Abstract
The absence of Klotho (KL) from mice causes the development of disorders associated with human aging and decreased longevity, whereas increased expression prolongs lifespan. With age, KL protein levels decrease, and keeping levels consistent may promote healthier aging and be disease-modifying. Using the KL promoter to drive expression of luciferase, we conducted a high-throughput screen to identify compounds that activate KL transcription. Hits were identified as compounds that elevated luciferase expression at least 30%. Following validation for dose-dependent activation and lack of cytotoxicity, hit compounds were evaluated further in vitro by incubation with opossum kidney and Z310 rat choroid plexus cells, which express KL endogenously. All compounds elevated KL protein compared with control. To determine whether increased protein resulted in an in vitro functional change, we assayed FGF23 (fibroblast growth factor 23) signalling. Compounds G–I augmented ERK (extracellular-signal-regulated kinase) phosphorylation in FGFR (fibroblast growth factor receptor)-transfected cells, whereas co-transfection with KL siRNA (small interfering RNA) blocked the effect. These compounds will be useful tools to allow insight into the mechanisms of KL regulation. Further optimization will provide pharmacological tools for in vivo studies of KL.
Keywords: choroid plexus, fibroblast growth factor 23 (FGF23), high-throughput screen, kidney, klotho, longevity
INTRODUCTION
Klotho (KL) is a type I transmembrane protein composed of two weakly homologous domains: KL1 and KL2. KL is shed from cell membranes [1] and has unique functions attributed to its transmembrane and shed forms. Transmembrane KL functions as the FGF23 (fibroblast growth factor 23) co-receptor, critical for proper calcium, phosphate and vitamin D homoeostasis [2]. Amelioration of homoeostatic dysregulation by dietary changes attenuates many, but not all, of the disorders observed in KL-null mice, emphasizing the importance of proper FGF23 signalling in vivo [3]. Shed KL functions as a sialidase to alter ion channel properties [TRPV (transient receptor potential vanilloid) 5/6 and ROMK1 (renal outer meduallary K+ channel 1)] [4,5]. KL shed from the membrane represses insulin/IGF-1 (insulin-like growth factor 1) signalling, up-regulating FOXO (forkhead box O) dephosphorylation and SOD2 (superoxide dismutase 2), an important scavenger of free radicals [6,7]. Similarly, shed KL represses Wnt signalling to prevent premature depletion of stem cells [8]. Modulation of KL also affects cellular senescence and apoptosis [7,9,10] and functions as a tumour suppressor in several cancers [1,11,12]. KL is protective in the cardiovascular system, where its absence causes a decrease in NO production resulting in vascular endothelial dysfunction [13–15]. Taken together, studies of the molecular mechanisms responsible for KL-mediated disorders reveal KL to be a protective molecule involved in numerous homoeostatic functions, antioxidant defence and proper cell cycling.
Absence of KL shortens lifespan in both mice and Caenorhabditis elegans [16,17]. In mice, the absence of KL induces a dramatic phenotype recapitulating many of the disorders commonly associated with human aging [17]. Although normal through development and adolescence, upon reaching adulthood, KL-deficient mice rapidly begin to deteriorate and die by no later than 4 months from the confluence of defects in multiple organ systems [17]. Phenotypically, these mice develop vascular calcification, infertility, emphysema, osteoporosis, skin atrophy, hair loss, thymic involution, osteopenia, motor neuron degeneration and cognitive impairment [2]. Although elimination of KL causes systemic dysfunction and shortened lifespan, overexpression of KL in mice extends lifespan ~20–30% and suppresses insulin signalling [18]. Exposure of KL-overexpressing animals to paraquat, which generates lethal doses of reactive oxygen species, revealed increased resistance to oxidative stress [7]. Resistance to stress has been attributed to KL-mediated FOXO dephosphorylation, a well-characterized antioxidant pathway [7,16].
Altering KL protein levels in humans results in disease or increased disease risk. The human equivalent of a knockout has not been observed, presumably due to lethality, especially in light of case studies reporting severe phenotypes from significant changes in KL protein levels. The missense mutation H193R decreased KL, resulting in severe tumoral calcinosis, hyperphosphataemia and hypercalcaemia [19]. Meanwhile, translocation and subsequent overexpression of the KL gene resulted in hypophosphataemic rickets and hyperparathyroidism [20]. Although these examples are extremely rare cases of severe KL dysregulation, a growing body of evidence suggests that even minor alterations in KL caused by polymorphisms can increase the risk of developing disease.
KL’s functions both as a co-receptor for FGF23 signalling and a sialidase implicate it as a molecule of critical importance in proper phosphate and vitamin D homoeostasis in the kidney and parathyroid [21]. Absence of KL from the kidney and parathyroid occur with chronic kidney disease progression and can result in development of secondary disorders [22–25]. The loss of transmembrane KL is reported in breast and cervical cancer and in lung cancer cell lines [11,12,26,27]. The magnitude of KL loss correlates with disease progression. Conversely, overexpression of the secreted alternative splice variant of KL correlates with progression and increased risk of death in ovarian cancer [28]. These results indicate that the tumour-suppressor activity of KL may depend on functions normally associated with transmembrane KL and that these functions are lost during disease progression. Although the loss of KL does not appear to be an initiating factor in disease development, loss of expression or alteration of normal expression does appear to coincide with disease progression/severity. As such, maintaining KL protein expression levels is anticipated to be beneficial and probably disease-modifying.
We determined previously that KL is decreased in the brain white matter of aged non-human primates, mice and rats [29]. Similar age-dependent decreases in protein expression occur in the heart and liver of rodents [30,31]. Although non-human primates do not develop neurodegenerative disorders, a subset does develop cognitive impairment, similar to human non-pathological brain aging. In humans, a polymorphism in KL is associated with decreased cognitive ability throughout life [32]. To study further the role of KL in normal physiology and in diseases, including neurodegenerative disorders, we sought to identify small-molecule modulators of KL expression. In order to accomplish this goal, an HTS (high-throughput screen) was developed using 1.8 kb of the KL promoter driving luciferase expression in a stably transfected HEK (human embryonic kidney)-293 cell line (KL5). This assay was then used to screen a compound library to identify molecules able to elevate KL transcription by >30%. After a series of validation steps, three compounds were identified that elevate both KL transcription and KL protein expression. All three compounds were also shown to enhance responsiveness to FGF23-mediated cell signalling, a KL-dependent process critical in homoeostasis. These compounds warrant further mechanistic evaluation in order to gain a better understanding of KL regulation. In addition, these compounds can also serve as starting points for further optimization in order to generate pharmacological tools to probe KL modulation in vivo.
EXPERIMENTAL
Cell culture
All cell lines were maintained under standard growth conditions and propagated in DMEM (Dulbecco’s modified Eagle’s medium) (4.5 g/ml glucose) containing 10% FBS (fetal bovine serum) (Atlanta Biologicals) and 1% penicillin/streptomycin (100 units/ml). Opossum kidney cells were a gift from Dr Steven Borkan (Boston University School of Medicine). Z310 rat choroid plexus cells were a gift from Dr Wei Zheng (Purdue University, West Lafayette, IN, U.S.A.) and were cultured in the presence of G418 (8 µg/ml). All cell culture solutions were obtained from Cellgro unless otherwise noted.
Stable cell line development
A 1.8 kb section of the human KL promoter was amplified using BD Advantage polymerase (Clontech) from human kidney genomic DNA (Clontech) using the primers 5′-GGGAAATGTGATACTCCATGTAGACGTAGC-3′ (forward) and 5′-GCTGCGCGGGAGCCAGGCTCCGGGGCCCCG-3′ (reverse). Owing to the high concentration of GC residues present in the promoter, addition of 1 M betaine (Sigma) was required in all PCRs. An expected, a 1.8 kb PCR product was produced and cloned into the pCR2.1 vector (Invitrogen). The promoter was subsequently subcloned into the pGL3 vector (Promega) using XhoI and HindIII restriction enzyme sites. The construct was confirmed by DNA sequencing. Stable cell lines were generated by co-transfection of the promoter construct and pcDNA3.1 (for G418 resistance) into HEK-293 cells with Lipofectamine™ 2000 (Invitrogen). Stable integrants were selected in G418-containing medium (800 µg/ml; Cellgro). Isolated colonies were evaluated for KL promoter-mediated luciferase expression according to the manufacturer’s instructions using the luciferase assay system (Promega). Further confirmation of construct integration into the genome was performed by PCR amplification of DNA isolated from HEK-293 or stable HEK-293KL lines using two primer pairs, the pair detailed above (to detect endogenous and the stably integrated KL promoter) and the forward primer described above paired with the GLprimer2 reverse primer (to detect only stably integrated KL promoter driving luciferase) (Promega). The fifth stable line (KL5) was selected for use in the HTS.
