Abstract
The ubiquitin conjugation system plays an important role in immune regulation; however, the ubiquitin-specific proteases (USPs) that carry out deubiquitination of cellular substrates are poorly understood. Here we show that in vivo knockdown of the deubiquitinating enzyme USP9X attenuates T-cell proliferation. In addition, naïve CD4+ T cells from USP9X knockdown chimeric mice display decreased cytokine production and T helper cell differentiation in vitro, which we confirmed in vivo by performing adoptive transfer of transgenic T cells and subsequent immunization. USP9X silencing in both a human T-cell line and mouse primary T cells reduced T-cell receptor (TCR) signaling-induced NF-κB activation. Mechanistically, USP9X interacts with Bcl10 of the Carma1-Bcl10-Malt1 (CBM) complex and removes the TCR-induced ubiquitin chain from Bcl10, which facilitates the association of Carma1 with Bcl0-Malt1. These results demonstrate that USP9X is a crucial positive regulator of the TCR signaling pathway and is required for T-cell function through the modulation of CBM complex formation.
Keywords: signal transduction, posttranslational modification
Protein ubiquitination is a fundamental mechanism for regulating various cellular processes, including signal transduction and transcriptional regulation (1). This process involves the attachment of ubiquitin to a target protein by a three-step enzymatic cascade, consisting of ubiquitin-activating (E1), ubiquitin-conjugating (E2), and ubiquitin ligase (E3) enzymes (2). The ubiquitin conjugation process is reversible, in which the attached ubiquitin chain can be removed by deubiquitinating enzymes (DUBs), consisting of members of a protease superfamily (3). Approximately 95 putative DUBs have been identified by a human genome database search, which can be subdivided into five subclasses based on their ubiquitin protease domains: ubiquitin-specific protease (USP), ubiquitin C-terminal hydrolase (UCH), otubain protease (OTU), Machado–Joseph disease protease (MJD), and JAB1/MPN/Mov34 metalloenzyme (JAMM) (4).
Although E3 ubiquitin ligases have been well studied in the immune system, the targets and physiological functions of most DUBs remain unknown. Previous studies have shown that the OTU domain-containing DUB A20 and the USP domain-containing DUB cylindromatosis protein negatively regulate NF-κB activation through deubiquitination of signaling molecules, such as RIP and TRAF2, in response to innate immune stimulation (5–7). In adaptive immunity, this is thought to involve ubiquitin-mediated regulation of key components of the T-cell receptor (TCR) signaling machinery, including the Carma1-Bcl10-Malt1 (CBM) complex (8) and Tak1 (9). How the deubiquitination system regulates the TCR-induced signaling pathway and T-cell function remains largely unclear, however.
TCR-induced NF-κB activation is critical for T lymphocyte activation (10). The CBM complex, a core signaling complex of the NF-κB activation process, regulates IκB kinase (IKK) complex activation after TCR ligation. On activation by the TCR, the scaffold protein Carma1 associates with the Bcl10-Malt1 complex in a signal-dependent manner after phosphorylation by protein kinase Cθ and is subsequently recruited into the lipid raft by phosphoinositide-dependent kinase 1 (11). This triggers TRAF6, which is thought to bind to Malt1 to promote K63-linked ubiquitination of Bcl10, thereby leading to ubiquitin-dependent activation of the IKK complex (12–14). Bcl10 is a caspase-recruitment domain (CARD)-containing adaptor protein that binds constitutively to Malt1 through both the carboxyl-terminal part of CARD motif and the adjoining 13 amino acids (107–119) (15–18). Even though Bcl10 ubiquitination is a pivotal mechanism in the regulation of NF-κB activation, our understanding of how the interaction among CBM complex components controls Bcl10 ubiquitination and its consequences on downstream IKK activation or, in reverse, whether Bcl10 ubiquitination regulates CBM complex formation remains limited. Furthermore, the involvement of DUBs in the deubiquitination of CBM complex in the TCR-induced NF-κB activation pathway is far from clear.
USP9X is a USP domain-containing DUB that was initially identified as a human homolog of Drosophila fat facets (faf) gene (19). Recent studies have reported that USP9X plays important role in the regulation of cell survival and TGF-β signaling by deubiquitinating myeloid leukemia cell differentiation protein 1 (Mcl1), Itch, and Smad4 (20–22); however, the function of USP9X in the immune system, particularly in T-cell regulation, remains unknown. In this study, we identified USP9X as a critical positive regulator of TCR signaling pathway and characterized the function of USP9X by genetic, molecular, and immunologic studies. Our study reveals an ubiquitin-dependent mechanism for regulation of the CBM complex by USP9X, leading to NF-κB activation in peripheral T cells, and thus suggests a possible function of USP9X in T-cell–mediated inflammatory responses.
Results
Phenotypic Analysis of USP9X Knockdown Chimeric Mice.
