Abstract
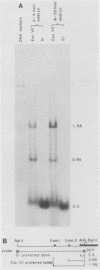
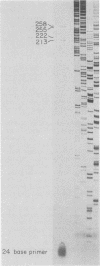
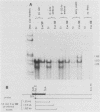
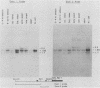
At least six mRNAs are made from the Drosophila melanogaster act5C gene. We investigated the structures of these RNAs in detail and determined that they are heterogeneous at both their 5' and 3' ends. At the 5' end there were two nonhomologous leader exons which were alternately spliced to the remainder of the gene. These leader exons mapped to 1.7 and 0.7 kilobases, respectively, upstream of a common splice acceptor site which was eight base pairs 5' to the translation initiator AUG. Exon 1 is 147 bases in length, while exon 2 is 111 bases. A consensus TATA sequence was found roughly 30 base pairs upstream from exon 1, but none was found in the analogous position upstream of exon 2. The transcript length diversity arose principally from the use of three polyadenylation sites. This gave rise to RNA molecules with 3'-untranslated regions of roughly 375, 655, and 945 base pairs. With two start sites and three termination sites, this gene has the potential to produce six different transcripts. All six possible transcripts were present in whole fly mRNA. Transcripts containing the two different leader exons were found in roughly the same relative quantities through development. In contrast, the various 3' ends were differentially represented through development.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker C. C., Herisse J., Courtois G., Galibert F., Ziff E. Messenger RNA for the Ad2 DNA binding protein: DNA sequences encoding the first leader and heterogenity at the mRNA 5' end. Cell. 1979 Oct;18(2):569–580. doi: 10.1016/0092-8674(79)90073-4. [DOI] [PubMed] [Google Scholar]
- Bantle J. A., Maxwell I. H., Hahn W. E. Specificity of oligo (dT)-cellulose chromatography in the isolation of polyadenylated RNA. Anal Biochem. 1976 May 7;72:413–427. doi: 10.1016/0003-2697(76)90549-2. [DOI] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. J Biol Chem. 1982 Mar 25;257(6):3018–3025. [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Benyajati C., Spoerel N., Haymerle H., Ashburner M. The messenger RNA for alcohol dehydrogenase in Drosophila melanogaster differs in its 5' end in different developmental stages. Cell. 1983 May;33(1):125–133. doi: 10.1016/0092-8674(83)90341-0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Spliced early mRNAs of simian virus 40. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1274–1278. doi: 10.1073/pnas.75.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Capetanaki Y. G., Ngai J., Flytzanis C. N., Lazarides E. Tissue-specific expression of two mRNA species transcribed from a single vimentin gene. Cell. 1983 Dec;35(2 Pt 1):411–420. doi: 10.1016/0092-8674(83)90174-5. [DOI] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Cheng H. L., Blattner F. R., Fitzmaurice L., Mushinski J. F., Tucker P. W. Structure of genes for membrane and secreted murine IgD heavy chains. Nature. 1982 Apr 1;296(5856):410–415. doi: 10.1038/296410a0. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Early P., Rogers J., Davis M., Calame K., Bond M., Wall R., Hood L. Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell. 1980 Jun;20(2):313–319. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
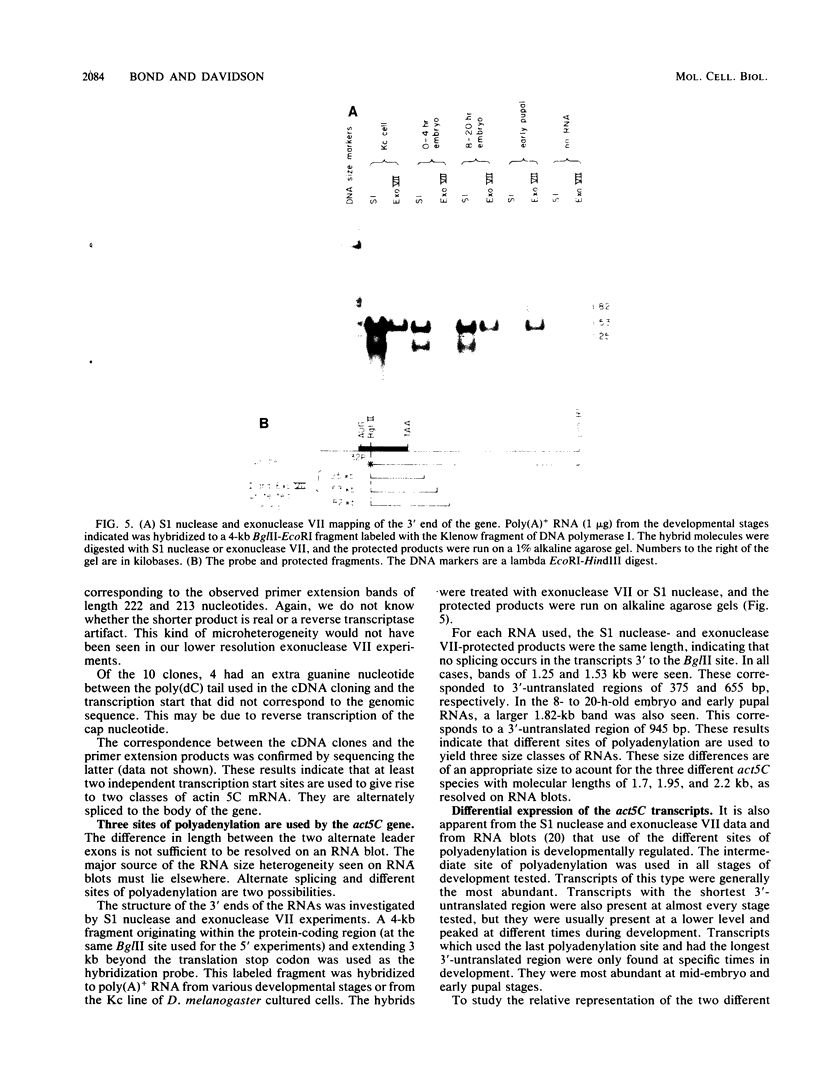
