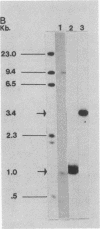
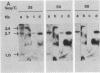
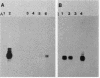
Abstract
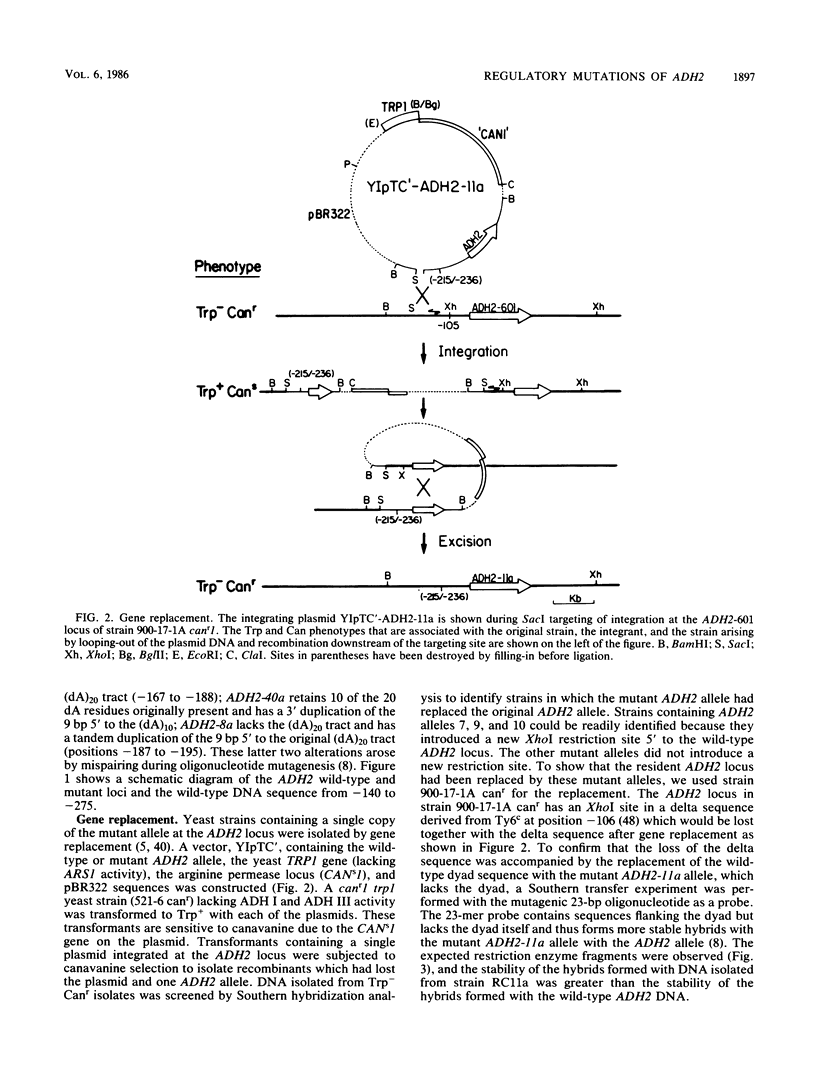
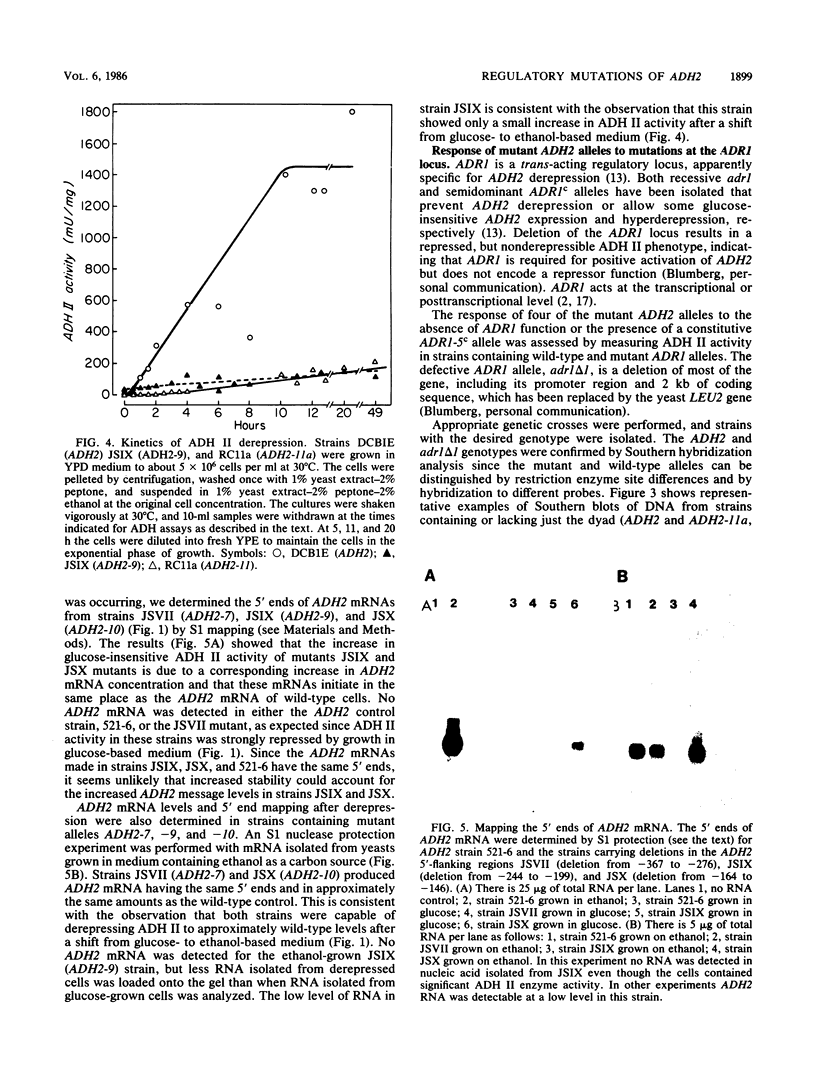
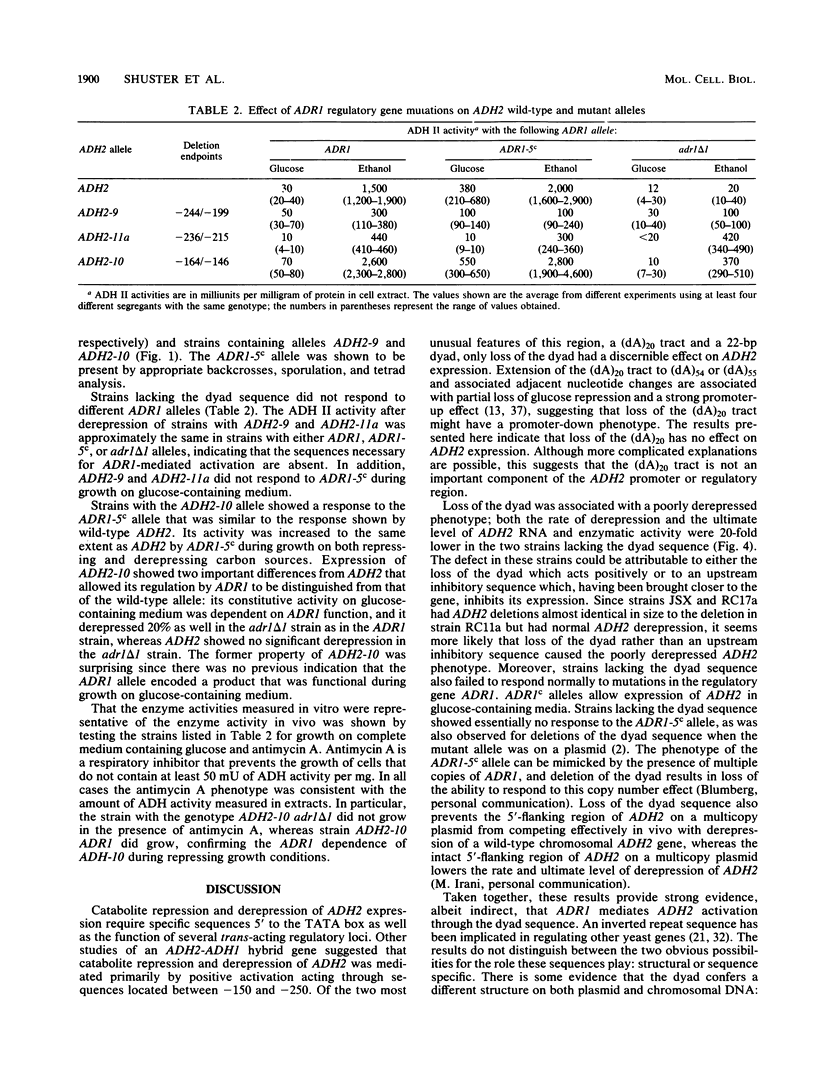
DNA sequence analysis of wild-type and mutant ADH2 loci suggested that two unusual features 5' of the promoter, a 22-base-pair perfect dyad sequence and a (dA)20 tract, were important for regulation of this gene (D. W. Russell, M. Smith, D. Cox, V. M. Williamson, and E. T. Young, Nature [London] 304:652-654, 1983). Oligonucleotide-directed mutagenesis was used to construct ADH2 genes lacking the 22-base-pair dyad or the (dA)20 tract (V.-L. Chan and M. Smith, Nucleic Acids Res. 12:2407-2419, 1984). These mutant genes and other ADH2 deletions constructed by BAL 31 endonuclease digestion were studied after replacing the wild-type chromosomal locus with the altered alleles by the technique of gene transplacement (T. L. Orr-Weaver, J. W. Szostak, and R. S. Rothstein, Proc. Natl. Acad. Sci. USA 78:6354-6358, 1981), using canavanine resistance as the selectable marker. Deletions lacking the dyad failed to derepress normally and did not respond to mutations at the ADR1 locus, which encodes a protein necessary to activate ADH2. Deletions of the (dA)20 tract did not have a detectable phenotype. A small deletion located just 3' to the (dA)20 tract (between positions -164 and -146) had a low amount of ADR1-dependent transcription during repressed growth conditions, indicating that the regulatory protein encoded by ADR1 is present in a potentially active form during repression and that alterations of a DNA sequence in the promoter region can unmask its latent activity.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beier D. R., Sledziewski A., Young E. T. Deletion analysis identifies a region, upstream of the ADH2 gene of Saccharomyces cerevisiae, which is required for ADR1-mediated derepression. Mol Cell Biol. 1985 Jul;5(7):1743–1749. doi: 10.1128/mcb.5.7.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier D. R., Young E. T. Characterization of a regulatory region upstream of the ADR2 locus of S. cerevisiae. Nature. 