Abstract
We provide an integrated dynamic view on a eukaryotic osmolyte system, linking signaling with regulation of gene expression, metabolic control and growth. Adaptation to osmotic changes enables cells to adjust cellular activity and turgor pressure to an altered environment. The yeast Saccharomyces cerevisiae adapts to hyperosmotic stress by activating the HOG signaling cascade, which controls glycerol accumulation. The Hog1 kinase stimulates transcription of genes encoding enzymes required for glycerol production (Gpd1, Gpp2) and glycerol import (Stl1) and activates a regulatory enzyme in glycolysis (Pfk26/27). In addition, glycerol outflow is prevented by closure of the Fps1 glycerol facilitator. In order to better understand the contributions to glycerol accumulation of these different mechanisms and how redox and energy metabolism as well as biomass production are maintained under such conditions we collected an extensive dataset. Over a period of 180 min after hyperosmotic shock we monitored in wild type and different mutant cells the concentrations of key metabolites and proteins relevant for osmoadaptation. The dataset was used to parameterize an ODE model that reproduces the generated data very well. A detailed computational analysis using time-dependent response coefficients showed that Pfk26/27 contributes to rerouting glycolytic flux towards lower glycolysis. The transient growth arrest following hyperosmotic shock further adds to redirecting almost all glycolytic flux from biomass towards glycerol production. Osmoadaptation is robust to loss of individual adaptation pathways because of the existence and upregulation of alternative routes of glycerol accumulation. For instance, the Stl1 glycerol importer contributes to glycerol accumulation in a mutant with diminished glycerol production capacity. In addition, our observations suggest a role for trehalose accumulation in osmoadaptation and that Hog1 probably directly contributes to the regulation of the Fps1 glycerol facilitator. Taken together, we elucidated how different metabolic adaptation mechanisms cooperate and provide hypotheses for further experimental studies.
Author Summary
Osmotic changes are common environmental challenges for cells, even in multi-cellular organisms, having led to sophisticated adaptation mechanisms. In order to adapt to hyperosmotic stress, yeast cells accumulate glycerol. This is achieved by short-term responses involving metabolic and transmembrane transport changes as well as long-term transcriptional responses. By integrating experimentation and simulation of a mathematical model we resolve the quantitative and temporal characteristics of different processes contributing to glycerol accumulation. We show that osmoadaptation prioritizes the redox and energy balance in glycolysis while rerouting flux from biomass to glycerol production. We further show that the glycerol accumulation network provides osmoadaptation with robustness by compensating for the loss of certain nodes and with the flexibility necessary for responding to different stress situations. Finally we provide novel insight into the roles of transport processes in glycerol accumulation and evidence that trehalose may play a role in yeast osmoadaptation. The present work provides for the first time an integrated dynamic view on a eukaryotic osmolyte system and links signaling with regulation of gene expression and metabolic control.
Introduction
Upon increase in external osmolarity, cells first shrink and subsequently recover volume by accumulating compatible solutes [1]. Different processes contribute to adaptation, establishing negative feedback loops for the regulation of osmotic pressure, cell volume, and turgor [2]–[5]. The yeast Saccharomyces cerevisiae employs glycerol as compatible solute for osmo-regulation. Upon hyperosmotic shock the High Osmolarity Glycerol (HOG) pathway is activated, resulting in phosphorylation of the stress-activated protein (SAP) kinase Hog1. Phosphorylated Hog1 stimulates expression of genes encoding enzymes involved in glycerol production and uptake. Hyperosmotic stress also leads to rapid closure of the glycerol facilitator Fps1 preventing glycerol outflow. The contributions to glycerol accumulation of the different processes and regulatory mechanisms – including central metabolism - have not been systematically studied in a quantitative and time-resolved manner.
In the absence of hyperosmotic stress, glycerol production is required for maintaining the redox balance [1], [6] and excess glycerol leaks out freely through the glycerol facilitator Fps1 [7], [8]. The glycerol production pathway starts with the reduction of the glycolytic intermediate di-hydroxyl-acetone phosphate (DHAP) to glycerol-3-phosphate (G3P) catalyzed by the NAD+-dependent glycerol-3-phospate dehydrogenase. This enzyme is encoded by two isogenes, where osmostress controls expression of GPD1 and cellular redox potential controls expression of GPD2 [6], [9]–[11]. Stimulated expression of GPD1 [12], [13] enhances glycerol production under hyperosmotic conditions. G3P is transformed to glycerol by the G3P phosphatases Gpp1/Rhr2 and Gpp2/Hor2 [6], [9], [10], [14]–[16]. Hog1 may also control the amount of DHAP available for glycerol production: it appears that Hog1 stimulates the 6-phosphofructo-2-kinase Pfk26, which, together with its isoform Pfk27, produces fructose-2,6-diphosphate (F26DP), an allosteric activator of the glycolytic enzyme phosphofructokinase (Pfk1) [17]. Glycerol transmembrane transport is facilitated by the aquaglyceroporin Fps1. Its rapid closure upon osmostress prevents glycerol outflow [18]. Active glycerol uptake is mediated by the glycerol-proton symporter Stl1, whose expression is strongly up-regulated upon osmoshock [19], [20] and down-regulated by glucose repression. Fig. 1 presents an overview of the known mechanisms involved in osmoadaptation.
Figure 1. Overview of osmoadaptation in S. cerevisiae.
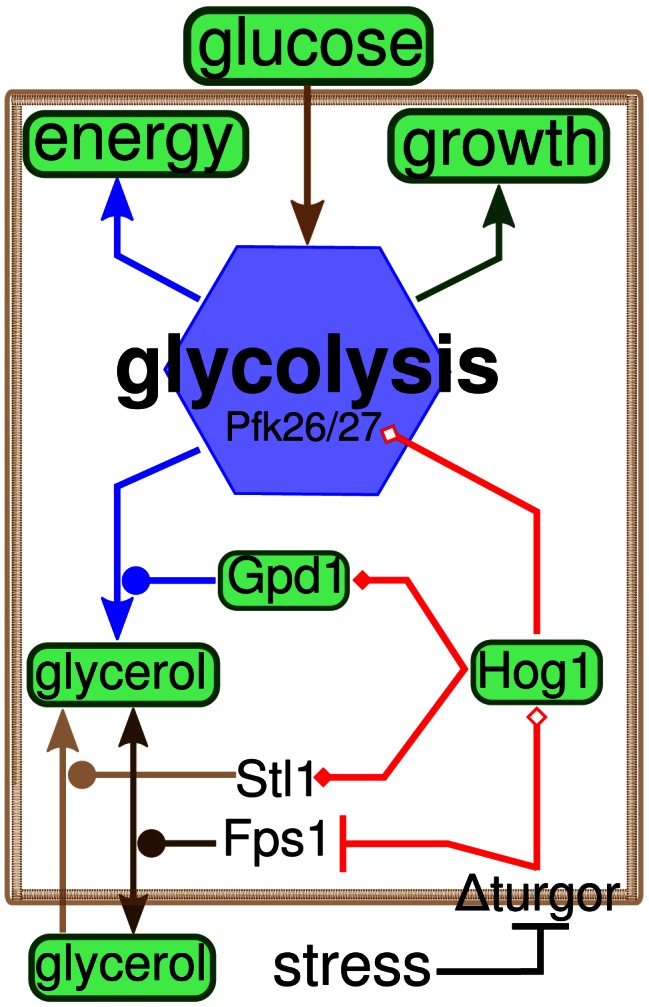
Arrows indicate mass flow, diamonds indicate positive regulation (empty: direct, filled: gene expression), circles indicate catalysis, bars indicate inhibition. Measured entities are highlighted green (for a complete list of experiments refer to SI). Processes are colored according to the different modules (transport: brown, glycolysis: blue, growth: green, adaptation: red).
Genetic tools to study osmoadaptation include a range of knockout mutants and several strains with alterations in regulatory nodes. The strain FPS1-Δ1 contains a mutation that prevents the stress dependent closure of Fps1; this strain produces glycerol but loses it by diffusion through Fps1 [8]. HOG1-att contains a tag that tethers Hog1 to the plasma membrane. Such cells are osmo-resistant but do not mount a Hog1-dependent gene expression response [21].
