Abstract
Multiple sequence changes that are simultaneously introduced in a single DNA transaction have a higher probability of altering gene function than do single base substitutions. DNA polymerase zeta (Pol ζ) has been shown to introduce such clustered mutations under specific selective and/or DNA damage-producing conditions. In this study, a forward mutation assay was used to determine the specificity of spontaneous mutations generated in Saccharomyces cerevisiae when either wild-type Pol ζ or a mutator Pol ζ variant (rev3-L979F) bypasses endogenous lesions. Mutagenesis in strains proficient for nucleotide excision repair (NER) was compared to mutagenesis in NER-deficient strains that retain unrepaired endogenous DNA lesions in the genome. Compared to NER-proficient strains, NER-deficient rad14Δ strains have elevated mutation rates that depend on Pol ζ. Rates are most strongly elevated for tandem base pair substitutions and clusters of multiple, closely spaced mutations. Both types of mutations depend on Pol ζ, but not on Pol η. Rates of each are further elevated in yeast strains bearing the rev3-979F allele. The results indicate that when Pol ζ performs mutagenic bypass of endogenous, helix-distorting lesions, it catalyzes a short track of processive, error-prone synthesis. We discuss the implications of this unique catalytic property of Pol ζ to its evolutionary conservation and possibly to multistage carcinogenesis.
Keywords: spontaneous mutagenesis, nucleotide excision repair, error-prone DNA synthesis translesion synthesis, multiple mutations
INTRODUCTION
Genome stability is threatened not only by exposure to environmental hazards, but also by endogenous sources of DNA damage created during cellular metabolism. Types of endogenous DNA damage include oxidized bases, alkylated bases, and abasic sites that are typically removed by repair systems such as nucleotide excision repair (NER) and base excision repair (BER). If not removed prior to replication, lesions that distort the DNA helix can stall replication fork progression, triggering cell cycle check-points and apoptosis. Replication fork stalling can be alleviated by two lesion-tolerance mechanisms: template switching and translesion DNA synthesis (TLS). Template switching makes use of an undamaged template, generally the complementary strand of the sister chromatid, and thus has a low mutagenic potential. During TLS, specialized DNA polymerases are recruited to directly bypass lesions. Depending on the lesion, the sequence context, and the polymerase(s) employed for bypass, TLS can be relatively accurate or mutagenic.
There are three TLS polymerases in Saccharomyces cerevisiae, polymerase zeta (Pol ζ, a heterodimer of Rev3 and Rev7 subunits), Rev1, and Pol η. Deleting REV3, REV7, or REV1 reduces the rate of spontaneous mutagenesis in yeast by 50–80% [Lawrence et al., 2000; Lawrence, 2004], suggesting that Pol ζ and Rev1 are responsible for TLS past endogenous DNA lesions. While Pol ζ can bypass some lesions without the need for a second polymerase [Haracska et al., 2003; Johnson et al., 2003; Garg et al., 2005; Stone et al., 2011], its ability to extend aberrant primer termini has lead to the suggestion that Pol ζ often extends aberrant termini resulting from nucleotide insertions opposite lesions by other TLS polymerases [reviewed by Lawrence et al., 2000; Prakash and Prakash, 2002; Lawrence, 2004; Gan et al., 2008; Waters et al., 2009]. Rev1 has a limited polymerase activity, inserting deoxycytidine triphosphate (dCTP) opposite damaged and undamaged bases [Nelson et al., 1996; Lawrence, 2002; Kim et al., 2011], and it is also thought to play a structural role in promoting TLS by Pol ζ [Haracska et al., 2001; Waters and Walker, 2006]. Pol η is responsible for efficient bypass of cis-syn cyclobutane pyrimidine dimers (CPDs) induced by ultraviolet light [Johnson et al., 1999; Yuan et al., 2000; McCulloch et al., 2004, 2007] and also it bypasses 8-oxoguanine, a common product of oxidation damage [Haracska et al., 2000; McCulloch et al., 2009]. While Pol η is relatively accurate during CPD bypass, it is less accurate than Pol ζ when copying undamaged templates in vitro [Matsuda et al., 2001; McCulloch et al., 2004; Zhong et al., 2006].
When copying undamaged DNA templates in vitro, Pol ζ and Pol η can both generate sequence changes involving multiple bases. For example, when copying undamaged DNA templates in vitro, Pol η creates tandem base pair substitutions (TBPSs) at a high rate [Matsuda et al., 2000, 2001], and yeast Pol ζ generates multiple, closely spaced sequence changes [Zhong et al., 2006; Stone et al., 2009]. Pol ζ-dependent complex mutations have also been reported in vivo. For example, human Pol ζ was implicated in forming complex mutations and TBPS induced by exposure of human cell lines to UV light [Gueranger et al., 2008] and during somatic hypermutation [Daly et al., 2012; Saribasak et al., 2012]. A Pol ζ-dependent increase in complex mutations was also observed in S. cerevisiae when NER was inactivated or when cells were exposed to UV light [Harfe and Jinks-Robertson, 2000; Minesinger and Jinks-Robertson, 2005; Abdulovic and Jinks-Robertson, 2006; Minesinger et al., 2006]. These complex mutations were composed of a selected frameshift mutation in combination with one or more base substitutions. They were completely dependent on Pol ζ but not Pol η, indicating that complex mutations are created during Pol ζ during bypass of DNA lesions normally subject to NER.