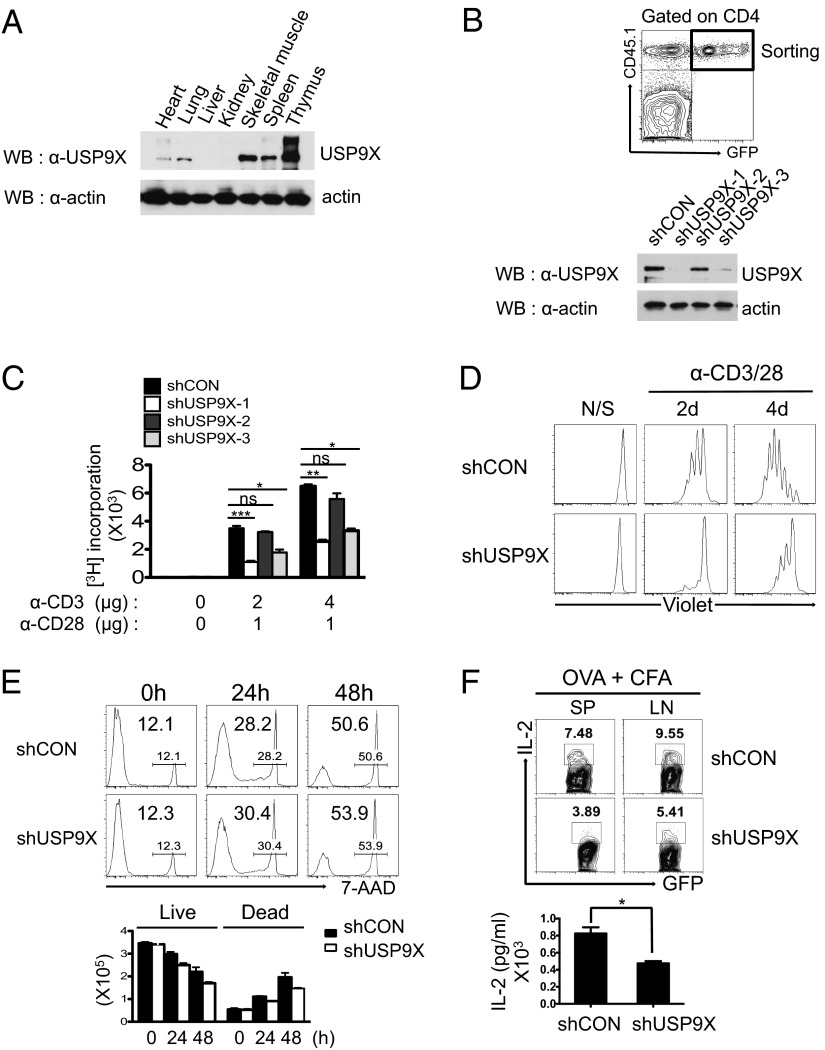
To begin to understand the biological function of USP9X in the immune system, we first examined the endogenous expression of USP9X by Western blot analysis in various mouse tissue extracts. USP9X was highly expressed in both the spleen and the thymus (Fig. 1A). This suggests that USP9X may play a role in the regulation of immune responses, especially T-cell function. We then generated shRNAs directed against USP9X and performed retroviral transduction in bone marrow from CD45.1+ mice and transplantation into lethally irradiated CD45.2+ recipient mice, to investigate the functional involvement of USP9X in T cells. Newly reconstituted CD4+ T cells (CD45.1+GFP+) were sorted from spleens of bone marrow chimeric mice, and the reduced USP9X expression was verified by immunoblotting in each USP9X shRNA-expressing CD4+ T cells (Fig. 1B).
Fig. 1.
Characterization of USP9X-deficient T cells from bone marrow chimeric mice. (A) Endogenous USP9X protein level was measured by immunoblotting with anti-USP9X antibody in various mouse tissues. The membrane was reprobed with anti-actin to reveal equivalent protein loading. (B) (Upper) Analysis of GFP expression in peripheral blood from bone marrow chimeric mice at 2 mo after reconstitution of USP9X shRNAs expressing bone marrow cells. (Lower) Immunoblot analysis of USP9X was performed in a sorted GFP+ CD4+ T-cell population. The data are representative of between 3 and 10 independent experiments. (C) CD4+ T cells were sorted from USP9X shRNAs expressing chimeric mice and stimulated with anti-CD3 + anti-CD28 for 36 h. Cell proliferation was measured by 3H-thymidine uptake. The data are compiled from between 3 and 10 independent experiments. Error bars indicate mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001, two-tailed unpaired t test. (D) Violet dye-labeled CD4+ T cells were stimulated with anti-CD3 + anti-CD28 for 2 and 4 d, after which cell division was analyzed by flow cytometry. The data are representative of three independent experiments. (E) Cell death of stimulated CD4+ T cells was examined by 7-AAD staining and analyzed by flow cytometry (Upper) and total cell counts (Lower). The data are representative of three independent experiments. (F) IL-2 production by OVA-specific CD4 T cells. CD4+ T cells from OT-II control or OT-II USP9X knockdown chimeric mice were adoptively transferred into WT C57BL/6J mice, followed by immunization with OVA plus CFA as an adjuvant. Splenocytes and lymph node cells obtained 6 d later were stimulated with OVA323–339 peptide for 24 h and analyzed by intracellular staining and flow cytometry (Upper) and ELISA (Lower). The data are representative of and compiled from two independent experiments (n = 2 per each experiment). Error bars indicate mean ± SD. *P < 0.05, two-tailed unpaired t test.
We next examined the effect of USP9X knockdown on T-cell function. shUSP9X-1– and shUSP9X-3–expressing CD4+ T cells, which exhibited efficient reduction of USP9X, showed less proliferation than control CD4+ T cells, whereas shUSP9X-2–expressing CD4+ T cells showed a moderate decrease in proliferation in response to anti-CD3/CD28 stimulation, as measured by 3H-thymidine incorporation (Fig. 1C). Thus, significant reductions in TCR stimulation-induced IFN-γ production were seen in both shUSP9X-1– and shUSP9X-3–expressing CD4+ T cells, as analyzed by intracellular cytokine staining and ELISA (Fig. S1 A and B). These results clearly show that T-cell proliferation and cytokine production are regulated by USP9X expression.
To further examine the role of USP9X in T-cell response, we selected shUSP9X-1, which demonstrated the most efficient knockdown of USP9X (termed “shUSP9X” hereinafter), and analyzed individual cell division of violet dye-labeled CD4+ T cells by flow cytometry. Consistent with the results of proliferation analysis, USP9X-deficient CD4+ T cells divided much more slowly compared with control CD4+ T cells under the same stimulation conditions (Fig. 1D). Thus, flow cytometry and total cell counts of 7-aminoactinomycin D (7-AAD) stained CD4+ T cells revealed that USP9X deficiency apparently had no effect on T-cell survival (Fig. 1E).
Despite the defective proliferation seen in USP9X knockdown CD4+ T cells, decreased expression of USP9X had no effect on the development of CD4 or CD8 single-positive or CD4/CD8 double-positive T cells in the thymus (Fig. S2A). Similarly, the frequency of CD4+ and CD8+ T cells, B-cell:T-cell ratios, and expression of CD44 and CD62L cell surface markers in CD4+ T cells in peripheral lymphoid tissues were quite similar in control and USP9X knockdown mice (Fig. S2 B–D).
To examine the potential effect of USP9X on IL-2 production, we generated ovalbumin (OVA)-specific, MHC class II restricted TCR (OT-II) transgenic control or OT-II USP9X knockdown chimeric mice and performed adoptive transfer and immunization experiments. The reduction of IL-2 production was observed upon stimulation with OVA323–339 peptide in USP9X knockdown OT-II T cells, as revealed by intracellular cytokine staining and ELISA (Fig. 1F). This defect was further confirmed by in vitro acute knockdown of USP9X in OT-II CD4 T cells (Fig. S3A). However, both control and USP9X deficient CD4+ T cells showed appropriate expression of CD69 and CD25 activation markers to a similar degree (Fig. S3B).
USP9X Is Required for Th Cell Differentiation.
