Abstract
The suprachiasmatic nucleus (SCN) coordinates circadian rhythms that adapt the individual to solar time. SCN pacemaking revolves around feedback loops in which expression of Period (Per) and Cryptochrome (Cry) genes is periodically suppressed by their protein products. Specifically, PER/CRY complexes act at E-box sequences in Per and Cry to inhibit their transactivation by CLOCK/BMAL1 heterodimers. To function effectively, these closed intracellular loops need to be synchronized between SCN cells and to the light/dark cycle. For Per expression, this is mediated by neuropeptidergic and glutamatergic extracellular cues acting via cAMP/calcium-responsive elements (CREs) in Per genes. Cry genes, however, carry no CREs, and how CRY-dependent SCN pacemaking is synchronized remains unclear. Furthermore, whereas reporter lines are available to explore Per circadian expression in real time, no Cry equivalent exists. We therefore created a mouse, B6.Cg-Tg(Cry1-luc)01Ld, carrying a transgene (mCry1-luc) consisting of mCry1 elements containing an E-box and E′-box driving firefly luciferase. mCry1-luc organotypic SCN slices exhibited stable circadian bioluminescence rhythms with appropriate phase, period, profile, and spatial organization. In SCN lacking vasoactive intestinal peptide or its receptor, mCry1 expression was damped and desynchronized between cells. Despite the absence of CREs, mCry1-luc expression was nevertheless (indirectly) sensitive to manipulation of cAMP-dependent signaling. In mPer1/2-null SCN, mCry1-luc bioluminescence was arrhythmic and no longer suppressed by elevation of cAMP. Finally, an SCN graft procedure showed that PER-independent as well as PER-dependent mechanisms could sustain circadian expression of mCry1. The mCry1-luc mouse therefore reports circadian mCry1 expression and its interactions with vasoactive intestinal peptide, cAMP, and PER at the heart of the SCN pacemaker.
Keywords: gene expression, period gene, adenylate cyclase, VPAC2, afterhours
The suprachiasmatic nucleus (SCN) of the hypothalamus is the principal circadian pacemaker in mammals, coordinating daily programs of gene expression across the body that ultimately underpin adaptation to day and night (1). SCN pacemaking revolves around an autoregulatory feedback loop in which expression of Period (Per) and Cryptochrome (Cry) genes is suppressed by their protein products on a daily basis. Specifically, PER/CRY complexes act at E-box regulatory sequences in Per and Cry to inhibit their transactivation by CLOCK/BMAL1 heterodimers (2). Supplementary feedback loops involving bZIP-family proteins [albumin D-element binding protein (Dbp) and nuclear factor, interleukin 3 regulated (Nfil3) acting at D-boxes] and REV-ERB family nuclear receptors (acting at RORE elements) confer additional robustness and amplitude (3), but E-box–mediated transcription is the principal determinant of SCN molecular pacemaking.
To generate a coherent circadian signal across the SCN, the intracellular loops are synchronized between cells by neuropeptidergic cues that act via signaling cascades triggered by cAMP and intracellular Ca2+ ([Ca2+]i) (4, 5). In addition, synchronization to the cycle of light and darkness is mediated by specialized glutamatergic retinal innervation of SCN neurons (6) that acts via [Ca2+]i (7). These cAMP/[Ca2+]i pathways converge upon cAMP/calcium-responsive elements (CREs) in the Per genes (8) to activate their expression (9). Thus, extracellular synchronizing stimuli gain access to the PER-dependent components of the feedback loops. Cry genes, however, carry no CREs as access points for extracellular information, raising the question of how CRY-dependent elements of the SCN pacemaker are synchronized. Consistent with a model of indirect regulation, reentrainment of daily Cry expression in the SCN of mice subjected to phase-advanced lighting cycles lags behind Per rhythms (10). Cry expression requires several days to resynchronize, but, importantly, the time course of mCry reentrainment parallels that of the rest–activity cycle, highlighting the contribution of Cry expression to the timing of behavior.
A comprehensive understanding of the SCN circadian pacemaker in toto requires, therefore, direct and dynamic analyses of Cry gene expression, but whereas various mouse lines (11, 12) have been powerful tools in examining Per expression, no reporter lines are available with which to characterize intra- and intercellular mechanisms governing mCry1 expression. We therefore generated a transgenic (Tg) mouse line, B6.Cg-Tg(Cry1-luc)01Ld, carrying a mCry1-luciferase bioluminescent reporter, using previously characterized Cry1 elements that contain an E-box and E′-box (13) with an overlapping D-box (14). We show that these sequences are sufficient to drive period- and phase-appropriate circadian cycles of mCry1 transcription consistent with E-box regulation in organotypic SCN and peripheral tissue cultures. Despite lacking CRE sequences, mCry1 circadian transcription in the SCN was nevertheless dependent upon signaling via vasoactive intestinal peptide (VIP) and its VPAC2 receptor, a positive regulator of adenylyl cyclase (AC). Moreover, direct pharmacological manipulation of cAMP levels, both positive and negative, disrupted circadian mCry1 expression. In the absence of mPER1 and mPER2 proteins, however, mCry1-luc transcription in the SCN was arrhythmic and no longer suppressed by elevated cAMP levels, highlighting a role for mPER1 and mPER2 as transducers in the circadian regulation of mCry1. Finally, by using an SCN slice grafting technique, we show that mPER1/mPER2-independent pathways could also sustain circadian expression of mCry1. Thus, parallel PER-dependent and PER-independent pathways direct circadian expression of mCRY1, the principal negative feedback regulator in the SCN.
Results
Period and Phase of SCN Circadian mCry1-luc Expression.
Following injection of mouse oocyte (strain B6CBAF1/OlaHsd) with linearized plasmid carrying −1,504 to +107 of mCry1 upstream of luciferase coding sequence (Fig. S1A), and subsequent embryo transfer, six transgenic founder mice were identified and crossed with nontransgenic mice to establish breeding lines. Of these, three lines produced offspring with a bioluminescent signal detectable in the SCN, and the line with the most intense SCN signal was selected for further analysis. All subsequent crosses (greater than six) were with mice on a C57BL/6 background to create the B6.Cg-Tg(Cry1-luc)01Ld (mCry1-luc) line, which bred true, and digital droplet PCR identified a single copy of the reporter construct at a single insertion.