In this study we integrate analysis of five distinct control mechanisms for glycerol accumulation and their link to glycolysis: (i) regulation of GPD1 expression by nuclear Hog1, (ii) activation of Pfk26 by cytosolic Hog1, (iii) regulation of glycerol transport through Fps1, (iv) volume and associated concentration changes of all cellular compounds, and (v) metabolic reconfiguration resulting in rerouting of fluxes. To study the interplay between these mechanisms, we employed an iterative approach of modeling and experimentation. This model is based on a far more extensive dataset than several previous models including experimental data on a range of metabolites as well as the Hog1 and Gpd1 proteins in wild type and different mutants. The interaction between the different glycerol accumulation processes can be considered as a network in which each node represents one means of control. We tested the robustness of this network to genetic perturbations by model simulation and experimentation.
This work for the first time provides an overview of the dynamics of a eukaryotic osmolyte system, integrating biophysical changes with signaling, control of gene expression, and regulation of metabolic flux. Specifically, we provide evidence that (i) rerouting of glycolytic flux from growth to glycerol production is a major effector of osmoadaptation, (ii) different glycerol accumulation control mechanisms can (partly) compensate for each other, providing robustness and flexibility, (iii) long-term adaptation may involve other osmolytes than glycerol and (iv) Fps1-mediated glycerol efflux is down-regulated by active Hog1.
Results
Model
Based on previous work [22], pilot experiments, and a previous model [2] we constructed a refined mathematical model (Supplementary Information (SI) and Fig. S10). We included carbon flux to biomass, the influence of Hog1 on glycolytic flux via Pfk26/27, and Stl1-mediated glycerol uptake. This model comprises the following modules:
Biophysical changes: changes in cell volume (basal solid volume, Vb, and osmotically active volume, Vos [23], [24]), cell surface area, osmotic pressure, and turgor pressure (where changes depend on Vos [2] and external osmolarity).
Glycolysis module: metabolic reactions from glucose uptake via phosphorylated intermediates to glycerol, trehalose, ethanol, and acetate. The module is based on models [2], [25] where reactions have been lumped to reduce the number of parameters (see Text S1 for details). The
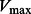 of the Pfk1 reaction is modulated by F26DP, which depends on the Hog1-regulated activity of Pfk26/27.
of the Pfk1 reaction is modulated by F26DP, which depends on the Hog1-regulated activity of Pfk26/27.Transport module: glucose, trehalose, ethanol, acetate transport, glycerol exchange via Fps1, and irreversible glycerol uptake via Stl1. We corrected rate laws of transport reactions for cell density increase in time-course experiments.
Biomass module: biomass production is necessary for describing the interplay between osmotic regulation and glycolysis, but also to maintain correct carbon balance. Biomass is measured as cell number or cell density. Upon osmoshock, Hog1 arrests cell cycle progression [26], [27] and hence proliferation. Hog1-activity affects metabolism by altering expression of metabolic genes [20], [28]–[30]. A Hog1-dependent drop in growth rate is included in the model (
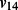 ).
).Adaptation module: Hog1 phosphorylation, GPD1, and STL1 mRNA, translation to Gpd1 and Stl1, activation of Pfk26/27, and closure of Fps1. Hog1 activity is correlated with volume changes and turgor [24]. The Hog1 signaling system is described with only two reactions.
In order to ensure that the available data support the model, which covers different major processes, metabolism is highly condensed for this specific study, compared to metabolic reconstructions suitable for steady state analyses. For example, we refrained from including redox balance dynamics since they are affected by cellular changes beyond the scope of this model, such as detailed biosynthetic pathways (see Text S1). We do not assume a steady state for the entire model because we consider external metabolites and cell density. Although most intracellular concentrations can be considered constant before osmotic stress, the increasing external concentrations of ethanol, acetate and glycerol and the decreasing external glucose level together with the increase in OD slightly affect model dynamics. Therefore, the model is not in steady state in a strict sense.
Models for the different mutant strains are generated by implementing known effects of the respective genetic perturbation (see Text S1). Besides these modifications, all models use the same parameter set.
Data set
To estimate model parameters and determine time scales, we measured intracellular and extracellular concentrations of glycerol, glucose, ethanol, acetate, and trehalose over 240 min for wild type and mutants lacking particular adaptation mechanisms (hog1Δ, gpd1Δ, pfk26Δ, pfk27Δ, pfk26/27Δ) or with altered regulation nodes (HOG1-att, FPS1-Δ1) (Fig. 2, SI). While external glucose is consumed, glycerol, ethanol, and acetate as well OD600 and cell numbers (SI, Fig. S7) increased over time. Internal glycerol (Fig. 2C) accumulates transiently, while trehalose shows two peaks at 45 min and 180 min (Fig. S7G). We also measured GPD1 mRNA concentration, Gpd1 concentration, and Hog1 phosphorylation (Fig. 2A–B, Fig. S13B). The following characteristics were noted.
Figure 2.
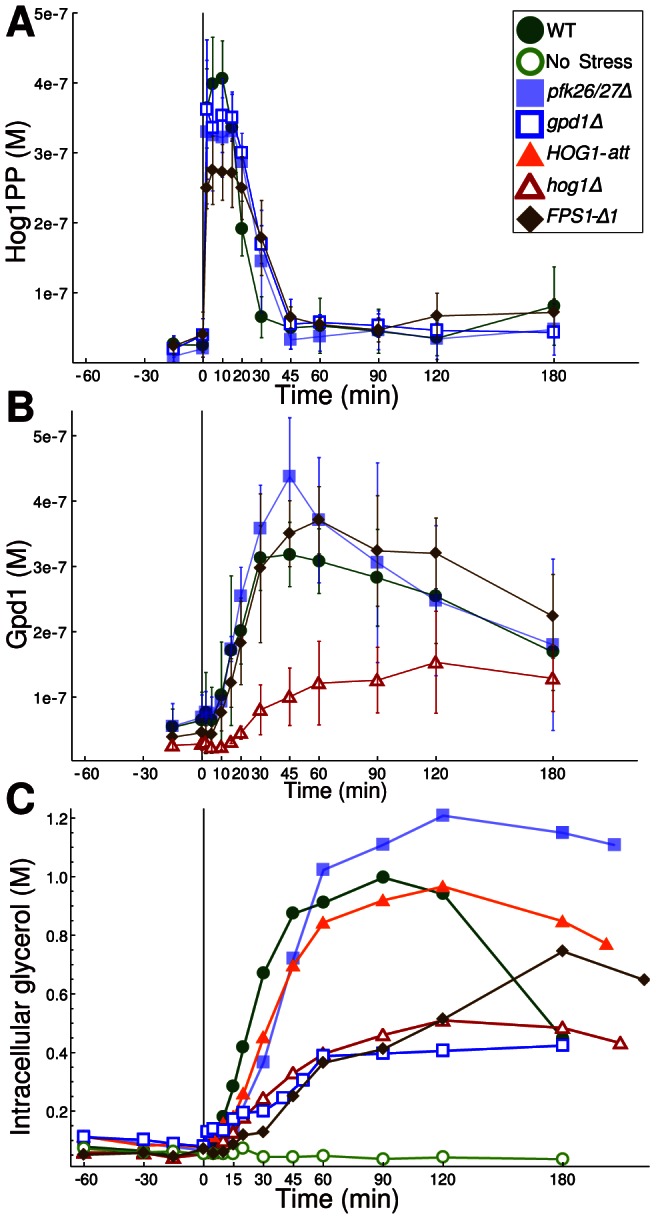
Time courses of (A) phosphorylated Hog1, (B) Gpd1, and (C) intracellular glycerol following hyperosmotic stress of 0.4 M NaCl at time point 0. The full dataset is provided in Datasets S1, S2, S3, S4, S5, S6, S7.
In the gpd1Δ mutant, the extracellular glycerol decreases, while the FPS1-Δ1 strain excretes large amounts of glycerol (Fig. S8).
Intracellular glycerol (Fig. 2C) builds up with a delay in the FPS1-Δ1 and hog1Δ mutants. In the pfk26/27Δ and Hog1-att [21] strains, intracellular glycerol remains high over the entire period.