The Egr-1 construct was a gift from Kathrin Kirsch (Boston University School of Medicine). Transfection of the transcription factor or control plasmid into the KL5 cell line was conducted according to the manufacturer’s instructions using Nanofect (Qiagen). Luciferase output was evaluated 48 h post-transfection using the Luciferase Assay System (Promega). Cell proliferation was also measured in parallel transfections via MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] assay according to the manufacturer’s instructions (Promega).
The KL5 stable cells were plated and, 24 h later, fresh medium containing compounds reported previously to alter KL mRNA or protein in cells were added at the following concentrations 0.05–1 µM Lipitor (Pfizer) [33,34], 0.05–1 µM vitamin D (Sigma) [3,35], 5–50 µM gentian root (Nature’s Answer) [36] or 100–500 µM troglitazone (Sigma) [37]. At 24 h after the addition of compounds, luciferase expression was measured as described above (Promega).
Compound library
The compound library consisting of approximately 150000 small molecules included compounds approved by the U.S. Food and Drug Administration. In addition, the compound library also contained purified natural products, and synthetic compound collections from Peakdale, Maybridge, Cerep, Bionet Research, Prestwick, Specs and Biospecs, ENAMINE, I.F. Lab, Chemical Diversity Labs, ChemBridge and from various academic institutions around the world. Compounds in the library are organized by number starting with the abbreviation LDN (Laboratory for Drug Discovery in Neurodegeneration number).
High-throughput compound screen for KL transcriptional activators
In order to utilize the 384-well format required for HTS, our screening platform (KL5 stable cell line) was miniaturized. Parameters tested for optimization include cell concentration, medium formulation, plate colour, detection reagent volume and time, and type of plate reader. We optimized conditions to minimize well-to-well and plate-to-plate variability in luciferase signal. For screening, 5000 KL5 cells per well were plated in complete medium without Phenol Red on to 384-well white plates (PerkinElmer) using a Multidrop 384 (Thermo Fisher Scientific). Cells were allowed to attach to the plate for 4 h before addition of compound to the medium overlaying the cells using a Biomek® NX system (Beckman Coulter). Compound source plates for the assay were prepared by spotting 0.4 µl of 1.67 mM compound in DMSO in each well of a 384-well plate. For all plates, columns 23 and 24 were spotted with DMSO as controls. Plates were sealed and stored at 4°C until the day of screening. Source plates were then brought to room temperature (22°C) and diluted in 60.4 µl of Phenol Red-free complete medium for an intermediate stock of 11 µM and 0.66% DMSO. Cells plated alone and treated with DMSO alone served as our baseline control (column 23). Although we are unaware of a compound that will highly activate KL transcription, a compound (LDN-22721, N-phenyl-1H-indole-2-carboxamide) identified during our validation studies was used as a positive control and evidence of successful activation on all plates (column 24). A total of 5 µl of compound or control was transferred from the intermediate plate to medium overlaying cells resulting in a final concentration of 1 µM compound and 0.06% DMSO. Cells were exposed to the compound overnight. Promoter activation was detected by the addition of SteadyLite to medium overlaying cells (PerkinElmer). Plates were read on an Envision plate reader (PerkinElmer) within 10 min of reagent addition.
Approximately 50 compound plates were tested each day of the HTS. Analysis of the quality of daily data was performed by determining the percentage CV (coefficient of variation) of wells containing cells treated with DMSO and wells containing cells treated with LDN-22721 controls. Any plate with a CV higher than 10% for either group was rescreened. Following screening/rescreening, hits were identified as any compound inducing greater than 30% activation above baseline (cells alone). A total of 414 compounds or ~0.27% of all compounds met or exceeded this threshold. Compounds identified as hits were selected for five-point dose–response curve retesting (2–0.02 µM). Hits were prepared and diluted into medium overlaying cells in a 384-well format in a Biomek® FX system (Beckman Coulter) for confirmation of activity.
Some 38 compounds that re-hit, produced dose-dependent activation and were deemed chemically tractable were ordered fresh from the original sources. Twelve-point dose–response assays were performed using similar methods to the full HTS. Compounds were serially diluted in DMSO and added into medium overlaying the cells. All compounds were tested from 30 µM to 0.0001 µM for both KL promoter activation and for general toxicity. Toxicity was measured according to the manufacturer’s instructions using Cell Titer Glo (Promega).
qPCR (quantitative PCR)
Z310 cells were treated with compounds G, H and I (3 µM) for 16 h. Total RNA was isolated using Qiagen’s RNeasy kit. The first strand cDNA was made with 2 µg of total RNA using Invitrogen SuperScript III reverse transcriptase. The qPCR experiments were performed with IQ SYBR Green (Bio-Rad Laboratories) detection on a Bio-Rad C1000 Thermal Cycler. Triplicates of 20 µl reaction volumes containing primers 5′-TCCATCTGGGACACTTTCACCCAT-3′ (forward) and 5′-TGTGTCGCGGTAGACGTTGTTGTA-3′ (reverse) and cDNA [100 ng for KL and 50 ng for housekeeping genes Hprt1 (hypoxanthine phosphoribosyltransferase 1) and Rpl13A (ribosomal protein L13A)] template were used for quantification. qPCR was carried out as follows: 1 cycle of 95°C for 10 min followed by 40 cycles of 95°C for 30 s, 55°C for 1 min and 72°C for 1 min. This was followed by a dissociation curve beginning at 55°C and increasing by 0.2 °C every 3 s, with SYBR Green fluorescence measured at every interval. Relative quantification of the difference between the control and drug-treated samples was done using RT2 Profiler PCR Array Data Analysis Template from Qiagen. KL gene expression was tested for statistical significance (P < 0.05) by ANOVA.
Western blotting
Validation of compound effects on endogenous protein was performed by incubating the top ten compounds with OK (opossum kidney) or Z310 (rat choroid plexus) cell lines. Compounds were diluted into fresh medium overlaying the cells at either 1 or 3 µM and left for ~18 h. Cell lysates were harvested in RIPA buffer [150 mM NaCl, 50 mM Tris/HCl (pH 7.5), 1% Triton X-100, 0.5% deoxycholic acid and 0.1% SDS, with fresh protease inhibitors added daily (Roche)]. BCA (bicinchoninic acid) protein assay was used to determine protein concentration and equal protein was loaded on to 10% Tris/glycine polyacrylamide gels. Proteins were transferred on to nitrocellulose (Millipore) for Western blotting. Nitrocellulose was blocked in 5% (w/v) non-fat dried skimmed milk powder before overnight incubation in primary antibody [in BSA/TBST [Tris-buffered saline with Tween 20 (50 mM Tris/HCl, pH 8, 150 mM NaCl and 0.05% Tween 20)] at room temperature. KL was detected using KM2076, a rat monoclonal antibody specific to the KL1 domain of KL (kindly provided by the Antibody Research Laboratories, Kyowa Hakko Kirin, Tokyo, Japan). Anti-β-tubulin or anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (Santa Cruz Biotechnology) antibodies were used to verify equal loading of protein. All washes were conducted in TBST. Relevant secondary antibodies were obtained from KPL. Antibody detection was accomplished using Immobilon (Millipore) or Super Signal Pico West Chemiluminescent (Pierce) reagents. Quantification of all bands was performed using ImageJ software (NIH).
FGF23 functional assay
FGFR1C plasmid was a gift from Dr Makoto Kuro-o (University of Texas Southwestern Medical School, Dallas, TX, U.S.A.). The KL plasmid was generated by our laboratory as described previously [38]. FGF23 and bFGF (basic fibroblast growth factor) were obtained from R&D Systems. Cells were transfected using Nanofect (Qiagen) according to the manufacturer’s instructions. After 24 h, compounds were added to medium overlaying the cells. After a further 24 h, cells were incubated in serum-free medium for 2 h and then medium was changed to fresh serum-free medium, containing 10 ng/ml FGF23 or 100 ng/ml bFGF. Cells were incubated for 20 min at 37°C. For experiments involving transfection of KL or control siRNA (small interfering RNA) (Santa Cruz Biotechnology) (pool of three to five target-specific siRNAs), cells were co-transfected with HiPerFect (Qiagen) according to the manufacturer’s instructions. siRNA effects were tested by co-transfection with a KL pcDNA3.1 plasmid into HEK-293 cells using HiPerFect according to the manufacturer’s instructions. Cells were lysed in RIPA buffer and processed for Western blotting as above. Antibodies against pERK [phospho-ERK (extracellular-signal-regulated kinase)] and total ERK were obtained from Cell Signaling Technology. Quantification of blots was performed using ImageJ software.
Statistics
Statistical significance was calculated using Graphpad Prism software version 5.0. Student’s t test or ANOVA with Dunnett’s post-test was used where indicated to determine whether the minimum threshold of 0.05 had been achieved between groups. For the HTS, Z′ calculations rely on both a maximum and minimum signal. Even though we identified an active compound that increased KL expression early in the HTS that we subsequently used as a positive control, the increase in signal with the positive control does not represent a maximum increase and therefore could not be used as the maximum for Z′ calculations. Instead, we relied on percentage CV of the basal luciferase activity as a reliable measure for assay robustness and reproducibility. The percentage CV was calculated as (S.D./mean) × 100. In addition, reproducibility of the positive control across a group of plates and assay days as measured by the mean and S.D. of the signal is included in the results.