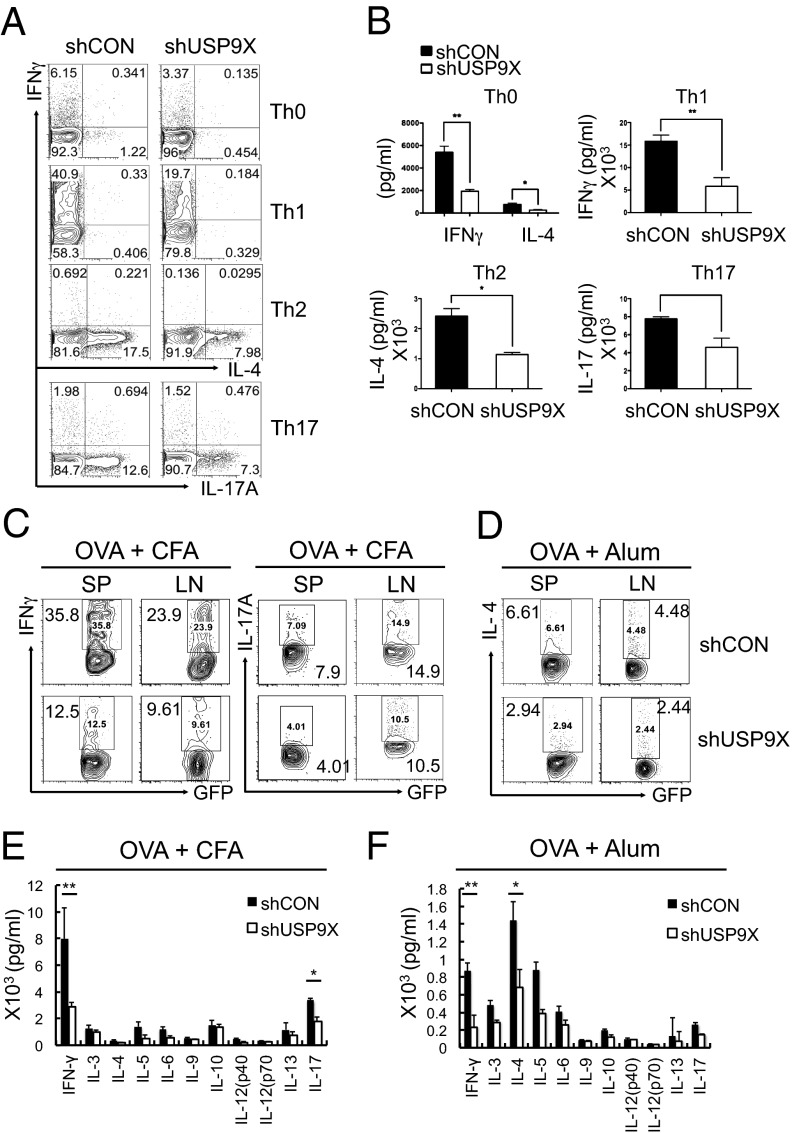
To determine whether USP9X knockdown affects the differentiation of naïve T cells into effector T helper (Th) cell subsets, we sorted control and USP9X-deficient naïve CD4+ T cells from chimeric mice and cultured them under nonpolarizing conditions for Th-neutral cells, or polarizing conditions for Th1, Th2, or Th17, with coincubation of appropriate cytokines and cytokine antibodies for 5 d, followed by restimulation with anti-CD3 and anti-CD28. Intracellular cytokine staining for IFN-γ, IL-4, and IL-17 showed that cytokine production by these Th subsets were decreased substantially under both nonpolarizing (neutral) and polarizing conditions (for Th1, Th2, and Th17) in USP9X knockdown T cells (Fig. 2A). This defect in Th cell differentiation was further confirmed by measuring cytokine secretion by ELISA; IFN-γ, IL-4, and IL-17 concentrations were markedly reduced in the culture supernatants of USP9X knockdown T cells (Fig. 2B). These results indicate that CD4+ T-cell differentiation into Th cells requires USP9X in vitro.
Fig. 2.
Defective Th cell differentiation of USP9X-deficient T cells. (A) Sorted GFP+CD4+CD62L+CD25− naïve CD4+ T cells from control (shCON) and USP9X shRNA (shUSP9X)-expressing chimeric mice were stimulated with anti-CD3 + anti-CD28 and cultured under different polarizing conditions for Th-neutral, Th1, Th2, or Th17 for 5 d. Cells producing IFN-γ, IL-4, and IL-17 were analyzed by intracellular cytokine staining (ICCS) at 6 h after restimulation with anti-CD3 + anti-CD28. The data are representative of five independent experiments. (B) IFN-γ, IL-4, and IL-17 production were measured by ELISA at 24 h after restimulation. The data are compiled from five independent experiments. Error bars indicate mean ± SD. *P < 0.05;**P < 0.01, two-tailed unpaired t test. (C and D) CD4+ T cells from OT-II control or OT-II USP9X knockdown chimeric mice were adoptively transferred into WT C57BL/6J mice, followed by immunization with OVA plus CFA (C) or alum (D) as adjuvants. Splenocytes and lymph node cells obtained 6 d later were stimulated and analyzed as described in Fig. 1F. The data are representative of three independent experiments. (E and F) Cytokine expression profile in C57BL/6J mice immunized as in C and D. CD4+ T cells obtained at 6 d after immunization were stimulated with OVA323–339 peptide for 48 h, and the culture supernatants were analyzed by a bioplex multicytokine assay. The data are compiled from three independent experiments. Error bars indicate mean ± SD. *P < 0.05; **P < 0.01, two-tailed unpaired t test.
Next, to investigate the biological relevance of USP9X in T cells, we examined the effect of USP9X knockdown using in vivo mouse models. We isolated CD4+ T cells from ovalbumin (OVA)-specific OT-II transgenic chimeric mice of control or USP9X knockdown group and adoptively transferred into WT C57BL/6J mice. These mice were then immunized with OVA peptide plus complete Freund’s adjuvant (CFA) as adjuvants 1 d after adoptive transfer, and examined for OVA-specific T cells by intracellular cytokine staining. OVA-induced IFN-γ– or IL-17A–producing T cells were greatly reduced in recipients of USP9X knockdown OT-II T cells (Fig. 2C). Similar reduction was observed in IL-4 production when recipient mice of USP9X knockdown OT-II T cells were immunized with OVA plus alum as adjuvants (Fig. 2D).