Sequence changes that are simultaneously introduced in a single DNA transaction are of particular interest because they have a higher probability of altering gene function than do single base substitutions. For this reason, we decided to obtain a comprehensive view of complex mutagenesis resulting from Pol ζ-dependent bypass of endogenous lesions, by determining the rate and specificity of mutagenesis using a forward mutation assay that does not require a selected frameshift mutation to score mutations resulting from TLS. The present study compares three yeast strains differing in REV3 status, wild-type REV3, a rev3Δ knockout incapable of Pol ζ-dependent TLS and having an anti-mutator phenotype [Lawrence, 2004], and a rev3-L979F allele that has the opposite effect of increasing the mutation rate [Sakamoto et al., 2007]. The rev3-L979F allele contains phenylalanine substituted for a conserved leucine near the polymerase active site of the catalytic Rev3 subunit [Sakamoto et al., 2007]. Purified L979F Pol ζ has lower fidelity than wild-type Pol ζ when copying undamaged DNA in vitro, and creates complex mutations at a rate that is 100–200 times higher than wild-type Pol ζ [Stone et al., 2009]. Also, L979F Pol ζ has increased capacity to bypass a variety of lesions, including abasic sites and UV-induced lesions [Stone et al., 2011]. These properties correlate with the observation that a rev3-L979F yeast strain has an elevated frequency of UV-induced mutagenesis, particularly with regard to complex mutations, a property that is further exacerbated by deleting RAD30, which encodes Pol η [Sakamoto et al., 2007].
In this study, the URA3 forward mutation reporter was used to examine the roles of Pol ζ and Pol η in mutagenic bypass of endogenous lesions in yeast. To obtain a comprehensive view of all possible mutations created as Pol ζ bypasses unrepaired endogenous lesions, we created NER-deficient strains by deleting RAD14, which encodes the protein that binds to sites of DNA damage during NER [Prakash and Prakash, 2000]. Spontaneous mutagenesis in NER-proficient strains was compared to mutagenesis in NER-deficient rad14Δ strains that retain unrepaired endogenous DNA lesions. We observe increased rates of TBPS and clustered mutations in the REV3 rad14Δ and rev3-L979F rad14Δ strains as compared to their NER-proficient parent strains. Both TBPS and clustered mutations were independent of Pol η status, but increased in rev3-L979F strains and decreased in rev3Δ strains. This suggests that the mutations result from bypass of endogenous DNA lesions that is dependent on Pol ζ, but not Pol η.
MATERIALS AND METHODS
Yeast Strains
All yeast strains used are listed in Supporting Information Table I and are derivatives of strain Δl(−2)l-7B-YUNI300 (MATa CAN1 his7-2 leu2-Δ::kanMX ura3-Δ trp1-289 ade2-1 lys2-ΔGG2899-2900) [Pavlov et al., 2002]. rad14::NAT strains were created by one-step transplacement with a PCR product generated by amplifying the natMX4 cassette from pAG25 [Goldstein and McCusker, 1999] with primers rad14::kan-F (5′ACTAGAAAAAGAGTTTGGATCTTCGTAGTGAAGGTATCGAACGTAACGCTCGTACGCTGCAGGTCGAC) and rad14::kan-R (5′AACACCTTATTATGACTTTCTTGTTATATTCTTATATACATAACCAACATATCGATGAATTCGAGCTCG). rad30::HYG strains were created by one-step transplacement with a PCR product generated by amplifying the hphMX4 cassette from pAG32 [Goldstein and McCusker, 1999] with primers rad30::kan-F (5′TAGCGCAGGCCTGCTCATTTTTGAACGGCTTTGATAAAACAAGACAAAGCCGTACGCTGCAGGTCGACGG) and rad30::kan-R (5′ATCAGGACGTTTTAGTTGCTGAAGCCATATAATTGTCTATTTGGAATAGGGAGCTCGTTTTCGACACTGG). rev1::loxP strains were created by Cre-lox disruption by first transforming strains with a PCR product generated by amplifying the loxP-flanked K. lactis URA3 cassette from pUG72 [Guldener et al., 1996] with primers rev1::KI-URA3-LOX-KO-F2 (5′ACAGATTTTCTCAAAATAAATCGATACTGCATTTCTAGGCATATCCAGCGCAGCTGAAGCTTCGTACGC) and rev1::KI-URA3-LOX-KO-R (5′GCAAACTGCGTGTTTACTGTATGCTGAAATGTTTTTTTTTTTTTAATTGCATAGGCCACTAGTGGATCTG). LoxP-mediated recombination was then used to remove URA3 from rev1::loxP-URA3 strains by expressing the Cre-recombinase from pSH63 [Guldener et al., 1996]. rev1::loxP strains that had lost pSH63 were then isolated. rev3::LEU2 strains were created by a one-step transplacement with a PCR product generated by amplifying LEU2 from genomic S. cerevisiae DNA with primers rev3::LEU2-F (5′ATGTCGAGGGAGTCGAACGACACAATACAGAGCGATACGGTTAGATCATCGCGCTATCGCACAGAATCAA) and rev3::LEU2-R (5′TTACCAATCATTTAGAGATATTAATGCTTCTTCCCTTTGAACAGATTGATTAGGAATCATAGTTTCATGAT). Strains carrying rev3-L979F allele were created via two-step transplacement as previously described [Sakamoto et al., 2007]. The URA3 reporter was inserted at the AGP1 locus in either Orientation 1 or Orientation 2, as previously described [Pavlov et al., 2002], to create agp1::URA3-OR1 and agp1::URA3-OR2 strains. All yeast strains were verified by PCR and/or sequencing.
Mutation Rates and Spectra
Fluctuation rate analyses were used to determine the rates of mutation of the CAN1, agp1::URA3-OR1, and agp1::URA3-OR2 forward reporters. For each strain, at least two experiments were performed, with each experiment consisting of at least 14 independent cultures split between two independent isogenic isolates. Liquid YPDA (1% yeast extract, 2% peptone, 2% dextrose,150 mg/L adenine sulfate) cultures were inoculated in parallel with single colonies and grown at 30°C for 36 hr to saturation (~2 × 108 cells/mL). Cells were washed with distilled water, diluted appropriately and then plated on nonselective media to determine cell count and synthetic media containing 5-flouroorotic acid (5-FOA) or canavanine to select for ura3 or can1 mutants, respectively. Mutation rates were determined via the method of the median using the Drake equation [Drake, 1991] and 95% confidence intervals were determined as described [Dixon and Massey, 1969]. Final rates for each strain were determined using the combined data from two or more experiments. To obtain mutation spectra, genomic DNA was purified from independently selected 5-FOAR isolates, and the URA3 ORF was amplified and sequenced using primers 5′CGCATATGTGGTGTTGAAGAAACATGAA and 5′GAAGCTCTAATTTGTGAGTTTAGTATACATGCA. The mutation rate and 95% confidence limits for each specific type of mutation were determined by multiplying the total mutation rate for that strain times the observed number of mutations of a specific type divided by the number of 5-FOAR isolates that were sequenced.
