In yeast, the localization of homologous recombination–associated proteins to heterochromatic regions of the genome is necessary for proper nuclear organization.
Abstract
The eukaryotic genome is highly organized in the nucleus, and this organization affects various nuclear processes. However, the molecular details of higher-order organization of chromatin remain obscure. In the present study, we show that the Saccharomyces cerevisiae silenced loci HML and HMR cluster in three-dimensional space throughout the cell cycle and independently of the telomeres. Long-range HML–HMR interactions require the homologous recombination (HR) repair pathway and phosphorylated H2A (γ-H2A). γ-H2A is constitutively present at silenced loci in unperturbed cells, its localization requires heterochromatin, and it is restricted to the silenced domain by the transfer DNA boundary element. SMC proteins and Scc2 localize to the silenced domain, and Scc2 binding requires the presence of γ-H2A. These findings illustrate a novel pathway for heterochromatin organization and suggest a role for HR repair proteins in genomic organization.
Introduction
Eukaryotic genomes are highly organized within the nucleus, and the genome of the budding yeast Saccharomyces cerevisiae is no exception (Taddei et al., 2010; Zimmer and Fabre, 2011): the 16 centromeres cluster at the spindle pole body (Jin et al., 1998, 2000), and the 32 telomeres localize at the nuclear periphery in five to eight clusters (Gotta et al., 1996; Laroche et al., 1998), whereas the ribosomal DNA (rDNA)-containing nucleolus localizes opposite the centromeres (Gotta et al., 1997). Some of the determinants for this organization are known (Maillet et al., 1996; Laroche et al., 1998, 2000; D’Ambrosio et al., 2008; Haeusler et al., 2008). Telomeric clustering is dependent on Esc1/Ku70, Rap1, and Sir4 proteins (Teixeira et al., 2002; Schober et al., 2008, 2009) as well as SUN proteins and membrane tethering (Chikashige et al., 2006; Antoniacci et al., 2007; Bupp et al., 2007; Grund et al., 2008; Mekhail and Moazed, 2010; Zimmer and Fabre, 2011), and loss of clustering results in a weakening of silencing. However, the identities of additional pathways and factors remain to be determined.
The silenced HML and HMR loci are located 10 and 20 kb from the telomeres on chromosome III. Although a large distance separates these loci, they are found in close proximity in three-dimensional space, clustering together at the nuclear periphery (Miele et al., 2009). Silencing at these loci initiate at silencers (Rine and Herskowitz, 1987; Pillus and Rine, 1989), which recruit the silent information regulator (Sir) proteins, leading to the formation of a heterochromatic domain (Rusche et al., 2003). At HMR, a tDNA acts as a barrier to the spread of silencing, whereas at HML, the CHA1 promoter mediates barrier function (Donze et al., 1999; Donze and Kamakaka, 2001; Oki and Kamakaka, 2005; Dhillon et al., 2009).
Although genomic organization affects proper gene regulation (Ahmed and Brickner, 2007; Brickner, 2009; Taddei et al., 2009), it may also play a role in other nuclear processes (Mekhail and Moazed, 2010; Nagai et al., 2010, 2011). DNA damage occurs spontaneously in cycling yeast cells, and >25% experience damage in each cell cycle (Lisby et al., 2001, 2003; Lisby and Rothstein, 2004). Damaged DNA is repaired to avoid chromosomal rearrangements (Branzei and Foiani, 2008; Lisby and Rothstein, 2009; van Attikum and Gasser, 2009; Polo and Jackson, 2011; Deem et al., 2012). In haploid yeast, the lesion is predominately repaired through the nonhomologous end joining (NHEJ) pathway in G1, whereas in G2, repair is primarily via homologous recombination (HR; Aylon and Kupiec, 2004). Upon detection of a double-strand break (DSB), the MRX (Mre11–Rad50–Xrs2) complex binds the break (Kinoshita et al., 2009; Stracker and Petrini, 2011), and in HR-mediated repair, histone H2A in nucleosomes are phosphorylated on Serine-129 (γ-H2A) by Tel1 or Mec1 (Flott et al., 2007; Polo and Jackson, 2011). Phosphorylation of H2A helps stabilize the binding of the repair machinery. H2A phosphorylation is followed by the recruitment of the resection machinery (Srs2 and Exo1) and then the late repair proteins (Rad51, Rad52, Rad54, and Rdh54 among others) are recruited to the single-stranded resected DNA (Heyer et al., 2006; Keogh et al., 2006; Wu, 2008; Mortensen et al., 2009). This leads to homology recognition, strand invasion, and repair (Sugawara et al., 2003). In addition, replication proteins and the structural maintenance of chromatin (SMC) proteins play a direct role in repair (Holmes and Haber, 1999; Unal et al., 2004; Wang et al., 2004; Cortés-Ledesma et al., 2007; De Piccoli et al., 2009; Bose and Gerton, 2010; Wood et al., 2010). The silencing proteins are also recruited to breaks and affect the kinetics of repair through an as yet unknown mechanism (Martin et al., 1999; Mills et al., 1999; Tamburini and Tyler, 2005).
Although spontaneous DNA breaks can occur anywhere in the genome as a consequence of environmental factors, they frequently occur at specific fragile sites during DNA replication (Deshpande and Newlon, 1996; Lopes et al., 2001; Mirkin and Mirkin, 2007; Schleker et al., 2009; Zegerman and Diffley, 2009; Branzei and Foiani, 2010). Beyond spontaneous damage, site-specific endonucleases also generate DSBs during mating-type switching. Mating-type switching involves gene conversion of the MAT locus, initiated by the generation of a DSB at this locus by the HO endonuclease in the G1 phase of the cell cycle. During HO-mediated gene conversion, the silenced HML or HMR loci physically interact with the MAT locus, and the HR repair machinery along with replication and SMC proteins use sequences at HM to repair the break at MAT (Haber, 1998; Holmes and Haber, 1999; Wang et al., 2004; Bystricky et al., 2009; Hicks et al., 2011).
In the current study, we aimed to identify the factors involved in long-range HML–HMR clustering and elucidate the mechanisms of heterochromatin clustering to identify potential mechanisms that underlie genome organization. Here, we show that silencing and HR repair proteins influence higher-order chromatin organization in the yeast nucleus. We show a role for silencers in clustering and also show that silencing plays a role in the phosphorylation of S129 of H2A at HMR. We further demonstrate that the tDNA insulator blocks the spread of this modified histone mark beyond the silenced domain. We show a novel role for the DSB repair machinery in this phenomenon and propose a model in which both silencing and repair proteins contribute to heterochromatic long-range interactions and genome organization by using specific DNA sequences and loci.
Results
HML and HMR cluster independent of the cell cycle and mating type
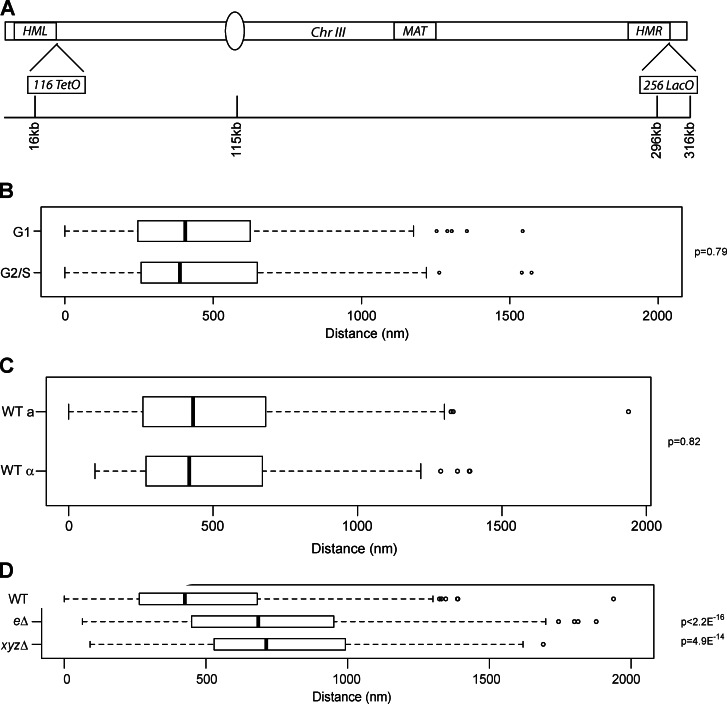
It has previously been shown using 3C and fluorescence imaging that HML and HMR physically interact and cluster together, and this association is dependent on the silencing proteins (Miele et al., 2009). To better understand this clustering, we initially inquired about the persistence of the association through the cell cycle. We used a strain with 256 copies of the LacO operator integrated near HMR and 128 copies of the TetO operator integrated near HML (Bystricky et al., 2005). The strain also contained the fusion proteins CFP-LacI and YFP-TetR (Fig. 1 A). A wide-field microscope was used to acquire optical slices of live, unperturbed, asynchronously growing cells to measure the distance between HML and HMR. Around 200 cells were analyzed in each strain, and comparisons between strains are presented as simple median distances, but it is important to consider the change in the distribution of distances, which is better reflected in the p-value between strains (Mann-Whitney U test; p-values are between a wild-type [WT] strain and the strain of interest unless otherwise stated). We measured the distances between the HM loci and simultaneously scored the asynchronously growing cells based on budding. The distances between HML and HMR were 399 nm for unbudded cells and 388 nm for budded cells (P = 0.79; Fig. 1 B). These data suggest that although the nuclear volume increases in G2, the two loci remain close to one another through the cell cycle. We also determined the distances between these two loci in strains of opposite mating type and did not observe any difference (Fig. 1 C).
Figure 1.
HML and HMR are in close three-dimensional proximity. (A) Schematic of budding yeast chromosome III with locations of TetO and LacO arrays. (B–D) Boxplots of the distance between TetR-YFP and CFP-LacI foci in asynchronously growing cells in G1 (n = 192) or G2/S (n = 136; B) in MATa (n = 153) and MATα cells (n = 152; C) and in an hmr-eΔ strain (n = 353) and an xyzΔ strain (n = 202; D). P-values were calculated using a Mann-Whitney U test (Wilcoxon test). Data are from all cells counted from at least two independent trials (see Materials and methods). The boxes represent the middle 50% of data points with the black lines showing the median of distances. Outliers are defined as distances >1.5 times the interquartile range (dashed lines) and are represented by open circles.
Long-range HML–HMR interactions are telomere independent
TEL-IIIR (telomere IIIR) and TEL-IIIL (telomere IIIL) interact with one another, forming a pseudocircular chromosome III (Bystricky et al., 2005; Therizols et al., 2010). Because HML and HMR are located ∼10 and 20 kb, respectively, from the telomeres, it was possible that the observed HM associations were in fact telomere–telomere interactions (Bystricky et al., 2005). To determine the role of the telomeres in HM clustering, we deleted the HMR-E silencer, such that the resulting strain does not silence HMR, though silencing in the rest of the genome, including HML, remains unaffected. Loss of silencing solely at HMR leads to a change in the long-range association between the HM loci in a large number of cells (Fig. 1 D), suggesting that silencing at HMR is necessary for HM clustering and that proximity of HMR to TEL-IIIR is not a sufficient driver for HM interactions. To test whether or not DNA sequence homology was playing a role in the HM clustering, we replaced the XYZ homology sequence present between the silencers at HMR with the HIS3 gene. The HIS3 gene within HMR was silenced (Fig. S1). Unexpectedly, the deletion of the XYZ sequence led to a reduction in the long-range associations between the HM loci (median = 713 nm; Fig. 1 D). This result suggests that HML and HMR reside in close proximity to one another over long distances through a mechanism that relies on silencing and sequence homology.
