Abstract
In many individuals suspected of the common cancer predisposition Lynch syndrome, variants of unclear significance (VUS), rather than an obviously pathogenic mutations, are identified in one of the DNA mismatch repair (MMR) genes. The uncertainty of whether such VUS inactivate MMR, and therefore are pathogenic, precludes targeted healthcare for both carriers and their relatives. To facilitate the identification of pathogenic VUS, we have developed an in cellulo genetic screen-based procedure for the large-scale mutagenization, identification, and cataloging of residues of MMR genes critical for MMR gene function. When a residue identified as mutated in an individual suspected of Lynch syndrome is listed as critical in such a reverse diagnosis catalog, there is a high probability that the corresponding human VUS is pathogenic. To investigate the applicability of this approach, we have generated and validated a prototypic reverse diagnosis catalog for the MMR gene MutS Homolog 2 (Msh2) by mutagenizing, identifying, and cataloging 26 deleterious mutations in 23 amino acids. Extensive in vivo and in vitro analysis of mutants listed in the catalog revealed both recessive and dominant-negative phenotypes. Nearly half of these critical residues match with VUS previously identified in individuals suspected of Lynch syndrome. This aids in the assignment of pathogenicity to these human VUS and validates the approach described here as a diagnostic tool. In a wider perspective, this work provides a model for the translation of personalized genomics into targeted healthcare.
Lynch syndrome is a prevalent, dominantly inherited predisposition to colon and other visceral cancers and is caused by heterozygosity for a deleterious defect in one of the DNA mismatch repair (MMR) genes MSH2, MSH6, MLH1, or PMS2 encoding the MutS Homologs 2 and 6, the MutL Homolog 1, and Postmeiotic Segregation increased 2, respectively. Inadvertent loss of the second, wild-type allele in somatic cells results in MMR deficiency. The consequent inability to remove accidental nucleotide misincorporations during DNA replication leads to the accumulation of spontaneous genomic mutations and the rapid development of cancer (1).
Lifelong preventive surveillance is an effective means to reduce cancer incidence and mortality in carriers of a pathogenic MMR gene mutation (2). In addition, carriers may benefit from both targeted chemoprevention (3) and chemotherapy (4). Unfortunately, in a significant fraction of all individuals suspected to have Lynch syndrome, a subtle alteration of one of the MMR genes is identified, such as a single amino acid alteration or a mutation of a putative splice site (5). The pathogenicity of these so-called variant(s) of unclear significance (VUS) often cannot be assessed because of the absence of conclusive clinical and segregation data and the absence of data on the effect of the mutation on gene function. It is believed that primary screening for MMR gene mutations in selected populations, a cost-effective approach to improve general health, will cause a further increase in the incidence of MMR gene VUS (6–8). The development and use of robust and accessible approaches to identify pathogenic MMR gene VUS will enable the implementation of targeted preventive and curative healthcare for carriers of pathogenic VUS. Meanwhile, unaffected relatives of such carriers can be liberated from the burden associated with the uncertain pathogenicity of the VUS while unwarranted inflow into preventive healthcare systems is reduced (9).
The genetics and biochemistry of the MMR pathway have been established in detail (10), which has enabled the development of in vitro assays to predict the pathogenicity of MMR gene VUS (11). Although these assays can be useful tools in the prediction of pathogenicity, they are often laborious and time consuming, whereas defects that specifically affect functions of the variant genes in vivo, such as splicing, intracellular localization, or stability, may escape detection. To circumvent these issues, we have taken an in vivo, genetic screen-based approach. This approach combines the large-scale mutagenesis and identification of critical amino acids of MMR genes, the analysis of the mutant cell lines and proteins, and the compilation of these data in an annotated reverse diagnosis catalog. Such a catalog enables us to rapidly see whether a VUS concerns a residue that is critical for gene function in vivo, and therefore aids in the assessment of pathogenicity. Here, we describe the construction and extensive validation of a prototypic reverse diagnosis catalog of the core MMR protein Msh2.
Results
Genetic Screen to Mutagenize and Map Critical MMR Gene Residues.