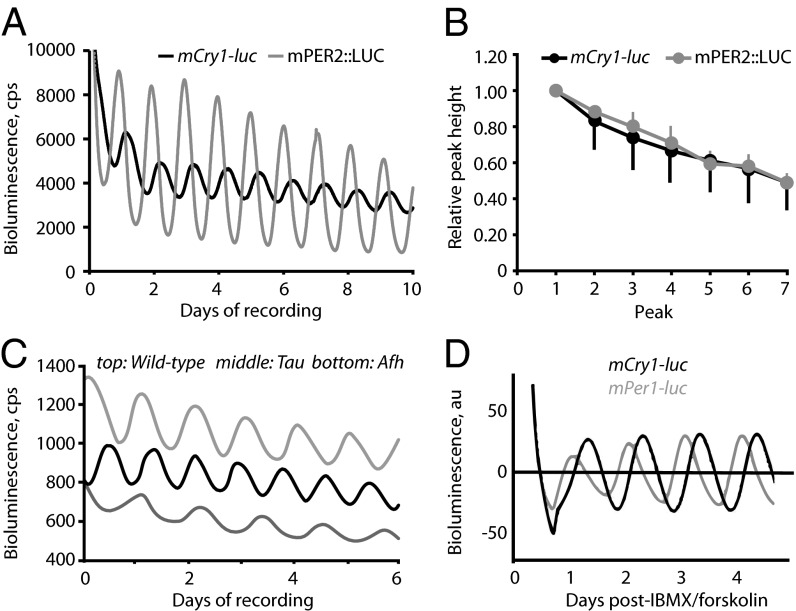
When recorded by photomultiplier array, all SCN slices carrying the reporter (n = 28) on an otherwise WT background exhibited stable bioluminescence rhythms (Fig. 1A) with periods within the circadian range (mean ± SEM, 24.21 ± 0.08 h) that were not significantly different from those of the widely used mPER2::LUC fusion protein reporter (12) (24.40 ± 0.04 h; n = 11; t test, P = 0.16), although they were more variable between slices than mPER2::LUC reports (range mCry1-luc, 22.89–24.99 h; mPER2::LUC, 24.13–24.59 h; SD, 0.42 vs. 0.12). mCry1-driven bioluminescence tended to be lower than that of mPER2::LUC SCN (2,940 ± 403 cps vs. 3,908 ± 521 cps; 75.2%) but not statistically so (t test, P = 0.19). The amplitude of the oscillation was, however, significantly smaller than that of mPER2::LUC (403 ± 49 cps vs. 2,377 ± 270 cps; t test, P < 0.01), and the relative amplitude error (RAE) was significantly higher in mCry1-luc slices (0.050 ± 0.004 vs. 0.033 ± 0.002; P < 0.01), indicative of lower cycle-to-cycle signal reproducibility. The relative decline of peak height (which arises from use of luciferin substrate) was calculated in a subset of recordings and followed the same exponential decay for both reporters (both r2 = 0.99; Fig. 1B). A one-phase exponential decay half-life showed no significant difference between reporters (mCry1-luc, 2.27 ± 0.69 d; mPER2::LUC, 3.00 ± 0.42 d; t test, P = 0.44; n = 9 and n = 6, respectively).
Fig. 1.
Circadian period and phase of mCry1-luc expression in SCN. (A) Representative bioluminescence recordings from mCry1-luc and mPER2::LUC SCN slices. (B) Relative decline in signal amplitude caused by substrate use from mCry1-luc and mPER2::LUC SCN (n = 10 for both; data normalized to initial value of 100%; mean ± SD). (C) Representative bioluminescence recordings from WT (gray), CK1εTau (black), and Fbxl3Afh (dark gray) mCry1-luc SCN. (D) Representative recordings of baseline-corrected bioluminescence from mCry1-luc and mPer1-luc SCN following washout of forskolin/IBMX. Note later phase of mCry1-luc.
Importantly, the mCry1-luc transgene faithfully reported the effects of two mutant alleles on SCN circadian period (Fig. 1C and Fig. S1B). The Tau allele of casein kinase 1ε (CK1εTau) allele dose-dependently accelerated pacemaking by approximately 2.2 h per copy (15), whereas the Afterhours allele of Fbxl3 (Fbxl3Afh) slowed the SCN by approximately 2.1 h per copy (16). Deceleration by the Fbxl3Afh mutation was also accompanied by a prolongation of the nadir of bioluminescence [WT, 6.40 ± 0.63 h; heterozygote, 8.13 ± 0.13 h; homozygote, 10.00 ± 0.32 h (mean ± SEM); n = 4, n = 3, and n = 4, respectively]. This reflects an extended phase of negative feedback on the E-boxes of the transgene caused by Afh-stabilized CRY proteins. Despite these differences in period, there was no difference in the quality of the rhythms reported by mCry1-luc (RAE, WT, 0.050 ± 0.004, n = 28; Tau homozygote, 0.054 ± 0.008, n = 6; Afh homozygote, 0.051 ± 0.007, n = 5).
In the SCN in vivo, expression of mCry1 mRNA peaks later in the circadian day, at approximately circadian time 10 to 12, whereas mPer1 mRNA levels peak earlier, at approximately circadian time 06 (17, 18). To determine the phase reported by the transgene in vitro, SCN from mCry1-luc and mPer1-luc mice were treated with forskolin and isobutyl-1-methylxanthine (IBMX) for 8 d and then transferred to fresh, drug-free medium. This treatment synchronizes previously unsynchronized SCN slices to a single-phase (19). This was evident within the separate groups of SCN carrying mCry1 or mPer1 reporters (Fig. S1C). Comparison between the groups, however, revealed a phase difference, such that the mCry1-reported rhythm was 5.3 ± 0.6 h delayed relative to the mPer1-luc cycle (P < 0.01, t test; n = 3 per reporter; Fig. 1D). Assuming that the time for translation and activation of luciferase is the same in the two reporters, the mCry1-luc peak is exactly in the phase predicted by the respective mRNA peaks (mCry1 delayed relative to mPer1 by 4–6 h). Therefore, as in cell culture (13), the mCry1-luc construct was sufficient to sustain the phase relationship observed in vivo. Thus, the circadian activation of the mCry1-luc transgene within the SCN slice of B6.Cg-Tg(Cry1-luc)01Ld mice is of appropriate period, profile, and phase to that observed in vivo, providing a high-fidelity tool with which to analyze E-box–mediated circadian control of mCry1 within the SCN. Finally, consistent with the earlier validation of the reporter in 3T3 cells (13), the majority of peripheral tissue explants from mCry1-luc mice exhibited circadian bioluminescence rhythms (Fig. S1D). As for SCN, the overall level of bioluminescence and circadian period were not different from corresponding mPER2::LUC explants, but amplitude and coherence were significantly lower (Tables S1 and S2). Finally, mCry1-luc MEFs exhibited clear circadian cycles of bioluminescence (Fig. S1E and Table S3) comparable to those of mPER2::LUC but with higher amplitude that progressively damped and were reactivated by medium change.