Statistical Analyses
To test whether the proportion of any particular mutation type observed in one strain was higher compared to that observed for another strain, one-tailed Fisher Exact tests were used to compare the observed number ura3 mutations of a particular type to the number of ura3 mutations observed that were not of that type. For each mutation type tested, the sequential P-value procedure was used to limit the false discovery rate [Benjamini and Hochberg, 1995]. For each set of comparisons, uncorrected P-values were determined first and then the Benjamini–Hochberg procedure was used with an initial value of α = 0.05 to determine which P-values were statistically significant. To test whether there was an increase in the length of mutation clusters in rev3-L979F strains, one-tailed, paired t-tests were used. GraphPad Prism 5.0 software was used for all of the aforementioned statistical tests.
Hotspots were identified as previously described [Nick McElhinny et al., 2008], with two exceptions. First, sites that exceeded expectations due to[Daly et al., 2012; Saribasak et al., 2012] random chance (binomial distribution, P ≥ 0.01) were considered significant for purposes of further calculations, but only those sites identified as outliers by the Dixon Q test (P ≥ 0.01) [Dixon, 1950] were defined as hotspots. Second, the number of detectable sites used in these calculations was binned by mutation type. For all single-base mutations, these numbers were taken from a database of known mutations in URA3 that result in resistance to 5-FOA (database available upon request). For TBPSs, the number of possible observable events was conservatively estimated as 1,346, which is the sum of all possible TBPS events containing at least one substitution found individually in the aforementioned database, all possible TBPS events yielding nonsense mutations and all observed TBPS events not covered by the preceding categories. The hotspot analysis was completed using a Microsoft Excel spreadsheet that was created by S.A.L. during this study (available upon request).
RESULTS AND DISCUSSION
Mutation Rates
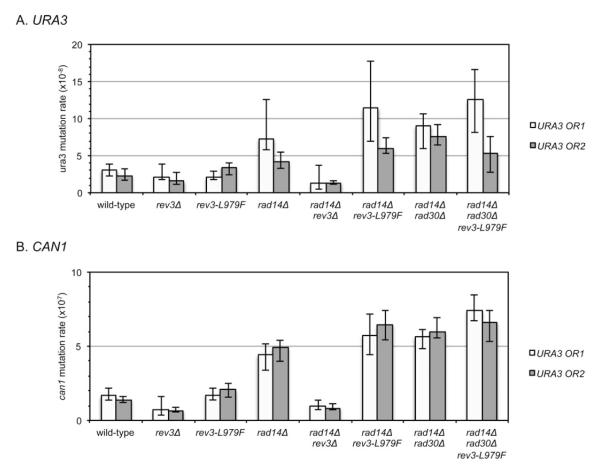
Mutation rates were determined for both the CAN1 and URA3 forward mutation reporters in isogenic strains of S. cerevisiae (Fig. 1, Supporting Information Tables II and III). The CAN1 and URA3 reporters detect any mutation that alters gene function by selecting for resistance to canavanine or 5-FOA, respectively. In order to determine whether there was a reported gene orientation bias, the URA3 target was inserted at the AGP1 locus on chromo-some III in each of two orientations relative to the ARS306 replication origin, as previously described [Pavlov et al., 2002]. There was no significant difference in mutation rates for the two orientations of URA3 for any genotype examined (Fig. 1, Supporting Information Tables II and III).
Fig. 1.
Overall spontaneous mutation rates. Mutation rates are shown for (A) ura3 and (B) can1 forward mutation reporters. Mutation rates from the URA3 Orientation 1 strain and the isogenic Orientation 2 strain are shown in white and gray, respectively. Error bars indicate 95% confidence intervals.
In the NER-deficient background, the spontaneous mutation rates for each of the reporters were about two-fold higher than those observed for the NER-proficient strains (Fig. 1, Supporting Information Tables II and III). The rev3Δ knockout was a strong anti-mutator in the NER-deficient background, but had only a weak effect in the NER-proficient background (Fig. 1, Supporting Information Tables II and III). In the NER-proficient background, the REV3 and rev3-L979F strains had similar mutation rates, whereas in the NER-deficient background, the rev3-L979F allele resulted in slightly increased mutation rates for each reporter (Fig. 1, Supporting Information Tables II and III). Deletion of RAD30 from REV3 or rev3-979F strains did not significantly affect the mutation rates in NER-proficient or NER-deficient backgrounds (Fig. 1, Supporting Information Tables II and III). In contrast, REV1 was required for Pol ζ-dependent mutagenesis both in NER-proficient and NER-deficient backgrounds (Supporting Information Tables II and III).
