase, reaction the glycosidic bond between nicotinamide and ribose is cleaved due to the nucleophylic attack of glutamate, aspartate residues, or the carboxy terminal of acceptor proteins (Bellocchi, et al. 2006.) (positively charged lysine residues were also shown to be PAR acceptors (Altmeyer, et al. 2009.)). Then the mono-ADPR units are bonded via an ester bond (Altmeyer, et al. 2009, Burzio, et al. 1979, Ogata, et al. 1980.). The ADPR moiety remains bound to the acceptor protein, while NAM is released in the reaction. Subsequently, the enzymes catalyze elongation and branching reactions using additional ADPR units from NAD+ leading to formation of branched polymers up to 200 ADPR units (Hayashi, et al. 1983.). The half-life of the polymer is estimated to be less than 1 min, it is rapidly degraded by poly(ADP-ribose) glycohydrolase (PARG) and ADP-ribosyl protein lyase (Kawaichi, et al. 1983, Ueda, et al. 1972.).
How are PARPs activated? The first known activator of PARP-1 was DNA strand breaks (Benjamin and Gill 1980.) and irregular DNA structures (Kun, et al. 2002.). PARP-1 binds to these structures through its zinc fingers that subsequently leads to its activation. To date PARP-1, -2 and -3 had been shown to be induced by DNA damage (Ame, et al. 1999, Boehler, et al. 2011, Menissier-de Murcia, et al. 1989, Rulten, et al. 2011.). The majority of DNA-induced PARP activity is covered by PARP-1 (85-90%), while PARP-2 is considered to be responsible for the rest (Schreiber, et al. 2002, Szanto, et al. 2011.). It seems that not all PARPs are active, or build polymers: PARP-13 is inactive, PARP-7, PARP-10 and PARP-16 perform only mono-ADP-ribosylation (Di Paola, et al. 2012, Kleine, et al. 2008, Leung, et al. 2012, Ma, et al. 2001.), while PARP activity of PARP-9 and PARP-13 is under debate.
PAR molecules may be introduced onto PARP-1 itself (autoPARylation), or onto other proteins (transPARylation). PARP-1 autoPARylation efficiently inhibits PARP-1 activity (Kawaichi, et al. 1981, Zahradka and Ebisuzaki 1982.) due to strong electrostatic repulsion between DNA and PAR. PARP-2 has also been reported to perform autoPARylation (Ame, et al. 1999.) suggesting the existence of a similar autoPARylation cycle as PARP-1. Inhibition of PARP-1 by autoPARylation seems an exquisite mechanism to avoid uncontrolled and excessive PARP-1 activity. The inhibitory effect of autoPARylation can be reverted by PARG that removes PAR polymers creating a reversible PARylation cycle for PARP-1 (Erdelyi, et al. 2009, Ying and Swanson 2000.). Indeed, inhibition, or deletion of PARG blocks PARP-1 in a PARylated state and therefore protect against PARP-1 mediated NAD+ and ATP depletion and the consequent cell death (Bakondi, et al. 2004, Erdelyi, et al. 2009, Ying and Swanson 2000.).
There are numerous posttranslational pathways through which the activity of PARP enzymes can be regulated. Reversible phosphorylation regulates PARP-1 (for a proteomic approach see (Gagne, et al. 2009.), for review, see Virág and Bürkle in this series) and tankyrases (Ha, et al. 2012, Li, et al. 2012, Yeh, et al. 2006.). PARP-1 is acetylated and activated by p300/CBP-association factor (PCAF) and p300 (Hassa, et al. 2005, Rajamohan, et al. 2009.), while deacyetylation by SIRT1 leads to radical decrease in PARP activity (Rajamohan, et al. 2009.). PARP-2 is acetylated by PCAF and GCN5L (Haenni, et al. 2008.). PIASy, a SUMO ligase physically interact and modify PARP-1 upon heat shock (Martin, et al. 2009.). PARP-1 can be mono-ADP-ribosylated and activated by PARP-3, or SIRT6 (Loseva, et al. 2010, Mao, et al. 2011.). PARP-1 activity seems to be linked to cellular calcium homeostasis (Bakondi, et al. 2003, Wyrsch, et al. 2012.).
PARP enzymes were related to numerous biological processes. The first function to be discovered for PARP-1 (and later PARP-2 and -3) was its involvement in DNA repair (Durkacz, et al. 1980, Purnell and Whish 1980.). Later, the involvement of these PARP enzymes and tankyrases in the maintenance of genomic integrity was evidenced (reviewed in this series by Valérie Schreiber and Francoise Dantzer). In our current understanding, under non-stress conditions the action of PARP-1 and -2 are not essential for efficient DNA repair (Allinson, et al. 2003, Bai, et al. 2011a, Bai, et al. 2011b, De Vos, et al. 2012.). However, deletion of PARP-1, PARP-2, or the application of PARP inhibitors leads to sensitization against DNA damaging agents (MNNG, ionizing radiation, etc.) (Menissier-de Murcia, et al. 1997, Menissier-de Murcia, et al. 2003, Wang, et al. 1995.). It seems that PAR polymers act as a scaffold matrix around DNA damage sites that other DNA repair enzymes bind to (Karras, et al. 2005, Mortusewicz, et al. 2007, Tartier, et al. 2003.). PARP-1 and -2 participate in the resolution of single strand breaks, base excision repair (Dantzer, et al. 2000, Schreiber, et al. 2002.) and double strand break repair (Langelier, et al. 2012, Szanto, et al. 2012.). PARP-1 has antirecombinogenic activity (Morrison, et al. 1997.) that consequently protects against retroviral infections (Ha, et al. 2001.).
Insufficient DNA repair on a longer timeline leads to either cell death, or to the accumulation of mutations, genomic instability that ultimately induce tumorigenic transformation. Indeed, the lack of PARP-1 enhanced the number of sister chromatid exchange events when challenged by DNA damaging agents (Menissier-de Murcia, et al. 1997, Schreiber, et al. 1995, Wang, et al. 1995.), however, to date, it seems that the lack of PARP-1, or -2 alone, under non-stress conditions, does not lead to tumorigenic transformation (Menissier-de Murcia, et al. 2003, Wang, et al. 1995.). It suggests that other parallel DNA repair pathways cope with DNA damage in the absence of PARPs. However, the simultaneous removal of PARP-1 and -2, or other DNA repair enzymes, such as ataxia-telangiectasia mutated (ATM) leads to embryonic lethality (Huber, et al. 2004, Menissier-de Murcia, et al. 2003.), or tumorigenic transformation as in the case of p53−/− PARP-1−/−, or p53−/− PARP-2−/− mice (Nicolas, et al. 2010, Tong, et al. 2001.).
The extent of PARP activation has major influence on the fate of the cell (reviewed in this series by László Virág, Agnieska Robaszkievicz, Jose Vargas and Javier Oliver). Obviously, if DNA damage is repairable, PARP activation contributes to the survival of the cell as discussed above. Unrepairable DNA damage induces apoptosis that is an energy intensive process that disposes of cells in a safe manner. Extensive PARP activation – as suggested by Berger and colleagues (Berger 1985.) – may markedly reduce cellular NAD+ content. NAD+ resynthesis through NMNAT and phosphoribosyl pyrophosphate synthetase (PPS) is energy consuming therefore reducing cellular ATP content. Further metabolic rearrangements encumber the replenishment of ATP: slowdown of glycolytic flow due to NAD+ loss (Ying, et al. 2002.), the reversal of the activity of F1/F0 ATPase (synthase activity shifts to ATPase activity) (Ha and Snyder 1999.) and the opening of mitochondrial transition pores (Virag, et al. 1998.). The lack of energy prevents the progression of the apoptotic program and turns cell death into necrosis (Leist, et al. 1997, Leist, et al. 1999.). The fact that PARP activation affect cell survival, moreover switches apoptosis into necrosis suggested that the application of PARP inhibitors may have beneficial effects in pathological states associated with oxidative stress (e.g. reperfusion injuries, or inflammatory pathologies) (Virag and Szabo 2002.).
PARP-1 is involved in transcriptional regulation at numerous levels (reviewed in this series by Lee Kraus and Michael Hottiger): it may modulate chromatin structure (de Murcia, et al. 1986, Quenet, et al. 2009.), bind to enhancer sequences, or promoters (Krishnakumar, et al. 2008.), act as a transcriptional cofactor (Oliver, et al. 1999.), or may promote chromatin insulation (Yu, et al. 2004.) leading to complex gene expression rearrangements (Frizzell, et al. 2009, Simbulan-Rosenthal, et al. 2000.). It is under debate whether the catalytic activation of PARP-1 is necessary in every transcriptional event (e.g. in the case of NFkB activation (Hassa, et al. 2001.)), however it seems that global and local NAD+ levels affect PARP action at transcription foci (Kraus 2008, Zhang, et al. 2012.). Besides, PARP-1, other PARP enzymes influence transcription (e.g PARP-2 (Szanto, et al. 2012.), or PARP-14 (Mehrotra, et al. 2011.)).
The above detailed biological functions of PARPs act jointly in complex physiological, or pathophysiological scenarios. PARP enzymes have major impact on inflammatory diseases (Bai and Virag 2012, Levaot, et al. 2011, Mehrotra, et al. 2012, Yelamos, et al. 2006.). PARPs influence the maturation and function of immune cells (Bai and Virag 2012.). PARP-1 is necessary for the appropriate activation of numerous proinflammatory transcription factors (e.g. NFAT, NFκB, AP-1, YY1, or sp1) that have key role in producing chemokines (Bai and Virag 2012, Oliver, et al. 1999.), cytokines, adhesion factors and other inflammatory mediators (matrix metalloproteinases, cyclooxyganse-2, or inducible NO synthase) (Virag and Szabo 2002.). Under inflammatory conditions oxidative stress is largely enhanced that leads to cell death that is diverged towards necrosis by PARP activation that further enhance the inflammatory response (Virag and Szabo 2002.). Vast amount of data had been assembled suggesting that inflammatory processes can be quenched by the application of PARP inhibitors (reviewed in (Bai and Virag 2012.)).
Recent data suggests the involvement of PARPs in metabolic regulation (Bai and Canto 2012.) that – similarly to the inflammatory role of PARPs – stem from multiple roots. As discussed above, prolonged PARP activation through depleting cellular NAD+ pools hamper cellular energy metabolism: glycolytic slowdown (Ying, et al. 2002.) and a rapid shutdown of mitochondrial function (Bai, et al. 2001, Bai, et al. 2007a, Cipriani, et al. 2005, Virag, et al. 1998.). Inversely, upon the lack, or inhibition of PARP-1, or -2 mitochondrial activity is not only preserved, but are further enhanced due to the activation SIRT1 (Bai, et al. 2011a, Bai, et al. 2011b.). It seems therefore that the level and activity of PARP-1, or -2 activity is in strong correlation with mitochondrial activity (Bai and Canto 2012.). PARPs are related to other metabolic processes by interacting with several metabotropic receptors (Bai and Canto 2012.) and by influencing energy intake (Asher, et al. 2010, Bai, et al. 2011b.). These metabolic changes together influence insulin and glucose sensitivity, adipogenesis and body weight (Bai, et al. 2011a, Bai, et al. 2011b, Bai, et al. 2007b, Erener, et al. 2012a, Erener, et al. 2012b, Mangerich, et al. 2010.). PARP-5a, -5b, -7 and -14 also seems to influence metabolism, however their action is yet blurry (Bai and Canto 2012.).
2. Levels of SIRT - PARP interaction
2.1. Interaction of PARPs and SIRTs through the common NAD+ substrate
As discussed in sections 1.2 and 1.3 both PARPs and SIRTs are NAD+ dependent enzymes that makes it likely that they may compete for the limiting NAD+ substrate. Most studies report 200-500 μM intracellular NAD+ concentrations, however the NAD+ levels in different compartments (mitochondria, nucleus, or cytosol) are still debated (Houtkooper, et al. 2010.). As discussed in detail in section 1.2, SIRT1 activity (and probably the activity of further members of the sirtuin family) is linked to fluctuations in NAD+ levels (Asher, et al. 2008, Canto, et al. 2012, Imai, et al. 2000, Nakahata, et al. 2008.) as the Km of SIRT1 falls in the range of physiological cellular NAD+ changes (Houtkooper, et al. 2010.).
The Km of PARP-1 towards NAD+ falls in the low micromolar range (20-60 μM) (Ame, et al. 1999, Mendoza-Alvarez and Alvarez-Gonzalez 1993.) suggesting that physiological fluctuations in NAD+ levels are unlikely to affect PARP-1 activity. In contrast, the Km of PARP-2 towards NAD+ is higher (around 130 μM) (Ame, et al. 1999.) that is comparable to the one of SIRT1 (Houtkooper, et al. 2010.). Also, PARP-1 had been described as an effective enzyme in NAD+ degradation (Ame, et al. 1999.) displaying high catalytic turnover when compared to SIRT1 (Bai and Canto 2012, Bai, et al. 2011b.). This is further highlighted by the fact that the maintenance of local NAD+ levels seems important upon PARP-1 activation. NMNAT-1 has been shown to recruit to sites of PARP-1 activation upon oxidative stress (Berger, et al. 2007.), or in transcriptional events (Zhang, et al. 2012.). It seems that NMNAT-1 recruitment does not only enhance local NAD+ availability, but activate PARP-1 in an NAD+-independent manner (Zhang, et al. 2012.). PARP-1 is responsible for the majority of PARP activity (section 1.3, (Schreiber, et al. 2002, Szanto, et al. 2011.)), while the rest is mostly covered by PARP-2.
The drop in NAD+ levels upon excessive DNA damage due to PARP activation is a long-known fact (Berger 1985.). Under such conditions NAD+ levels may drop to 20-30% of the original that is likely to rate limit sirtuin enzymes (Houtkooper, et al. 2010.). SIRT1 activity is largely reduced under these conditions (Pillai, et al. 2006, Qin, et al. 2012, Rajamohan, et al. 2009.) that might be followed by decreased SIRT1 expression (Qin, et al. 2012.). It is logical to assume that the activity of other nuclear sirtuins will drop under these conditions, however it is not known whether extranuclear sirtuins would respond to these insults, or would remain intact.
When the biochemical changes upon deletion, or inhibition of PARP-1 were analyzed we found that NAD+ leveles were induced (20-100% as a function of cell model, or tissue) in animal and cellular models (Bai, et al. 2011b.). It is likely that PARP-1 activity is a major activity in NAD+ degradation and consequently in NAD+ turnover (Houtkooper, et al. 2010.), therefore the lack of PARP-1 activity elevates NAD+ levels. That induction is translated into higher SIRT1 activity and better metabolic performance (Bai, et al. 2011b.).
As previously mentioned, the affinity of PARP-2 to NAD+ and the rate of NAD+ degradation is similar to SIRT1, therefore it is unlikely that these enzymes could limit NAD+ for one another. In line that we were unable to detect differences NAD+ levels of PARP-2+/+ and −/− cells and tissues under non-stress and oxidative stress conditions (Bai, et al. 2011a, Szanto, et al. 2011.)
A particularity on the interaction between PARP-1 and PARP-2 with sirtuins is that these two models seem to specifically target SIRT1: neither cytoplasmic SIRT2, nor mitochondrial SIRT3 activities were increased by the absence of PARP-1 or PARP-2 (Bai, et al. 2011a, Bai, et al. 2011b.). In the case of PARP-2, the nature of this selectivity is clearer, as it roots on the direct regulation of the SIRT1 promoter (discussed in the following section). The case of PARP-1 is a bit more complicated, as the modulation of NAD+ levels could potentially impact on all sirtuins. The reasons for the specificity might be that the changes in NAD+ levels promoted the reduction of PARP-1 activity could be restricted to the nucleus (Bai, et al. 2011b.). This is logical, as PARP-1 is predominantly a nuclear protein. Another possible explanation is that different sirtuins might have different windows of sensitivity for NAD+. Confirming this, recent efforts from the Denu lab have demonstrated that SIRT6 binds to NAD+, even in the absence of acetylated substrate, at a Kd around 27 μM, which is a concentration far lower than that of intracellular NAD+ content (Pan, et al. 2011.). This means that NAD+ might rarely be rate-limiting. Hence, SIRT6 activity might not act as an NAD+ sensor and, rather, other regulatory mechanisms, such as specific protein binding or post-translational modifications, determine SIRT6 activity. In fact, it is conceivable that NAD+ could just be permissive for certain sirtuins, and that the true switch for their activity is found in changes in their protein interactions or post-translational modifications. This might explain why only a subset of sirtuins (such as SIRT1 in the PARP-1 KO mice) is responsive to fluctuations in NAD+. Of note, also recent studies have highlighted how post-translational modifications might change the affinity of sirtuins for NAD+, therefore enhancing or blocking their sensing capabilities. This is the case actually for SIRT1, as phosphorylation by PKA during fasting enhances the sensitivity of SIRT1 for NAD+ (Gerhart-Hines, et al. 2011.).
2.2. Post-translational modifications
While SIRT1 and PARP activities might influence each other through the competition for a limited NAD+ pool, other events, such as their interaction with different proteins and the impact of diverse post-translational modifications, act also as key determinants.
A first crucial possibility would be the cross-action of both activities, i.e: that PARPs could PARylate SIRT1 and, conversely that SIRT1 could deacetylate PARPs. Very little information exists on whether SIRT1 could be a substrate for PARylation. However, SIRT1 is not PARylated in C2C12 myotubes when PARP activity is triggered by exposure to genotoxic hydrogen peroxide concentrations (Bai, et al. 2011b.). This suggests that endogenous SIRT1 might not be a direct PARylation target, even though additional scenarios of enhanced PARP activity will have to be tested in order to solidify this conclusion.
Conversely, it could be hypothesized that PARP-1 might be targeted by SIRT1 deacetylase activity. In line with this hypothesis, reduction of PARP activity is observed upon SIRT1 activation (Kolthur-Seetharam, et al. 2006.). Given the relatively high Km and low Vmax of sirtuins, it is unlikely that sirtuin activity ever rate-limits NAD+ availability for PARP-1, characterized by a 5-fold lower Km and much stronger Vmax of PARP-1 then the one of SIRT1 (Houtkooper, et al. 2010.) and section 2.1. Clues to our understanding on how SIRT1 might impact on PARP activity were given when PARP-1 was identified to be an acetylated protein (Hassa, et al. 2005, Rajamohan, et al. 2009.) and section 1.3. In cardiomyocytes, PARP-1 acetylation was increased by mechanical stress, phenylephrine or angiotensin-II (Rajamohan, et al. 2009.). This increase in PARP-1 acetylation was coupled to enhanced catalytic activity and was enough to trigger PARP-1 activation in the absence of DNA damage (Rajamohan, et al. 2009.).
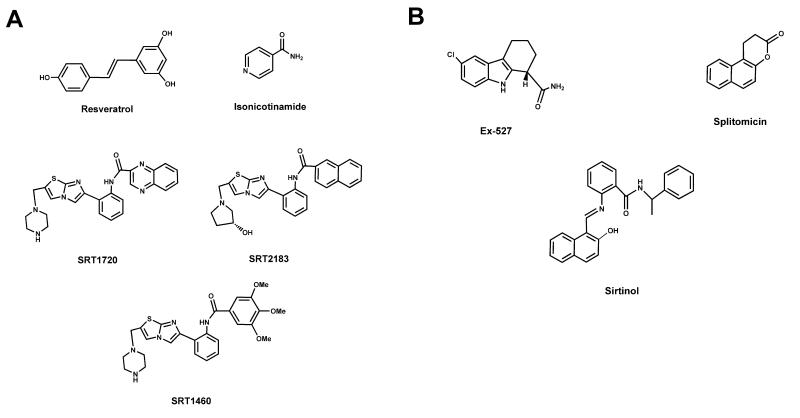
Following the discovery that PARP-1 activity is influenced by its acetylation status, Rajamohan and colleagues demonstrated that SIRT1 could directly deacetylate PARP-1. Overexpression of SIRT1 or treatment with resveratrol, as a SIRT1 agonist, both led to the deacetylation of PARP-1 in cell cultured models (Rajamohan, et al. 2009.). Finally, the authors also demonstrated that SIRT1-mediated deacetylation blocks PARP-1 catalytic activity (Rajamohan, et al. 2009.). Altogether, these observations set a scenario in which enhanced SIRT1 activity would reduce PARP-1 activity via direct deacetylation. However, if PARP-1 activity is prompted through DNA damage, this will reduce NAD+ availability, hence blocking the ability of SIRT1 to retain PARP-1 in a deacetylated (low activity) state.
The direct influence of sirtuins on PARP activity was further reinforced when trying to elucidate why SIRT6 deficient mice display genomic instability. Remarkably, it was found that SIRT6, but not other nuclear sirtuins, is directly recruited to the sites of DNA double-strand breaks and enhances the efficiency of non-homologous end joining and homologous recombination after paraquat treatment (Mao, et al. 2011.). In these experiments, PARP-1 was found to be a mono-ADP-ribosylation substrate for SIRT6 (Mao, et al. 2011.). Both proteins bind to each other, and the binding is somehow potentiated by DNA damage. SIRT6 overexpression did not stimulate DNA repair in PARP-1 knock-out cells, indicating that PARP-1 is required to mediate the effects of SIRT6 (Mao, et al. 2011.). Key experiments demonstrated that while PARP-1 can be mono-ADP-ribosylated in at least 6 sites, only K521 is the only one affected by SIRT6. Of note, SIRT6 did not seem to affect the acetylation status of PARP-1 (Mao, et al. 2011.).
As mentioned in previous chapters, it will be of crucial interest to understand in which scenarios sirtuins might be selectively activated and how this is molecularly channelled. Illustrating this point, the cases above show how SIRT1 and SIRT6 exert theoretically opposite effects on PARP-1 activity (inhibition and activation, respectively). Hence, it should be expected that the docking of specific sirtuins to DNA locations or differential protein interaction might crucially determine sirtuin activity. An example of the latter case can be found in Deleted in Breast Cancer-1 (DBC-1), a protein that can selectively bind the catalytic domain of SIRT1, negatively regulating its activity (Kim, et al. 2008, Zhao, et al. 2008.). Upon genotoxic stress, a condition that triggers PARP-1 activation, DBC-1 is phosphorylated by ATM at Thr454, creating a second binding site for SIRT1 (Yuan, et al. 2012, Zannini, et al. 2012.). This leads to enhanced binding between SIRT1 and DBC-1, hence abolishing SIRT1 activity (Yuan, et al. 2012, Zannini, et al. 2012.). This would provide a very elegant mechanism for shutting down SIRT1 and relieve the inhibition of PARP-1 exerted via deacetylation, while promoting simultaneously PARP-1 activation via SIRT6 mediated mono-ADP-ribosylation.
Acetylated residues have also been identified in other PARP enzymes, such as PARP-2 (Haenni, et al. 2008.). However, whether the acetylation status of these residues is modulated by sirtuins is not currently clear. Similarly, the identification of mitochondrial PARP activity (Du, et al. 2003, Lai, et al. 2008, Pankotai, et al. 2009.), opens a whole new world for possible direct cross-regulation between PARP enzymes with mitochondrial sirtuins (SIRT3-5).
2.3. SIRT - PARP interaction through the regulation of gene expression
Marked changes in SIRT1 expression is capable of influencing metabolic and energetic balance. In humans, SIRT1 mRNA levels and certain SNPs in the SIRT1 gene correlated well with enhanced energy expenditure, insulin sensitivity (Rutanen, et al. 2010.), insulin secretion (Dong, et al. 2011.), or predisposition to obesity (Clark, et al. 2012, Zillikens, et al. 2009a.).