The pfk26/27Δ mutant displays slower glucose consumption (Fig. S7B).
Trehalose dynamics at 180 min indicate that it may accumulate further in wild type; pfk26/27Δ and HOG1-att strains do not display a late increase of trehalose (Fig. S7G).
Hog1 phosphorylation (Fig. 2A) is slightly prolonged in cells lacking Gpd1 or Pfk26/27 or expressing constitutively open Fps1.
The lowest Gpd1 levels are observed for the hog1Δ mutant (Fig. 2B).
Simulation of osmoadaptation
The model was fitted to experimental data (Figs. S12, S13, S14). In general, model fitting can follow two different paradigms: (i) either fit the model to part of the data and use the rest of the data to test model predictions or (ii) use all data for fitting to obtain the model best describing the observations. Since our quantitative time-resolved data of various compounds for wild type and different mutants comprehensively elucidates the potential network dynamics, we decided to eventually use all data from all yeast strains and conditions to parameterize the final model presented here. To reproduce the Gpd1-mediated increase of glycerol production, dimerization of Gpd1, as reported for human Gpd [21], [31], [32], was implemented. We further implemented a partial Hog1-independent increase of GPD1 mRNA to reproduce hog1Δ data, which is in accordance with experimental observations [21], [22]. To fit gpd1Δ mutant data, we incorporated a small osmo-dependent increase in GPD2 transcription. Including regulation of biomass production [30] significantly improved model behavior (SI): A rapid drop in cell growth, dependent on stress and cell volume, is necessary to provide sufficient carbon for glycerol production and reproduce experimental data on cell density. To reproduce experimental data for intracellular glycerol in hog1Δ and HOG1-att strains, we included a negative regulation of open Fps1 by active Hog1.
Model analysis and predictions
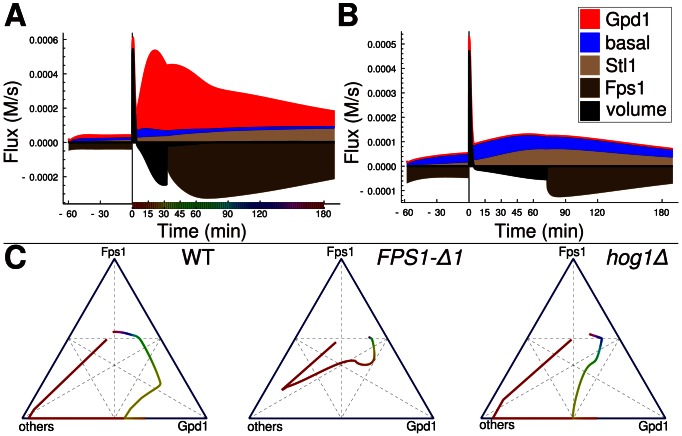
To characterize the contribution of different processes to osmoadaptation (Fig. 3), we plotted the absolute fluxes to and from glycerol for wild type and gpd1Δ over time (Fig. 3A and B) and relative contributions of fluxes in ternary plots (Fig. 3C). We distinguish Fps1-reliant, Gpd1-reliant, and other contributions (‘Others’) to net glycerol flux in each strain. For wild type, immediate adaptation is influenced mainly by changes in volume (part of ‘Others’) and closure of Fps1. Within approximately 15 min after stress, glycerol production becomes the predominant contribution. After 30 min, Fps1 reopens and glycerol levels decrease. Open Fps1 in FPS1-Δ1 results in constitutive glycerol efflux. Although the model underestimates the glycerol production for this mutant, it clearly shows that the distribution of fluxes remains at a state of sustained glycerol production. Sustained glycerol production occurs also in the hog1Δ mutant, but due to reduced Hog1-dependent GPD1 transcription its level remains low until 60 min after stress. Subsequently, the system moves to a state similar to that observed for the FPS1-Δ1 strain.
Figure 3. Contribution of glycerol accumulation mechanisms in different strains.
(A, B) Absolute fluxes towards glycerol as well as relative contributions of specific mechanisms differ between wild type WT (A) and gpd1Δ (B). (C) Changes in relative contributions of Fps1, Gpd1, and other effects (basal glycerol production, uptake through Stl1, effects of volume change) over time are depicted for WT, fps1-Δ1 and hog1Δ. Colors in (C) indicate time as shown on the x-axis in (A).
Although the relative composition of fluxes approaches pre-stress values in wild-type, we do not observe perfect adaptation [33] (Fig. 4). For a more comprehensive analysis, the complete state of cells has to be considered, including changes in reactions or pathways indirectly affecting glycerol concentrations, e.g. changes in biomass production. To do so, we employed scaled time-dependent response coefficients (RCs), 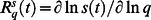 for a compound concentration
for a compound concentration 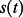 and a parameter q
[34]. RCs express the relative change in
and a parameter q
[34]. RCs express the relative change in  given a small change in q, serving as quantitative measure for the effect of a parameter perturbation on a time course taking all direct and indirect effects into account. Positive or negative values indicate that the time course increases or decreases upon parameter increase, respectively. Scaled RCs (Fig. 4B–C) show that regulation of Pfk26/27 does not have a pronounced effect on intracellular glycerol (Fig. 4B), which is in accordance with experimental data for pfk26/27Δ (Fig. 2C). Additionally, other reactions downstream of Pfk1 are positively affected while glycolytic reactions upstream of Pfk1 (trehalose and biomass production) are reduced. This indicates that Pfk26/27 might be part of a rerouting mechanism of metabolic flux ensuring that the influx to each branch of glycolysis is adjusted to demands and that ATP-production downstream of pyruvate is maintained during osmoadaptation.
given a small change in q, serving as quantitative measure for the effect of a parameter perturbation on a time course taking all direct and indirect effects into account. Positive or negative values indicate that the time course increases or decreases upon parameter increase, respectively. Scaled RCs (Fig. 4B–C) show that regulation of Pfk26/27 does not have a pronounced effect on intracellular glycerol (Fig. 4B), which is in accordance with experimental data for pfk26/27Δ (Fig. 2C). Additionally, other reactions downstream of Pfk1 are positively affected while glycolytic reactions upstream of Pfk1 (trehalose and biomass production) are reduced. This indicates that Pfk26/27 might be part of a rerouting mechanism of metabolic flux ensuring that the influx to each branch of glycolysis is adjusted to demands and that ATP-production downstream of pyruvate is maintained during osmoadaptation.
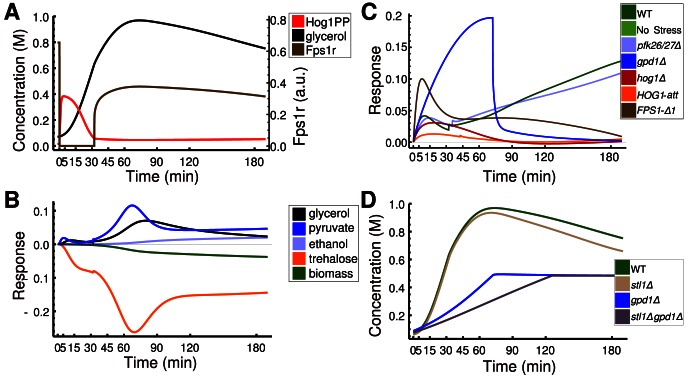
Figure 4. Model analysis with time-dependent response coefficients.
A: Model simulation for phosphorylated Hog1, intracellular glycerol, and abundance of open Fps1. B: Effect of small changes in Pfk26/27 activation on different model variables as expressed by normalized response coefficient indicates that Pfk26/27 contributes to a rerouting of flux towards lower glycolysis. C: Response of intracellular glycerol concentration to perturbations in Stl1 gene expression as measured by normalized response coefficients in different strains indicates a specific time- and context-dependent role of Stl1in osmoadaptation. D: Simulation of genetic perturbations of Stl1 results in time courses as expected from C: in wild type, deletion of STL1 affects intracellular glycerol levels only at later time points. In gpd1Δ background, the effect of additional deletion of STL1 is early and transient.