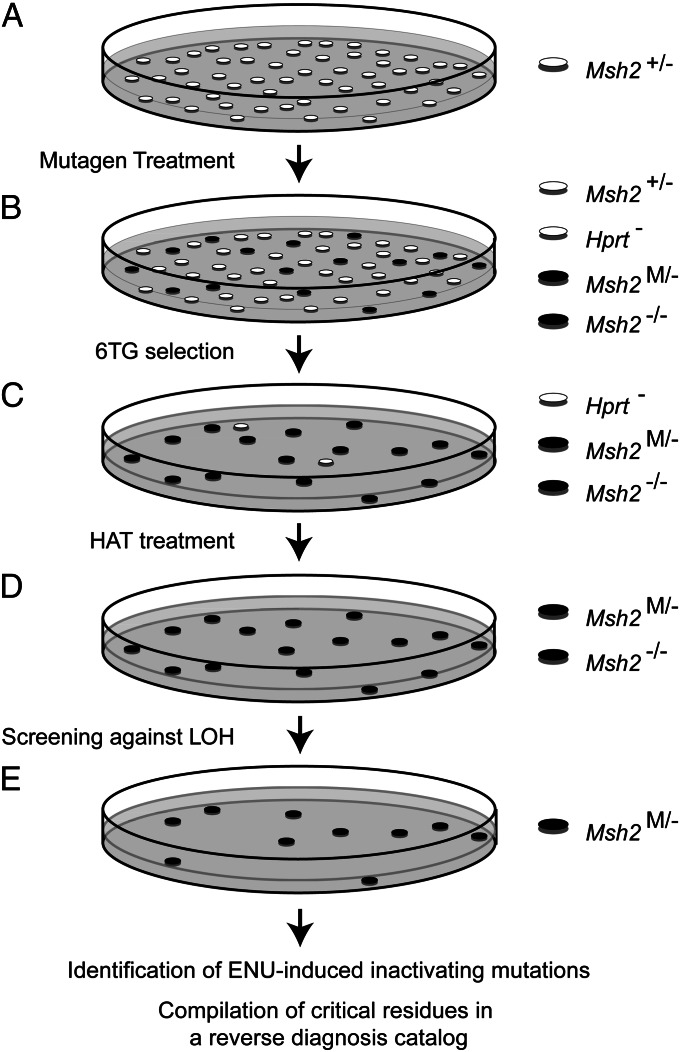
We argue that residues, which are critical for MMR gene function in vivo, might be identified by a three-step procedure: treatment of cultured cells with a wide-spectrum mutagen to randomly induce genetic variants; isolation of clones that have lost MMR consequent to mutation of a critical amino acid; and identification of the causal mutations and their compilation in a reverse diagnosis catalog. Mouse embryonic stem cells (mESCs) are a suitable cell type for such a screen, as ES cells are primary diploid cells, although they are immortal. Furthermore, mESCs that are monoallelic for each of the MMR genes are available (12, 13), which requires the inactivation of only the single, wild-type allele by a mutagenic event. Importantly, mouse and human MMR genes are >95% identical or homologous at the amino acid level.
To generate a pilot reverse diagnosis catalog, we used an mESC line that is monoallelic for Msh2 (Msh2+/−) (12). This line was treated with the powerful, wide-spectrum, and relatively nontoxic point mutagen N-ethyl-N-nitrosourea (ENU; Fig. 1A) (15, 16). To isolate clones that have become MMR-deficient because of a mutagenic event at the monoallelic MMR gene, we subsequently exposed cells to the nucleotide analog 6-thioguanine (6TG; Fig. 1B), to which MMR-deficient cells are tolerant (14, 16). Then, unwanted clones that had acquired 6TG tolerance because of mutational inactivation of the X-linked yypoxanthine phosphorybosyl transferase (Hprt) gene, rather than loss of MMR, were killed by culture in a hypoxanthine-aminopterin-thymidine-supplement-containing medium (Fig. 1C). In the remaining clones, the single Msh2 allele might be lost either by an ENU-induced substitution at a critical residue (Msh2M/− in Fig. 1D) or by loss of heterozygosity (Msh2−/− in Fig. 1D) (14). The latter clones were discarded after screening with an Msh2 allele-specific PCR. The remaining cell clones are expected to harbor an ENU-induced substitution that inactivates a critical residue of the monoallelic Msh2 gene. Importantly, when wild-type (Msh2+/+) ES cells were subjected to these treatments, no surviving clones were obtained, indicating that this procedure does not result in clones with a simultaneous mutational inactivation of both alleles of any of the MMR genes.
Fig. 1.
A genetic screen to identify critical MMR gene residues. (A) A mESC line, hemizygous for Msh2 (Msh2+/−), is subjected to mutagenic treatment with ENU. Preexisting, inadvertent, Msh2−/− cells (14) are not displayed here. (B) Cells that have acquired low-level 6TG tolerance, by loss of heterozygosity at Msh2 (LOH, Msh2−/−), by an ENU-induced loss-of-function mutation at a critical residue in the monoallelic Msh2 gene (Msh2M/−), or by a deleterious mutation at the monoallelic Hprt gene (Hprt-) are selected using two brief 6TG selections. (C) Unwanted Hprt-deficient clones are eliminated using HAT-supplemented medium. (D) Unwanted clones that have lost the wild-type Msh2 allele by LOH are excluded, using an allele-specific PCR. (E) The inactivating mutation in the remaining clones is identified by sequence analysis. A reverse diagnosis catalog is compiled that lists critical residues at Msh2.
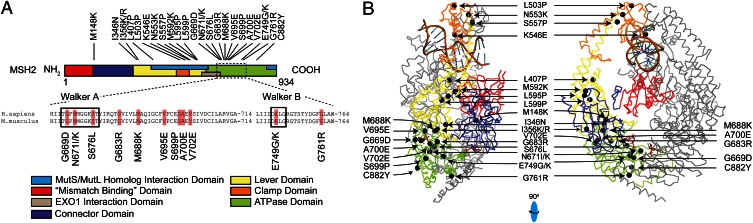
To identify ENU-induced amino acid substitutions in Msh2 in the selected clones, we sequenced the complete Msh2 ORF of clones that had retained expression of full-length Msh2 cDNA, as judged by reverse transcription–PCR (Table 1). This resulted in the identification of an amino acid substitution in monoallelic Msh2 in 41 clones. These substitutions alter 23 different amino acids, of which 3 amino acids were represented by two different substitutions in individual clones. These residues were then compiled in a prototypic reverse diagnosis catalog (Table 2 and Table S1). Mapping these amino acids to the MSH2 crystal structure revealed a clustering around the lever, connector, and ATPase domains of the protein, indicating the relative importance of these domains (Fig. 2 A and B) (18). Conversely, only a single deleterious mutation was found in the N-terminal region of Msh2 (Fig. 2A). Multivariate analysis of protein-polymorphisms-MMR (MAPP-MMR) (19), an alignment-based algorithm, predicted 25 of 26 Msh2 mutants in the reverse diagnosis catalog to be deleterious and one to be borderline deleterious, a result that cross-validates both MAPP-MMR and our reverse diagnosis screen (Table 2).