Spatiotemporal Control of SCN mCry1 Circadian Activation.
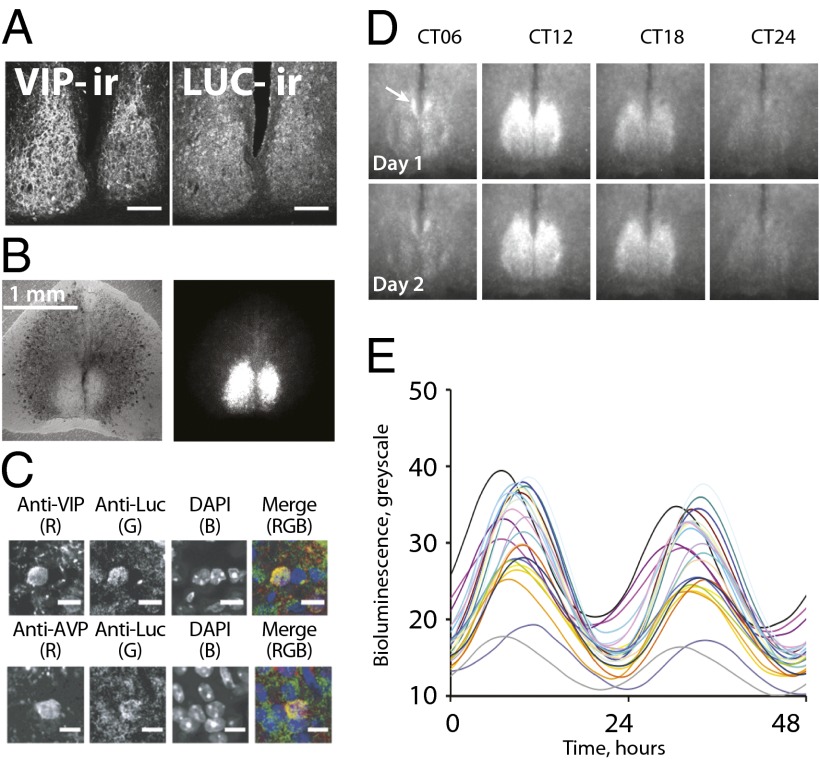
Immunofluorescent staining for luciferase and CCD imaging revealed mCry1-luc activation across the SCN (Fig. 2 A and B), with expression in arginine vasopressin (AVP)- and VIP-positive neurons (Fig. 2C), which are markers, respectively, of shell and core SCN subdivisions. A distinctive feature of the circadian behavior of mPer-based reporters is their spatiotemporal “wave” by which bioluminescent or fluorescent activity progresses across the SCN from the dorsomedial shell to the ventrolateral core (11, 12, 20). It is not known whether this dynamic is a unique feature of mPer expression, possibly reflecting CRE-mediated activation of Per by interneuronal cues, or a conserved aspect of the complete molecular cycle. If the latter, the wave would be common to all components of the pacemaker, including mCry1. Analysis of regional mCry1-luc bioluminescence highlighted a wave emanating from a leading edge in the dorsomedial lip of the SCN (Fig. 2D, Fig. S2, and Movie S1). The activity in this region was advanced by approximately 2.30 ± 0.13 h relative to the remainder of the SCN [assessed by fast Fourier transform (FFT)]. Similarly, bioluminescence rhythms of individual cells were clearly synchronous but with a distribution of phases (Fig. 2E and Fig. S2), and, when assigned into phase groups, cells in the dorsomedial lip were advanced by between 1.16 ± 0.28 h and 3.77 ± 0.15 h relative to laterally and ventrally placed cells, respectively. The mCry1-luc reporter therefore demonstrates that the spatiotemporal wave across the SCN circuitry is not exclusively a CRE-dependent phenomenon. Rather, it is a feature of both components of the negative feedback axis, mPer and mCry; and may arise from temporally and spatially sequential activation of E-box–mediated transcription.
Fig. 2.
Spatiotemporal control of mCry1 circadian activation across the SCN circuit. (A) Confocal photomicrographs (magnification of 20×) reveal mCry1-driven luciferase immunoreactivity (-ir) across the SCN, delineated by VIP expression. (Scale bar: 100 μm.) (B) Organotypic SCN slice viewed in phase (Left) or as mCry1-luc bioluminescence (Right). (C) High-power (magnification of 63×) confocal images reveal expression of mCry1-driven luciferase in both VIP- (Upper) and AVP-immunoreactive (Lower) SCN neurons. (Scale bar: 10 μm.) (D) Serial frames from a representative slice demonstrate circadian expression of mCry1 across SCN over 2 d. Arrow indicates phase-leading dorsomedial lip. (E) Representative plots of circadian bioluminescence recorded from individual SCN cells by CCD.
Circadian Regulation of mCry1 by VIP/VPAC2 Signaling.