RESULTS
Development and validation of a KL HTS platform
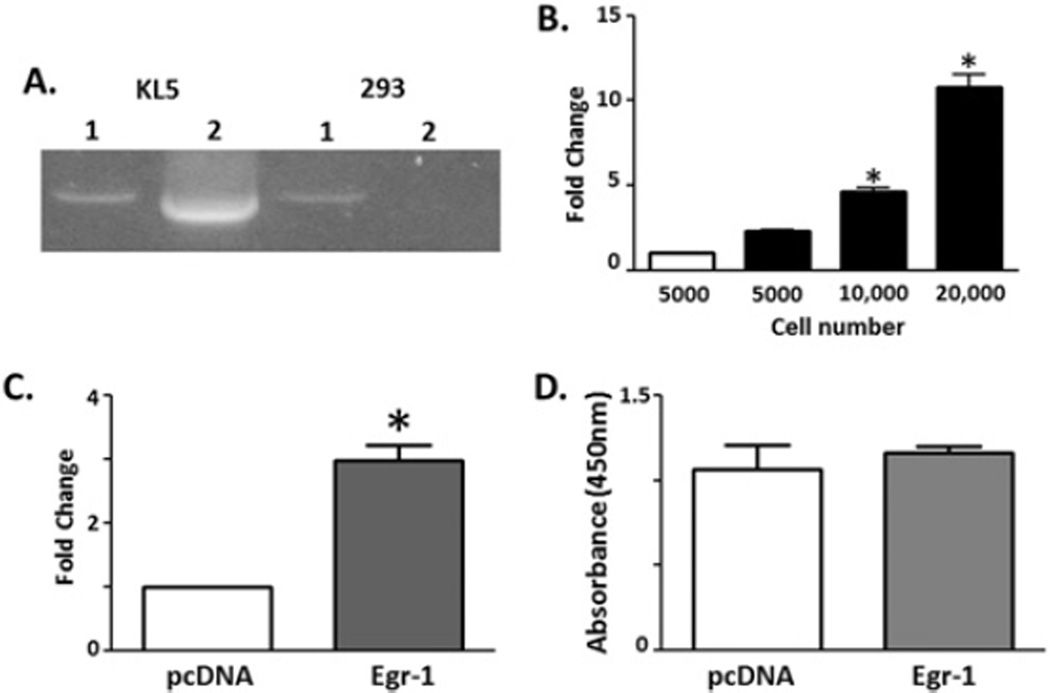
We cloned the 1.8 kb of DNA immediately upstream of the KL translation start site into the pGL3 luciferase reporter vector. Stable cell lines were then generated by co-transfection of the KL reporter with pcDNA3.1 (providing G418 resistance) into HEK-293 cells. Individual colonies were expanded and tested for stable integration of the KL promoter reporter construct and colony 5 (KL5) was selected for further studies. PCR primers used to clone the construct detect the KL promoter and will amplify both the endogenous KL promoter and the promoter associated with the reporter construct (Figure 1A, PCR product 1), while primer set 2 using the GL2 reverse primer located in the luciferase gene will only detect the promoter when associated with the reporter construct (Figure 1A, PCR product 2). Similarly, KL5 cells were plated in increasing concentrations and tested for luciferase gene product to determine whether HEK-293 cells could endogenously activate the KL promoter reporter construct. HEK-293 cells alone (Figure 1B, white bar) do not produce luciferase; however, KL5 cells showed luciferase expression in proportion to their cell concentration (Figure 1B).
Figure 1. HTS platform development.
Stable HEK-293 cells expressing the KL promoter construct driving reporter gene (firefly luciferase) expression were generated. (A) Candidate stable cell line 5 (KL5) and the parental HEK-293 cell line were evaluated for integration of the construct by PCR using different reverse primers generated from within the KL promoter (1) or within the luciferase gene (2). (B) The KL5 line was screened for baseline luciferase gene expression by plating increasing numbers of cells and detecting luminescence (*P < 0.05, ANOVA, compared with 5000 KL5 cells, four independent wells; results are means ± S.E.M.). (C) Egr-1 was transfected into the KL5 cell line to determine whether the signal could be modulated. Reporter gene activity was compared with and without Egr-1 transfection (*P < 0.05, ANOVA, n = 8; results are means ± S.E.M.). (D) Cell proliferation was measured using the MTS assay. Cell proliferation was not different between mock and Egr-1-transfected cells (ANOVA, n = 8; results are means ± S.E.M.).
Previous papers have reported compounds that are able to alter the amount of KL mRNA or protein in cells. However, incubation of KL5 cells in the medium containing Lipitor [33,34], active vitamin D [3,35], gentian root extract [36] or troglitazone [37] did not increase the KL reporter expression (see Supplementary Figure S1 at http://www.BiochemJ.org/bj/441/bj4410453add.htm). As such, compounds identified previously either must stabilize mRNA/protein post-transcriptionally or else require promoter elements not present in our 1.8 kb KL promoter construct for transcriptional effects. KL is activated by the Egr-1 transcription factor [39]. To determine whether increased luciferase expression could be induced by activation of the KL promoter, Egr-1 transcription factor or pcDNA3.1 (control) plasmids were transfected into KL5 cells. At 48 h after transfection a ~2-fold increase in luciferase was detected (Figure 1C). This increase could not be accounted for by increased cell proliferation (Figure 1D, MTS assay).
KL high-throughput screen
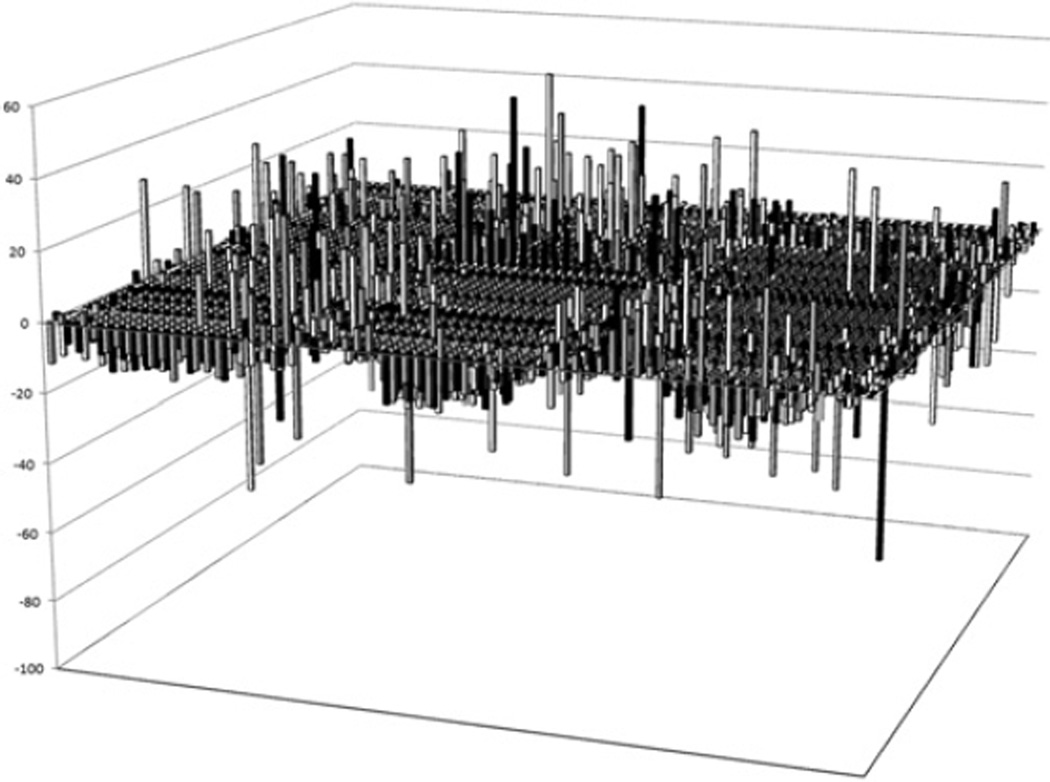
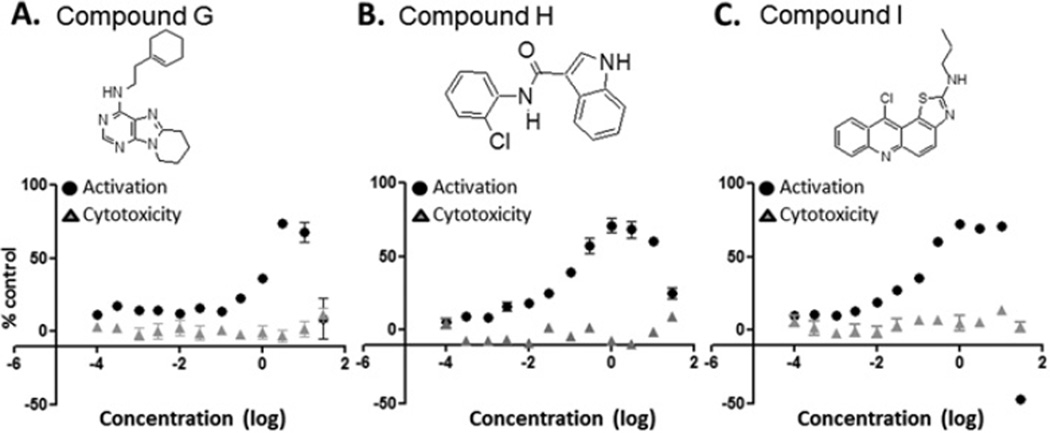
The assay was miniaturized to a 384-well format and an initial series of compound plates (Prestwick collection) were tested to determine the number of hits anticipated in the HTS. During this phase of assay validation, a compound, LDN-22721, was identified as being able to elevate luciferase expression by ~30%. LDN-22721 was subsequently included on all plates as a positive control. The performance of the compound was very reproducible across the HTS. Over a representative 20 plates, DMSO control wells, which served as the background, produced an average signal of 190693 ± 7201 light units (mean ± S.D.) with CV = 3.8%. For the LDN-22721 control, the average signal was 275856 ± 8799 light units (mean ± S.D.) with CV = 3.2%. Although initial testing identified LDN-22721, the HTS produced more appealing compounds during the validation phases and therefore LDN-22721 was used only as a positive control for the HTS. Approximately 150000 compounds were tested at a concentration of 1 µM for the ability to increase luciferase expression as a marker of KL promoter activation. The statistics for the control wells were examined on each plate daily and any plate with a CV higher than 10% was rescreened; as such, four plates were rescreened. After the screen, hits were identified as any compound