We use RCs to compare the role of Stl1 in different experiments (Fig. 4C): apparently the contribution of Stl1, though negligible in most strains until 90 min after stress, is significant in gpd1Δ. In silico predictions show an early decrease in intracellular glycerol accumulation in the gpd1Δ stl1Δ double mutant compared with gpd1Δ but a late decrease in the stl1Δ mutant compared with wild type (Fig. 4D). The different roles of Stl1 in wild type and gpd1Δ cells highlight context specificity in osmoadaptation.
The regulation of biomass production significantly contributes to glycerol accumulation (Fig. 5, Fig. S15B). Rerouting of glycolytic flux can be assumed to result from Hog1-mediated cell-cycle arrest and glycolytic regulation. The cost of maintaining a certain cell volume by producing more glycerol is compensated by a decrease in growth rate. Changes in doubling times before and after stress are plotted in Fig. 5A. The observed decrease in growth rate is similar for wild type, pfk26/27Δ, and HOG1-att strains. This indicates that the main contribution of Hog1 activity to osmoadaptation is not the transcriptional activation of GPD1, which is absent in the HOG1-att strain. Instead, control of cytosolic or membrane-bound targets constitutes the main contributions of Hog1. A strong growth rate drop is observed in FPS1-Δ1 and hog1Δ, while this drop is relatively low in the gpd1Δ mutant. This supports the idea that a Hog1-effect on Fps1 is mainly responsible for the prominent role of Hog1 in osmoadaptation. Fig. 5B shows model simulations of the relative carbon fluxes from glycolysis to glycerol or biomass production, respectively, for wild type and different mutants before 20 min and 90 min after stress induction, indicating a trade-off of cellular adaptation versus growth.
Figure 5. Effect of salt stress on growth rate.
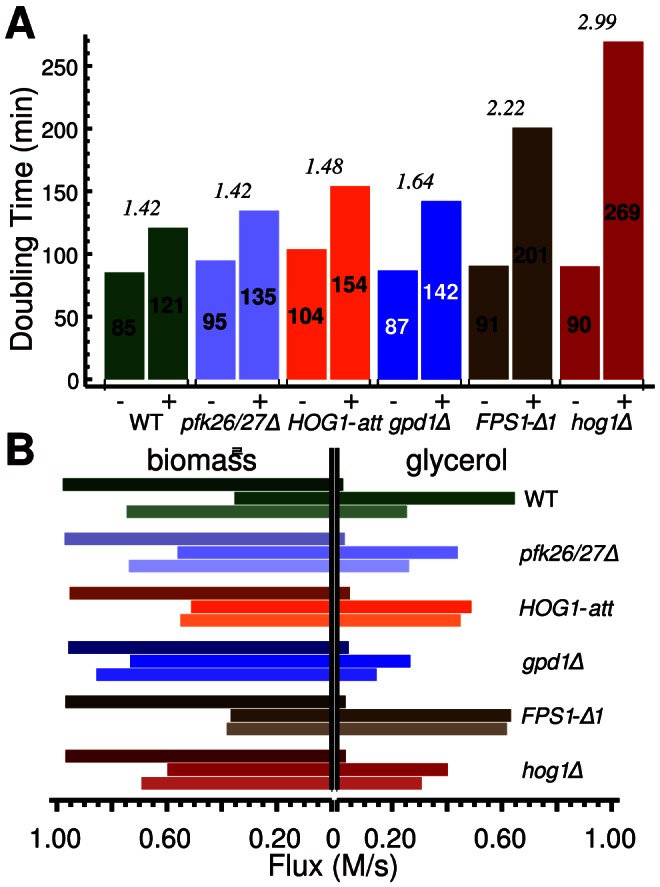
A: In vivo doubling times (−: before, +: after addition of 0.4 M NaCl) strongly differ between strains. B: Model simulations of the flux towards biomass production (left) and glycerol production (right) in the different strains at 0, 20, and 90 minutes after osmotic upshift to 0.4 M NaCl indicate a link between insufficient glycerol accumulation and a prolonged decrease in growth rate.
The model is fitted to data covering a period until 180 min after stimulation. However, cellular processes beyond the scope of the model may interfere with the adaptation process at later stages, hence model results and hypotheses derived from those may be less reliable beyond 120 min.
Discussion
For the first time we present a data-driven analysis of the dynamic regulation of a eukaryotic osmolyte system mediating adaptation to hyperosmotic stress that integrates different regulatory layers. Our mathematical model, supported by a detailed dedicated dataset, provides novel insight into the quantitative contributions of regulatory processes underlying glycerol accumulation during the yeast osmostress response.
The main conclusions are:
The Hog1 SAPK mediates regulation of glycolysis via indirect activation of the enzyme Pfk1 and increased expression of glycerol-producing enzymes.
Pfk1 activation serves stabilization of glycolytic flux as much as increased glycerol accumulation.
Glycerol accumulation occurs at the expense of biomass production.
Glycerol accumulation is achieved by different fluxes comprising basal (Gpd2-dependent) and Hog1-regulated (Gpd1-dependent) production, glycerol exchange over the plasma membrane as well as concentration increase through volume loss; these fluxes contribute to varying extent during different periods of adaptation.
The parameter set was fitted to reproduce data from seven different strains and experimental observations on 14 of 29 model variables, providing the model with an unprecedented coverage [35]. This coverage was achieved employing coordinated model reduction and experimentation with the intention to maintain all regulatory nodes while lumping metabolites and proteins for which no experimental data was gathered.
Deleting individual nodes of the glycerol accumulation network (Figs. 1, S10) results in slightly prolonged HOG pathway activation. For FPS1-Δ1, this prolongation is less pronounced than in previous data [2], which is probably due to different copy numbers of the expression construct. In contrast to Dihazi et al. [17], we found that only the double deletion mutant, pfk26Δpfk27Δ shows prolonged Hog1 phosphorylation. The mutation causes a transient delay of glycerol accumulation (Fig. 2C).
We hypothesize that, like human Gpd1 [32], also yeast Gpd1 dimerizes since this assumption results in significantly better fits to experimental data.
It has been shown that Hog1 interacts with and controls Fps1 under certain conditions [36]–[38], although the exact mechanism of Hog1-dependent control of Fps1 remains elusive. Experimental observations (e.g. [8]), as well as the simulations shown here and elsewhere [5], are consistent with a Hog1-dependent down-regulation of glycerol flux through Fps1 under osmostress. In hog1Δ cells the glycerol efflux is elevated while membrane-attached Hog1 results in a very strict regulation.
In the gpd1Δ mutant the glycerol production rate is diminished. Our experimental data show that in this strain the extracellular glycerol concentration decreases over time, indicating glycerol uptake through the Stl1 glycerol-proton symporter. We determined the role of Stl1 in glycerol accumulation using time varying response coefficient analysis [34]. Our simulations indicate that the stl1Δgpd1Δ mutant adapts even worse than the gpd1Δ mutant and does not show a decrease in the extracellular glycerol concentration. This indicates that osmostress response can overrule glucose repression of Stl1, which is supported by experimental data from Ferreira et al [19] showing strongly stimulated STL1 gene expression under osmostress in mutants unable to produce glycerol as well as growth phenotypes of such mutants lacking in addition Stl1.
The presented model underestimates the production of glycerol for FPS1-Δ1. This may have two reasons. First, FPS1-Δ1 cells adapt to the imposed genetic perturbation prior to stress, which can, for example, lead to an up-regulation of glycerol production. Second, the proposed interaction of the modified protein with Hog1 or other potential regulators [38] may be perturbed. Thus, the model indicates that FPS1-Δ1 has global effects beyond the expected de-regulation of glycerol efflux that need to be investigated further.
In adapted yeast cells the intracellular glycerol level diminishes. We observed an increase in intracellular trehalose concentrations coinciding with the decline of intracellular glycerol (Fig. S7G). In the FPS1-Δ1 strain, which is unable to accumulate intracellular glycerol, the intracellular trehalose concentration is permanently high. Although the combination of high trehalose and glycerol concentrations has a synergistic osmoprotective effect [39], the observed trehalose concentrations are too small to significantly contribute to intracellular osmotic pressure. Hence, trehalose may have more specific roles in osmoadaptation by protecting proteins and membranes [40], [41].
We do not observe perfect adaptation in the sense defined previously [33]. Although cell volume returns to its original values, other variables remain perturbed in adapted cells at constant high external osmolarity, such as Gpd1, trehalose or glycerol concentration, and especially growth rate.