The activity of the SIRT1 promoter had been shown to be controlled by several transcription factors, such as CREB (cAMP response element-binding protein), ChREBP (carbohydrate response element binding protein) (Noriega, et al. 2011.), FOXOs (forkhead box transcription factor O), p53 (Nemoto, et al. 2004.), HIC1 (hypermethylated in cancer 1) (Chen, et al. 2005, Zhang, et al. 2007.), PPARs (peroxisome proliferator-activated receptors) (Han, et al. 2010.) and c-Myc (Yuan, et al. 2009.). Most of these transcription factors integrate nutritional signal (Nemoto, et al. 2004, Noriega, et al. 2011.). We have described the presence of PARP-2 on the SIRT1 promoter and provided evidence that PARP-2 acts as a suppressor of SIRT1 transcription (Bai, et al. 2011a, Szanto, et al. 2011.).
PARP-2 binds to DNA in the proximal region of the SIRT1 promoter (−1 to −91 region of the mouse SIRT1 promoter) (Bai, et al. 2011a.). This region is on one hand directly adjacent to the region where FOXOs bind (−91 to −202 region of the mouse SIRT1 promoter) (Nemoto, et al. 2004.), while on the other it’s sequence is highly conserved among mammals and shows conservation when compared to the distantly related sequence of the promoter of SIRT1 in Xenopus (Bai, et al. 2011a.). Depletion of PARP-2 enhanced the activity of the SIRT1 promoter that translated into higher SIRT1 mRNA and protein levels in skeletal and smooth muscle, liver, brown adipose tissue and pancreas as shown in murine and cellular models (Bai, et al. 2011a, Szanto, et al. 2011.). Interestingly, although in brown adipose tissue SIRT1 protein levels are enhanced the induction of mitochondrial activity was not detected (Bai, et al. 2011a.) suggesting yet unknown tissue-specific mechanisms that limit the phenotypical manifestation of PARP-2, or SIRT1 action. Likewise, tissue specific gene expression changes alter the effects of SIRT1 induction in PARP-2−/− mice, wherein in contrast to pancreatic SIRT1 overxpression that ameliorates β cell function (Moynihan, et al. 2005.) PARP-2 deficiency hampers β cell expansion leading to pancreatic dysfunction (Bai, et al. 2011a.) (discussed in detail in section 3.1.4).
Alterations in NAD+ levels upon the depletion of PARP-2 were minor or negligible in cellular models and inconsistent in in vivo experiments (Bai, et al. 2011a, Schreiber, et al. 2002, Szanto, et al. 2011.). That suggest that activation of SIRT1 upon PARP-2 depletion seems to rely primarily on transcriptional effects and unlikely on activation through enhanced NAD+ availability. PARP-2 seems specific for the SIRT1 promoter, as the depletion of PARP-1 did not alter promoter activity (Bai, et al. 2011a.). To date, no further direct regulation of other sirtuin genes by PARPs has been clearly evaluated.
3. Physiological processes influenced by SIRT - PARP interaction
3.1. Metabolism
Experiments in cell lines and animal models have shown that sirtuins act as key regulators of oxidative metabolism and global metabolic homeostasis. The multiple levels of interaction between PARP enzymes and sirtuins (see section 2), predict, therefore, that the modulation of PARP activity could also have a strong impact on energy metabolism.
While many of the original studies showed a negative correlation of PARP activity and sirtuin activity in situations of supraphysiological oxidative stress or DNA damage, it is worth mentioning that this relation has recently been found also in physiological scenarios. For example, PARP activity is largely increased upon high-fat feeding, when SIRT1 activity is lower (Bai, et al. 2011b.). Oppositely, PARP activity is lower in muscle after an overnight fast, where enhanced SIRT1 activity is observed (Bai, et al. 2011b.). A recent report has also highlighted how higher PARP activity is observed in aged rodent tissues, leading to decreased NAD+ content and limiting SIRT1 activity, even though SIRT1 protein content is higher (Braidy, et al. 2011.). All these observations indicate how genetical and physiological variations in PARP activity might have a large impact on sirtuin activity, and, consequently, on global metabolism.
3.1.1. PARP - SIRT1 interactions in food intake behavior
PARP-1 null C57Bl/6 mice display a clear metabolic phenotype, characterized by lower body weight gain upon ageing and high-fat feeding. Strikingly, this happens despite the increased food intake observed in PARP-1 knockout mice (Bai, et al. 2011b, Devalaraja-Narashimha and Padanilam 2010.). Moreover, recent data indicates that PARP-1 plays role in the regulation of the circadian entrainment of feeding behavior and body temperature cycles (Asher, et al. 2010.). Interestingly, also SIRT1 is a key regulator of the core circadian clock molecular machinery (Asher, et al. 2008, Nakahata, et al. 2008.). The regulation of NAD+ bioavailability might constitute an attractive mechanism tying the circadian fluctuations of PARP-1 and SIRT1 activities. Essentially, the expression levels of Nampt, the critical rate limiting enzyme in the mammalian NAD+ salvaging pathway, display a robust diurnal oscillation, with a peak around the beginning of the dark period in mice, in line with the maximal peak for the circadian fluctuation of SIRT1 activity (Nakahata, et al. 2009, Ramsey, et al. 2009.). SIRT1 negatively regulates CLOCK:BMAL-1 transcriptional activity, which is a key positive controller of Nampt expression (Nakahata, et al. 2009, Ramsey, et al. 2009.). Hence, the activation of SIRT1 shuts down Nampt expression. This will likely promote a decrease in NAD+ levels low enough to limit SIRT1. It is likely that PARP-1 activity could also rise simultaneously, as the decrease in SIRT1 activity should lead to increased PARP-1 acetylation and activity. This would further limit NAD+ availability for SIRT1, completely shutting down its activity. Once SIRT1 activity is low enough, CLOCK:BMAL-1 activity will be increased, and Nampt expression will be slowly recovered, reaching full circle.
3.1.2. PARP - SIRT1 interaction in the regulation of energy expenditure
A key element driving the metabolic phenotype of the PARP-1 knock-out mice is their enhanced energy expenditure (Bai, et al. 2011b.). This effect likely derives, at least in part, from a potentiation in SIRT1 activity and the activation of key transcriptional metabolic regulators, such as the transcriptional coactivator PGC-1α (Rodgers, et al. 2005.). It has been shown that PGC-1α activation is linked to enhanced mitochondrial biogenesis and a more oxidative profile of skeletal muscle fibers (Lin, et al. 2002.). Another key downstream effector of SIRT1 contributing to the regulation of oxidative metabolism is the FOXO family of transcription factors. FOXOs are deacetylated by SIRT1 (Brunet, et al. 2004.), prompting their activation and the transcriptional activation of genes linked to lipid oxidation and stress resistance (Banks, et al. 2011, Gross, et al. 2008.). It was therefore reassuring to see that, consistent with SIRT1 activation, mice where PARP activity is impaired, either by genetic or pharmacological means, show a marked deacetylation of PGC-1α and FOXO1 in a key metabolic tissue such as skeletal muscle (Bai, et al. 2011b.). Consistent with the activation of gene programs related to mitochondrial biogenesis, the muscles from PARP-1 deficient mice displayed a large increase in mitochondrial content and an enhanced oxidative profile of their muscle fibers (Bai, et al. 2011b.).
Another key tissue influencing whole body energy expenditure is the brown adipose tissue (BAT), which has a key role in thermogenesis. As seen in muscle, the BAT from PARP-1 deficient mice is characterized by increased NAD+ content and SIRT1 activity, as manifested in the deacetylation and activation of PGC-1α (Bai, et al. 2011b.). This leads to a marked increase in mitochondrial content in the BAT of PARP-1 deficient mice (Bai, et al. 2011b.). Physiologically, this renders the PARP-1−/− mice with a stronger ability to maintain body temperature when exposed to cold compared to their wild-type littermates.
In agreement with the observations in mice, the knock-down of PARP-1 in cultured HEK293 or inhibition of PARP activity, using PJ34 (a pan-PARP inhibitor), in C2C12 myotubes is enough to drive an increase in mitochondrial gene expression and O2 consumption (Bai, et al. 2011b.). Noteworthy, the simultaneous knock-down of SIRT1 largely prevented the increase in cellular respiration triggered by the reduction of PARP activity (Bai, et al. 2011b.). Importantly, when analyzing the expression of a panel of genes related to oxidative metabolism in response to PARP inhibition, it was clear that SIRT1 only participated in the regulation of certain subsets, but not all (Bai, et al. 2011b.). This indicates that reductions in PARP activity leads to a plethora of effects, and that SIRT1 solely controls a few contributing to enhanced mitochondrial respiration and energy expenditure.
The evaluation of PARP-2 deficient mice further consolidated the link between sirtuins and PARP enzymes on energy expenditure. PARP-2 mice also display resistance against high-fat diet-induced obesity, linked to increased energy expenditure and an enhanced oxidative profile of skeletal muscle (Bai, et al. 2011a.). As mentioned in section 2.3, defects in PARP-2 expression also enhance SIRT1 activity through enhancing SIRT1 expression. As PARP-2 is a repressor of the SIRT1 promoter, PARP-2 deletion relieves the repression on the SIRT1 promoter and enhances SIRT1 mRNA and protein levels (Bai, et al. 2011a, Szanto, et al. 2011.). Experiments in C2C12 myotubes demonstrated that the knock-down of PARP-2 triggered mitochondrial gene expression in a SIRT1 dependent fashion.
3.1.3. PARP - SIRT1 interaction in the regulation of fat deposition
This leaner phenotype of PARP-1−/− and PARP-2−/− mice can be explained, at least in part, due to their enhanced energy expenditure when compared to wild-type littermates. However, another attractive mechanism by which PARP deficiency might impact on body weight relies on the direct regulation of fat deposition in white adipose tissues (WAT). Indeed, PARP-1 and -2 deficient mice present a largely reduced size of their WAT depots (Bai, et al. 2011a, Bai, et al. 2011b, Bai, et al. 2007b.). PPARγ is a nuclear receptor that is mainly expressed in white adipose tissue and plays key roles in adipocyte differentiation, lipid synthesis and storage (Heikkinen, et al. 2007.). The lower fat deposition in the PARP-1 and PARP-2 knock-out mice, therefore, might be explained by affecting the activity of PPARγ. PARP-1 and -2 had been shown to physically interact with PPARγ (Bai, et al. 2007b, Miyamoto, et al. 1999.) and were already correlated with WAT tissue mass (Bai, et al. 2007b, Erener, et al. 2012a.). PARylation can be observed in differentiating 3T3-L1 preadipocyte cells and in subcutaneous adipose tissue (Gehl, et al. 2012, Janssen and Hilz 1989.), likely consequent to PARP-1 activation (Erener, et al. 2012a, Janssen and Hilz 1989, Simbulan-Rosenthal, et al. 1996, Smulson, et al. 1995.). Indeed, PARP-1 is recruited to PPARγ target genes in a PAR-dependent manner, allowing a sustained expression of PPARγ and its target genes (Erener, et al. 2012a.). Also PARP-2 can contribute to the adipogenic program, as the lack of PARP-2 hampers the adipocytic differentiation of embryonic fibroblasts and 3T3-L1 cells (Bai, et al. 2007b.). PARP-2 binds to the same sites on promoters as PPARγ and apparently acts as positive cofactor (Bai, et al. 2007b.). Interestingly, the expression of some adipokines, such as leptin or adiponectin, is regulated by PARP-1 and -2 (Bai, et al. 2007b, Erener, et al. 2012a.),
Besides regulation through direct physical interaction the higher SIRT1 activity in PARP-1 and PARP-2 deficient models might have a key role. SIRT1 is known to decrease PPARγ transcriptional activity through direct interaction and docking of transcriptional co-repressors, such as NCoR and SMART (Picard, et al. 2004.) and more recently, PPARγ has been identified as a deacetylation target for SIRT1 (Qiang, et al. 2012.). Activation of PPARγ through TZD decreased PPARγ acetylation levels on K268 and K293 by prompting the binding of SIRT1 (Qiang, et al. 2012.). The deacetylation of PPARγ at these two residues allows the recruitment of the transcriptional coactivator PRDM16 and promotes adipokine production and an upregulation of BAT-like gene expression (Qiang, et al. 2012.). Physiologically, the deacetylation of PPARγ is also triggered by cold exposure and blunted when mice are fed a high-fat diet (Qiang, et al. 2012.). This way, SIRT1-induced deacetylation of PPARγ will promote a brown-like phenotype of the WAT, enhancing energy expenditure, lowering fat deposition and favoring insulin sensitivity. Of note, PARP activity is enhanced upon high-fat feeding, which could limit SIRT1 activity and compromise PPARγ deacetylation that is in line with the enhanced insulin sensitivity and lower fat storage of PARP-1 and PARP-2 null mice, even if browning effects have never been closely examined. Of note, the regulation of SIRT1 alone might not be enough to directly impact on PPARγ activity, as the binding of both proteins seems to be ligand-dependent. This is in line with previous observations suggesting that SIRT1 in vivo does not deacetylate its substrates in an undiscriminated manner. Rather, substrates might be primed for deacetylation via different means, such as conformational changes upon ligand binding or through the modulation of other post-translational modifications. Altogether, it seems clear that both SIRT1-dependent and -independent mechanisms might contribute to the lower PPARγ activity and the blunted fat deposition in PARP-1 and PARP-2 knock-out mice (Bai, et al. 2011a, Bai, et al. 2011b.).
An interesting question is whether reduced WAT depots could potentially lead to ectopic lipid deposition. A recent report identified increased fat deposition in the livers of PARP-1−/− mice when fed a HFD (Erener, et al. 2012b.). PARP-1 is poorly expressed in the liver and global deletion of PARP-1 does not seem to have a major influence on hepatic expression of mitochondrial and lipid oxidation genes (Bai, et al. 2011b.), which might create a permissive scenario for lipid deposition. However, it is difficult to match this observation with the notion that PARP-1 deficiency dampens PPARγ activity and with the lower body weight of PARP-1−/− mice. Similarly, the possible activation of sirtuins would be theoretically at odds with a predisposition for lipid accumulation. Further evaluation of these models or the generation of tissue-specific deletions will be required to clarify this apparent discrepancy.
Another apparent discrepancy lies in the fact that PARP-1 deletion on an SV129 background renders the mice susceptible to obesity (Devalaraja-Narashimha and Padanilam 2010.). It is to be noted that the SV129 background is less suited for metabolic studies than C57Bl/6J mice (Champy, et al. 2008.) that may provide a plausible explanation for the misalignment of observations. Illustrating this latter point, the pharmacological inhibition of PARP activity in diverse human and murine cell types prompts an increase in oxygen consumption and mitochondrial biogenesis, very much in line with the results obtained in C57Bl/6J mice (Bai, et al. 2011b, Modis, et al. 2012.). Furthermore, the expression of an additional copy of PARP-1 in mice leads to enhanced adiposity, perfectly mirroring once more the data obtained in the C57Bl6/J mice (Mangerich, et al. 2010.). The convergent results of these genetic, physiological, pharmacological and in vitro studies clearly support that a reduction in PARP activity would result in the enhancement of energy expenditure and prevention against HFD-induced body weight gain. The particular reasons by which the deletion of PARP-1 in the SV129 rendered an opposite phenotype are still elusive. Analyses on disturbances on NAD+ and sirtuin activity might bring some light into this question and will warrant further investigation.
3.1.4. PARP - sirtuin interaction in whole body glucose metabolism
The large influence of PARP enzymes on highly metabolic tissues, such as muscle and brown adipose tissue, predicts that PARP enzymes should have a major impact on whole body glucose homeostasis. PARP-1 and PARP-2 deficient mice displayed increased glucose clearance in response to an insulin tolerance test compared to their wild-type littermates (Bai, et al. 2011a, Bai, et al. 2011b.). This is likely to be consequent to increased insulin-stimulated muscle glucose uptake, as skeletal muscle accounts for ~80% of the whole body glucose disposal in insulin-stimulated conditions (DeFronzo, et al. 1985.) involving many different factors. For example, oxidative muscle fibers are generally more insulin sensitive than glycolytic fibers (Hom and Goodner 1984.), and, as described above, PARP-1, or -2 deficiency is linked to a higher oxidative profile of muscle fibers. A second key factor is that PARP-1 and -2 deficient mice are leaner due to enhanced energy expenditure and have an impaired ability to accumulate fat (Bai, et al. 2011a, Bai, et al. 2011b.). This grants the muscle protection against the chronic deposition of lipid species that could be detrimental for an efficient insulin signaling (Petersen and Shulman 2006.). Finally, the higher mitochondrial content in oxidative fibers gives the organism a greater potential to obtain energy from fatty acids, also contributing to the prevention of fat deposition. While no causality links can yet be established between these observations and sirtuin activation, there are a number of interesting correlations. Notably, most transgenic and pharmacological approaches aimed to increase SIRT1 activity also lead to enhanced insulin sensitivity (Banks, et al. 2008, Canto, et al. 2012, Feige, et al. 2008, Lagouge, et al. 2006, Pfluger, et al. 2008.). The link between other sirtuins and insulin sensitivity is not so well established. While SIRT3 might be protective against insulin resistance and obesity upon high-fat feeding (Hirschey, et al. 2011.), most evidences do not support the activation of SIRT3 or any other non-nuclear sirtuin upon PARP inhibition (Bai, et al. 2011a, Bai, et al. 2011b.). However, we cannot rule out that other nuclear sirtuins could contribute to the insulin-sensitizing effect of reducing PARP activity. In this sense, it should be pointed out that mice overexpressing SIRT6 are not more insulin sensitive (Kanfi, et al. 2010.). These observations points towards SIRT1 as a very likely mediator of the insulin sensitizing effects of PARP deficiency. It will be important in the future to evaluate whether PARP inhibition can render insulin-sensitization in SIRT1-deficient tissues.
The enhanced insulin sensitivity of PARP-1 and PARP-2 deficient mice should theoretically be aligned with a better glucose tolerance. While this is certainly the case in PARP-1−/− mice (Bai, et al. 2011b.), it was surprising to observe that PARP-2−/− mice became markedly glucose intolerant upon high fat feeding (Bai, et al. 2011a.). This glucose intolerance was rooted in pancreatic β-cell dysfunction. Upon high-fat feeding, the pancreatic β-cell mass increases in order to compensate for peripheral insulin resistance (Buteau and Accili 2007.). In the PARP-2−/− mice, however, this hyperplasic response is largely impaired, resulting in a blunted ability to release insulin upon a glucose load. This way, PARP-2−/− mice have a lower average β-cell islet size and pancreatic insulin content (Bai, et al. 2011a.). The molecular mechanism by which PARP-2 deletion impairs β-cell proliferation might be consequent to a constitutive SIRT1 activation, which leads to FOXO1 deacetylation and activation. FOXO1 is a well-known repressor of Pdx-1, a key regulator for β-cell proliferation and development (Bai, et al. 2011a, Buteau and Accili 2007.). An interesting question, yet unresolved, is why this is not observed in the PARP-1−/− mice. PARP-1 is also expressed in the β-cell and might have also have a critical role on its functionality. A role for PARP-1 in β-cells was first unravelled by studies showing how PARP inhibitors improved diabetes mellitus in partially depancreatized rats (Yonemura, et al. 1988, Yonemura, et al. 1984.). These studies revealed that PARP inhibitors allowed more efficient β-cell regeneration after pancreatectomy and a faster normalization of blood glucose. Posterior studies reinforced such concept by demonstrating that mice lacking PARP-1 are completely resistant to the development of diabetes upon streptozocin (Burkart, et al. 1999b.). When compared to their wild-type littermates, PARP-1−/− mice remained normoglycemic and maintained normal pancreatic insulin content and islet morphology (Bai, et al. 2011a, Burkart, et al. 1999b.). Parallel research demonstrated that PARP-1 is actually a master controller of β-cell death upon exposure to nitric oxide or oxygen radical generating compounds and that the inhibition of PARP-1 allows to retain β-cell survival in such circumstances (Burkart, et al. 1999a.). Also important for pancreatic β-cell function, PARP inhibition prevents the detrimental effects of glucotoxicity on insulin promoter activity and biosynthesis (Ye, et al. 2006.). Despite the major effects of PARP-1 in situations of toxicity for β-cells, PARP-1 ablation does not seem to have a major impact on the endocrine pancreas β-cell function in the basal state (Bai, et al. 2011a.) suggesting that the protective phenotype of PARP-1 inhibition is only effective under β cell stress. Given the above observations on the PARP deficient mice models, would the inhibition of PARP activity be beneficial or detrimental for β-cell function? A priori one would argue that in the absence of β-cell toxicity PARP activity would be detrimental, as supported by the marked β-cell dysfunction in PARP-2−/− mice and the lack of phenotype in the PARP-1−/− mice. However, in situations of β-cell toxicity, PARP inhibitors might prevent β-cell death and allow a better regeneration of the tissue.
Overall, the ability of PARP enzymes to modulate SIRT1 activity and the fact that human studies have provided evidence on the connection between SIRT1 gene expression and insulin sensitivity (Rutanen, et al. 2010, Zillikens, et al. 2009b.), further strengthens the promising possibility of modulating PARP activity in the management of metabolic disease. In this sense, preliminary evidence indicates that short term pharmacological inhibition of PARP activity is able to decrease plasma glucose, triglyceride and free fatty acid levels (Bai, et al. 2011b.). However, more protracted treatments will be required to evaluate the feasibility of engaging long-term inhibition of PARP activity and whether this might have a negative impact on DNA damage and chromosome maintenance. Furthermore, while the regulation of SIRT1 activity might help granting insulin sensitivity in the PARP deficient models, this does not exclude that other mechanism yet to be found might be equally important in explaining this phenotype. Amongst them, it will be important to evaluate possible direct PARylation targets that could influence glucose homeostasis.
3.2. Interplay between PARPs and SIRTs in oxidative stress response
Reactive species are partially reduced, highly reactive molecules (radicals and non-radicals). Most reactive species can be classified as reactive oxygen intermediates (ROI), or reactive nitrogen intermediates (RNI) depending on the central atom. The major source of ROIs in unstimulated cells is the leakage of mitochondrial electron transport chain that creates superoxide anion that serves as a parent molecule of downstream ROIs, such as hydrogen peroxide, or hydroxyl radical. Besides the mitochondrial origin, ROIs can stem from the oxidation of certain xenobiotics, activated neutrophils and macrophages, or from xanthine oxidase (Ray, et al. 2012, Virag and Szabo 2002.).
The parent molecule of RNIs is nitrogen monoxide (NO) that is formed by nitrogen monoxide synthase enzymes termed neuronal (nNOS), endothelial (eNOS) and inducible (iNOS) (Forstermann and Sessa 2012, Nathan 1992.). While NO is an important signaling molecule and considered cytoprotective (Pacher, et al. 2007.), its downstream derivatives such as peroxynitrite (formed in reaction with superoxide (Pacher, et al. 2007.)), or nitroxyl radical (formed by NO reduction (Bai, et al. 2001.)) are cytotoxic. In oxidative stress-related diseases the balance between free radical species and antioxidant defence systems is hampered. Free radicals, in abundance, damage lipids, DNA and proteins, or may react with protein-bound metals affecting all vital components of cells and tissues (Pacher, et al. 2007.). Various enzyme systems are on duty to cope with free radical-induced damage and hence to protect against free radical-provoked diseases, among them PARPs and SIRTs play prominent roles.