causing at least a 30% increase in luciferase expression. Some 414 compounds or 0.27% of all compounds met this threshold of activation. A representative sampling of 4416 compounds are shown (Figure 2). All 414 compounds were retested at five concentrations (2–0.02 µM) to validate the initial identification as a hit and to obtain an initial indication of dose-dependent response. A total of 200 compounds were again identified as hits. These compounds were prioritized on the basis of dose-response and chemical tractability (i.e. lack of reactive functional groups, molecular mass < 500 Da, cLogP < 5, synthetic feasibility, etc.). Compounds that had been identified in other luciferase-based HTS campaigns conducted at the Laboratory for Drug Discovery in Neurodegeneration were removed from further consideration. Of 50 compounds prioritized for further characterization, 38 were reordered from original sources. Fresh stocks were utilized to ensure that the initial hit was not a side product of freeze–thaw breakdown or some other contaminating by-product specific to an individual lot. Fresh stocks were applied to the HTS platform at 12 concentrations (30–0.0001 µM). Luciferase expression and cell cytotoxicity assays using Cell Titer Glo were performed in parallel to examine both activation and toxicity. None of the compounds tested showed evidence of toxicity at any of the concentrations examined. The Cell Titer Glo assay is another luciferase-based assay and therefore also should have eliminated any compounds that worked directly on luciferase. Of 38 compounds tested in 12-point dose–response assays, 30 revealed dose-dependent profiles. These were prioritized on the basis of dose-response and compound structure for additional validation. The structure, dose-response and toxicity profile of the top three compounds are shown in Figure 3 (Figure 3A: Compound G, N-[2-(1-cyclohexen-1-yl)ethyl]-6,7,8,9-tetrahydropyrido[1,2-e]purin-4-amine; Figure 3B: Compound H, N-(2-chlorophenyl)-1H-indole-3-carboxamide; Figure 3C: Compound I, 2-(1-propyl)amino-11-chlorothiazolo[5,4-a]acridine). Table 1 shows the primary HTS percentage activation above baseline at 1 µM and 12-point dose-response EC50 values for the top nine compounds (A–I). Most compounds show an activation of 30–40% and a sub-micromolar EC50.
Figure 2. High-throughput screen.
Following optimization and miniaturization, ~150 000 compounds were tested for the ability to increase luciferase signal. Results are percentage activation above DMSO-treated KL5 cells (baseline) for a representative 4416 compounds from the primary HTS.
Figure 3. Compound structure, activation and toxicity.
Three compounds identified in the HTS were validated further and found to be active across a range of concentrations and not show evidence of overt toxicity. (A) Compound G, N-[2-(1-cyclohexen-1-yl)ethyl]-6,7,8,9-tetrahydropyrido[1,2-e]purin-4-amine. (B) Compound H, N-(2-chlorophenyl)-1H-indole-3-carboxamide. (C) Compound I, 2-(1-propyl)amino-11-chlorothiazolo[5,4-a]acridine. Activation (black circles) and cytotoxicity (grey triangles) are reported as the mean ± S.E.M. percentage of the relevant DMSO control for two independent experiments with each point being run in quadruplicate in each assay.
Table 1.
Candidate compound activation and EC50 values
| Compound | Activation (%) | EC50 (µM) |
|---|---|---|
| A | 42 | 0.29 |
| B | 31 | 1.97 |
| C | 32 | 0.75 |
| D | 33 | 0.31 |
| E | 35 | 6.14 |
| F | 31 | 3.01 |
| G | 50 | 50.17 |
| H | 32 | 0.05 |
| I | 30 | 0.02 |
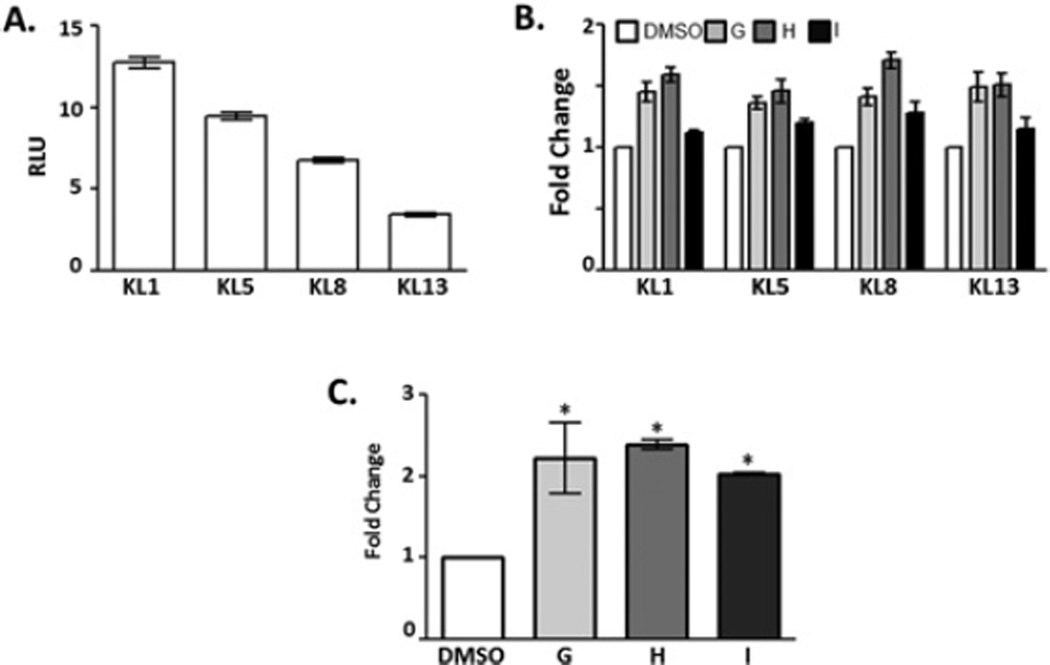
The KL5 cell line was one of several lines of HEK-293 cell determined to stably express the KL promoter driving luciferase expression. To determine whether the effects of compounds G–I were specific to the single stable clone used in the HTS, three additional lines stably expressing higher (KL1) and lower (KL8 and KL13) levels of luciferase were evaluated (Figure 4A). When compounds G–I were applied to any of four stable cell lines, the fold change in luciferase expression was consistent (Figure 4B). These results demonstrate that luciferase expression is the result of compounds G–I influencing the KL promoter and are not a result of integration site-specific off-target effects. Activation of the reporter construct, although specific, does not examine whether compounds will affect the actual level of KL mRNA and protein in a cell. DMSO or compounds G–I were applied to the medium overlaying Z310 rat choroid plexus cells and the relative expression of KL mRNA determined by qPCR. All three compounds induced a significant increase in KL mRNA relative to DMSO-treated cells (Figure 4C).
Figure 4. Compound effects across stable lines and on mRNA.