Table 1.
Generation of the pilot Msh2 reverse diagnosis catalog described here
| Clone characteristics | No. |
| Total no. of 6TG and HAT-tolerant clones | 1,483 |
| Clones that did not display LOH at Msh2* | 426 |
| Clones analyzed for Msh2 mutations† | 81 |
| Clones with a single, unique, amino acid substitution‡ | 41 (26) |
| Mutated amino acids | 23 |
| Other mutations identified in Msh2¶ | 10 |
Loss of heterozygosity was assessed by allele-specific PCR.
Only full-length cDNAs were sequenced.
Six substitutions were found in more than a single individual clone. Between parentheses are unique substitutions.
Comprises nonsense, frameshift or silent mutations, tandem mutations, or small in-frame deletions.
Table 2.
Msh2 mutants generated in this study
| Mutation | cDNA | MAPP-MMR* | LOVD† | |
| M148K | c.443 | T > A | 3.75 | — |
| I346N | c.1037 | T > A | 18.3 | — |
| I356K | c.1067 | T > A | 14.66 | — |
| I356R | c.1067 | T > G | 16.34 | — |
| L407P | c.1220 | T > C | 18.56 | L407L |
| L503P | c.1508 | T > C | 14.91 | L503P |
| K546E | c.1636 | A > G | 23.7 | — |
| N553K | c.1659 | C > A | 24.75 | — |
| S557P | c.1669 | T > C | 9.71 | T557P |
| M592K | c.1775 | T > A | 8.21 | M592V |
| L595P | c.1784 | T > C | 7.68 | — |
| L599P | c.1796 | T > C | 17.42 | — |
| G669D | c.2006 | G > A | 35.86 | G669D/R |
| N671I | c.2012 | A > T | 22.05 | |
| N671K | c.2013 | T > A | 25.31 | N671K/Y |
| S676L | c.2027 | C > T | 28.64 | S676P |
| G683R | c.2047 | G > A | 44 | G683R/W |
| M688K | c.2063 | T > A | 32.5 | M688R/I |
| V695E | c.2084 | T > A | 30.37 | — |
| S699P | c.2095 | T > C | 5.97 | — |
| A700E | c.2099 | C > A | 37.91 | — |
| V702E | c.2105 | T > A | 33.26 | V702G |
| E749K | c.2245 | G > A | 19.98 | |
| E749G | c.2246 | A > G | 28.23 | E749K |
| G761R | c.2281 | G > A | 40.89 | — |
| C822Y | c.2465 | G > A | 6.48 | — |
Boldface represents the mutants included in the validation panel.
In silico MAPP-MMR prediction of pathogenicity. Scores above 4.55 are predicted to be pathogenic, variant M148K is borderline pathogenic (calculated from http://mappmmr.blueankh.com/Impact.php).
A mutation at the same residue identified in a suspected Lynch syndrome patient, as reported in the Leiden Open Variant Database (LOVD). Bold, identical substitution found in reverse diagnosis catalog and LOVD, allowing to assess pathogenicity to the patient VUS.
Fig. 2.
Critical Msh2 residues mapped in this study. (A) Schematic representation of all identified deleterious amino acid substitutions in Msh2. The colored protein domains are as described (18). It should be noted that the term “mismatch binding domain” is a misnomer, as rather than Msh2, the homologous domain in Msh6 binds the mismatch. (Middle) An alignment of the Walker A and Walker B ATPase domains of human and mouse Msh2, highlighting residues that were mutated in the screen described here (orange boxes). Numbers reflect amino acid numbering. (B) Mapping of critical Msh2 residues mutated in our screen to the crystal structure of the human MSH2/MSH6 heterodimer. In color, MSH2 (see Fig. 2A for legends); in gray, MSH6. Mutated residues are shown as black spheres. The MutS/MutL and Exonuclease 1 (EXO1) interaction domains are not indicated in the crystal structure. (Right) Mutants S699P and V695E are obscured.
To investigate the applicability of our approach in the identification of pathogenic human MMR gene variants, we aligned the Leiden Open Variation Database, which lists human MSH2 variants, with the reverse diagnosis catalog. This revealed that of the 23 residues listed as critical in the catalog, 10 were previously identified as mutated in humans. Moreover, in 6 of these 10 cases, human VUS were identical to an ENU-induced deleterious amino acid substitution (Table 2). This result provides strong support for the applicability of reverse diagnosis catalogs to identify pathogenic human MMR gene variants.
Msh2-Mutant Cell Lines Display Phenotypic Hallmarks of MMR Deficiency.
To validate the genetic screen experimentally, we assembled a validation panel consisting of 13 of the 26 mutant cell lines. These substitutions were chosen because all of these residues are represented in the Leiden Open Variation Database, either as an identical or different substitution or as a silent mutation (Table 2).