To further investigate the in vivo function of USP9X in T cells, we examined the cytokine profiles in T cells of the immunized mice using a bioplex multicytokine assay. Marked reductions were found in the levels of IFN-γ, IL-4, IL-5, IL-6, IL-13, and IL-17 production in USP9X knockdown T cells from OVA + CFA (Fig. 2E) or alum (Fig. 2F) immunized mice. These results demonstrate that USP9X is necessary for the differentiation of Th1, Th2, and Th17 cells in vivo.
USP9X Regulates NF-κB Activation During TCR Signaling.
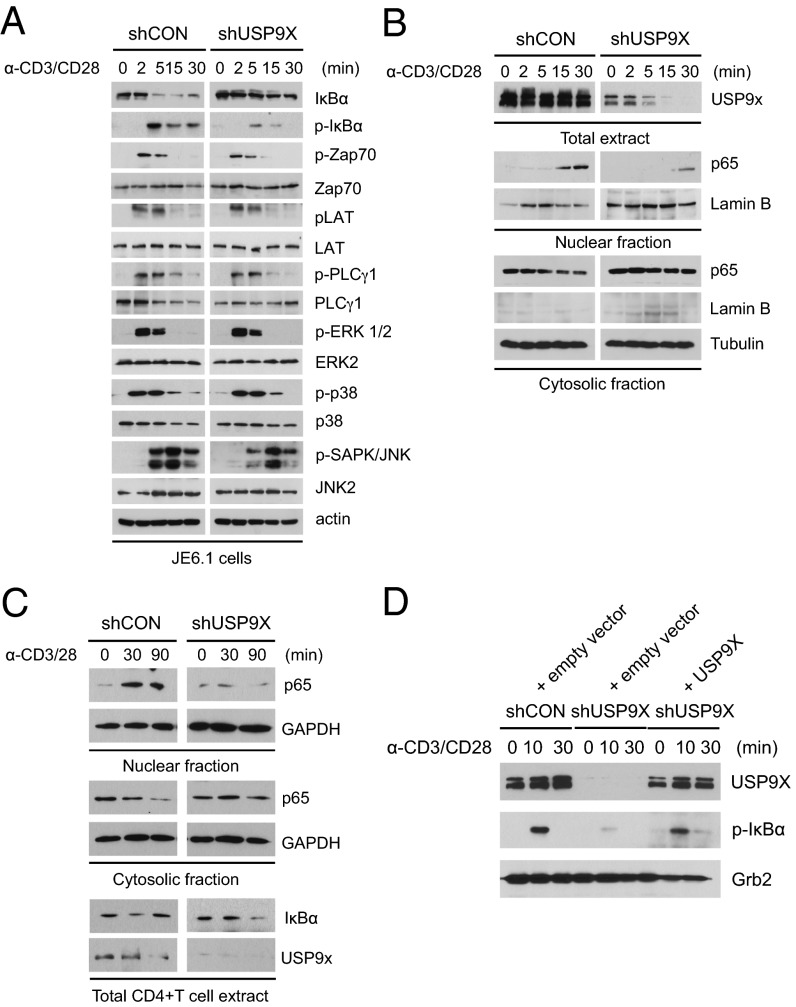
To define the molecular mechanism of USP9X in TCR signaling event, we established a Jurkat E6.1 (JE6.1) cell line with stable expression of control or USP9X shRNA using lentiviral transduction, and analyzed TCR-induced activation of both upstream and downstream signaling molecules. The phosphorylation of Zap-70, LAT, PLCγ1, ERK1/2, and p38 were quite similar between control and USP9X knockdown JE6.1 cells, with the exception of c-Jun N-terminal kinase (JNK), which seemed to be slightly less phosphorylated in USP9X knockdown cells (Fig. 3A). Notably, loss of USP9X in JE6.1 cells was associated with marked impairment of both phosphorylation and degradation of IκBα. Consistent with this result, translocation of the NF-κB subunit p65 into the nucleus was reduced in response to CD3/CD28 stimulation (Fig. 3B), demonstrating that USP9X is required for NF-κB activation by modulating upstream signaling molecules of the NF-κB pathway, but not TCR proximal or other downstream signaling events, such as Erk or p38 activation.
Fig. 3.
Impaired TCR/CD28-induced NF-κB activation in USP9X-deficient T cells. (A) Analysis of the phosphorylation status of TCR signaling proteins. Control and USP9X shRNA-expressing JE6.1 cells were stimulated with anti-CD3 + anti-CD28 (α-CD3/28) for the indicated times, after which the cell lysates were subjected to immunoblotting with the indicated antibodies. Anti-actin blot was used as a loading control. (B) Immunoblot analysis of translocation of p65 to the nucleus in control and USP9X shRNA-expressing JE6.1 cells treated as in A. After stimulation, cell lysates were separated into nuclear and cytosolic fractions. (C) Immunoblot analysis of translocation of p65 to the nucleus in mouse CD4+ T cells. Sorted Naïve CD4+ T cells from control and USP9X knockdown chimeric mice were stimulated with anti-CD3 + anti-CD28 for the indicated times, and nuclear fractionation was performed as in B. The protein content of IκBα was detected in the total cell lysates (Lower). The data are representative of three independent experiments. (D) Immunoblot analysis of phosphorylation of IκBα in USP9X shRNA-expressing JTAg cells reconstituted with USP9X. USP9X shRNA-expressing JTAg cells were transfected with empty vector or USP9X and CD28 and stimulated with anti-CD3 + anti-CD28 for the indicated times, after which the cell lysates were subjected to immunoblotting with the indicated antibodies. The data are representative of two independent experiments.
To further confirm the effect of USP9X knockdown on NF-κB activation in mouse primary T cells, we purified naïve CD4+ T cells from control or USP9X knockdown chimeric mice and assessed the nuclear translocation of p65 on stimulation. Consistent with the results in JE6.1 cells, USP9X deficiency resulted in decreased translocation of p65 on stimulation in mouse primary T cells (Fig. 3C). In addition, degradation of IκBα was inhibited in USP9X knockdown mouse T cells after 30 min of stimulation, and the subsequent reexpression of IκBα was also reduced after 90 min of stimulation (Fig. 3C, Lower).
We next performed reconstitution of USP9X in USP9X knockdown Jurkat cell line carrying SV40 large T antigen (JTAg) cells to confirm the effect of USP9X on NF-κB activation. Impaired phosphorylation of IκBα in USP9X knockdown JTAg cells was restored by the expression of full-length USP9X in response to anti-CD3/CD28 stimulation (Fig. 3D). These results suggest that UPS9X regulates the NF-κB signaling pathway in T cells.
USP9X Binds to Bcl10 and Regulates CBM Complex Formation by Modulating Bcl10 Ubiquitination.