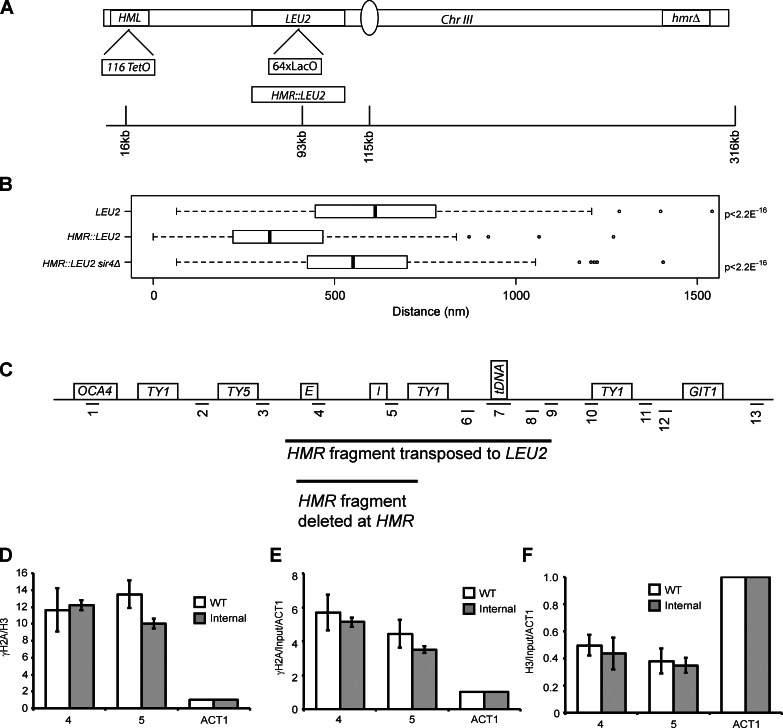
In a second set of experiments, we moved the HMR locus away from TEL-IIIR. We inserted a LacO array at the LEU2 gene (∼80 kb from HML) and measured the distance between the LEU2 locus and HML (Fig. 2 A). Our analysis indicates that the LEU2 locus resides at a median distance of 612 nm from HML. When a 3.5-kb HMR-containing fragment was inserted at LEU2, the distance between HML and HMR::LEU2 greatly decreased in a large population of cells (median = 321 nm, P < 2.2 × 10−16), showing that clustering of HML and LEU2::HMR is more frequent than between HML and LEU2 (Fig. 2). These data again show that HM clustering is independent of its proximity to TEL-IIIR. Moreover, this association between HML and HMR at LEU2 was dependent on Sir4 (median = 551 nm, P < 2.2 × 10−16 in a sir4Δ strain compared with HMR::LEU2), comparable to the locus lacking HMR (P = 0.03 compared with LEU2). Recently, it has been shown that LacO arrays bind LacI, which leads to ectopic recruitment of silencing proteins by the array (Dubarry et al., 2011). The results in Fig. 2 argue against this phenomenon being the sole basis for the long-range HM association.
Figure 2.
Role of telomeres in HML and HMR proximity. (A) Schematic of chromosome III constructs for internal HMR analysis. A 64× LacO array integrated at the LEU2 gene with or without a 3.3-kb fragment of HMR, a 128× TetO array at HML, and an hmrΔ at its native locus. (B) Boxplots of the distance between TetR-YFP and CFP-LacI foci in a given strain: LEU2 (n = 228), HMR::LEU2 (n = 274), and HMR::LEU2 sir4Δ (n = 209). The boxes represent the middle 50% of data points with the black lines showing the median of distances. Outliers are defined as distances >1.5 times the interquartile range (dashed lines) and are represented by open circles. (C) A schematic of the HMR locus. (D–F) ChIP of ratio γ-H2A/H3 (D), γ-H2A (E), and H3 (F) compared with the native locus (Fig. 6 C). Error bars represent standard deviation from the mean for n = 6 (WT) and n = 4 (internal).
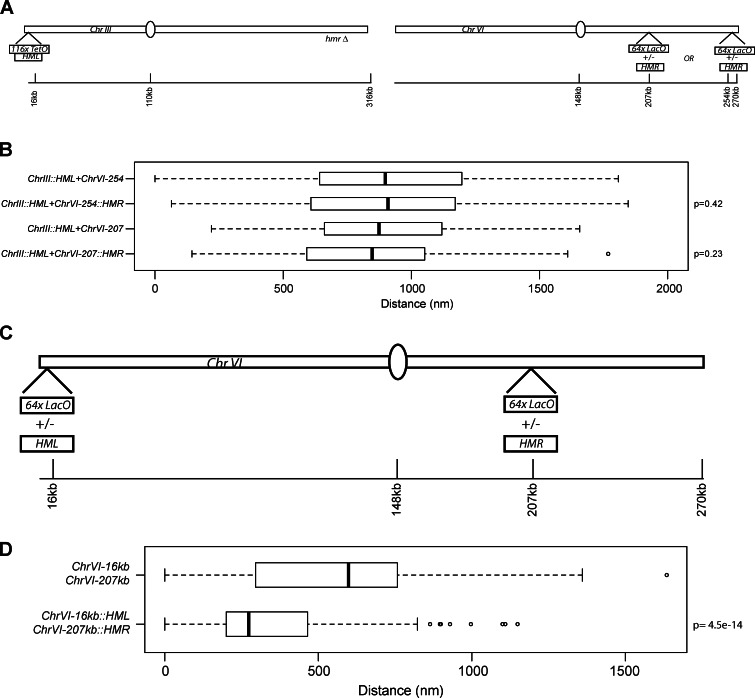
Next, we performed an experiment in which HMR was moved to different locations on chromosome VI to test whether or not HMR can also drive an interchromosomal association. A LacO array with or without the 3.3-kb HMR fragment was integrated ∼15 kb from the ChrVIR telomere or internally (∼60 kb from the centromeres on VIR; Fig. 3 A). Clustering between HML on chromosome III and HMR on chromosome VI were assayed. We did not observe any significant association between HMR on chromosome VI and HML at its native site on chromosome III (Fig. 3 B). This is not totally unexpected given the observation that HR is less efficient for loci present on two different chromosomes as on the same chromosome (Lichten and Haber, 1989).
Figure 3.
HML and HMR proximity when placed on chromosome VI. (A) Schematic of chromosome III and chromosome VI. A 64× LacO array was placed in one of two positions on the right arm of chromosome VI with or without HMR. (B) Boxplots of the distances between the TetO and LacO arrays in each strain (ChrIII::HML + ChrVI-254kb, n = 178; ChrIII::HML + ChrVI-254kb::HMR, n = 159; ChrIII::HML + ChrVI-207kb, n = 109; and ChrIII::HML + ChrVO-207kb::HMR, n = 133). (C) Schematic of chromosome VI. (D) Boxplots of the distances between the TetO and LacO arrays in each strain (ChrVI-16kb + ChrVI-207kb, n = 160; and ChrVI-16kb::HML + ChrVI-207kb::HMR, n = 188). The boxes represent the middle 50% of data points with the black lines showing the median of distances. Outliers are defined as distances >1.5 times the interquartile range (dashed lines) and are represented by open circles.
This result raised the possibility that the association observed between HML and HMR may be specific for chromosome III. To address this, we asked whether HML and HMR could cluster together when both loci were located on chromosome VI. In the strain in which HMR was integrated 60 kb from centromere VI, we integrated a LacO array ∼16 kb from telomere VIL with or without HML (Fig. 3 C). In the absence of HML at TEL-VIL, the HMR locus did not cluster with TEL-VIL, but in the presence of HML at TEL-VIL, the HMR locus clustered with HML in a large number of cells (Fig. 3 D). Clustering occurred even though HML and HMR were not located equidistant from the centromeres. These data show that the clustering of the two HM loci is not chromosome III specific but a specific property of these two loci.
DNA DSB repair proteins contribute to HML–HMR clustering in the nucleus
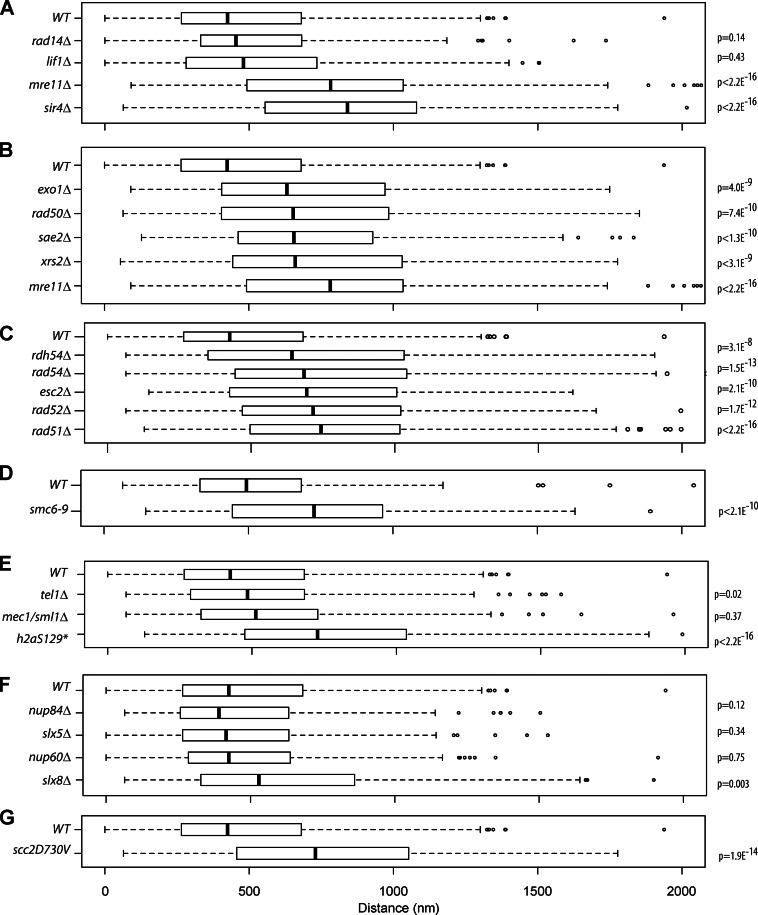
We next wished to know the identities of the proteins that play a role in long-range HM association. We performed a directed screen in nonperturbed, asynchronously growing cells for mutants in histone modifications, chromatin remodelers, and nuclear transport. However, none of these mutants affected the long-range association between HML and HMR (Fig. S1 C). Because telomere clustering utilizes the NHEJ/HR-mediated repair protein Ku, we screened DNA repair proteins. These included Rad14, in the nucleotide excision repair pathway, Lif1, a member of the NHEJ pathway that is not thought to play a major role in the HR repair pathway, and Mre11, which plays a role in the HR repair pathway. Loss of proteins specific to nucleotide excision repair and NHEJ did not have a significant effect on HM distances (median = 454 nm, P = 0.14; median = 480 nm, P = 0.43, respectively; vs. 425 nm for WT). However, loss of the HR protein Mre11 (median = 782 nm, P < 2.2 × 10−16) led to a significant increase in HM distances similar to those seen with the loss of silencing proteins (sir4Δ: median = 840 nm, P = 2.2 × 10−16; Fig. 4 A). A more detailed analysis of the distribution pattern indicates that in ∼60% of WT cells, HML and HMR are in close proximity, whereas in mre11Δ cells, these two loci are in close proximity in <25% of the cells, indicating that the change in median distribution is a result of a change in a large number of cells in the population and not simply caused by a change in a few outliers (Fig. S2 A).
Figure 4.