1982 Dec 23;300(5894):724–728. doi: 10.1038/300724a0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Spliced early mRNAs of simian virus 40. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1274–1278. doi: 10.1073/pnas.75.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Brown P. A., Szostak J. W. Yeast vectors with negative selection. Methods Enzymol. 1983;101:278–290. doi: 10.1016/0076-6879(83)01021-6. [DOI] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Carnevali F., Caserta M., Di Mauro E. Transitions in topological organization of supercoiled DNA domains as a potential regulatory mechanism. J Biol Chem. 1984 Oct 25;259(20):12633–12643. [PubMed] [Google Scholar]
- Chan V. L., Smith M. In vitro generation of specific deletions in DNA cloned in M13 vectors using synthetic oligodeoxyribonucleotides: mutants in the 5'-flanking region of the yeast alcohol dehydrogenase II gene. Nucleic Acids Res. 1984 Mar 12;12(5):2407–2419. doi: 10.1093/nar/12.5.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriacy M. Genetics of alcohol dehydrogenase in Saccharomyces cerevisiae. II. Two loci controlling synthesis of the glucose-repressible ADH II. Mol Gen Genet. 1975;138(2):157–164. doi: 10.1007/BF02428119. [DOI] [PubMed] [Google Scholar]
- Ciriacy M. Isolation and characterization of further cis- and trans-acting regulatory elements involved in the synthesis of glucose-repressible alcohol dehydrogenase (ADHII) in Saccharomyces cerevisiae. Mol Gen Genet. 1979 Nov;176(3):427–431. doi: 10.1007/BF00333107. [DOI] [PubMed] [Google Scholar]
- Ciriacy M. Isolation and characterization of yeast mutants defective in intermediary carbon metabolism and in carbon catabolite derepression. Mol Gen Genet. 1977 Jul 20;154(2):213–220. doi: 10.1007/BF00330840. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Jacobs H. T., Britten R. J. Very short repeats and coordinate induction of genes. Nature. 1983 Feb 10;301(5900):468–470. doi: 10.1038/301468a0. [DOI] [PubMed] [Google Scholar]
- Davis R. W., Thomas M., Cameron J., St John T. P., Scherer S., Padgett R. A. Rapid DNA isolations for enzymatic and hybridization analysis. Methods Enzymol. 1980;65(1):404–411. doi: 10.1016/s0076-6879(80)65051-4. [DOI] [PubMed] [Google Scholar]