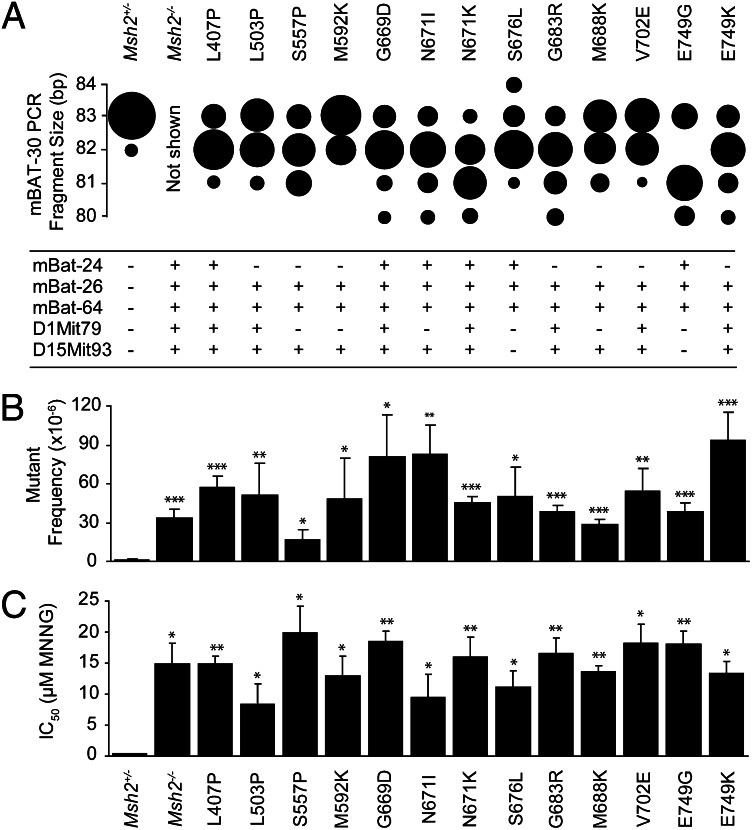
Microsatellite instability is a diagnostic hallmark of cancer in Lynch syndrome (1). We measured the size of six different microsatellites in 40–50 subclones of each line from the validation panel. Microsatellites were shortened in the large majority of the subclones of each cell line, in support of MMR deficiency (Fig. 3A). The microsatellite size reductions were less pronounced in the mESC lines than in Lynch syndrome-associated cancers (1). This likely is a consequence of the absence of iterative cycles of expansion and clonal selection, which characterizes carcinogenesis. In addition to microsatellite instability, MMR defects are characterized by the accumulation of spontaneous nucleotide substitutions. Indeed, all cell lines from the validation panel displayed high frequencies of spontaneous Hprt mutants, mimicking the Msh2-disrupted control mESC line (Fig. 3B).
Fig. 3.
Analysis of the validation panel. (A) (Upper) Microsatellite instability analysis on mononucleotide microsatellite mBAT-30. The size of each sphere is proportional to the relative number of subclones with the indicated mBAT-30 PCR fragment length. All mutants from the validation panel display microsatellite instability (P < 0.05 compared with the Msh2+/− line). Microsatellite size shifts in the Msh2−/− control line are not displayed, as in this line the number of cell divisions before subcloning was much higher than in the lines of the validation panel, which makes a quantitative comparison impossible. (Lower) Microsatellite instability analysis on mononucleotide repeats mBAT-24, mBAT-26, and mBAT-64, and on dinucleotide repeats D1Mit79 and D15Mit93. +, MSI (P < 0.05 compared with the Msh2+/− line); −, no MSI. (B) Assessment of spontaneous mutator phenotypes at the genomic Hprt gene in Msh2-mutant mESC lines from the validation panel. Bars represent Hprt mutant frequencies ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 compared with the parental Msh2+/− line. (C) Tolerance to the methylating drug N-methyl-N′-nitro-N-nitrosoguanidine in Msh2-mutant mESC lines. Bars represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 compared with the parental Msh2+/− line.
MMR-deficient cells have acquired tolerance of DNA-methylating drugs, a class of agents that includes several chemotherapeutic drugs (10). We tested our validation panel of cell lines for tolerance of the prototypic methylating drug N-methyl-N′-nitro-N-nitrosoguanidine. Indeed, all Msh2-mutant cell lines proved as tolerant as the Msh2-disrupted control line (Fig. 3C). Cumulatively, these results demonstrate that all cell lines from the validation panel have acquired all phenotypic hallmarks of MMR deficiency.
MMR Deficiency Is Caused by the ENU-Induced Mutations in Msh2.
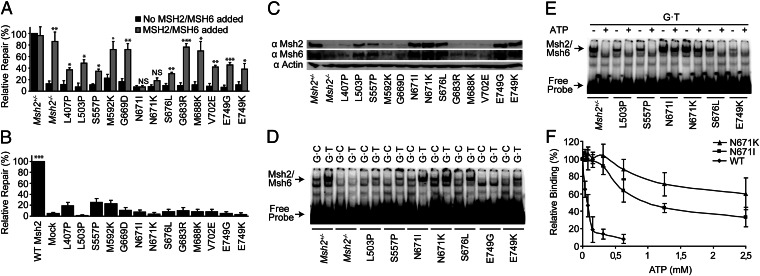
To provide conclusive evidence of the causality of the Msh2 mutations for MMR deficiency, we followed a biochemical approach, using an MMR assay that measures the ability of cell extracts to repair a defined G⋅T mismatch (20). MMR activity was lost in extracts from all lines from the validation panel (Fig. 4A, black bars). To determine causality of the Msh2 mutations for MMR loss, we investigated whether the addition of recombinant purified wild-type human MSH2/MSH6 rescued the MMR defect in extracts from each mutant cell line. Indeed, the defect was significantly complemented in 11 cell extracts, confirming that the MMR deficiency is caused by inactivation of Msh2/Msh6, rather than by inadvertent loss of one of the other (biallelic) MMR genes (Fig. 4B, gray bars). Only in two of the extracts, each derived from a cell line with a different substitution at Msh2 residue N671, was the MMR activity not significantly restored by wild-type MSH2/MSH6 (Fig. 4A and see following).
Fig. 4.
