SUMMARY
The genetics of complex disease produce alterations in the molecular interactions of cellular pathways whose collective effect may become clear through the organized structure of molecular networks. To characterize molecular systems associated with late-onset Alzheimer’s disease (LOAD), we constructed gene regulatory networks in 1647 post-mortem brain tissues from LOAD patients and non-demented subjects, and demonstrate that LOAD reconfigures specific portions of the molecular interaction structure. Through an integrative network-based approach, we rank-ordered these network structures for relevance to LOAD pathology, highlighting an immune and microglia-specific module dominated by genes involved in pathogen phagocytosis, containing TYROBP as a key regulator and up-regulated in LOAD. Mouse microglia cells over-expressing intact or truncated TYROBP revealed expression changes that significantly overlapped the human brain TYROBP network. Thus the causal network structure is a useful predictor of response to gene perturbations and presents a novel framework to test models of disease mechanisms underlying LOAD.
INTRODUCTION
Complex diseases such as late-onset Alzheimer’s disease (LOAD) arise from the downstream interplay of DNA sequence variants and non-genetic factors that act through molecular networks to confer disease risk (Schadt, 2009). Despite decades of intensive research, the causal chain of mechanisms behind LOAD remains elusive. In fact, there are no effective disease modifying or preventive therapies and the only available treatment remains symptomatic; meanwhile the incidence of LOAD is expected to double by 2050 (Brookmeyer et al., 2007). Progress in LOAD research is fundamentally limited by our reliance on mouse models of severe familial/early-onset Alzheimer’s disease: therefore our primary knowledge of LOAD is in actuality based on the downstream effects of three rare mutations in APP, PSEN1 and PSEN2 (Bertram et al., 2010). While such mouse models are necessary and helpful, the cognitive deficits in these transgenic mice are less severe than those in humans and they do not exhibit equivalent neurodegeneration, which is the most accurate clinical marker of cognitive disease progression in humans. Correspondingly, attrition rates from early discovery to late drug development have been very high (Schafer and Kolkhof, 2008).
In contrast to the plethora of potential disease mechanisms detected in humans with LOAD, the search for LOAD-modifying interventions has focused primarily on compounds targeting the amyloid-β pathway. Both biological risk factors, often related to vascular health and psychosocial factors (Cechetto et al., 2008; Qiu et al., 2010), as well as genetic susceptibility play a critical role in the underlying pathophysiology of LOAD (Bertram et al., 2010). APOE is still the best validated suceptibility gene accounting for at least 30% of the genetic variance in LOAD (Corder et al., 1993). Several additional genetic risk loci for LOAD have been identified via genome-wide association studies (GWAS) that seem to cluster in patterns suggesting immunity (CLU, CR1, CD33, EPHA1, MS4A4A/MS4A6A), lipid processing (APOE, ABCA7) and endocytosis (PICALM, BIN1, CD2AP) as important causal biological processes (Bettens et al., 2013). More recently, low-frequency missense variants in APP and TREM2 were found to confer strong protection or elevated risk of LOAD (Guerreiro et al., 2013; Jonsson et al., 2012; Jonsson et al., 2013). However, the overall contribution of these new common and low-frequency variants to the heritability of LOAD is very small, suggesting that a large fraction of the genetic variance beyond the APOE risk still remains hidden. Can we clarify the pathology of LOAD by zooming out to the pathway level to search for emergent risk of many genomic contributions? If so, how can we identify the key causal genes in these pathways?
In light of the complexity and elusiveness of LOAD pathogenesis, new approaches are needed to boost the probability of identifying causal genes and pathways. Recently, we have leveraged the molecular network structure that is reflected in genotypic and gene expression data to uncover biologically meaningful gene modules involved in the development of complex disease (Chen et al., 2008; Emilsson et al., 2008). Targeting such causal networks in ways that restore them to a normal state has been proposed as a path to treat disease (Schadt et al., 2009), but this potential has never been realized for LOAD. However, the complexity of these networks makes it difficult to distinguish the causal from correlated disease effects or how the causal regulators propagate their effects. To better address this, we constructed molecular networks based on whole-genome gene expression profiling and genotyping data in 1647 autopsied brain tissues from hundreds of LOAD patients and non-demented subjects. We identified numerous modules of distinct functional categories and cellular specificity, many showing a massive remodeling effect in the LOAD brain. Next we applied an integrative network-based approach to rank-order these modules for relevance to LOAD pathology and used a Bayesian inference to identify the key causal regulators of these remodeled networks. For instance, we identified, eight causal regulators of the top-ranked immune/microglia module including TYROBP (aka DAP12) as the highest ranking in terms of regulatory strength and differential expression in LOAD brains. We demonstrate through mouse microglia cells over-expressing intact or truncated dominant-negative TYROBP that downstream expression changes significantly overlapped the human TYROBP brain network. This study presents many of the network advantages useful in identifying and prioritizing pathways and gene targets involved in the pathophysiology of LOAD.
RESULTS
Leveraging a Systems Approach to LOAD
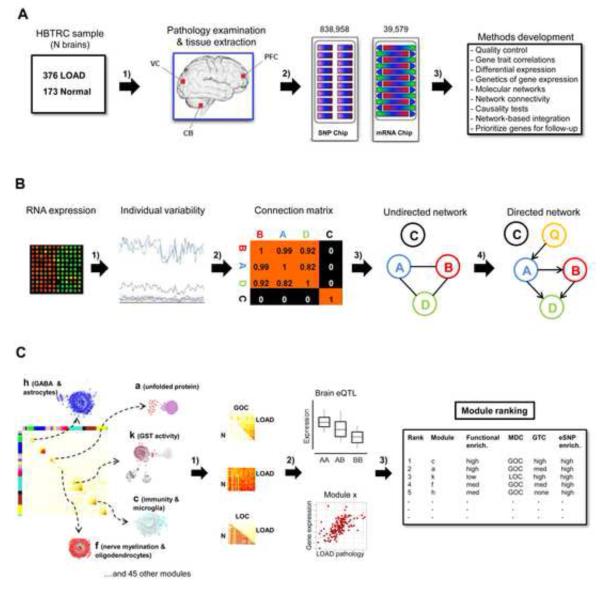
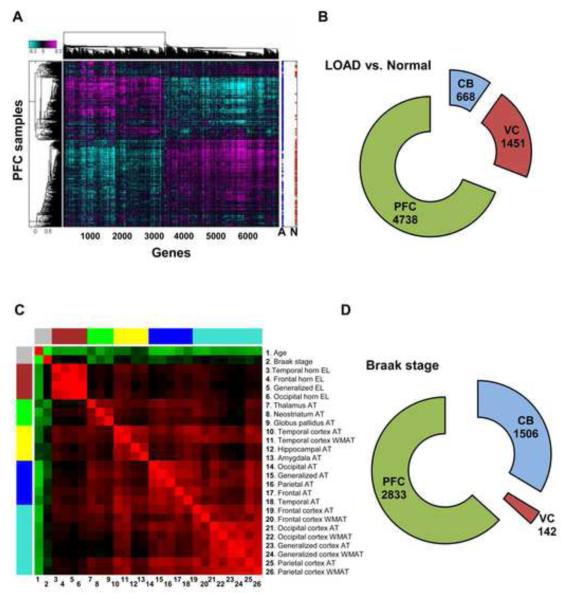
We developed and applied an integrative network-based approach to identify modules of genes associated with neurodegenerative disease (Figure 1A-C). We processed 1647 autopsied tissues from dorsolateral prefrontal cortex (PFC), visual cortex (VC) and cerebellum (CB) in 549 brains of 376 LOAD patients and 173 non-demented healthy controls (Figure 1A). All subjects were diagnosed at intake and each brain underwent extensive LOAD-related pathology examination. We note that the known APOE genotype exposure was confirmed in the HBTRC sample, showing an odds ratio of 3.74 per copy ε4 allele (P=4.1e-13). Each tissue sample was profiled for 39,579 transcripts representing 25,242 known and 14,337 predicted gene expression traits and each subject genotyped for 838,958 unique SNPs (Figure 1A). Unless otherwise noted, gene expression analyses were adjusted for age and sex, postmortem interval (PMI) in hours, sample pH and RNA integrity number (RIN). In the overall cohort of LOAD and non-demented brains the mean±SD for sample PMI, pH and RIN were 17.8±8.3, 6.4±0.3 and 6.8±0.8, respectively. Extensive analysis of the effect of covariates on gene expression variation in LOAD and non-demented brains was carried out, as shown in Figure S1 and described in the Extended Experimental Procedures. Here, we used a robust linear regression model for covariate corrections in all our gene expression analyses (Experimental Procedures). Results of traditional differential expression analysis demonstrate that subsets of genes were up- or down-regulated in LOAD (Figure 2A). Consistent with the known progression and regional severity of LOAD pathology (Braak and Braak, 1991), we observed the PFC region contained the greatest number of differentially expressed genes (Figure 2B). Figure 2C summarizes the clustering or co-linearity of the various LOAD pathology traits and age within the HBTRC cohort, resulting in distinct groups of clinical pathology and age as separate clusters. For instance, the number of significant correlations of expression traits to neuropathology like Braak stage within the LOAD patient group was highest in the PFC region (Figure 2D). Given these observations and the fact that PFC is more commonly affected in LOAD than CB and VC (Braak and Braak, 1991), a particular attention was paid to this region in our strategy to rank-order modules for relevance to LOAD. These massive datasets were the basis of further method development with the aim to identify and rank-order network modules and gene targets associated with LOAD pathology (Figure 1A-C). Results of these various analysis steps are discussed in the sections that follow, and a more detailed description of methods and statistical procedures is found in the Extended Experimental Procedures.