As reactive species cause DNA damage and consequently induce PARP activation in the effort to restore DNA integrity (Virag and Szabo 2002.). As discussed previously (section 1.3), PARP activation has pleiotropic effects (induction of necrosis, mitochondrial damage, proinflammatory actions, reprogramming of gene expression) that worsen free radical-mediated pathologies (Modis, et al. 2012, Szanto, et al. 2012, Virag and Szabo 2002.). The idea that PARP inactivation may provide protection against free radical production-mediated PARP activation and the consequent cell, or tissue stress is almost 15 years old (Szabo, et al. 1998.) and it had been successfully tested in a plethora of animal and cellular models (reviewed in (Virag and Szabo 2002.)).
In contrast to PARP activation the induction of SIRT1 protects against oxidative stress. The protective effects of SIRT induction involve numerous mechanisms.
SIRT1 modify numerous components of the cell cycle coordination machinery (e.g. p53 and FOXO) upon oxidative injury that leads to cell cycle arrest and suppression of apoptosis (Brunet, et al. 2004, Han, et al. 2008, Luo, et al. 2001.);
SIRT1 activation induces antioxidant defence systems, such as manganese superoxide dismutase (MnSOD) (Danz, et al. 2009.), or catalase (Hasegawa, et al. 2008.).;
Mitochondrial biogenesis is hampered upon oxidative stress that is restored upon SIRT1 induction (Danz, et al. 2009, Szanto, et al. 2011.);
In oxidative stress SIRT1 induction induces autophagy (Alcendor, et al. 2007.);
SIRT1 itself is redox sensitive, its protective actions can be hampered under oxidative stress conditions by carbonylation of SIRT1 (Caito, et al. 2010a.) and the disregulation of thiol redox balance (Caito, et al. 2010b.) both impairing SIRT1 activity. Recent data by Caito and colleagues (Caito, et al. 2010b.) have shown the redox-dependent phosphorylation of SIRT1 that may have a role in the oxidant-mediated reduction of SIRT1 activity.
Apparently SIRT1 and PARP activation have opposing characteristics under oxidative stress conditions. As PARP activation seems to be a key factor of cellular and tissue damage under oxidative stress (Pacher and Szabo 2008.) these observations prompted research to understand PARP – SIRT interaction under pathological scenarios. The interaction of SIRT1 and PARPs had been associated with pathologies of the cardiovascular system, the central nervous system and the liver in the gastrointestinal tract (summarized in Table 1).
Table 1. Occasions of SIRT1 – PARP interaction in oxidative stress-related pathologies.
| Organ system | Disease | Partners | Models | Mechanism | Effect | Ref. |
|---|---|---|---|---|---|---|
| Cardiovascular system |
Angiotensin-induced cardiac hypertrophy |
PARP-1 SIRT1 |
PARP-1−/− mice, primary cardiomyocytes treated with resveratrol, SIRT1 siRNA, or by SIRT1 overexpression. |
PARP-1 activation inhibits SIRT1 through NAD+ depletion. |
Both SIRT1 activation and PARP inhibition protect against angiotensin II-mediated and oxidant-mediated cardiomyocyte cell death. |
(Pillai, et al. 2006.) |
| Heart failure (aortic banding model) |
PARP-1 SIRT1 |
Cardiomyocytes overexpressing PARP-1, SIRT1, or treated with resveratrol, sirtinol, SIRT1 siRNA |
PARP-1 limits NAD+ levels and hence SIRT1 activity through limiting NAD+ for SIRT1 action and probably by repressing SIRT1 expression. |
SIRT1 induction protect against oxidant-induced cardiomyocytes cell death. |
(Pillai, et al. 2005.) | |
| Shear stress on endothelial cells |
PARP-1 SIRT1 |
HUVEC cells undergoing shear stress treated with ABT888 and PARP-1 siRNA |
Shear stress decrease NAD+ levels, SIRT1 activity and expression that is reverted by PARP-1 depletion, or inhibition. |
Proinflammatory conditions and cell death provoked by shear stress is reduced. |
(Qin, et al. 2012.) | |
| DOX-induced vascular damage |
PARP-2 SIRT1 |
PARP-2 knockout mice and PARP-2 knockdown MOVAS cells |
SIRT1 promoter is released from suppression upon PARP-2 depletion that induces mitochondrial biogenesis. |
Vascular protection against DOX damage upon SIRT1 induction after PARP-2 depletion. |
(Szanto, et al. 2011.) | |
| Central nervous system |
Trophic deprivation and oxidant mediated neuronal cell death |
PARP-1 SIRT1 |
Near-pure cortical neuronal cell cultures from PARP-1−/− mice. Same neurons treated with fisetin, resveratrol and sirtinol. |
PARP-1 activation inhibits SIRT1 through NAD+ depletion. Protective effect of SIRT1 activation is not explained. |
Both SIRT1 activation and PARP inhibition protect against neurotoxicity. |
(Sheline, et al. 2010.) |
| Glutamate/NMDA neurotoxicity |
PARP-1 SIRT1 |
Dissociated cerebral cortical cell cultures from embryonic rat treated with sirtinol and resveratrol. |
NMDA treatment reduces SIRT1 activation probably due to PARP activation and NAD+ depletion. Protective effect of SIRT1 activation is not explained. |
Reduction of cell death upon SIRT1 activation and PARP inhibition. |
(Liu, et al. 2008.) (Liu, et al. 2009.) |
|
| Gastrointestinal tract |
Glucose toxicity on hepatocytes |
PARP-1 SIRT1 |
HepG2 cells treated with PJ34 and PARP-1 siRNA. |
PARP-1 activation decreases SIRT1 activity that is restored by PARP inhibition probably through conservation of NAD+ levels. |
Glucose toxicity is reverted by PARP inhibition. |
(Pang, et al. 2011.) |
Abbreviations in text.
Fistein and resveratrol are SIRT1 activators, sirtinol is a SIRT1 inhibitor, 3-amino-benzamide (3AB) and PJ34 are PARP inhibitors.
The pathological states listed are characterized by increased production of reactive species either from an internal source like angiotensin II (AngII), N-Methyl-D-aspartic acid (NMDA) receptor activation, high glucose levels, pressure overload in the heart, shear stress on endothelial cells and from an external noxa such as oxidants, or doxorubicin treatment (Moncada and Bolanos 2006, Pacher, et al. 2003, Pacher, et al. 2002b, Qin, et al. 2012, Soriano, et al. 2001, Wilson 1990.) that lead to PARP activation contributing to cellular and organ dysfunction (Moncada and Bolanos 2006, Pacher, et al. 2002a, Pillai, et al. 2006, Soriano, et al. 2001, Szabo, et al. 2004.). In the case of trophic deprivation, heart failure, AngII, NMDA and glucose toxicity extensive PARP activation decreased SIRT1 activity (Liu, et al. 2009, Pang, et al. 2011, Pillai, et al. 2006, Pillai, et al. 2005, Qin, et al. 2012, Sheline, et al. 2010.) through reducing NAD+ levels that compromised SIRT1 activation and hence the protective effects of SIRT1 activation (Figure 4). This hypothesis is further underlind by the fact that preventing NAD+ depletion was protective in most of these pathologies (Houtkooper, et al. 2010, Liu, et al. 2008.). As discussed in section 1.3 and 2.1, enhanced PARP activation depletes cellular NAD+ (Berger 1985.) that limits substrate availability for SIRT1 (Kolthur-Seetharam, et al. 2006.). On the other hand SIRT1 induction limits PARP-1 through deacetylating and hence inactivating it (Rajamohan, et al. 2009.) (Figure 4).
Figure 4. The molecular level interactions between SIRT1 PARP-1 and -2 under oxidative stress conditions.
The pathways enhancing oxidative stress-mediated tissue damage are in red, in turn, protective pathways are in green.
Oxidative stress induces PARP-1 that through the depletion of NAD+ pools inhibits SIRT1. That pathway seems to participate in the tissue damage inflicted by PARP activation.
On the contrary, SIRT1 activation by pharmacological agents (e.g. resveratrol, fistein) or by induction of its expression (e.g. PARP-2 ablation) leads to SIRT1-mediated deacetylation and inactivation of PARP-1 that seems a crucial pathway in the cytoprotective action of SIRT1 activation.
Alternative, non-NAD+ dependent pathways may also exist between SIRT1 and PARP-1. It would be easy to speculate that PARP-1 could inhibit SIRT1 activity through direct PARylation of SIRT1, although we were unable to show SIRT1 PARylation under oxidative stress ((Bai, et al. 2011b.) and section 2.2). It is also plausible that other transcription factors are PARylated (e.g. c-fos/c-jun PARylation upon angiotensin II treatment (Huang, et al. 2009.)) that modify oxidative stress sensitivity of cells, however that field is largely unexplored.
Kolthur-Seetharam and colleagues (Kolthur-Seetharam, et al. 2006.) have identified a possible alternative connection between SIRT1 and PARP-1 that may influence oxidative stress-induced cell death. PARylation has been shown to induce the nuclear translocation of apoptosis inducing factor (AIF) in certain cell lines contributing to cell death (Yu, et al. 2002.). In the absence of SIRT1 enhanced PARylation was observed that is further accentuated upon oxidative damage (Kolthur-Seetharam, et al. 2006.) (Figure 4). In line with that observation in SIRT1−/− cells nuclear translocation of AIF is induced that may contribute to cell death.
The depletion of PARP-2 protected partially the vasculature against doxorubicin toxicity (Szanto, et al. 2011.). The protective enhancement of SIRT1 activity upon PARP-2 depletion relies on the induction of the SIRT1 promoter (Szanto, et al. 2011.) (Figure 4). Importantly, PARP-2 depletion and the consequent enhancement of SIRT1 activity did not reduce PARP-1 activation under oxidative stress conditions (Szanto, et al. 2011.).
Apparently PARP-1 and SIRT1 under oxidative conditions regulate the activity of each other through various mechanisms. SIRT1 induction leads to protection against oxidative damage, while PARP-1 activation is a detrimental consequence of oxidative stress. There is a large overlap between the oxidative stress-mediated pathologies that are corrected by SIRT1 induction (Chong, et al. 2012, Chung, et al. 2010.), or PARP inhibition (Virag and Szabo 2002.) due to joint regulation of key enzymes involved in these pathologies (e.g. matrix metalloproteinase activation in cardiovascular and dermatological diseases (Bai, et al. 2009, Bai, et al. 2004, Brunyanszki, et al. 2010, Choi, et al. 2005, Lee, et al. 2010, Nakamaru, et al. 2009, Ohguchi, et al. 2010, Pacher, et al. 2002a.) suggesting that there are way more pathologies (e.g. diabetic complications, stroke, multiple sclerosis, inflammation) where PARPs and SIRTs collaborate on the outcome.
3.3. PARP – SIRT interaction in the maintenance of genomic stability
PARP-1, -2, -3 and tankyrases are involved in DNA repair and the maintenance of genomic integrity, as discussed in section 1.3, SIRT1 and SIRT6 had been shown to be involved in DNA repair (Mostoslavsky, et al. 2006, Oberdoerffer, et al. 2008.). The interaction between PARylation and SIRT1 in DNA repair events were first suggested by Zhang in 2003 (Zhang 2003.). First experimental evidence for that interaction in chromatin remodelling was presented by Tulin and co-workers (Tulin, et al. 2006.) showing that PARG and SIR2 co-localized in cell nuclei of Drosophyla larvae and localization of SIR2 was dependent on PARG expression suggestive of the involvement of PAR levels (Tulin, et al. 2006.).
The apparent functional convergence of SIRT1 and PARP activation in DNA repair and genomic maintenance was assessed in detail by El-Ramy and colleagues (El Ramy, et al. 2009.) by studying the double deletion of PARP-1 and SIRT1 in mice. The deletion of SIRT1 induced early postnatal lethality, chromosomal and DNA repair defects as expected from previous studies (Cheng, et al. 2003, McBurney, et al. 2003, Oberdoerffer, et al. 2008.). Although, the concurrent deletion of PARP-1 did not prevent postnatal lethality, it did influence DNA repair defects (El Ramy, et al. 2009.).
The absence of SIRT1 have led to telomere dysfunction, the spreading of heterochromatic regions, rearranged nucleolar architecture by increasing the number of nucleoli and hampered mitotic cell division by inducing the number of mitotic divisions, however also enhanced the number of aberrant divisions, the incidence of unequal distribution of chromosomes between daughter cells and the extent of DNA damage associated with mitosis (occurrence of micronuclei) (El Ramy, et al. 2009.). PARP-1 had been shown to act as an actor in maintaining genome stability under genotoxic stress and to protect against the above features (De Vos, et al. 2012.), however unexpectedly it was the deletion of PARP-1 that was protective against the SIRT1-induced genome instability except for maintenance of telomere integrity (El Ramy, et al. 2009.). Yet the exact molecular mechanism of the above detailed phenomenon is unknown.
Among sirtuins, besides SIRT1, SIRT6 is involved in the regulation of DNA repair events (Mostoslavsky, et al. 2006.). The absence of SIRT6 enhances the sensitivity of fibroblasts to ionizing radiation and brings about genomic instability (increased number of chromosome fragmentation, centromere default, abnormal metaphase, chromosome translocations) (Mostoslavsky, et al. 2006.). SIRT6 translocate to the damage sites where it promotes DNA repair (Mao, et al. 2011, Mostoslavsky, et al. 2006.). SIRT6 has pivotal role in the appropriate function of base excision and double strand break repair (Mao, et al. 2011, Mostoslavsky, et al. 2006.). SIRT6 mono-ADP-ribosylate PARP-1 on K521, whereby the activity of PARP-1 is induced that contribute to successful resolution of double strand breaks, however the deacetylase activity of SIRT6 has equal contribution to DNA repair as its mono-ADP-ribosyl transferase activity (Mao, et al. 2011.) suggesting unknown parallel DNA repair pathways influenced by SIRT6.
It remains a question whether other PARPs involved in genome maintenance (PARP-2, -3, or tankyrases) would similarly interact with SIRT1 in DNA repair events, or whether the interaction of SIRT1 and PARP-1 would take place also in the resolution of double and single strand breaks. It is plausible that the interaction of SIRTs and PARPs in the maintenance of genomic integrity has prominent role in senescence, apoptosis, cell cycle regulation and tumorigenesis.
3.4. PARP - sirtuin interaction in ageing
Higher PARP activation capacity had been associated with successful ageing in mammals (Burkle, et al. 1994.) and in humans (Muiras, et al. 1998.). It was postulated that the benefit of the higher PARylation capacity is more reliable DNA repair that prevents the occurrence of DNA damage-associated diseases (e.g. neoplasms). Although, as of yet the lack of PARP-1 has not been associated with spontaneous tumorigenesis without tumorigenic challenge in vivo unless other DNA repair enzymes (e.g. p53) were removed too (Nicolas, et al. 2010, Tong, et al. 2001.). Recent data, however validate that concept: the hPARP-1 mouse strain, that overexpresses an extra copy of PARP-1, are protected against neoplastic diseases (Mangerich, et al. 2010.). However the incidence of other age-related pathologies increase upon PARP-1 overexpression: obesity, glucose intolerance and certain inflammatory pathologies (Mangerich, et al. 2010.) suggesting that the previous model might need to be refined.
Aging is associated with the dysregulation of the oxidative balance enhancing oxidative stress. Upon ageing PARylation capacity increases (Braidy, et al. 2011, Massudi, et al. 2012.) that upon stress puts heavy burden on NAD+ homeostasis. Indeed, lower NAD+ levels were detected in aged animals and humans that coincided with lower SIRT1 activity despite the induction of SIRT1 expression (Braidy, et al. 2011, Massudi, et al. 2012.). As decrease in SIRT1 activity leads to decrease in mitochondrial biogenesis it is tempting to speculate that the crosstalk between PARP-1 and SIRT1 might be a cause of the age-associated loss of mitochondrial function and vice versa, influencing that angle could be exploited to combat age-associated loss of mitochondrial function. The dysregulation of the PARP-1 – NAD+ - SIRT1 balance may stay behind some of the pathologies observed in the hPARP-1 mice (Mangerich, et al. 2010.). As hampered mitochondrial biogenesis is a hallmark of ageing and it is probably a major cause of several age-associated metabolic and central nerve system diseases (Lopez-Lluch, et al. 2008.) fine tuning of the PARP-1 – SIRT1 interaction may prove to be a successful strategy to counteract these diseases and provide longer healthspan (Canto and Auwerx 2011b.).
4. Pharmacology of NAD+, SIRT1 and PARPs
4.1. NAD+ modulating agents
The idea that PARPs and sirtuins may be affected by NAD+ metabolism, and that in turn, activities of these enzymes modulate NAD+ levels is cells has indicated that NAD+ metabolism itself is an interesting target for pharmacological modulation. It is important to bear in mind that NAD+ itself and associated metabolites (Figure 5) are active forms of Vitamin B3, whose levels are sensitive to pharmacologic interventions featuring different forms of Vitamin B3. Importantly, as was discussed in section 1.1, different vitamin forms are metabolized through distinct pathways, indicating that each has unique pharmacologic and metabolic properties. This has been noted particularly for nicotinic acid, which has remarkable anti-lipogenic effects, as well as the ability to lower LDL cholesterol and raise HDL cholesterol. These effects are not found for pharmacologic use of NAM. Thus, different precursors to NAD+ have distinct therapeutic as well as nutritional significance.
Figure 5. Compounds that modulate NAD+ concentrations or modulate GPR109A.
4.1.1. Niacin
The distinctive effects of NA (niacin) supplementation are thought to include systemic NAD+ increase, as revealed by supplementation studies of NAM or NA in rats (Jackson, et al. 1995.). These authors supplemented each of the two Vitamin B3 compounds in 30, 100, 500, and 1000 mg/kg for three weeks and then tissue NAD+ was analyzed by HPLC. Animals fed at the highest dose NA experienced 1.44 (packed RBC), 1.54 (liver), 1.62 (heart), 1.12 (lung), and 1.88 (kidney) increased NAD+ contents (measured as fold over control), whereas NAM at the highest dose yielded 1.44 (packed RBC), 1.47 (liver), 1.20 (heart), 1.18 (lung), and 1.03 (kidney) increases in NAD+ contents (measured as fold over control). It is apparent from these data that NA generally provides greater NAD+ enhancements than NAM in mammalian tissues. This observation has been rationalized by observations that NAM and NA are metabolized to NAD+ differently, and that NA production is typically low in mammalian tissues, enabling enhanced biosynthesis of NAD+ when NA is provided via a pharmacologic route. NA administration enhances NAD+ availability, therefore is protective in PARP-mediated pathologies (Benavente, et al. 2009, Hageman and Stierum 2001, Weidele, et al. 2010.).
NA is distinct versus NAM in its ability to bind a G-protein coupled receptor (GPR109A, or HM74A) (Soga, et al. 2003, Tunaru, et al. 2003, Wise, et al. 2003.), which increases vasodilation and leads to uncomfortable flushing (Benyo, et al. 2005.). This effect appears to be mediated by release of prostaglandins D2 and E2 from epidermal Langerhans cells (Benyo, et al. 2006.). In fact, three distinct receptors for NA have been identified in mammalian cells. The high affinity receptor has a measured affinity to NA of 63-250 nM (Soga, et al. 2003, Wise, et al. 2003.). The knockout of the murine homologue PUMA-G appears to ablate many of the effects of NA in lipid lowering (Tunaru, et al. 2003.). Some authors have argued that GPR109A activation is responsible for the entirety of the anti-lipidemic effects of NA (Wanders and Judd 2011.). This view is in part supported by the effects of a NA mimic acipomox, which has been used extensively in Europe for treatment of hyperlipidemias. Acipomox does not have an apparent pathway for stimulation of systemic NAD+ increase, suggesting it parces the non-NAD+ component in NAs effects. However, acipomox has been reported to exhibit smaller effects than NA in lipid modulation (Seed, et al. 1993.), and acipomox did not cause statistically significant increase of HDL in one study (Seed, et al. 1993.).
The latter result suggests that some of NA effects could act through NAD+ metabolism, and through NAD+ modulation of signal transducers (such as sirtuins and PARPs) that can impinge on lipid metabolism. Even those who are strong advocates of GPR109A as responsible for mediating NA’s effects acknowledge that some of NA effects are unclear, such as how exactly NA causes the magnitudes of lipid modulations as well as how NA causes increases in HDL levels (Wanders and Judd 2011.). It is important to bear in mind that NAD+ metabolism could play key, but yet to be determined roles, in these effects.
Niacin’s lipid altering effects, were first reported in 1955 (Altschul, et al. 1955.) and niacin has been widely used clinically since that time for modulating serum lipids. As such, niacin supplementation therapy is the oldest known therapy for modulating cholesterol and lipids, and predates the development of the statin drugs. High dose niacin is still widely prescribed as a treatment for lipidemia, in the form of Niaspan (Abbott laboratories), for treating elevated cholesterol and for raising HDL levels. Niacin increases HDL cholesterol, better than any other known pharmacologic agent, and decreases LDL and VLDL cholesterol (Capuzzi, et al. 2000.). It also can decrease serum fatty acids. Data on its effectiveness indicate that niacin reduces long-term cardiac disease mortality compared with untreated patients (Berge and Canner 1991.). Because, niacin can profoundly increase HDL cholesterol, high dose niacin was evaluated as a supplement to statin therapy (simvistatin) for possible benefit in a large 3414 patient study called AIM-HIGH (www.aimhigh.com). This study was halted in 2011 after no benefit was found in the Niaspn treated arm of the study as measured by rates of cardiovascular events. There was a slightly increased risk of stroke in the Niaspan arm (28 events versus 12 events) which prompted the study to be terminated.
4.1.2. Nicotinamide
Nicotinamide does not behave like NA as a serum cholesterol modulation agent (Altschul, et al. 1955.). It also cannot activate the GPR109A receptor (Wise, et al. 2003.). Not surprisingly, NAM does not activate flushing in people. However, as already discussed, data suggests that NAM is not particularly effective for increasing NAD+ levels in tissues, suggesting it has limited potential as an NAD+ enhancement agent. Nevertheless, NAM can increase liver NAD+ contents substantially (Jackson, et al. 1995.). NAM is a known inhibitor of several ADP-ribosyl transferase enzymes, including sirtuins (Jackson, et al. 2003, Sauve and Schramm 2003.) and PARPs (Preiss, et al. 1971, Virag and Szabo 2002.), causing its effects on NAD+ metabolism to be in part due to decreasing rates of NAD+ turnover.
The ability of NAM to attenuate rate of depletion of NAD+ by inhibition of PARP probably accounts for some of the protective effects identified for NAM in models of disease (Virag and Szabo 2002.). NAM has been shown to provide protection in stroke and hypoxic neural injury (Feng, et al. 2006, Liu, et al. 2009.), fetal alcohol syndrome (Ieraci and Herrera 2006.) and in acute neurotrauma (Hoane, et al. 2003, Hoane, et al. 2006a, Hoane, et al. 2006b, Hoane, et al. 2006c.). NAM also provides protection against streptozotocin induced β cell loss in the pancreas (Lazarus and Shapiro 1973.). A large clinical study using NAM to treat prevent Type I diabetes in Europe failed to show a clinical benefit (Gale, et al. 2004.), but reinforced the safety of longterm high dose NAM administration to healthy patients.