- Falkenthal S., Parker V. P., Davidson N. Developmental variations in the splicing pattern of transcripts from the Drosophila gene encoding myosin alkali light chain result in different carboxyl-terminal amino acid sequences. Proc Natl Acad Sci U S A. 1985 Jan;82(2):449–453. doi: 10.1073/pnas.82.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Fiers W., Contreras R., Haegemann G., Rogiers R., Van de Voorde A., Van Heuverswyn H., Van Herreweghe J., Volckaert G., Ysebaert M. Complete nucleotide sequence of SV40 DNA. Nature. 1978 May 11;273(5658):113–120. doi: 10.1038/273113a0. [DOI] [PubMed] [Google Scholar]
- Flavell A. J., Cowie A., Legon S., Kamen R. Multiple 5' terminal cap structures in late polyoma virus RNA. Cell. 1979 Feb;16(2):357–371. doi: 10.1016/0092-8674(79)90012-6. [DOI] [PubMed] [Google Scholar]
- Fyrberg E. A., Bond B. J., Hershey N. D., Mixter K. S., Davidson N. The actin genes of Drosophila: protein coding regions are highly conserved but intron positions are not. Cell. 1981 Apr;24(1):107–116. doi: 10.1016/0092-8674(81)90506-7. [DOI] [PubMed] [Google Scholar]
- Fyrberg E. A., Kindle K. L., Davidson N., Kindle K. L. The actin genes of Drosophila: a dispersed multigene family. Cell. 1980 Feb;19(2):365–378. doi: 10.1016/0092-8674(80)90511-5. [DOI] [PubMed] [Google Scholar]
- Fyrberg E. A., Mahaffey J. W., Bond B. J., Davidson N. Transcripts of the six Drosophila actin genes accumulate in a stage- and tissue-specific manner. Cell. 1983 May;33(1):115–123. doi: 10.1016/0092-8674(83)90340-9. [DOI] [PubMed] [Google Scholar]
- Gerlinger P., Krust A., LeMeur M., Perrin F., Cochet M., Gannon F., Dupret D., Chambon P. Multiple initiation and polyadenylation sites for the chicken ovomucoid transcription unit. J Mol Biol. 1982 Dec 5;162(2):345–364. doi: 10.1016/0022-2836(82)90531-9. [DOI] [PubMed] [Google Scholar]
- Ghosh P. K., Reddy V. B., Swinscoe J., Choudary P. V., Lebowitz P., Weissman S. M. The 5'-terminal leader sequence of late 16 S mRNA from cells infected with simian virus 40. J Biol Chem. 1978 May 25;253(10):3643–3647. [PubMed] [Google Scholar]
- Ghosh P. K., Reddy V. B., Swinscoe J., Lebowitz P., Weissman S. M. Heterogeneity and 5'-terminal structures of the late RNAs of simian virus 40. J Mol Biol. 1978 Dec 25;126(4):813–846. doi: 10.1016/0022-2836(78)90022-0. [DOI] [PubMed] [Google Scholar]
- Grez M., Land H., Giesecke K., Schütz G., Jung A., Sippel A. E. Multiple mRNAs are generated from the chicken lysozyme gene. Cell. 1981 Sep;25(3):743–752. doi: 10.1016/0092-8674(81)90182-3. [DOI] [PubMed] [Google Scholar]
- Hagenbüchle O., Schibler U., Petrucco S., Van Tuyle G. C., Wellauer P. K. Expression of mouse Amy-2a alpha-amylase genes is regulated by strong pancreas-specific promoters. J Mol Biol. 1985 Sep 20;185(2):285–293. doi: 10.1016/0022-2836(85)90404-8. [DOI] [PubMed] [Google Scholar]
- Hagenbüchle O., Tosi M., Schibler U., Bovey R., Wellauer P. K., Young R. A. Mouse liver and salivary gland alpha-amylase mRNAs differ only in 5' non-translated sequences. Nature. 1981 Feb 19;289(5799):643–646. doi: 10.1038/289643a0. [DOI] [PubMed] [Google Scholar]
- Hanauer A., Levin M., Heilig R., Daegelen D., Kahn A., Mandel J. L. Isolation and characterization of cDNA clones for human skeletal muscle alpha actin. Nucleic Acids Res. 1983 Jun 11;11(11):3503–3516. doi: 10.1093/nar/11.11.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilig R., Perrin F., Gannon F., Mandel J. L., Chambon P. The ovalbumin gene family: structure of the X gene and evolution of duplicated split genes. Cell. 1980 Jul;20(3):625–637. doi: 10.1016/0092-8674(80)90309-8. [DOI] [PubMed] [Google Scholar]