Figure 1. Sample Processing and the Integrative Network-based Approach.
(A) 549 brains were collected through the Harvard Brain Tissue Resource Center (HBTRC) from 376 LOAD patients and 173 non-demented subjects and tissues extracted from three brain regions, the commonly affected PFC in LOAD, and the less affected VC and CB 1). Each brain went through extensive neuropathology examination, and all tissues were profiled for 39,579 transcripts and every subject genotyped for 838,958 SNPs 2). These datasets were the basis of the method development in the present study 3). (B) From the microarray RNA expression data we identified gene expression traits showing individual variability in gene expression traits as per brain region 1). Next we computed the co-regulation (connectivity) strength between genes, defined the appropriate connectivity threshold 2), and performed hierarchical clustering analysis to construct the undirected co-expression network 3). Finally, we used brain eSNPs (Q) as causal anchors in the construction of directed Bayesian networks to infer a causal relationship between nodes in the network 4). A variant of the underlying causality scoring process here can be applied to relationships among thousands of nodes to infer genome-scale networks. (C) Comparison of LOAD and non-demented networks was performed to explore any effect on the molecular interaction structure associated with the disease. Differentially connected modules in LOAD were investigated for their functional organization 1), module relevance to clinical outcome as well as the enrichment of brain eSNPs 2). Modules were rank-ordered (this figure does not show the true rank order) for their strength of the functional enrichment, module correlation to neuropathology and eSNP enrichment 3). See also Figure S1.
Figure 2. Differential Gene Expression in LOAD Brains and Expression Correlation to Braak stage.
(A) The heat-plot shows the genes (n=6457), absolute mean-log ratio >1.5 for each profile, which most significantly differentiate disease status in PFC. The legend to the right shows the arrangement of samples with blue points denoting LOAD (A), and red points denoting non-demented state (N). (B) The number of differentially expressed genes in LOAD compared with controls per brain region using Bonferroni adjusted P<0.05 by correcting for the number of probes tested (P ≤ 2.46×10−7). (C) Clustering analysis where the rows and columns represent age and 25 LOAD pathology traits are arranged in a symmetric fashion and sorted by the hierarchical clustering tree of the correlation matrix. The color intensity signifies the correlation strength between two traits (red positive and green negative). AT, atrophy; WMAT, white matter atrophy; EL, enlargement. (D) Number of genes showing significant expression correlation to Braak stage as measured per brain region using Bonferroni adjusted P<0.05 by correcting for the number of probes tested (P ≤ 2.46×10−7). See also Table S1.
Remodeling of the Molecular Interaction Structure in LOAD Brains
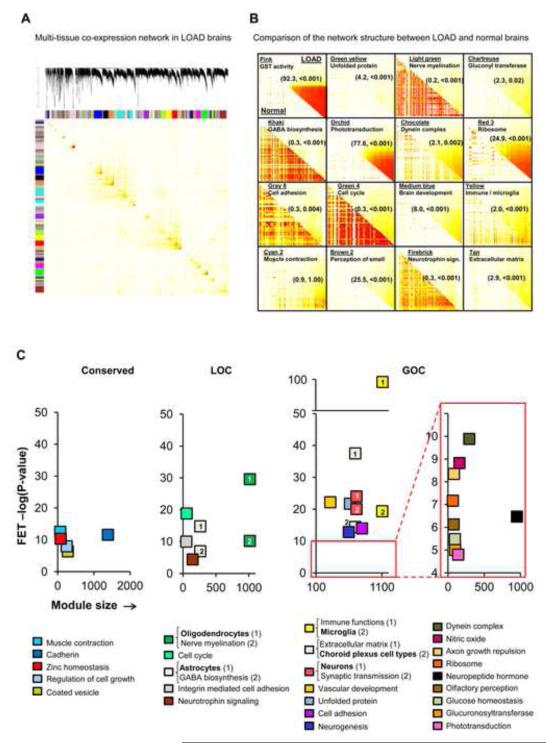
For simultaneously capturing the intra- and inter-regional gene-gene interactions in the LOAD or non-demented state, we constructed multi-tissue co-expression networks consisting of the top one-third (n=13,193) of the most variable gene expression traits per brain region in individuals donating tissues from all three regions (Extended Experimental Procedures). The multi-tissue co-expression network in LOAD brains indicated strong structurally segregated regions of the human brain molecular interactome (Figure 3A), generating 111 modules in the LOAD network, each containing between 30 and 1446 gene members (Figure 3A), while the network generated from non-demented samples has 89 modules ranging in size from 30 to 2278 genes. Figure 3B highlights a direct comparison of the two topological overlap matrices corresponding to the LOAD or non-demented associated network for a subset of 16 modules, demonstrating that LOAD reconfigures specific portions of the molecular interaction structure. To analytically detect and quantify this network reorganization across the demented and non-demented states, we developed a metric we refer to as modular differential connectivity (MDC) (Extended Experimental Procedures). MDC is the ratio of the average connectivity for any pair of module sharing genes in LOAD compared to that of the same genes in the non-demented state, and is a continuous measure ranging from 0 to infinity. This module-centric measure of differential connectivity between the two states is therefore fundamentally different from the gene-centric analysis of previous studies that applied hard cutoffs (Mani et al., 2008). Given the nature of the co-expression network analysis, MDC > 1 indicates gain of connectivity (GOC) or enhanced co-regulation between genes, while MDC < 1 indicates loss of connectivity (LOC) or reduced co-regulation between genes. In extreme cases where MDC >>1, e.g. the glutathione transferase (GST) module (Figure 3B), or MDC <<1, e.g. the nerve myelination module (Figure 3B), the corresponding genes do not form a coherent cluster in the non-demented state or LOAD, respectively. Thus, new modules are created in LOAD while in other cases a portion of the network is completely disrupted. The statistical significance of the MDC metrics was computed through the false discovery rate (FDR) procedure as described in Extended Experimental Procedures. Based on 10% FDR, 54% of all modules showed GOC while 4.5% of modules exhibited LOC. The structure of the remaining 41.5% of the modules were found to be conserved across the LOAD and non-demented states by this MDC measure. We note a negligible overlap of only 6% between signatures of differential connectivity and standard differential gene expression in LOAD brains, implying that the observed disruption in co-regulation of genes reflects a previously untapped marker of LOAD neuropathology.