DNA DSB repair proteins contribute to HM long-range interactions. (A) Boxplots of the distance between TetR-YFP and CFP-LacI foci in a given strain. Strains containing deletions in rad14Δ (n = 186), lif1Δ (n = 143), mre11Δ (n = 443), and sir4Δ (n = 134). (B) Strains containing deletions in early HR repair proteins (exo1Δ, n = 214; rad50Δ, n = 414; sae2Δ, n = 161; xrs2Δ, n = 123; and mre11Δ, the mre11Δ data are the same as in A and are simply shown for easy comparison). (C) Strains containing deletions in downstream HR repair proteins (rdh54Δ, n = 215; rad54Δ, n = 223; esc2Δ, n = 167; rad52Δ, n = 163; and rad51Δ, n = 676). (D) WT and a temperature-sensitive smc6-9 strain after 2 h at 37°C. (E) The checkpoint proteins tel1Δ and mec1Δ/sml1Δ and a H2A mutant (tel1Δ, n = 143; mec1Δ/sml1Δ, n = 184; and hta1-129*Δ hta2-129*Δ, n = 437). (F) Nuclear pore proteins and ubiquitin pathway proteins (nup84Δ, n = 215; slx5Δ, n = 196; slx8Δ, n = 215; and nup60Δ, n = 210). (G) A point mutant scc2D730V. The WT data in all panels (except D) are the same as in A and are simply shown for easy comparison. The boxes represent the middle 50% of data points with the black lines showing the median of distances. Outliers are defined as distances >1.5 times the interquartile range (dashed lines) and are represented by open circles.
We then tested mutants in the early HR pathway—Mre11, Rad50, Xrs2, Sae2, and Exo1—all of which adversely affected the proximity of HML to HMR to varying degrees (exo1Δ: 632 nm, P = 4.0 × 10−9; rad50Δ: 654 nm, P = 7.4 × 10−10; sae2Δ: 656 nm, P = 1.3 × 10−10; xrs2Δ: 661 nm, P = 3.1 × 10−9; and mre11Δ: 782 nm, P < 2.2 × 10−16; Fig. 4 B). Some of the early HR repair proteins (specifically Mre11 and Ku) are thought to bind the telomeres in the absence of any damage, but late repair protein association with telomeres in unperturbed cells is not known; therefore, we investigated whether late repair proteins also affected HM clustering. Surprisingly, members of the late HR response pathway—Rad51, Rad52, Rad54, and Rdh54—all affected long-range HM association, as their loss led to an increase in the distance between the HM loci in large numbers of cells (rdh54Δ: 642nm, P = 3.1 × 10−8; rad54Δ: 684 nm, P = 1.5 × 10−13; rad52Δ: 716 nm, P = 1.7 × 10−12; and rad51Δ: 743 nm, P < 2.2 × 10−16; p-values are from WT; Fig. 4 C).
We also tested mutants in SMC proteins and discovered that mutations in these proteins lead to loss of HM interactions as well (Fig. 4, D and G). These results suggest that all of these proteins either directly or indirectly contribute to long-range heterochromatin clustering.
Because the HM loci cluster throughout the cell cycle, it is unlikely that mere cell cycle delay (some DSB repair mutants spend more time in G2) is sufficient to explain the loss of HM interactions. However, we analyzed the distances between the HM loci in cells in G1 or S/G2. We performed this analysis in strains lacking Mre11, Rad51, or mutants in γ-H2A. There was no difference in HM clustering as a function of cell cycle phase for these strains, though we did observe a statistically insignificant cell cycle–dependent difference for strains lacking Rad51 (Fig. S2 B).
We next asked whether there was a mating type–specific effect on HM clustering in these mutants. We measured distances between HML and HMR in both MATa and MATα cells. Once again, we did not observe any difference in the clustering of the HM loci as a function of mating type, either in the WT cells or in cells lacking Mre11 or Rad51 (Fig. S2 C).
The checkpoint proteins Tel1 and Mec1 also did not have an effect on HM association when deleted individually (median for tel1Δ: 513 nm, P = 0.015; mec1Δ/sml1Δ: 484 nm, P = 0.37; Fig. 4 E). The mec1, tel1, and sml1 mutant strains have senescent phenotypes caused by shortened telomeres (Ritchie et al., 1999), so these strains were not tested.
Tel1 and Mec1 perform some redundant functions, including the phosphorylation of H2A (γ-H2A), and so we assayed an H2A mutant that could not be phosphorylated (Downs et al., 2000). Strains that cannot phosphorylate H2A at Ser129 show a significant increase in HM distances (median = 727 nm, P < 2.2 × 10−16; Fig. 4 E), showing that γ-H2A was necessary for long-range HM clustering.
Some DSB repair occurs at the nuclear periphery and nuclear pore, and SUMO-targeted ubiquitin ligase proteins Nup84, Slx5, and Slx8 affect this repair (Nagai et al., 2008; Oza et al., 2009). However, loss of these proteins or another pore protein, Nup60 (which localizes at the HMR boundary; Ruben et al., 2011), did not affect HML–HMR interactions significantly (Fig. 4 F). These data suggest that the SUMO-targeted ubiquitin ligase pathway is not playing a major role in HM interactions.
It was possible that the separation of the HM loci in the mutants was caused by changes in nuclear size. We therefore measured the diameter of the nucleus in WT and various mutants. To assay nuclear periphery, we used a strain containing an HDEL-dsRed fusion protein, which incorporates into lumenal membranes and marks the nuclear periphery (Madrid et al., 2006; Ruben et al., 2011). Measurements of nuclear diameter in WT and mutants show that the nuclear diameter changes slightly in some of the mutants as compared with WT cells (Fig. S2 D). We inquired whether the variation in diameter affected the distance between HML and HMR. The median distance between HML and HMR was divided by the median nuclear diameter and plotted relative to WT cells. The small differences in the nuclear diameter of the mutants did not alter our interpretation that HML–HMR clustering was disrupted in these mutants, and the two loci were further apart in the nucleus of the mutants compared with WT cells (Fig. S2 E). In conclusion, these data show that HR repair proteins are directly or indirectly involved in HM clustering, but not all DNA repair proteins are necessary.
Do DSB repair proteins play a role in peripheral nuclear localization of HMR?
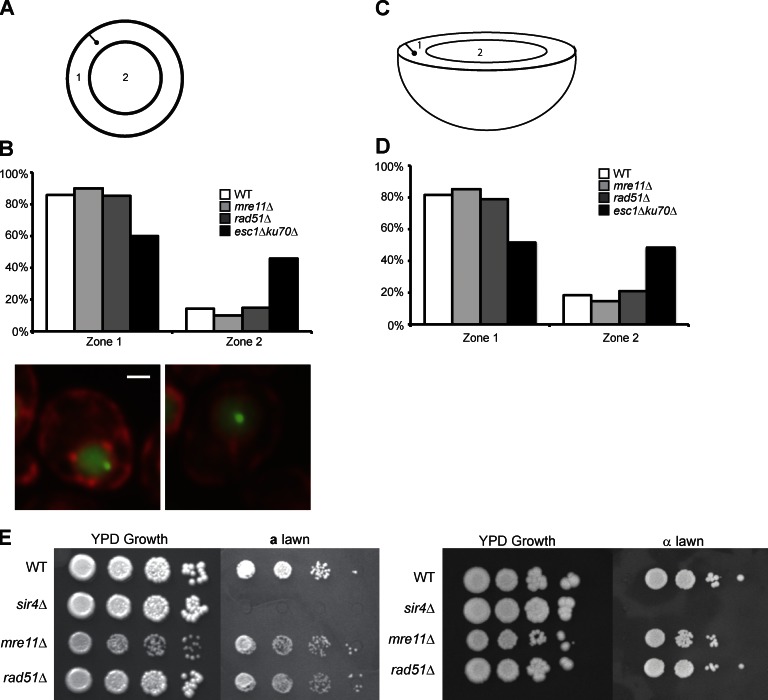
Because DNA DSB repair proteins play a role in HM clustering, we investigated their ability to affect the organization of these loci in the nucleus. One possibility could be that HML and HMR don’t cluster in the mutants because they are no longer tethered to the nuclear periphery where they normally could come into contact with each other (because of the restricted area they occupy). To assay nuclear peripheral localization, we marked HMR with a LacO array and the periphery with HDEL-dsRed fusion. The distance between HMR and the nuclear periphery was measured as was the diameter of the nucleus going through the GFP focus. These distances allow one to divide the nucleus into two sections of equal surface area and assign a locus to either the interior zone or the peripheral zone (Fig. 5 A). Strains lacking HR proteins Mre11 and Rad51 did not exhibit a marked loss of peripheral localization, whereas a previously published esc1Δ/ku70Δ control did (Fig. 5 B; reproduced from Ruben et al., 2011).
Figure 5.
DSB repair proteins play a role in silencing and localization. (A) Schematic of a cell’s nucleus showing two zones of equal surface area and a GFP focus. (B) Graph of the percentage of cells in each of two zones in a given strain for three independent trials the combined data for the three trials is shown: WT (n = 132); mre11Δ (n = 122), P = 0.27; rad51Δ (n = 134), P = 0.90; and ku70Δ/esc1Δ (n = 197), P = 5.8 × 10−7. Data shown are for all three trials combined, and p-values were determined by χ2 test compared with WT (see Materials and methods). The ku70Δ/esc1Δ data were previously published (Ruben et al., 2011) and are shown here for ease of comparison. Representative images of the two zones are provided under the graph. Bar, 1,000 nm. (C) Schematic of a cell’s nucleus showing two zones of equal volume and a GFP focus. (D) Graph of the percentage of cells in each of two zones of equal volume in a given strain using the data from B: WT (n = 132); mre11Δ (n = 122), P = 0.46; rad51Δ (n = 134), P = 0.58; and ku70Δ/esc1Δ (n = 197), P = 2.7 × 10−8. The ku70Δ/esc1Δ data were previously published (Ruben et al., 2011) and are shown here for ease of comparison. (E) 10-fold dilutions of α strains on a YPD plate (growth control) or an a lawn to assay silencing at HMR (left) or 10-fold dilutions of a strains on a YPD plate (growth control) or an α lawn to assay silencing at HML (right).
These data were also analyzed by dividing the nucleus into two zones of equal volume and the conclusions remain unchanged (Fig. 5, C and D). This finding shows that although Mre11 and Rad51 are not necessary for the peripheral localization of HMR, mutations in these proteins still lead to a loss in HM clustering, suggesting that they do not simply function by bringing both loci in close proximity via tethering to the nuclear periphery.
DSB repair proteins are not necessary for HM silencing
Ku is a protein involved in both NHEJ- and the HR-mediated repair, and mutations in Ku affect both clustering and telomeric gene silencing (Maillet et al., 2001). It was therefore possible that loss of HM silencing in strains lacking HR repair proteins led to loss of long-range interactions. To test silencing, we built both a and α strains that were deleted for a DSB repair protein or Sir4 but contained the TetO and LacO arrays. WT and deletion strains were serially diluted in a 10-fold series and spotted on a or α tester lawns and assayed for growth. If the HM loci are derepressed, the haploid cell is functionally diploid and will not mate with the tester strain. Growth signifies that the strain is able to mate and therefore has a functionally silenced HM locus. An a strain gives information on the state of silencing of the HML-α locus and an α strain allows the testing of silencing at the HMRa locus. Loss of the DSB proteins tested, Mre11 and Rad51, did not lead to a mating defect on either a or α lawns, whereas in strains lacking Sir4, HML and HMR were completely derepressed (Fig. 5 E). Therefore, the strains that were deficient in DSB repair had functional silencing at both HML and HMR, suggesting that DSB repair proteins do not mediate their effects on HM clustering simply through the disruption of silencing at these loci. This is consistent with our demonstration that long-range interactions are lost upon deletion of the XYZ homology sequences (Fig. 1 D) even though under these conditions, silencing is not perturbed (Fig. S1). These results also indicate that the HR repair protein–mediated effects are not caused by pseudodiploid formation via loss of silencing. Interestingly, we find that the repair proteins when they are artificially tethered to a locus have the ability to recruit Sir proteins and silence genes at this locus (Fig. S1 B). Loss of these repair proteins does not affect silencing probably because Sir protein recruitment by ORC/Rap1/Abf1 bound to the native silencers is very robust.