Glycolysis serves three major fluxes resulting in the production of ethanol plus acetate, biomass, and glycerol. Although active Hog1 leads to cell cycle arrest [26], [27], potentially decreasing carbon flux towards biomass, a role of glycolysis in the osmostress response has not been considered in detail. Data presented by Dihazi et al. [17] as well as recent gene expression data [42] indicate that glycolytic flux may play a role in osmoadaptation. Our preliminary theoretical work [22] indicated that Pfk26/Pfk27 participate in maintaining the flux towards pyruvate upon adaptation to hyper-osmotic conditions. Glucose consumption and ethanol production rates are similar in all strains studied here, regardless which node of the glycerol accumulation network was removed. The pfk26/27Δ strain is the only exception: the slightly lower ethanol production and glucose consumption observed in this mutant indicate that Pfk26/27 influence glycolysis rather than osmotic adaptation. We conclude that under hyper-osmotic stress part of the carbon flux, which is normally directed towards biomass production, is used for increasing glycerol production. One of the elements in this metabolic prioritization mechanism is Pfk26/27, although it does not seem to be the only factor. The ratio between the production of glycerol and pyruvate (resulting in the formation of ethanol and acetate) is crucial for a proper energy and redox balance. In all strains, the flux towards pyruvate remains unaffected by osmoadaptation while flux towards biomass decreases. Apparently, energy production and redox balance are maintained while growth is temporarily stopped for faster adaptation. This pattern is likely to hold for a wide variety of stresses and adaptation mechanisms.
The relative contribution of each individual glycerol accumulation mechanism depends on the environmental conditions and the physiological state of the cell (Fig. 3A). In their natural environment, cells have to cope with combinations of stresses demanding for dynamic modulation of response mechanisms. The different mechanisms contributing to osmoadaptation might make the process robust but also allow choosing the ‘cheapest’ adaptation strategy under different conditions.
Response coefficient analysis is an excellent tool to elucidate this kind of interdependences. We found that introducing an individual deletion into the glycerol accumulation network is compensated. Our experimental data show that the adaptation takes longer for single knock-out strains, compared to wild type. The activation of the remaining glycerol accumulation mechanisms is prolonged (active component of the compensation) and the remaining mechanisms become more important (passive compensation component).
Minimalistic models of biological processes, such as the osmoadaptation model presented by Mettetal et al. [3] are of great value for understanding general principles underlying a given process. With a sufficient degree of generalization, conclusions drawn from minimalistic models can be applicable for characterizing other cellular events. This approach neglects the individual mechanisms contributing to a cellular function. On the other extreme, the approach of hierarchical control analysis [43] measures the effects of changes on a cellular process in an enormous amount of detail. This approach is crucial for understanding the role of individual components and events. Our work attempts to bridge between the two paradigms, which can be viewed as an intermediate stage of different magnifications of the same picture.
The principles of osmoregulation are conserved from yeast to mammalian cells. The mammalian stress-activated protein kinase p38 is a homolog of Hog1. Like Hog1 it plays a critical role in mounting the adaptive response to stress by controlling metabolism, gene expression and cell cycle progression [44]. Hence, the approaches employed in the present work as well as the conclusions drawn may have consequences for studies on mammalian cells as well.
By integration of experimental data for different strains into a medium-size model and reliably estimating its parameters, we were able to achieve a better understanding of the contribution of individual players to a cellular response – osmoadaptation – in a quantitative and time-resolved manner. Moreover, our analysis has revealed the trade-off between growth control and glycerol accumulation in the adaptation process.
Materials and Methods
Experimental methods
Yeast strains and culturing
Strains used in this work originate from W303-1A (MATa leu2-3/112 ura3-1 trp1-1 his3-11/15 ade2-1 can1-100 GAL SUC2 mal0) [45]. The strains include: wild type, gpd1Δ, pfk26Δpfk27Δ, fps1-Δ1 (kindly provided by Markus Tamás, University of Gothenburg), stl1Δ, hog1Δ, HOG1-att (kindly provided by Jeremy Thorner, University of California at Berkeley). A complete list of strains used in this study is presented in Table 1. Yeast cultures were grown until mid-exponential phase (OD600 = 0.7–1.0) in YPD medium (Yeast Peptone D-glucose; 1% yeast extract (Bacto), 2% peptone (Bacto), 2% glucose), then NaCl was added from a stock solution of 5 M in water to a final concentration of 0.4 M at t = 0 min.
Table 1. Yeast strains used in this study.
| Strain name | Strain genotype | Origin |
| W303-1a | MATa leu23/112 ura31 trp11 his311/15 ade21 can1100 GAL SUC2 | Thomas and Rothstein (1989) [45] |
| YMR84 | W303-1A with gpd1Δ::URA3 | Martijn Rep (Amsterdam)* |
| YSH1583 | W303-1A with pfk26Δ::KanMX | This study |
| YSH1585 | W303-1A with pfk27Δ::KanMX | This study |
| YSH1586 | W303-1A with pfk26Δ::KanMX pfk27Δ::KanMX | This study |
| YMT101 | W303-1A with fsp1Δ::LEU2 Ylp-URA3-fps1-Δ1 | Tamas et al., (1999) [8] |
| YSH2293 | W303-1A with stl1Δ::KanMX | This study |
| YSH444 | W303-1A with hog1Δ::TRP1 | Albertyn et al,. (1994) [12] |
| W303-1A with hog1-att | This study |
*Strain YMR84 was kindly provided by Martijn Rep (Amsterdam) and contains a replacement of the GPD1 upstream region (−883 to +91) by the URA3 gene. The strains was generated using a PCR approach and does not express the GPD1 gene product.
Western blot analysis
Samples of 1 ml were collected at the indicated time points, sedimented and frozen in liquid nitrogen after removing the supernatant. Proteins were extracted by boiling for 10 min in extraction buffer (100 mM Tris-HCl pH 6.8, 20% glycerol, 200 mM DTT, 4% SDS, 10 mM NaF, 0.1 mM Na3V04 (sodium orthovanadate), protease inhibitor (Complete EDTA-free Protease Inhibitor Cocktail tablets, Roche), and 20 mM mercapto-ethanol). The extracts were claried by centrifugation (13 000 rpm in 4°C for 10 min). For each sample 40 µg of protein was separated by electrophoresis on a 10% polyacrylamide gel (SDS-PAGE) and transferred (semi-dry) to a nitrocellulose membrane (Hybond-ECL, Amersham). Membranes were blocked with Odyssey Blocking Buffer (Li-Cor Biosciences) and incubated sequentially with primary antibodies: first primary antibody - phospho-p38 MAPK (Thr180/Tyr182) monoclonal rabbit antibody (Cell Signalling), 1∶1000 in Odyssey Blocking Buffer with TBST (1∶1000), overnight at 4°C; second primary antibody - yC20 total Hog1 polyclonal goat antibody (Santa Cruz Biotechnology Inc.), 1∶2000 in Odyssey Blocking Buffer with TBST (1∶1000), 1 h at room temperature, third primary antibody - rabbit polyclonal antisera Gpd1-A (Innovagen), 1∶2000 in Odyssey Blocking Buffer with TBST (1∶1000), 1 h at room temperature, and simultaneously with secondary antibodies: donkey anti-goat IR Dye 680, 1∶12 500 and donkey anti-rabbit IR Dye 800CW 1∶12500 (Li-Cor Biosciences), in Odyssey Blocking Buffer with TBST (1∶1000), for 45 min at room temperature.
The membranes were scanned using Odyssey Infrared Imaging System (Li-Cor Biosciences) and quantified using Multi Gauge 3.0 (FujiFilm) software.