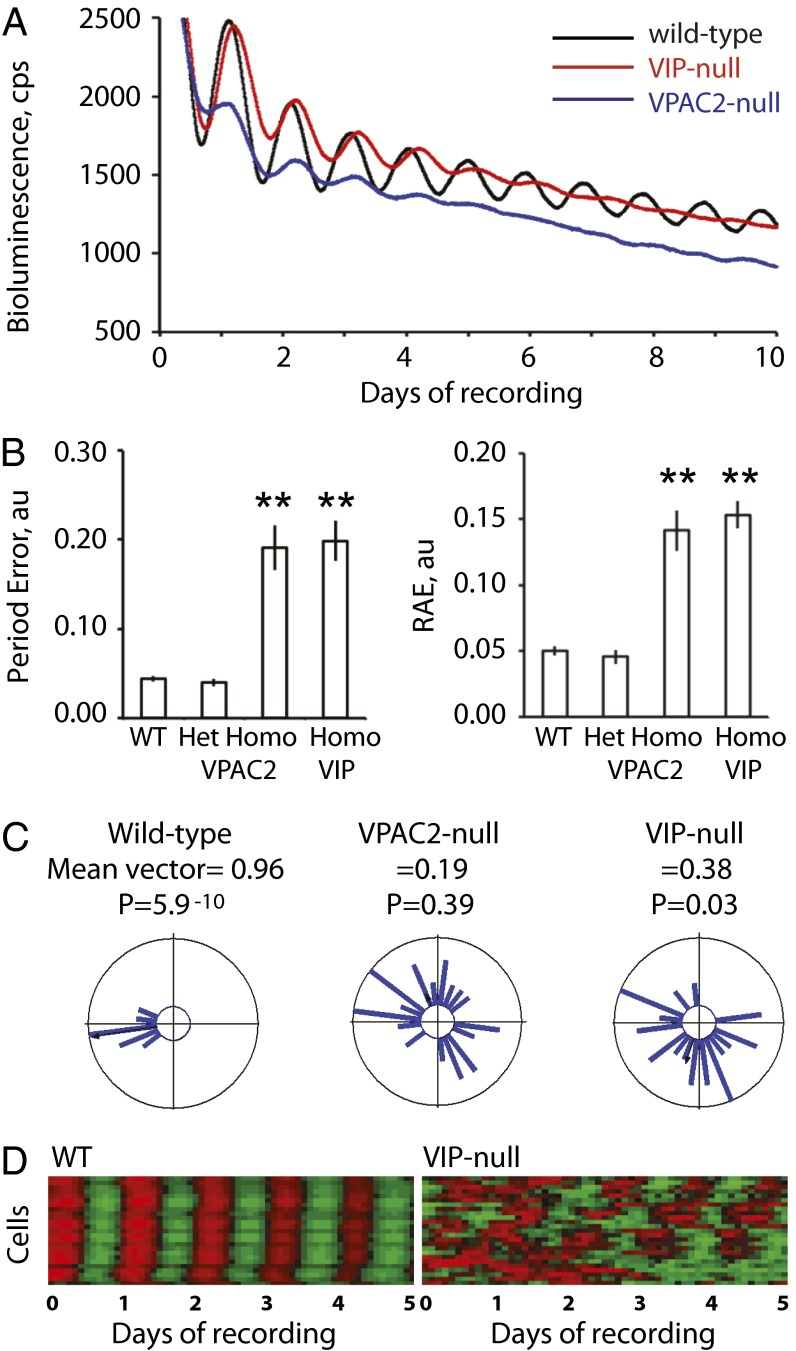
The neuropeptide VIP and its receptor, VPAC2, are necessary to maintain the amplitude and synchrony of mPer-reported molecular pacemaking (21, 22). The VPAC2 receptor stimulates cAMP synthesis, thereby maintaining the amplitude and synchrony of Per expression via CREs in the mPer genes. The degree to which the loss of VIP/VPAC2 signaling compromises other components of the molecular pacemaker is unknown. If E-box–directed circadian expression of mCry1 is independent of cAMP/mPER proteins, loss of VIP/VPAC2 signaling would not affect mCry1-luc rhythms. To test this directly, mCry1-luc mice were crossed onto VIP- or VPAC2-null backgrounds (Fig. 3A). Mean bioluminescence levels were comparable between WT and VPAC2-null heterozygous slices (2,940 ± 403 cps, n = 28; 2,759 ± 463 cps, n = 11). Bioluminescence tended to be lower in VPAC2-null homozygous slices (2,010 ± 399, n = 9; P > 0.05) and was significantly lower in VIP-null slices (622 ± 120, n = 7; P < 0.01). FFT analysis was nevertheless able to define periodicity in the recordings from VPAC2-null SCN (24.27 ± 0.40 h) that was not significantly different from WT (24.21 ± 0.09 h) or VPAC2-null heterozygotes (23.92 ± 0.13 h). The period of VIP-null SCN was, however, statistically longer than WT (25.18 ± 0.61; P < 0.05). More significantly, there was a rapid damping of the circadian pattern in VIP- and VPAC2-null SCN, such that the oscillation progressively lost definition, and errors of period and relative amplitude were greater (Fig. 3B).
Fig. 3.
Circadian regulation of mCry1 by interneuronal VIP/VPAC2 signaling. (A) Representative bioluminescence recordings from WT, VPAC2-null, or VIP-null mCry1-luc SCN. (B) Group data (mean ± SEM) for period error (Left) and RAE (Right) of mCry1-luc bioluminescence rhythms from WT and VPAC2- or VIP-null SCN. (**P < 0.01 vs. WT by ANOVA and post-hoc Bonferroni correction.) (C) Rayleigh plots (n = 25) of cellular bioluminescence rhythms from representative WT and VPAC2- and VIP-null SCN. (D) Raster plots of cellular mCry1-luc bioluminescence rhythms from representative WT and VIP-null SCN recorded by CCD over 5 d.
To determine the cause of these disorganized rhythms, mCry1-luc emission was imaged with CCD cameras. In WT SCN, cellular rhythms were highly synchronous, as revealed by the Rayleigh analysis (Fig. 3 C and D): the small spread in phases reflecting the spatiotemporal wave. In contrast, cellular rhythms in VPAC2- and VIP-null slices were poorly synchronized, with Rayleigh plots having low or null statistical significance and reduced mean vectors. This loss of synchrony mirrors that shown with mPer-based reporters. To test this further, VPAC2-null, mCry1-luc SCN slices were treated with forskolin (20 μM), which is known to transiently restore circadian organization to Per expression in VPAC2-null SCN (21). Forskolin triggered a transient increase in bioluminescence levels, associated at the cellular level with resynchronized rhythmic expression (Fig. S3A). Thus, mCry1 expression was perturbed by upstream deficiencies in VIP/VPAC2 and compromised cAMP signals.
Indirect Regulation of SCN mCry1 by cAMP Signaling.