γ-H2A is enriched at HM loci and their surrounding chromosomal regions
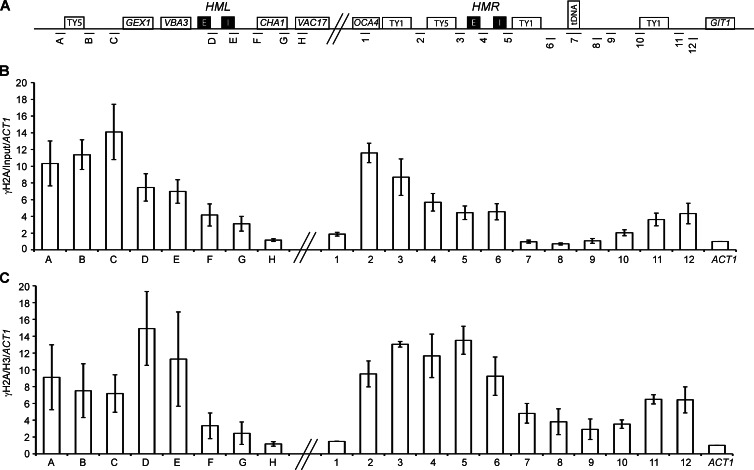
To determine the role of HR repair proteins in HM clustering, we inquired whether or not the HM loci were enriched for chromatin marks normally found at DNA damage sites. Phosphorylation of H2A is an early damage-specific chromatin mark. We therefore performed chromatin immunoprecipitation (IP; ChIP) followed by quantitative PCR (qPCR) to map the distribution of γ-H2A in asynchronously growing, unperturbed cells. All primer pairs were unique with similar amplification efficiencies and did not generate primer–dimer artifacts (Table S2 and Fig. 6 A). We found significant enrichment of this histone modification at both HMR and HML (Fig. 6 B). Enrichment was found in heterochromatic amplicons containing the silencers and at regions previously shown to bind Sir proteins, but this mark reached background levels at amplicons that were not silenced (amplicons 1 and H). Interestingly, this mark extends up to the TY1 and TY5 long terminal repeats and suggests an accumulation of this modification at repetitive elements, which are also sites of replication stress. (Deshpande and Newlon, 1996; Wang et al., 2001; Ivessa et al., 2002; Lemoine et al., 2005; Admire et al., 2006; Azvolinsky et al., 2009).
Figure 6.
γ-H2A is enriched at HM loci and their surrounding chromosomal regions. (A) Schematic of the HML and HMR loci on chromosome III with amplicons used in ChIP-qPCR. Schematic is not to scale. (B) ChIP mapping of γ-H2A. Data are presented as the mean enrichment of IP/input further normalized to an ACT1 amplicon for six IPs from three independent cross-links. Error bars are standard deviation from the mean. (C) γ-H2A enrichment normalized to H3 enrichment to consider nucleosome occupancy. All amplicons normalized to the ACT1 locus.
Because the silencers and the tDNA are depleted of histones, we performed an H3 IP using a polyclonal H3 antibody on the same cross-linked material and normalized the γ-H2A distribution data for histone occupancy (Fig. 6 C and Fig. S3). The data from γ-H2A versus γ-H2A/H3 show two different things: in the first instance, γ-H2A enrichment shows whether or not there is enrichment of γ-H2A at the locus compared with the control ACT1 locus, whereas in the second set of data, γ-H2A/H3 enrichment shows whether or not there is enrichment of γ-H2A on a per nucleosome basis compared with the ACT1 control locus. These latter data reveal an even higher enrichment of γ-H2A at the nucleosome-depleted silencers, suggesting that a high proportion of the few nucleosomes found (in a population of cells) at the silencers and boundary element are phosphorylated on H2AS129 (Fig. 6 C).
Because γ-H2A is enriched at telomeres, we were concerned that the γ-H2A at HMR was simply caused by a gradient of this modification originating at the telomeres and encompassing the HM loci. We therefore also analyzed the presence of γ-H2A when HMR was inserted at the LEU2 locus on chromosome III by ChIP (Fig. 2, C–F). We still observed an enrichment of this mark at this internal HMR locus, suggesting that its presence was not caused by the proximity of HMR to the telomeres.
The HMR-E silencer is required for γ-H2A localization at HMR
We have shown that γ-H2A localizes to silenced chromatin, and the distribution of γ-H2A is very similar to that observed for the Sir proteins. This raised the possibility that silenced loci were persistently being damaged in most cells in the population.
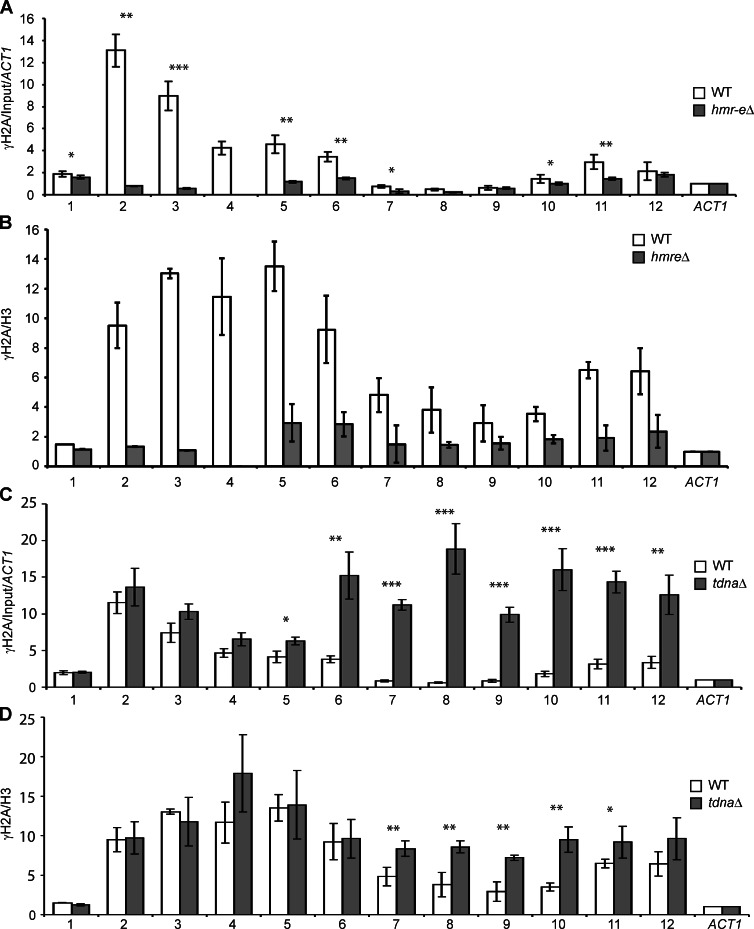
Because γ-H2A is enriched at telomeres (Szilard et al., 2010; Kitada et al., 2011) and the HM loci (Fig. 6), we aimed to determine whether the silenced HM loci recruit γ-H2A. We performed γ-H2A and H3 ChIP in a strain in which the HMR-E silencer has been deleted. In this strain, silencing at HML remains unaffected. When the E silencer is deleted, γ-H2A enrichment across the entire HMR locus, including the HMR-I silencer and the amplicons distal to the telomere end, was lost (Fig. 7, A and B). Loss of an HMR-E silencer does not affect γ-H2A enrichment at HML (Fig. S3 D), consistent with the hypothesis that loss of γ-H2A at HMR is a specific reaction to the loss of heterochromatin at HMR and that loss of the HM interaction does not relieve γ-H2A recruitment at HML.
Figure 7.
The heterochromatin silencer is necessary for γ-H2A enrichment at HMR, and the tDNA insulator restricts the spread of γ-H2A from HMR. (A) ChIP-qPCR plots of γ-H2A enrichment at HMR in an hmr-eΔ strain. (B) γ-H2A/H3 enrichment at HMR in the hmr-eΔ strain. (C) γ-H2A enrichment at HMR in a tT(AUG)CΔ strain (D) γ-H2A/H3 enrichment at HMR in the tT(AUG)CΔ strain. Plots are a mean of three cross-links and six IPs. All amplicons normalized to the ACT1 locus. WT data are the same as in Fig 6 and are included for ease of comparison. Error bars are standard deviation from the mean. P-values by t test are assigned as ***, P < 0.001; **, P < 0.01; and *, P < 0.05.
One of the major mechanisms of spontaneous break formation in cells occurs during replication when replication forks pause or stall at sites in the genome (Aguilera and Gómez-González, 2008; Heyer et al., 2010). We therefore asked whether the HM loci were sites of replication fork pausing, which could then play a role in loading repair proteins. To determine the sites of replication fork pause, we performed ChIP experiments with DNA polymerase, Pol-ε, in asynchronously growing, unperturbed cells. Enrichment of Pol-ε at a locus under these conditions is indicative of the protein spending more time (paused) at that locus (Azvolinsky et al., 2006, 2009). These data show a small enrichment of Pol-ε at HMR and HML though there is more enrichment at the boundaries of the silenced loci (Fig. S3 B). These data suggest that the silenced loci are sites of subtle replication stress, though the boundaries show more fork pausing.
Because there was some pausing of the replication machinery at the tDNA insulator, we next asked whether the tDNA insulator at HMR was required for the recruitment of γ-H2A. Therefore, we assayed a strain in which the tDNA was deleted. In a tdnaΔ, the formerly nucleosome-depleted region incorporated histones (amplicons 6–12; Fig. S3 C). To our surprise, loss of the tDNA did not lead to a decrease in γ-H2A but, on the contrary, leads to a marked increase in γ-H2A enrichment especially in the formerly nucleosome-depleted region as seen in amplicons 7–10 (Fig. 7, C and D). When one normalizes γ-H2A levels to H3 to take into account nucleosome occupancy differences between the WT strain and the tdnaΔ strain at the HMR locus (Dhillon et al., 2009), the tdnaΔ and the regions immediately surrounding the tdnaΔ still showed a smaller, but significant, increase in γ-H2A enrichment on a per nucleosome basis (Fig. 7 D). These data are very similar to those observed for Sir protein distribution upon loss of the tDNA boundary element (Oki and Kamakaka, 2005; Dhillon et al., 2009) and suggest that the tDNA is playing a role in directly or indirectly blocking the spread of γ-H2A but not in the recruitment of this protein and that γ-H2A localization at HMR is directly coupled with silenced chromatin.
The data suggest that the silencer or silenced chromatin is playing a role in the phosphorylation of γ-H2A at the HM loci, or alternatively, silent chromatin stabilizes γ-H2A by preventing the eviction of this modified histone and its subsequent dephosphorylation by the Pph3 phosphatase (Keogh et al., 2006). The latter possibility is consistent with the observation that histone exchange is reduced at silenced loci (Dion et al., 2007; Rufiange et al., 2007). Importantly, these data are inconsistent with the hypothesis that γ-H2A is solely enriched at HM as a result of spreading from the telomeres. Furthermore, the tDNA boundary element does not recruit γ-H2A but instead acts as a barrier to the spread of γ-H2A.