4.1.3. Tryptophan
Tryptophan is a precursor of NAD+ via degradation through the oxidative kyurenine pathway. The value of tryptophan as an NAD+ precursor is limited. It has been estimated that only 1 of 67 mg of tryptophan is shunted into NAD+ synthesis in human females. This indicates that 1 mg of niacin is equivalent to 67 mg of tryptophan for NAD+ biosynthesis (Fukuwatari, et al. 2004.). Nevertheless defects in tryptophan metabolism to NAD+ have been suggested in some diseases, such as neurodegenerative disorders, through accumulation of quinolinic acid (Schwarcz, et al. 1983.). Inhibition of a key enzyme in this pathway, kynurenine 3-monooxygenase provides relief in a transgenic model of Alzheimers disease (Zwilling, et al. 2011.).
4.1.4. Nicotinamide Riboside
Limited data exists for the use of NR as an NAD+ precursor in human or mammalian cell types. Sauve and co-workers were first to establish that NR can be broadly useful for increasing NAD+ levels in a variety of mammalian cell types (Yang, et al. 2007b.). NR was found to be protective in a model of optic neuritis by direct injection into the eye (Shindler, et al. 2007.). More recently, NR was shown to increase NAD+ levels in muscle and other tissues when administered to mice in food (Canto, et al. 2012.). NR was able to stimulate mitochondrial biogenesis, increase insulin sensitivity, lower cholesterol, and reduce weight gain in mice fed a high fat diet (Canto, et al. 2012.). NR supplementation was able to provide in vivo activation of sirtuins SIRT1 and SIRT3 (Canto, et al. 2012.). The beneficial effects observed in vivo, in combination with high levels of NAD+ enhancements on cells (Yang, et al. 2007b.) suggests this compound could be of interest for nutritional supplements and for possible therapeutic formulations.
4.2. Sirtuin modulating agents
The quest for sirtuin activators (Figure 6) began short after the finding that Sir2 could modulate replicative lifespan in yeast. These efforts culminated when, in 2003, Howitz and collaborators identified resveratrol and a few other polyphenols, including quercetin and piceatannol, as natural compounds that could directly bind and enhance SIRT1 activity (Howitz, et al. 2003.). A number of studies have subsequently shown that resveratrol treatment leads to enhanced SIRT1 activity in diverse cells, tissues and organisms (Baur 2010.). In line with this, resveratrol has been shown to promote calorie-restriction like health benefits in numerous organisms. Notably, resveratrol largely prevented the onset of diet-induced obesity and metabolic disease upon high-feeding, ultimately protecting against the lifespan curbing associated with high caloric intake (Baur, et al. 2006, Lagouge, et al. 2006.). The same study demonstrated that resveratrol also improved mitochondrial function and fatty acid oxidation (Lagouge, et al. 2006.). In line with the possible inhibitory role of SIRT1 on PARP activity, it has been reported that resveratrol-treatment can lead to reductions in PARP activity (Kolthur-Seetharam, et al. 2006.).
Figure 6. Examples of compounds that (A) activate and (B) inhibit SIRT1 or other sirtuins.
A major caveat in the identification of resveratrol as a direct SIRT1 activator relies in the use of a fluorescently labelled substrate in the original screening. These results were questioned by convincing evidence demonstrating that the nonphysiological fluorescent “Fluor de Lys” substrate can lead to artifactual results (Borra, et al. 2005, Kaeberlein, et al. 2005.). This suggested that the actions of resveratrol on SIRT1 might be indirect, igniting a new quest for the possible early drivers of resveratrol action. A likely candidate to initiate the metabolic actions of resveratrol is the AMPK. Many works have reported how resveratrol treatment leads to AMPK activation (see (Canto and Auwerx 2011a.) for review). Elegant studies by the Hardie lab demonstrated that AMP-insensitive forms of AMPK are resistant to activation by resveratrol, clearly indicating that AMPK activation in response to resveratrol relies on an AMP/ATP imbalance. In line with this hypothesis, resveratrol has been shown to interfere with the mitochondrial respiratory chain (Zini, et al. 1999.), providing a likely mechanism by which resveratrol might affect AMP/ATP balances and activate AMPK. The link between AMPK and SIRT1 activation is provided at least by a couple of different mechanisms. First, AMPK activation is followed by an increase in NAD+ levels. On an initial phase this increase is powered by increased fatty acid oxidation flux (Canto, et al. 2009.), which is later sustained by enhanced Nampt expression (Canto, et al. 2009, Fulco, et al. 2008.). A second mechanism recently proposed relies on the direct phosphorylation of SIRT1 by AMPK, which would disrupt the ability of DBC-1 to interact and inhibit SIRT1 (Nin, et al. 2012.). Another layer of complexity is provided by the possible impact of SIRT1 in AMPK activity. Indeed, some labs have shown that SIRT1 might also influence AMPK activity (Hou, et al. 2008, Lan, et al. 2008, Price, et al. 2012, Suchankova, et al. 2009.). This would create a positive feedback loop between both signaling events to amplify the signal. The fact that AMPK is activated by resveratrol in SIRT1 defective cells and that defective AMPK activity compromises resveratrol-induced SIRT1 activation (Canto, et al. 2010, Dasgupta and Milbrandt 2007, Um, et al. 2010.), indicates that AMPK might be the initial trigger of this signal loop. This, however, might largely depend on the dose of resveratrol used (Price, et al. 2012.). In any case, a clear implication of this relationship is that AMPK activation should have similar silencing effects on PARP activity as SIRT1 activation. Surprisingly, however, the only study directly studying a possible relation between AMPK and PARPs described how AMPK could phosphorylate PARP-1 in vitro and enhance its activity in certain scenarios (Walker, et al. 2006.). These observations are in apparent contradiction with the metabolic observations in animal models, which indicate that AMPK rather mimics the transcriptional adaptations promoted by decreased PARP activity (Bai, et al. 2011a, Bai, et al. 2011b, Canto and Auwerx 2010.). These discrepancies might be explained by the very different contexts and degrees of PARP and AMPK activation reached in the diverse experimental settings. In this sense, it is common that the conditions used in cell culture-based experiments activate these enzymes to supra-physiological levels. Therefore, the possible existence of interaction and phosphorylation events between AMPK and PARP-1 requires further confirmation and evaluation to validate its physiological relevance.
Following the footsteps of resveratrol, a more recent and ambitious screening for SIRT1 activators provided a new collection of compounds, amongst which N-[2-[3-(piperazin-1-ylmethyl)imidazo[2,1-b][1,2]thiazol-6-yl]phenyl]quinoxaline-2-carboxamide (SRT1720, Figure 6) has been the one receiving most attention. SRT1720 is structurally different from resveratrol and acts as a more potent and efficient SIRT1 activator (Milne, et al. 2007.). As expected from a SIRT1 activator, the treatment of mice with SRT1720 enhanced oxidative metabolism and mitochondrial biogenesis, leading to protection against obesity and benefits on glucose homeostasis (Feige, et al. 2008, Milne, et al. 2007.). However, it was recently pointed out that the screen that identified SRT1720 as a direct SIRT1 agonist had similar flaws as the one that identified resveratrol, namely: confusing results derived from the use of the “Fluor de Lys” fluorescent moiety (Huber, et al. 2010, Pacholec, et al. 2010.). Therefore, a shadow of doubt stands on whether SRT1720 can be truly considered a direct SIRT1 agonist. Of note, the much higher potency of SRT1720 as a SIRT1 activator found in the in vitro screen was not translated in vivo (Feige, et al. 2008.), suggesting a rather indirect activation of SIRT1 or poor bioavailability. Whatever the case, no study to this data has clearly evaluated any possible impact of SRT1720 on PARP activity.
4.3. Agents modulating poly(ADP-ribosyl)ation
Most PARP activity in cells is driven by PARP-1, while the rest is mostly covered by PARP-2 both under non-stress conditions and upon oxidative injury (Bai, et al. 2011a, Schreiber, et al. 2002, Szanto, et al. 2011.). PARP activation – as discussed in section 1.3 – can be triggered by DNA damage. PARP activation is induced voluntarily mostly under experimental conditions, by oxidants (e.g hydrogen peroxide, peroxynitrite, radical donor compounds, certain cytostatic drugs), or by DNA alkylating agents (e.g. MNNG, or streptozotocin), or ionizing radiation (Schreiber, et al. 2006.). Besides DNA damage, PARP-1 can be activated by posttranslational modifications (see section 1.3 and (Gibson and Kraus 2012.)), however it remains a question to be answered whether pharmacological modulation of these signaling pathways can efficiently modulate cellular PARP activity.
The first compound identified as PARP inhibitor was benzamide (IC50 = 22 μM) (Shall 1975.) and its 3-substituted versions (3-amino-benzamide is used the most frequently (IC50 = 33 μM)) that are simple analogues of NAM (IC50 = 210 μM) that is a by-product of the PARylation reaction. Benzamide and its analogues are applied in cellular assays in the millimolar range that may lead to aspecific interactions (Milam and Cleaver 1984, Milam, et al. 1986.). Interestingly, certain dietary compounds (theophylline, caffeine and certain dietary flavonoids) that possess pleiotropic biological effects were also shown to inhibit PARP activity in cellular models in the submillimolar range (>100 μM) (Geraets, et al. 2007, Geraets, et al. 2006, Moonen, et al. 2005.).
The careful modification of the backbone provided by nicotinamide and benzamide gave rise to current cutting edge PARP inhibitors (elegantly reviewed in (Ferraris 2010.) and (Curtin 2006.)) (Figure 7). These inhibitors bind to the NAM-binding pocket of PARP-1, hence act as competitive inhibitors of NAD+-binding (Curtin 2006, Ferraris 2010, Jagtap and Szabo 2005.) with IC50 values in the low nanomolar range (1-5 nM) (Javle and Curtin 2012.). Targeting NAD+ binding, to date, seems the only successful way to design specific PARP inhibitors, as an alternative approach to inhibit PARP-1 via impeding DNA binding through impairing the zinc fingers was not a viable strategy (Liu, et al. 2012.). PARG inhibitors would provide another alternative indirect approach for PARP inhibition (Erdelyi, et al. 2009, Ying and Swanson 2000.), however the specificity of tannin derivatives used as PARG inhibitors had been questioned (Erdelyi, et al. 2005.).
Figure 7. Compounds that inhibit PARPs as discussed in the text.
Current PARP inhibitors are considered pan-PARP inhibitors, as they inhibit both PARP-1 and PARP-2 (Wahlberg, et al. 2012.). There is a current quest for the design of PARP-1/-2 specific inhibitors, however to date the best achieved specificity is 60-fold higher affinity towards PARP-2 than PARP-1 (Moroni, et al. 2009.), or ~10 fold preference towards PARP-1 than PARP-2 (Ferraris 2010.) in in vitro assays. These inhibitors are unlikely to act as highly selective agents in an in vivo setting. It seems that due to the large structural similarity between the PARP-1 and -2 catalytic domains (Oliver, et al. 2004.), the design of a highly specific inhibitor is a difficult task to accomplish (reviewed in (Szanto, et al. 2012.)).
The application of PARP inhibitors seems a promising strategy in a large number of physiological and pathophysiological states. Experimental evidence suggest the applicability of these compounds provide means to combat oxidative stress related diseases such as reperfusion injuries (gut, eye, kidney, myocardium), stroke, neurotrauma, inflammatory pathologies, shock, or diabetes and its consequent complications ((Bai and Canto 2012, Virag and Szabo 2002.) and in the present series a review by Nicola Curtin and Csaba Szabó). Several PARP inhibitors are currently making their ways through the different phases of clinical trials for the treatment of different solid and lymphoblastoid neoplasias as single agents or in combination (reviewed in (Javle and Curtin 2011.) and in the present series a review by Nicola Curtin and Csaba Szabó). Importantly, in clinical studies PARP inhibitors were reported to have good tolerability that further ensure the applicability of these agents (Fong, et al. 2009.).
PARP inhibitors were protective as shown in multiple models of oxidative stress and ageing, through guarding cellular NAD+ levels by preventing excessive PARP activation and therefore inducing SIRT1 activity (Braidy, et al. 2011, Liu, et al. 2009, Massudi, et al. 2012, Pang, et al. 2011, Pillai, et al. 2006, Pillai, et al. 2005, Qin, et al. 2012, Sheline, et al. 2010.). PARP inhibitor treatment of C2C12 myotubes, or C57/Bl5J mice induced NAD+ levels and SIRT1 activity (Bai, et al. 2011b.) that further consolidates the importance of the NAD+ link between PARP-1 and SIRT1. PARP inhibitor treatment did not alter NAD+ levels and sirtuin activity in other compartments such as cytosol, or mitochondria evidenced by the lack of SIRT2 and SIRT3 activation (Bai, et al. 2011b.).
Yet the effects of longer PARP inhibitor treatment on NAD+ homeostasis and SIRT1 activity has not been studied, moreover the chronic applicability of PARP inhibitors raise concerns, due to reduction of DNA repair capacity and consequently to enhanced genomic instability that requires further studies.
5. Concluding remarks, perspectives
NAD+ is a cofactor, or substrate of numerous enzymes that suggest widespread influence for NAD+ over a plethora of cellular functions (Houtkooper and Auwerx 2012, Houtkooper, et al. 2010.). Out of these enzymes we focused the subject of our review on two major NAD+-dependent enzyme families: PARPs and sirtuins.
The interaction of SIRT and PAPR enzymes has multiple layers. Both enzyme families consist of multiple members that show different affinities towards NAD+. Obviously, as both enzymes are NAD+-dependent in some occasions they may limit NAD+ availability for one another. Furthermore, sirtuin and PARP enzymes interact through modifying acetylation levels, or in some cases these enzymes may interact through mutual regulation of gene expression. These interactions were described to modulate a series of biological processes ranging from metabolism, oxidative stress-mediated diseases through DNA repair to ageing. Apparently, the appropriate balance between sirtuin and PARP activity is crucial to adequately regulate these processes. In some instances these balances are disturbed (e.g in ageing (Braidy, et al. 2011, Massudi, et al. 2012.)) that presumably contributes to these pathologies.
Both SIRT1 and PARPs can be modulated pharmacologically (Feige, et al. 2008, Fong, et al. 2009, Jagtap and Szabo 2005, Lagouge, et al. 2006.) enabling the modulation of both ends of this molecular seesaw. Importantly, there is ample data that not only the genetical, but pharmacological modulation of PARP, or SIRT1 activity affects the other partner (Bai, et al. 2011b, Liu, et al. 2009, Liu, et al. 2008, Pang, et al. 2011, Pillai, et al. 2006, Rajamohan, et al. 2009.) suggesting that it is possible to appropriately arrange the SIRT-PARP balance by applying pharmacological agents.
It should be noted that the activity of SIRTs and PARPs can be modulated through cellular NAD+ levels (Chambon, et al. 1963, Imai, et al. 2000.) suggesting the applicability of NAD+ precursors, such as NR, NA, or NAM. These agents impact profoundly on pathologies involving SIRTs or PARPs (Canto, et al. 2012, Jackson, et al. 1995, Schwarcz, et al. 1983, Virag and Szabo 2002.) making them likely tools to fine tune PARP and SIRT enzymes, however their exact mode of action on both enzyme families had not been sufficiently mapped. The physiological and pathophysiological processes that govern NAD+ levels are summarized on Figure 8.
Figure 8. Physiological and pathophysiological processes modulating NAD+ levels.
Physiolgical and pathological processes (in blue boxes) that enhance NAD+ content, or availability are marked by green arrow, while those ones that lead to NAD+ degradation are in red.
It is very likely that the occasions of sirtuin – PARP interaction is way more widespread than demonstrated experimentally - as discussed in section 3.2 - suggesting that the circle of pathological states, where PARP-SIRT disbalance takes place can be enlarged and better defined. In fact, this notion raises the question whether other sirtuins, or PARPs may interact. It will be also important to distinguish between the gene-specific effects and the side effects of the available pharmacological agents (Antolin, et al. 2012, Nicolescu, et al. 2009, Pacholec, et al. 2010.) and to clearly define effects that are PARP, or SIRT independent in these pathologies (Bai, et al. 2011b.). Our current knowledge on the SIRT – PARP interaction represent only the tip of the iceberg and the field is expanding that will warrant further research in that field.
Acknowledgements
This work was supported by Bolyai fellowship to PB, grants from the National Innovation Office (TéT_09-2010-0023, Baross program Seahorse grant), TÁMOP-4.2.1./B-09/KONV-2010-0007, TÁMOP-4.2.2/A-11/1/KONV-2012-0025, TÁMOP-4.2.2/B-10/1-2010-0024, OTKA PD83473, K105872 and Medical and Health Science Center (Mecenatura Mec-8/2011).
Vitae
Carles Cantó is a metabolism and diabetes specialist at the Nestlé Institute of Health Sciences S. A. in Lausanne, Switzerland. His research field involves the study of metabolic sensing and adaptation, including sirtuins and NAD+ signaling.
Anthony A. Sauve is an associate professor of Pharmacology Weill Cornell Medical College, New York, NY, USA. His research involves the study of NAD+ signaling and ADP-ribosyltransferases.
Peter Bai is an associate professor at the University of Debrecen, Hungary. His research focuses on the metabolic properties of PARP enzymes and the role of PARPs in oxidative stress response.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure
P. B. and A. A. S. declare no conflict of interest. C.C. is an employee of the Nestlé Institute of Health Sciences S.A. and declares no financial interests related to the work discussed in this review.