Figure 3. Multi-Tissue Gene Co-expression Network in LOAD Brains.
(A) The topological overlap matrix (TOM) plot corresponds to the LOAD multi-tissue co-expression network. The rows and columns represent the same set of the top one-third (13,193) of the most variably expressed genes in each of the three brain tissues and states, expressed in a symmetric fashion and sorted by the hierarchical clustering tree of the LOAD network. (B) Individual TOM covariance matrices of 15 differentially connected and one conserved modules in LOAD (the upper right triangle of each module) versus that in the non-demented state (the lower left triangle of each module). Differential connectivity (MDC) and FDR estimate is specified in each panel in parenthesis (MDC, FDR). (C) Significant (FET P-value after correcting for number of modules and functional categories/pathways tested) enrichment of functional categories in conserved modules (left most panel), LOC modules (center panel) or GOC modules (right most panel). The y-axis represents the −log(P-value) of enrichment while the x-axis denotes the number of genes per module. Each module contains at least 50 unique gene symbols. See also Table S1.
Functional Organization of the Network and Its Relevance to LOAD Pathology
As observed in previous network-based studies (Chen et al., 2008; Emilsson et al., 2008; Zhang and Horvath, 2005), we find that the brain gene expression is organized into modules of distinct functional categories (Figure 3C). Over-representation of canonical pathways and biological processes in modules was measured through Fisher’s exact test (FET), corrected for number of modules and functional categories tested. Figure 3C highlights significant over-representation of functional categories in modules showing GOC, LOC or conserved connectivity and containing at least 50 genes. The multi-factorial basis of LOAD neuropathology involves biological processes active in both the CNS and the metabolic and vascular peripheral system that have often progressed silently for many years (Huang and Mucke, 2012; Murray et al., 2011). In fact, we find that multiple functional categories are highly enriched in the LOAD-associated modules, including the immune response, unfolded protein, vascular system, extracellular matrix, neurogenesis (brain development), glucose homeostasis, synaptic transmission and olfactory sensory perception categories in the GOC modules (Figure 3C), while the LOC modules are enriched for genes involved in nerve myelination, cell-cycle, γ-aminobutyricacid (GABA) metabolism, and neurotrophin signaling (Figure 3C). Many of these functional categories have previously been implicated in LOAD and/or CNS-related function (Ansari and Scheff, 2010; Cechetto et al., 2008; Dodel et al., 2003; Luchsinger, 2008; Morawski et al., 2012; Schiffman et al., 2002), again reinforcing the complex multi-factorial basis of the underlying pathophysiology. The functional categories enriched in the conserved modules included “muscle contraction” (actin-related system), coated vesicle, cadherin and zinc ion metabolism (Figure 3C).
CNS cell-type specific gene signatures, from the Allen Brain Atlas (http://www.brain-map.org/) were enriched in distinct network modules as previously observed (Oldham et al., 2008): neurons in the synaptic transmission modules (11 fold, P=3.7e-24), astrocytes in GABA biosynthesis module (22 fold, P=1.5e-15), oligodendrocytes in the nerve myelination module (30 fold, P=2.5e-30), choroid plexus cell types in the extracellular matrix module (35 fold, P=3.9e-15) and microglia signatures responding to amyloid-β treatment (Walker et al., 2006) in the immune module (10 fold, P=4.5e-20) (Figure 3C). In contrast to the GOC and LOC modules, conserved modules were not enriched for any CNS specific cell types (Figure 3C). Pathways enriched in the brain modules and not previously implicated in LOAD, may therefore represent novel disease mechanisms including, for instance, the glucuronosyl transferase activity and the dynein complex (Figure 3C). Moreover, the comprehensive representation of gene-gene interactions in the LOAD-associated networks can uncover novel gene members in pathways already implicated in LOAD, thus allowing us to work out a known pathologic mechanism in more detail than ever before. In summary, the immune module shows the statistically most significant functional enrichment of all modules (Figure 3C), and as such may have a more comprehensive representation of its respective pathway members.
Table S1, available online in the Supplementary Data, contains extensive information regarding the functional enrichment and gene membership of modules containing at least 50 unique gene symbols. We highlight some specific findings of interest from Figure 3C: (1) the enrichment of pathways related to olfactory sensory perception in a LOAD-associated module is of interest given the processing of olfactory function is affected in subjects who are genetically at risk of developing LOAD long before the symptoms of dementia are manifested (Schiffman et al., 2002); (2) the APOE transcript is located in the LOC module enriched for astrocyte signatures and GABA metabolism, consistent with the observation that astrocytes are the major source of APOE in the CNS (Boyles et al., 1985). The close connectivity of APOE and GABA metabolism in the brain network, may therefore have some bearing on the observation that GABA interneuron dysfunction is particularly severe in APOE4 carriers (Li et al., 2009); (3) the previously identified macrophage enriched metabolic network (MEMN) in peripheral tissues and strongly supported as causal for a number of metabolic and vascular traits related to obesity, diabetes and heart disease (Chen et al., 2008; Emilsson et al., 2008), is remarkably enriched within the brain immune/microglia module (3.9 fold, P= 2.4e-46). This is of interest given the strong epidemiological evidence for metabolic- and vascular-based exposure on LOAD (Huang and Mucke, 2012; Murray et al., 2011); (4) The postsynaptic density proteome in the human neo-cortex of 748 proteins over-represented with risk loci known to underlie cognitive, affective and motor phenotypes (Bayes et al., 2011), was significantly enriched in the synaptic transmission module (3 fold, P = 1.6e-32). It is still unclear how and which of these different biological processes mentioned above interact to affect LOAD, however it is likely that only few downstream mechanisms on which many diverse effects converge are causally related to LOAD (Huang and Mucke, 2012; Murray et al., 2011). The accumulated data show a strikingly coherent organization of molecular processes in the LOAD-associated network.