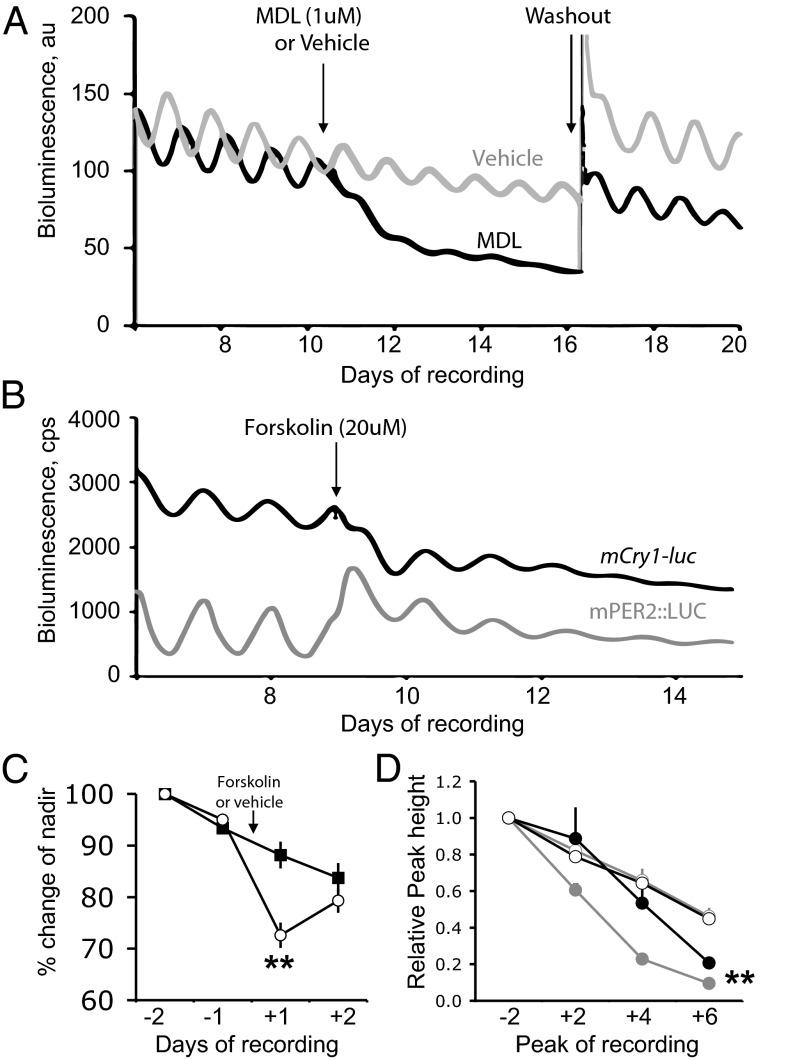
To test directly the dependence of mCry1 expression on cAMP, SCNs were treated with MDL-12,330A (1 μM), which inhibits AC and reversibly suppresses cAMP levels in the SCN, thereby curtailing CRE activation and mPer1/mPer2 expression (19). Vehicle-treated slices (n = 5) displayed the typical progressive decline in bioluminescence levels, falling by 73.8 ± 3.8% during the 5 d (Fig. 4A). The amplitude of the oscillation also decreased to 69 ± 11%. With addition of MDL, however, there was a dramatic and significant decrease in bioluminescence to 45.6 ± 1.7% of pretreatment level (n = 7; P < 0.01, t test) and amplitude to 22 ± 3% (t test, P < 0.01). The period of oscillation was not affected (vehicle, 23.75 ± 0.26 h; vs. MDL, 24.32 ± 0.26 h), but the coherence was reduced by MDL (RAE before, 0.038 ± 0.006; after, 0.096 ± 0.014; P < 0.01, t test) and not by vehicle (RAE before, 0.026 ± 0.002; after, 0.028 ± 0.002). After MDL washout, the SCN reestablished circadian rhythms comparable to those seen before treatment. Thus, inhibition of AC reversibly suppressed the absolute level of mCry1 activation, damped the amplitude, and reduced coherence of the oscillation. Circadian mCry1-luc expression therefore requires competent cAMP/CRE signaling.
Fig. 4.
Indirect regulation of mCry1 expression in the SCN by cAMP-mediated signaling. (A) Representative bioluminescence plots from mCry1-luc SCN treated with vehicle or AC inhibitor MDL-12,330A for 5 d and then subjected to medium change. Plots normalized to 100% at time of treatment. (B) Representative bioluminescence plots from mCry1-luc and mPER2::LUC SCN treated with forskolin. (C) Effect of forskolin (closed) or vehicle (open) on nadir of bioluminescence of mCry1-luc SCN (mean ± SEM, n = 7 for both treatments). (D) Effect of forskolin (closed circles) or vehicle (open circles) on relative peak height of bioluminescence of mPER2::LUC (gray) or mCry1-luc (black) SCN (mean ± SEM, n = 3 for all conditions). (**P < 0.01 vs. vehicle-treated SCN, ANOVA and post-hoc Bonferroni correction.)
To explore further the role of cAMP/CRE signaling in mCry1 expression, SCNs from mPER2::LUC or mCry1-luc mice were treated with forskolin (20 μM) to activate AC. In mPER2::LUC slices, forskolin acutely induced PER2 protein (relative change in bioluminescence over 24 h following treatment, mPER2::LUC vehicle, 0.92 ± 0.02; forskolin, 1.37 ± 0.08; n = 6; P < 0.0.01, t test; Fig. 4B). No such acute effect was apparent in mCry1-luc slices (vehicle, 0.90 ± 0.02; forskolin, 0.86 ± 0.02; n = 7). Rather, mCry1-luc activity was suppressed on the first nadir after addition of forskolin (P < 0.01, vehicle vs. forskolin, paired t test; n = 7; Fig. 4C). This occurred subsequent to the surge in mPER2 protein levels in the parallel mPER2::LUC slices. The elevated mPER2::LUC expression progressively damped with prolonged treatment (Fig. 4 B and D). Comparable damping was seen in the mCry1-luc oscillation, such that, after six cycles, the peak amplitude for both reporters was significantly suppressed relative to vehicle-treated slices (ANOVA, P < 0.01; n = 3 per group; Fig. 4D). Circadian regulation of mCry1 expression is therefore indirectly dependent on cAMP-dependent signaling. Although mCry1-luc did not share the acute CRE-mediated sensitivity of mPER2::LUC to altered cAMP levels, the acute increase and long-term damping of mPER expression arising from elevated cAMP levels fed forward to dysregulate E-box–dependent mCry1-luc rhythms.
Circadian Control of mCry1-luc Expression by mPER1/mPER2.