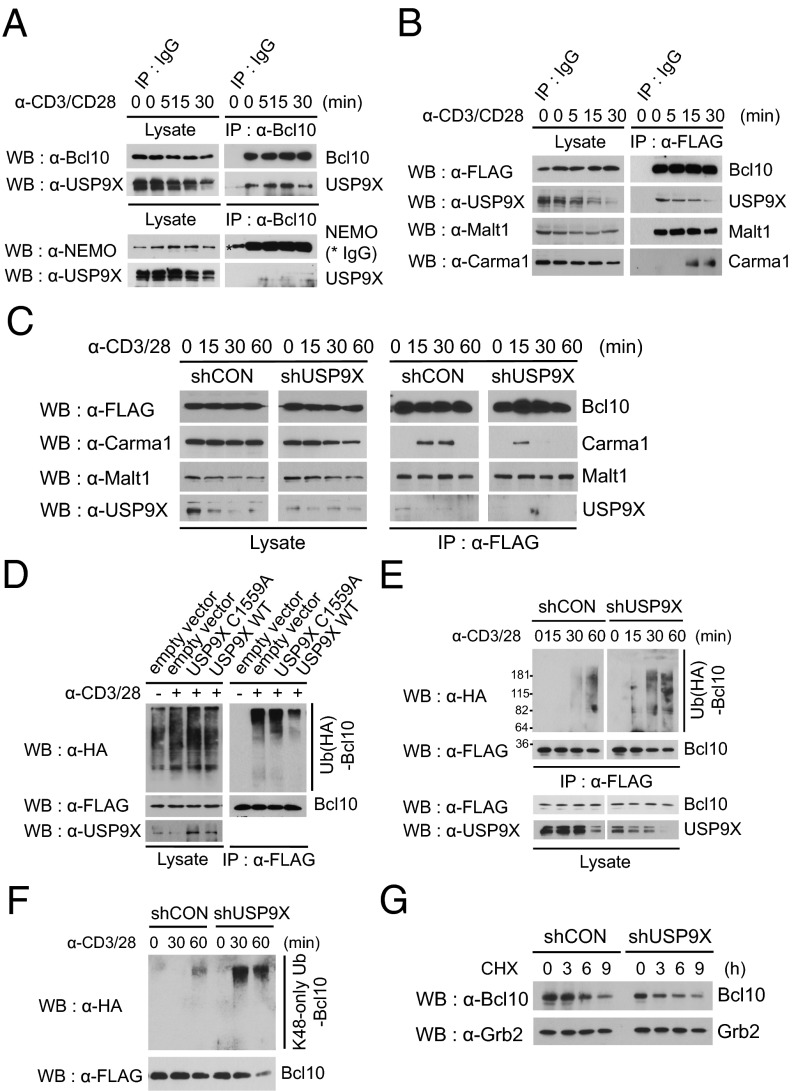
We investigated the specific mechanism through which USP9X regulates NF-κB signaling. The CBM complex is a critical mediator of TCR/CD28-triggered NF-κB activation in T cells (10, 23). Given that CBM complex-mediated NF-κB essential modulator (NEMO) ubiquitination is a major regulatory mechanism of the NF-κB activation process, we reasoned that USP9X deubiquitinase activity may act on either the CBM complex or NEMO. To test this hypothesis, we performed coimmunoprecipitation experiments in JE6.1 cells. Endogenous Bcl10 or NEMO was immunoprecipitated with anti-Bcl10 or NEMO antibodies, and coprecipitation of USP9X was monitored by immunoblotting with anti-USP9X antibody. USP9X was coimmunoprecipitated with Bcl10 constitutively in a TCR stimulation-independent manner (Fig. 4A, Upper); however, no apparent association was observed between USP9X and NEMO under either resting or stimulated conditions (Fig. 4A, Lower).
Fig. 4.
USP9X is required for CBM complex formation. (A) Interaction between endogenous USP9X and Bcl10. JE6.1 cells were stimulated with anti-CD3 + anti-CD28 for the indicated times, and endogenous Bcl10 or NEMO was immunoprecipitated with antibodies against Bcl10 or NEMO, respectively. The immunoprecipitates were separated by SDS/PAGE and immunoblotted with the indicated antibodies. (B) Interaction between USP9X and the CBM complex. JE6.1 cells expressing FLAG-Bcl10 were stimulated as in A, after which the FLAG-Bcl10 was immunoprecipitated with anti-FLAG antibody. The immunoprecipitates were then subjected to SDS/PAGE and immunoblotted with the indicated antibodies. (C) JE6.1 cells stably expressing both FLAG-Bcl10 and control or USP9X shRNA were stimulated with anti-CD3 + anti-CD28 for the indicated times, after which FLAG-Bcl10 was immunoprecipitated with anti-FLAG antibody. The immunoprecipitates were subjected to SDS/PAGE and immunoblotted with the indicated antibodies. (D) TCR-induced ubiquitination of Bcl10 in WT USP9X and catalytically inactive mutant (C1559A)-overexpressing cells. JTAg cells were transiently transfected with expression constructs for CD28, FLAG-Bcl10, HA ubiquitin, and USP9X WT or USP9X C1559A mutant. At 48 h after transfection, cells were left untreated or were stimulated with anti-CD3 + anti-CD28 for 20 min. Cell lysates were generated by adding SDS lysis buffer (2% SDS) and immunoprecipitated with anti-FLAG antibody. The immunoprecipitates were subjected to SDS/PAGE and analyzed by immunoblotting with the indicated antibodies. (E) TCR-induced ubiquitination of Bcl10 in control and USP9X shRNA-expressing cells. JTAg cells stably expressing control or USP9X shRNA were transfected with FLAG-Bcl10, HA ubiquitin, and CD28 and stimulated with anti-CD3 + anti-CD28 for the indicated times, and then analyzed as in D. The data are representative of three independent experiments. (F) TCR-induced K48-linked ubiquitination of Bcl10 in control and USP9X-shRNA expressing cells. JTAg cells stably expressing control or USP9X shRNA were transfected with FLAG-Bcl10, HA-K48–only ubiquitin, and CD28; stimulated with anti-CD3 + anti-CD28 for the indicated times; and then analyzed as in D. The data are representative of three independent experiments. (G) Stability of Bcl10 in JE6.1 cells expressing USP9X shRNA. Cells were stimulated with anti-CD3 + anti-CD28 and cycloheximide (50 μg/mL) was added for the indicated times. The cell lysates were subjected to immunoblotting with the indicated antibodies. Anti-Grb2 blot was used as a loading control. The data are representative of two independent experiments.