References
- Alcendor RR, Gao S, Zhai P, Zablocki D, Holle E, Yu X, Tian B, Wagner T, Vatner SF, Sadoshima J. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ Res. 2007;100:1512–1521. doi: 10.1161/01.RES.0000267723.65696.4a. [DOI] [PubMed] [Google Scholar]
- Allinson SL, Dianova, Dianov GL. Poly(ADP-ribose) polymerase in base excision repair: always engaged, but not essential for DNA damage processing. Acta Biochim Pol. 2003;50:169–179. [PubMed] [Google Scholar]
- Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009;37:3723–3738. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul R, Hoffer A, Stephen JD. Influence of nicotinic acid on serum cholesterol in man. Arch Biochem Biophys. 1955;54:558–559. doi: 10.1016/0003-9861(55)90070-9. [DOI] [PubMed] [Google Scholar]
- Alvarez-Gonzalez R, Mendoza-Alvarez H. Dissection of ADP-ribose polymer synthesis into individual steps of initiation, elongation, and branching. Biochimie. 1995;77:403–407. doi: 10.1016/0300-9084(96)88153-3. [DOI] [PubMed] [Google Scholar]
- Ame JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, Muller S, Hoger T, Menissier-de Murcia J, de Murcia G. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J.Biol.Chem. 1999;274:17860–17868. doi: 10.1074/jbc.274.25.17860. [DOI] [PubMed] [Google Scholar]
- Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- Antolin AA, Jalencas X, Yelamos J, Mestres J. Identification of Pim Kinases as Novel Targets for PJ34 with Confounding Effects in PARP Biology. ACS Chem Biol. 2012 doi: 10.1021/cb300317y. DOI: 10.1021/cb300317y. [DOI] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- Asher G, Reinke H, Altmeyer M, Gutierrez-Arcelus M, Hottiger MO, Schibler U. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell. 2010;142:943–953. doi: 10.1016/j.cell.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Bai P, Bakondi E, Szabo E, Gergely P, Szabo C, Virag L. Partial protection by poly(ADP-ribose) polymerase inhibitors from nitroxyl-induced cytotoxity in thymocytes. Free Radic.Biol.Med. 2001;31:1616–1623. doi: 10.1016/s0891-5849(01)00756-0. [DOI] [PubMed] [Google Scholar]
- Bai P, Canto C. The role of PARP-1 and PARP-2 enzymes in metabolic regulation and disease. Cell Metab. 2012;16:290–295. doi: 10.1016/j.cmet.2012.06.016. [DOI] [PubMed] [Google Scholar]
- Bai P, Canto C, Brunyanszki A, Huber A, Szanto M, Cen Y, Yamamoto H, Houten SM, Kiss B, Oudart H, Gergely P, Menissier-de Murcia J, Schreiber V, Sauve AA, Auwerx J. PARP-2 regulates SIRT1 expression and whole-body energy expenditure. Cell Metab. 2011a;13:450–460. doi: 10.1016/j.cmet.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai P, Canto C, Oudart H, Brunyanszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, Schoonjans K, Schreiber V, Sauve AA, Menissier-de Murcia J, Auwerx J. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab. 2011b;13:461–468. doi: 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai P, Hegedus C, Erdelyi K, Szabo E, Bakondi E, Gergely S, Szabo C, Virag L. Protein tyrosine nitration and poly(ADP-ribose) polymerase activation in N-methyl-N-nitro-N-nitrosoguanidine-treated thymocytes: implication for cytotoxicity. Toxicol.Lett. 2007a;170:203–213. doi: 10.1016/j.toxlet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Bai P, Hegedus C, Szabo E, Gyure L, Bakondi E, Brunyanszki A, Gergely S, Szabo C, Virag L. Poly(ADP-ribose) polymerase mediates inflammation in a mouse model of contact hypersensitivity. J Invest Dermatol. 2009;129:234–238. doi: 10.1038/jid.2008.196. [DOI] [PubMed] [Google Scholar]
- Bai P, Houten SM, Huber A, Schreiber V, Watanabe M, Kiss B, de Murcia G, Auwerx J, Menissier-de Murcia J. Poly(ADP-ribose) polymerase-2 controls adipocyte differentiation and adipose tissue function through the regulation of the activity of the retinoid X receptor/peroxisome proliferator-activated receptor-gamma heterodimer. J.Biol.Chem. 2007b;282:37738–37746. doi: 10.1074/jbc.M701021200. [DOI] [PubMed] [Google Scholar]
- Bai P, Mabley JG, Liaudet L, Virag L, Szabo C, Pacher P. Matrix metalloproteinase activation is an early event in doxorubicin-induced cardiotoxicity. Oncol.Rep. 2004;11:505–508. [PubMed] [Google Scholar]
- Bai P, Virag L. Role of poly(ADP-ribose) polymerases in the regulation of inflammatory processes. FEBS Lett. 2012;586:3771–3777. doi: 10.1016/j.febslet.2012.09.026. [DOI] [PubMed] [Google Scholar]
- Bakondi E, Bai P, Erdelyi K, Szabo C, Gergely P, Virag L. Cytoprotective effect of gallotannin in oxidatively stressed HaCaT keratinocytes: the role of poly(ADP-ribose) metabolism. Exp.Dermatol. 2004;13:170–178. doi: 10.1111/j.0906-6705.2004.0150.x. [DOI] [PubMed] [Google Scholar]
- Bakondi E, Gonczi M, Szabo E, Bai P, Pacher P, Gergely P, Kovacs L, Hunyadi J, Szabo C, Csernoch L, Virag L. Role of intracellular calcium mobilization and cell-density-dependent signaling in oxidative-stress-induced cytotoxicity in HaCaT keratinocytes. J.Invest Dermatol. 2003;121:88–95. doi: 10.1046/j.1523-1747.2003.12329.x. [DOI] [PubMed] [Google Scholar]
- Banks AS, Kim-Muller JY, Mastracci TL, Kofler NM, Qiang L, Haeusler RA, Jurczak MJ, Laznik D, Heinrich G, Samuel VT, Shulman GI, Papaioannou VE, Accili D. Dissociation of the glucose and lipid regulatory functions of FoxO1 by targeted knockin of acetylation-defective alleles in mice. Cell Metab. 2011;14:587–597. doi: 10.1016/j.cmet.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks AS, Kon N, Knight C, Matsumoto M, Gutierrez-Juarez R, Rossetti L, Gu W, Accili D. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008;8:333–341. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber MF, Michishita-Kioi E, Xi Y, Tasselli L, Kioi M, Moqtaderi Z, Tennen RI, Paredes S, Young NL, Chen K, Struhl K, Garcia BA, Gozani O, Li W, Chua KF. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature. 2012;487:114–118. doi: 10.1038/nature11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur JA. Resveratrol, sirtuins, and the promise of a DR mimetic. Mech Ageing Dev. 2010;131:261–269. doi: 10.1016/j.mad.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell EL, Emerling BM, Ricoult SJ, Guarente L. SirT3 suppresses hypoxia inducible factor 1alpha and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011;30:2986–2996. doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellocchi D, Costantino G, Pellicciari R, Re N, Marrone A, Coletti C. Poly(ADP-ribose)-polymerase-catalyzed hydrolysis of NAD+: QM/MM simulation of the enzyme reaction. ChemMedChem. 2006;1:533–539. doi: 10.1002/cmdc.200500061. [DOI] [PubMed] [Google Scholar]
- Bembenek ME, Kuhn E, Mallender WD, Pullen L, Li P, Parsons T. A fluorescence-based coupling reaction for monitoring the activity of recombinant human NAD synthetase. Assay Drug Dev Technol. 2005;3:533–541. doi: 10.1089/adt.2005.3.533. [DOI] [PubMed] [Google Scholar]
- Benavente CA, Jacobson MK, Jacobson EL. NAD in skin: therapeutic approaches for niacin. Curr Pharm Des. 2009;15:29–38. doi: 10.2174/138161209787185760. [DOI] [PubMed] [Google Scholar]
- Benjamin RC, Gill DM. Poly(ADP-ribose) synthesis in vitro programmed by damaged DNA. A comparison of DNA molecules containing different types of strand breaks. J Biol Chem. 1980;255:10502–10508. [PubMed] [Google Scholar]
- Benyo Z, Gille A, Bennett CL, Clausen BE, Offermanns S. Nicotinic acid-induced flushing is mediated by activation of epidermal langerhans cells. Mol Pharmacol. 2006;70:1844–1849. doi: 10.1124/mol.106.030833. [DOI] [PubMed] [Google Scholar]
- Benyo Z, Gille A, Kero J, Csiky M, Suchankova MC, Nusing RM, Moers A, Pfeffer K, Offermanns S. GPR109A (PUMA-G/HM74A) mediates nicotinic acid-induced flushing. J Clin Invest. 2005;115:3634–3640. doi: 10.1172/JCI23626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berge KG, Canner PL. Coronary drug project: experience with niacin. Coronary Drug Project Research Group. Eur J Clin Pharmacol. 1991;40(Suppl 1):S49–51. [PubMed] [Google Scholar]
- Berger F, Lau C, Ziegler M. Regulation of poly(ADP-ribose) polymerase 1 activity by the phosphorylation state of the nuclear NAD biosynthetic enzyme NMN adenylyl transferase 1. Proc.Natl.Acad.Sci.U.S.A. 2007;104:3765–3770. doi: 10.1073/pnas.0609211104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger NA. Poly(ADP-ribose) in the cellular response to DNA damage. Radiat Res. 1985;101:4–15. [PubMed] [Google Scholar]
- Bieganowski P, Brenner C. The reported human NADsyn2 is ammonia-dependent NAD synthetase from a pseudomonad. J Biol Chem. 2003;278:33056–33059. doi: 10.1074/jbc.M302276200. [DOI] [PubMed] [Google Scholar]
- Bieganowski P, Brenner C. Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell. 2004;117:495–502. doi: 10.1016/s0092-8674(04)00416-7. [DOI] [PubMed] [Google Scholar]
- Bobrowska A, Donmez G, Weiss A, Guarente L, Bates G. SIRT2 ablation has no effect on tubulin acetylation in brain, cholesterol biosynthesis or the progression of Huntington’s disease phenotypes in vivo. PLoS One. 2012;7:e34805. doi: 10.1371/journal.pone.0034805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehler C, Gauthier LR, Mortusewicz O, Biard DS, Saliou JM, Bresson A, Sanglier-Cianferani S, Smith S, Schreiber V, Boussin F, Dantzer F. Poly(ADP-ribose) polymerase 3 (PARP3), a newcomer in cellular response to DNA damage and mitotic progression. Proc Natl Acad Sci U S A. 2011;108:2783–2788. doi: 10.1073/pnas.1016574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan KL, Brenner C. Nicotinic acid, nicotinamide, and nicotinamide riboside: a molecular evaluation of NAD+ precursor vitamins in human nutrition. Annu Rev Nutr. 2008;28:115–130. doi: 10.1146/annurev.nutr.28.061807.155443. [DOI] [PubMed] [Google Scholar]
- Bordone L, Motta MC, Picard F, Robinson A, Jhala US, Apfeld J, McDonagh T, Lemieux M, McBurney M, Szilvasi A, Easlon EJ, Lin SJ, Guarente L. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 2006;4:e31. doi: 10.1371/journal.pbio.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borra MT, Smith BC, Denu JM. Mechanism of human SIRT1 activation by resveratrol. J Biol Chem. 2005;280:17187–17195. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- Braidy N, Guillemin GJ, Mansour H, Chan-Ling T, Poljak A, Grant R. Age related changes in NAD+ metabolism oxidative stress and Sirt1 activity in wistar rats. PLoS One. 2011;6:e19194. doi: 10.1371/journal.pone.0019194. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Brunyanszki A, Hegedus C, Szanto M, Erdelyi K, Kovacs K, Schreiber V, Gergely S, Kiss B, Szabo E, Virag L, Bai P. Genetic Ablation of PARP-1 Protects Against Oxazolone-Induced Contact Hypersensitivity by Modulating Oxidative Stress. J Invest Dermatol. 2010;130:2629–2637. doi: 10.1038/jid.2010.190. [DOI] [PubMed] [Google Scholar]
- Burgos ES, Schramm VL. Weak coupling of ATP hydrolysis to the chemical equilibrium of human nicotinamide phosphoribosyltransferase. Biochemistry. 2008;47:11086–11096. doi: 10.1021/bi801198m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkart V, Blaeser K, Kolb H. Potent beta-cell protection in vitro by an isoquinolinone-derived PARP inhibitor. Horm Metab Res. 1999a;31:641–644. doi: 10.1055/s-2007-978813. [DOI] [PubMed] [Google Scholar]
- Burkart V, Wang ZQ, Radons J, Heller B, Herceg Z, Stingl L, Wagner EF, Kolb H. Mice lacking the poly(ADP-ribose) polymerase gene are resistant to pancreatic beta-cell destruction and diabetes development induced by streptozocin. Nat Med. 1999b;5:314–319. doi: 10.1038/6535. [DOI] [PubMed] [Google Scholar]
- Burkle A, Muller M, Wolf I, Kupper JH. Poly(ADP-ribose) polymerase activity in intact or permeabilized leukocytes from mammalian species of different longevity. Mol Cell Biochem. 1994;138:85–90. [PubMed] [Google Scholar]
- Burnett C, Valentini S, Cabreiro F, Goss M, Somogyvari M, Piper MD, Hoddinott M, Sutphin GL, Leko V, McElwee JJ, Vazquez-Manrique RP, Orfila AM, Ackerman D, Au C, Vinti G, Riesen M, Howard K, Neri C, Bedalov A, Kaeberlein M, Soti C, Partridge L, Gems D. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477:482–485. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burzio LO, Riquelme PT, Koide SS. ADP ribosylation of rat liver nucleosomal core histones. J Biol Chem. 1979;254:3029–3037. [PubMed] [Google Scholar]
- Buteau J, Accili D. Regulation of pancreatic beta-cell function by the forkhead protein FoxO1. Diabetes Obes Metab. 2007;9(Suppl 2):140–146. doi: 10.1111/j.1463-1326.2007.00782.x. [DOI] [PubMed] [Google Scholar]
- Caito S, Hwang JW, Chung S, Yao H, Sundar IK, Rahman I. PARP-1 inhibition does not restore oxidant-mediated reduction in SIRT1 activity. Biochem Biophys Res Commun. 2010a;392:264–270. doi: 10.1016/j.bbrc.2009.12.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caito S, Rajendrasozhan S, Cook S, Chung S, Yao H, Friedman AE, Brookes PS, Rahman I. SIRT1 is a redox-sensitive deacetylase that is post-translationally modified by oxidants and carbonyl stress. Faseb J. 2010b;24:3145–3159. doi: 10.1096/fj.09-151308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Auwerx J. Caloric restriction, SIRT1 and longevity. Trends Endocrinol Metab. 2009;20:325–331. doi: 10.1016/j.tem.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Auwerx J. AMP-activated protein kinase and its downstream transcriptional pathways. Cell Mol Life Sci. 2010;67:3407–3423. doi: 10.1007/s00018-010-0454-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Auwerx J. Calorie restriction: is AMPK a key sensor and effector? Physiology (Bethesda) 2011a;26:214–224. doi: 10.1152/physiol.00010.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Auwerx J. Interference between PARPs and SIRT1: a novel approach to healthy ageing? Aging (Albany NY) 2011b;3:543–547. doi: 10.18632/aging.100326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Auwerx J. Targeting sirtuin 1 to improve metabolism: all you need is NAD+? Pharmacol Rev. 2012;64:166–187. doi: 10.1124/pr.110.003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Houtkooper RH, Pirinen E, Youn DY, Oosterveer MH, Cen Y, Fernandez-Marcos PJ, Yamamoto H, Andreux PA, Cettour-Rose P, Gademann K, Rinsch C, Schoonjans K, Sauve AA, Auwerx J. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 2012;15:838–847. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Jiang LQ, Deshmukh AS, Mataki C, Coste A, Lagouge M, Zierath JR, Auwerx J. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab. 2010;11:213–219. doi: 10.1016/j.cmet.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capuzzi DM, Morgan JM, Brusco OA, Jr., Intenzo CM. Niacin dosing: relationship to benefits and adverse effects. Curr Atheroscler Rep. 2000;2:64–71. doi: 10.1007/s11883-000-0096-y. [DOI] [PubMed] [Google Scholar]
- Cen Y, Sauve AA. Transition state of ADP-ribosylation of acetyllysine catalyzed by Archaeoglobus fulgidus Sir2 determined by kinetic isotope effects and computational approaches. J Am Chem Soc. 2010;132:12286–12298. doi: 10.1021/ja910342d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambon P, Weill JD, Mandel P. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem.Biophys.Res.Commun. 1963;11:39–43. doi: 10.1016/0006-291x(63)90024-x. 39-43. [DOI] [PubMed] [Google Scholar]
- Champy MF, Selloum M, Zeitler V, Caradec C, Jung B, Rousseau S, Pouilly L, Sorg T, Auwerx J. Genetic background determines metabolic phenotypes in the mouse. Mamm Genome. 2008;19:318–331. doi: 10.1007/s00335-008-9107-z. [DOI] [PubMed] [Google Scholar]
- Chen D, Bruno J, Easlon E, Lin SJ, Cheng HL, Alt FW, Guarente L. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22:1753–1757. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci U S A. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KC, Ryu OH, Lee KW, Kim HY, Seo JA, Kim SG, Kim NH, Choi DS, Baik SH, Choi KM. Effect of PPAR-alpha and -gamma agonist on the expression of visfatin, adiponectin, and TNF-alpha in visceral fat of OLETF rats. Biochem.Biophys.Res.Commun. 2005;336:747–753. doi: 10.1016/j.bbrc.2005.08.203. [DOI] [PubMed] [Google Scholar]
- Chong ZZ, Shang YC, Wang S, Maiese K. SIRT1: new avenues of discovery for disorders of oxidative stress. Expert Opin Ther Targets. 2012;16:167–178. doi: 10.1517/14728222.2012.648926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Yao H, Caito S, Hwang JW, Arunachalam G, Rahman I. Regulation of SIRT1 in cellular functions: role of polyphenols. Arch Biochem Biophys. 2010;501:79–90. doi: 10.1016/j.abb.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipriani G, Rapizzi E, Vannacci A, Rizzuto R, Moroni F, Chiarugi A. Nuclear poly(ADP-ribose) polymerase-1 rapidly triggers mitochondrial dysfunction. J.Biol.Chem. 2005;280:17227–17234. doi: 10.1074/jbc.M414526200. [DOI] [PubMed] [Google Scholar]
- Clark SJ, Falchi M, Olsson B, Jacobson P, Cauchi S, Balkau B, Marre M, Lantieri O, Andersson JC, Jernas M, Aitman TJ, Richardson S, Sjostrom L, Wong HY, Carlsson LM, Froguel P, Walley AJ. Association of sirtuin 1 (SIRT1) gene SNPs and transcript expression levels with severe obesity. Obesity (Silver Spring) 2012;20:178–185. doi: 10.1038/oby.2011.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colabroy KL, Begley TP. The pyridine ring of NAD is formed by a nonenzymatic pericyclic reaction. J Am Chem Soc. 2005;127:840–841. doi: 10.1021/ja0446395. [DOI] [PubMed] [Google Scholar]
- Costford SR, Bajpeyi S, Pasarica M, Albarado DC, Thomas SC, Xie H, Church TS, Jubrias SA, Conley KE, Smith SR. Skeletal muscle NAMPT is induced by exercise in humans. Am J Physiol Endocrinol Metab. 2010;298:E117–126. doi: 10.1152/ajpendo.00318.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin N. PARP inhibitors and cancer therapy. 2006:218–223. [Google Scholar]
- Dantzer F, de La RG, Menissier-de Murcia J, Hostomsky Z, de Murcia G, Schreiber V. Base excision repair is impaired in mammalian cells lacking Poly(ADP-ribose) polymerase-1. Biochemistry. 2000;39:7559–7569. doi: 10.1021/bi0003442. [DOI] [PubMed] [Google Scholar]
- Danz ED, Skramsted J, Henry N, Bennett JA, Keller RS. Resveratrol prevents doxorubicin cardiotoxicity through mitochondrial stabilization and the Sirt1 pathway. Free Radic Biol Med. 2009;46:1589–1597. doi: 10.1016/j.freeradbiomed.2009.03.011. [DOI] [PubMed] [Google Scholar]
- Dasgupta B, Milbrandt J. Resveratrol stimulates AMP kinase activity in neurons. Proc Natl Acad Sci U S A. 2007;104:7217–7222. doi: 10.1073/pnas.0610068104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Murcia G, Huletsky A, Lamarre D, Gaudreau A, Pouyet J, Daune M, Poirier GG. Modulation of chromatin superstructure induced by poly(ADP-ribose) synthesis and degradation. J.Biol.Chem. 1986;261:7011–7017. [PubMed] [Google Scholar]
- de Murcia G, Schreiber V, Molinete M, Saulier B, Poch O, Masson M, Niedergang C, Menissier de MJ. Structure and function of poly(ADP-ribose) polymerase. Mol.Cell Biochem. 1994;138:15–24. doi: 10.1007/BF00928438. [DOI] [PubMed] [Google Scholar]
- De Vos M, Schreiber V, Dantzer F. The diverse roles and clinical relevance of PARPs in DNA damage repair: current state of the art. Biochem Pharmacol. 2012;84:137–146. doi: 10.1016/j.bcp.2012.03.018. [DOI] [PubMed] [Google Scholar]
- DeFronzo RA, Gunnarsson R, Bjorkman O, Olsson M, Wahren J. Effects of insulin on peripheral and splanchnic glucose metabolism in noninsulin-dependent (type II) diabetes mellitus. J Clin Invest. 1985;76:149–155. doi: 10.1172/JCI111938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devalaraja-Narashimha K, Padanilam BJ. PARP1 deficiency exacerbates diet-induced obesity in mice. J Endocrinol. 2010;205:243–252. doi: 10.1677/JOE-09-0402. [DOI] [PubMed] [Google Scholar]
- Di Paola S, Micaroni M, Di Tullio G, Buccione R, Di Girolamo M. PARP16/ARTD15 Is a Novel Endoplasmic-Reticulum-Associated Mono-ADP-Ribosyltransferase That Interacts with, and Modifies Karyopherin-ss1. PLoS One. 2012;7:e37352. doi: 10.1371/journal.pone.0037352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Guo T, Traurig M, Mason CC, Kobes S, Perez J, Knowler WC, Bogardus C, Hanson RL, Baier LJ. SIRT1 is associated with a decrease in acute insulin secretion and a sex specific increase in risk for type 2 diabetes in Pima Indians. Mol Genet Metab. 2011;104:661–665. doi: 10.1016/j.ymgme.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet-Chabeaud G, Godon C, Brutesco C, de Murcia G, Kazmaier M. Ionising radiation induces the expression of PARP-1 and PARP-2 genes in Arabidopsis. Mol.Genet.Genomics. 2001;265:954–963. doi: 10.1007/s004380100506. [DOI] [PubMed] [Google Scholar]
- Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, He B, Chen W, Zhang S, Cerione RA, Auwerx J, Hao Q, Lin H. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Zhang X, Han YY, Burke NA, Kochanek PM, Watkins SC, Graham SH, Carcillo JA, Szabo C, Clark RS. Intra-mitochondrial poly(ADP-ribosylation) contributes to NAD+ depletion and cell death induced by oxidative stress. J Biol Chem. 2003;278:18426–18433. doi: 10.1074/jbc.M301295200. [DOI] [PubMed] [Google Scholar]
- Durkacz BW, Omidiji O, Gray DA, Shall S. (ADP-ribose)n participates in DNA excision repair. Nature. 1980;283:593–596. doi: 10.1038/283593a0. [DOI] [PubMed] [Google Scholar]
- El Ramy R, Magroun N, Messadecq N, Gauthier LR, Boussin FD, Kolthur-Seetharam U, Schreiber V, McBurney MW, Sassone-Corsi P, Dantzer F. Functional interplay between Parp-1 and SirT1 in genome integrity and chromatin-based processes. Cell Mol Life Sci. 2009;66:3219–3234. doi: 10.1007/s00018-009-0105-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdelyi K, Bai P, Kovacs I, Szabo E, Mocsar G, Kakuk A, Szabo C, Gergely P, Virag L. Dual role of poly(ADP-ribose) glycohydrolase in the regulation of cell death in oxidatively stressed A549 cells. Faseb J. 2009;23:3553–3563. doi: 10.1096/fj.09-133264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdelyi K, Kiss A, Bakondi E, Bai P, Szabo C, Gergely P, Erdodi F, Virag L. Gallotannin inhibits the expression of chemokines and inflammatory cytokines in A549 cells. Mol.Pharmacol. 2005;68:895–904. doi: 10.1124/mol.105.012518. [DOI] [PubMed] [Google Scholar]
- Erener S, Hesse M, Kostadinova R, Hottiger MO. Poly(ADP-ribose)polymerase-1 (PARP1) controls adipogenic gene expression and adipocyte function. Mol Endocrinol. 2012a;26:79–86. doi: 10.1210/me.2011-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erener S, Mirsaidi A, Hesse M, Tiaden AN, Ellingsgaard H, Kostadinova R, Donath MY, Richards PJ, Hottiger MO. ARTD1 deletion causes increased hepatic lipid accumulation in mice fed a high-fat diet and impairs adipocyte function and differentiation. Faseb J. 2012b;26:2631–2638. doi: 10.1096/fj.11-200212. [DOI] [PubMed] [Google Scholar]