Mutation Spectra
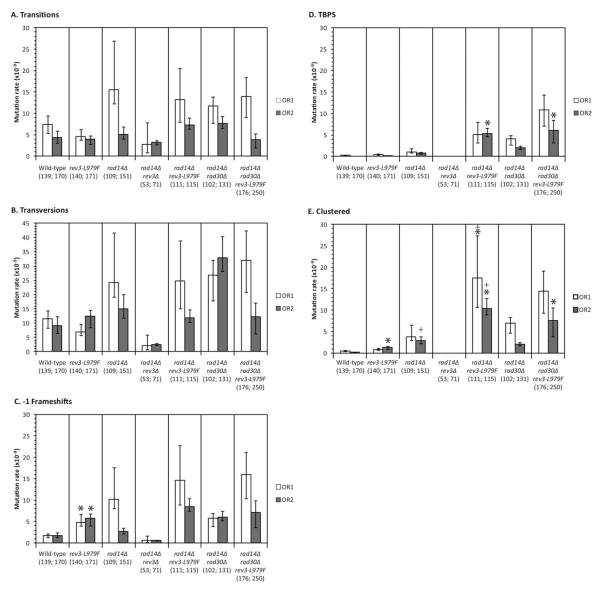
The URA3 ORF of independent 5-FOAR isolates from NER-proficient and NER-deficient strains was sequenced (Supporting Information Figs. 1–8, and Supporting Information Tables IV–VIII). For the NER-proficient wild-type strains [Nick McElhinny et al., 2010], the mutation spectra with URA3 in Orientations 1 and 2 are composed of 80–85% base substitutions, 8–13% single base frameshifts, and 7% multi-base mutations that are primarily large duplications and deletions. Compared to NER-proficient RAD14 strains, NER-deficient rad14Δ strains encoding wild-type or L979F Pol ζ (but not rev3Δ strains) had small increases in the rates of single base substitutions and frameshifts (Figs. 2A–2C). Greater increases were observed in rates for mutations involving multiple sequence changes (Figs. 2D and 2E). Multiple mutations from each isolate occurred within short patches, and were classified as either TBPSs or clustered mutations. A few 5-FOAR isolates contained two or three widely spaced mutations in the URA3 ORF (see Supporting Information Tables IV and V, “≥2 ura3 mutations”). The four main mutation types observed are considered individually below.
Fig. 2.
Rates for each mutation type. Mutation spectra were determined by sequencing independently isolated 5-FOAR derivatives from each indicated genotype. The numbers in parenthesis under the genotypes indicate the total number of ura3 mutations examined for URA3 Orientation 1 and Orientation 2, respectively. Mutation rates are shown for (A) transitions, (B) transversions, (C) −1 frameshifts, (D) TBPS, and (E) clustered mutations (multiple mutations with <10 base pairs between each adjacent mutation). Genotypes are indicated at the bottom. Mutation rates from the URA3 Orientation 1 strain and the isogenic Orientation 2 strain are shown in white and gray, respectively. Mutation rates and error bars for each mutation type were calculated as indicated in the Materials and Methods. One-tailed Fisher exact tests were used to compare the number of mutations of a particular type to all other mutations in a pair of strains. Asterisks (*) indicate that a statistically significant increase is observed in the rev3-L979F strain when compared to the isogenic REV3 strain using the same test (P value = 0.0027). Plus signs (+) indicate that a statistically significant increase is observed for the rad14Δ strain when compared to the isogenic RAD14 strain (P values ≤ 0.0006).
Single Base Mutations
In our previous study using the CAN1 reporter gene, we observed an elevated rate of C:G to G:C substitutions in a rev3-L979F strain compared to wild-type [Sakamoto et al., 2007]. This normally rare type of mutation has been suggested to result from bypass of modified guanines commonly induced by endogenous oxidative stress [Minnick et al., 1994; Minesinger et al., 2006; Neeley and Essigmann, 2006]. In the present study, the rate of C:G to G:C substitutions in the rev3-L979F strain is slightly decreased in Orientation 1 of the URA3 gene when compared to wild-type (Supporting Information Table IV). This may reflect a difference in the reporter gene used or an unknown difference in the strain backgrounds used in the two studies. Nonetheless, consistent with our original study, the rate of C:G to G:C substitutions in the rev3-L979F strain is slightly elevated in URA3 Orientation 2 (Supporting Information Table V). In the present study, we also observe a significantly elevated rate of C:G to G:C mutations in the rad14Δ strain compared to the wild-type strain (Supporting Information Table V), including one hotspot for C:G to G:C changes at URA3 base pair 356 (Supporting Information Fig. 4). This supports the notion that NER-deficiency elevates the level of unrepaired, modified guanines that are mutagenic when bypassed by Pol ζ. This idea is supported by decreases in the rate of C:G to G:C mutations in the rad14Δ rev3Δ strains (Supporting Information Tables IV and V). In fact, a more general 2- to 4-fold bias exists for transversions over transitions in the REV3 and rev3-L979F strains, regardless of NER status (Figs. 2A and 2B), whereas equal numbers of transitions and transversions are observed in the rad14Δ rev3Δ strains lacking Pol ζ activity. This suggests that Pol ζ bypass of unrepaired spontaneous lesions preferentially favors formation of mismatches that result in transversions.
There is a strong bias for −1 deletions over +1 insertions in all of the REV3 and rev3-L979F strains examined in this study (Supporting Information Tables IV and V), indicating that Pol ζ does generate single base deletion mismatches in vivo. Disruption of NER resulted in 2- to 6-fold increases in the rates of −1 deletions (Fig. 2C), and these increases were not apparent in the rad14Δ rev3Δ strain, indicating some single base deletions are generated during TLS by Pol ζ. The rate of −1 deletions in the NER-deficient background was slightly exacerbated by the rev3-L979F allele (Fig. 2C), but the increase was not statistically significant.
Tandem Base Pair Substitutions
TBPSs observed in this study were primarily tandem double substitutions (boxed in Figs. 3 and 4). There were also rare tandem triple base substitutions (Figs. 3B, 3D, and 4A). Compared to NER-proficient strains, the rates of TBPSs in NER-deficient strains were elevated from 3- to 42-fold in REV3 and rev3-L979F strains, respectively (Fig. 2D). In contrast, no such mutations were recovered from rad14Δ rev3Δ strains lacking Pol ζ (Supporting Information Tables IV and V). This indicates that the vast majority of TBPSs result from Pol ζ-dependent bypass of endogenous lesions that would normally be removed by NER. In the rad14Δ background, the rev3-L979F allele significantly elevated TBPS events when compared to the otherwise isogenic REV3 strain (Fig. 3D). Deleting RAD30 had no significant effect on the level of TBPSs in the rad14Δ or rad14Δ rev3-L979F backgrounds.