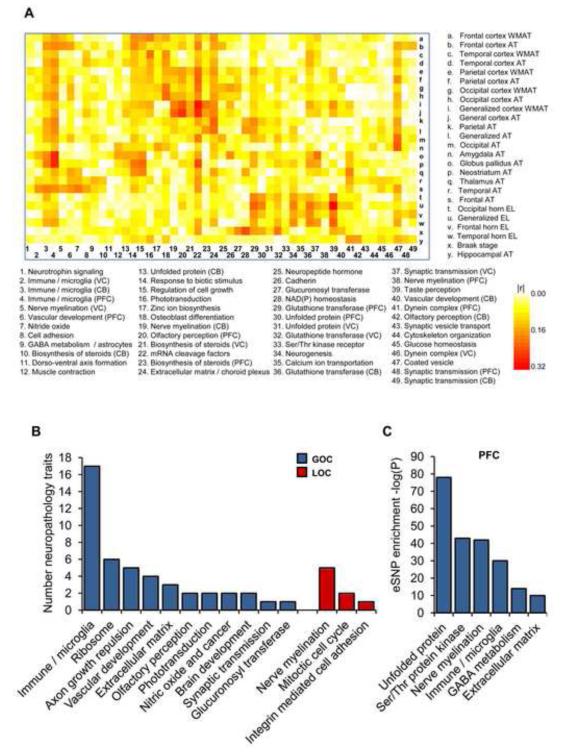
The co-expression network structure, its changes between non-demented and LOAD brains, and the genetic loci responsible for the expression co-variation behind these networks, collectively reflect molecular processes associated with LOAD. By linking the network modules to clinical outcome or LOAD neuropathology via a multiple regression analysis (Extended Experimental Procedures), we can infer key molecular processes associated with LOAD. A covariance matrix of the average expression correlation (∣r∣) between 49 modules, comprised of at least 100 probes, and 25 LOAD-related traits is shown in Figure 4A. We performed principal component analysis (PCA) to estimate the module-trait correlation and used the FDR method to assess the significance (see Extended Experimental Procedures). Of all modules, the immune/microglia showed correlation to the greatest number of LOAD-related neuropathology traits (Figure 4B). Expression of the PFC immune/microglia module correlated to atrophy levels in multiple brain regions, including frontal cortex (r=0.27, FDR=0.018), parietal (r=0.20, FDR=0.016), temporal (r=0.19, FDR=0.022) and neostriatum regions (r=0.28, FDR=3.3e-09) as well as ventricular enlargement (r=0.17, FDR=0.031). Several modules, however, showed correlation to a more restricted type of neuropathology, including the modules characteristic for the glucuronosyl transferase correlated to Braak stage (r=0.18, FDR=9.8e-05), NAD(P) homeostasis to Braak stage (r=0.25, FDR=1.4e-07), neurogenesis to ventricular enlargement (r=0.19, FDR=5.1e-05) and GST to ventricular enlargement (r=0.22, FDR=4e-06). The significance of functional enrichment in modules and the number of neuropathology traits correlated with modules were considered important criteria in rank-ordering modules for their potential to affect LOAD.
Figure 4. Module Relevance to LOAD Pathology and Enrichment of Brain eSNPs.
(A) A heat-map of the correlations (∣r∣) between 49 module principal components (PCs) and 25 LOAD-related neuropathology traits. These modules contain at least 100 probes. AT, atrophy; WMAT, white matter atrophy; EL, enlargement. (B) Number of significant module-dependent correlation to LOAD related neuropathology of all differentially connected modules with at least 100 members and showing significant correlation to at least single neuropathology trait (see Extended Experimental Procedures). The total number of traits associated with a module was used to rank-order modules for relevance to LOAD pathology. (C) We tested the enrichment of brain eSNPs in the differentially connected modules of the multi-tissue co-expression network in LOAD as per brain region. Here we present a significant enrichment of brain eSNPs in many of the PFC modules. We used the FET analysis to access the significance of the overlap between each module and cis eSNPs, correcting for the number of modules tested. See also Figure S2 and Table S1.
Bayesian Networks and the Immune Module as an Effector in LOAD
Causal probabilistic Bayesian networks were constructed and used as an alternative approach to delineate potential regulatory mechanisms. In order to establish a causal relationship or dependency between nodes in the network, we constructed a directed probabilistic Bayesian network through the application of brain cis expression (e)SNPs as causal anchors. Because cis eSNPs are in LD with causal variants that affect the expression levels of a neighboring gene or they are the causal variant themselves, they serve as an excellent source of natural perturbation to infer causal relationships among genes and between genes and higher order phenotypes like disease (Chen et al., 2008; Emilsson et al., 2008). We detected a total of 11,318 unique cis eSNPs-transcripts in the three brain regions, at FDR of 10% (Figure S2A), which is the largest number of brain eSNP-transcripts detected to date in a single study (Webster et al., 2009). The methodology to identify cis and trans eSNPs is detailed in Extended Experimental Procedures, while Table S1 lists all cis and trans acting eSNPs detected in the present study at FDR of 10%. There was between 70 and 80% sharing of cis eSNP-transcripts between different brain regions and 37% overlapped all brain regions (Figure S2A). Importantly, we find a variable and often strong enrichment of brain eSNPs in many of the LOAD-associated modules compared to all probes on the array, suggesting the possibility that these variants determine the differential connectivity observed in LOAD. For instance, in the PFC region (Figure 4C) there were five modules showing significant enrichment for cis eSNPs including the unfolded protein (3.8 fold, P=3.8e-81), nerve myelination (2.5 fold, P=2.9e-40), immune function (2.2 fold, P=4.3e-30), GABA metabolism (2.7 fold, P=2.3e-13) and extracellular matrix (1.6 fold, P=2.3e-07) modules (Figure 4C). The enrichment of cis eSNPs in the differentially connected LOAD modules in the VC and CB regions is shown in Figure S2B. For the present study, however, a particular attention was paid to the cis eSNPs for their applicability as priors in the construction of Bayesian networks (Extended Experimental Procedures and schematic Figure S3).
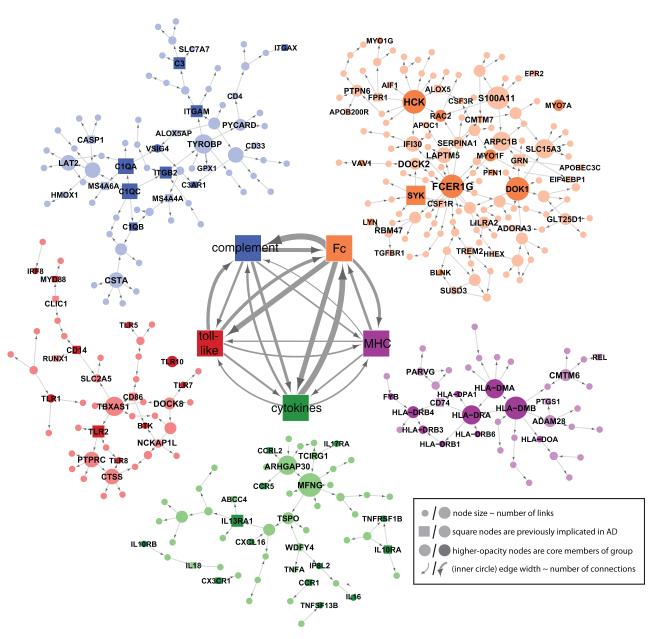
We constructed Bayesian networks for each co-expression module. While many of the LOAD-associated network modules are of potential interest, the reconstruction of the Bayesian network for the immune/microglia module is highlighted given it has the strongest disease association based on clinical covariates and network-associated properties: (1) significant differential connectivity of the cortex specific immune modules in LOAD (MDC between 49 and 100% GOC at FDR < 0.001); (2) the immune/microglia module showed the most significant enrichment of functional categories; (3) the highest degree of gene expression correlation to several measures of LOAD neuropathology; (4) the PFC version of the module was highly enriched for brain eSNPs. To increase the predictive power of inflammation related regulatory networks, we further built up the directed Bayesian network for the inflammation modules derived from the individual brain regions. Figure 5 highlights the interactions within and between the five predominant immunologic families in the PFC-based putative microglia module. To generate this roadmap to the complex structure of the immune/microglia module, genes which were not direct members of one of these five core pathways were assigned to the family with which they have the greatest number of causal interactions. The immune module was dissected into five families representing functional immune pathways that were labeled according to their main function as ‘Complement’, ‘Fc’ for Fc-receptors, ‘MHC’ for major histocompatibility complex, ‘Cytokines’ for cytokines/chemokines and ‘Toll-like’ for toll-like receptors (Figure 5).