Owing to the technical obstacle of demonstrating endogenous CBM complex in JE6.1 cells, to further examine whether USP9X binds to Bcl10 in the CBM complex including Malt1 and Carma1, we established a JE6.1 cell line with stable expression of FLAG-Bcl10 using lentiviral transduction and immunoprecipitated Bcl10 with anti-FLAG beads. Consistent with previous studies (15, 16), Bcl10 interacted with Malt1 constitutively, but with Carma1 transiently after 15 min of stimulation with anti-CD3/CD28. Importantly, USP9X coimmunoprecipitated not only with Bcl10, but also with the CBM complex under stimulated conditions (Fig. 4B).
To further confirm that USP9X is biophysically a part of the CBM complex, we performed a purification and elution experiment using anti-FLAG beads, followed by silver staining and immunoblotting of the eluted protein mixture. USP9X was indeed detected in the eluted samples under both resting and stimulated conditions, whereas Carma1 was detected only in the stimulated sample (Fig. S4A). Furthermore, USP9X was coimmunoprecipitated with Myc-Malt (Fig. S4B). Taken together, these findings indicate that USP9X participates in the CBM complex through the interaction with Bcl10.
Next, to assess the role of USP9X on either the formation and/or the functional regulation of the CBM complex, we established JE6.1 cell lines stably expressing both FLAG-Bcl10 and control or USP9X shRNA using lentiviral transduction, and performed coimmunoprecipitation experiments in these cell lines. Knockdown of USP9X did not affect coprecipitation of Malt1 with FLAG-Bcl10 (Fig. 4C); however, T-cell stimulation-induced interaction of Carma1 with Bcl10 was reduced after 30 min of treatment in the USP9X knockdown T cells (Fig. 4C), suggesting that USP9X is required for CBM complex formation. We then examined whether the deubiquitinating enzymatic activity of USP9X is directly involved in the removal of polyubiquitin chains from Bcl10. JTAg cells were transiently transfected with CD28, FLAG-Bcl10, HA ubiquitin, and USP9X or its C1559A mutant, which contains an active site cysteine-to-alanine mutation at the 1559 position (24). Overexpression of the WT USP9X reduced ubiquitination of Bcl10 on TCR stimulation (Fig. 4D). However, the catalytically inactive USP9X C1559A mutant was not effective in removing the ubiquitin chain formation in Bcl10. Rather, it appeared to function as a dominant negative to enhance Bcl10 ubiquitination (Fig. 4D).
We then examined whether knockdown of USP9X expression could cause an increase in stimulation-induced Bcl10 ubiquitination by the loss of its deubiquitination. To test this possibility, we performed an in vivo ubiquitination assay in JTAg cell lines stably expressing control or USP9X shRNA. Both JTAg cells lines were transiently transfected with CD28, FLAG-Bcl10, and HA ubiquitin; left untreated or stimulated with anti-CD3/CD28; and then lysed in 2% SDS buffer and boiled, followed by FLAG-Bcl10 immunoprecipitation with anti-FLAG beads. Polyubiquitin chains conjugated to Bcl10 were detected by an anti-HA antibody. In USP9X shRNA-expressing cells, stimulation-induced Bcl10 ubiquitination was markedly augmented compared with control cells (Fig. 4E); however, USP9X knockdown did not affect the ubiquitination status of Malt1 under the same conditions (Fig. S5). Furthermore, although we observed an increase in lysine 48 (K48)-linked ubiquitination of Bcl10 in USP9X knockdown cells by transfection with HA-K48–only ubiquitin (Fig. 4F), Bcl10 stability the after inhibition of new protein synthesis with cycloheximide was similar in control and USP9X shRNA-expressing JE6.1 cells (Fig. 4G), likely related to nonproteasomal degradation of Bcl10 on TCR stimulation, as reported previously (25, 26). Indeed, TCR-induced Bcl10 degradation was not stabilized by MG132 treatment (Fig. S6A), and the K48-linked ubiquitination of Malt1-associated Bcl10 was not altered in the presence of MG132, as revealed by blotting with ubiquitin-K48 linkage-specific antibody (Fig. S6B). Consistent with the result of ubiqutination with K48-only ubiquitin (Fig. 4F), K48-linked Bcl10 ubiquitination was increased in USP9X knockdown cells, whereas K63-linked ubiquitination of Bcl10 was similar in control and USP9X knockdown T cells (Fig. S6C). Collectively, these results indicate that USP9X acts as a deubiquitinating enzyme for Bcl10 and functionally regulates TCR-triggered CBM complex formation.
Discussion
In the present study, using both in vivo and in vitro genetic and molecular approaches, we have characterized the role of the deubiquitinating enzyme USP9X as an important regulator of T-cell proliferation, cytokine production, and Th differentiation by controlling the TCR-induced NF-κB signaling pathway. At the molecular level, a constitutive interaction is observed between the CBM complex component Bcl10 and USP9X that is required for both binding of Carma1 to Bcl10-Malt1 and deubiquitination of Bcl10. Thus, the defect of USP9X-mediated CBM complex formation leads to impaired NF-κB activation in USP9X-ablated T cells. Based on these findings, we propose that USP9X acts as a positive regulator of the TCR-CD28 signaling pathway by modulating CBM complex assembly and subsequent TCR/CD28-induced NF-κB activation.
Ubiquitination is a key mechanism that operates at different stages of the sequential events in the CBM complex-mediated NF-κB activation pathway. K63-linked ubiquitination of Bcl10 triggered by TCR stimulation has been known to recruit IKK and subsequently activate NF-κB (12, 13). In addition to its nonproteolytic function in ubiqutination, E3 ligase (e.g., neural precursor cell expressed, developmentally downregulated 4; Itch; βTRCP)-mediated Bcl10 ubiquitination has been reported to induce Bcl10 degradation (26, 27), although which ubiquitin linkage is involved remains elusive. A more recent study suggested that K63-linked ubiqutination of Bcl10 is involved not only in NF-κB activation, but also in Bcl10 degradation by autophagy in effector T cells (25). In addition to K63-linked ubiquitination, K48-linked ubiquitination of Bcl10 has been reported, although the role of K48-linked ubiquitination on the regulation of Bcl10 remains unclear (13). We observed that USP9X plays an important role as a positive regulator in this process by interacting with and regulating the CBM complex. Interestingly, the constitutive Bcl10-Malt1 complex interacts with USP9X without TCR stimulation, suggesting that USP9X is a newly identified component of the Bcl10-Malt1 complex. Further molecular mechanistic studies in USP9X-deficient T cells revealed that binding of Carma1 to the Bcl10-Malt1 complex was reduced on TCR stimulation, accompanied by an increase in concurrent Bcl10 ubiquitination, which collectively leads to impaired NF-κB activation. Although the exact mechanisms by which USP9X modulates CBM complex formation are not clear, we suggest that USP9X maintains the integrity of the Bcl10-Malt1 complex by deubiquitinating Bcl10 that is conjugated with K48-linked or other lysine-linked ubiquitin chains, which may inhibit the accessibility of Carma1 to the Bcl10-Malt1 complex. Thus, TCR/CD28-induced oscillatory degradation of USP9X, which was revealed by the present study, proposes that USP9X is critically correlated to TCR-induced signaling, including Bcl10 regulation. Further study is needed to gain more insight into the mechanisms of USP9X function and regulation during TCR-induced signaling transduction.