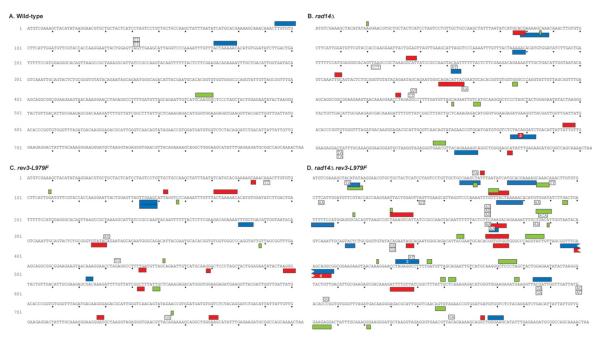
Fig. 3.
Spectra of clustered and TBPS mutations observed in wild-type, rev3-L979F, rad14Δ, and rad14Δ rev3-L979F strains. The sequence of the URA3 coding sequence is shown in black text with dots marking every 10 bases. Mutations from the URA3 Orientation 1 strain are shown above the URA3 reference sequence and mutations from the isogenic URA3 Orientation 2 strain are shown below. Boxed letters indicate TBPS with the mutant sequence indicated within the box. Filled boxes indicate clustered mutations with the length of the box extending from the first to the last mutated base in the mutation cluster. Blue boxes indicate cbps, clustered mutations composed of only base substitutions; green boxes indicate clustered mutations that include ≥1 insertions; and red boxes indicate clustered mutations that include ≥1 deletions. A white number within a box indicates the number of identical clustered mutations observed. Mutation spectra for the entire URA3 ORF and the sequences of clustered mutations are shown in the Supporting Information.
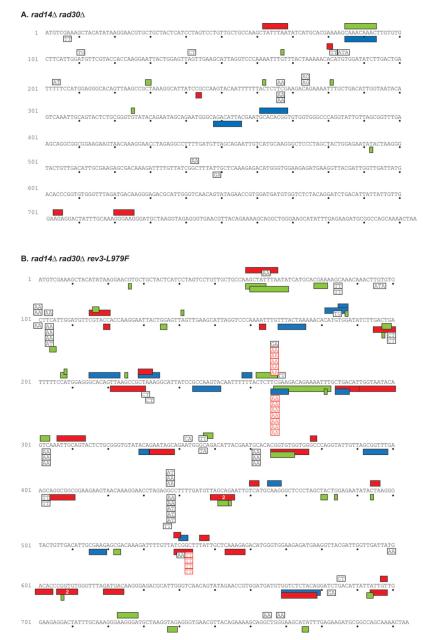
Fig. 4.
Spectra of clustered and TBPS mutations observed in rad14Δ rad30Δ and rad14Δ rad30Δ rev3-L979F strains. All text and symbol are as designated in Figure 3. Red boxes and text indicate TBPS mutations considered to be at hotspots. Mutation spectra for the entire URA3 ORF and the sequences of clustered mutations are shown in the Supporting Information.
Closer examination of TBPS events from all rad14Δ strains (Figs. 3 and 4, Supporting Information Table VII) reveals two interesting features. First, TBPS events commonly occur at specific locations. For example in the rad14Δ rad30Δ rev3-L979F strains (Fig. 4B), three or more TBPS events were observed at three TC:AG locations and at four GC:CG locations (Fig. 4). Two of these sites are statistically significant hotspots (see Methods) for TBPSs (indicated by red text in Fig. 4). Multiple TBPS events were also observed at these same locations in the rad14Δ rev3-L979F and rad14Δ rad30Δ strains (Figs. 3 and 4). This specificity indicates that formation of TBPSs is highly sequence context-dependent, which could be due to the probability of initial lesion formation, the probability of mutagenic bypass, or both.
The second feature of interest is that the vast majority of TBPSs represent but a small subset of the 64 possible types of TBPSs. For example, among 61 TBPSs observed in the rad14Δ rad30Δ rev3-L979F strains (Fig. 4B), 23 occurred at TC:AG dinucleotides and 25 occurred at GC:CG dinucleotides, despite the fact that these two dinucleotides are not over-represented in the URA3 ORF (Table I). Moreover, most of these TBPS events involved insertion of A and/or T bases (Table I). While Table I illustrates this finding using the TBPS events from the rad14Δ rad30Δ rev3-L979F strains, it is also the case for TBPS from the other rad14Δ strains, regardless of whether they are REV3, rev3-L979F, or rad30Δ (Figs. 3 and 4, Supporting Information Table VII). The data therefore indicate that spontaneous TBPSs are due to endogenous TLS that requires Pol ζ but is independent of Pol η, and that TBPSs are due to bypass of a lesion (or a variety of lesions) that is usually removed by NER. While the lesions responsible for the mutagenesis are unknown, the TBPS hotspots suggest that some locations in the genome may be more prone to lesion formation than others. In addition, the high rates of TBPS events seen here suggest that L979F Pol ζ, and to a lesser extent wild-type Pol ζ, can create, and then extend, primers containing two, and sometimes even three, consecutive terminal mismatches.
TABLE I.