- Denis C. L., Ciriacy M., Young E. T. A positive regulatory gene is required for accumulation of the functional messenger RNA for the glucose-repressible alcohol dehydrogenase from Saccharomyces cerevisiae. J Mol Biol. 1981 Jun 5;148(4):355–368. doi: 10.1016/0022-2836(81)90181-9. [DOI] [PubMed] [Google Scholar]
- Denis C. L. Identification of new genes involved in the regulation of yeast alcohol dehydrogenase II. Genetics. 1984 Dec;108(4):833–844. doi: 10.1093/genetics/108.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis C. L., Young E. T. Isolation and characterization of the positive regulatory gene ADR1 from Saccharomyces cerevisiae. Mol Cell Biol. 1983 Mar;3(3):360–370. doi: 10.1128/mcb.3.3.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue T. F., Daves R. S., Lucchini G., Fink G. R. A short nucleotide sequence required for regulation of HIS4 by the general control system of yeast. Cell. 1983 Jan;32(1):89–98. doi: 10.1016/0092-8674(83)90499-3. [DOI] [PubMed] [Google Scholar]
- Federoff H. J., Eccleshall T. R., Marmur J. Carbon catabolite repression of maltase synthesis in Saccharomyces carlsbergensis. J Bacteriol. 1983 Oct;156(1):301–307. doi: 10.1128/jb.156.1.301-307.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giniger E., Varnum S. M., Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985 Apr;40(4):767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters: positive and negative elements. Cell. 1984 Apr;36(4):799–800. doi: 10.1016/0092-8674(84)90028-x. [DOI] [PubMed] [Google Scholar]
- Guarente L., Yocum R. R., Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol. 1967 May;93(5):1662–1670. doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper J. E., Broach J. R., Rowe L. B. Regulation of the galactose pathway in Saccharomyces cerevisiae: induction of uridyl transferase mRNA and dependency on GAL4 gene function. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2878–2882. doi: 10.1073/pnas.75.6.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston G. C., Pringle J. R., Hartwell L. H. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp Cell Res. 1977 Mar 1;105(1):79–98. doi: 10.1016/0014-4827(77)90154-9. [DOI] [PubMed] [Google Scholar]