Biochemical characterization of Msh2-mutant mESC lines from the validation panel. (A) In vitro repair of a G·T mismatch in mutant cell extracts in the absence (black bars) and presence (gray bars) of purified wild-type human MSH2/MSH6. Bars represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, when comparing repair efficiencies in the presence and absence of recombinant MSH2/MSH6 for each extract. NS, not significant. (B) MMR activity in in vitro recreated mutant humanized MSH2 proteins. These proteins were tested for the repair of a G·T mismatch after addition to an MSH2/MSH6-deficient human cell extract. Mock, mock Msh2/Msh6 expression. Bars represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 compared with the mock reaction. (C) Western blot analysis of total lysates from mutant ES cell extracts. Actin, β-actin loading control. (D) Binding of control and mutant Msh2/Msh6 cell extracts to a G·T mismatch within a double-stranded oligonucleotide probe in an electrophoretic mobility shift assay. Msh2/Msh6 indicates the Msh2/Msh6·DNA complex. (E) ATP-induced mismatch release of Msh2-mutant cell extracts was tested in an electrophoretic mobility shift assay. ATP (1.25 mM) was added after allowing the proteins to bind to the probe. (F) Quantification of ATP-induced mismatch release of Msh2/Msh6, Msh2-N671I/Msh6, or Msh2-N671K/Msh6. The values represent the average from three independent experiments. Error bars represent SEM.
We wanted to directly prove causality of the mutated Msh2 amino acids for loss of MMR. To this aim, we used a wild-type human MSH2 cDNA as a template to mimic the 13 mouse Msh2 mutants from the validation panel by site-directed mutagenic PCR in the human gene. This was followed by in vitro transcription and translation to recreate MSH2 mutant proteins (20, 21). The resulting proteins were heterodimerized with wild-type MSH6 and tested for their ability to restore MMR in extracts from the MSH2/MSH6-deficient human cancer cell line LoVo (22). None of the 13 mutant MSH2/MSH6 human proteins complemented the LoVo cell extract (Fig. 4B). This result provides direct and conclusive evidence for the causality of each of the ENU-induced amino acid substitutions in Msh2 for MMR deficiency, also in the human homolog. Therefore, this result provides strong support for pathogenicity of the human mutants.
MMR Deficiency Is Caused by Loss of Protein Expression or by Specific Biochemical Defects.
To provide biochemical insights into the MMR defects in the cell lines, we investigated levels of mutant Msh2 proteins in the validation panel by Western blotting. This revealed that 6 of the cell lines had lost expression of Msh2 (Fig. 4C), suggesting defective protein folding and/or stability. The heterodimeric partner of Msh2, Msh6, was also destabilized in these low-protein-level Msh2 mutants (Fig. 4C), consistent with previous results (13). By analyzing levels of mutant Msh2 proteins in fractionated cell extracts, we excluded intracellular mislocalization as a cause for the MMR defect of the seven Msh2 mutants that had retained significant protein levels (Fig. S1).
To obtain insights into the biochemical defects of the mutants that had retained protein expression, we tested binding to G⋅T mismatched probes in extracts from these cell lines, using an electrophoretic mobility shift assay. With the exception of mutants N671I and N671K, binding to both the matched and mismatched probes was significantly reduced, suggesting a general defect in DNA binding in those mutants (Fig. 4D).
Substitutions at Residue N671 of Msh2 Confer a Dominant-Negative Phenotype.
After mismatch binding by Msh2/Msh6, ATP binding to the distal ATPase domains of the protein (Fig. 2) results in the conversion of the mismatch-bound form of Msh2/Msh6 into a repair-competent DNA-embracing clamp that releases the mismatch (10). We tested ATP-induced mismatch release in cell extracts from all 6 mutants that had retained mismatch binding. ATP released the protein from the mismatch in most of these extracts, indicating that neither ATP binding nor intramolecular signaling is affected (Fig. 4E). In contrast, extracts from both mutants at residue N671 were defective in ATP-induced release from the mismatch (Fig. 4 E and F). Indeed, N671 is a highly conserved residue at the Walker A (ATP-binding) motif of the ATPase domain (Fig. 2A) (10). It is not unlikely that the normal mismatch binding of the N671I and N671K mutants, combined with the defect in ATP-induced mismatch release, precludes binding of wild-type Msh2/Msh6. This result is consistent with the inability of wild-type MSH2/MSH6 to restore MMR activity to extracts from both mutant cell lines, as discussed earlier (Fig. 4A). Complementary to this, we show that the addition of both N671 mutant proteins, but not the recessive G683R mutant protein, to a HeLa cell extract inhibits MMR activity (Fig. S2). On the basis of these results, we conclude that the N671I and N671K substitutions confer dominant-negative phenotypes to Msh2.
Discussion
Genetic Screens for the Identification of Pathogenic MMR Gene Variants.
The absence of well-established procedures to classify VUS in genes associated with cancer predisposition is a bottleneck in the translation of personalized genomics into targeted healthcare. To aid in resolving this problem in Lynch syndrome, we have developed a genetic screening procedure to mutagenize and identify critical MMR gene residues. This results in reverse diagnosis catalogs that list critical gene residues, together with the associated mutant cell lines. Given the causal relation of MMR deficiency for Lynch syndrome, VUS that are listed as deleterious in the reverse diagnosis catalog have a high probability of pathogenicity.