Figure 5. The Bayesian Brain Immune and Microglia Module.
A module that correlates with multiple LOAD clinical covariates and is enriched for immune functions and pathways related to microglia activity (PFC module shown). [Inner networks] The PFC module is enriched in genes which can be classified as members of the complement cascade, ‘Complement’, toll-like receptor signaling, ‘Toll-like’, chemokines/cytokines, ‘Chemokine’, and the major histocompatibility complex, ‘MHC’, or Fc receptor system, ‘Fc’. The direction and strength of interactions between these pathways are collected across all gene-gene causal relationships that span different pathways. The minimum line width corresponds to a single interaction (MHC to toll-like) and scales linearly to a maximum of 17 interactions (Fc to Complement). [Outer networks] Each color-coded group of genes consists of the core members of the different families and genes that are causally related to a given family. Core family members of each pathway are shaded darkly, while square nodes in any family denote literature-supported nodes (at least two PubMed abstracts implicating the gene or final protein complex in LOAD or a model of LOAD). Labeled nodes are either highly connected in the original network, literature-implicated LOAD genes or core members of one of the five immune families. Node size is proportional to connectivity in the module. See also Figures S5.
Highlighting the Microglia Pathway with TYROBP as Causal Regulator
The Bayesian inference enabled us to compute the causal regulators of the differential connectivity in individual modules, defined as the genes controlling many downstream nodes in the respective network (see Extended Experimental Procedures). The causal regulators of the highest scoring immune/microglia module were rank-ordered based on the number of downstream nodes, i.e. the power of regulating other genes, as well as differential expression in LOAD brains. Here, we used a combined score as , where, gji is the discriminant value of a j in the case i, and is defined as (Duda et al., 2000). In comparison to the average gene/node in a given network the causal regulators are expected to have a stronger effect on the clinical outcome as they direct the expression of a significant portion of the network module they reside in. The size of the gene membership for the different regional specific immune modules ranges from 386 in CB to 1108 in the PFC, with 247 of the genes in the CB detected in all regions (P=1e-19). The identity of the key causal regulators is somewhat variable across each brain regional version of the microglia module of which CTSC, HCK, TYROBP, SERPINA1, S100A11, LY86, DOCK2 and FCER1G were common to all immune modules, regardless of brain region. Through the combined ranking score based on regulatory strength and differential expression in PFC of LOAD brains, TYROBP scored the highest (Figure S4A). Table 1 lists the 20 top ranking PFC modules and their respective key causal regulators. Expression of TYROBP is restricted to cells involved in the innate immunity including the microglial cells in the brain (Schleinitz et al., 2009). Here, TYROBP was significantly up-regulated in LOAD brains in the HBTRC sample (1.18 fold, P = 0.028) and the direction of this effect was replicated (1.17 fold, P = 5.1e-05) in an independent multi-center cohorts study (see Extended Experimental Procedures and Figure S4B). Additionally, we observed a progression of TYROBP expression changes across mild cognitive impairment (MCI) in the replication study (Figure S4B). Estimating what constitutes a “large” or “small” change in gene expression levels is challenging in microarray analyses. We note, however, that TYROBP was the 124th most differentially-expressed probe out of 48,803 probes assayed in the replication study cohort. Moreover, TYROBP was more differentially expressed in LOAD brains than the classical markers of microglia AIF1, and CD68, indicating there was not a relative down-regulation of TYROBP despite elevated microgliosis in LOAD brains (Perry et al., 2010).
Table 1. Top 20 Modules in PFC Ranked for Relevance to LOAD Pathology.
This table lists the top 20 rank-ordered modules consisting of at least 50 genes from PFC, or if majority of genes are from PFC in mixed modules with a total of 50 genes or more. See also Table S1 and Figure S4.
| Module | Rank | Top Functional & Cellular Category |
N PFC Genes |
MDC | Highlighted Causal Regulatorsa |
|---|---|---|---|---|---|
| Yellow | 1 | Immune & microglia | 1102 | 1,49 | TYROBP, DOCK2, FCER1G |
| Pink | 2 | Glutathione transferase | 113 | 92,67 | GSTA4, ABCC2, TIMELESS |
| Gray 1 | 3 | Cell junction | 51 | 0,82b | ACBD5, LMAN1, MLL3S |
| Seashell | 4 | Coated vesicle | 278 | 1,29b | KIFAP3, PCTK2, SNCA |
| Red 3 | 5 | Ribosome | 50 | 24,93 | RPS27, RPS18, PCBP2 |
| Green yellow | 6 | Unfolded protein | 721 | 4,50 | STIP1, HSPA1A, DOPEY1 |
| Red | 7 | Nerve myelination & oligodendrocytes |
987 | 0,68 | ENPP2, PSEN1, GAB2 |
| Gold 2 | 8 | Axon growth repulsion | 80 | 3,27 | TUBB4, ACTL9, ACTG1 |
| Tan | 9 | Extracellular matrix & choroid plexus cells |
700 | 2,88 | SLC22A2, AGTR1, ZIC2 |
| Gold 3 | 10 | Dynein complex | 67 | 12,12 | TEKT1, FANK1, HYDIN |
| Light yellow | 11 | mRNA cleavage | 96 | 6,01 | MED6, STATIP1, SFRS3 |
| Brown 2 | 12 | Olfactory perception | 77 | 25,51 | PPP2R5A, C1ORF143, RNASE11 |
| Dark cyan | 13 | Steroid biosynthesis | 110 | 1,39b | LAMP2, P2RX7, MID1IP1 |
| Khaki | 14 | GABA biosynthesis & astrocytes |
267 | 0,29 | GJA1, STON2, CST3 |
| Grey 60 | 15 | Ser/Thr kinase receptor | 495 | 4,64 | CREBBP, ABCC11, MDGA1 |
| Purple | 16 | Synaptic transmission & neurons |
805 | 1,22 | SNAP91, BSN, GLS |
| Green 4 | 17 | Cell cycle | 50 | 0,33 | DTL, UBE2C, BUB1 |
| Honey dew | 18 | Muscle contraction | 128 | 1,10b | RFX4, DGCR6, AQP4 |
| Red 2 | 19 | Zinc homeostasis | 83 | 1,17b | MT1M, MT1JP, MT1P3 |
| Beige | 20 | Glucose homeostasis | 95 | 12,64 | AMPD1, EGR2, PDGFB |
Selected set of maximum three causal regulators per module
MDC FDR > 10% and therefore not considered significant
The majority of the common causal regulators were located either in the ‘Fc’ pathway and associated/clustered genes (HCK, SERPINA1, S100A11, DOCK2 and FCER1G) or the ‘Complement’ pathway (TYROBP) in the immune/microglia network (Figure 5). Recent reports (we note that our submission predates these reports) show a striking association of a low-frequency DNA variant in TREM2 to LOAD (Guerreiro et al., 2013; Jonsson et al., 2013). More specifically, TREM2 is known to associate and signal via TYROBP, the key regulator of the immune/microglia network activated in LOAD. Thus our data-driven network-based approach places both TREM2 and TYROBP in a gene network that literally unifies them with previous top GWAS risk loci including MS4A4A, MS4A6A and CD33 (Figure 5). These new results provide exciting convergent evidence for the specific microglia network that we had directly implicated as activated in LOAD, and reinforce the potential causality of this pathway in LOAD pathology. In fact, the dissection of the immune/microglia module into distinct families and key causal regulators point towards an important function of the microglia pathways involving genes of the ‘Complement’ and/or ‘Fc’ network clusters. Figure S5 (genes marked in red) highlight many of the key genes in the pathogen phagocytosis pathways found in the immune/microglia module. It is noteable how comprehensive representation of a specific signal transduction pathways is observed within the two immune families of this module. The strategic network position of TYROBP as a causal regulator of many genes mirrors its bottleneck position in several microglia activation signaling cascades. Extrapolating from this data-driven interaction, it is possible that TYROBP may be associated with neuronal pruning activity of the complement system that may be reawakened in LOAD via amyloid-β and Tau aggregates (Stevens, 2007; Perry, 2010). In this manner, the network structure can become a data-driven hypothesis generator for novel disease-relevant interactions.