(A) Multiple cell lines were generated by transfection and stable selection of the KL promoter/luciferase construct into the genome of HEK-293 cells. KL5 cells were selected for use in the HTS because of their intermediate expression of luciferase. Additional stable lines were evaluated for luciferase expression. The KL1 line expresses higher levels of luciferase, whereas KL8 and KL13 lines express lower levels of luciferase. (B) In concurrent experiments, the ability of compounds G–I was evaluated in each independent stable cell line. A concentration of 1 µM DMSO or compound G (light grey), H (dark grey) and I (black) was applied to the medium overlaying the selected stable cell lines. Results are the mean ± S.E.M. fold change relative to DMSO-treated cells of the relevant cell line for three independent experiments with each sample run in triplicate. (C) Compound effects on KL mRNA were validated in Z310 cells. Z310 cells were incubated in DMSO or 3 µM compound G–I, mRNA was isolated, and the relative expression level was determined by qPCR. Results are relative levels of expression (2−ΔΔCt) as fold change compared with DMSO-treated control (*P < 0.03, ANOVA, n = 2).
Compound effects on endogenous KL protein
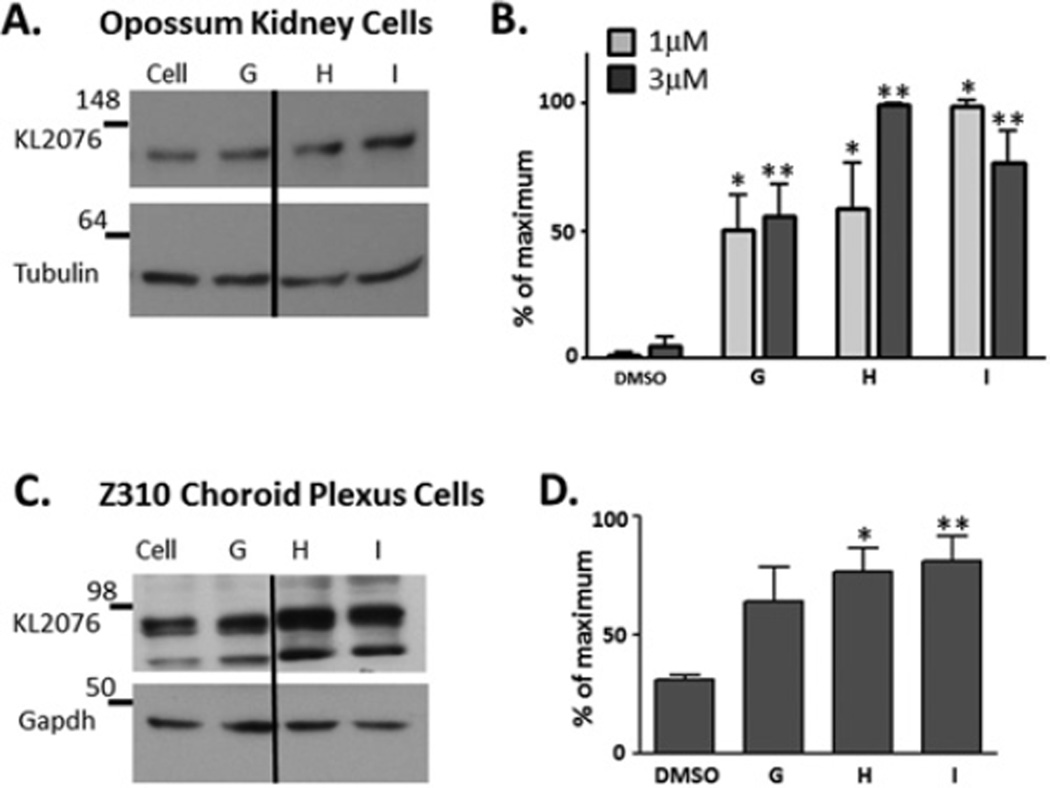
As protein levels are the ultimate target for increasing transcriptional activation, we next determined whether compounds G–I modulated KL protein expression in cells endogenously expressing KL protein at high levels. We utilized two types of cell lines, an opossum kidney cell line (OK) and a rat choroid plexus (Z310) cell line, as these two tissues express the highest levels of KL in mammals. Compounds G–I were tested on OK cells at 1 and 3 µM (Figures 5A and 5B), and all compounds significantly increased KL protein at both concentrations utilized (Figures 5A and 5B). Z310 cells were incubated in the presence of compounds G–I at 1 and 3 µM (Figures 5C and 5D). Z310 cells did not show evidence of increased protein expression at 1 µM, but did at 3 µM (Figures 5C and 5D). Results for both cell lines were normalized to loading control protein.
Figure 5. Effects of compounds G–I on endogenous KL protein.
Compound effects on the KL protein were validated in two cell lines that endogenously express high levels of KL protein. (A and B) OK cells were treated with 1 µM (grey bars) or 3 µM (black bars) compound G–I. At 24 h after compound addition, cell lysates were subjected to Western blotting and semi-quantitative densitometry analysis. Each sample was normalized to β-tubulin as a control. Results are mean ± S.E.M. percentages of the maximum activation compared with the DMSO control. Each sample was independently repeated in three to five experiments (*P < 0.001, ANOVA, compared with 1 µM DMSO control, **P < 0.001, ANOVA, compared with 3 µM DMSO control). Dividing lines indicate where the image was spliced together; all samples were run on the same gel. (C and D) A similar analysis was conducted, using rat choroid plexus cells (Z310), but 3 µM, not 1 µM, was required to increase protein expression in Z310 cells. Z310 cells did not express β-tubulin and so GAPDH was used to normalize for protein expression. Each sample was repeated in five independent experiments and results are means ± S.E.M. (*P <0.01, **P < 0.001, ANOVA). Dividing lines indicate where the image was spliced together; all samples were run on the same gel. Molecular masses are indicated in kDa.
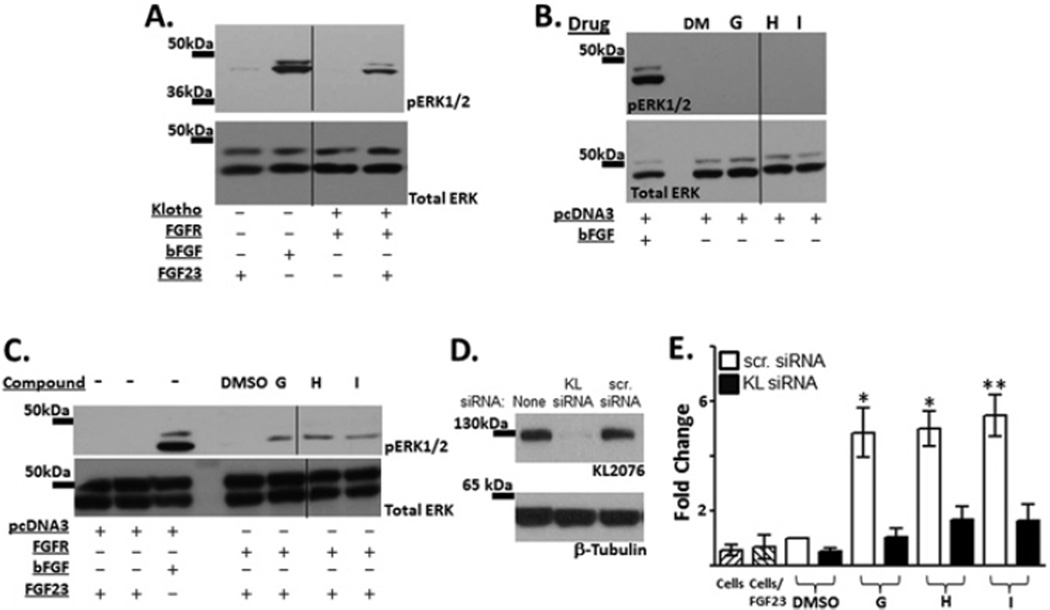
Increasing KL expression should induce functional consequences for cellular processes requiring KL protein. One of the most well defined pathways of KL action is its role as a co-receptor with FGFR (fibroblast growth factor receptor) in transducing FGF23 signalling. If the compounds elevate KL expression, increased FGF23 signalling should be observed. We verified previous reports that upon transfection with KL and FGFR1 plasmid constructs, FGF23 signalling increased in HEK-293 cells (Figure 6A). In all experiments, bFGF was used to verify that ERK phosphorylation occurs in the HEK-293 cells in response to an outside stimulus.
Figure 6. FGF23 functional assay.