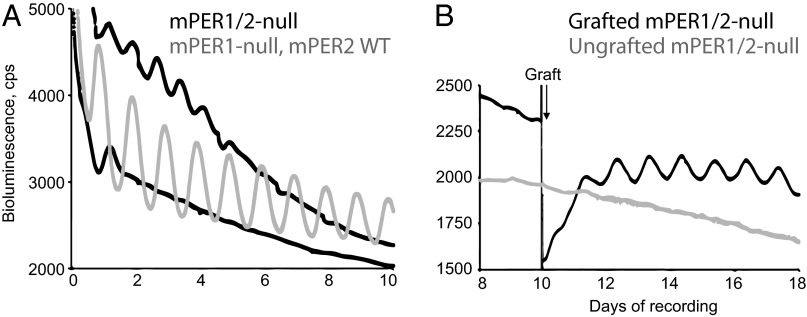
To test the presumed role of PER proteins in the indirect control of mCry1 by intercellular cues, the Cry1-luc reporter mouse was crossed to an mPER1/mPER2-null background. These mice are behaviorally arrhythmic (23) and, at the level of the SCN, molecular pacemaking was severely compromised (Fig. 5A). In some slices (n = 4), no significant rhythm of mCry1-luc expression was detectable by FFT. In others (n = 6), a weak, short period oscillation (20.36 ± 1.81 h) persisted for four or five cycles. The oscillation had low amplitude [35.39 ± 17.63 vs. 403 ± 49 (WT)] and was poorly organized (RAE, 0.361 ± 0.175; vs. WT, 0.050 ± 0.004). Thus, E-box–mediated circadian mCry1 expression in the SCN is dependent on mPER1/2.
Fig. 5.
Circadian control over mCry1 expression by mPER1 and mPER2. (A) Representative bioluminescence plots from mPER1/2-null mCry1-luc SCN, one immediately arrhythmic and the other with transient short period cycles. A reference plot from a representative mPER1-null mPER2 WT slice is included. (B) Representative bioluminescence plots from mPER1/2-null mCry1-luc SCN that did or did not receive a WT SCN graft.
To test the presumed role of PER proteins as transducers of cAMP signals onto mCry1 expression, mPER1/2-null SCNs were treated with forskolin (20 μM). In contrast to the suppression of mCry1-driven bioluminescence observed in WT slices, forskolin caused a small (ca. 10%) increase in emission in mPER1/2-null SCN over the subsequent 24 h (Fig. S3B). In the absence of PER proteins, this brief activation of mCry1-luc was not accompanied by the transient restoration of circadian oscillation noted earlier with the PER-proficient VIP-null SCN. The weak response was not a consequence of the mPER1/2-null background per se, because, in mPER1/2-null SCN carrying a mPer1-luc reporter, forskolin triggered a >50% increase in bioluminescence that persisted for several days (Fig. S3B). Thus, cAMP-dependent signals can be activated in mPER1/2-null SCN, so the absence of cAMP-dependent suppression of mCry1 was likely caused by the absence of cAMP-inducible PER proteins to act upon E/E′-boxes of the mCry1 reporter.
Having demonstrated the contribution of PER proteins to circadian/cAMP control of mCry1 expression, we tested whether mPER1/2 are the only transducers for mCry1. To address this, we applied our SCN graft procedure (4), which previously highlighted VPAC2-dependent and -independent mechanisms for synchronization of circadian mPER2 expression. To confirm that SCN grafts can restore circadian mCry1-luc expression, VIP-null slices were recorded by PMT for approximately 10 d, by which time mCry1 rhythms had damped. A WT SCN was then grafted onto the mutant host. As seen previously with the mPER2::LUC reporter, the graft restored more coherent and higher high-amplitude mCry1-driven bioluminescence in the host SCN (Fig. S3C), confirming the earlier conclusion that extracellular cues are able, indirectly, to control mCry1 circadian expression. We then tested the effect of grafting onto mPER1/2-null SCN. As noted earlier, these slices were arrhythmic before grafting. In slices that did not receive a graft (n = 3), residual rhythms were of very low amplitude (3.39 ± 0.62) and poorly organized (RAE, 0.288 ± 0.037; Fig. 5B). Forty-eight hours after grafting of a WT SCN onto the host, clear circadian rhythmicity was restored (period, 23.64 ± 0.31 h; n = 6). Graft-driven rhythmicity was of higher amplitude (39.06 ± 5.29) and better organized (RAE, 0.109 ± 0.043) than in nongrafted slices, and continued for the duration of recording (>10 d; all P < 0.01). Thus, in the absence of mPER1 and mPER2, circadian expression of mCry1 can be restored by extracellular cues. Importantly, the resulting oscillations were of lower amplitude than the spontaneous rhythms of WT SCN noted earlier (403 ± 49), and were less well defined (WT RAE, 0.050 ± 0.004). Nevertheless, our results reveal the contribution of PER-independent as well as PER-dependent mechanisms to circadian expression of mCry1.
Discussion
mCRY1 is the principal negative regulator within the feedback loops that define circadian time (2, 17), so description of the dynamics of its expression in the SCN is central to understanding the timekeeping mechanism. This study therefore developed a bioluminescent reporter mouse with which to monitor E-box–dependent circadian regulation of mCry1 to extend our view of the molecular clockwork beyond the existing Per-based constructs. We then used the mouse to investigate the mechanisms that direct circadian expression of mCry1, in particular to determine how mCry1 expression is synchronized within and between SCN neurons. The B6.Cg-Tg(Cry1-luc)01Ld line provided a high-fidelity, real-time report of SCN mCry1 expression, with appropriate phase, period, and profile. CCD imaging showed that a coordinated spatiotemporal wave of expression is a feature of both components of the negative feedback axis: Cry as well as Per. Despite lacking CRE sequences, mCry1 expression was dependent on VIP/VPAC2 interneuronal cues and intraneuronal cAMP signaling, highlighting indirect circadian regulation. Moreover, mPER1 and mPER2 proteins were necessary transducers for circadian and cAMP-mediated regulation of mCry1. By using an SCN graft procedure, we showed that PER-independent as well as PER-dependent mechanisms sustain circadian expression of mCry1. The B6.Cg-Tg(Cry1-luc)01Ld mouse therefore provides a unique view upon mCry1 expression and its interaction with PER at the heart of the circadian pacemaker.