- Feige JN, Lagouge M, Canto C, Strehle A, Houten SM, Milne JC, Lambert PD, Mataki C, Elliott PJ, Auwerx J. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8:347–358. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- Feng Y, Paul IA, LeBlanc MH. Nicotinamide reduces hypoxic ischemic brain injury in the newborn rat. Brain Res Bull. 2006;69:117–122. doi: 10.1016/j.brainresbull.2005.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Marcos PJ, Jeninga EH, Canto C, Harach T, de Boer VC, Andreux P, Moullan N, Pirinen E, Yamamoto H, Houten SM, Schoonjans K, Auwerx J. Muscle or liver-specific Sirt3 deficiency induces hyperacetylation of mitochondrial proteins without affecting global metabolic homeostasis. Sci Rep. 2012;2:425. doi: 10.1038/srep00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraris DV. Evolution of poly(ADP-ribose) polymerase-1 (PARP-1) inhibitors. From concept to clinic. J Med Chem. 2010;53:4561–4584. doi: 10.1021/jm100012m. [DOI] [PubMed] [Google Scholar]
- Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O’Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JH, de Bono JS. Inhibition of Poly(ADP-Ribose) Polymerase in Tumors from BRCA Mutation Carriers. N Engl J Med. 2009;361(2):123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006;20:1075–1080. doi: 10.1101/gad.1399706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstermann U, Sessa WC. Nitric oxide synthases: regulation and function. Eur Heart J. 2012;33:829–837. doi: 10.1093/eurheartj/ehr304. 837a-837d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French JB, Cen Y, Vrablik TL, Xu P, Allen E, Hanna-Rose W, Sauve AA. Characterization of nicotinamidases: steady state kinetic parameters, classwide inhibition by nicotinaldehydes, and catalytic mechanism. Biochemistry. 2010;49:10421–10439. doi: 10.1021/bi1012518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frizzell KM, Gamble MJ, Berrocal JG, Zhang T, Krishnakumar R, Cen Y, Sauve AA, Kraus WL. Global analysis of transcriptional regulation by poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase in MCF-7 human breast cancer cells. J Biol Chem. 2009;284:33926–33938. doi: 10.1074/jbc.M109.023879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuwatari T, Ohta M, Kimtjra N, Sasaki R, Shibata K. Conversion ratio of tryptophan to niacin in Japanese women fed a purified diet conforming to the Japanese Dietary Reference Intakes. J Nutr Sci Vitaminol (Tokyo) 2004;50:385–391. doi: 10.3177/jnsv.50.385. [DOI] [PubMed] [Google Scholar]
- Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, Sartorelli V. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell. 2008;14:661–673. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne JP, Moreel X, Gagne P, Labelle Y, Droit A, Chevalier-Pare M, Bourassa S, McDonald D, Hendzel MJ, Prigent C, Poirier GG. Proteomic investigation of phosphorylation sites in poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase. J Proteome Res. 2009;8:1014–1029. doi: 10.1021/pr800810n. [DOI] [PubMed] [Google Scholar]
- Galassi L, Di Stefano M, Brunetti L, Orsomando G, Amici A, Ruggieri S, Magni G. Characterization of human nicotinate phosphoribosyltransferase: Kinetic studies, structure prediction and functional analysis by site-directed mutagenesis. Biochimie. 2012;94:300–309. doi: 10.1016/j.biochi.2011.06.033. [DOI] [PubMed] [Google Scholar]
- Gale EA, Bingley PJ, Emmett CL, Collier T. European Nicotinamide Diabetes Intervention Trial (ENDIT): a randomised controlled trial of intervention before the onset of type 1 diabetes. Lancet. 2004;363:925–931. doi: 10.1016/S0140-6736(04)15786-3. [DOI] [PubMed] [Google Scholar]
- Gehl Z, Bai P, Bodnar E, Emri G, Remenyik E, Nemeth J, Gergely P, Virag L, Szabo E. Poly(ADP-ribose) in the skin and in melanomas. Histol Histopathol. 2012;27:651–659. doi: 10.14670/HH-27.651. [DOI] [PubMed] [Google Scholar]
- Geraets L, Moonen HJ, Brauers K, Wouters EF, Bast A, Hageman GJ. Dietary flavones and flavonoles are inhibitors of poly(ADP-ribose)polymerase-1 in pulmonary epithelial cells. J Nutr. 2007;137:2190–2195. doi: 10.1093/jn/137.10.2190. [DOI] [PubMed] [Google Scholar]
- Geraets L, Moonen HJ, Wouters EF, Bast A, Hageman GJ. Caffeine metabolites are inhibitors of the nuclear enzyme poly(ADP-ribose)polymerase-1 at physiological concentrations. Biochem Pharmacol. 2006;72:902–910. doi: 10.1016/j.bcp.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Gerhart-Hines Z, Dominy JE, Jr., Blattler SM, Jedrychowski MP, Banks AS, Lim JH, Chim H, Gygi SP, Puigserver P. The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD(+) Mol Cell. 2011;44:851–863. doi: 10.1016/j.molcel.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gholson RK, Ueda I, Ogasawara N, Henderson LM. The Enzymatic Conversion of Quinolinate to Nicotinic Acid Mononucleotide in Mammalian Liver. J Biol Chem. 1964;239:1208–1214. [PubMed] [Google Scholar]
- Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- Gilley J, Coleman MP. Endogenous Nmnat2 is an essential survival factor for maintenance of healthy axons. PLoS Biol. 2010;8:e1000300. doi: 10.1371/journal.pbio.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giralt A, Villarroya F. SIRT3, a pivotal actor in mitochondrial functions: metabolism, cell death and aging. Biochem J. 2012;444:1–10. doi: 10.1042/BJ20120030. [DOI] [PubMed] [Google Scholar]
- Goodwin PM, Lewis PJ, Davies MI, Skidmore CJ, Shall S. The effect of gamma radiation and neocarzinostatin on NAD and ATP levels in mouse leukaemia cells. Biochim Biophys Acta. 1978;543:576–582. doi: 10.1016/0304-4165(78)90312-4. [DOI] [PubMed] [Google Scholar]
- Gross DN, van den Heuvel AP, Birnbaum MJ. The role of FoxO in the regulation of metabolism. Oncogene. 2008;27:2320–2336. doi: 10.1038/onc.2008.25. [DOI] [PubMed] [Google Scholar]
- Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 2000;14:1021–1026. [PubMed] [Google Scholar]
- Ha GH, Kim HS, Go H, Lee H, Seimiya H, Chung DH, Lee CW. Tankyrase-1 function at telomeres and during mitosis is regulated by Polo-like kinase-1-mediated phosphorylation. Cell Death Differ. 2012;19:321–332. doi: 10.1038/cdd.2011.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha HC, Juluri K, Zhou Y, Leung S, Hermankova M, Snyder SH. Poly(ADP-ribose) polymerase-1 is required for efficient HIV-1 integration. Proc.Natl.Acad.Sci.U.S.A. 2001;98:3364–3368. doi: 10.1073/pnas.051633498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha HC, Snyder SH. Poly(ADP-ribose) polymerase is a mediator of necrotic cell death by ATP depletion. Proc Natl Acad Sci U S A. 1999;96:13978–13982. doi: 10.1073/pnas.96.24.13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenni SS, Hassa PO, Altmeyer M, Fey M, Imhof R, Hottiger MO. Identification of lysines 36 and 37 of PARP-2 as targets for acetylation and auto-ADP-ribosylation. Int.J.Biochem.Cell Biol. 2008;40:2274–2283. doi: 10.1016/j.biocel.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Hageman GJ, Stierum RH. Niacin, poly(ADP-ribose) polymerase-1 and genomic stability. Mutat Res. 2001;475:45–56. doi: 10.1016/s0027-5107(01)00078-1. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Hallows WC, Albaugh BN, Denu JM. Where in the cell is SIRT3?--functional localization of an NAD+-dependent protein deacetylase. Biochem J. 2008;411:e11–13. doi: 10.1042/BJ20080336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Zhou R, Niu J, McNutt MA, Wang P, Tong T. SIRT1 is regulated by a PPAR{gamma}-SIRT1 negative feedback loop associated with senescence. Nucleic Acids Res. 2010;38:7458–7471. doi: 10.1093/nar/gkq609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MK, Song EK, Guo Y, Ou X, Mantel C, Broxmeyer HE. SIRT1 regulates apoptosis and Nanog expression in mouse embryonic stem cells by controlling p53 subcellular localization. Cell Stem Cell. 2008;2:241–251. doi: 10.1016/j.stem.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harden A, Young WJ. The alcoholic ferment of yeast-juice. Proceedings of the Royal Society of London Series B-Containing Papers of a Biological Character. 1906;77:405–420. [Google Scholar]
- Hasegawa K, Wakino S, Yoshioka K, Tatematsu S, Hara Y, Minakuchi H, Washida N, Tokuyama H, Hayashi K, Itoh H. Sirt1 protects against oxidative stress-induced renal tubular cell apoptosis by the bidirectional regulation of catalase expression. Biochem.Biophys.Res.Commun. 2008;372:51–56. doi: 10.1016/j.bbrc.2008.04.176. [DOI] [PubMed] [Google Scholar]
- Hassa PO, Covic M, Hasan S, Imhof R, Hottiger MO. The enzymatic and DNA binding activity of PARP-1 are not required for NF-kappa B coactivator function. J.Biol.Chem. 2001;276:45588–45597. doi: 10.1074/jbc.M106528200. [DOI] [PubMed] [Google Scholar]
- Hassa PO, Haenni SS, Buerki C, Meier NI, Lane WS, Owen H, Gersbach M, Imhof R, Hottiger MO. Acetylation of poly(ADP-ribose) polymerase-1 by p300/CREB-binding protein regulates coactivation of NF-kappaB-dependent transcription. J Biol Chem. 2005;280:40450–40464. doi: 10.1074/jbc.M507553200. [DOI] [PubMed] [Google Scholar]
- Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Tanaka M, Shimada T, Miwa M, Sugimura T. Size and shape of poly(ADP-ribose): examination by gel filtration, gel electrophoresis and electron microscopy. Biochem Biophys Res Commun. 1983;112:102–107. doi: 10.1016/0006-291x(83)91803-x. [DOI] [PubMed] [Google Scholar]
- Heikkinen S, Auwerx J, Argmann CA. PPARgamma in human and mouse physiology. Biochim Biophys Acta. 2007;1771:999–1013. doi: 10.1016/j.bbalip.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka M, Inoue T, Toda T, Kimura N, Shirayoshi Y, Kamitani H, Watanabe T, Ohama E, Tahimic CG, Kurimasa A, Oshimura M. Proteomics-based identification of differentially expressed genes in human gliomas: down-regulation of SIRT2 gene. Biochem Biophys Res Commun. 2003;309:558–566. doi: 10.1016/j.bbrc.2003.08.029. [DOI] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV, Jr., Alt FW, Kahn CR, Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Jing E, Grueter CA, Collins AM, Aouizerat B, Stancakova A, Goetzman E, Lam MM, Schwer B, Stevens RD, Muehlbauer MJ, Kakar S, Bass NM, Kuusisto J, Laakso M, Alt FW, Newgard CB, Farese RV, Jr., Kahn CR, Verdin E. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol Cell. 2011;44:177–190. doi: 10.1016/j.molcel.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoane MR, Akstulewicz SL, Toppen J. Treatment with vitamin B3 improves functional recovery and reduces GFAP expression following traumatic brain injury in rats. J Neurotrauma. 2003;20:1189–1199. doi: 10.1089/089771503770802871. [DOI] [PubMed] [Google Scholar]
- Hoane MR, Gilbert DR, Holland MA, Pierce JL. Nicotinamide reduces acute cortical neuronal death and edema in the traumatically injured brain. Neurosci Lett. 2006a;408:35–39. doi: 10.1016/j.neulet.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Hoane MR, Kaplan SA, Ellis AL. The effects of nicotinamide on apoptosis and blood-brain barrier breakdown following traumatic brain injury. Brain Res. 2006b;1125:185–193. doi: 10.1016/j.brainres.2006.10.019. [DOI] [PubMed] [Google Scholar]
- Hoane MR, Tan AA, Pierce JL, Anderson GD, Smith DC. Nicotinamide treatment reduces behavioral impairments and provides cortical protection after fluid percussion injury in the rat. J Neurotrauma. 2006c;23:1535–1548. doi: 10.1089/neu.2006.23.1535. [DOI] [PubMed] [Google Scholar]
- Hom FG, Goodner CJ. Insulin dose-response characteristics among individual muscle and adipose tissues measured in the rat in vivo with 3[H]2-deoxyglucose. Diabetes. 1984;33:153–159. doi: 10.2337/diab.33.2.153. [DOI] [PubMed] [Google Scholar]
- Honjo T, Nishizuka Y, Hayaishi O. Diphtheria toxin-dependent adenosine diphosphate ribosylation of aminoacyl transferase II and inhibition of protein synthesis. J Biol Chem. 1968;243:3553–3555. [PubMed] [Google Scholar]
- Hottiger MO, Hassa PO, Luscher B, Schuler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Hou X, Xu S, Maitland-Toolan KA, Sato K, Jiang B, Ido Y, Lan F, Walsh K, Wierzbicki M, Verbeuren TJ, Cohen RA, Zang M. SIRT1 regulates hepatocyte lipid metabolism through activating AMP-activated protein kinase. J Biol Chem. 2008;283:20015–20026. doi: 10.1074/jbc.M802187200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Auwerx J. Exploring the therapeutic space around NAD+ J Cell Biol. 2012;199:205–209. doi: 10.1083/jcb.201207019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Canto C, Wanders RJ, Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev. 2010;31:194–223. doi: 10.1210/er.2009-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- Huang D, Wang Y, Yang C, Liao Y, Huang K. Angiotensin II promotes poly(ADP-ribosyl)ation of c-Jun/c-Fos in cardiac fibroblasts. J Mol Cell Cardiol. 2009;46:25–32. doi: 10.1016/j.yjmcc.2008.10.019. [DOI] [PubMed] [Google Scholar]
- Huber A, Bai P, Menissier-de Murcia J, de Murcia G. PARP-1, PARP-2 and ATM in the DNA damage response: functional synergy in mouse development. DNA Repair (Amst) 2004;3:1103–1108. doi: 10.1016/j.dnarep.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Huber JL, McBurney MW, Distefano PS, McDonagh T. SIRT1-independent mechanisms of the putative sirtuin enzyme activators SRT1720 and SRT2183. Future Med Chem. 2010;2:1751–1759. doi: 10.4155/fmc.10.257. [DOI] [PubMed] [Google Scholar]
- Ieraci A, Herrera DG. Nicotinamide protects against ethanol-induced apoptotic neurodegeneration in the developing mouse brain. PLoS Med. 2006;3:e101. doi: 10.1371/journal.pmed.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ijichi H, Ichiyama A, Hayaishi O. Studies on the biosynthesis of nicotinamide adenine dinucleotide. 3. Comparative in vivo studies on nicotinic acid, nicotinamide, and quinolinic acid as precursors of nicotinamide adenine dinucleotide. J Biol Chem. 1966;241:3701–3707. [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Ivy JM, Klar AJ, Hicks JB. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:688–702. doi: 10.1128/mcb.6.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MD, Schmidt MT, Oppenheimer NJ, Denu JM. Mechanism of nicotinamide inhibition and transglycosidation by Sir2 histone/protein deacetylases. J Biol Chem. 2003;278:50985–50998. doi: 10.1074/jbc.M306552200. [DOI] [PubMed] [Google Scholar]
- Jackson TM, Rawling JM, Roebuck BD, Kirkland JB. Large supplements of nicotinic acid and nicotinamide increase tissue NAD+ and poly(ADP-ribose) levels but do not affect diethylnitrosamine-induced altered hepatic foci in Fischer-344 rats. J Nutr. 1995;125:1455–1461. doi: 10.1093/jn/125.6.1455. [DOI] [PubMed] [Google Scholar]
- Jagtap P, Szabo C. Poly(ADP-ribose) polymerase and the therapeutic effects of its inhibitors. Nat.Rev.Drug Discov. 2005;4:421–440. doi: 10.1038/nrd1718. [DOI] [PubMed] [Google Scholar]
- Janssen OE, Hilz H. Differentiation of 3T3-L1 pre-adipocytes induced by inhibitors of poly(ADP-ribose) polymerase and by related noninhibitory acids. Eur.J.Biochem. 1989;180:595–602. doi: 10.1111/j.1432-1033.1989.tb14686.x. [DOI] [PubMed] [Google Scholar]
- Javle M, Curtin NJ. The role of PARP in DNA repair and its therapeutic exploitation. Br J Cancer. 2011;105:1114–1122. doi: 10.1038/bjc.2011.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javle M, Curtin NJ. The potential for poly (ADP-ribose) polymerase inhibitors in cancer therapy. Ther Adv Med Oncol. 2012;3:257–267. doi: 10.1177/1758834011417039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Wang S, Xiao M, Lin Y, Zhou L, Lei Q, Xiong Y, Guan KL, Zhao S. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol Cell. 2011;43:33–44. doi: 10.1016/j.molcel.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing E, Gesta S, Kahn CR. SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab. 2007;6:105–114. doi: 10.1016/j.cmet.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, McDonagh T, Heltweg B, Hixon J, Westman EA, Caldwell SD, Napper A, Curtis R, DiStefano PS, Fields S, Bedalov A, Kennedy BK. Substrate-specific activation of sirtuins by resveratrol. J Biol Chem. 2005;280:17038–17045. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW. Sir2 and calorie restriction in yeast: a skeptical perspective. Ageing Res Rev. (3rd) 2007;6:128–140. doi: 10.1016/j.arr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, Bar-Joseph Z, Cohen HY. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- Kanfi Y, Peshti V, Gil R, Naiman S, Nahum L, Levin E, Kronfeld-Schor N, Cohen HY. SIRT6 protects against pathological damage caused by diet-induced obesity. Aging Cell. 2010;9:162–173. doi: 10.1111/j.1474-9726.2009.00544.x. [DOI] [PubMed] [Google Scholar]
- Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, Bycroft M, Ladurner AG. The macro domain is an ADP-ribose binding module. Embo J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz J, Tayek JA. Gluconeogenesis and the Cori cycle in 12-, 20-, and 40-h-fasted humans. Am J Physiol. 1998;275:E537–542. doi: 10.1152/ajpendo.1998.275.3.E537. [DOI] [PubMed] [Google Scholar]
- Kawaichi M, Oka J, Zhang J, Ueda K, Hayaishi O. Properties of poly(ADP-ribose) synthetase and ADP-ribosyl histone splitting enzyme. Princess Takamatsu Symp. 1983;13:121–128. [PubMed] [Google Scholar]
- Kawaichi M, Ueda K, Hayaishi O. Multiple autopoly(ADP-ribosyl)ation of rat liver poly(ADP-ribose) synthetase. Mode of modification and properties of automodified synthetase. J Biol Chem. 1981;256:9483–9489. [PubMed] [Google Scholar]
- Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, Vassilopoulos A, Ozden O, Park SH, Singh KK, Abdulkadir SA, Spitz DR, Deng CX, Gius D. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010a;17:41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Xiao C, Wang RH, Lahusen T, Xu X, Vassilopoulos A, Vazquez-Ortiz G, Jeong WI, Park O, Ki SH, Gao B, Deng CX. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab. 2010b;12:224–236. doi: 10.1016/j.cmet.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- Kleine H, Poreba E, Lesniewicz K, Hassa PO, Hottiger MO, Litchfield DW, Shilton BH, Luscher B. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell. 2008;32:57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Koch-Nolte F, Haag F, Guse AH, Lund F, Ziegler M. Emerging roles of NAD+ and its metabolites in cell signaling. Sci Signal. 2009;2:mr1. doi: 10.1126/scisignal.257mr1. [DOI] [PubMed] [Google Scholar]
- Kolthur-Seetharam U, Dantzer F, McBurney MW, de Murcia G, Sassone-Corsi P. Control of AIF-mediated cell death by the functional interplay of SIRT1 and PARP-1 in response to DNA damage. Cell Cycle. 2006;5:873–877. doi: 10.4161/cc.5.8.2690. [DOI] [PubMed] [Google Scholar]
- Kraus WL. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr Opin Cell Biol. 2008;20:294–302. doi: 10.1016/j.ceb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, Kraus WL. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319:819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- Kun E, Kirsten E, Ordahl CP. Coenzymatic activity of randomly broken or intact double-stranded DNAs in auto and histone H1 trans-poly(ADP-ribosylation), catalyzed by poly(ADP-ribose) polymerase (PARP I) J.Biol.Chem. 2002;277:39066–39069. doi: 10.1074/jbc.C200410200. [DOI] [PubMed] [Google Scholar]
- Kurnasov O, Goral V, Colabroy K, Gerdes S, Anantha S, Osterman A, Begley TP. NAD biosynthesis: identification of the tryptophan to quinolinate pathway in bacteria. Chem Biol. 2003;10:1195–1204. doi: 10.1016/j.chembiol.2003.11.011. [DOI] [PubMed] [Google Scholar]
- Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol Improves Mitochondrial Function and Protects against Metabolic Disease by Activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Lai Y, Chen Y, Watkins SC, Nathaniel PD, Guo F, Kochanek PM, Jenkins LW, Szabo C, Clark RS. Identification of poly-ADP-ribosylated mitochondrial proteins after traumatic brain injury. J Neurochem. 2008;104:1700–1711. doi: 10.1111/j.1471-4159.2007.05114.x. [DOI] [PubMed] [Google Scholar]
- Lan F, Cacicedo JM, Ruderman N, Ido Y. SIRT1 modulation of the acetylation status, cytosolic localization, and activity of LKB1. Possible role in AMP-activated protein kinase activation. J Biol Chem. 2008;283:27628–27635. doi: 10.1074/jbc.M805711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier MF, Planck JL, Roy S, Pascal JM. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science. 2012;336:728–732. doi: 10.1126/science.1216338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier MF, Servent KM, Rogers EE, Pascal JM. A third zinc-binding domain of human poly(ADP-ribose) polymerase-1 coordinates DNA-dependent enzyme activation. J Biol Chem. 2008;283:4105–4114. doi: 10.1074/jbc.M708558200. [DOI] [PubMed] [Google Scholar]
- Lau C, Niere M, Ziegler M. The NMN/NaMN adenylyltransferase (NMNAT) protein family. Front Biosci. 2009;14:410–431. doi: 10.2741/3252. [DOI] [PubMed] [Google Scholar]
- Lazarus SS, Shapiro SH. Influence of nicotinamide and pyridine nucleotides on streptozotocin and alloxan-induced pancreatic B cell cytotoxicity. Diabetes. 1973;22:499–506. doi: 10.2337/diab.22.7.499. [DOI] [PubMed] [Google Scholar]
- Lee JS, Park KY, Min HG, Lee SJ, Kim JJ, Choi JS, Kim WS, Cha HJ. Negative regulation of stress-induced matrix metalloproteinase-9 by Sirt1 in skin tissue. Exp Dermatol. 2010;19:1060–1066. doi: 10.1111/j.1600-0625.2010.01129.x. [DOI] [PubMed] [Google Scholar]
- Leist M, Single B, Castoldi AF, Kuhnle S, Nicotera P. Intracellular adenosine triphosphate (ATP) concentration: a switch in the decision between apoptosis and necrosis. J Exp Med. 1997;185:1481–1486. doi: 10.1084/jem.185.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist M, Single B, Naumann H, Fava E, Simon B, Kuhnle S, Nicotera P. Inhibition of mitochondrial ATP generation by nitric oxide switches apoptosis to necrosis. Exp Cell Res. 1999;249:396–403. doi: 10.1006/excr.1999.4514. [DOI] [PubMed] [Google Scholar]
- Lepiniec L, Babiychuk E, Kushnir S, Van Montagu M, Inze D. Characterization of an Arabidopsis thaliana cDNA homologue to animal poly(ADP-ribose) polymerase. FEBS Lett. 1995;364:103–108. doi: 10.1016/0014-5793(95)00335-7. [DOI] [PubMed] [Google Scholar]