Structure of Causal Networks Guides Differential Expression in a Distance-dependent Manner
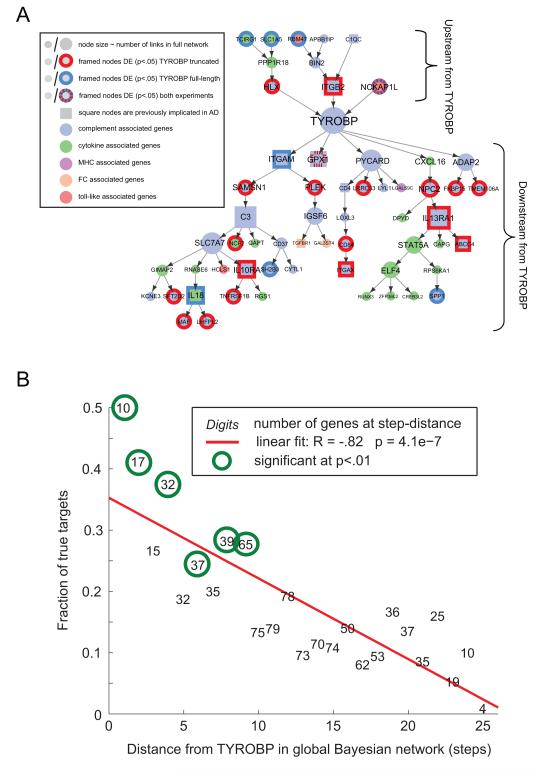
To test our prediction that TYROBP can direct LOAD-associated gene networks, we contrast both the molecular function and genome-wide effects of TYROBP with those predicted by the structure of causal networks inferred from human LOAD brains. For this, microglia cells derived from mouse embryonic stem cells were genetically modified by lentiviral vectors to over-express either full length or a truncated version of Tyrobp which lacks both intracellular ITAM motifs (Extended Experimental Procedures and Figure S6). To assess the genome-wide gene expression changes in response to the perturbation of Tyrobp, we derived gene expression data from the RNA sequencing of mouse microglia cell lines over-expressing (1) vehicle, (2) the full length Tyrobp or (3) dominant negative truncated Tyrobp. We identified 2638 and 3415 differentially expressed genes for the over-expression of full length Tyrobp and truncated Tyrobp, respectively (Table S1), at FDR < 2.5%. Roughly one-third (858 to 1092) of these genes are found in the most variable gene set in the brain dataset used for the network reconstruction. The PFC variant of the human immune/microglia module was highly enriched for genes which are differentially expressed in the full length or truncated Tyrobp experiments (P < 1e-15) (Figure 6A). We projected results of RNA sequencing experiments onto a large Bayesian brain network of ~8000 nodes that contains the microglia module as well as many other modules. In this large network, we could track differential expression of genes which are predicted to be downstream of TYROBP at various network path distances (Figure 6B). The highest predictive power for differential expression is in the primary neighborhood of the perturbed gene, and this power decreases for genes which are farther away in the network. The enrichment for differentially expressed genes in the network neighborhood of TYROBP and strong negative correlation between the fraction of confirmed targets and path distance (r= −0.82, P=4.e-07) (Figure 6B), show that our causal network structure is a significant and useful predictor of response to gene perturbations, even in a challenging cross-species setting. Thus both the structure and direction of links in these causal networks provide significant information on the effects of complex signal transduction mechanisms.
Figure 6. Structure of Causal Networks Guides Differential Expression in a Distance-dependent Manner.
A) Within the microglia module, we show all genes which receive direct or indirect causal inputs to/from TYROBP. Genes which were differentially expressed in either full-length or truncated Tyrobp experiments are circled (P-value<0.05, n=4/4/4 for control/truncated/full-length RNA sequenced samples). Possible reasons for differentially expressed (DE) of predicted upstream genes are mouse-human network differences, network inaccuracy, or presence of feedback loops, which are not represented in a Bayesian framework. (B) We mapped results of RNA sequencing experiments onto a large Bayesian network of ~8000 nodes that contains the microglia module as well as many other modules. In this large network, we could track differential expression of genes which are predicted to be downstream of TYROBP at various network distances (link distances). There was a strong negative correlation (r= −0.82, P=4.e-07) between the differentially expressed genes in the microglia and the path distance from TYROBP in the brain immune network. See also Figure S6 and Table S1.
The inferred network structure has significant predictive power for nodes which are several links away from TYROBP. We studied the enrichment of functional categories in the gene sets responding to the Tyrobp perturbation experiments, and applied Bonferroni-corrected P-values for statistical significance (Extended Experimental Procedures). Approximately 99% of the differentially expressed genes from the microglia over-expressing intact Tyrobp were down-regulated compared to the control vehicle. This set was enriched for genes involved in RNA metabolism (P=6.2e-05) and cell-cycle mitosis (P=2.7e-03). In the microglia cells over-expressing the dominant negative truncated Tyrobp, 2856 up-regulated genes were enriched for the vacuole/autophagy (P=1.7e-08) and mitochondrion (P=4.6e-04), while 559 genes involved in histone assemply (P=1.6e-31) were down-regulated. Moreover, the Tyrobp regulatory effect reflects a degree of symmetry as 658 genes, related to the vacuole/autophagy (P=5e-03), were down-regulated by active Tyrobp but up-regulated in cells expressing dominant negative truncated Tyrobp. These findings are of interest because they link the far down-stream effects of TYROPB to known molecular pathology in LOAD, such as abnormalities in the cell-cycle, mitochondrion and autophagy (Coskun et al., 2004; Webber et al., 2005). The accumulated data suggest that TYROBP may be a therapeutic target in prevention of neuronal damage in LOAD.
DISCUSSION
The construction of gene regulatory networks in a large sampling of human brain specimens has revealed many facets of the molecular interaction structure in LOAD, when compared to that in non-demented brains. A comprehensive characterization of gene network connectivity, its regulation and association to disease can provide critical insights into the underlying mechanisms, and identify genes that may serve as effective targets for therapeutic intervention. For instance, targeting genes that are the most central (highly connected) may be more effective in disrupting disease-related networks for the purpose of therapy, but that could be at the cost of more adverse effects. In summary, the utility of network-based approaches to complex disease includes: (1) elucidating the biological function and molecular context of a particular set of causal genes, (2) establishing a framework to map interaction between genes and network modules, (3) providing an objective filter for rank-ordering genes based on connectivity or other network features, (4) defining dynamic changes and corresponding causal regulators of the altered network structure associated with disease condition, (5) identifying modules and pathways causally related to disease, and (6) revealing tissue-to-tissue interactions that can aid in the identification of key target tissues for disease (Dobrin et al., 2009). The present study utilizes many of these network advantages to highlight and prioritize pathways and gene targets causally related to LOAD.