KL and FGFR function as co-receptors for FGF23 signalling. (A) Assay validation. bFGF stimulates ERK phosphorylation, indicating that cells express ERK and are able to transduce a signal from the cell surface. Upon transfection of KL and FGFR into HEK-293 cells, FGF23 signalling induces ERK phosphorylation. Shown is a representative blot of pERK and total ERK. (B) HEK-293 cells transfected with control plasmid were treated with compounds G–I for 24 h and processed similarly to the FGF23 assay, although no protein stimulus was added to the medium. Although phosphorylation was detected in bFGF controls, no pERK was detected as a result of compound stimulation alone. (C) HEK-293 cells were transfected with FGFR1c or control plasmid pcDNA3.1. Following transfection, KL-activating compounds G–I were added to the medium overlaying cells (1 µM) for 24 h. FGF23 was subsequently added to the medium for 20 min to determine whether increased KL protein would result in elevated pERK signalling. Increases in pERK were observed in all of the compound-treated wells. Shown is a blot representative of three independent experiments. (D) HEK-293 cells were transfected with a plasmid expressing KL alone or in combination with KL siRNA or a scrambled control siRNA. Although no effects were observed on β-tubulin control proteins, KL siRNA reduced KL expression to nearly undetectable levels. (E) HEK-293 cells were co-transfected with FGFR and either KL or scrambled control siRNA. Cells were then exposed to compound and processed for FGF23 signalling as above. Fold change of pERK expression was normalized to total ERK in the cell and quantified across multiple experiments. All three compounds tested showed elevated levels of pERK (white bars, G–I) unless transfected with KL siRNA which blocked FGF23 signalling (black bars, G–I). Hatched bars represent cells alone or cells treated with FGF23 as controls for background levels of pERK in HEK-293 cells. Each compound was tested in three independent experiments and results are means ± S.E.M. (*P < 0.01, **P < 0.001, ANOVA). Dividing lines indicate where the image was spliced together; all samples were run on the same gel. Molecular masses are indicated in kDa.
Cells were transfected with control plasmid before exposure to compounds G–I to determine whether the compounds alone would elevate pERK (Figure 6B). Although the cells were responsive to outside stimulus (bFGF), exposure to compounds G–I did not induce an increase in pERK signalling. Next we transfected HEK-293 cells with FGFR, the co-receptor with KL for FGF23 signalling, before compound exposure. Cells were then stimulated by addition of FGF23-containing medium, and pERK levels were evaluated compared with DMSO controls. Compounds G–I increased FGF23 signalling (Figure 6C).
Upon co-transfection with a KL-expressing plasmid, KL siRNA reduced KL expression, whereas control siRNA did not (Figure 6D). To determine whether the effect of compounds G–I was specific to KL, we co-transfected KL siRNA or control siRNA with the FGFR, exposed cells first to compounds and then to FGF23 (Figure 6E). Whereas control siRNA had no effect on increased FGF23 signalling induced by compounds G–I (Figure 6E, white bars), the presence of KL siRNA blocked FGF23 signalling (Figure 6E, black bars). These results indicate that the compounds elevate KL protein expression in the cells to a functionally relevant level and that the effects are induced via KL.
DISCUSSION
KL protein decreases with age and during progression of age-related diseases [25–27,29,30]. We have identified three compounds that increase KL expression, resulting in increased protein in kidney (OK) and increased mRNA and protein in choroid plexus (Z310) cell lines. This elevation in KL protein has functional consequences for the cell as is evident by increased responsiveness to FGF23 stimulation resulting in FGFR-dependent ERK phosphorylation. Whereas both kidney and choroid plexus cells up-regulated KL protein as detected by Western blotting, kidney cells appeared to be more sensitive and responded at a lower compound concentration than choroid plexus cells. It is possible that choroid plexus, which has not been studied as intensely as kidney, could regulate KL in a different way for unique and presently unappreciated functions in brain.
Our HTS utilized 1.8 kb of the KL promoter driving luciferase expression and, as such, our assumption is that compounds identified in the HTS activate transcription by interacting with the KL promoter or influencing transcription factors involved in transcriptional activation or repression. qPCR showing increased KL mRNA are consistent with this idea, however, we cannot rule out post-transcriptional and post-translational effects that could stabilize the protein and be unrelated to a direct effect on transcription also contributing. As such, further optimization of compound structure and studies into the exact mechanism of each compound are necessary to determine whether compounds G–I are potential novel therapeutics.
Our attempts to determine whether compounds reported previously to elevate KL protein levels in the cell would do so via transcriptional activation in our cell line indicate that these compounds act through alternative mechanisms [3,33–37]. This was not entirely unexpected since most of the compounds were not tested previously in transcription assays and could have other mechanisms of action. However, gentian root extract was identified as an activator of KL transcription in an HTS similar to ours using the KL promoter and a panel of traditional Chinese medicines [36]. Although gentian root in the previous study was extracted in the laboratory [36], our preparation was from a commercial source. Nonetheless, this previous study highlights the possibility that there may be natural compounds that result in KL elevation and it would be interesting to test whether natural dietary supplements that claim to promote healthy aging have an effect on KL transcription. Use of the oral sorbent AST-120 increased expression of KL in kidney of rats with chronic renal failure [40]. Although the mechanism of AST-120 action is not directly on KL transcription, the downstream effect of its use appears to promote increased transcription. AST-120 reduces toxic levels of indoxyl sulfate, lowering the oxidative stress load. These results indicate that loss of KL correlates with disease progression and it will be important to determine whether elevating KL is disease-modifying.
Three common KL polymorphisms repeatedly show disease association in genetic studies. The G395A KL polymorphism is located in the KL promoter. This polymorphism is found across ethnic groups and its status has been associated with increased risk of hypertension [41], knee osteoarthritis [42] and hand osteoarthritis [43] in women, lower bone mineral density in women [44], cardioembolic stroke [45] and potentially an increased risk of coronary artery disease, although reports conflict regarding this latter condition [46–50]. In healthy adults, G395A was associated with high fasting glucose [49] and high blood pressure [48]. Decreased protein interaction with DNA is observed in EMSAs (electrophoretic mobility-shift assays) using the G395A polymorphism; however, the exact transcription factor or factors presumed to cause a decrease in KL transcription are presently not known [44]. Depending on the mechanism by which compounds G–I elevate KL transcription, differential effects in G395A carriers may be observed.
Decreased levels of KL protein have been observed in aged non-human primate brains during the time of life when cognitive impairment develops [29]. Also, a recent study found that methylation of the KL promoter resulted in decreased KL protein in the most invasive cervical carcinoma consistent with its inactivation during disease progression [26]. If decreased KL protein with age and in disease is the result of epigenetic silencing of KL, use of compounds G–I to increase transcription alone may not be sufficient for disease modification. Particularly in applications for cancer, the contribution of epigenetic modifications will be critical to assess. Knowledge of the exact mechanism of action as well as investigation of the KL promoter methylation status in target populations will potentially be required for effective therapeutic use of these compounds or optimized analogues.
Decreased expression of KL with age and during disease indicates that loss of KL contributes to an environment where disease progression can occur, worsening the primary deficit and spawning secondary conditions. The further development of compounds identified in the present study, which elevate KL transcription, seems warranted in order to determine whether increasing KL protein alone can be disease-modifying. The multiple dysfunctions noted in the KL-knockout mice and subsequent studies that have analysed individual organ systems indicate that KL is a critical protein in homoeostatic functions and a protective protein suppressing insulin signalling and enhancing antioxidative defences. Modulation and mechanistic evaluation of these compounds could unveil a novel therapeutic target for numerous human disorders.
The identification of three compounds with functionally validated effects on KL protein expression will require further intensive evaluation and optimization before testing in animal models of disease is warranted. Since the HTS was based on a 1.8 kb stretch of the KL promoter driving a reporter gene, the exact target and mechanism of the compounds is currently unknown. Compounds could interact with the DNA, but, more likely, they could affect the ability of transcription factors to interact with or activate the KL promoter. Although changes in the expression of KL were not accompanied by changes in the control proteins, it may be expected that these compounds would modulate other proteins, and may have off-target effects in vivo. Optimization to KL-specific activation and determination of the exact molecular target of each compound is required to understand the mechanism of activation and to evaluate further the potential use of this therapeutic strategy. Work is ongoing to examine whether chemical modifications of compounds G–I will result in improved KL activation. Optimized compounds will then be used to define the mechanism of action for each compound of interest.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ross Stein for his insightful discussions and leadership at the Laboratory for Drug Discovery in Neurodegeneration at the initial stages of this work.
FUNDING
This work was funded by the National Institutes of Health (NIH)/National Institute on Aging (NIA) [grant numbers P01 AG-00001 and ADDF/ISOA (to C.R.A.), and 5T32AG000115-24 and K99/R00 AG034989-01 (to G.D.K.)]. M.M.H., E.R.S., G.D.C. and M.A.G. thank the Harvard NeuroDiscovery Center for financial support.
Abbreviations used
- bFGF
basic fibroblast growth factor
- CV
coefficient of variation
- ERK
extracellular-signal-regulated kinase
- FGF23
fibroblast growth factor 23
- FGFR
fibroblast growth factor receptor
- FOXO
forkhead box O
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HEK
human embryonic kidney
- HTS
high-throughput screen
- KL
klotho
- MTS
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
- pERK
phospho-ERK
- qPCR
quantitative PCR
- siRNA
small interfering RNA
- TBST
Tris-buffered saline with Tween 20
Footnotes
AUTHOR CONTRIBUTION
Gwendalyn King generated the cell line, conducted the HTS and validation, FGF23 assay, Western blotting experiments, and wrote and edited the paper before submission. CiDi Chen cloned the KL promoter and conducted qPCR experiments. Mickey Huang participated in the HTS and validation. Ella Zeldich conducted FGF23 assays. Patricia Brazee conducted qPCR experiments. Eli Schuman optimized robotics and assay design for the HTS. Maxime Robin provided a previously unpublished compound. Gregory Cuny conducted compound assessment, oversaw the HTS and validation and edited the paper before submission. Marcie Glicksman oversaw all aspects of the HTS and validation and edited the paper before submission. Carmela Abraham oversaw all aspects of the project from conception and edited the paper before submission.