Matrix of TBPS Mutations from rad14Δ rad30Δ rev3-L979F Strains
| ura3 Mutant Sequence | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | Total | % | ||
| ura3 Reference Sequence |
AA (74) | - | 1 | 1 | 2 | ||||||||||||||
| AC (46) | - | 0 | 0 | ||||||||||||||||
| AG (66) | - | 0 | 0 | ||||||||||||||||
| AT (60) | - | 1 | 1 | 2 | |||||||||||||||
| CA (55) | . | 0 | 0 | ||||||||||||||||
| CC (21) | - | 0 | 0 | ||||||||||||||||
| CG (21) | 1 | 1 | 2 | ||||||||||||||||
| CT (37) | - | 0 | 0 | ||||||||||||||||
| GA (63) | 3 | - | 1 | 4 | 7 | ||||||||||||||
| GC (43) | 10 | 5 | - | 1 | 9 | 25 | 41 | ||||||||||||
| GG (59) | 1 | - | 1 | 2 | 3 | ||||||||||||||
| GT (47) | - | 1 | 1 | 2 | |||||||||||||||
| TA (54) | 1 | - | 1 | 2 | |||||||||||||||
| TC (24) | 19 | 1 | 3 | - | 23 | 38 | |||||||||||||
| TG (66) | 1 | - | 1 | 2 | |||||||||||||||
| TT (67) | 1 | - | 1 | 2 | |||||||||||||||
| Total | 33 | 0 | 0 | 6 | 1 | 0 | 0 | 5 | 3 | 0 | 0 | 0 | 1 | 0 | 1 | 11 | 61 | ||
| % | 54 | 0 | 0 | 10 | 2 | 0 | 0 | 8 | 5 | 0 | 0 | 0 | 2 | 0 | 2 | 18 | |||
Tandem base substitutions (TBPS) from the rad14Δ rad30Δ rev3-L979F URA3 orientation 1 and orientation 2 strains and were summed. The left column lists each type of TBPS, with the total number in of each dinucleotide present in URA3 coding strand in parenthesis. Because the strand that was erroneously copied is not known, all mutations could be complementary to what is shown here. For example, AA → CT changes could also have been notated as TT → GA.
Clustered Mutations
For this study, clustered mutations are defined as two or more mutations separated by less than 10 nucleotides. This class excludes TBPSs, as well as deletions, insertions, and duplications of two or more adjacent bases (Supporting Information Table VIII). The clustered mutation class is quite diverse (Supporting Information Table VI). Clustered mutations contain from 2 to 16 sequence changes, separated by distances ranging from 1 to 20 base pairs. All types of single base changes (substitutions, insertions, and deletions) were observed within the clusters. Clustered mutations were classified as “cbps” if composed of only base substitutions (blue boxes in Figs. 3 and 4), “cins” if composed of one or more substitution and insertions (green boxes), and “cdel” if composed of one or more substitutions and deletions (red boxes).
The rates of clustered mutations were elevated 3- to 14-fold in NER-deficient strains compared to NER-proficient strains (Fig. 2E). Deletion of RAD30 did not affect the rate of clustered mutations in any genetic background examined, suggesting that Pol η is not required for their creation. Clustered mutations were observed in every strain examined except the rad14Δ rev3Δ strain, indicating that they are Pol ζ-dependent. The rev3-L979F allele significantly elevated the rate of clustered mutations compared to the corresponding REV3 strain in RAD14, rad14Δ, and rad14Δ rad30Δ backgrounds for URA3 Orientation 2 and in the rad14Δ background for URA3 Orientation 1 (Fig. 2E). Additionally, mutation clusters (TBPS events plus clustered mutations) span significantly longer overall distances in each of the rev3-L979F strains compared to their isogenic REV3 counterparts (P values <0.05; Supporting Information Tables VI and VII). These data are in agreement with our previous observations that the rev3-L979F mutation exacerbates the tendency of Pol ζ to create mutation clusters [Sakamoto et al., 2007; Stone et al., 2009].
As with the TBPS events, the data suggest that spontaneous clustered mutations are generated during bypass of lesions that would normally be removed from DNA by the NER machinery and this bypass is Pol ζ-dependent, but Pol η-independent. This observation is consistent with previous studies from the Jinks-Roberston group, in which spontaneous complex mutations were detected in a lys2ΔA746 frameshift reporter assay that selects primarily for +1 and rarely −2 frameshifts [Harfe and Jinks-Robertson, 2000; Minesinger and Jinks-Robertson, 2005; Minesinger et al., 2006]. The clustered mutations observed in lys2ΔA746 studies almost always contained a +1 frameshift and tended to occur at one of three hotspots. While we did observe some TBPS hotspots in this study, we did not find any hotspots for clustered mutations. This could be due to the two different sequence contexts; the lys2ΔA746 reporter has a reversion window of ~150 bases, while the URA3 reporter used in this study is 804 bases.
Because our study used a forward mutation reporter, we were able to observe clustered mutations composed of only base substitutions, in addition to clustered mutations that involve a frameshift. Of the 210 clustered mutations observed in all strains used in this study, 36% include at least one deletion, 30% include at least one insertion, and 34% contain only base substitutions (Figs. 3 and 4, Supporting Information Table VI). There is a significant overrepresentation of insertions observed within clustered mutations compared to +1 insertions observed as single-base mutations. For example, 22 of the 48 complex mutations from the rad14Δ rad30Δ rev3-L979F URA3 Orientation 2 strain contained at least one insertion, compared to 4 insertions among 157 single-base mutations (P < 0.0001; Supporting Information Table V). The overrepresentation of insertions within clustered mutations is independent of the NER status of the strain or whether the strain is REV3 or rev3-L979F. There were no overrepresentations for deletions or for any type of base substitution observed as part of clustered mutations or TBPS events, compared to single mutations for any strain studied (Supporting Information Tables IV–VI).
Implications
This study shows that Pol ζ-dependent, Pol η-independent bypass of unrepaired endogenous lesions could be a significant source of multiple, clustered, simultaneously introduced sequence changes. TBPSs and clustered mutations like those observed in this study have the potential to change multiple amino acids in one mutagenic DNA synthesis reaction. Compared to single base changes that may alter only a single amino acid, multiple amino acid changes resulting from multiple closely-spaced base substitutions provide a more diverse pool of mutant alleles upon which selection could act to more rapidly jump fitness barriers [Smith, 1970] and to increase survival in changing environments, in a manner similar to that discussed for multiple mutations spread over hundreds to thousands of base pairs [i.e., so-called “mutation showers” reviewed in Drake, 2007].