The screens are based on the mutagenic treatment of a mESC line, which is monoallelic for one of the MMR genes, followed by selection of clones that have acquired a deleterious substitution of a critical residue of the wild-type allele. This procedure mimics the somatic inactivation of the wild-type allele and the subsequent clonal outgrowth of MMR-deficient tumor cells that precedes cancer development in Lynch syndrome (23). Therefore, the mutant cell lines can be used as models for cancer cell lines from patients carrying the corresponding mutation. The generation of reverse diagnosis catalogs is not restricted to Msh2, as in addition, cells deficient for the other core MMR genes are tolerant to 6TG (17), and monoallelic mESC lines for these genes are available. A comprehensive reverse diagnosis catalog (Table 2 and Table S1), linear gene map (Fig. 2A), or structural representation (Fig. 2B and Fig. S3), annotated with functional data, has to be generated only once for each MMR gene and will aid in assessing pathogenicity to human MMR gene VUS both now and in the future.
In the prototypic screen presented here, we have identified 26 cell lines, each carrying a deleterious Msh2 amino acid substitution, in total unveiling 23 amino acids that are critical for Msh2 function in vivo. Ten of these 23 amino acids have previously been identified as mutated in humans (Table 2). In 6 of these mutated amino acids, the amino acid substitutions in the individual and in the reverse diagnosis catalog were identical (Table 2).
To validate the screen, we have extensively tested a panel consisting of 13 of the mutants, including those that were also identified as VUS in humans. All mutant cell lines from the validation panel display all hallmarks of MMR deficiency (Table S1). With the exception of the dominant-negative substitutions at residue N671 (see following), the biochemical MMR defect was rescued by wild-type human MSH2/MSH6 in extracts from all mutant mouse ES cell lines. This demonstrates that the mutagenic treatment has not inadvertently inactivated any of the other MMR genes (Fig. 4A). Finally, we have directly proven causality of the ENU-induced Msh2 substitutions in these 13 cell lines for the loss of MMR in a human setting (Fig. 4B). Taken together, these experiments validate the use of the mouse cell-based genetic screen presented here to identify critical MMR gene residues, to use these data for the assignment of pathogenicity to human MMR gene VUS, and to investigate in vivo phenotypes of the mutant cell lines as models for tumor cell lines.
Our pilot screen yielded only a single inactivating substitution at the N terminus of Msh2. This may have different causes, including a bias in the mutagenicity of ENU, a bias in the recovery of N-terminal deleterious mutations [e.g., resulting from degradation of mRNA (the clones analyzed here have stable cDNAs)], and an intrinsically low density of critical residues in the N-terminal part of Msh2. In support, deletion of the N-terminal 133 amino acids of Msh2 hardly affects MMR in Saccharomyces cerevisiae (24). In the Human Gene Mutation Database (http://www.hgmd.cf.ac.uk/ac/index.php), presumed deleterious missense variants are ubiquitous in the N-terminal part of MSH2. Possibly, many of these N-terminal substitutions represent hypomorphic mutations, whereas our screen selects for complete loss of function. Alternatively, it cannot be excluded that many of these N-terminal (and other) substitutions in MSH2 are classified incorrectly, as the criteria for their assignment are unclear. In this light, it is noteworthy to mention that the MMR Gene Unclassified Variants Database (www.mmruv.info) only reports a stretch of loss-of-function VUS located between V161 and C199 and then from C333 onward, based on functional assays.
Dominant-Negative Phenotypes of Msh2 Variants and Cancer Predisposition.
The extensive analysis of the mutants generated here has allowed the identification of dominant-negative phenotypes of Msh2 mutants N671K and N671I. In families suspected of Lynch syndrome, both identical (N671K) and nonidentical (N671Y) substitutions at MSH2 N671 have been described (19, 25). The identification of dominant-negative mutations is of particular relevance, as they may confer a disease phenotype already in a heterozygous state. This may include the accelerated onset and expanded tropism of cancer. In fact, a family with a MSH2 N671Y mutation displays both an extended spectrum (colon, ovary, breast, esophagus) and a young average age of onset of cancer (37 y) (25). In addition to substitutions N671I and N671K, several other mutants obtained in our screen have retained Msh2 protein expression (Fig. 4C). From a diagnostic point of view, this is a significant observation, as loss of protein expression frequently is used to direct MMR gene sequencing in individuals suspected of Lynch syndrome (1). Therefore, even when immunostaining of a tumor is positive for the presence of all MMR proteins, the patient should be analyzed for VUS.
Approaches to Generate Comprehensive Reverse Diagnosis Catalogs.
In this study, we have focused on the identification of ENU-induced mutants that had retained full-length Msh2 cDNA. For this reason, Msh2 mutants that cause defects at the RNA level will have escaped identification. Indeed, in many of the clones, full-length cDNA or protein was not detected, and these clones were discarded (Table 1). In the future, deleterious mutations that disrupt transcription, splicing, or mRNA stability (26–28) may be identified by genomic, rather than cDNA, sequencing.
Analysis of the ENU-induced mutation spectrum at Msh2 in the clones generated here reveals substitutions at both A⋅T and G⋅C base pairs (Fig. S4), supporting the notion that ENU is a wide-spectrum mutagen (15). However, as mutagenesis by ENU may be not fully random, the mutagenesis and ensuing identification of all critical gene residues may be hindered. Higher saturation of the reverse diagnosis catalog may be achieved by the use of drugs with a complementary mutagenic activity, such as 4-nitroquinoline N-oxide (29) or 7,12-dimethylbenz[a]anthracene (30).
Integration of Reverse Diagnosis Catalogs in Lynch Syndrome Diagnostics.