Metabolite measurements
Samples of 1 ml were collected at the indicated time points. Three types of samples were collected for each time point: intracellular - centrifuged cell pellet without supernatant, extracellular - pure supernatant, removed from intracellular samples, total - 1 ml cells in medium, and frozen in liquid nitrogen. Extracellular samples did not require further processing; total extracts were boiled for 10 min and cleared by centrifugation, cell pellets were extracted with sterile water by boiling for 10 min and cleared by centrifugation. The concentrations of glucose, trehalose, glycerol, acetate, succinate, pyruvate and ethanol were measured by high performance liquid chromatography (DIONEX) with an Aminex HPX-87H ion exchange column (Bio-Rad, Hercules, USA). An isocratic condition was performed with 5 mM H2SO4 as mobile phase at flow rate of 0.6 ml/min and oven temperature of 65°C. Glucose, trehalose, glycerol and ethanol were quantified by a refraction index detector (Waters 410 Differential Refractometer Millipore, CA, USA) and acetate, succinate and pyruvate by ultraviolet-1 visible light absorbance detector (Waters 486 Tunable Absorbance Detector set at 210 nm, Millipore, CA, USA).
Data processing and additional data
Experimental data was processed to account for cell density increase and the reliability of HPLC measurements was assessed by comparison with enzyme assay quantification. For a full description of data processing and additional data, see Text S1.
Mathematical modeling
System dynamics were described by ordinary differential equations (ODEs). A complete list of model equations and parameter estimation procedures is provided in SI. Time-dependent response coefficients have been calculated as described in [34]. Temporal simulations were performed with Mathematica7 (Wolfram Research. Mathematica edition: Version 7.0, 2008). Parameter estimation was done with PottersWheel [46] and SBML-PET [47].
Supporting Information
Raw and processed metabolite measurements for different strains, stress 0.4 M NaCl added at t = 0.
(XLS)
Overview of experimental data as used in fitting.
(XLS)
Comparison of intracellular trehalose measurements by HPLC and enzyme assay, stress 0.4 M NaCl added at t = 0.
(XLS)
Western Blot data for different strains and stress strengths.
(XLS)
Intracellular glycerol quantification by enzyme assay in different strains for different stresses added at t = 0.
(XLS)
Intracellular trehalose quantification by enzyme assay in different strains, stress 0.4 M NaCl added at t = 0.
(XLS)
Northern Blot results for CTT1, GRE2, STL1, 18S in different strains.
(XLS)
Cell density and optical density in stressed (0.4 M NaCl added at t = 0) and control experiments (A) and normalized to the values at t = 0 (B). Experimental setup as explained in main text.
(PDF)
Plot of OD versus cell density values as obtained from control and stressed time course experiments and the function for computing cell density from OD.
(PDF)
Measured (blue) and inferred (pink) intracellular glycerol for different experiments. Errors in the inferred values due to inconsistencies in measurements are visible in FPS1-Δ1, hog1Δ and WT1.
(PDF)
Comparison of glycerol time courses for WT1 and WT4. (A) intracellular glycerol, (B) extracellular glycerol.
(PDF)
Comparison of enzyme assay (blue) and HPLC (pink) intracellular glycerol quantifications. Underlying data is given in Supplemental Dataset S2.
(PDF)
Comparison of intracellular trehalose levels obtained with different methods. Enzyme assay data is in g D-glucose/l per µg protein/ml, HPLC data in mol/l is normalized to the enzyme assay value at t = 30. Underlying data is given in Supplemental Dataset S3.
(PDF)
Experimental data as used for model fitting. Measured entities are indicated on y-axes. Shown are representative experiments for each strain. Stress of 0.4 M NaCl is added at t = 0. A: intracellular glucose, B: extracellular glucose, C: Optical density, D: intracellular ethanol, E: extracellular ethanol, F: extracellular acetate, G: intracellular trehalose, H: extracellular trehalose, I: extracellular glycerol.
(PDF)
Experimental data for extracellular glycerol (A,B) and Gpd1 (C) following addition of 0.4 M NaCl at t = 0. Close inspection reveals a decrease of extracellular glycerol in gpd1Δ strain. The decrease intracellular glycerol in gpd1Δ is on a similar timescale as the transcriptionally regulated increase of Gpd1 in other strains, indicating that a transcriptionally regulated mechanism is responsible for this decrease as well.
(PDF)
Effects of HOG1-att (Hog1 attached to the plasma membrane) compared to wild-type. Under unstressed conditions, inactive Hog1 (dark red stars) is localized throughout the cell, residual active Hog1 (light red stars) is localized in the nucleus. Possible Hog1 interaction partners are depicted: Fps1 (brown triangles), Pfk26/27 (blue spirals) and genes (black waves). In wild-type, osmoadaptation leads to active Hog1 translocating to the nucleus to stimulate transcription. In HOG1-att, transcriptional regulation is abolished and cytosolic Hog1-concentration is reduced while possible interactions with membrane-bound proteins are increased.
(PDF)
Model topology in SBGN syntax. The different modules are color coded (red: adaptation, yellow: biophysical, brown: transport, blue: glycolytic, green: growth). Measured entities are indicated by a green background. Perturbations to the model (as stress [NaCl] or different mutations [I: hog1Δ, II: pfk26/27Δ, III: HOG1-att, IV: gpd1Δ, V: FPS1-Δ1 ]).
(PDF)
Changes in cell density in a batch culture experiment. Cells covered by an ODE model highlighted in yellow.
(PDF)
Agreement between main model variables and experimental data. Concentrations of intracellular glycerol (dashed) and phosphorylated Hog1 in different strains (A: WT, B: pfk26/27Δ, C: HOG1-att, D: FPS1-Δ1, E: gpd1Δ, F: hog1Δ) following hyperosmotic stress of 0.4 M NaCl at t = 0.
(PDF)
Experimental data and simulated model variables, stress 0.4 M NaCl added at t = 0. A: phosphorylated Hog1, B: GPD1mRNA, C: Gpd1, D: cell volume, E: abundance of open Fps1, F: intracellular trehalose. Concentrations in A and B are scaled as described in text.
(PDF)
Simulation of model variables, stress 0.4 M NaCl added at t = 0. A: extracellular glycerol, B: intracellular glycerol, C: extracellular trehalose, D: extracellular glycerol, E: extracellular ethanol, F: extracellular acetate.
(PDF)
Model Simulations and scaled response coefficients for models of different strains. A,D,G: Model variables (solid line: phosphorylated Hog1, dashed line: intracellular glycerol, dotted: abundance of open Fps1) for wild-type, gpd1Δ and hog1Δ, respectively. B,E,H: scaled response coefficients of osmoshock dependent parameters on intracellular glycerol for wild-type, gpd1Δ and hog1Δ, respectively. C,F,I: scaled response coefficients of glycolytic parameters on intracellular pyruvate for wild-type, gpd1Δ and hog1Δ, respectively.
(PDF)
Annotated model of osmoadaptation in wild type.
(XML)
Annotated model of osmoadaptation in pfk26/27Δ.
(XML)
Annotated model of osmoadaptation in HOG1-att.
(XML)
Annotated model of osmoadaptation in FPS1-Δ1.
(XML)
Annotated model of osmoadaptation in gpd1Δ.
(XML)
Annotated model of osmoadaptation in hog1Δ.
(XML)
Optical density (OD) and cell density (CD) for control and stressed WT cultures. OD in arbitrary units, cell density in 106 cells/ml.
(PDF)
Materials and Methods, data processing, and details concerning modeling, parameter estimation, and response coefficients.