The SMC proteins mediate long-range association of HML and HMR
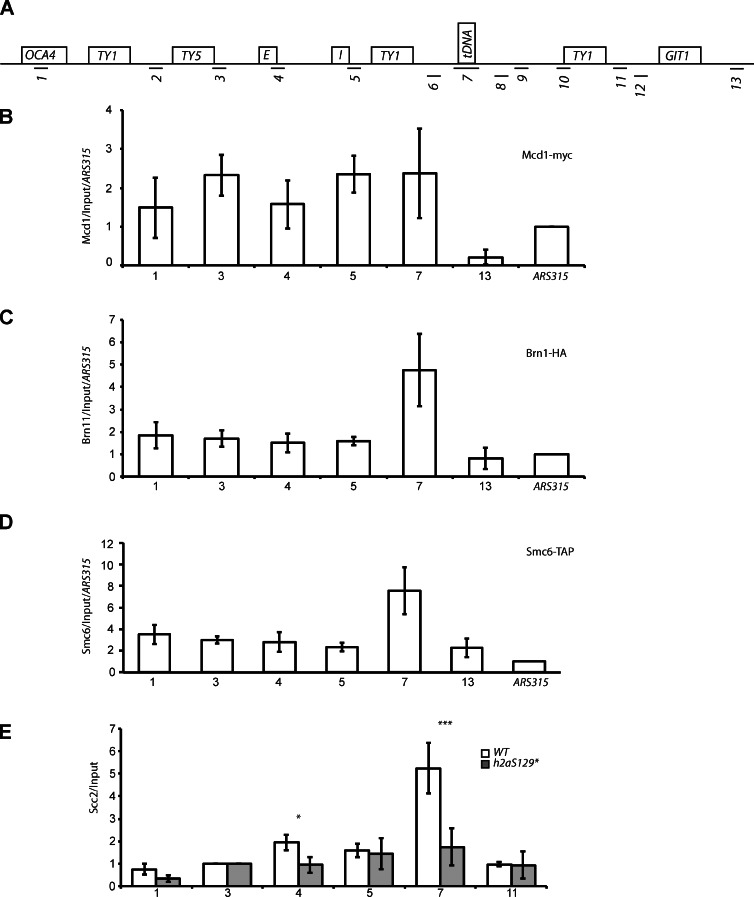
We next asked the question of how does γ-H2A mediate the long-range clustering of HML and HMR. We reasoned that the SMC proteins might play a role in this process given their centrality in long-range chromatin dynamics (De Piccoli et al., 2009; Hudson et al., 2009; Bose and Gerton, 2010; Wood et al., 2010). We mapped the distribution of a subunit of the cohesin complex, Myc-Mcd1 (Fig. 8 B), as well as a subunit of the condensin complex, HA-Brn1 (Fig. 8 C), and a subunit of the repair SMCs, tandem affinity peptide (TAP)–Smc6 (Fig. 8 D). This mapping showed that the condensin and repair SMCs were primarily localized to the boundary of the silenced HMR domain (Fig. 8, C and D) and is consistent with genomic data showing that these marks localize to tDNAs in both S. cerevisiae and Schizosaccharomyces pombe (Ampatzidou et al., 2006; D’Ambrosio et al., 2008; Haeusler et al., 2008; Pebernard et al., 2008; Iwasaki et al., 2010), whereas Mcd1 was modestly distributed across the entire silenced domain (Fig. 8 B) also consistent with previous results (Glynn et al., 2004; Dubey and Gartenberg, 2007; Kogut et al., 2009; Ocampo-Hafalla and Uhlmann, 2011).
Figure 8.
SMC proteins are enriched at HMR and Scc2 enrichment requires γH2A. (A) Schematic of chromosome III qPCR amplicons. (B–D) ChIP-qPCR enrichment of Mcd1-myc (cohesin; B), Brn1-HA (condensin; C), and SMC6-TAP (reparin; D). (E) ChIP-qPCR plots of Scc2 in a WT and htaS129* mutant. Plots are a mean of at least two independent cross-links and four IPs. Error bars are standard deviation from the mean. P-values by t test are assigned as ***, P < 0.001; and *, P < 0.05.
Scc2 helps loads SMC proteins (Uhlmann and Nasmyth, 1998; Ciosk et al., 2000), and we therefore mapped the distribution of Scc2 in WT yeast cells (Fig. 8 E). ChIP showed the enrichment of this protein at the tDNA boundary element as well.
If Scc2 was mediating long-range clustering of the HM loci, loss of Scc2 should lead to a separation of the HM loci. The allele of SCC2, scc2D730V, which does not affect telomere clustering but exhibits a condensin-loading defect at tDNAs (Gard et al., 2009), shows a significant loss of HM interactions (median distance = 730 nm, P = 1.88 × 10−14; Fig. 4 G), suggesting that this protein plays a role in HM clustering.
We finally inquired about the role of γ-H2A in the binding of the SMC proteins to the HMR domain. We mapped Scc2 in a γ-H2A mutant and found that in the absence of this histone modification, the SMC loader Scc2 was no longer enriched at the tDNA boundary (Fig. 8 E). In summary, we have shown that γ-H2A stabilizes the binding of Scc2 and possibly the SMC proteins to the HM loci, which then likely mediate long-range association between the two loci.
Discussion
HML and HMR are located on opposite ends of chromosome III but are in close three-dimensional space in the yeast nucleus. and this clustering in the WT yeast nucleus is most likely caused by direct interactions between the silencers (Miele et al., 2009). In this study, we aimed to elucidate the factors that contribute to this long-range clustering and identify mutants in which these associations are lost. We have uncovered a network of cis-elements and trans-factors that influence the long-range association.
Heterochromatic HML–HMR clustering occurs independently of the telomeres
Our results indicate a direct mechanism underlying HML–HMR association. If long-range associations between the HM loci were being mediated by the localization of Sir proteins and Esc1/Ku at the telomeres, deletion of the HMR-E silencer should have no effect on these interactions. However, loss of silencing at HMR, when silencing at HML and the telomeres remains unaffected, was sufficient to abrogate long-range interactions (Figs. 1 and 4). Moving HMR to an internal locus was sufficient to form long-range interactions (Fig. 2), and these associations are not restricted to chromosome III because we observe similar clustering when both HML and HMR are moved to chromosome VI (Fig. 3). We do not observe clustering when HML resides on chromosome III and HMR resided on chromosome VI. One possibility is that interchromosomal interactions are rare and thus not detected, or alternatively, HM interactions only occur within individual chromosome territories. We tend to favor the former possibility based on the observation that HR between loci occurs but is less efficient when the loci are on different chromosomes (Lichten and Haber, 1989).
It has been shown that telomere clustering is dependent on chromosome arm length, in which arms of similar length are found in the same telomere cluster (Taddei et al., 2010; Therizols et al., 2010). Despite the fact that chromosome III has arms of different lengths (IIIR is twice as long as IIIL), TEL-IIIR and TEL-IIIL cluster together (Therizols et al., 2010), but our results suggest that the telomere clustering is indirect and is driven primarily by the HML–HMR association. Loss of silencing at HMR (hmr-eΔ) results in the separation of HML (TEL-IIIL) and HMR (TEL-IIIR), but these loci still localize to the nuclear periphery, consistent with the proposition that HML and HMR drive telomere IIIL–IIIR interactions, and upon the loss of these interactions, the fluorescence foci separate, as the two telomeres now migrate to sites at the nuclear periphery dependent on chromosome arm length. Additionally, the HM loci clustered when present on chromosome VI despite being placed at positions that conferred differences in arm lengths.
HR repair proteins affect long-range HML–HMR association
We showed that most members of the HR repair pathway tested affected HM long-range interactions, including early and late HR repair proteins as well as the SMC proteins. Neither a nucleotide excision repair protein nor a NHEJ protein affected HM interactions, suggesting that the HR pathway contributed specifically to HM interactions.
Although it is not clear whether these mutants affect HM clustering directly or indirectly, we did not see silencing defects at either HML or HMR in HR-deficient strains, and therefore, although HR-deficient strains lead to a dispersal of some telomeric Sir proteins (Maillet et al., 2001), that does not appear to be the case at the HM loci. In addition, Nup60 mutants also result in a small decrease in silencing at HMR (Ruben et al., 2011), which does not affect HML–HMR interactions. Furthermore, in an XYZ delete strain the long-range interactions are lost, but the silencers and silencing are unperturbed. It is therefore unlikely that the loss of HR proteins leads to the loss of HM clustering solely via the dispersal of Sir proteins from silenced loci.
Two very recent studies described an increase in chromosome mobility upon DNA damage (Dion et al., 2012; Miné-Hattab and Rothstein, 2012). Both studies showed an increase in mobility of a break point, dependent on Mec1, Rad51, and Rad54. It is possible that HR repair proteins influence the search mechanism after damage by increasing mobility genome wide, and it is also possible that the loss of long-range HM interactions in unperturbed cells in our study is a reflection of this role of these proteins, but our study did not directly examine mobility, and future experiments will be necessary to shed light on this issue.
γ-H2A and long-range clustering of HML and HMR
γ-H2A, a mark for DNA damage, which acts as a chromatin scaffold for repair proteins to stably bind was highly enriched at both HML and HMR in logarithmically growing unperturbed yeast cells and is consistent with recent observations in S. cerevisiae that showed that γ-H2A is present at subtelomeric heterochromatin in undamaged cells (Szilard et al., 2010; Kitada et al., 2011). γ-H2A localization to silenced heterochromatin is not restricted to budding yeast and is also found at the silent mating-type locus, centromeres, telomeres, and rDNA in S. pombe (Rozenzhak et al., 2010). The observation that this histone mark is present at heterochromatin from both of these yeasts despite the different proteins involved in the formation of heterochromatin in these two yeasts suggests that the localization of this modification to heterochromatin may be conserved across other species as well.
Our localization experiments of γ-H2A are in apparent contrast to another study (Kim et al., 2007), but the differences between Kim et al. (2007) and our results are simply a result of the manner of normalization and presentation of γ-H2A distribution patterns. Kim et al. (2007) presented γ-H2A distribution across the HMR domain as fold difference between undamaged and damaged cells. Because HMR already possessed γ-H2A in undamaged cells, there was no observable change in this protein after damage.
How is γ-H2A recruited to heterochromatic regions? One possibility is that replication fork pausing at or in the vicinity of silenced domains leads to the modification of this histone by Mec1 and/or Tel1. Monitoring DNA Pol-ε indicates pausing at HMR. Others have also shown, using 2D gel analysis, that HM silencers and tDNAs are sites of fork pausing/stalling (Deshpande and Newlon, 1996; Wang et al., 2001; Ivessa et al., 2002; Lemoine et al., 2005; Admire et al., 2006; Azvolinsky et al., 2009). Although we see increased pausing at these sites, loss of the tDNA boundary element does not lead to loss of γ-H2A from HMR, arguing against the tDNA being solely involved in the recruitment of γ-H2A. The recruitment and spreading of γ-H2A at HMR requires the HMR-E silencer and is probably a function of silencing. Whether it is also a function of replication pausing at the silenced domains is unclear. Our observation suggests that silencing either aid in the modification of H2A by recruiting a kinase (Mec1 or Tel1) or stabilizes the modification after it is laid down by blocking the eviction of the modified histone and its subsequent dephosphorylation by the Pph3 phosphatase (Keogh et al., 2006). Consistent with this latter possibility is the observation that silenced chromatin has lower histone turnover (Dion et al., 2007).
Interestingly, γ-H2A was necessary for long-range HM association, suggesting a possible link between proteins involved in HR repair and long-range heterochromatin clustering. Replication pausing-induced γ-H2A could trigger the recruitment of the HR repair pathway, leading to homology-based clustering (because sequence homology between HML and HMR is necessary for HM clustering). After replication, the packaging of the domain with Sir proteins would reduce histone exchange, preventing eviction of γ-H2A and thus maintaining the clustering of the HM loci. We also show that mutations in the SMC loader Scc2 affect long-range heterochromatin clustering and that γ-H2A is required for the binding/stabilization of Scc2 to the silenced chromatin boundary. Furthermore, all three classes of SMC proteins localize to the silenced chromatin domain (Fig. 8) or to the boundary of this domain, and numerous laboratories have shown that the binding of SMC proteins to chromatin requires Scc2 (Ciosk et al., 2000; Lengronne et al., 2004; Lindroos et al., 2006; Pebernard et al., 2008; Kogut et al., 2009). Thus, one simple model for the mechanism by which γ-H2A functions in long-range clustering of the HM loci would be via the stabilization of binding of the SMC proteins to these loci and possibly other repair proteins. Studies have mapped Ku (Patterson and Fox, 2008; Vandre et al., 2008; Bystricky et al., 2009) to the silenced HM loci.
Why do HML–HMR cluster and why is this dependent on the HR repair proteins?
Telomeric heterochromatin is a reservoir for repair proteins, and mutations in Ku, Mre11, and Tel1 affect clustering of telomeres and clustering of silencing proteins at the telomeres (Martin et al., 1999; Hiraga et al., 2008). Ku, Scc2, and the SMC proteins also map to the silenced HM loci in unperturbed cells, and these proteins play key roles in HR repair (Aguilera and Gómez-González, 2008; Onn et al., 2008; De Piccoli et al., 2009; Hudson et al., 2009; Heyer et al., 2010). It is therefore possible that the HM loci are in part reservoirs for repair proteins in undamaged cells. Further studies will help shed light on the interrelationships between the HR repair proteins, HM-bound Ku, and the long-range clustering of HM loci.