Although overall bioluminescence levels were not different between mCry1-luc and mPER2::LUC SCN, the amplitude of oscillation was smaller in mCry1-luc tissue, as noted in vivo (17). Furthermore, consistent with the previous validation of the mCry1 sequence for circadian reporting in fibroblasts (13), the B6.Cg-Tg(Cry1-luc)01Ld reporter was useful for the analysis of circadian expression in peripheral tissues and MEFs, albeit less well defined than the mPER2::LUC reporter. Importantly, the principal circadian elements of the mCry1-luc reporter are the E-box and E′-box (13), which provide a readout for activation within the central circadian feedback loop. In addition, the E′-box incorporates two overlapping D-box motifs (14), which may moderately tune its activity in terms of phase and amplitude. The reporter does not, however, contain the intronic RORE sequences, which have been shown in cell-based assays to be capable of conferring a marked phase delay of approximately 4 h to the oscillation (14). The E-boxes, on the contrary, were sufficient to recapitulate in the SCN slice a phase of circadian mCry1 expression, relative to mPer1, that replicated their in vivo expression. Thus, whereas the intronic ROREs are reported as necessary to determine mCry1 phase in cells (14) and peripheral tissues (24; but see ref. 13), they were clearly not required for appropriate phase control in the SCN. The effects of Fbxl3Afh, in which CRY protein degradation is impaired, further emphasized the fidelity of the reporter as a readout of E-box–mediated gene expression. mCry1-luc reported the molecular signature of the Afh mutant mice: prolonged negative feedback onto the E/E′-boxes arising from stabilized mCRY.
The reporter provided explicit insights into the role of interneuronal communication in driving the core clockwork. Previous analyses have relied on Per-based reporters, but these carry both E-boxes and CREs, so activity arising from acute interneuronal signals (via CREs) would confound analysis of sustained circadian cycles (via E-boxes). CCD recording revealed that mCry1 activation followed a spatiotemporal wave that was initiated at the dorsomedial lip of the SCN and processed ventrally and laterally (11). Although this wave is initiated by localized cAMP-dependent signals (25), consistent with actions on Per expression, the present study shows that the wave is shared by other components of the feedback loop. Future studies with the mCry1-luc mouse will seek to identify the dependence of the wave on PER-dependent signals. Thus, different spatially specified neuronal populations of the SCN encode temporally specific oscillations, adding a further layer of circuit-level complexity to the control of E-box–dependent SCN outputs. The role of interneuronal cues in specifying this phase structure was evident in the progressive damping of circadian mCry1 expression and loss of cellular synchrony in VIP- and VPAC2-null SCN. Previous in situ hybridization assays of steady-state mCry1 expression in the VPAC2-null SCN showed little change in mean expression level but a loss of rhythmicity (26). The reporter now shows that loss of cellular synchrony of mCry1 expression is the cause of this phenotype. It also shows that VPAC2-dependent signaling has access into the core clockwork beyond the CREs of Per, as highlighted by the restoration of mCry1 rhythms in mPER1/2-null SCN by grafts of WT SCN. Importantly, this restoration demonstrates that the paracrine mechanisms that drive circadian-incompetent SCN (4) really can address the entire oscillatory system, rather than simply stimulating the Per-based reporters via their CREs but having no downstream consequences.
Sensitivity to manipulations of AC/cAMP revealed an indirect pathway whereby VIP/VPAC2 and other interneuronal signals can direct mCry1 rhythms. The serial responses of mPER2::LUC and mCry1 to forskolin provided correlative evidence that PER proteins act as transducers in such a pathway, their elevation subsequently suppressing E-box activation of mCry1. Evidence of a necessary role was seen in mPER1/2-null SCN: mCry1 expression damped rapidly and activation of cAMP signaling no longer suppressed mCry1. It should be noted, however, that the mPER1 and mPER2 proteins are not the sole means of extracellular control over circadian mCry1 expression because a WT graft restored circadian expression of mCry1 in mPER1/2-null SCN. Although we did not specifically address any contribution from PER3, this is an unlikely mediator of this effect because it is neither necessary nor sufficient for circadian function; it does not carry CREs and is not acutely regulated by extracellular cues (23). If PER-independent regulation of mCry1 is mediated by other components of the feedback loop, DBP, acting via the D-boxes (14), is a candidate, although, as with mPer3, it does not carry CREs. In contrast, DEC-1, a factor that suppresses CLOCK/BMAL1 activity at E-boxes, does carry CREs (27). Alternatively, metabolic, redox-dependent cues may play a role. It is now well established that the transcriptional loops are intermeshed with cycles of cellular metabolism (28), and the SCN slice in culture expresses a marked circadian cycle of superoxidation of peroxiredoxin proteins (29). Whether such metabolic cues are capable of driving the core loop in the absence of PER proteins remains to be determined, but there is growing evidence that cytosolic processes can cycle independently of, and influence the behavior of, the transcriptional feedback loops (30). Transient circadian cycles were evident in mPER1/2-null SCN in the present study, and have also been reported in BMAL1- (31) and CRY1/2-null SCN (4), indicative of ongoing cytosolic oscillations. Circadian control of mCry1 by the SCN graft in the mPER1/2-null SCN may involve such “cytoscillators” (30). In conclusion, therefore, the creation and subsequent analysis of the B6.Cg-Tg(Cry1-luc)01Ld mouse has provided a unique perspective on inter- and intracellular pathways (Fig. S4) that act on the fulcrum of the core circadian pacemaker: E-box–mediated expression of the principal negative regulator, mCRY1 (2).
Materials and Methods
All animal work was licensed under the UK Animals (Scientific Procedures) Act of 1986 with local ethical approval. To create transgenic mice we used a mCry1 sequence (−1,504 to +107; Fig. S1A) equivalent to ovine sequence previously shown to sustain circadian bioluminescence when transfected as a plasmid into fibroblasts (13). The transgenic fragment was excised by using EcoR1 and Sal1 and injected into the pronuclei of fertilized eggs (B6CBAF1/OlaHsd strain). Six potential founder mice were identified by PCR analysis, whereby a 250-bp band from the luciferase transgene was amplified with primers 5′ CTT CAG AAA CGT GAG GTG CCG 3′ and 5′ AGC GTA AGT GAT GTC CAC CTC G 3′. One line was selected for further use and crossed with WT and circadian mutant lines from our in-house colonies all with a C57Bl6 background C57Bl6 (more than six back-crosses to C57Bl6). Copy number was determined by QX100 Droplet Digital PCR (Bio-Rad). SCN organotypic slices were routinely made from 5- to 12-d-old pups, and bioluminescence recorded after at least 7 d in culture by using photomultiplier arrays or CCD camera as described previously (21). Luciferin-EF (Promega) was added at an initial concentration of 0.1 μM. Peripheral tissue explants were recorded immediately under PMTs, and primary fibroblasts were prepared from mCry1-luc and mPER2::LUC embryos (embryonic day 13–14) and maintained in DMEM supplemented with 10% (vol/vol) FBS before plating in 35-mm dishes at ∼1 × 106 cells per dish in Hepes-buffered medium (20). Data were analyzed by using IPLab and Oriana software (4) and the FFT–nonlinear least squares algorithm, accessed through the BioDare database (www.biodare.ed.ac.uk). The first 24 h of data were discarded to avoid experimental artifacts. For single-cell analysis, CCD images were converted to 8-bit image stacks by using ImageJ (National Institutes of Health) and analyzed by using Semiautomated Routines for Functional Image Analysis plug-in component within Igor-Pro (Wavemetrics) data analysis software. Single cells were identified by the automated region of interest (ROI) selection tool. Bioluminescence intensity in grayscale values was extracted from all ROIs above 0.4 threshold and 6 μm diameter, to select for “cell-like” ROIs. Bioluminescence intensity units were generated for each ROI through the image stack and visualized in a raster plot. Immunofluorescent staining with goat anti-luciferase (1:1,000; Promega), rabbit anti–[Arg8]-vasopressin, and guinea pig anti-VIP (1:1,000; Bachem) and graft cocultures were conducted as described previously (4, 20). Drugs (forskolin, IBMX, and MDL-12,330A) were obtained from Sigma-Aldrich.