REFERENCES
- 1.Chen CD, Podvin S, Gillespie E, Leeman SE, Abraham CR. Insulin stimulates the cleavage and release of the extracellular domain of Klotho by ADAM10 and ADAM17. Proc. Natl. Acad. Sci. U.S.A. 2007;104:19796–19801. doi: 10.1073/pnas.0709805104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuro-o M. Klotho and aging. Biochim. Biophys. Acta. 2009;1790:1049–1058. doi: 10.1016/j.bbagen.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morishita K, Shirai A, Kubota M, Katakura Y, Nabeshima Y, Takeshige K, Kamiya T. The progression of aging in klotho mutant mice can be modified by dietary phosphorus and zinc. J. Nutr. 2001;131:3182–3188. doi: 10.1093/jn/131.12.3182. [DOI] [PubMed] [Google Scholar]
- 4.Cha SK, Ortega B, Kurosu H, Rosenblatt KP, Kuro OM, Huang CL. Removal of sialic acid involving Klotho causes cell-surface retention of TRPV5 channel via binding to galectin-1. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9805–9810. doi: 10.1073/pnas.0803223105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cha SK, Hu MC, Kurosu H, Kuro-o M, Moe O, Huang CL. Regulation of renal outer medullary potassium channel and renal K+ excretion by Klotho. Mol. Pharmacol. 2009;76:38–46. doi: 10.1124/mol.109.055780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Utsugi T, Ohno T, Ohyama Y, Uchiyama T, Saito Y, Matsumura Y, Aizawa H, Itoh H, Kurabayashi M, Kawazu S, et al. Decreased insulin production and increased insulin sensitivity in the klotho mutant mouse, a novel animal model for human aging. Metab. Clin. Exp. 2000;49:1118–1123. doi: 10.1053/meta.2000.8606. [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto M, Clark JD, Pastor JV, Gurnani P, Nandi A, Kurosu H, Miyoshi M, Ogawa Y, Castrillon DH, Rosenblatt KP, Kuro-o M. Regulation of oxidative stress by the anti-aging hormone klotho. J. Biol. Chem. 2005;280:38029–38034. doi: 10.1074/jbc.M509039200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu H, Fergusson MM, Castilho RM, Liu J, Cao L, Chen J, Malide D, Rovira II, Schimel D, Kuo CJ, et al. Augmented Wnt signaling in a mammalian model of accelerated aging. Science. 2007;317:803–806. doi: 10.1126/science.1143578. [DOI] [PubMed] [Google Scholar]
- 9.de Oliveira RM. Klotho RNAi induces premature senescence of human cells via a p53/p21 dependent pathway. FEBS Lett. 2006;580:5753–5758. doi: 10.1016/j.febslet.2006.09.036. [DOI] [PubMed] [Google Scholar]
- 10.Ikushima M, Rakugi H, Ishikawa K, Maekawa Y, Yamamoto K, Ohta J, Chihara Y, Kida I, Ogihara T. Anti-apoptotic and anti-senescence effects of Klotho on vascular endothelial cells. Biochem. Biophys. Res. Commun. 2006;339:827–832. doi: 10.1016/j.bbrc.2005.11.094. [DOI] [PubMed] [Google Scholar]
- 11.Chen B, Wang X, Zhao W, Wu J. Klotho inhibits growth and promotes apoptosis in human lung cancer cell line A549. J. Exp. Clin. Cancer Res. 2010;29:99. doi: 10.1186/1756-9966-29-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolf I, Levanon-Cohen S, Bose S, Ligumsky H, Sredni B, Kanety H, Kuro-o M, Karlan B, Kaufman B, Koeffler HP, Rubinek T. Klotho: a tumor suppressor and a modulator of the IGF-1 and FGF pathways in human breast cancer. Oncogene. 2008;27:7094–7105. doi: 10.1038/onc.2008.292. [DOI] [PubMed] [Google Scholar]
- 13.Nagai R, Saito Y, Ohyama Y, Aizawa H, Suga T, Nakamura T, Kurabayashi M, Kuro-o M. Endothelial dysfunction in the klotho mouse and downregulation of klotho gene expression in various animal models of vascular and metabolic diseases. Cell. Mol. Life Sci. 2000;57:738–746. doi: 10.1007/s000180050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saito Y, Nakamura T, Ohyama Y, Suzuki T, Iida A, Shiraki-Iida T, Kuro-o M, Nabeshima Y, Kurabayashi M, Nagai R. In vivo klotho gene delivery protects against endothelial dysfunction in multiple risk factor syndrome. Biochem. Biophys. Res. Commun. 2000;276:767–772. doi: 10.1006/bbrc.2000.3470. [DOI] [PubMed] [Google Scholar]
- 15.Saito Y, Yamagishi T, Nakamura T, Ohyama Y, Aizawa H, Suga T, Matsumura Y, Masuda H, Kurabayashi M, Kuro-o M, et al. Klotho protein protects against endothelial dysfunction. Biochem. Biophys. Res. Commun. 1998;248:324–329. doi: 10.1006/bbrc.1998.8943. [DOI] [PubMed] [Google Scholar]
- 16.Chateau MT, Araiz C, Descamps S, Galas S. Klotho interferes with a novel FGF-signalling pathway and insulin/Igf-like signalling to improve longevity and stress resistance in Caenorhabditis elegans. Aging. 2010;2:567–581. doi: 10.18632/aging.100195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuro-o M, Matsumura Y, Aizawa H, Kawaguchi H, Suga T, Utsugi T, Ohyama Y, Kurabayashi M, Kaname T, Kume E, et al. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature. 1997;390:45–51. doi: 10.1038/36285. [DOI] [PubMed] [Google Scholar]
- 18.Kurosu H, Yamamoto M, Clark JD, Pastor JV, Nandi A, Gurnani P, McGuinness OP, Chikuda H, Yamaguchi M, Kawaguchi H, et al. Suppression of aging in mice by the hormone Klotho. Science. 2005;309:1829–1833. doi: 10.1126/science.1112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ichikawa S, Imel EA, Kreiter ML, Yu X, Mackenzie DS, Sorenson AH, Goetz R, Mohammadi M, White KE, Econs MJ. A homozygous missense mutation in human KLOTHO causes severe tumoral calcinosis. J. Clin. Invest. 2007;117:2684–2691. doi: 10.1172/JCI31330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brownstein CA, Adler F, Nelson-Williams C, Iijima J, Li P, Imura A, Nabeshima Y, Reyes-Mugica M, Carpenter TO, Lifton RP. A translocation causing increased α-klotho level results in hypophosphatemic rickets and hyperparathyroidism. Proc. Natl. Acad. Sci. U.S.A. 2008;105:3455–3460. doi: 10.1073/pnas.0712361105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torres PU, Prie D, Molina-Bletry V, Beck L, Silve C, Friedlander G. Klotho: an antiaging protein involved in mineral and vitamin D metabolism. Kidney Int. 2007;71:730–737. doi: 10.1038/sj.ki.5002163. [DOI] [PubMed] [Google Scholar]
- 22.Komaba H, Koizumi M, Fukagawa M. Parathyroid resistance to FGF23 in kidney transplant recipients: back to the past or ahead to the future? Kidney Int. 2010;78:953–955. doi: 10.1038/ki.2010.283. [DOI] [PubMed] [Google Scholar]
- 23.Krajisnik T, Olauson H, Mirza MA, Hellman P, Akerstrom G, Westin G, Larsson TE, Bjorklund P. Parathyroid Klotho and FGF-receptor 1 expression decline with renal function in hyperparathyroid patients with chronic kidney disease and kidney transplant recipients. Kidney Int. 2010;78:1024–1032. doi: 10.1038/ki.2010.260. [DOI] [PubMed] [Google Scholar]
- 24.Bjorklund P, Krajisnik T, Akerstrom G, Westin G, Larsson TE. Type I membrane klotho expression is decreased and inversely correlated to serum calcium in primary hyperparathyroidism. J. Clin. Endocrinol. Metab. 2008;93:4152–4157. doi: 10.1210/jc.2008-0564. [DOI] [PubMed] [Google Scholar]
- 25.Koh N, Fujimori T, Nishiguchi S, Tamori A, Shiomi S, Nakatani T, Sugimura K, Kishimoto T, Kinoshita S, Kuroki T, Nabeshima Y. Severely reduced production of klotho in human chronic renal failure kidney. Biochem. Biophys. Res. Commun. 2001;280:1015–1020. doi: 10.1006/bbrc.2000.4226. [DOI] [PubMed] [Google Scholar]