- Lutstorf U., Megnet R. Multiple forms of alcohol dehydrogenase in Saccharomyces cerevisiae. I. Physiological control of ADH-2 and properties of ADH-2 and ADH-4. Arch Biochem Biophys. 1968 Sep 10;126(3):933–944. doi: 10.1016/0003-9861(68)90487-6. [DOI] [PubMed] [Google Scholar]
- Mahler H. R., Lin C. C. Exogenous adenosine 3': 5'-monophosphate can release yeast from catabolite repression. Biochem Biophys Res Commun. 1978 Aug 14;83(3):1039–1047. doi: 10.1016/0006-291x(78)91500-0. [DOI] [PubMed] [Google Scholar]
- Martinez-Arias A., Yost H. J., Casadaban M. J. Role of an upstream regulatory element in leucine repression of the Saccharomyces cerevisiae leu2 gene. Nature. 1984 Feb 23;307(5953):740–742. doi: 10.1038/307740b0. [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Uno I., Oshima Y., Ishikawa T. Isolation and characterization of yeast mutants deficient in adenylate cyclase and cAMP-dependent protein kinase. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2355–2359. doi: 10.1073/pnas.79.7.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakis E. S., Bartley W. Changes in the enzyme activities of Saccharomyces cerevisiae during aerobic growth on different carbon sources. Biochem J. 1965 Oct;97(1):284–297. doi: 10.1042/bj0970284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell D. W., Smith M., Cox D., Williamson V. M., Young E. T. DNA sequences of two yeast promoter-up mutants. Nature. 1983 Aug 18;304(5927):652–654. doi: 10.1038/304652a0. [DOI] [PubMed] [Google Scholar]
- Russell D. W., Smith M., Williamson V. M., Young E. T. Nucleotide sequence of the yeast alcohol dehydrogenase II gene. J Biol Chem. 1983 Feb 25;258(4):2674–2682. [PubMed] [Google Scholar]
- Sarokin L., Carlson M. Upstream region required for regulated expression of the glucose-repressible SUC2 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Dec;4(12):2750–2757. doi: 10.1128/mcb.4.12.2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S., Davis R. W. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz L. D. Transcriptional role of yeast deoxyribonucleic acid dependent ribonucleic acid polymerase III. Biochemistry. 1978 Feb 21;17(4):750–758. doi: 10.1021/bi00597a031. [DOI] [PubMed] [Google Scholar]