Among all DNA polymerases studied to date, the unique ability of Pol ζ to generate multiple closely spaced point mutations may be relevant to the evolutionary conservation of Pol ζ. As recently pointed out by Roberts et al. [2012], there are likely to be positive selective advantages to mutagenesis that can produce locally high mutation densities without overloading the rest of the genome, thereby reducing the chance of deleterious mutations that could obscure an advantageous change. For example, in pathogenic unicellular eukaryotes, multiple mutations introduced by Pol ζ may provide the mutations upon which selection can act to allow survival in the face of host defense mechanisms and/or drug treatments. In mammals, two recent studies demonstrate that DNA synthesis errors by mouse Pol ζ strongly contribute to somatic hypermutation of immunoglobulin genes [Daly et al., 2012; Saribasak et al., 2012]. Both studies report a high frequency of TBPSs in the hyper-variable region of immunoglobulin genes generated during affinity maturation. Conversely, mutagenesis can have adverse consequences, for example, in initiating and promoting multistage carcinogenesis [Loeb, 2011]. Recent studies demonstrate that mutagenic TLS by Pol ζ during replication of damaged, single-stranded DNA results in clustered, strand-specific hypermutation [Roberts et al., 2012]. During such a transaction, TBPS and clustered mutations like those observed herein have the potential to contribute more rapidly than single base substitutions to multistage carcinogenesis.
Supplementary Material
ACKNOWLEDGMENTS
Authors thank Amy Abdulovic-Cui, Mercedes Arana, and Alan B. Clark for helpful advice and reviewing the manuscript prior to submission. They also acknowledge the NIEHS Molecular Genetics Core Facility for performing all of the DNA sequencing.
Grant sponsor: Division of Intramural Research of the National Institute of Environmental Health Sciences, National Institutes of Health; Grant number: Project Z01 ES065070.
Footnotes
AUTHOR CONTRIBUTIONS JES and TAK designed the study; JES performed experiments and collected data; JES, SAL, and TAK analyzed results; JES prepared figures and tables; JES and TAK wrote the manuscript; all authors approved the final manuscript.
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- Abdulovic AL, Jinks-Robertson S. The in vivo characterization of translesion synthesis across UV-induced lesions in Saccharomyces cerevisiae: Insights into Pol zeta- and Pol eta-dependent frameshift mutagenesis. Genetics. 2006;172:1487–1498. doi: 10.1534/genetics.105.052480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300. [Google Scholar]
- Daly J, Bebenek K, Watt DL, Richter K, Jiang C, Zhao ML, Ray M, McGregor WG, Kunkel TA, Diaz M. Altered Ig hypermutation pattern and frequency in complementary mouse models of DNA polymerase zeta activity. J Immunol. 2012;188:5528–5537. doi: 10.4049/jimmunol.1102629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J. Analysis of extreme values. Annual Math Stat. 1950;21:488–506. [Google Scholar]
- Dixon WJ, Massey FJ. Introduction to Statistical Analysis. McGraw-Hill; New York: 1969. [Google Scholar]
- Drake JW. A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci USA. 1991;88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JW. Mutations in clusters and showers. Proc Natl Acad Sci USA. 2007;104:8203–8204. doi: 10.1073/pnas.0703089104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan GN, Wittschieben JP, Wittschieben BO, Wood RD. DNA polymerase zeta (pol zeta) in higher eukaryotes. Cell Res. 2008;18:174–183. doi: 10.1038/cr.2007.117. [DOI] [PubMed] [Google Scholar]
- Garg P, Stith CM, Majka J, Burgers PM. Proliferating cell nuclear antigen promotes translesion synthesis by DNA polymerase zeta. J Biol Chem. 2005;280:23446–23450. doi: 10.1074/jbc.C500173200. [DOI] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Gueranger Q, Stary A, Aoufouchi S, Faili A, Sarasin A, Reynaud CA, Weill JC. Role of DNA polymerases eta, iota and zeta in UV resistance and UV-induced mutagenesis in a human cell line. DNA Repair (Amst) 2008;7:1551–1562. doi: 10.1016/j.dnarep.2008.05.012. [DOI] [PubMed] [Google Scholar]
- Guldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase eta. Nat Genet. 2000;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- Haracska L, Unk I, Johnson RE, Johansson E, Burgers PM, Prakash S, Prakash L. Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites. Genes Dev. 2001;15:945–954. doi: 10.1101/gad.882301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Prakash S, Prakash L. Yeast DNA polymerase zeta is an efficient extender of primer ends opposite from 7,8-dihydro-8-Oxoguanine and O6-methylguanine. Mol Cell Biol. 2003;23:1453–1459. doi: 10.1128/MCB.23.4.1453-1459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, Jinks-Robertson S. DNA polymerase zeta introduces multiple mutations when bypassing spontaneous DNA damage in Saccharomyces cerevisiae. Mol Cell. 2000;6:1491–1499. doi: 10.1016/s1097-2765(00)00145-3. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Pol eta. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Yu SL, Prakash S, Prakash L. Yeast DNA polymerase zeta (zeta) is essential for error-free replication past thymine glycol. Genes Dev. 2003;17:77–87. doi: 10.1101/gad.1048303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Mudrak SV, Jinks-Robertson S. The dCMP transferase activity of yeast Rev1 is biologically relevant during the bypass of endogenously generated AP sites. DNA Repair (Amst) 2011;10:1262–1271. doi: 10.1016/j.dnarep.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CW. Cellular roles of DNA polymerase zeta and Rev1 protein. DNA Repair (Amst) 2002;1:425–435. doi: 10.1016/s1568-7864(02)00038-1. [DOI] [PubMed] [Google Scholar]
- Lawrence CW. Cellular functions of DNA polymerase zeta and Rev1 protein. Adv Protein Chem. 2004;69:167–203. doi: 10.1016/S0065-3233(04)69006-1. [DOI] [PubMed] [Google Scholar]