It is conceivable that, even when performed at a large scale and using multiple mutagens, the genetic screens proposed here may not be fully saturating, resulting in the absence of critical residues from the reverse diagnosis catalog. In other instances, the substitution identified in an individual, although at the same amino acid, may be different from that listed in the catalog. Such variants may correctly be assessed using cell-free MMR assays (20, 21). Conversely, cell-free assays may not correctly identify pathogenic variants when the mutation specifically affects in vivo characteristics of the variant protein, such as its stability or intracellular localization, and these may rely on reverse diagnosis catalogs for identification. We envision that reverse diagnosis catalogs and cell-free activity assays may become important components of an integrated diagnostic procedure that also includes independent data sources such as segregation and pathological and bioinformatics data (11). This will benefit targeted healthcare for individuals suspected to have Lynch syndrome but will also provide a paradigm for the management of gene variants in any genetic disease that has a defined underlying cellular and biochemical phenotype in the upcoming era of personalized genomics.
Materials and Methods
Determination of genomic mutator phenotypes, microsatellite instability analysis, methylation tolerance assays, and other procedures are described in the SI Materials and Methods.
Genetic Screens to Identify Critical Msh2 Residues.
The outline of the generation of a reverse diagnosis catalog for Msh2 is described in Fig. 1. In a P90 dish on irradiated feeder cells in mESC culture medium (Gibco), 5 × 106 Msh2+/− mESCs (12) were seeded. To induce mutations, cells either were incubated in medium containing 0.2 mg/mL ENU (Sigma-Aldrich) for 2 h (14) or were mock-treated. Cells were propagated for 1 wk to allow loss of wild Msh2. Then, cells were seeded at a density of 2 × 106 cells per T90, followed by double treatment, with a 7-d interval, with 40 μM 6TG (Sigma-Aldrich) for 4 h. Inadvertent Hprt-deficient cells were eliminated by culturing the clones for 1 wk in hypoxanthine-aminopterin-thymidine (HAT)-supplemented medium (Gibco). Surviving clones were picked and expanded in 96-well plates. Next, clones that had lost the wild-type Msh2 allele by loss of heterozygosity, rather than by mutational inactivation, were discarded after their identification by allele-specific PCR (14). In the remaining clones, the status of the Msh2 mRNA was analyzed by reverse transcriptase-PCR analysis. The Msh2 ORF of full-length cDNA clones was sequenced as described (24).
Statistical Analyses.
For experiments measuring microsatellite instability (Fig. 2A), mutator phenotypes (Fig. 2B), and methylation tolerance (Fig. 2C), P values were calculated by comparing with the Msh2+/− ES cell line, using a one-tailed t test assuming unequal variance. For the cell-free MMR complementation assay (Fig. 4A), P values were calculated for every extract individually, comparing repair efficiencies in the absence or presence of exogenous MSH2/MSH6, using a one-tailed t test assuming unequal variance. For the cell-free MMR assay (Fig. 4B), P values were calculated compared with the mock using a one-tailed t test assuming unequal variance.
Supplementary Material
Acknowledgments
We thank Jacob G. Jansen and Piya Temviriyanukul for critically reading the manuscript, Joyce H. G. Lebbink (Erasmus Medical Center) and Meindert H. Lamers (Medical Research Council) for help with crystal structure modeling, and Flora Groothuizen and Titia Sixma (The Netherlands Cancer Institute) for recombinant wild-type MSH2/MSH6. This work was funded by European Union Grant FP6-018754.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220537110/-/DCSupplemental.
References
- 1.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348(10):919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 2.Lagerstedt Robinson K, et al. Lynch syndrome (hereditary nonpolyposis colorectal cancer) diagnostics. J Natl Cancer Inst. 2007;99(4):291–299. doi: 10.1093/jnci/djk051. [DOI] [PubMed] [Google Scholar]
- 3.Burn J, et al. CAPP2 Investigators Long-term effect of aspirin on cancer risk in carriers of hereditary colorectal cancer: An analysis from the CAPP2 randomised controlled trial. Lancet. 2011;378(9809):2081–2087. doi: 10.1016/S0140-6736(11)61049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hewish M, Lord CJ, Martin SA, Cunningham D, Ashworth A. Mismatch repair deficient colorectal cancer in the era of personalized treatment. Nat Rev Clin Oncol. 2010;7(4):197–208. doi: 10.1038/nrclinonc.2010.18. [DOI] [PubMed] [Google Scholar]
- 5.Peltomäki P, Vasen HFA. Mutations associated with HNPCC predisposition — Update of ICG-HNPCC/INSiGHT mutation database. Dis Markers. 2004;20(4-5):269–276. doi: 10.1155/2004/305058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dinh TA, et al. Health benefits and cost-effectiveness of primary genetic screening for Lynch syndrome in the general population. Cancer Prev Res (Phila) 2011;4(1):9–22. doi: 10.1158/1940-6207.CAPR-10-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnetson RA, et al. Classification of ambiguous mutations in DNA mismatch repair genes identified in a population-based study of colorectal cancer. Hum Mutat. 2008;29(3):367–374. doi: 10.1002/humu.20635. [DOI] [PubMed] [Google Scholar]