- Leung A, Todorova T, Ando Y, Chang P. Poly(ADP-ribose) regulates post-transcriptional gene regulation: in the cytoplasm. RNA Biol. 2012;9:5. doi: 10.4161/rna.19899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levaot N, Voytyuk O, Dimitriou I, Sircoulomb F, Chandrakumar A, Deckert M, Krzyzanowski PM, Scotter A, Gu S, Janmohamed S, Cong F, Simoncic PD, Ueki Y, La Rose J, Rottapel R. Loss of Tankyrase-mediated destruction of 3BP2 is the underlying pathogenic mechanism of cherubism. Cell. 2011;147:1324–1339. doi: 10.1016/j.cell.2011.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Yamauchi Y, Kamakura M, Murayama T, Goshima F, Kimura H, Nishiyama Y. Herpes simplex virus requires poly(ADP-ribose) polymerase activity for efficient replication and induces extracellular signal-related kinase-dependent phosphorylation and ICP0-dependent nuclear localization of tankyrase 1. J Virol. 2012;86:492–503. doi: 10.1128/JVI.05897-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, Lowell BB, Bassel-Duby R, Spiegelman BM. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- Liu D, Gharavi R, Pitta M, Gleichmann M, Mattson MP. Nicotinamide Prevents NAD(+) Depletion and Protects Neurons Against Excitotoxicity and Cerebral Ischemia: NAD(+) Consumption by SIRT1 may Endanger Energetically Compromised Neurons. Neuromolecular Med. 2009;11:28–42. doi: 10.1007/s12017-009-8058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Pitta M, Mattson MP. Preventing NAD(+) depletion protects neurons against excitotoxicity: bioenergetic effects of mild mitochondrial uncoupling and caloric restriction. Ann N Y Acad Sci. 2008;1147:275–282. doi: 10.1196/annals.1427.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Shi Y, Maag DX, Palma JP, Patterson MJ, Ellis PA, Surber BW, Ready DB, Soni NB, Ladror US, Xu AJ, Iyer R, Harlan JE, Solomon LR, Donawho CK, Penning TD, Johnson EF, Shoemaker AR. Iniparib nonselectively modifies cysteine-containing proteins in tumor cells and is not a bona fide PARP inhibitor. Clin Cancer Res. 2012;18:510–523. doi: 10.1158/1078-0432.CCR-11-1973. [DOI] [PubMed] [Google Scholar]
- Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV, Jr., Weissman S, Verdin E, Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard DB, Pletcher SD, Canto C, Auwerx J. Ageing: longevity hits a roadblock. Nature. 2011;477:410–411. doi: 10.1038/477410a. [DOI] [PubMed] [Google Scholar]
- Lopez-Lluch G, Irusta PM, Navas P, de Cabo R. Mitochondrial biogenesis and healthy aging. Exp Gerontol. 2008;43:813–819. doi: 10.1016/j.exger.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loseva O, Jemth AS, Bryant HE, Schuler H, Lehtio L, Karlberg T, Helleday T. PARP-3 is a mono-ADP-ribosylase that activates PARP-1 in the absence of DNA. J Biol Chem. 2010;285:8054–8060. doi: 10.1074/jbc.M109.077834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Luthi-Carter R, Taylor DM, Pallos J, Lambert E, Amore A, Parker A, Moffitt H, Smith DL, Runne H, Gokce O, Kuhn A, Xiang Z, Maxwell MM, Reeves SA, Bates GP, Neri C, Thompson LM, Marsh JL, Kazantsev AG. SIRT2 inhibition achieves neuroprotection by decreasing sterol biosynthesis. Proc Natl Acad Sci U S A. 2010;107:7927–7932. doi: 10.1073/pnas.1002924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Baldwin KT, Renzelli AJ, McDaniel A, Dong L. TCDD-inducible poly(ADP-ribose) polymerase: a novel response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Biochem Biophys Res Commun. 2001;289:499–506. doi: 10.1006/bbrc.2001.5987. [DOI] [PubMed] [Google Scholar]
- Mangerich A, Herbach N, Hanf B, Fischbach A, Popp O, Moreno-Villanueva M, Bruns OT, Burkle A. Inflammatory and age-related pathologies in mice with ectopic expression of human PARP-1. Mech Ageing Dev. 2010;131:389–404. doi: 10.1016/j.mad.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, Seluanov A, Gorbunova V. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin N, Schwamborn K, Schreiber V, Werner A, Guillier C, Zhang XD, Bischof O, Seeler JS, Dejean A. PARP-1 transcriptional activity is regulated by sumoylation upon heat shock. Embo J. 2009;28:3534–3548. doi: 10.1038/emboj.2009.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massudi H, Grant R, Braidy N, Guest J, Farnsworth B, Guillemin GJ. Age-Associated Changes In Oxidative Stress and NAD(+) Metabolism In Human Tissue. PLoS One. 2012;7:e42357. doi: 10.1371/journal.pone.0042357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazen A, Menissier-de MJ, Molinete M, Simonin F, Gradwohl G, Poirier G, de MG. Poly(ADP-ribose)polymerase: a novel finger protein. Nucleic Acids Res. 1989;17:4689–4698. doi: 10.1093/nar/17.12.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBurney MW, Yang X, Jardine K, Hixon M, Boekelheide K, Webb JR, Lansdorp PM, Lemieux M. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meder VS, Boeglin M, de Murcia G, Schreiber V. PARP-1 and PARP-2 interact with nucleophosmin/B23 and accumulate in transcriptionally active nucleoli. J.Cell Sci. 2005;118:211–222. doi: 10.1242/jcs.01606. [DOI] [PubMed] [Google Scholar]
- Mehrotra P, Hollenbeck A, Riley JP, Li F, Patel RJ, Akhtar N, Goenka S. Poly (ADP-ribose) polymerase 14 and its enzyme activity regulates T(H)2 differentiation and allergic airway disease. J Allergy Clin Immunol. 2012;2012:25. doi: 10.1016/j.jaci.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra P, Riley JP, Patel R, Li F, Voss L, Goenka S. PARP-14 functions as a transcriptional switch for Stat6-dependent gene activation. J Biol Chem. 2011;286:1767–1776. doi: 10.1074/jbc.M110.157768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Alvarez H, Alvarez-Gonzalez R. Poly(ADP-ribose) polymerase is a catalytic dimer and the automodification reaction is intermolecular. J Biol Chem. 1993;268:22575–22580. [PubMed] [Google Scholar]
- Menissier-de Murcia J, Molinete M, Gradwohl G, Simonin F, de Murcia G. Zinc-binding domain of poly(ADP-ribose)polymerase participates in the recognition of single strand breaks on DNA. J.Mol.Biol. 1989;210:229–233. doi: 10.1016/0022-2836(89)90302-1. [DOI] [PubMed] [Google Scholar]
- Menissier-de Murcia J, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M, Walztinger C, Chambon P, de Murcia G. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc.Natl.Acad.Sci.U.S.A. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menissier-de Murcia J, icoul M, Tartier L, Niedergang C, Huber A, Dantzer F, Schreiber V, Ame JC, Dierich A, LeMeur M, Sabatier L, Chambon P, de Murcia G. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 2003;22:2255–2263. doi: 10.1093/emboj/cdg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milam KM, Cleaver JE. Inhibitors of poly(adenosine diphosphate-ribose) synthesis: effect on other metabolic processes. Science. 1984;223:589–591. doi: 10.1126/science.6420886. [DOI] [PubMed] [Google Scholar]
- Milam KM, Thomas GH, Cleaver JE. Disturbances in DNA precursor metabolism associated with exposure to an inhibitor of poly(ADP-ribose) synthetase. Exp Cell Res. 1986;165:260–268. doi: 10.1016/0014-4827(86)90550-1. [DOI] [PubMed] [Google Scholar]
- Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, Bemis JE, Xie R, Disch JS, Ng PY, Nunes JJ, Lynch AV, Yang H, Galonek H, Israelian K, Choy W, Iffland A, Lavu S, Medvedik O, Sinclair DA, Olefsky JM, Jirousek MR, Elliott PJ, Westphal CH. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T, Kakizawa T, Hashizume K. Inhibition of nuclear receptor signalling by poly(ADP-ribose) polymerase. Mol.Cell Biol. 1999;19:2644–2649. doi: 10.1128/mcb.19.4.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modis K, Gero D, Erdelyi K, Szoleczky P, Dewitt D, Szabo C. Cellular bioenergetics is regulated by PARP1 under resting conditions and during oxidative stress. Biochem Pharmacol. 2012;83:633–643. doi: 10.1016/j.bcp.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncada S, Bolanos JP. Nitric oxide, cell bioenergetics and neurodegeneration. J Neurochem. 2006;97:1676–1689. doi: 10.1111/j.1471-4159.2006.03988.x. [DOI] [PubMed] [Google Scholar]
- Moonen HJ, Geraets L, Vaarhorst A, Bast A, Wouters EF, Hageman GJ. Theophylline prevents NAD+ depletion via PARP-1 inhibition in human pulmonary epithelial cells. Biochem Biophys Res Commun. 2005;338:1805–1810. doi: 10.1016/j.bbrc.2005.10.159. [DOI] [PubMed] [Google Scholar]
- Moroni F, Formentini L, Gerace E, Camaioni E, Pellegrini-Giampietro DE, Chiarugi A, Pellicciari R. Selective PARP-2 inhibitors increase apoptosis in hippocampal slices but protect cortical cells in models of post-ischaemic brain damage. Br J Pharmacol. 2009;157:854–862. doi: 10.1111/j.1476-5381.2009.00232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison C, Smith GC, Stingl L, Jackson SP, Wagner EF, Wang ZQ. Genetic interaction between PARP and DNA-PK in V(D)J recombination and tumorigenesis. Nat Genet. 1997;17:479–482. doi: 10.1038/ng1297-479. [DOI] [PubMed] [Google Scholar]
- Mortusewicz O, Ame JC, Schreiber V, Leonhardt H. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 2007;35:7665–7675. doi: 10.1093/nar/gkm933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Moynihan KA, Grimm AA, Plueger MM, Bernal-Mizrachi E, Ford E, Cras-Meneur C, Permutt MA, Imai S. Increased dosage of mammalian Sir2 in pancreatic beta cells enhances glucose-stimulated insulin secretion in mice. Cell Metab. 2005;2:105–117. doi: 10.1016/j.cmet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Muiras ML, Muller M, Schachter F, Burkle A. Increased poly(ADP-ribose) polymerase activity in lymphoblastoid cell lines from centenarians. J Mol Med (Berl) 1998;76:346–354. doi: 10.1007/s001090050226. [DOI] [PubMed] [Google Scholar]
- Murray-Zmijewski F, Slee EA, Lu X. A complex barcode underlies the heterogeneous response of p53 to stress. Nat Rev Mol Cell Biol. 2008;9:702–712. doi: 10.1038/nrm2451. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Guarente L. Sirtuins at a glance. J Cell Sci. 2011;124:833–838. doi: 10.1242/jcs.081067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamaru Y, Vuppusetty C, Wada H, Milne JC, Ito M, Rossios C, Elliot M, Hogg J, Kharitonov S, Goto H, Bemis JE, Elliott P, Barnes PJ, Ito K. A protein deacetylase SIRT1 is a negative regulator of metalloproteinase-9. Faseb J. 2009;23:2810–2819. doi: 10.1096/fj.08-125468. [DOI] [PubMed] [Google Scholar]
- Nasrin N, Wu X, Fortier E, Feng Y, Bare OC, Chen S, Ren X, Wu Z, Streeper RS, Bordone L. SIRT4 regulates fatty acid oxidation and mitochondrial gene expression in liver and muscle cells. J Biol Chem. 2010;285:31995–32002. doi: 10.1074/jbc.M110.124164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C. Nitric oxide as a secretory product of mammalian cells. Faseb J. 1992;6:3051–3064. [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- Nicolas L, Martinez C, Baro C, Rodriguez M, Baroja-Mazo A, Sole F, Flores JM, Ampurdanes C, Dantzer F, Martin-Caballero J, Aparicio P, Yelamos J. Loss of poly(ADP-ribose) polymerase-2 leads to rapid development of spontaneous T-cell lymphomas in p53-deficient mice. Oncogene. 2010;29:2877–2883. doi: 10.1038/onc.2010.11. [DOI] [PubMed] [Google Scholar]
- Nicolescu AC, Holt A, Kandasamy AD, Pacher P, Schulz R. Inhibition of matrix metalloproteinase-2 by PARP inhibitors. Biochem Biophys Res Commun. 2009;387:646–650. doi: 10.1016/j.bbrc.2009.07.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nin V, Escande C, Chini CC, Giri S, Camacho-Pereira J, Matalonga J, Lou Z, Chini EN. Role of deleted in breast cancer 1 (DBC1) protein in SIRT1 deacetylase activation induced by protein kinase A and AMP-activated protein kinase. J Biol Chem. 2012;287:23489–23501. doi: 10.1074/jbc.M112.365874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizuka Y, Ueda K, Honjo T, Hayaishi O. Enzymic adenosine diphosphate ribosylation of histone and poly adenosine diphosphate ribose synthesis in rat liver nuclei. J Biol Chem. 1968;243:3765–3767. [PubMed] [Google Scholar]
- Noriega LG, Feige JN, Canto C, Yamamoto H, Yu J, Herman MA, Mataki C, Kahn BB, Auwerx J. CREB and ChREBP oppositely regulate SIRT1 expression in response to energy availability. EMBO Rep. 2011;12:1069–1076. doi: 10.1038/embor.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, Wright SM, Mills KD, Bonni A, Yankner BA, Scully R, Prolla TA, Alt FW, Sinclair DA. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata N, Ueda K, Kagamiyama H, Hayaishi O. ADP-ribosylation of histone H1. Identification of glutamic acid residues 2, 14, and the COOH-terminal lysine residue as modification sites. J Biol Chem. 1980;255:7616–7620. [PubMed] [Google Scholar]
- Ohguchi K, Itoh T, Akao Y, Inoue H, Nozawa Y, Ito M. SIRT1 modulates expression of matrix metalloproteinases in human dermal fibroblasts. Br J Dermatol. 2010;163:689–694. doi: 10.1111/j.1365-2133.2010.09825.x. [DOI] [PubMed] [Google Scholar]
- Oliver AW, Ame JC, Roe SM, Good V, de Murcia G, Pearl LH. Crystal structure of the catalytic fragment of murine poly(ADP-ribose) polymerase-2. Nucleic Acids Res. 2004;32:456–464. doi: 10.1093/nar/gkh215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver FJ, Menissier-de MJ, Nacci C, Decker P, Andriantsitohaina R, Muller S, de La RG, Stoclet JC, de MG. Resistance to endotoxic shock as a consequence of defective NF-kappaB activation in poly (ADP-ribose) polymerase-1 deficient mice. EMBO J. 1999;18:4446–4454. doi: 10.1093/emboj/18.16.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortolan E, Vacca P, Capobianco A, Armando E, Crivellin F, Horenstein A, Malavasi F. CD157, the Janus of CD38 but with a unique personality. Cell Biochem Funct. 2002;20:309–322. doi: 10.1002/cbf.978. [DOI] [PubMed] [Google Scholar]
- Otto H, Reche PA, Bazan F, Dittmar K, Haag F, Koch-Nolte F. In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs) BMC Genomics. 2005;6:139. doi: 10.1186/1471-2164-6-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacher P, Beckman JS, Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacher P, Liaudet L, Bai P, Mabley JG, Kaminski PM, Virag L, Deb A, Szabo E, Ungvari Z, Wolin MS, Groves JT, Szabo C. Potent metalloporphyrin peroxynitrite decomposition catalyst protects against the development of doxorubicin-induced cardiac dysfunction. Circulation. 2003;107:896–904. doi: 10.1161/01.cir.0000048192.52098.dd. [DOI] [PubMed] [Google Scholar]
- Pacher P, Liaudet L, Bai P, Virag L, Mabley JG, Hasko G, Szabo C. Activation of poly(ADP-ribose) polymerase contributes to development of doxorubicin-induced heart failure. J.Pharmacol.Exp.Ther. 2002a;300:862–867. doi: 10.1124/jpet.300.3.862. [DOI] [PubMed] [Google Scholar]
- Pacher P, Liaudet L, Mabley J, Komjati K, Szabo C. Pharmacologic inhibition of poly(adenosine diphosphate-ribose) polymerase may represent a novel therapeutic approach in chronic heart failure. J.Am.Coll.Cardiol. 2002b;40:1006–1016. doi: 10.1016/s0735-1097(02)02062-4. [DOI] [PubMed] [Google Scholar]
- Pacher P, Szabo C. Role of the peroxynitrite-poly(ADP-ribose) polymerase pathway in human disease. Am J Pathol. 2008;173:2–13. doi: 10.2353/ajpath.2008.080019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacholec M, Bleasdale JE, Chrunyk B, Cunningham D, Flynn D, Garofalo RS, Griffith D, Griffor M, Loulakis P, Pabst B, Qiu X, Stockman B, Thanabal V, Varghese A, Ward J, Withka J, Ahn K. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J Biol Chem. 2010;285:8340–8351. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan PW, Feldman JL, Devries MK, Dong A, Edwards AM, Denu JM. Structure and biochemical functions of SIRT6. J Biol Chem. 2011;286:14575–14587. doi: 10.1074/jbc.M111.218990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang J, Gong H, Xi C, Fan W, Dai Y, Zhang TM. Poly(ADP-ribose) polymerase 1 is involved in glucose toxicity through SIRT1 modulation in HepG2 hepatocytes. J Cell Biochem. 2011;112:299–306. doi: 10.1002/jcb.22919. [DOI] [PubMed] [Google Scholar]
- Pankotai E, Lacza Z, Muranyi M, Szabo C. Intra-mitochondrial poly(ADP-ribosyl)ation: potential role for alpha-ketoglutarate dehydrogenase. Mitochondrion. 2009;9:159–164. doi: 10.1016/j.mito.2009.01.013. [DOI] [PubMed] [Google Scholar]
- Park SJ, Ahmad F, Philp A, Baar K, Williams T, Luo H, Ke H, Rehmann H, Taussig R, Brown AL, Kim MK, Beaven MA, Burgin AB, Manganiello V, Chung JH. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell. 2012;148:421–433. doi: 10.1016/j.cell.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen KF, Shulman GI. Etiology of insulin resistance. Am J Med. 2006;119:S10–16. doi: 10.1016/j.amjmed.2006.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleiderer G. Isoenzymes of NAD and NADP dependent dehydrogenases. Vitam Horm. 1970;28:195–211. doi: 10.1016/s0083-6729(08)60894-8. [DOI] [PubMed] [Google Scholar]
- Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci U S A. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai JB, Gupta M, Rajamohan SB, Lang R, Raman J, Gupta MP. Poly(ADP-ribose) polymerase-1-deficient mice are protected from angiotensin II-induced cardiac hypertrophy. Am.J.Physiol Heart Circ.Physiol. 2006;291:H1545–H1553. doi: 10.1152/ajpheart.01124.2005. [DOI] [PubMed] [Google Scholar]
- Pillai JB, Isbatan A, Imai S, Gupta MP. Poly(ADP-ribose) polymerase-1-dependent cardiac myocyte cell death during heart failure is mediated by NAD+ depletion and reduced Sir2alpha deacetylase activity. J.Biol.Chem. 2005;280:43121–43130. doi: 10.1074/jbc.M506162200. [DOI] [PubMed] [Google Scholar]
- Pittelli M, Formentini L, Faraco G, Lapucci A, Rapizzi E, Cialdai F, Romano G, Moneti G, Moroni F, Chiarugi A. Inhibition of nicotinamide phosphoribosyltransferase: cellular bioenergetics reveals a mitochondrial insensitive NAD pool. J Biol Chem. 2010;285:34106–34114. doi: 10.1074/jbc.M110.136739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss J, Handler P. Biosynthesis of diphosphopyridine nucleotide. I. Identification of intermediates. J Biol Chem. 1958a;233:488–492. [PubMed] [Google Scholar]
- Preiss J, Handler P. Biosynthesis of diphosphopyridine nucleotide. II. Enzymatic aspects. J Biol Chem. 1958b;233:493–500. [PubMed] [Google Scholar]
- Preiss J, Schlaeger R, Hilz H. Specific inhibition of poly adpribose polymerase by thymidine and nicotinamide in HeLa cells. FEBS Lett. 1971;19:244–246. doi: 10.1016/0014-5793(71)80524-0. [DOI] [PubMed] [Google Scholar]
- Price NL, Gomes AP, Ling AJ, Duarte FV, Martin-Montalvo A, North BJ, Agarwal B, Ye L, Ramadori G, Teodoro JS, Hubbard BP, Varela AT, Davis JG, Varamini B, Hafner A, Moaddel R, Rolo AP, Coppari R, Palmeira CM, de Cabo R, Baur JA, Sinclair DA. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 2012;15:675–690. doi: 10.1016/j.cmet.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purnell MR, Whish WJ. Novel inhibitors of poly(ADP-ribose) synthetase. Biochem J. 1980;185:775–777. doi: 10.1042/bj1850775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang L, Wang L, Kon N, Zhao W, Lee S, Zhang Y, Rosenbaum M, Zhao Y, Gu W, Farmer SR, Accili D. Brown Remodeling of White Adipose Tissue by SirT1-Dependent Deacetylation of Ppargamma. Cell. 2012;150:620–632. doi: 10.1016/j.cell.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, Yang T, Ho L, Zhao Z, Wang J, Chen L, Zhao W, Thiyagarajan M, MacGrogan D, Rodgers JT, Puigserver P, Sadoshima J, Deng H, Pedrini S, Gandy S, Sauve AA, Pasinetti GM. Neuronal SIRT1 activation as a novel mechanism underlying the prevention of Alzheimer disease amyloid neuropathology by calorie restriction. J Biol Chem. 2006;281:21745–21754. doi: 10.1074/jbc.M602909200. [DOI] [PubMed] [Google Scholar]
- Qin WD, Wei SJ, Wang XP, Wang J, Wang WK, Liu F, Gong L, Yan F, Zhang Y, Zhang M. Poly(ADP-ribose) Polymerase 1 Inhibition Protects Against Low Shear Stress Induced Inflammation. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbamcr.2012.10.013. dx.doi.org/10.1016/j.bbamcr.2012.1010.1013. [DOI] [PubMed] [Google Scholar]
- Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010;12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- Quenet D, El Ramy R, Schreiber V, Dantzer F. The role of poly(ADP-ribosyl)ation in epigenetic events. Int J Biochem Cell Biol. 2009;41:60–65. doi: 10.1016/j.biocel.2008.07.023. [DOI] [PubMed] [Google Scholar]
- Rajamohan SB, Pillai VB, Gupta M, Sundaresan NR, Konstatin B, Samant S, Hottiger MO, Gupta MP. SIRT1 promotes cell survival under stress by deacetylation-dependent deactivation of poly (ADP-ribose) polymerase 1. Mol Cell Biol. 2009;26:4116–4129. doi: 10.1128/MCB.00121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna R, Edwards JS, McCulloch A, Palsson BO. Flux-balance analysis of mitochondrial energy metabolism: consequences of systemic stoichiometric constraints. Am J Physiol Regul Integr Comp Physiol. 2001;280:R695–704. doi: 10.1152/ajpregu.2001.280.3.R695. [DOI] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, Takahashi JS, Imai S, Bass J. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray PD, Huang BW, Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal. 2012;24:981–990. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- Revollo JR, Korner A, Mills KF, Satoh A, Wang T, Garten A, Dasgupta B, Sasaki Y, Wolberger C, Townsend RR, Milbrandt J, Kiess W, Imai S. Nampt/PBEF/Visfatin regulates insulin secretion in beta cells as a systemic NAD biosynthetic enzyme. Cell Metab. 2007;6:363–375. doi: 10.1016/j.cmet.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimachi R, Bruzzi de Carvahlo F, Orellano-Jimenez C, Cotton F, Vincent JL, De Backer D. Lactate/pyruvate ratio as a marker of tissue hypoxia in circulatory and septic shock. Anaesth Intensive Care. 2012;40:427–432. doi: 10.1177/0310057X1204000307. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci U S A. 2007;104:12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongvaux A, Galli M, Denanglaire S, Van Gool F, Dreze PL, Szpirer C, Bureau F, Andris F, Leo O. Nicotinamide phosphoribosyl transferase/pre-B cell colony-enhancing factor/visfatin is required for lymphocyte development and cellular resistance to genotoxic stress. J Immunol. 2008;181:4685–4695. doi: 10.4049/jimmunol.181.7.4685. [DOI] [PubMed] [Google Scholar]
- Rongvaux A, Shea RJ, Mulks MH, Gigot D, Urbain J, Leo O, Andris F. Pre-B-cell colony-enhancing factor, whose expression is up-regulated in activated lymphocytes, is a nicotinamide phosphoribosyltransferase, a cytosolic enzyme involved in NAD biosynthesis. Eur J Immunol. 2002;32:3225–3234. doi: 10.1002/1521-4141(200211)32:11<3225::AID-IMMU3225>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Rothgiesser KM, Erener S, Waibel S, Luscher B, Hottiger MO. SIRT2 regulates NF-kappaB dependent gene expression through deacetylation of p65 Lys310. J Cell Sci. 2010;123:4251–4258. doi: 10.1242/jcs.073783. [DOI] [PubMed] [Google Scholar]