(PDF)
Funding Statement
This work was supported by grants from the European Commission 7th Framework Programme: UNICELLSYS (Contract No. 201142, to SH, JN, and EK), CELLCOMPUT (Contract No. 043310 to SH, EK), the Chalmers Foundation (JN), and a grant from the German Research Foundation: IRTG 1360 “Genomics and Systems Biology of Molecular Networks” to EK. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Yancey PH (2005) Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J Exp Biol 208: 2819–2830. [DOI] [PubMed] [Google Scholar]
- 2. Klipp E, Nordlander B, Kruger R, Gennemark P, Hohmann S (2005) Integrative model of the response of yeast to osmotic shock. Nat Biotechnol 23: 975–982. [DOI] [PubMed] [Google Scholar]
- 3. Mettetal JT, Muzzey D, Gomez-Uribe C, van Oudenaarden A (2008) The frequency dependence of osmo-adaptation in Saccharomyces cerevisiae. Science 319: 482–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zi Z, Liebermeister W, Klipp E (2010) A quantitative study of the Hog1 MAPK response to fluctuating osmotic stress in Saccharomyces cerevisiae. PLoS One 5: e9522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schaber J, Baltanas R, Bush A, Klipp E, Colman-Lerner A (2012) Modelling reveals novel roles of two parallel signalling pathways and homeostatic feedbacks in yeast. Mol Syst Biol 8: 622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ansell R, Granath K, Hohmann S, Thevelein JM, Adler L (1997) The two isoenzymes for yeast NAD+-dependent glycerol 3-phosphate dehydrogenase encoded by GPD1 and GPD2 have distinct roles in osmoadaptation and redox regulation. EMBO J 16: 2179–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Luyten K, Albertyn J, Skibbe WF, Prior BA, Ramos J, et al. (1995) Fps1, a yeast member of the MIP family of channel proteins, is a facilitator for glycerol uptake and efflux and is inactive under osmotic stress. EMBO J 14: 1360–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tamas MJ, Luyten K, Sutherland FC, Hernandez A, Albertyn J, et al. (1999) Fps1p controls the accumulation and release of the compatible solute glycerol in yeast osmoregulation. Mol Microbiol 31: 1087–1104. [DOI] [PubMed] [Google Scholar]
- 9. Albertyn J, Hohmann S, Prior BA (1994) Characterization of the osmotic-stress response in Saccharomyces cerevisiae: osmotic stress and glucose repression regulate glycerol-3-phosphate dehydrogenase independently. Curr Genet 25: 12–18. [DOI] [PubMed] [Google Scholar]
- 10. Påhlman AK, Granath K, Ansell R, Hohmann S, Adler L (2001) The yeast glycerol 3-phosphatases Gpp1p and Gpp2p are required for glycerol biosynthesis and differentially involved in the cellular responses to osmotic, anaerobic, and oxidative stress. J Biol Chem 276: 3555–3563. [DOI] [PubMed] [Google Scholar]
- 11. Valadi A, Granath K, Gustafsson L, Adler L (2004) Distinct intracellular localization of Gpd1p and Gpd2p, the two yeast isoforms of NAD+-dependent glycerol-3-phosphate dehydrogenase, explains their different contributions to redox-driven glycerol production. J Biol Chem 279: 39677–39685. [DOI] [PubMed] [Google Scholar]
- 12. Albertyn J, Hohmann S, Thevelein JM, Prior BA (1994) GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol Cell Biol 14: 4135–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rep M, Albertyn J, Thevelein JM, Prior BA, Hohmann S (1999) Different signalling pathways contribute to the control of GPD1 gene expression by osmotic stress in Saccharomyces cerevisiae. Microbiology 145 Pt 3: 715–727. [DOI] [PubMed] [Google Scholar]
- 14. Norbeck J, Påhlman AK, Akhtar N, Blomberg A, Adler L (1996) Purification and characterization of two isoenzymes of DL-glycerol-3-phosphatase from Saccharomyces cerevisiae. Identification of the corresponding GPP1 and GPP2 genes and evidence for osmotic regulation of Gpp2p expression by the osmosensing mitogen-activated protein kinase signal transduction pathway. J Biol Chem 271: 13875–13881. [DOI] [PubMed] [Google Scholar]
- 15. Hirayama T, Maeda T, Saito H, Shinozaki K (1995) Cloning and characterization of seven cDNAs for hyperosmolarity-responsive (HOR) genes of Saccharomyces cerevisiae. Mol Gen Genet 249: 127–138. [DOI] [PubMed] [Google Scholar]
- 16. Remize F, Barnavon L, Dequin S (2001) Glycerol export and glycerol-3-phosphate dehydrogenase, but not glycerol phosphatase, are rate limiting for glycerol production in Saccharomyces cerevisiae. Metab Eng 3: 301–312. [DOI] [PubMed] [Google Scholar]
- 17. Dihazi H, Kessler R, Eschrich K (2004) High osmolarity glycerol (HOG) pathway-induced phosphorylation and activation of 6-phosphofructo-2-kinase are essential for glycerol accumulation and yeast cell proliferation under hyperosmotic stress. J Biol Chem 279: 23961–23968. [DOI] [PubMed] [Google Scholar]
- 18. Tamas MJ, Karlgren S, Bill RM, Hedfalk K, Allegri L, et al. (2003) A short regulatory domain restricts glycerol transport through yeast Fps1p. J Biol Chem 278: 6337–6345. [DOI] [PubMed] [Google Scholar]
- 19. Ferreira C, van Voorst F, Martins A, Neves L, Oliveira R, et al. (2005) A member of the sugar transporter family, Stl1p is the glycerol/H+ symporter in Saccharomyces cerevisiae. Mol Biol Cell 16: 2068–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rep M, Krantz M, Thevelein JM, Hohmann S (2000) The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J Biol Chem 275: 8290–8300. [DOI] [PubMed] [Google Scholar]
- 21. Westfall PJ, Patterson JC, Chen RE, Thorner J (2008) Stress resistance and signal fidelity independent of nuclear MAPK function. Proc Natl Acad Sci U S A 105: 12212–12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kuhn C, Petelenz E, Nordlander B, Schaber J, Hohmann S, et al. (2008) Exploring the impact of osmoadaptation on glycolysis using time-varying response-coefficients. Genome Inform 20: 77–90. [PubMed] [Google Scholar]
- 23. Schaber J, Klipp E (2008) Short-term volume and turgor regulation in yeast. Essays Biochem 45: 147–159 doi: 10.1042/BSE0450147 [DOI] [PubMed] [Google Scholar]
- 24. Schaber J, Adrover MA, Eriksson E, Pelet S, Petelenz-Kurdziel E, et al. (2010) Biophysical properties of Saccharomyces cerevisiae and their relationship with HOG pathway activation. Eur Biophys J 39: 1547–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Teusink B, Passarge J, Reijenga CA, Esgalhado E, van der Weijden CC, et al. (2000) Can yeast glycolysis be understood in terms of in vitro kinetics of the constituent enzymes? Testing biochemistry. Eur J Biochem 267: 5313–5329. [DOI] [PubMed] [Google Scholar]
- 26. Clotet J, Escote X, Adrover MA, Yaakov G, Gari E, et al. (2006) Phosphorylation of Hsl1 by Hog1 leads to a G2 arrest essential for cell survival at high osmolarity. Embo J 25: 2338–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Escote X, Zapater M, Clotet J, Posas F (2004) Hog1 mediates cell-cycle arrest in G1 phase by the dual targeting of Sic1. Nat Cell Biol 6: 997–1002. [DOI] [PubMed] [Google Scholar]
- 28. Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, et al. (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, et al. (2001) Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell 12: 323–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nordlander B, Krantz M, Hohmann S (2008) Hog1-mediated Metabolic Adjustments Following Hyperosmotic Shock in the Yeast Saccharomyces cerevisiae. In: Posas F, Nebreda AR, editors. Stress-Activated Protein Kinases. Berlin/Heidelberg: Springer. pp. 141–158.