Supplementary Material
Acknowledgments
We thank Dr. Tomasz Zielinski for expert assistance with BioDare, Mr. P. Margiotta for assistance with figures, and LMB Biomedical Services Group for animal care. This work was supported by Medical Research Council (United Kingdom). BioDare is supported by Biotechnology and Biological Sciences Research Council and Engineering and Physical Sciences Research Council Awards BB/D019621, BB/F59011/1, and BB/F005237/1 to SynthSys and ROBuST (University of Edinburgh).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220894110/-/DCSupplemental.
References
- 1.Hastings MH, Reddy AB, Maywood ES. A clockwork web: Circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4(8):649–661. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- 2.Koike N, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338(6105):349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ueda HR, et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37(2):187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 4.Maywood ES, Chesham JE, O’Brien JA, Hastings MH. A diversity of paracrine signals sustains molecular circadian cycling in suprachiasmatic nucleus circuits. Proc Natl Acad Sci USA. 2011;108(34):14306–14311. doi: 10.1073/pnas.1101767108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.An S, Irwin RP, Allen CN, Tsai C, Herzog ED. Vasoactive intestinal polypeptide requires parallel changes in adenylate cyclase and phospholipase C to entrain circadian rhythms to a predictable phase. J Neurophysiol. 2011;105(5):2289–2296. doi: 10.1152/jn.00966.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Güler AD, et al. Melanopsin cells are the principal conduits for rod-cone input to non-image-forming vision. Nature. 2008;453(7191):102–105. doi: 10.1038/nature06829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hastings MH, et al. Entrainment of the circadian clock. Prog Brain Res. 1996;111:147–174. doi: 10.1016/s0079-6123(08)60406-9. [DOI] [PubMed] [Google Scholar]
- 8.Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc Natl Acad Sci USA. 2002;99(11):7728–7733. doi: 10.1073/pnas.102075599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shigeyoshi Y, et al. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell. 1997;91(7):1043–1053. doi: 10.1016/s0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- 10.Reddy AB, Field MD, Maywood ES, Hastings MH. Differential resynchronisation of circadian clock gene expression within the suprachiasmatic nuclei of mice subjected to experimental jet lag. J Neurosci. 2002;22(17):7326–7330. doi: 10.1523/JNEUROSCI.22-17-07326.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamaguchi S, et al. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science. 2003;302(5649):1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- 12.Yoo SH, et al. PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101(15):5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fustin JM, O’Neill JS, Hastings MH, Hazlerigg DG, Dardente H. Cry1 circadian phase in vitro: wrapped up with an E-box. J Biol Rhythms. 2009;24(1):16–24. doi: 10.1177/0748730408329267. [DOI] [PubMed] [Google Scholar]
- 14.Ukai-Tadenuma M, et al. Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell. 2011;144(2):268–281. doi: 10.1016/j.cell.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 15.Meng QJ, et al. Setting clock speed in mammals: The CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron. 2008;58(1):78–88. doi: 10.1016/j.neuron.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Godinho SI, et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316(5826):897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- 17.Kume K, et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98(2):193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 18.Field MD, et al. Analysis of clock proteins in mouse SCN demonstrates phylogenetic divergence of the circadian clockwork and resetting mechanisms. Neuron. 2000;25(2):437–447. doi: 10.1016/s0896-6273(00)80906-x. [DOI] [PubMed] [Google Scholar]
- 19.O’Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science. 2008;320(5878):949–953. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hastings MH, Reddy AB, McMahon DG, Maywood ES. Analysis of circadian mechanisms in the suprachiasmatic nucleus by transgenesis and biolistic transfection. Methods Enzymol. 2005;393:579–592. doi: 10.1016/S0076-6879(05)93030-9. [DOI] [PubMed] [Google Scholar]
- 21.Maywood ES, et al. Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr Biol. 2006;16(6):599–605. doi: 10.1016/j.cub.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 22.Atkinson SE, et al. Cyclic AMP signaling control of action potential firing rate and molecular circadian pacemaking in the suprachiasmatic nucleus. J Biol Rhythms. 2011;26(3):210–220. doi: 10.1177/0748730411402810. [DOI] [PubMed] [Google Scholar]
- 23.Bae K, et al. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30(2):525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 24.Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421(6919):177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- 25.Doi M, et al. Circadian regulation of intracellular G-protein signalling mediates intercellular synchrony and rhythmicity in the suprachiasmatic nucleus. Nat Commun. 2011;2:327. doi: 10.1038/ncomms1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harmar AJ, et al. The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell. 2002;109(4):497–508. doi: 10.1016/s0092-8674(02)00736-5. [DOI] [PubMed] [Google Scholar]
- 27.Honma S, et al. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419(6909):841–844. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- 28.Bass J. Circadian topology of metabolism. Nature. 2012;491(7424):348–356. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- 29.Edgar RS, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485(7399):459–464. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hastings MH, Maywood ES, O’Neill JS. Cellular circadian pacemaking and the role of cytosolic rhythms. Curr Biol. 2008;18(17):R805–R815. doi: 10.1016/j.cub.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 31.Ko CH, et al. Emergence of noise-induced oscillations in the central circadian pacemaker. PLoS Biol. 2010;8(10):e1000513. doi: 10.1371/journal.pbio.1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.