In this study, we emphasize that USP9X regulates T-cell function by modulating the TCR-induced NF-κB activation pathway. Previous studies have demonstrated that USP9X has different target substrates in different cell types. One study showed that USP9X promotes cell survival by stabilizing Mcl1 in a deubiquitination-dependent manner in human lymphoma (20), whereas a more recent report showed that USP9X expression is inversely linked with pancreatic cancer by acting as a tumor suppressor (21). It seems that USP9X does not play a role in T-cell survival, as revealed in the present study. Nevertheless, these observations, together with our findings regarding the role of USP9X in T-cell regulation, raise the possibility that regulation of USP9X activity may be prognostically and therapeutically important in the management of human lymphomas and T-cell–mediated inflammatory diseases. Thus, further studies to determine the precise mechanism by which USP9X activity is regulated during T-cell activation may aid in the design of therapeutic approaches for human diseases.
Materials and Methods
Retroviral Transduction and Bone Marrow Reconstitution.
C57BL/6 mice were obtained from Jackson Laboratory, and B6 SJL (CD45.1) mice were obtained from Taconic. All mice were housed in specific pathogen-free conditions. The animal experiment protocols were approved by members of the Institutional Animal Care and Use Committee of the La Jolla Institute for Allergy and Immunology. To generate bone marrow chimeric mice expressing USP9X shRNAs (shUSP9X-1: 5′- TGCTGTTGACAGTGAGCGCGGTGCTAATCTCATTAAAGAATAGTGAAGCCACAGATGTATTCTTTAATGAGATTAGCACCTTGCCTACTGCCTCGGA-3′; shUSP9X-2: 5′- TGCTGTTGACAGTGAGCGCTTCCAGTGTGTCATACTATACTAGTGAAGCCACAGATGTAGTATAGTATGACACACTGGAAATGCCTACTGCCTCGGA-3′; shUSP9X-3: 5′- TGCTGTTGACAGTGAGCGAAGGGATGATGTGTTTGGATATTAGTGAAGCCACAGATGTAATATCCAAACACATCATCCCTGTGCCTACTGCCTCGGA-3′), Plat-E cells were transfected with 3 µg of LMP vector with 9 µL of TransIT-LT1 (Mirus), and the culture supernatant containing retrovirus was collected after 48 h. Mature T-cell–depleted bone marrow cells from B6 SJL (CD45.1) mice were cultured for 24 h in IL-3 (10 ng/mL), IL-6 (10 ng/mL), and stem cell factor (100 ng/mL) (all from Peptrotech) containing complete DMEM before initial retroviral infection. Mature T-cell–depleted bone marrow cells were infected with retrovirus together with 5 µg/mL polybrene by centrifuging cells at 420 × g for 60 min at room temperature. At 2 d after infection, retrovirally transduced bone marrow cells were injected into lethally irradiated (900 rad) C57BL/6 recipient mice. Recipient mice were killed at 8 wk after reconstitution and analyzed as described below.
Cell Proliferation and Division Analysis.
Purified CD4+ T cells (2 × 105 cells/200 μL) were plated in 96-well tissue culture plates with the indicated concentrations of plate-bound anti-CD3 (clone 145-2C11; BioLegend) and soluble anti-CD28 (clone 37.1; Bio-Xell). Proliferation of the last 12 h of a 48-h culture was detected by addition of 1 μCi/mL of 3H-thymidine, and cell-incorporated radiation was monitored by a β-plate counter. Data are presented as the mean value from triplicate wells. Cell division was analyzed by prelabeling T cells with 5 μM Cell Trace Violet (Molecular Probes) and stimulating them at a concentration of 2 × 106/mL with plate-coated anti-CD3 (2 μg/mL) and soluble anti-CD28 (1 μg/mL) for 72 h. Violet intensity was measured by flow cytometry.
Antibodies.
Antibodies to phospho-IκBα, p65, phospho-PLCγ, phospho-Zap70, Zap70, phospho-LAT (Ser473), LAT, phospho-Erk1/2, phospho-p38, p38, phospho-JNK, JNK2, Carma1, Ub-K48, and Ub-K63 were purchased from Cell Signaling Technology. Antibodies to IκBα, Erk2, Lamin B, Malt1, Bcl10, Grb2, ubiquitin, HA, and Myc were purchased from Santa Cruz Biotechnology. Anti-USP9X was purchased from Novagen, anti-FLAG was purchased from Sigma-Aldrich, and anti-actin was purchased from Millipore.
Multicytokine Assay.
Supernatants were collected and diluted for cytokine detection. Cytokines were detected with a multiplex cytokine kit (Bio-Rad) according to the manufacturer’s instructions.
Adoptive Transfer and Immunization.
CD4+ T cells from OT-II control or OT-II USP9X knockdown chimeric mice were isolated, and 1 × 106 cells were injected retro-orbitally into WT C57BL/6 mice. The next day, the recipient mice were immunized with OVA (50 μg, grade V; Sigma-Aldrich) emulsified in CFA (BD Diagnostics) or alum (Pierce) by s.c. injection. At 6 d after immunization, cells were collected from spleen and inguinal lymph nodes and cultured with OVA323–339 peptide (10 μg/mL; AnaSpec) for 8 h at 37 °C in the presence of Golgi Stop (BD Biosciences). The intracellular cytokine profiles were analyzed by flow cytometry.
Establishment of Stable Jurkat E6.1 Cell Line by Lentiviral Transduction.