- Sledziewski A., Young E. T. Chromatin conformational changes accompany transcriptional activation of a glucose-repressed gene in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1982 Jan;79(2):253–256. doi: 10.1073/pnas.79.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Negative control at a distance mediates catabolite repression in yeast. 1985 Oct 31-Nov 6Nature. 317(6040):822–824. doi: 10.1038/317822a0. [DOI] [PubMed] [Google Scholar]
- Taguchi A. K., Ciriacy M., Young E. T. Carbon source dependence of transposable element-associated gene activation in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Jan;4(1):61–68. doi: 10.1128/mcb.4.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West R. W., Jr, Yocum R. R., Ptashne M. Saccharomyces cerevisiae GAL1-GAL10 divergent promoter region: location and function of the upstream activating sequence UASG. Mol Cell Biol. 1984 Nov;4(11):2467–2478. doi: 10.1128/mcb.4.11.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson V. M., Cox D., Young E. T., Russell D. W., Smith M. Characterization of transposable element-associated mutations that alter yeast alcohol dehydrogenase II expression. Mol Cell Biol. 1983 Jan;3(1):20–31. doi: 10.1128/mcb.3.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson V. M., Young E. T., Ciriacy M. Transposable elements associated with constitutive expression of yeast alcohol dehydrogenase II. Cell. 1981 Feb;23(2):605–614. doi: 10.1016/0092-8674(81)90156-2. [DOI] [PubMed] [Google Scholar]
- Young T., Williamson V., Taguchi A., Smith M., Sledziewski A., Russell D., Osterman J., Denis C., Cox D., Beier D. The alcohol dehydrogenase genes of the yeast, Saccharomyces cerevisiae: isolation, structure, and regulation. Basic Life Sci. 1982;19:335–361. doi: 10.1007/978-1-4684-4142-0_26. [DOI] [PubMed] [Google Scholar]
- Zitomer R. S., Montgomery D. L., Nichols D. L., Hall B. D. Transcriptional regulation of the yeast cytochrome c gene. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3627–3631. doi: 10.1073/pnas.76.8.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]