- Hunkapiller M., Kent S., Caruthers M., Dreyer W., Firca J., Giffin C., Horvath S., Hunkapiller T., Tempst P., Hood L. A microchemical facility for the analysis and synthesis of genes and proteins. Nature. 1984 Jul 12;310(5973):105–111. doi: 10.1038/310105a0. [DOI] [PubMed] [Google Scholar]
- Lai E. C., Roop D. R., Tsai M. J., Woo S. L., O'Malley B. W. Heterogeneous initiation regions for transcription of the chicken ovomucoid gene. Nucleic Acids Res. 1982 Sep 25;10(18):5553–5567. doi: 10.1093/nar/10.18.5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeMeur M. A., Galliot B., Gerlinger P. Termination of the ovalbumin gene transcription. EMBO J. 1984 Dec 1;3(12):2779–2786. doi: 10.1002/j.1460-2075.1984.tb02209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeMeur M., Glanville N., Mandel J. L., Gerlinger P., Palmiter R., Chambon P. The ovalbumin gene family: hormonal control of X and Y gene transcription and mRNA accumulation. Cell. 1981 Feb;23(2):561–571. doi: 10.1016/0092-8674(81)90152-5. [DOI] [PubMed] [Google Scholar]
- Lee M. G., Lewis S. A., Wilde C. D., Cowan N. J. Evolutionary history of a multigene family: an expressed human beta-tubulin gene and three processed pseudogenes. Cell. 1983 Jun;33(2):477–487. doi: 10.1016/0092-8674(83)90429-4. [DOI] [PubMed] [Google Scholar]
- Malek L. T., Eschenfeldt W. H., Munns T. W., Rhoads R. E. Heterogeneity of the 5' terminus of hen ovalbumin messenger ribonucleic acid. Nucleic Acids Res. 1981 Apr 10;9(7):1657–1673. doi: 10.1093/nar/9.7.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McDonell M. W., Simon M. N., Studier F. W. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J Mol Biol. 1977 Feb 15;110(1):119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- Milcarek C., Hall B. Cell-specific expression of secreted versus membrane forms of immunoglobulin gamma 2b mRNA involves selective use of alternate polyadenylation sites. Mol Cell Biol. 1985 Oct;5(10):2514–2520. doi: 10.1128/mcb.5.10.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima Y., Fujii-Kuriyama Y., Muramatsu M., Ogata K. Alternative transcription and two modes of splicing results in two myosin light chains from one gene. Nature. 1984 Mar 22;308(5957):333–338. doi: 10.1038/308333a0. [DOI] [PubMed] [Google Scholar]
- Nevins J. R., Wilson M. C. Regulation of adenovirus-2 gene expression at the level of transcriptional termination and RNA processing. Nature. 1981 Mar 12;290(5802):113–118. doi: 10.1038/290113a0. [DOI] [PubMed] [Google Scholar]
- Parker C. S., Topol J. A Drosophila RNA polymerase II transcription factor contains a promoter-region-specific DNA-binding activity. Cell. 1984 Feb;36(2):357–369. doi: 10.1016/0092-8674(84)90229-0. [DOI] [PubMed] [Google Scholar]
- Parker V. P., Falkenthal S., Davidson N. Characterization of the myosin light-chain-2 gene of Drosophila melanogaster. Mol Cell Biol. 1985 Nov;5(11):3058–3068. doi: 10.1128/mcb.5.11.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnes J. R., Robinson R. R., Seidman J. G. Multiple mRNA species with distinct 3' termini are transcribed from the beta 2-microglobulin gene. 1983 Mar 31-Apr 6Nature. 302(5907):449–452. doi: 10.1038/302449a0. [DOI] [PubMed] [Google Scholar]