There is also increasing evidence that repair proteins bind chromatin at specific sites that are not damaged but are more prone to damage. In mammalian cells, γ-H2A and SMC5/6 are enriched at repetitive elements, such as short-interspersed nuclear elements and tDNAs in the absence of damage (Barlow et al., 2013), whereas mammalian Rad51 binds sites of replication fork pausing before DNA break formation by a process that does not trigger HR repair unless forks collapse (Petermann et al., 2010). In yeast, silenced chromatin, Ty elements, and tRNA genes are all sites of replication stress (Deshpande and Newlon, 1996; Wang et al., 2001; Ivessa et al., 2002; Lemoine et al., 2005; Admire et al., 2006; Azvolinsky et al., 2009) that could trigger a HR repair response in the absence of fork collapse and be bound by γ-H2A.
Repetitive DNA sequences localize to specific nuclear compartments, and it has been suggested that this reduces deleterious recombination (Torres-Rosell et al., 2007; Aguilera and Gómez-González, 2008; Lukas et al., 2011). It is therefore possible that during replication stress, repair proteins phosphorylate H2A at heterochromatic sites. The phosphorylation is stabilized by heterochromatin formation after replication, which then stabilizes long-range capture of homologous sequences by the SMC proteins resulting in clustering.
It should also be pointed out that mating-type switching is used by haploid yeast to switch the genetic information at the MAT locus using the donor information that is stored at the cryptic mating-type loci HML and HMR (Haber, 1998, 2012). Switching initiates by the formation of a HO-induced DSB at the MAT locus during the G1 phase of the cell cycle, and the HM donor locus moves to the nuclear interior, where it interacts with MAT (Bystricky et al., 2009) mediated by the HR repair proteins, including γ-H2A. Although G1 is the phase of the cell cycle when yeast cells use NHEJ to repair DSBs (Symington and Gautier, 2011), for switching, yeast cells use the HR machinery, and mutations in the HR repair proteins disrupt mating-type switching (Haber, 1998, 2012). In this context, having the silenced HML and HMR loci poised for HR-mediated repair could favor HR-mediated recombination and mating-type switching even under conditions that typically favor NHEJ mechanisms. The lack of HR marks/proteins at MAT before the HO-induced break precludes MAT from interacting with HM outside of mating-type switching (Szilard et al., 2010); instead, HM interact with each other at the nuclear periphery, where they are secluded from the MAT locus, thus precluding premature/deleterious switching. This model is analogous to what is observed at the rDNA locus (Torres-Rosell et al., 2007) and is also consistent with observations that HR mutants that affect HM clustering also affect mating-type switching (Haber, 1998, 2012), but testing this model will require the isolation of separation-of-function alleles in repair proteins.
In summary, these data show that a novel and diverse set of pathways contribute to heterochromatic organization in the budding yeast nucleus, where both the HR pathway and the silencing pathway contribute to HM interactions. γ-H2A is found at heterochromatin, where it helps facilitate HM long-range clustering via the SMC proteins.
Materials and methods
Yeast strains and primers
Yeast strains and primer sequences can be found in Table S1 and S2, respectively. Some yeast strains and plasmids were provided by K. Bystricky (Laboratoire de Biologie Moleculaire Eucaryote, Toulouse, France), L. Aragon (Medical Research Council Clinical Sciences Centre and Imperial College London Mill Hill, London, England, UK), J. Gerton (Stowers Institute for Medical Research, Kansas City, MO), M. Gartenberg (University of Medicine and Dentistry of New Jersey and Robert Woods Johnson Medical School, New Brunswick, NJ), R. Rothstein (Columbia University, New York, NY), and S. Jackson (Cambridge University, Cambridge, England, UK).
ChIP
ChIPs were performed as previously described (Dhillon et al., 2009; Valenzuela et al., 2009). In brief, 300 ml of asynchronously dividing cells were grown in YPD (yeast extract, peptone, and dextrose) to an OD of 2.2–2.4 and fixed in 1% formaldehyde for 15 min followed by quenching in glycine for 12–15 min, spun down, washed in PBS, spun down, and frozen. Cells were thawed and lysed by agitation with glass beads. Chromatin was sheared to a mean length of 300 bp by sonication. 950 µl of cleared lysates was incubated with primary antibody for 3 h at 4°C and then added to preblocked protein A/G–Sepharose beads overnight with mixing at 4°C. IPs were performed in duplicate for each cross-link reaction. Chromatin was eluted with 10% Chelex 100 (Bio-Rad Laboratories) and treated with proteinase K. Eluted chromatin was quantified by PicoGreen (Invitrogen). For qPCR, equal amounts of input DNA and IP DNA (100–200 pg) were added to each reaction. Reactions were performed in technical duplicate. Enrichment is reported as IP/input followed by normalization to another locus (γ-H2A and H3: ACT1). To account for differential histone density, some data are reported as γ-H2A/H3, in which the ratio of the means of the two IPs for each mark within a fix is taken, allowing for a standard deviation. For these experiments, the n is reported as the number of cross-links rather than the number of IPs as in other experiments. Error bars shown are standard deviations. Pol-ε ChIP cells were performed as described in this paper with the following changes: cells were grown to 1.0 OD, only input DNA was quantified and used at 40 pg, and 4 µl of IP material was used in qPCR reactions.
Quantitative mating assays
Quantitative mating assays were performed as previously described (Ruben et al., 2011). In brief, 1 OD of a or α lawn was plated on YMD plates. Serial dilutions of tester strains (1–0.001 ODs) are spotted on mating lawns. Mating is only able to occur when the a and α genes at HMR and HML are repressed. Mating leads to complementation of auxotrophic markers in a diploid and growth on YMD plates. When necessary, strains were kept under plasmid selection. Plates were incubated at 30°C and photographed at 24–72 h.
Antibodies
Antibodies used were from the following sources and used at the following dilutions or concentrations: antiphospho–histone H2A (Ser129), yeast specific (catalogue no. 07–745, lot no. 30559; EMD Millipore) at 5 µl, anti–histone H3 (source) at 5 µg, and monoclonal anti–myc 9E11 (Abcam) at 5 µl.
Image acquisition and manipulation
Microscopy was performed on live cells for all experiments. Cells were grown exponentially in YMD (yeast minimal dextrose) + amino acids (Leu, Ura, Trp, Lys, Ade, and His) to an OD600nm of ∼0.6. Cells were rinsed in YMD + amino acids before imaging and placed on YMD + amino acids and 1.5% agarose patches on slides, covered, and imaged. Images were acquired on an inverted wide-field microscope (Xi70; Olympus) with precise stage (DeltaVision; Applied Precision) using a camera (CoolSNAP HQ2; Photometrics). Optical image stacks of 20 images were acquired with a step size of 200 nm for 400–500 ms in the appropriate wavelength channel (CFP/YFP/GFP/mCherry). A 100×/1.4 NA oil objective was used. Acquisition software softWoRx 3.7.1 (Applied Precision) was used for image acquisition and analysis. All images were taken at 25°C. Cropping of images was performed in Photoshop (Adobe).
For distance analysis, the distances between HML (yellow) and HMR (cyan) were calculated in nanometers using the measure tool in three dimensions. The measured distances were loaded into R software, and the data were plotted as a boxplot. The box represents the middle 50% of data points with the black line showing the median of distances. Outliers are defined as distances >1.5 times the interquartile range and are represented by open circles. Data presented are the sum of at least two independent strains and trials.
For zone analysis, 200-nm optical slices were taken on live cells, and only the 10 middle planes of the nucleus were assayed. Images were acquired in the GFP and mCherry channels. For the position of the GFP focus in relation to the HDEL-dsRed, marked nuclear envelopes were determined as a percentage of fluorescent foci in one of two concentric nuclear zones of equal surface in the plane bearing the brightest GFP-LacI focus. Three independent trials were performed for each strain, and strains were scored in a blind manner by measuring the distance between the GFP spot (array) and the nuclear membrane (s2p) and the diameter of the nucleus (p2p) in nanometers. A ratio of (s2p/p2p) × 2 was calculated and used for assigning to one of two zones, either the peripheral zone (zone 1) or the interior zone (zone 2), of approximately equal surface area (Fig. 5 B) or volume (Fig. 5 D) as previously described. P-values were determined by χ2 test (Hediger et al., 2002; Ruben et al., 2011).
Three independent trials were performed for each strain, and strains were scored in a blind manner by measuring the distance between the GFP spot (array) and the nuclear membrane (s2p) and the diameter of the nucleus (p2p) in nanometers. A ratio of (s2p/p2p) × 2 was calculated and used for assigning to one of two zones of approximately equal surface area.
Online supplemental material
Fig. S1 shows silencing controls and representative two-dot assay images. Fig. S2 shows cell cycle, mating type, and nuclear diameter controls. Fig. S3 shows ChIP controls used in this study. Table S1 shows strains. Table S2 shows oligonucleotides. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201211105/DC1.
Supplementary Material
Acknowledgments
We would like to thank S. Gasser, J. Gerton, J. Haber, M. Gartenberg, N. Dhillon, J. Raab, G. Hartzog, and N. Bhalla for experimental suggestions, advice, and comments during the progression of this research and comments on the manuscript.
This work was supported by grants from the National Institutes of Health to R.T. Kamakaka (GM078068) and to J.G. Kirkland (T32-GM008646) and a President’s Dissertation Year Fellowship to J.G. Kirkland.
