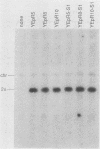
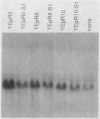
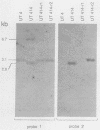
Abstract
The coding sequences of genes in the yeast Saccharomyces cerevisiae show a preference for 25 of the 61 possible coding triplets. The degree of this biased codon usage in each gene is positively correlated to its expression level. Highly expressed genes use these 25 major codons almost exclusively. As an experimental approach to studying biased codon usage and its possible role in modulating gene expression, systematic codon replacements were carried out in the highly expressed PGK1 gene. The expression of phosphoglycerate kinase (PGK) was studied both on a high-copy-number plasmid and as a single copy gene integrated into the chromosome. Replacing an increasing number (up to 39% of all codons) of major codons with synonymous minor ones at the 5' end of the coding sequence caused a dramatic decline of the expression level. The PGK protein levels dropped 10-fold. The steady-state mRNA levels also declined, but to a lesser extent (threefold). Our data indicate that this reduction in mRNA levels was due to destabilization caused by impaired translation elongation at the minor codons. By preventing translation of the PGK mRNAs by the introduction of a stop codon 3' and adjacent to the start codon, the steady-state mRNA levels decreased dramatically. We conclude that efficient mRNA translation is required for maintaining mRNA stability in S. cerevisiae. These findings have important implications for the study of the expression of heterologous genes in yeast cells.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Backman K., Ptashne M., Gilbert W. Construction of plasmids carrying the cI gene of bacteriophage lambda. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4174–4178. doi: 10.1073/pnas.73.11.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Chen C. Y., Oppermann H., Hitzeman R. A. Homologous versus heterologous gene expression in the yeast, Saccharomyces cerevisiae. Nucleic Acids Res. 1984 Dec 11;12(23):8951–8970. doi: 10.1093/nar/12.23.8951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeddel D. V., Shepard H. M., Yelverton E., Leung D., Crea R., Sloma A., Pestka S. Synthesis of human fibroblast interferon by E. coli. Nucleic Acids Res. 1980 Sep 25;8(18):4057–4074. doi: 10.1093/nar/8.18.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M., Gautier C. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 1982 Nov 25;10(22):7055–7074. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M., Grantham R. Polypeptide elongation and tRNA cycling in Escherichia coli: a dynamic approach. FEBS Lett. 1980 Jun 30;115(2):151–155. doi: 10.1016/0014-5793(80)81155-0. [DOI] [PubMed] [Google Scholar]
- Grantham R., Gautier C., Gouy M. Codon frequencies in 119 individual genes confirm consistent choices of degenerate bases according to genome type. Nucleic Acids Res. 1980 May 10;8(9):1893–1912. doi: 10.1093/nar/8.9.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitzeman R. A., Chen C. Y., Hagie F. E., Patzer E. J., Liu C. C., Estell D. A., Miller J. V., Yaffe A., Kleid D. G., Levinson A. D. Expression of hepatitis B virus surface antigen in yeast. Nucleic Acids Res. 1983 May 11;11(9):2745–2763. doi: 10.1093/nar/11.9.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitzeman R. A., Clarke L., Carbon J. Isolation and characterization of the yeast 3-phosphoglycerokinase gene (PGK) by an immunological screening technique. J Biol Chem. 1980 Dec 25;255(24):12073–12080. [PubMed] [Google Scholar]
- Hitzeman R. A., Hagie F. E., Hayflick J. S., Chen C. Y., Seeburg P. H., Derynck R. The primary structure of the Saccharomyces cerevisiae gene for 3-phosphoglycerate kinase. Nucleic Acids Res. 1982 Dec 11;10(23):7791–7808. doi: 10.1093/nar/10.23.7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol. 1981 Sep 25;151(3):389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J Mol Biol. 1982 Jul 15;158(4):573–597. doi: 10.1016/0022-2836(82)90250-9. [DOI] [PubMed] [Google Scholar]