Our network-based integrative analysis not only highlighted the immune/microglia module as the molecular system most strongly associated with the pathophysiology of LOAD, but identified the key network regulators including TYROBP. In a separate in vitro study we have found that the microglia expressed TYROBP is directly involved in amyloid-β turnover and neuronal damage (unpublished results). Of interest, mutations in TYROBP or TREM2 cause Nasu-Hakola disease (Bianchin et al., 2010), a rare Mendelian disease characterized by bone reabsorption dysfunction and chronic inflammatory neurodegeneration leading to death in the fourth or fifth decade of life. The exact pathomechanism underlying Nasu-Hakola disease is still unclear, but it was hypothesized that failure of proper microglial clearance is causal for the lethal effect of neurodegeneration. Thus dysfunctional immune/microglia pathways might not be unique to LOAD. To test the generalization of this concept, we explored the connection of the immune/microglia module to Huntington disease (HD), another neurodegenerative disease. HD pathology, caused by expanded alleles of a variable stretch of trinucleotide (CAG) repeat length in HTT (The Huntington’s Disease Collaborative Research Group, 1993), features astrogliosis and neurodegeneration of the striatum, prefrontal cortex and hippocampus. We constructed molecular networks in the PFC from 194 HD patients genotyped for CAG allele size (see Extended Experimental Procedures) and found that the PFC version of the immune/microglia module was well conserved between LOAD and HD in terms of gene annotation (75% overlap, P-value <1e-300). This module, however, did not show any alteration in connectivity in HD brains compared to the disease-free controls used in our LOAD study. Moreover, through a PCA we did not detect any gene expression correlation of the HD brain immune/microglia module to expanded CAG repeat length (r=-0.05, FDR=56%), a key biomarker for predicting HD severity (Gusella and MacDonald, 2006). Thus based on the comparison to HD, the disease-related effect of the immune/microglia module appears to be specific to LOAD (and possibly Nasu-Hakola disease).
Immune activation in LOAD may have multi-faceted activity: long-term use of non-steroid anti-inflammatory drugs (NSAIDs) before onset of the disease decreases risk (Etminan et al., 2003), and microglia effector function via interfering with reactive oxygen production, cytokines and complement cascade members have been postulated to damage healthy neurons and synapses (Cameron and Landreth, 2010). Close association and positive feedback between amyloid-β and microglia (Meyer-Luehmann et al., 2008) further clouds the cause and effect relationships of inflammation to disease progression. Without a causal framework for these observations, it is difficult to find optimal molecular targets that direct LOAD inflammation. Therefore, we integrated clinical factors with whole-genome genotype and molecular trait data to identify a network module containing several microglia signaling cascades functionally related to the reactive oxygen burst during pathogen phagocytosis. We highlight the causal regulator TYROBP that exerts control over multiple genes within this module and pathways involved in LOAD, thus validating our network structure and its relevance to LOAD pathology. This approach appears to offer novel insights for drug discovery programs that can affect neurodegenerative diseases, such as LOAD.
EXPERIMENTAL PROCEDURES
Raw gene expression data together with information related to demographics, disease state and technical covariates are available via the GEO database (GEO accession number GSE44772; GSE44768, GSE44770 and GSE44771). A brief description of key methods and sample description is provided below while complete details are found in the Extended Experimental Procedures.
Data Sets and Sample Processing
We compiled six disease- and tissue-specific gene expression datasets consisting of 1647 postmortem specimens from three brain regions (PCF (BA9), VC (BA17) and CB) in LOAD and non-demented subjects recruited through the Harvard Brain Tissue Resource Center (HBTRC). Each subject was diagnosed at intake and via extensive neuropathology examination. Tissues were profiled on a custom-made Agilent 44K array of 40,638 DNA probes and each subject genotyped for 838,958 SNPs.
Molecular Networks and Causal Regulators
We constructed both multi-tissue and single tissue co-expression networks from the top one-third (n=13,193) of the most variably expressed genes in each tissue and condition. We computed the module differential connectivity (MDC) in LOAD brains as: , where, kij is the connectivity between two genes i and j in a given network, and assessed the statistical significance through the FDR method. We constructed causal probabilistic Bayesian networks from individual co-expression modules and used brain cis eSNPs as priors to infer directionality between nodes (see Figure S3). For this, we identified 11,318 unique cis eSNPs-transcripts at FDR of 10% (Extended Experimental Procedures), all listed in Table S1. The Bayesian inference allowed us to compute the causal regulators of the differential connectivity in individual modules by examining the number of N-hob downstream nodes.
Mouse Microglia Cultivation, Cell Transduction and RNA sequencing
Genome-wide gene expression of mRNA from cultivated microglia cells over-expressing intact or genetically modified TYROBP was sequenced using TruSeq Kit for RNA capture and HiSeq 2000 for the sequencing. Read mapping was done using the TopHat (Trapnell et al., 2009) RNA-seq aligner.
Supplementary Material
HIGHLIGHTS.
▶ Systems approach to LOAD based on large-scale human brain tissue sampling
▶ Molecular networks show strong remodeling effect in LOAD brains
▶ Integrative network-based analysis implicates the immune/microglia network in LOAD
▶ TYROBP is key causal regulator within the immune/microglia module
ACKNOWLEDGMENTS
H.N. and L-G.B. were supported by the Hertie-Foundation and the Deutsche Forschungsgemeinschaft (FOR1336, SFB704, KFO177). H.N. is a member of the DFG-funded excellence cluster ImmunoSensation. We thank Jessica Schumacher and Rita Hass for technical assistance. We are grateful to the HBTRC for the generous gift of human postmortem brain samples. The authors are also grateful to the participants in the Religious Orders Study and the Memory and Aging Project. This work is supported by the National Institutes of Health (R01AG034504, R01 AG030146, P30 AG10161, R01 AG17917, R01 AG15819, K08 AG034290, P30 AG10161, R01 AG11101 and NS032765), the Illinois Department of Public Health and start-up funding from the University of Miami, Miller School of Medicine. JM, AAP, CZ, TX, RD, EF, SM, MN, CM and DJS are employees and shareholders of Merck & Co., Inc. JRL is an employee and shareholder of Novartis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION Supplemental Information includes Supplementary Data with one table, Extended Experimental Procedures with six figures and can be found with this article online at doi:xx
REFERENCES
- Ansari MA, Scheff SW. Oxidative stress in the progression of Alzheimer disease in the frontal cortex. J Neuropathol Exp Neurol. 2010;69:155–167. doi: 10.1097/NEN.0b013e3181cb5af4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayes A, van de Lagemaat LN, Collins MO, Croning MD, Whittle IR, Choudhary JS, Grant SG. Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat Neurosci. 2011;14:19–21. doi: 10.1038/nn.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, Lill CM, Tanzi RE. The genetics of Alzheimer disease: back to the future. Neuron. 2010;68:270–281. doi: 10.1016/j.neuron.2010.10.013. [DOI] [PubMed] [Google Scholar]
- Bettens K, Sleegers K, Van Broeckhoven C. Genetic insights in Alzheimer’s disease. Lancet Neurol. 2013;12:92–104. doi: 10.1016/S1474-4422(12)70259-4. [DOI] [PubMed] [Google Scholar]