Footnotes
Abbreviations used in this paper:
- ChIP
- chromatin IP
- DSB
- double-strand break
- HR
- homologous recombination
- IP
- immunoprecipitation
- NHEJ
- nonhomologous end joining
- qPCR
- quantitative PCR
- rDNA
- ribosomal DNA
- Sir
- silent information regulator
- SMC
- structural maintenance of chromatin
- TAP
- tandem affinity peptide
- WT
- wild type
References
- Admire A., Shanks L., Danzl N., Wang M., Weier U., Stevens W., Hunt E., Weinert T. 2006. Cycles of chromosome instability are associated with a fragile site and are increased by defects in DNA replication and checkpoint controls in yeast. Genes Dev. 20:159–173 10.1101/gad.1392506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A., Gómez-González B. 2008. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 9:204–217 10.1038/nrg2268 [DOI] [PubMed] [Google Scholar]
- Ahmed S., Brickner J.H. 2007. Regulation and epigenetic control of transcription at the nuclear periphery. Trends Genet. 23:396–402 10.1016/j.tig.2007.05.009 [DOI] [PubMed] [Google Scholar]
- Ampatzidou E., Irmisch A., O’Connell M.J., Murray J.M. 2006. Smc5/6 is required for repair at collapsed replication forks. Mol. Cell. Biol. 26:9387–9401 10.1128/MCB.01335-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniacci L.M., Kenna M.A., Skibbens R.V. 2007. The nuclear envelope and spindle pole body-associated Mps3 protein bind telomere regulators and function in telomere clustering. Cell Cycle. 6:75–79 10.4161/cc.6.1.3647 [DOI] [PubMed] [Google Scholar]
- Aylon Y., Kupiec M. 2004. DSB repair: the yeast paradigm. DNA Repair (Amst.). 3:797–815 10.1016/j.dnarep.2004.04.013 [DOI] [PubMed] [Google Scholar]
- Azvolinsky A., Dunaway S., Torres J.Z., Bessler J.B., Zakian V.A. 2006. The S. cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes Dev. 20:3104–3116 10.1101/gad.1478906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azvolinsky A., Giresi P.G., Lieb J.D., Zakian V.A. 2009. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol. Cell. 34:722–734 10.1016/j.molcel.2009.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow J.H., Faryabi R.B., Callén E., Wong N., Malhowski A., Chen H.T., Gutierrez-Cruz G., Sun H.W., McKinnon P., Wright G., et al. 2013. Identification of early replicating fragile sites that contribute to genome instability. Cell. 152:620–632 10.1016/j.cell.2013.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose T., Gerton J.L. 2010. Cohesinopathies, gene expression, and chromatin organization. J. Cell Biol. 189:201–210 10.1083/jcb.200912129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D., Foiani M. 2008. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 9:297–308 10.1038/nrm2351 [DOI] [PubMed] [Google Scholar]
- Branzei D., Foiani M. 2010. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 11:208–219 10.1038/nrm2852 [DOI] [PubMed] [Google Scholar]
- Brickner J.H. 2009. Transcriptional memory at the nuclear periphery. Curr. Opin. Cell Biol. 21:127–133 10.1016/j.ceb.2009.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bupp J.M., Martin A.E., Stensrud E.S., Jaspersen S.L. 2007. Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J. Cell Biol. 179:845–854 10.1083/jcb.200706040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystricky K., Laroche T., van Houwe G., Blaszczyk M., Gasser S.M. 2005. Chromosome looping in yeast: telomere pairing and coordinated movement reflect anchoring efficiency and territorial organization. J. Cell Biol. 168:375–387 10.1083/jcb.200409091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystricky K., Van Attikum H., Montiel M.D., Dion V., Gehlen L., Gasser S.M. 2009. Regulation of nuclear positioning and dynamics of the silent mating type loci by the yeast Ku70/Ku80 complex. Mol. Cell. Biol. 29:835–848 10.1128/MCB.01009-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y., Tsutsumi C., Yamane M., Okamasa K., Haraguchi T., Hiraoka Y. 2006. Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell. 125:59–69 10.1016/j.cell.2006.01.048 [DOI] [PubMed] [Google Scholar]
- Ciosk R., Shirayama M., Shevchenko A., Tanaka T., Toth A., Shevchenko A., Nasmyth K. 2000. Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell. 5:243–254 10.1016/S1097-2765(00)80420-7 [DOI] [PubMed] [Google Scholar]
- Cortés-Ledesma F., de Piccoli G., Haber J.E., Aragón L., Aguilera A. 2007. SMC proteins, new players in the maintenance of genomic stability. Cell Cycle. 6:914–918 10.4161/cc.6.8.4107 [DOI] [PubMed] [Google Scholar]
- D’Ambrosio C., Schmidt C.K., Katou Y., Kelly G., Itoh T., Shirahige K., Uhlmann F. 2008. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 22:2215–2227 10.1101/gad.1675708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deem A.K., Li X., Tyler J.K. 2012. Epigenetic regulation of genomic integrity. Chromosoma. 121:131–151 10.1007/s00412-011-0358-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Piccoli G., Torres-Rosell J., Aragón L. 2009. The unnamed complex: what do we know about Smc5-Smc6? Chromosome Res. 17:251–263 10.1007/s10577-008-9016-8 [DOI] [PubMed] [Google Scholar]
- Deshpande A.M., Newlon C.S. 1996. DNA replication fork pause sites dependent on transcription. Science. 272:1030–1033 10.1126/science.272.5264.1030 [DOI] [PubMed] [Google Scholar]
- Dhillon N., Raab J., Guzzo J., Szyjka S.J., Gangadharan S., Aparicio O.M., Andrews B., Kamakaka R.T. 2009. DNA polymerase epsilon, acetylases and remodellers cooperate to form a specialized chromatin structure at a tRNA insulator. EMBO J. 28:2583–2600 10.1038/emboj.2009.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion M.F., Kaplan T., Kim M., Buratowski S., Friedman N., Rando O.J. 2007. Dynamics of replication-independent histone turnover in budding yeast. Science. 315:1405–1408 10.1126/science.1134053 [DOI] [PubMed] [Google Scholar]
- Dion V., Kalck V., Horigome C., Towbin B.D., Gasser S.M. 2012. Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat. Cell Biol. 14:502–509 10.1038/ncb2465 [DOI] [PubMed] [Google Scholar]
- Donze D., Kamakaka R.T. 2001. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 20:520–531 10.1093/emboj/20.3.520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D., Adams C.R., Rine J., Kamakaka R.T. 1999. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 13:698–708 10.1101/gad.13.6.698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs J.A., Lowndes N.F., Jackson S.P. 2000. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature. 408:1001–1004 10.1038/35050000 [DOI] [PubMed] [Google Scholar]
- Dubarry M., Loïodice I., Chen C.L., Thermes C., Taddei A. 2011. Tight protein-DNA interactions favor gene silencing. Genes Dev. 25:1365–1370 10.1101/gad.611011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey R.N., Gartenberg M.R. 2007. A tDNA establishes cohesion of a neighboring silent chromatin domain. Genes Dev. 21:2150–2160 10.1101/gad.1583807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flott S., Alabert C., Toh G.W., Toth R., Sugawara N., Campbell D.G., Haber J.E., Pasero P., Rouse J. 2007. Phosphorylation of Slx4 by Mec1 and Tel1 regulates the single-strand annealing mode of DNA repair in budding yeast. Mol. Cell. Biol. 27:6433–6445 10.1128/MCB.00135-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard S., Light W., Xiong B., Bose T., McNairn A.J., Harris B., Fleharty B., Seidel C., Brickner J.H., Gerton J.L. 2009. Cohesinopathy mutations disrupt the subnuclear organization of chromatin. J. Cell Biol. 187:455–462 10.1083/jcb.200906075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn E.F., Megee P.C., Yu H.G., Mistrot C., Unal E., Koshland D.E., DeRisi J.L., Gerton J.L. 2004. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2:E259 10.1371/journal.pbio.0020259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M., Laroche T., Formenton A., Maillet L., Scherthan H., Gasser S.M. 1996. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol. 134:1349–1363 10.1083/jcb.134.6.1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M., Strahl-Bolsinger S., Renauld H., Laroche T., Kennedy B.K., Grunstein M., Gasser S.M. 1997. Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J. 16:3243–3255 10.1093/emboj/16.11.3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grund S.E., Fischer T., Cabal G.G., Antúnez O., Pérez-Ortín J.E., Hurt E. 2008. The inner nuclear membrane protein Src1 associates with subtelomeric genes and alters their regulated gene expression. J. Cell Biol. 182:897–910 10.1083/jcb.200803098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J.E. 1998. Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 32:561–599 10.1146/annurev.genet.32.1.561 [DOI] [PubMed] [Google Scholar]
- Haber J.E. 2012. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics. 191:33–64 10.1534/genetics.111.134577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler R.A., Pratt-Hyatt M., Good P.D., Gipson T.A., Engelke D.R. 2008. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev. 22:2204–2214 10.1101/gad.1675908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger F., Neumann F.R., Van Houwe G., Dubrana K., Gasser S.M. 2002. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr. Biol. 12:2076–2089 10.1016/S0960-9822(02)01338-6 [DOI] [PubMed] [Google Scholar]
- Heyer W.D., Li X., Rolfsmeier M., Zhang X.P. 2006. Rad54: the Swiss Army knife of homologous recombination? Nucleic Acids Res. 34:4115–4125 10.1093/nar/gkl481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer W.D., Ehmsen K.T., Liu J. 2010. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 44:113–139 10.1146/annurev-genet-051710-150955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks W.M., Yamaguchi M., Haber J.E. 2011. Real-time analysis of double-strand DNA break repair by homologous recombination. Proc. Natl. Acad. Sci. USA. 108:3108–3115 10.1073/pnas.1019660108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga S., Botsios S., Donaldson A.D. 2008. Histone H3 lysine 56 acetylation by Rtt109 is crucial for chromosome positioning. J. Cell Biol. 183:641–651 10.1083/jcb.200806065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes A.M., Haber J.E. 1999. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell. 96:415–424 10.1016/S0092-8674(00)80554-1 [DOI] [PubMed] [Google Scholar]
- Hudson D.F., Marshall K.M., Earnshaw W.C. 2009. Condensin: Architect of mitotic chromosomes. Chromosome Res. 17:131–144 10.1007/s10577-008-9009-7 [DOI] [PubMed] [Google Scholar]
- Ivessa A.S., Zhou J.Q., Schulz V.P., Monson E.K., Zakian V.A. 2002. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 16:1383–1396 10.1101/gad.982902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki O., Tanaka A., Tanizawa H., Grewal S.I., Noma K. 2010. Centromeric localization of dispersed Pol III genes in fission yeast. Mol. Biol. Cell. 21:254–265 10.1091/mbc.E09-09-0790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q., Trelles-Sticken E., Scherthan H., Loidl J. 1998. Yeast nuclei display prominent centromere clustering that is reduced in nondividing cells and in meiotic prophase. J. Cell Biol. 141:21–29 10.1083/jcb.141.1.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q.W., Fuchs J., Loidl J. 2000. Centromere clustering is a major determinant of yeast interphase nuclear organization. J. Cell Sci. 113:1903–1912 [DOI] [PubMed] [Google Scholar]
- Keogh M.C., Kim J.A., Downey M., Fillingham J., Chowdhury D., Harrison J.C., Onishi M., Datta N., Galicia S., Emili A., et al. 2006. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature. 439:497–501 10.1038/nature04384 [DOI] [PubMed] [Google Scholar]
- Kim J.A., Kruhlak M., Dotiwala F., Nussenzweig A., Haber J.E. 2007. Heterochromatin is refractory to γ-H2AX modification in yeast and mammals. J. Cell Biol. 178:209–218 10.1083/jcb.200612031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E., van der Linden E., Sanchez H., Wyman C. 2009. RAD50, an SMC family member with multiple roles in DNA break repair: how does ATP affect function? Chromosome Res. 17:277–288 10.1007/s10577-008-9018-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T., Schleker T., Sperling A.S., Xie W., Gasser S.M., Grunstein M. 2011. γH2A is a component of yeast heterochromatin required for telomere elongation. Cell Cycle. 10:293–300 10.4161/cc.10.2.14536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogut I., Wang J., Guacci V., Mistry R.K., Megee P.C. 2009. The Scc2/Scc4 cohesin loader determines the distribution of cohesin on budding yeast chromosomes. Genes Dev. 23:2345–2357 10.1101/gad.1819409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche T., Martin S.G., Gotta M., Gorham H.C., Pryde F.E., Louis E.J., Gasser S.M. 1998. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr. Biol. 8:653–656 10.1016/S0960-9822(98)70252-0 [DOI] [PubMed] [Google Scholar]
- Laroche T., Martin S.G., Tsai-Pflugfelder M., Gasser S.M. 2000. The dynamics of yeast telomeres and silencing proteins through the cell cycle. J. Struct. Biol. 129:159–174 10.1006/jsbi.2000.4240 [DOI] [PubMed] [Google Scholar]