To generate Jurkat E6.1(JE6.1) cells stably expressing USP9X shRNA, USP9X shRNA (USP9X: 5′- TGCTGTTGACAGTGAGCGCGGTGCTAATCTCATTAAAGAATAGTGAAGCCACAGATGTATTCTTTAATGAGATTAGCACCTTGCCTACTGCCTCGGA-3′) was subcloned into pGIPz lentiviral expression vector. Then 293T cells were transfected with 1 µg of pGIPz vector and 5 μg of viral packaging mix (Sigma-Aldrich) with 9 µL of TransIT-LT1 (Mirus). After 48 h, the culture supernatant containing lentivirus was collected. JE6.1 cells were infected with lentivirus together with 5 µg/mL polybrene by centrifuging cells at 420 × g for 60 min at room temperature. Then lentivirus-transduced cells were selected with puromycin (1 μg/mL). One week after selection, the cells were analyzed for GFP expression by FACS. To generate JE6.1 cells stably expressing FLAG-Bcl10, full-length human Bcl10 cDNA was subcloned into pLENTI6/V5-DEST lentiviral expression vector (Invitrogen) in frame with an N-terminal 3× FLAG epitope. Lentivirus-transduced cells were selected with blasticidin (5 μg/mL).
Supplementary Material
Acknowledgments
We thank J. Lopez and C. Elly for mouse breeding, and Y. Harada for technical help. Y.P. is supported in part by a fellowship from National Research Foundation of Korea. This work is supported by National Institutes of Health Grants R01 AI62969 and R01 AI78272, from the National Institute of Allergy and Infectious Diseases.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1221925110/-/DCSupplemental.
References
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 3.Nijman SM, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123(5):773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Komander D, Clague MJ, Urbé S. Breaking the chains: Structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10(8):550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 5.Wertz IE, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430(7000):694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 6.Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature. 2003;424(6950):797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- 7.Trompouki E, et al. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature. 2003;424(6950):793–796. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- 8.Düwel M, et al. A20 negatively regulates T cell receptor signaling to NF-kappaB by cleaving Malt1 ubiquitin chains. J Immunol. 2009;182(12):7718–7728. doi: 10.4049/jimmunol.0803313. [DOI] [PubMed] [Google Scholar]
- 9.Reiley WW, et al. Deubiquitinating enzyme CYLD negatively regulates the ubiquitin-dependent kinase Tak1 and prevents abnormal T cell responses. J Exp Med. 2007;204(6):1475–1485. doi: 10.1084/jem.20062694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schulze-Luehrmann J, Ghosh S. Antigen-receptor signaling to nuclear factor kappa B. Immunity. 2006;25(5):701–715. doi: 10.1016/j.immuni.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 11.Lee KY, D’Acquisto F, Hayden MS, Shim JH, Ghosh S. PDK1 nucleates T cell receptor-induced signaling complex for NF-kappaB activation. Science. 2005;308(5718):114–118. doi: 10.1126/science.1107107. [DOI] [PubMed] [Google Scholar]
- 12.Sun L, Deng L, Ea CK, Xia ZP, Chen ZJ. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol Cell. 2004;14(3):289–301. doi: 10.1016/s1097-2765(04)00236-9. [DOI] [PubMed] [Google Scholar]
- 13.Wu CJ, Ashwell JD. NEMO recognition of ubiquitinated Bcl10 is required for T cell receptor-mediated NF-kappaB activation. Proc Natl Acad Sci USA. 2008;105(8):3023–3028. doi: 10.1073/pnas.0712313105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oeckinghaus A, et al. Malt1 ubiquitination triggers NF-kappaB signaling upon T-cell activation. EMBO J. 2007;26(22):4634–4645. doi: 10.1038/sj.emboj.7601897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langel FD, et al. Multiple protein domains mediate interaction between Bcl10 and MALT1. J Biol Chem. 2008;283(47):32419–32431. doi: 10.1074/jbc.M800670200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lucas PC, et al. Bcl10 and MALT1, independent targets of chromosomal translocation in malt lymphoma, cooperate in a novel NF-kappa B signaling pathway. J Biol Chem. 2001;276(22):19012–19019. doi: 10.1074/jbc.M009984200. [DOI] [PubMed] [Google Scholar]
- 17.Gaide O, et al. CARMA1 is a critical lipid raft-associated regulator of TCR-induced NF-kappa B activation. Nat Immunol. 2002;3(9):836–843. doi: 10.1038/ni830. [DOI] [PubMed] [Google Scholar]
- 18.Uren AG, et al. Identification of paracaspases and metacaspases: Two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol Cell. 2000;6(4):961–967. doi: 10.1016/s1097-2765(00)00094-0. [DOI] [PubMed] [Google Scholar]
- 19.Wood SA, et al. Cloning and expression analysis of a novel mouse gene with sequence similarity to the Drosophila fat facets gene. Mech Dev. 1997;63(1):29–38. doi: 10.1016/s0925-4773(97)00672-2. [DOI] [PubMed] [Google Scholar]
- 20.Schwickart M, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463(7277):103–107. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- 21.Pérez-Mancera PA, et al. Australian Pancreatic Cancer Genome Initiative The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature. 2012;486(7402):266–270. doi: 10.1038/nature11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dupont S, et al. FAM/USP9X, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell. 2009;136(1):123–135. doi: 10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- 23.Thome M, Charton JE, Pelzer C, Hailfinger S. Antigen receptor signaling to NF-kappaB via CARMA1, BCL10, and MALT1. Cold Spring Harb Perspect Biol. 2010;2(9):a003004. doi: 10.1101/cshperspect.a003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Al-Hakim AK, et al. Control of AMPK-related kinases by USP9X and atypical Lys(29)/Lys(33)-linked polyubiquitin chains. Biochem J. 2008;411(2):249–260. doi: 10.1042/BJ20080067. [DOI] [PubMed] [Google Scholar]
- 25.Paul S, Kashyap AK, Jia W, He YW, Schaefer BC. Selective autophagy of the adaptor protein Bcl10 modulates T cell receptor activation of NF-κB. Immunity. 2012;36(6):947–958. doi: 10.1016/j.immuni.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scharschmidt E, Wegener E, Heissmeyer V, Rao A, Krappmann D. Degradation of Bcl10 induced by T-cell activation negatively regulates NF-kappa B signaling. Mol Cell Biol. 2004;24(9):3860–3873. doi: 10.1128/MCB.24.9.3860-3873.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lobry C, Lopez T, Israël A, Weil R. Negative feedback loop in T cell activation through IkappaB kinase-induced phosphorylation and degradation of Bcl10. Proc Natl Acad Sci USA. 2007;104(3):908–913. doi: 10.1073/pnas.0606982104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.