- Lawrence CW, Gibbs PE, Murante RS, Wang XD, Li Z, McManus TP, McGregor WG, Nelson JR, Hinkle DC, Maher VM. Roles of DNA polymerase zeta and Rev1 protein in eukaryotic mutagenesis and translesion replication. Cold Spring Harb Symp Quant Biol. 2000;65:61–69. doi: 10.1101/sqb.2000.65.61. [DOI] [PubMed] [Google Scholar]
- Loeb LA. Human cancers express mutator phenotypes: Origin, consequences and targeting. Nat Rev Cancer. 2011;11:450–457. doi: 10.1038/nrc3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Bebenek K, Masutani C, Hanaoka F, Kunkel TA. Low fidelity DNA synthesis by human DNA polymerase-eta. Nature. 2000;404:1011–1013. doi: 10.1038/35010014. [DOI] [PubMed] [Google Scholar]
- Matsuda T, Bebenek K, Masutani C, Rogozin IB, Hanaoka F, Kunkel TA. Error rate and specificity of human and murine DNA polymerase eta. J Mol Biol. 2001;312:335–346. doi: 10.1006/jmbi.2001.4937. [DOI] [PubMed] [Google Scholar]
- McCulloch SD, Kokoska RJ, Masutani C, Iwai S, Hanaoka F, Kunkel TA. Preferential cis-syn thymine dimer bypass by DNA polymerase eta occurs with biased fidelity. Nature. 2004;428:97–100. doi: 10.1038/nature02352. [DOI] [PubMed] [Google Scholar]
- McCulloch SD, Wood A, Garg P, Burgers PM, Kunkel TA. Effects of accessory proteins on the bypass of a cis-syn thymine-thymine dimer by Saccharomyces cerevisiae DNA polymerase eta. Biochemistry. 2007;46:8888–8896. doi: 10.1021/bi700234t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch SD, Kokoska RJ, Garg P, Burgers PM, Kunkel TA. The efficiency and fidelity of 8-oxo-guanine bypass by DNA polymerases delta and eta. Nucleic Acids Res. 2009;37:2830–2840. doi: 10.1093/nar/gkp103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minesinger BK, Jinks-Robertson S. Roles of RAD6 epistasis group members in spontaneous pol zeta-dependent translesion synthesis in Saccharomyces cerevisiae. Genetics. 2005;169:1939–1955. doi: 10.1534/genetics.104.033894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minesinger BK, Abdulovic AL, Ou TM, Jinks-Robertson S. The effect of oxidative metabolism on spontaneous Pol zeta-dependent translesion synthesis in Saccharomyces cerevisiae. DNA Repair (Amst) 2006;5:226–234. doi: 10.1016/j.dnarep.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Minnick DT, Pavlov YI, Kunkel TA. The fidelity of the human leading and lagging strand DNA replication apparatus with 8-oxodeoxyguanosine triphosphate. Nucleic Acids Res. 1994;22:5658–5664. doi: 10.1093/nar/22.25.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neeley WL, Essigmann JM. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem Res Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC. Thymine-thymine dimer bypass by yeast DNA polymerase zeta. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Kissling GE, Kunkel TA. Differential correction of lagging-strand replication errors made by DNA polymerases {alpha} and {delta} Proc Natl Acad Sci USA. 2010;107:21070–21075. doi: 10.1073/pnas.1013048107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov YI, Newlon CS, Kunkel TA. Yeast origins establish a strand bias for replicational mutagenesis. Mol Cell. 2002;10:207–213. doi: 10.1016/s1097-2765(02)00567-1. [DOI] [PubMed] [Google Scholar]
- Prakash S, Prakash L. Nucleotide excision repair in yeast. Mutat Res. 2000;451:13–24. doi: 10.1016/s0027-5107(00)00037-3. [DOI] [PubMed] [Google Scholar]
- Prakash S, Prakash L. Translesion DNA synthesis in eukaryotes: A one- or two-polymerase affair. Genes Dev. 2002;16:1872–1883. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- Roberts SA, Sterling J, Thompson C, Harris S, Mav D, Shah R, Klimczak LJ, Kryukov GV, Malc E, Mieczkowski PA, Resnick MA, Gordenin DA. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell. 2012;46:424–435. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto AN, Stone JE, Kissling GE, McCulloch SD, Pavlov YI, Kunkel TA. Mutator alleles of yeast DNA polymerase zeta. DNA Repair (Amst) 2007;6:1829–1838. doi: 10.1016/j.dnarep.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribasak H, Maul RW, Cao Z, Yang WW, Schenten D, Kracker S, Gearhart PJ. DNA polymerase zeta generates tandem mutations in immunoglobulin variable regions. J Exp Med. 2012;209:1075–1081. doi: 10.1084/jem.20112234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM. Natural selection and the concept of a protein space. Nature. 1970;225:563–564. doi: 10.1038/225563a0. [DOI] [PubMed] [Google Scholar]
- Stone JE, Kissling GE, Lujan SA, Rogozin IB, Stith CM, Burgers PM, Kunkel TA. Low-fidelity DNA synthesis by the L979F mutator derivative of Saccharomyces cerevisiae DNA polymerase zeta. Nucleic Acids Res. 2009;37:3774–3787. doi: 10.1093/nar/gkp238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JE, Kumar D, Binz SK, Inase A, Iwai S, Chabes A, Burgers PM, Kunkel TA. Lesion bypass by S. cerevisiae Pol zeta alone. DNA Repair. 2011;10:826–834. doi: 10.1016/j.dnarep.2011.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Walker GC. The critical mutagenic translesion DNA polymerase Rev1 is highly expressed during G(2)/M phase rather than S phase. Proc Natl Acad Sci USA. 2006;103:8971–8976. doi: 10.1073/pnas.0510167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Minesinger BK, Wiltrout ME, D’Souza S, Woodruff RV, Walker GC. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev. 2009;73:134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan F, Zhang Y, Rajpal DK, Wu X, Guo D, Wang M, Taylor JS, Wang Z. Specificity of DNA lesion bypass by the yeast DNA polymerase eta. J Biol Chem. 2000;275:8233–8239. doi: 10.1074/jbc.275.11.8233. [DOI] [PubMed] [Google Scholar]
- Zhong X, Garg P, Stith CM, Nick McElhinny SA, Kissling GE, Burgers PM, Kunkel TA. The fidelity of DNA synthesis by yeast DNA polymerase zeta alone and with accessory proteins. Nucleic Acids Res. 2006;34:4731–4742. doi: 10.1093/nar/gkl465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.