- Lemoine F.J., Degtyareva N.P., Lobachev K., Petes T.D. 2005. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell. 120:587–598 10.1016/j.cell.2004.12.039 [DOI] [PubMed] [Google Scholar]
- Lengronne A., Katou Y., Mori S., Yokobayashi S., Kelly G.P., Itoh T., Watanabe Y., Shirahige K., Uhlmann F. 2004. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 430:573–578 10.1038/nature02742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten M., Haber J.E. 1989. Position effects in ectopic and allelic mitotic recombination in Saccharomyces cerevisiae. Genetics. 123:261–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroos H.B., Ström L., Itoh T., Katou Y., Shirahige K., Sjögren C. 2006. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol. Cell. 22:755–767 10.1016/j.molcel.2006.05.014 [DOI] [PubMed] [Google Scholar]
- Lisby M., Rothstein R. 2004. DNA damage checkpoint and repair centers. Curr. Opin. Cell Biol. 16:328–334 10.1016/j.ceb.2004.03.011 [DOI] [PubMed] [Google Scholar]
- Lisby M., Rothstein R. 2009. Choreography of recombination proteins during the DNA damage response. DNA Repair (Amst.). 8:1068–1076 10.1016/j.dnarep.2009.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M., Rothstein R., Mortensen U.H. 2001. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl. Acad. Sci. USA. 98:8276–8282 10.1073/pnas.121006298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M., Antúnez de Mayolo A., Mortensen U.H., Rothstein R. 2003. Cell cycle-regulated centers of DNA double-strand break repair. Cell Cycle. 2:477–483 10.4161/cc.2.5.483 [DOI] [PubMed] [Google Scholar]
- Lopes M., Cotta-Ramusino C., Pellicioli A., Liberi G., Plevani P., Muzi-Falconi M., Newlon C.S., Foiani M. 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 412:557–561 10.1038/35087613 [DOI] [PubMed] [Google Scholar]
- Lukas J., Lukas C., Bartek J. 2011. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat. Cell Biol. 13:1161–1169 10.1038/ncb2344 [DOI] [PubMed] [Google Scholar]
- Madrid A.S., Mancuso J., Cande W.Z., Weis K. 2006. The role of the integral membrane nucleoporins Ndc1p and Pom152p in nuclear pore complex assembly and function. J. Cell Biol. 173:361–371 10.1083/jcb.200506199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet L., Boscheron C., Gotta M., Marcand S., Gilson E., Gasser S.M. 1996. Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev. 10:1796–1811 10.1101/gad.10.14.1796 [DOI] [PubMed] [Google Scholar]
- Maillet L., Gaden F., Brevet V., Fourel G., Martin S.G., Dubrana K., Gasser S.M., Gilson E. 2001. Ku-deficient yeast strains exhibit alternative states of silencing competence. EMBO Rep. 2:203–210 10.1093/embo-reports/kve044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S.G., Laroche T., Suka N., Grunstein M., Gasser S.M. 1999. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell. 97:621–633 10.1016/S0092-8674(00)80773-4 [DOI] [PubMed] [Google Scholar]
- Mekhail K., Moazed D. 2010. The nuclear envelope in genome organization, expression and stability. Nat. Rev. Mol. Cell Biol. 11:317–328 10.1038/nrm2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele A., Bystricky K., Dekker J. 2009. Yeast silent mating type loci form heterochromatic clusters through silencer protein-dependent long-range interactions. PLoS Genet. 5:e1000478 10.1371/journal.pgen.1000478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills K.D., Sinclair D.A., Guarente L. 1999. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell. 97:609–620 10.1016/S0092-8674(00)80772-2 [DOI] [PubMed] [Google Scholar]
- Miné-Hattab J., Rothstein R. 2012. Increased chromosome mobility facilitates homology search during recombination. Nat. Cell Biol. 14:510–517 10.1038/ncb2472 [DOI] [PubMed] [Google Scholar]
- Mirkin E.V., Mirkin S.M. 2007. Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev. 71:13–35 10.1128/MMBR.00030-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen U.H., Lisby M., Rothstein R. 2009. Rad52. Curr. Biol. 19:R676–R677 10.1016/j.cub.2009.06.001 [DOI] [PubMed] [Google Scholar]
- Nagai S., Dubrana K., Tsai-Pflugfelder M., Davidson M.B., Roberts T.M., Brown G.W., Varela E., Hediger F., Gasser S.M., Krogan N.J. 2008. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 322:597–602 10.1126/science.1162790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai S., Heun P., Gasser S.M. 2010. Roles for nuclear organization in the maintenance of genome stability. Epigenomics. 2:289–305 10.2217/epi.09.49 [DOI] [PubMed] [Google Scholar]
- Nagai S., Davoodi N., Gasser S.M. 2011. Nuclear organization in genome stability: SUMO connections. Cell Res. 21:474–485 10.1038/cr.2011.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocampo-Hafalla M.T., Uhlmann F. 2011. Cohesin loading and sliding. J. Cell Sci. 124:685–691 10.1242/jcs.073866 [DOI] [PubMed] [Google Scholar]
- Oki M., Kamakaka R.T. 2005. Barrier function at HMR. Mol. Cell. 19:707–716 10.1016/j.molcel.2005.07.022 [DOI] [PubMed] [Google Scholar]
- Onn I., Heidinger-Pauli J.M., Guacci V., Unal E., Koshland D.E. 2008. Sister chromatid cohesion: a simple concept with a complex reality. Annu. Rev. Cell Dev. Biol. 24:105–129 10.1146/annurev.cellbio.24.110707.175350 [DOI] [PubMed] [Google Scholar]
- Oza P., Jaspersen S.L., Miele A., Dekker J., Peterson C.L. 2009. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev. 23:912–927 10.1101/gad.1782209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson E.E., Fox C.A. 2008. The Ku complex in silencing the cryptic mating-type loci of Saccharomyces cerevisiae. Genetics. 180:771–783 10.1534/genetics.108.091710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pebernard S., Schaffer L., Campbell D., Head S.R., Boddy M.N. 2008. Localization of Smc5/6 to centromeres and telomeres requires heterochromatin and SUMO, respectively. EMBO J. 27:3011–3023 10.1038/emboj.2008.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E., Orta M.L., Issaeva N., Schultz N., Helleday T. 2010. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell. 37:492–502 10.1016/j.molcel.2010.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillus L., Rine J. 1989. Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell. 59:637–647 10.1016/0092-8674(89)90009-3 [DOI] [PubMed] [Google Scholar]
- Polo S.E., Jackson S.P. 2011. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 25:409–433 10.1101/gad.2021311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rine J., Herskowitz I. 1987. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 116:9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie K.B., Mallory J.C., Petes T.D. 1999. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 19:6065–6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenzhak S., Mejía-Ramírez E., Williams J.S., Schaffer L., Hammond J.A., Head S.R., Russell P. 2010. Rad3 decorates critical chromosomal domains with gammaH2A to protect genome integrity during S-Phase in fission yeast. PLoS Genet. 6:e1001032 10.1371/journal.pgen.1001032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruben G.J., Kirkland J.G., MacDonough T., Chen M., Dubey R.N., Gartenberg M.R., Kamakaka R.T. 2011. Nucleoporin mediated nuclear positioning and silencing of HMR. PLoS ONE. 6:e21923 10.1371/journal.pone.0021923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufiange A., Jacques P.E., Bhat W., Robert F., Nourani A. 2007. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol. Cell. 27:393–405 10.1016/j.molcel.2007.07.011 [DOI] [PubMed] [Google Scholar]
- Rusche L.N., Kirchmaier A.L., Rine J. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72:481–516 10.1146/annurev.biochem.72.121801.161547 [DOI] [PubMed] [Google Scholar]
- Schleker T., Nagai S., Gasser S.M. 2009. Posttranslational modifications of repair factors and histones in the cellular response to stalled replication forks. DNA Repair (Amst.). 8:1089–1100 10.1016/j.dnarep.2009.04.010 [DOI] [PubMed] [Google Scholar]
- Schober H., Kalck V., Vega-Palas M.A., Van Houwe G., Sage D., Unser M., Gartenberg M.R., Gasser S.M. 2008. Controlled exchange of chromosomal arms reveals principles driving telomere interactions in yeast. Genome Res. 18:261–271 10.1101/gr.6687808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober H., Ferreira H., Kalck V., Gehlen L.R., Gasser S.M. 2009. Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev. 23:928–938 10.1101/gad.1787509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker T.H., Petrini J.H. 2011. The MRE11 complex: starting from the ends. Nat. Rev. Mol. Cell Biol. 12:90–103 10.1038/nrm3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N., Wang X., Haber J.E. 2003. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell. 12:209–219 10.1016/S1097-2765(03)00269-7 [DOI] [PubMed] [Google Scholar]
- Symington L.S., Gautier J. 2011. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45:247–271 10.1146/annurev-genet-110410-132435 [DOI] [PubMed] [Google Scholar]
- Szilard R.K., Jacques P.E., Laramée L., Cheng B., Galicia S., Bataille A.R., Yeung M., Mendez M., Bergeron M., Robert F., Durocher D. 2010. Systematic identification of fragile sites via genome-wide location analysis of gamma-H2AX. Nat. Struct. Mol. Biol. 17:299–305 10.1038/nsmb.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A., Van Houwe G., Nagai S., Erb I., van Nimwegen E.J., Gasser S.M. 2009. The functional importance of telomere clustering: global changes in gene expression result from SIR factor dispersion. Genome Res. 19:611–625 10.1101/gr.083881.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A., Schober H., Gasser S.M. 2010. The budding yeast nucleus. Cold Spring Harb. Perspect. Biol. 2:a000612 10.1101/cshperspect.a000612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburini B.A., Tyler J.K. 2005. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol. Cell. Biol. 25:4903–4913 10.1128/MCB.25.12.4903-4913.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira M.T., Dujon B., Fabre E. 2002. Genome-wide nuclear morphology screen identifies novel genes involved in nuclear architecture and gene-silencing in Saccharomyces cerevisiae. J. Mol. Biol. 321:551–561 10.1016/S0022-2836(02)00652-6 [DOI] [PubMed] [Google Scholar]
- Therizols P., Duong T., Dujon B., Zimmer C., Fabre E. 2010. Chromosome arm length and nuclear constraints determine the dynamic relationship of yeast subtelomeres. Proc. Natl. Acad. Sci. USA. 107:2025–2030 10.1073/pnas.0914187107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Rosell J., Sunjevaric I., De Piccoli G., Sacher M., Eckert-Boulet N., Reid R., Jentsch S., Rothstein R., Aragón L., Lisby M. 2007. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat. Cell Biol. 9:923–931 10.1038/ncb1619 [DOI] [PubMed] [Google Scholar]
- Uhlmann F., Nasmyth K. 1998. Cohesion between sister chromatids must be established during DNA replication. Curr. Biol. 8:1095–1101 10.1016/S0960-9822(98)70463-4 [DOI] [PubMed] [Google Scholar]
- Unal E., Arbel-Eden A., Sattler U., Shroff R., Lichten M., Haber J.E., Koshland D. 2004. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell. 16:991–1002 10.1016/j.molcel.2004.11.027 [DOI] [PubMed] [Google Scholar]
- Valenzuela L., Dhillon N., Kamakaka R.T. 2009. Transcription independent insulation at TFIIIC-dependent insulators. Genetics. 183:131–148 10.1534/genetics.109.106203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H., Gasser S.M. 2009. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 19:207–217 10.1016/j.tcb.2009.03.001 [DOI] [PubMed] [Google Scholar]
- Vandre C.L., Kamakaka R.T., Rivier D.H. 2008. The DNA end-binding protein Ku regulates silencing at the internal HML and HMR loci in Saccharomyces cerevisiae. Genetics. 180:1407–1418 10.1534/genetics.108.094490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Ira G., Tercero J.A., Holmes A.M., Diffley J.F., Haber J.E. 2004. Role of DNA replication proteins in double-strand break-induced recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 24:6891–6899 10.1128/MCB.24.16.6891-6899.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Vujcic M., Kowalski D. 2001. DNA replication forks pause at silent origins near the HML locus in budding yeast. Mol. Cell. Biol. 21:4938–4948 10.1128/MCB.21.15.4938-4948.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A.J., Severson A.F., Meyer B.J. 2010. Condensin and cohesin complexity: the expanding repertoire of functions. Nat. Rev. Genet. 11:391–404 10.1038/nrg2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. 2008. Wrestling off RAD51: a novel role for RecQ helicases. Bioessays. 30:291–295 10.1002/bies.20735 [DOI] [PubMed] [Google Scholar]
- Zegerman P., Diffley J.F. 2009. DNA replication as a target of the DNA damage checkpoint. DNA Repair (Amst.). 8:1077–1088 10.1016/j.dnarep.2009.04.023 [DOI] [PubMed] [Google Scholar]
- Zimmer C., Fabre E. 2011. Principles of chromosomal organization: lessons from yeast. J. Cell Biol. 192:723–733 10.1083/jcb.201010058 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.