- 8.Heinen CD. Genotype to phenotype: Analyzing the effects of inherited mutations in colorectal cancer families. Mutat Res. 2010;693(1-2):32–45. doi: 10.1016/j.mrfmmm.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castells A, Castellví-Bel S, Balaguer F. Concepts in familial colorectal cancer: Where do we stand and what is the future? Gastroenterology. 2009;137(2):404–409. doi: 10.1053/j.gastro.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 10.Hsieh P, Yamane K. DNA mismatch repair: Molecular mechanism, cancer, and ageing. Mech Ageing Dev. 2008;129(7-8):391–407. doi: 10.1016/j.mad.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rasmussen LJ, et al. Pathological assessment of mismatch repair gene variants in Lynch syndrome: Past, present, and future. Hum Mutat. 2012;33(12):1617–1625. doi: 10.1002/humu.22168. [DOI] [PubMed] [Google Scholar]
- 12.de Wind N, Dekker M, Berns A, Radman M, te Riele H. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell. 1995;82(2):321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 13.de Wind N, et al. HNPCC-like cancer predisposition in mice through simultaneous loss of Msh3 and Msh6 mismatch-repair protein functions. Nat Genet. 1999;23(3):359–362. doi: 10.1038/15544. [DOI] [PubMed] [Google Scholar]
- 14.Borgdorff V, van Hees-Stuivenberg S, Meijers CM, de Wind N. Spontaneous and mutagen-induced loss of DNA mismatch repair in Msh2-heterozygous mammalian cells. Mutat Res. 2005;574(1-2):50–57. doi: 10.1016/j.mrfmmm.2005.01.021. [DOI] [PubMed] [Google Scholar]
- 15.Russell WL, et al. Specific-locus test shows ethylnitrosourea to be the most potent mutagen in the mouse. Proc Natl Acad Sci USA. 1979;76(11):5818–5819. doi: 10.1073/pnas.76.11.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jansen JG, et al. Marked differences in the role of O6-alkylguanine in hprt mutagenesis in T-lymphocytes of rats exposed in vivo to ethylmethanesulfonate, N-(2-hydroxyethyl)-N-nitrosourea, or N-ethyl-N-nitrosourea. Cancer Res. 1995;55(9):1875–1882. [PubMed] [Google Scholar]
- 17.Glaab WE, et al. Resistance to 6-thioguanine in mismatch repair-deficient human cancer cell lines correlates with an increase in induced mutations at the HPRT locus. Carcinogenesis. 1998;19(11):1931–1937. doi: 10.1093/carcin/19.11.1931. [DOI] [PubMed] [Google Scholar]
- 18.Warren JJ, et al. Structure of the human MutSalpha DNA lesion recognition complex. Mol Cell. 2007;26(4):579–592. doi: 10.1016/j.molcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 19.Chao EC, et al. Accurate classification of MLH1/MSH2 missense variants with multivariate analysis of protein polymorphisms-mismatch repair (MAPP-MMR) Hum Mutat. 2008;29(6):852–860. doi: 10.1002/humu.20735. [DOI] [PubMed] [Google Scholar]
- 20.Drost M, et al. A cell-free assay for the functional analysis of variants of the mismatch repair protein MLH1. Hum Mutat. 2010;31(3):247–253. doi: 10.1002/humu.21180. [DOI] [PubMed] [Google Scholar]
- 21.Drost M, et al. A rapid and cell-free assay to test the activity of lynch syndrome-associated MSH2 and MSH6 missense variants. Hum Mutat. 2012;33(3):488–494. doi: 10.1002/humu.22000. [DOI] [PubMed] [Google Scholar]
- 22.Liu B, et al. Mismatch repair gene defects in sporadic colorectal cancers with microsatellite instability. Nat Genet. 1995;9(1):48–55. doi: 10.1038/ng0195-48. [DOI] [PubMed] [Google Scholar]
- 23.Lu SL, et al. Loss or somatic mutations of hMSH2 occur in hereditary nonpolyposis colorectal cancers with hMSH2 germline mutations. Jpn J Cancer Res. 1996;87(3):279–287. doi: 10.1111/j.1349-7006.1996.tb00218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee SD, Surtees JA, Alani E. Saccharomyces cerevisiae MSH2-MSH3 and MSH2-MSH6 complexes display distinct requirements for DNA binding domain I in mismatch recognition. J Mol Biol. 2007;366(1):53–66. doi: 10.1016/j.jmb.2006.10.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharp A, Pichert G, Lucassen A, Eccles D. RNA analysis reveals splicing mutations and loss of expression defects in MLH1 and BRCA1. Hum Mutat. 2004;24(3):272. doi: 10.1002/humu.9267. [DOI] [PubMed] [Google Scholar]
- 26.Tournier I, et al. A large fraction of unclassified variants of the mismatch repair genes MLH1 and MSH2 is associated with splicing defects. Hum Mutat. 2008;29(12):1412–1424. doi: 10.1002/humu.20796. [DOI] [PubMed] [Google Scholar]
- 27.Betz B, et al. Comparative in silico analyses and experimental validation of novel splice site and missense mutations in the genes MLH1 and MSH2. J Cancer Res Clin Oncol. 2010;136(1):123–134. doi: 10.1007/s00432-009-0643-z. [DOI] [PubMed] [Google Scholar]
- 28.Arnold S, et al. Classifying MLH1 and MSH2 variants using bioinformatic prediction, splicing assays, segregation, and tumor characteristics. Hum Mutat. 2009;30(5):757–770. doi: 10.1002/humu.20936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryu J, et al. Mutation spectrum of 4-nitroquinoline N-oxide in the lacI transgenic Big Blue Rat2 cell line. Mutat Res. 1999;445(1):127–135. doi: 10.1016/s1383-5718(99)00136-9. [DOI] [PubMed] [Google Scholar]
- 30.Manjanatha MG, et al. Molecular analysis of lacI mutations in Rat2 cells exposed to 7,12-dimethylbenz[a]anthracene: Evidence for DNA sequence and DNA strand biases for mutation. Mutat Res. 1996;372(1):53–64. doi: 10.1016/s0027-5107(96)00162-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.