- Ruf A, Mennissier de MJ, de MG, Schulz GE. Structure of the catalytic fragment of poly(AD-ribose) polymerase from chicken. Proc.Natl.Acad.Sci.U.S.A. 1996;93:7481–7485. doi: 10.1073/pnas.93.15.7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulten SL, Fisher AE, Robert I, Zuma MC, Rouleau M, Ju L, Poirier G, Reina-San-Martin B, Caldecott KW. PARP-3 and APLF function together to accelerate nonhomologous end-joining. Mol Cell. 2011;41:33–45. doi: 10.1016/j.molcel.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Rutanen J, Yaluri N, Modi S, Pihlajamaki J, Vanttinen M, Itkonen P, Kainulainen S, Yamamoto H, Lagouge M, Sinclair DA, Elliott P, Westphal C, Auwerx J, Laakso M. SIRT1 mRNA expression may be associated with energy expenditure and insulin sensitivity. Diabetes. 2010;59:829–835. doi: 10.2337/db09-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyanarayana U, Rao BS. Dietary tryptophan level and the enzymes of tryptophan NAD pathway. Br J Nutr. 1980;43:107–113. doi: 10.1079/bjn19800070. [DOI] [PubMed] [Google Scholar]
- Sauve AA. NAD+ and vitamin B3: from metabolism to therapies. J Pharmacol Exp Ther. 2008;324:883–893. doi: 10.1124/jpet.107.120758. [DOI] [PubMed] [Google Scholar]
- Sauve AA, Schramm VL. Sir2 regulation by nicotinamide results from switching between base exchange and deacetylation chemistry. Biochemistry. 2003;42:9249–9256. doi: 10.1021/bi034959l. [DOI] [PubMed] [Google Scholar]
- Sauve AA, Schramm VL. SIR2: The biochemical mechanism of NAD(+)-dependent protein deacetylation and ADP-ribosyl enzyme intermediates. Current Medicinal Chemistry. 2004;11:807–826. doi: 10.2174/0929867043455675. [DOI] [PubMed] [Google Scholar]
- Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annu Rev Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- Schreiber V, Ame JC, Dolle P, Schultz I, Rinaldi B, Fraulob V, Menissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J.Biol.Chem. 2002;277:23028–23036. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat.Rev.Mol.Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- Schreiber V, Hunting D, Trucco C, Gowans B, Grunwald D, de Murcia G, Menissier-de Murcia J. A dominant-negative mutant of human poly(ADP-ribose) polymerase affects cell recovery, apoptosis, and sister chromatid exchange following DNA damage. Proc.Natl.Acad.Sci.U.S.A. 1995;92:4753–4757. doi: 10.1073/pnas.92.11.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber V, Molinete M, Boeuf H, de Murcia G, Menissier-de Murcia J. The human poly(ADP-ribose) polymerase nuclear localization signal is a bipartite element functionally separate from DNA binding and catalytic activity. EMBO J. 1992;11:3263–3269. doi: 10.1002/j.1460-2075.1992.tb05404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarcz R, Whetsell WO, Jr., Mangano RM. Quinolinic acid: an endogenous metabolite that produces axon-sparing lesions in rat brain. Science. 1983;219:316–318. doi: 10.1126/science.6849138. [DOI] [PubMed] [Google Scholar]
- Schweiger M, Hennig K, Lerner F, Niere M, Hirsch-Kauffmann M, Specht T, Weise C, Oei SL, Ziegler M. Characterization of recombinant human nicotinamide mononucleotide adenylyl transferase (NMNAT), a nuclear enzyme essential for NAD synthesis. FEBS Lett. 2001;492:95–100. doi: 10.1016/s0014-5793(01)02180-9. [DOI] [PubMed] [Google Scholar]
- Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol. 2002;158:647–657. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Schumacher B, Lombard DB, Xiao C, Kurtev MV, Gao J, Schneider JI, Chai H, Bronson RT, Tsai LH, Deng CX, Alt FW. Neural sirtuin 6 (Sirt6) ablation attenuates somatic growth and causes obesity. Proc Natl Acad Sci U S A. 2010;107:21790–21794. doi: 10.1073/pnas.1016306107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed M, O’Connor B, Perombelon N, O’Donnell M, Reaveley D, Knight BL. The effect of nicotinic acid and acipimox on lipoprotein(a) concentration and turnover. Atherosclerosis. 1993;101:61–68. doi: 10.1016/0021-9150(93)90102-z. [DOI] [PubMed] [Google Scholar]
- Shall S. Proceedings: Experimental manipulation of the specific activity of poly(ADP-ribose) polymerase. J Biochem. 1975;77:2p. [PubMed] [Google Scholar]
- Sheline CT, Cai AL, Zhu J, Shi C. Serum or target deprivation-induced neuronal death causes oxidative neuronal accumulation of Zn2+ and loss of NAD+ Eur J Neurosci. 2010;32:894–904. doi: 10.1111/j.1460-9568.2010.07372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt FW, Denu JM, Jacobson MP, Verdin E. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 2010;12:654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y, Hasegawa S, Fujimura S, Sugimura T. Solubilization of enzyme forming ADPR polymer from NAD. Biochem Biophys Res Commun. 1967;29:80–83. doi: 10.1016/0006-291x(67)90544-x. [DOI] [PubMed] [Google Scholar]
- Shindler KS, Ventura E, Rex TS, Elliott P, Rostami A. SIRT1 activation confers neuroprotection in experimental optic neuritis. Invest Ophthalmol Vis Sci. 2007;48:3602–3609. doi: 10.1167/iovs.07-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D, Squire M, Nasmyth KA. Characterization of two genes required for the position-effect control of yeast mating-type genes. EMBO J. 1984;3:2817–2823. doi: 10.1002/j.1460-2075.1984.tb02214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simbulan-Rosenthal CM, Ly DH, Rosenthal DS, Konopka G, Luo R, Wang ZQ, Schultz PG, Smulson ME. Misregulation of gene expression in primary fibroblasts lacking poly(ADP-ribose) polymerase. Proc.Natl.Acad.Sci.U.S.A. 2000;97:11274–11279. doi: 10.1073/pnas.200285797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simbulan-Rosenthal CM, Rosenthal DS, Hilz H, Hickey R, Malkas L, Applegren N, Wu Y, Bers G, Smulson ME. The expression of poly(ADP-ribose) polymerase during differentiation-linked DNA replication reveals that it is a component of the multiprotein DNA replication complex. Biochemistry. 1996;35:11622–11633. doi: 10.1021/bi953010z. [DOI] [PubMed] [Google Scholar]
- Skidmore CJ, Davies MI, Goodwin PM, Halldorsson H, Lewis PJ, Shall S, Zia’ee AA. The involvement of poly(ADP-ribose) polymerase in the degradation of NAD caused by gamma-radiation and N-methyl-N-nitrosourea. Eur J Biochem. 1979;101:135–142. doi: 10.1111/j.1432-1033.1979.tb04225.x. [DOI] [PubMed] [Google Scholar]
- Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- Smulson ME, Kang VH, Ntambi JM, Rosenthal DS, Ding R, Simbulan CM. Requirement for the expression of poly(ADP-ribose) polymerase during the early stages of differentiation of 3T3-L1 preadipocytes, as studied by antisense RNA induction. J.Biol.Chem. 1995;270:119–127. doi: 10.1074/jbc.270.1.119. [DOI] [PubMed] [Google Scholar]
- Soga T, Kamohara M, Takasaki J, Matsumoto S, Saito T, Ohishi T, Hiyama H, Matsuo A, Matsushime H, Furuichi K. Molecular identification of nicotinic acid receptor. Biochem Biophys Res Commun. 2003;303:364–369. doi: 10.1016/s0006-291x(03)00342-5. [DOI] [PubMed] [Google Scholar]
- Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, Tanokura M, Denu JM, Prolla TA. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143:802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano FG, Virag L, Szabo C. Diabetic endothelial dysfunction: role of reactive oxygen and nitrogen species production and poly(ADP-ribose) polymerase activation. J.Mol.Med. 2001;79:437–448. doi: 10.1007/s001090100236. [DOI] [PubMed] [Google Scholar]
- Suchankova G, Nelson LE, Gerhart-Hines Z, Kelly M, Gauthier MS, Saha AK, Ido Y, Puigserver P, Ruderman NB. Concurrent regulation of AMP-activated protein kinase and SIRT1 in mammalian cells. Biochem Biophys Res Commun. 2009;378:836–841. doi: 10.1016/j.bbrc.2008.11.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Dai C, Xie J, Hu X. Biochemical issues in estimation of cytosolic free NAD/NADH ratio. PLoS One. 2012;7:e34525. doi: 10.1371/journal.pone.0034525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo C, Pacher P, Zsengeller Z, Vaslin A, Komjati K, Benko R, Chen M, Mabley JG, Kollai M. Angiotensin II-mediated endothelial dysfunction: role of poly(ADP-ribose) polymerase activation. Mol.Med. 2004;10:28–35. doi: 10.2119/2004-00001.szabo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo C, Virag L, Cuzzocrea S, Scott GS, Hake P, O’Connor MP, Zingarelli B, Salzman A, Kun E. Protection against peroxynitrite-induced fibroblast injury and arthritis development by inhibition of poly(ADP-ribose) synthase. Proc.Natl.Acad.Sci.U.S.A. 1998;95:3867–3872. doi: 10.1073/pnas.95.7.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szanto M, Brunyánszki A, Kiss B, Nagy L, Gergely P, Virag L, Bai P. Poly(ADP-ribose) polymerase-2: emerging transcriptional roles of a DNA repair protein. Cellular and Molecular Life Sciences. 2012;69:4079–4092. doi: 10.1007/s00018-012-1003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szanto M, Rutkai I, Hegedus C, Czikora A, Rozsahegyi M, Kiss B, Virag L, Gergely P, Toth A, Bai P. Poly(ADP-ribose) polymerase-2 depletion reduces doxorubicin-induced damage through SIRT1 induction. Cardiovasc Res. 2011;92:430–438. doi: 10.1093/cvr/cvr246. [DOI] [PubMed] [Google Scholar]
- Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem. 2007;282:6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- Tartier L, Spenlehauer C, Newman HC, Folkard M, Prise KM, Michael BD, Menissier-de MJ, de MG. Local DNA damage by proton microbeam irradiation induces poly(ADP-ribose) synthesis in mammalian cells. Mutagenesis. 2003;18:411–416. doi: 10.1093/mutage/geg015. [DOI] [PubMed] [Google Scholar]
- Tempel W, Rabeh WM, Bogan KL, Belenky P, Wojcik M, Seidle HF, Nedyalkova L, Yang T, Sauve AA, Park HW, Brenner C. Nicotinamide riboside kinase structures reveal new pathways to NAD+ PLoS Biol. 2007;5:e263. doi: 10.1371/journal.pbio.0050263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari M, Quan LT, O’Rourke K, Desnoyers S, Zeng Z, Beidler DR, Poirier GG, Salvesen GS, Dixit VM. Yama/CPP32 beta, a mammalian homolog of CED-3, is a CrmA-inhibitable protease that cleaves the death substrate poly(ADP-ribose) polymerase. Cell. 1995;81:801–809. doi: 10.1016/0092-8674(95)90541-3. [DOI] [PubMed] [Google Scholar]
- Tong WM, Hande MP, Lansdorp PM, Wang ZQ. DNA strand break-sensing molecule poly(ADP-Ribose) polymerase cooperates with p53 in telomere function, chromosome stability, and tumor suppression. Mol Cell Biol. 2001;21:4046–4054. doi: 10.1128/MCB.21.12.4046-4054.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulin A, Naumova NM, Menon AK, Spradling AC. Drosophila poly(ADP-ribose) glycohydrolase mediates chromatin structure and SIR2-dependent silencing. Genetics. 2006;172:363–371. doi: 10.1534/genetics.105.049239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunaru S, Kero J, Schaub A, Wufka C, Blaukat A, Pfeffer K, Offermanns S. PUMA-G and HM74 are receptors for nicotinic acid and mediate its anti-lipolytic effect. Nat Med. 2003;9:352–355. doi: 10.1038/nm824. [DOI] [PubMed] [Google Scholar]
- Ueda K, Oka J, Naruniya S, Miyakawa N, Hayaishi O. Poly ADP-ribose glycohydrolase from rat liver nuclei, a novel enzyme degrading the polymer. Biochem Biophys Res Commun. 1972;46:516–523. doi: 10.1016/s0006-291x(72)80169-4. [DOI] [PubMed] [Google Scholar]
- Um JH, Park SJ, Kang H, Yang S, Foretz M, McBurney MW, Kim MK, Viollet B, Chung JH. AMP-activated protein kinase-deficient mice are resistant to the metabolic effects of resveratrol. Diabetes. 2010;59:554–563. doi: 10.2337/db09-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakhrusheva O, Braeuer D, Liu Z, Braun T, Bober E. Sirt7-dependent inhibition of cell growth and proliferation might be instrumental to mediate tissue integrity during aging. J Physiol Pharmacol. 2008a;59(Suppl 9):201–212. [PubMed] [Google Scholar]
- Vakhrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, Braun T, Bober E. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res. 2008b;102:703–710. doi: 10.1161/CIRCRESAHA.107.164558. [DOI] [PubMed] [Google Scholar]
- van der Veer E, Ho C, O’Neil C, Barbosa N, Scott R, Cregan SP, Pickering JG. Extension of human cell lifespan by nicotinamide phosphoribosyltransferase. J Biol Chem. 2007;282:10841–10845. doi: 10.1074/jbc.C700018200. [DOI] [PubMed] [Google Scholar]
- van der Veer E, Nong Z, O’Neil C, Urquhart B, Freeman D, Pickering JG. Pre-B-cell colony-enhancing factor regulates NAD+-dependent protein deacetylase activity and promotes vascular smooth muscle cell maturation. Circ Res. 2005;97:25–34. doi: 10.1161/01.RES.0000173298.38808.27. [DOI] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Vinitsky A, Grubmeyer C. A new paradigm for biochemical energy coupling. Salmonella typhimurium nicotinate phosphoribosyltransferase. J Biol Chem. 1993;268:26004–26010. [PubMed] [Google Scholar]
- Virag L, Salzman AL, Szabo C. Poly(ADP-ribose) synthetase activation mediates mitochondrial injury during oxidant-induced cell death. J.Immunol. 1998;161:3753–3759. [PubMed] [Google Scholar]
- Virag L, Szabo C. The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol.Rev. 2002;54:375–429. doi: 10.1124/pr.54.3.375. [DOI] [PubMed] [Google Scholar]
- Viswanathan M, Guarente L. Regulation of Caenorhabditis elegans lifespan by sir-2.1 transgenes. Nature. 2011;477:E1–2. doi: 10.1038/nature10440. [DOI] [PubMed] [Google Scholar]
- Wahlberg E, Karlberg T, Kouznetsova E, Markova N, Macchiarulo A, Thorsell AG, Pol E, Frostell A, Ekblad T, Oncu D, Kull B, Robertson GM, Pellicciari R, Schuler H, Weigelt J. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat Biotechnol. 2012;30:283–288. doi: 10.1038/nbt.2121. [DOI] [PubMed] [Google Scholar]
- Walker JW, Jijon HB, Madsen KL. AMP-activated protein kinase is a positive regulator of poly(ADP-ribose) polymerase. Biochem Biophys Res Commun. 2006;342:336–341. doi: 10.1016/j.bbrc.2006.01.145. [DOI] [PubMed] [Google Scholar]
- Wanders D, Judd RL. Future of GPR109A agonists in the treatment of dyslipidaemia. Diabetes Obes Metab. 2011;13:685–691. doi: 10.1111/j.1463-1326.2011.01400.x. [DOI] [PubMed] [Google Scholar]
- Wang ZQ, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, Wagner EF. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 1995;9:509–520. doi: 10.1101/gad.9.5.509. [DOI] [PubMed] [Google Scholar]
- Warburg O, Christian W. Pyridine as a functional group of dehydrated enzymes. Biochemische Zeitschrift. 1936;286:142–142. [Google Scholar]
- Weidele K, Kunzmann A, Schmitz M, Beneke S, Burkle A. Ex vivo supplementation with nicotinic acid enhances cellular poly(ADP-ribosyl)ation and improves cell viability in human peripheral blood mononuclear cells. Biochem Pharmacol. 2010;80:1103–1112. doi: 10.1016/j.bcp.2010.06.010. [DOI] [PubMed] [Google Scholar]
- Williamson DH, Lund P, Krebs HA. The redox state of free nicotinamide-adenine dinucleotide in the cytoplasm and mitochondria of rat liver. Biochem J. 1967;103:514–527. doi: 10.1042/bj1030514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SK. Role of oxygen-derived free radicals in acute angiotensin II--induced hypertensive vascular disease in the rat. Circ Res. 1990;66:722–734. doi: 10.1161/01.res.66.3.722. [DOI] [PubMed] [Google Scholar]
- Wise A, Foord SM, Fraser NJ, Barnes AA, Elshourbagy N, Eilert M, Ignar DM, Murdock PR, Steplewski K, Green A, Brown AJ, Dowell SJ, Szekeres PG, Hassall DG, Marshall FH, Wilson S, Pike NB. Molecular identification of high and low affinity receptors for nicotinic acid. J Biol Chem. 2003;278:9869–9874. doi: 10.1074/jbc.M210695200. [DOI] [PubMed] [Google Scholar]
- Wyrsch P, Blenn C, Bader J, Althaus FR. Cell Death and Autophagy under Oxidative Stress: Roles of Poly(ADP-Ribose) Polymerases and Ca2+ Mol Cell Biol. 2012;32:3541–3553. doi: 10.1128/MCB.00437-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Sauve AA. Vitamin B3, the nicotinamide adenine dinucleotides and aging. Mech Ageing Dev. 2010;131:287–298. doi: 10.1016/j.mad.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Yamada K, Toribe Y, Yanagihara K, Mano T, Akagi M, Suzuki Y. Diagnostic accuracy of blood and CSF lactate in identifying children with mitochondrial diseases affecting the central nervous system. Brain Dev. 2012;34:92–97. doi: 10.1016/j.braindev.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, de Cabo R, Sauve AA, Sinclair DA. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007a;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Chan NY, Sauve AA. Syntheses of nicotinamide riboside and derivatives: effective agents for increasing nicotinamide adenine dinucleotide concentrations in mammalian cells. J Med Chem. 2007b;50:6458–6461. doi: 10.1021/jm701001c. [DOI] [PubMed] [Google Scholar]
- Ye DZ, Tai MH, Linning KD, Szabo C, Olson LK. MafA expression and insulin promoter activity are induced by nicotinamide and related compounds in INS-1 pancreatic beta-cells. Diabetes. 2006;55:742–750. doi: 10.2337/diabetes.55.03.06.db05-0653. [DOI] [PubMed] [Google Scholar]
- Yeh TY, Sbodio JI, Chi NW. Mitotic phosphorylation of tankyrase, a PARP that promotes spindle assembly, by GSK3. Biochem Biophys Res Commun. 2006;350:574–579. doi: 10.1016/j.bbrc.2006.09.080. [DOI] [PubMed] [Google Scholar]
- Yelamos J, Monreal Y, Saenz L, Aguado E, Schreiber V, Mota R, Fuente T, Minguela A, Parrilla P, de Murcia G, Almarza E, Aparicio P, Menissier-de Murcia J. PARP-2 deficiency affects the survival of CD4+CD8+ double-positive thymocytes. EMBO J. 2006;25:4350–4360. doi: 10.1038/sj.emboj.7601301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying W, Chen Y, Alano CC, Swanson RA. Tricarboxylic acid cycle substrates prevent PARP-mediated death of neurons and astrocytes. J Cereb Blood Flow Metab. 2002;22:774–779. doi: 10.1097/00004647-200207000-00002. [DOI] [PubMed] [Google Scholar]
- Ying W, Swanson RA. The poly(ADP-ribose) glycohydrolase inhibitor gallotannin blocks oxidative astrocyte death. Neuroreport. 2000;11:1385–1388. doi: 10.1097/00001756-200005150-00007. [DOI] [PubMed] [Google Scholar]
- Yonemura Y, Takashima T, Matsuda Y, Miwa K, Sugiyama K, Miyazaki I, Yamamoto H, Okamoto H. Induction of islet B-cell regeneration in partially pancreatectomized rats by poly(ADP-ribose) synthetase inhibitors. Int J Pancreatol. 1988;3:73–82. doi: 10.1007/BF02788225. [DOI] [PubMed] [Google Scholar]
- Yonemura Y, Takashima T, Miwa K, Miyazaki I, Yamamoto H, Okamoto H. Amelioration of diabetes mellitus in partially depancreatized rats by poly(ADP-ribose) synthetase inhibitors. Evidence of islet B-cell regeneration. Diabetes. 1984;33:401–404. doi: 10.2337/diab.33.4.401. [DOI] [PubMed] [Google Scholar]
- Yoshino J, Mills KF, Yoon MJ, Imai S. Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 2011;14:528–536. doi: 10.1016/j.cmet.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- Yu W, Ginjala V, Pant V, Chernukhin I, Whitehead J, Docquier F, Farrar D, Tavoosidana G, Mukhopadhyay R, Kanduri C, Oshimura M, Feinberg AP, Lobanenkov V, Klenova E, Ohlsson R. Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat Genet. 2004;36:1105–1110. doi: 10.1038/ng1426. [DOI] [PubMed] [Google Scholar]
- Yuan J, Luo K, Liu T, Lou Z. Regulation of SIRT1 activity by genotoxic stress. Genes Dev. 2012;26:791–796. doi: 10.1101/gad.188482.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Minter-Dykhouse K, Lou Z. A c-Myc-SIRT1 feedback loop regulates cell growth and transformation. J Cell Biol. 2009;185:203–211. doi: 10.1083/jcb.200809167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahradka P, Ebisuzaki K. A shuttle mechanism for DNA-protein interactions. The regulation of poly(ADP-ribose) polymerase. Eur J Biochem. 1982;127:579–585. [PubMed] [Google Scholar]
- Zannini L, Buscemi G, Kim JE, Fontanella E, Delia D. DBC1 phosphorylation by ATM/ATR inhibits SIRT1 deacetylase in response to DNA damage. J Mol Cell Biol. 2012;2012:26. doi: 10.1093/jmcb/mjs035. [DOI] [PubMed] [Google Scholar]
- Zhang J. Are poly(ADP-ribosyl)ation by PARP-1 and deacetylation by Sir2 linked? Bioessays. 2003;25:808–814. doi: 10.1002/bies.10317. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Wang SY, Fleuriel C, Leprince D, Rocheleau JV, Piston DW, Goodman RH. Metabolic regulation of SIRT1 transcription via a HIC1:CtBP corepressor complex. Proc Natl Acad Sci U S A. 2007;104:829–833. doi: 10.1073/pnas.0610590104. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang T, Berrocal JG, Yao J, DuMond ME, Krishnakumar R, Ruhl DD, Ryu KW, Gamble MJ, Kraus WL. Regulation of poly(ADP-ribose) polymerase-1-dependent gene expression through promoter-directed recruitment of a nuclear NAD+ synthase. J Biol Chem. 2012;287:12405–12416. doi: 10.1074/jbc.M111.304469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–590. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, D’Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, Clish CB, Vaitheesvaran B, Iliopoulos O, Kurland I, Dor Y, Weissleder R, Shirihai OS, Ellisen LW, Espinosa JM, Mostoslavsky R. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillikens MC, van Meurs JB, Rivadeneira F, Amin N, Hofman A, Oostra BA, Sijbrands EJ, Witteman JC, Pols HA, van Duijn CM, Uitterlinden AG. SIRT1 genetic variation is related to BMI and risk of obesity. Diabetes. 2009a;58:2828–2834. doi: 10.2337/db09-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillikens MC, van Meurs JB, Sijbrands EJ, Rivadeneira F, Dehghan A, van Leeuwen JP, Hofman A, van Duijn CM, Witteman JC, Uitterlinden AG, Pols HA. SIRT1 genetic variation and mortality in type 2 diabetes: interaction with smoking and dietary niacin. Free Radic Biol Med. 2009b;46:836–841. doi: 10.1016/j.freeradbiomed.2008.12.022. [DOI] [PubMed] [Google Scholar]
- Zini R, Morin C, Bertelli A, Bertelli AA, Tillement JP. Effects of resveratrol on the rat brain respiratory chain. Drugs Exp Clin Res. 1999;25:87–97. [PubMed] [Google Scholar]
- Zwilling D, Huang SY, Sathyasaikumar KV, Notarangelo FM, Guidetti P, Wu HQ, Lee J, Truong J, Andrews-Zwilling Y, Hsieh EW, Louie JY, Wu T, Scearce-Levie K, Patrick C, Adame A, Giorgini F, Moussaoui S, Laue G, Rassoulpour A, Flik G, Huang Y, Muchowski JM, Masliah E, Schwarcz R, Muchowski PJ. Kynurenine 3-monooxygenase inhibition in blood ameliorates neurodegeneration. Cell. 2011;145:863–874. doi: 10.1016/j.cell.2011.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]