- Iserentant D., Fiers W. Secondary structure of mRNA and efficiency of translation initiation. Gene. 1980 Apr;9(1-2):1–12. doi: 10.1016/0378-1119(80)90163-8. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E. W. Proteinase mutants of Saccharomyces cerevisiae. Genetics. 1977 Jan;85(1):23–33. doi: 10.1093/genetics/85.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lawn R. M., Adelman J., Bock S. C., Franke A. E., Houck C. M., Najarian R. C., Seeburg P. H., Wion K. L. The sequence of human serum albumin cDNA and its expression in E. coli. Nucleic Acids Res. 1981 Nov 25;9(22):6103–6114. doi: 10.1093/nar/9.22.6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losson R., Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas M. T., Chen C. Y., Hitzeman R. A., Riggs A. D. Active human-yeast chimeric phosphoglycerate kinases engineered by domain interchange. Science. 1986 Aug 15;233(4765):788–790. doi: 10.1126/science.3526552. [DOI] [PubMed] [Google Scholar]
- Mellor J., Dobson M. J., Roberts N. A., Kingsman A. J., Kingsman S. M. Factors affecting heterologous gene expression in Saccharomyces cerevisiae. Gene. 1985;33(2):215–226. doi: 10.1016/0378-1119(85)90096-4. [DOI] [PubMed] [Google Scholar]
- Mellor J., Dobson M. J., Roberts N. A., Tuite M. F., Emtage J. S., White S., Lowe P. A., Patel T., Kingsman A. J., Kingsman S. M. Efficient synthesis of enzymatically active calf chymosin in Saccharomyces cerevisiae. Gene. 1983 Sep;24(1):1–14. doi: 10.1016/0378-1119(83)90126-9. [DOI] [PubMed] [Google Scholar]
- Pedersen S. Escherichia coli ribosomes translate in vivo with variable rate. EMBO J. 1984 Dec 1;3(12):2895–2898. doi: 10.1002/j.1460-2075.1984.tb02227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M., Lilley R., Little S., Emtage J. S., Yarranton G., Stephens P., Millican A., Eaton M., Humphreys G. Codon usage can affect efficiency of translation of genes in Escherichia coli. Nucleic Acids Res. 1984 Sep 11;12(17):6663–6671. doi: 10.1093/nar/12.17.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S., Davis R. W. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schottel J. L., Sninsky J. J., Cohen S. N. Effects of alterations in the translation control region on bacterial gene expression: use of cat gene constructs transcribed from the lac promoter as a model system. Gene. 1984 May;28(2):177–193. doi: 10.1016/0378-1119(84)90255-5. [DOI] [PubMed] [Google Scholar]
- Scopes R. K. 3-phosphoglycerate kinase of baker's yeast. Methods Enzymol. 1975;42:134–138. doi: 10.1016/0076-6879(75)42106-1. [DOI] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuite M. F., Dobson M. J., Roberts N. A., King R. M., Burke D. C., Kingsman S. M., Kingsman A. J. Regulated high efficiency expression of human interferon-alpha in Saccharomyces cerevisiae. EMBO J. 1982;1(5):603–608. doi: 10.1002/j.1460-2075.1982.tb01215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varenne S., Lazdunski C. Effect of distribution of unfavourable codons on the maximum rate of gene expression by an heterologous organism. J Theor Biol. 1986 May 7;120(1):99–110. doi: 10.1016/s0022-5193(86)80020-0. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Yarus M., Folley L. S. Sense codons are found in specific contexts. J Mol Biol. 1985 Apr 20;182(4):529–540. doi: 10.1016/0022-2836(85)90239-6. [DOI] [PubMed] [Google Scholar]
- Zitomer R. S., Hall B. D. Yeast cytochrome c messenger RNA. In vitro translation and specific immunoprecipitation of the CYC1 gene product. J Biol Chem. 1976 Oct 25;251(20):6320–6326. [PubMed] [Google Scholar]
- Zitomer R. S., Montgomery D. L., Nichols D. L., Hall B. D. Transcriptional regulation of the yeast cytochrome c gene. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3627–3631. doi: 10.1073/pnas.76.8.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis using M13-derived vectors: an efficient and general procedure for the production of point mutations in any fragment of DNA. Nucleic Acids Res. 1982 Oct 25;10(20):6487–6500. doi: 10.1093/nar/10.20.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]