- 31. Rep M, Reiser V, Gartner U, Thevelein JM, Hohmann S, et al. (1999) Osmotic stress-induced gene expression in Saccharomyces cerevisiae requires Msn1p and the novel nuclear factor Hot1p. Mol Cell Biol 19: 5474–5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ou X, Ji C, Han X, Zhao X, Li X, et al. (2006) Crystal structures of human glycerol 3-phosphate dehydrogenase 1 (GPD1). J Mol Biol 357: 858–869. [DOI] [PubMed] [Google Scholar]
- 33. Muzzey D, Gomez-Uribe CA, Mettetal JT, van Oudenaarden A (2009) A systems-level analysis of perfect adaptation in yeast osmoregulation. Cell 138: 160–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ingalls BP, Sauro HM (2003) Sensitivity analysis of stoichiometric networks: an extension of metabolic control analysis to non-steady state trajectories. J Theor Biol 222: 23–36. [DOI] [PubMed] [Google Scholar]
- 35. Kuhn C, Klipp E (2012) Zooming in on yeast osmoadaptation. Adv Exp Med Biol 736: 293–310. [DOI] [PubMed] [Google Scholar]
- 36. Mollapour M, Piper PW (2007) Hog1 Mitogen-Activated Protein Kinase Phosphorylation Targets the Yeast Fps1 Aquaglyceroporin for Endocytosis, Thereby Rendering Cells Resistant to Acetic Acid. Mol Cell Biol 27: 6446–6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thorsen M, Di Y, Tängemo C, Morillas M, Ahmadpour D, et al. (2006) The MAPK Hog1p Modulates Fps1p-dependent Arsenite Uptake and Tolerance in Yeast. Mol Biol Cell 17: 4400–4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Beese SE, Negishi T, Levin DE (2009) Identification of positive regulators of the yeast fps1 glycerol channel. PLoS Genet 5: e1000738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Davis DJ, Burlak C, Money NP (2000) Osmotic pressure of fungal compatible osmolytes. Mycological Research 104: 800–804. [Google Scholar]
- 40. Pagnotta SE, McLain SE, Soper AK, Bruni F, Ricci MA (2010) Water and trehalose: how much do they interact with each other? J Phys Chem B 114 14: 4904–4908. [DOI] [PubMed] [Google Scholar]
- 41. Pereira CS, Hunenberger PH (2008) Effect of trehalose on a phospholipid membrane under mechanical stress. Biophys J 95: 3525–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nordlander B, Krantz M, Hohmann S (2008) Hog1-mediated Metabolic Adjustments Following Hyperosmotic Shock in the Yeast Saccharomyces cerevisiae. Stress-Activated Protein Kinases. Berlin/Heidelberg: Springer pp. 141–158.
- 43. Bruggeman FJ, de Haan J, Hardin H, Bouwman J, Rossell S, et al. (2006) Time-dependent hierarchical regulation analysis: deciphering cellular adaptation. Syst Biol (Stevenage) 153: 318–322. [DOI] [PubMed] [Google Scholar]
- 44. Duch A, de Nadal E, Posas F (2012) The p38 and Hog1 SAPKs control cell cycle progression in response to environmental stresses. FEBS Lett 586: 2925–2931. [DOI] [PubMed] [Google Scholar]
- 45. Thomas BJ, Rothstein RJ (1989) Elevated recombination rates in transcriptionally active DNA. Cell 56: 619–630. [DOI] [PubMed] [Google Scholar]
- 46. Maiwald T, Timmer J (2008) Dynamical modeling and multi-experiment fitting with PottersWheel. Bioinformatics 24: 2037–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zi Z, Klipp E (2006) SBML-PET: a Systems Biology Markup Language-based parameter estimation tool. Bioinformatics 22: 2704–2705. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Raw and processed metabolite measurements for different strains, stress 0.4 M NaCl added at t = 0.
(XLS)
Overview of experimental data as used in fitting.
(XLS)
Comparison of intracellular trehalose measurements by HPLC and enzyme assay, stress 0.4 M NaCl added at t = 0.
(XLS)
Western Blot data for different strains and stress strengths.
(XLS)
Intracellular glycerol quantification by enzyme assay in different strains for different stresses added at t = 0.
(XLS)
Intracellular trehalose quantification by enzyme assay in different strains, stress 0.4 M NaCl added at t = 0.
(XLS)
Northern Blot results for CTT1, GRE2, STL1, 18S in different strains.
(XLS)
Cell density and optical density in stressed (0.4 M NaCl added at t = 0) and control experiments (A) and normalized to the values at t = 0 (B). Experimental setup as explained in main text.
(PDF)
Plot of OD versus cell density values as obtained from control and stressed time course experiments and the function for computing cell density from OD.
(PDF)
Measured (blue) and inferred (pink) intracellular glycerol for different experiments. Errors in the inferred values due to inconsistencies in measurements are visible in FPS1-Δ1, hog1Δ and WT1.
(PDF)
Comparison of glycerol time courses for WT1 and WT4. (A) intracellular glycerol, (B) extracellular glycerol.
(PDF)
Comparison of enzyme assay (blue) and HPLC (pink) intracellular glycerol quantifications. Underlying data is given in Supplemental Dataset S2.
(PDF)
Comparison of intracellular trehalose levels obtained with different methods. Enzyme assay data is in g D-glucose/l per µg protein/ml, HPLC data in mol/l is normalized to the enzyme assay value at t = 30. Underlying data is given in Supplemental Dataset S3.
(PDF)
Experimental data as used for model fitting. Measured entities are indicated on y-axes. Shown are representative experiments for each strain. Stress of 0.4 M NaCl is added at t = 0. A: intracellular glucose, B: extracellular glucose, C: Optical density, D: intracellular ethanol, E: extracellular ethanol, F: extracellular acetate, G: intracellular trehalose, H: extracellular trehalose, I: extracellular glycerol.
(PDF)
Experimental data for extracellular glycerol (A,B) and Gpd1 (C) following addition of 0.4 M NaCl at t = 0. Close inspection reveals a decrease of extracellular glycerol in gpd1Δ strain. The decrease intracellular glycerol in gpd1Δ is on a similar timescale as the transcriptionally regulated increase of Gpd1 in other strains, indicating that a transcriptionally regulated mechanism is responsible for this decrease as well.
(PDF)
Effects of HOG1-att (Hog1 attached to the plasma membrane) compared to wild-type. Under unstressed conditions, inactive Hog1 (dark red stars) is localized throughout the cell, residual active Hog1 (light red stars) is localized in the nucleus. Possible Hog1 interaction partners are depicted: Fps1 (brown triangles), Pfk26/27 (blue spirals) and genes (black waves). In wild-type, osmoadaptation leads to active Hog1 translocating to the nucleus to stimulate transcription. In HOG1-att, transcriptional regulation is abolished and cytosolic Hog1-concentration is reduced while possible interactions with membrane-bound proteins are increased.
(PDF)
Model topology in SBGN syntax. The different modules are color coded (red: adaptation, yellow: biophysical, brown: transport, blue: glycolytic, green: growth). Measured entities are indicated by a green background. Perturbations to the model (as stress [NaCl] or different mutations [I: hog1Δ, II: pfk26/27Δ, III: HOG1-att, IV: gpd1Δ, V: FPS1-Δ1 ]).
(PDF)
Changes in cell density in a batch culture experiment. Cells covered by an ODE model highlighted in yellow.
(PDF)
Agreement between main model variables and experimental data. Concentrations of intracellular glycerol (dashed) and phosphorylated Hog1 in different strains (A: WT, B: pfk26/27Δ, C: HOG1-att, D: FPS1-Δ1, E: gpd1Δ, F: hog1Δ) following hyperosmotic stress of 0.4 M NaCl at t = 0.
(PDF)
Experimental data and simulated model variables, stress 0.4 M NaCl added at t = 0. A: phosphorylated Hog1, B: GPD1mRNA, C: Gpd1, D: cell volume, E: abundance of open Fps1, F: intracellular trehalose. Concentrations in A and B are scaled as described in text.
(PDF)
Simulation of model variables, stress 0.4 M NaCl added at t = 0. A: extracellular glycerol, B: intracellular glycerol, C: extracellular trehalose, D: extracellular glycerol, E: extracellular ethanol, F: extracellular acetate.
(PDF)
Model Simulations and scaled response coefficients for models of different strains. A,D,G: Model variables (solid line: phosphorylated Hog1, dashed line: intracellular glycerol, dotted: abundance of open Fps1) for wild-type, gpd1Δ and hog1Δ, respectively. B,E,H: scaled response coefficients of osmoshock dependent parameters on intracellular glycerol for wild-type, gpd1Δ and hog1Δ, respectively. C,F,I: scaled response coefficients of glycolytic parameters on intracellular pyruvate for wild-type, gpd1Δ and hog1Δ, respectively.
(PDF)
Annotated model of osmoadaptation in wild type.
(XML)
Annotated model of osmoadaptation in pfk26/27Δ.
(XML)
Annotated model of osmoadaptation in HOG1-att.
(XML)
Annotated model of osmoadaptation in FPS1-Δ1.
(XML)
Annotated model of osmoadaptation in gpd1Δ.
(XML)
Annotated model of osmoadaptation in hog1Δ.
(XML)
Optical density (OD) and cell density (CD) for control and stressed WT cultures. OD in arbitrary units, cell density in 106 cells/ml.
(PDF)
Materials and Methods, data processing, and details concerning modeling, parameter estimation, and response coefficients.
(PDF)