- Bianchin MM, Martin KC, de Souza AC, de Oliveira MA, Rieder CR. Nasu-Hakola disease and primary microglial dysfunction. Nat Rev Neurol. 2010;6:193–201. doi: 10.1038/nrneurol.2010.17-c1. [DOI] [PubMed] [Google Scholar]
- Boyles JK, Pitas RE, Wilson E, Mahley RW, Taylor JM. Apolipoprotein E associated with astrocytic glia of the central nervous system and with nonmyelinating glia of the peripheral nervous system. J Clin Invest. 1985;76:1501–1513. doi: 10.1172/JCI112130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Brookmeyer R, Johnson E, Ziegler-Graham K, Arrighi HM. Forecasting the global burden of Alzheimer’s disease. Alzheimers Dement. 2007;3:186–191. doi: 10.1016/j.jalz.2007.04.381. [DOI] [PubMed] [Google Scholar]
- Cameron B, Landreth GE. Inflammation, microglia, and Alzheimer’s disease. Neurobiology of disease. 2010;37:503–509. doi: 10.1016/j.nbd.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cechetto DF, Hachinski V, Whitehead SN. Vascular risk factors and Alzheimer’s disease. Expert Rev Neurother. 2008;8:743–750. doi: 10.1586/14737175.8.5.743. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, Zhang C, Lamb J, Edwards S, Sieberts SK, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Coskun PE, Beal MF, Wallace DC. Alzheimer’s brains harbor somatic mtDNA control-region mutations that suppress mitochondrial transcription and replication. Proc Natl Acad Sci U S A. 2004;101:10726–10731. doi: 10.1073/pnas.0403649101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrin R, Zhu J, Molony C, Argman C, Parrish ML, Carlson S, Allan MF, Pomp D, Schadt EE. Multi-tissue coexpression networks reveal unexpected subnetworks associated with disease. Genome Biol. 2009;10:R55. doi: 10.1186/gb-2009-10-5-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodel R, Hampel H, Du Y. Immunotherapy for Alzheimer’s disease. Lancet Neurol. 2003;4:215–220. doi: 10.1016/s1474-4422(03)00349-1. [DOI] [PubMed] [Google Scholar]
- Duda RO, Hart PE, Stork DG. Pattern classification. 2nd edn John Wily & Sons, Inc; New York: 2000. [Google Scholar]
- Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, Carlson S, Helgason A, Walters GB, Gunnarsdottir S, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- Etminan M, Gill S, Samii A. Effect of non-steroidal anti-inflammatory drugs on risk of Alzheimer’s disease: systematic review and meta-analysis of observational studies. BMJ (Clinical research ed) 2003;327:128. doi: 10.1136/bmj.327.7407.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C, Kauwe JS, Younkin S, et al. TREM2 variants in Alzheimer’s disease. N Engl J Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusella JF, MacDonald ME. Huntington’s disease: seeing the pathogenic process through a genetic lens. Trends Biochem Sci. 2006;31:533–540. doi: 10.1016/j.tibs.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Huang Y, Mucke L. Alzheimer mechanisms and therapeutic strategies. Cell. 2012;148:1204–1222. doi: 10.1016/j.cell.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson T, Atwal JK, Steinberg S, J. S, Jonsson PV, S. B, Stefansson H, Sulem P, Gudbjartsson D, Maloney J, et al. A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. 2012 doi: 10.1038/nature11283. LID - 10.1038/nature11283 [doi]. Nature doi: 10.1038. [DOI] [PubMed] [Google Scholar]
- Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J, Levey AI, Lah JJ, et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N Engl J Med. 2013;368:107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GF, Bien-Ly N, Andrews-Zwilling Y, Xu Q, Bernardo A, Ring K, Halabisky B, Deng C, Mahley RW, Huang Y. GABAergic interneuron dysfunction impairs hippocampal neurogenesis in adult apolipoprotein E4 knockin mice. Cell Stem Cell. 2009;5:634–645. doi: 10.1016/j.stem.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchsinger JA. Adiposity, hyperinsulinemia, diabetes and Alzheimer’s disease: an epidemiological perspective. Eur J Pharmacol. 2008;585:119–129. doi: 10.1016/j.ejphar.2008.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani KM, Lefebvre C, Wang K, Lim WK, Basso K, Dalla-Favera R, Califano A. A systems biology approach to prediction of oncogenes and molecular perturbation targets in B-cell lymphomas. Molecular systems biology. 2008;4:169. doi: 10.1038/msb.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Luehmann M, Spires-Jones TL, Prada C, Garcia-Alloza M, de Calignon A, Rozkalne A, Koenigsknecht-Talboo J, Holtzman DM, Bacskai BJ, Hyman BT. Rapid appearance and local toxicity of amyloid-beta plaques in a mouse model of Alzheimer’s disease. Nature. 2008;451:720–724. doi: 10.1038/nature06616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawski M, Bruckner G, Jager C, Seeger G, Matthews RT, Arendt T. Involvement of perineuronal and perisynaptic extracellular matrix in Alzheimer’s disease neuropathology. Brain Pathol. 2012;22:547–561. doi: 10.1111/j.1750-3639.2011.00557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray IV, Proza JF, Sohrabji F, Lawler JM. Vascular and metabolic dysfunction in Alzheimer’s disease: a review. Exp Biol Med (Maywood) 2011;236:772–782. doi: 10.1258/ebm.2011.010355. [DOI] [PubMed] [Google Scholar]
- Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, Geschwind DH. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry V, Nicoll J, Holmes C. Microglia in neurodegenerative disease. Nat Rev Neurol. 2010;4:193–201. doi: 10.1038/nrneurol.2010.17. [DOI] [PubMed] [Google Scholar]
- Qiu C, Xu W, Fratiglioni L. Vascular and psychosocial factors in Alzheimer’s disease: epidemiological evidence toward intervention. J Alzheimers Dis. 2010;20 doi: 10.3233/JAD-2010-091663. [DOI] [PubMed] [Google Scholar]
- Schadt EE. Molecular networks as sensors and drivers of common human diseases. Nature. 2009;461:218–223. doi: 10.1038/nature08454. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Friend SH, Shaywitz DA. A network view of disease and compound screening. Nat Rev Drug Discov. 2009;4:286–295. doi: 10.1038/nrd2826. [DOI] [PubMed] [Google Scholar]
- Schafer S, Kolkhof P. Failure is an option: learning from unsuccessful proof-of-concept trials. Drug Discov Today. 2008;21-22:913–916. doi: 10.1016/j.drudis.2008.03.026. [DOI] [PubMed] [Google Scholar]
- Schiffman SS, Graham BG, Sattely-Miller EA, Zervakis J, Welsh-Bohmer K. Taste, smell and neuropsychological performance of individuals at familial risk for Alzheimer’s disease. Neurobiol Aging. 2002;23:397–404. doi: 10.1016/s0197-4580(01)00337-2. [DOI] [PubMed] [Google Scholar]
- Schleinitz N, Chiche L, Guia S, Bouvier G, Vernier J, Morice A, Houssaint E, Harle JR, Kaplanski G, Montero-Julian FA, et al. Pattern of DAP12 expression in leukocytes from both healthy and systemic lupus erythematosus patients. PloS one. 2009;4:e6264. doi: 10.1371/journal.pone.0006264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Huntington’s Disease Collaborative Research Group. H. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DG, Link J, Lue LF, Dalsing-Hernandez JE, Boyes BE. Gene expression changes by amyloid beta peptide-stimulated human postmortem brain microglia identify activation of multiple inflammatory processes. Journal of leukocyte biology. 2006;79:596–610. doi: 10.1189/jlb.0705377. [DOI] [PubMed] [Google Scholar]
- Webber KM, Raina AK, Marlatt MW, Zhu X, Prat MI, Morelli L, Casadesus G, Perry G, Smith MA. The cell cycle in Alzheimer disease: a unique target for neuropharmacology. Mech Ageing Dev. 2005;126:1019–1025. doi: 10.1016/j.mad.2005.03.024. [DOI] [PubMed] [Google Scholar]
- Webster JA, Gibbs JR, Clarke J, Ray M, Zhang W, Holmans P, Rohrer K, Zhao A, Marlowe L, Kaleem M, et al. Genetic control of human brain transcript expression in Alzheimer disease. American journal of human genetics. 2009;84:445–458. doi: 10.1016/j.ajhg.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:12. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.