- 26.Lee J, Jeong DJ, Kim J, Lee S, Park JH, Chang B, Jung SI, Yi L, Han Y, Yang Y, et al. The anti-aging gene KLOTHO is a novel target for epigenetic silencing in human cervical carcinoma. Mol. Cancer. 2010;9:109. doi: 10.1186/1476-4598-9-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolf I, Laitman Y, Rubinek T, Abramovitz L, Novikov I, Beeri R, Kuro OM, Koeffler HP, Catane R, Freedman LS, et al. Functional variant of KLOTHO: a breast cancer risk modifier among BRCA1 mutation carriers of Ashkenazi origin. Oncogene. 2010;29:26–33. doi: 10.1038/onc.2009.301. [DOI] [PubMed] [Google Scholar]
- 28.Lu L, Katsaros D, Wiley A, de la Longrais IA, Puopolo M, Yu H. Klotho expression in epithelial ovarian cancer and its association with insulin-like growth factors and disease progression. Cancer Invest. 2008;26:185–192. doi: 10.1080/07357900701638343. [DOI] [PubMed] [Google Scholar]
- 29.Duce JA, Podvin S, Hollander W, Kipling D, Rosene DL, Abraham CR. Gene profile analysis implicates Klotho as an important contributor to aging changes in brain white matter of the rhesus monkey. Glia. 2008;56:106–117. doi: 10.1002/glia.20593. [DOI] [PubMed] [Google Scholar]
- 30.Shih PH, Yen GC. Differential expressions of antioxidant status in aging rats: the role of transcriptional factor Nrf2 and MAPK signaling pathway. Biogerontology. 2007;8:71–80. doi: 10.1007/s10522-006-9033-y. [DOI] [PubMed] [Google Scholar]
- 31.Nabeshima Y. Ectopic calcification in Klotho mice. Clin. Calcium. 2002;12:1114–1117. [PubMed] [Google Scholar]
- 32.Deary IJ, Harris SE, Fox HC, Hayward C, Wright AF, Starr JM, Whalley LJ. KLOTHO genotype and cognitive ability in childhood and old age in the same individuals. Neurosci. Lett. 2005;378:22–27. doi: 10.1016/j.neulet.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 33.Kuwahara N, Sasaki S, Kobara M, Nakata T, Tatsumi T, Irie H, Narumiya H, Hatta T, Takeda K, Matsubara H, Hushiki S. HMG-CoA reductase inhibition improves anti-aging klotho protein expression and arteriosclerosis in rats with chronic inhibition of nitric oxide synthesis. Int. J. Cardiol. 2008;123:84–90. doi: 10.1016/j.ijcard.2007.02.029. [DOI] [PubMed] [Google Scholar]
- 34.Narumiya H, Sasaki S, Kuwahara N, Irie H, Kusaba T, Kameyama H, Tamagaki K, Hatta T, Takeda K, Matsubara H. HMG-CoA reductase inhibitors up-regulate anti-aging klotho mRNA via RhoA inactivation in IMCD3 cells. Cardiovasc. Res. 2004;64:331–336. doi: 10.1016/j.cardiores.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 35.Tsujikawa H, Kurotaki Y, Fujimori T, Fukuda K, Nabeshima Y. Klotho, a gene related to a syndrome resembling human premature aging, functions in a negative regulatory circuit of vitamin D endocrine system. Mol. Endocrinol. 2003;17:2393–2403. doi: 10.1210/me.2003-0048. [DOI] [PubMed] [Google Scholar]
- 36.Xu ZL, Gao H, Ou-Yang KQ, Cai SX, Hu YH. Establishment of a cell-based assay to screen regulators for Klotho gene promoter. Acta Pharmacol. Sin. 2004;25:1165–1170. [PubMed] [Google Scholar]
- 37.Yamagishi T, Saito Y, Nakamura T, Takeda S, Kanai H, Sumino H, Kuro-o M, Nabeshima Y, Kurabayashi M, Nagai R. Troglitazone improves endothelial function and augments renal klotho mRNA expression in Otsuka Long–Evans Tokushima Fatty (OLETF) rats with multiple atherogenic risk factors. Hypertens. Res. 2001;24:705–709. doi: 10.1291/hypres.24.705. [DOI] [PubMed] [Google Scholar]
- 38.Kurosu H, Ogawa Y, Miyoshi M, Yamamoto M, Nandi A, Rosenblatt KP, Baum MG, Schiavi S, Hu MC, Moe OW, Kuro-o M. Regulation of fibroblast growth factor-23 signaling by klotho. J. Biol. Chem. 2006;281:6120–6123. doi: 10.1074/jbc.C500457200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi BH, Kim CG, Lim Y, Lee YH, Shin SY. Transcriptional activation of the human Klotho gene by epidermal growth factor in HEK293 cells; role of Egr-1. Gene. 2010;450:121–127. doi: 10.1016/j.gene.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 40.Adijiang A, Niwa T. An oral sorbent, AST-120, increases Klotho expression and inhibits cell senescence in the kidney of uremic rats. Am. J. Nephrol. 2010;31:160–164. doi: 10.1159/000264634. [DOI] [PubMed] [Google Scholar]
- 41.Wang HL, Xu Q, Wang Z, Zhang YH, Si LY, Li XJ, Yang QH, Xiao H. A potential regulatory single nucleotide polymorphism in the promoter of the Klotho gene may be associated with essential hypertension in the Chinese Han population. Clin. Chim. Acta. 2010;411:386–390. doi: 10.1016/j.cca.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 42.Tsezou A, Furuichi T, Satra M, Makrythanasis P, Ikegawa S, Malizos KN. Association of KLOTHO gene polymorphisms with knee osteoarthritis in Greek population. J. Orthop. Res. 2008;26:1466–1470. doi: 10.1002/jor.20634. [DOI] [PubMed] [Google Scholar]
- 43.Zhang F, Zhai G, Kato BS, Hart DJ, Hunter D, Spector TD, Ahmadi KR. Association between KLOTHO gene and hand osteoarthritis in a female Caucasian population. Osteoarthritis Cartilage. 2007;15:624–629. doi: 10.1016/j.joca.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 44.Kawano K, Ogata N, Chiano M, Molloy H, Kleyn P, Spector TD, Uchida M, Hosoi T, Suzuki T, Orimo H, et al. Klotho gene polymorphisms associated with bone density of aged postmenopausal women. J. Bone Miner. Res. 2002;17:1744–1751. doi: 10.1359/jbmr.2002.17.10.1744. [DOI] [PubMed] [Google Scholar]
- 45.Kim Y, Kim JH, Nam YJ, Kong M, Kim YJ, Yu KH, Lee BC, Lee C. Klotho is a genetic risk factor for ischemic stroke caused by cardioembolism in Korean females. Neurosci. Lett. 2006;407:189–194. doi: 10.1016/j.neulet.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 46.Imamura A, Okumura K, Ogawa Y, Murakami R, Torigoe M, Numaguchi Y, Murohara T. Klotho gene polymorphism may be a genetic risk factor for atherosclerotic coronary artery disease but not for vasospastic angina in Japanese. Clin. Chim. Acta. 2006;371:66–70. doi: 10.1016/j.cca.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 47.Jo SH, Kim SG, Choi YJ, Joo NR, Cho GY, Choi SR, Kim EJ, Kim HS, Kim HJ, Rhim CY. KLOTHO gene polymorphism is associated with coronary artery stenosis but not with coronary calcification in a Korean population. Int. Heart. J. 2009;50:23–32. doi: 10.1536/ihj.50.23. [DOI] [PubMed] [Google Scholar]
- 48.Rhee EJ, Oh KW, Yun EJ, Jung CH, Lee WY, Kim SW, Baek KH, Kang MI, Park SW. Relationship between polymorphisms G395A in promoter and C1818T in exon 4 of the KLOTHO gene with glucose metabolism and cardiovascular risk factors in Korean women. J. Endocrinol. Invest. 2006;29:613–618. doi: 10.1007/BF03344160. [DOI] [PubMed] [Google Scholar]
- 49.Shimoyama Y, Taki K, Mitsuda Y, Tsuruta Y, Hamajima N, Niwa T. KLOTHO gene polymorphisms G-395A and C1818T are associated with low-density lipoprotein cholesterol and uric acid in Japanese hemodialysis patients. Am. J. Nephrol. 2009;30:383–388. doi: 10.1159/000235686. [DOI] [PubMed] [Google Scholar]
- 50.Kim Y, Jeong SJ, Lee HS, Kim EJ, Song YR, Kim SG, Oh JE, Lee YK, Seo JW, Yoon JW, et al. Polymorphism in the promoter region of the klotho gene (G-395A) is associated with early dysfunction in vascular access in hemodialysis patients. Korean. J. Intern. Med. 2008;23:201–207. doi: 10.3904/kjim.2008.23.4.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.