- Perlman D., Halvorson H. O., Cannon L. E. Presecretory and cytoplasmic invertase polypeptides encoded by distinct mRNAs derived from the same structural gene differ by a signal sequence. Proc Natl Acad Sci U S A. 1982 Feb;79(3):781–785. doi: 10.1073/pnas.79.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochownik E. V. Relationship between an enhancer element in the human antithrombin III gene and an immunoglobulin light-chain gene enhancer. 1985 Aug 29-Sep 4Nature. 316(6031):845–848. doi: 10.1038/316845a0. [DOI] [PubMed] [Google Scholar]
- Reddy V. B., Thimmappaya B., Dhar R., Subramanian K. N., Zain B. S., Pan J., Ghosh P. K., Celma M. L., Weissman S. M. The genome of simian virus 40. Science. 1978 May 5;200(4341):494–502. doi: 10.1126/science.205947. [DOI] [PubMed] [Google Scholar]
- Sanchez F., Tobin S. L., Rdest U., Zulauf E., McCarthy B. J. Two Drosophila actin genes in detail. Gene structure, protein structure and transcription during development. J Mol Biol. 1983 Feb 5;163(4):533–551. doi: 10.1016/0022-2836(83)90111-0. [DOI] [PubMed] [Google Scholar]
- Sasavage N. L., Smith M., Gillam S., Woychik R. P., Rottman F. M. Variation in the polyadenylylation site of bovine prolactin mRNA. Proc Natl Acad Sci U S A. 1982 Jan;79(2):223–227. doi: 10.1073/pnas.79.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U., Hagenbüchle O., Wellauer P. K., Pittet A. C. Two promoters of different strengths control the transcription of the mouse alpha-amylase gene Amy-1a in the parotid gland and the liver. Cell. 1983 Jun;33(2):501–508. doi: 10.1016/0092-8674(83)90431-2. [DOI] [PubMed] [Google Scholar]
- Setzer D. R., McGrogan M., Nunberg J. H., Schimke R. T. Size heterogeneity in the 3' end of dihydrofolate reductase messenger RNAs in mouse cells. Cell. 1980 Nov;22(2 Pt 2):361–370. doi: 10.1016/0092-8674(80)90346-3. [DOI] [PubMed] [Google Scholar]
- Setzer D. R., McGrogan M., Schimke R. T. Nucleotide sequence surrounding multiple polyadenylation sites in the mouse dihydrofolate reductase gene. J Biol Chem. 1982 May 10;257(9):5143–5147. [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin S. L., Zulauf E., Sánchez F., Craig E. A., McCarthy B. J. Multiple actin-related sequences in the Drosophila melanogaster genome. Cell. 1980 Jan;19(1):121–131. doi: 10.1016/0092-8674(80)90393-1. [DOI] [PubMed] [Google Scholar]
- Tosi M., Young R. A., Hagenbüchle O., Schibler U. Multiple polyadenylation sites in a mouse alpha-amylase gene. Nucleic Acids Res. 1981 May 25;9(10):2313–2323. doi: 10.1093/nar/9.10.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterman R. D., Lynch K. R., Nakhasi H. L., Dolan K. P., Hamilton J. W., Cohn D. V., Feigelson P. Cloning and sequence of several alpha 2u-globulin cDNAs. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3478–3482. doi: 10.1073/pnas.78.6.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M., Stephenson P. Role of the conserved AAUAAA sequence: four AAUAAA point mutants prevent messenger RNA 3' end formation. Science. 1984 Nov 30;226(4678):1045–1051. doi: 10.1126/science.6208611. [DOI] [PubMed] [Google Scholar]
- Young R. A., Hagenbüchle O., Schibler U. A single mouse alpha-amylase gene specifies two different tissue-specific mRNAs. Cell. 1981 Feb;23(2):451–458. doi: 10.1016/0092-8674(81)90140-9. [DOI] [PubMed] [Google Scholar]
- Zehner Z. E., Paterson B. M. Characterization of the chicken vimentin gene: single copy gene producing multiple mRNAs. Proc Natl Acad Sci U S A. 1983 Feb;80(4):911–915. doi: 10.1073/pnas.80.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]