Abstract
Retinal endothelial cells line the arborizing microvasculature that supplies and drains the neural retina. The anatomical and physiological characteristics of these endothelial cells are consistent with nutritional requirements and protection of a tissue critical to vision. On the one hand, the endothelium must ensure the supply of oxygen and other nutrients to the metabolically active retina, and allow access to circulating cells that maintain the vasculature or survey the retina for the presence of potential pathogens. On the other hand, the endothelium contributes to the blood-retinal barrier that protects the retina by excluding circulating molecular toxins, microorganisms, and pro-inflammatory leukocytes. Features required to fulfill these functions may also predispose to disease processes, such as retinal vascular leakage and neovascularization, and trafficking of microbes and inflammatory cells. Thus, the retinal endothelial cell is a key participant in retinal ischemic vasculopathies that include diabetic retinopathy and retinopathy of prematurity, and retinal inflammation or infection, as occurs in posterior uveitis. Using gene expression and proteomic profiling, it has been possible to explore the molecular phenotype of the human retinal endothelial cell and contribute to understanding of the pathogenesis of these diseases. In addition to providing support for the involvement of well-characterized endothelial molecules, profiling has the power to identify new players in retinal pathologies. Findings may have implications for the design of new biological therapies. Additional progress in this field is anticipated as other technologies, including epigenetic profiling methods, whole transcriptome shotgun sequencing, and metabolomics, are used to study the human retinal endothelial cell.
Keywords: retina, endothelial cell, molecular profiling, posterior uveitis, autoimmune uveitis, infectious uveitis, diabetic retinopathy, retinopathy of prematurity
1. Introduction
Diseases involving the retinal vasculature, including 2 ischemic vasculopathies (i.e., diabetic retinopathy and retinopathy of prematurity) and various posterior forms of uveitis, are important causes of blindness in both industrialized countries and developing nations. Diabetic retinopathy affects approximately one-third of all persons who suffer from diabetes mellitus (Kempen et al., 2004b), a disease that is expected to affect 300 million people worldwide by 2025 (King et al., 1998). Retinopathy of prematurity accounts for up to one-third of childhood blindness, particularly in countries with intermediate infant mortality rates (Gilbert et al., 1997). Uveitis is a relatively uncommon disease, but due to an often substantial impact on vision at a relatively earlier age, its socioeconomic impact is roughly equivalent to that of diabetic retinopathy (Suttorp-Schulten and Rothova, 1996).
Retinal microvessels are complex structures, to which multiple cell populations contribute. Microvascular dysfunction is associated with retinal ischemia and neovascularization in diabetic retinopathy and retinopathy of prematurity, and leukocyte or microbial trafficking and potentiation of retinal inflammation in posterior uveitis. In this review, we focus on the critical participation of the retinal vascular endothelial cell in these pathological processes and highlight elements of the endothelial molecular phenotype that may predispose the retina to involvement in the stated diseases. We introduce our subject with a discussion of relevant anatomy and physiology, as well as descriptions of the model systems that are used to study the basic disease mechanisms.
2. Clinical significance of the retinal vascular diseases
2.1 Posterior uveitis
Inflammations that involve the intraocular tissues are termed uveitis. This large group of diseases is classified anatomically into anterior uveitis (primarily involving the anterior chamber), intermediate uveitis (primarily involving the vitreous), posterior uveitis (primarily involving the retina or choroid), and panuveitis (involving the anterior chamber, vitreous, and retina or choroid) (Bloch-Michel and Nussenblatt, 1987). Up to 10% of blindness in Western nations has been attributed to uveitis (Nussenblatt, 1990; Suttorp-Schulten and Rothova, 1996). In developing countries, the figure may be as high as 25% (London et al., 2010). Although 3 United States population-based studies report different patterns of age-stratified incidence, all show that more cases of uveitis begin during the working years than at any other period in life (Darrell et al., 1962; Gritz and Wong, 2004; Suhler et al., 2008). As a result, uveitis exacts an annual cost on society equivalent to that of diabetic retinopathy (Suttorp-Schulten and Rothova, 1996). In particular, approximately 50% of individuals with uveitis affecting the posterior segment of the eye suffer vision loss (Rothova et al., 1996).
Posterior uveitis is actually a diverse group of diseases with varied etiologies, including both autoimmune and infectious entities (Rodriguez et al., 1996; Suhler et al., 2008). Autoimmune uveitis may occur as part of a systemic inflammatory disease or be confined to the eye. Isolated autoimmune uveitis may take the form of a specific ocular syndrome or, if characteristic clinical features are not present, is often termed “idiopathic.” Large case series of posterior uveitis from the United States (Rodriguez et al., 1996) and Germany (Jakob et al., 2009) found that approximately 22% and 6% of cases were associated with systemic diseases and approximately 15% and 25% of cases were due to specific ocular syndromes, in the respective countries. In these same series, roughly 50% and 41% of cases were caused by infections. In developing nations, the percentage of cases caused by infections is even higher than in industrialized countries (London et al., 2010). Systemic inflammatory diseases identified in the 2 series included sarcoidosis, Behcet’s disease, and multiple sclerosis. Ocular syndromes identified in the 2 series included birdshot retinochoroidopathy and a number of other conditions characterized by multiple inflammatory lesions in the retina and/or choroid, commonly grouped as the “white dot syndromes” (Quillen et al., 2004). Although bacterial, viral and fungal infections were observed, by far the most common infectious cause of posterior uveitis found in the United States and Germany was ocular toxoplasmosis—infection of the retina with the parasite Toxoplasma gondii—accounting for one quarter of total cases of posterior uveitis in both studies. Clinical aspects of ocular toxoplasmosis have been extensively described by Holland (Holland, 2003, 2004).
Management of uveitis depends on etiology. Since the 1950s, systemic corticosteroids have been used as first-line treatment for patients with non-infectious posterior uveitis (Gordon, 1956). High-dose corticosteroid therapy is frequently effective, but the multitude of metabolic side effects (Stanbury and Graham, 1998) necessitates the use of corticosteroid-sparing agents for the long-term treatment most patients require. Immunosuppressive drugs, including antimetabolites, T-cell inhibitors, and alkylating agents, are the most frequently used corticosteroid-sparing agents (Jabs et al., 2000). Unfortunately, half of patients with inflammatory eye disease who are treated with corticosteroid-sparing agents will eventually stop treatment, primarily due to lack of efficacy or adverse effects (Baker et al., 2006). This is an incentive for clinicians to consider recently developed biologic agents or locally delivered corticosteroids for patients with posterior uveitis.
A biologic is defined as “a therapy based on a contemporary understanding of the disease biology and usually produced by molecular (recombinant DNA or monoclonal) technology” (James T. Rosenbaum, American Academy of Ophthalmology Annual Meeting, Uveitis Subspecialty Day, 2005). Multiple publications describe the effectiveness of biologic agents in patients with recalcitrant posterior uveitis, including drugs directed against lymphocyte markers, tumor necrosis factor alpha, and interleukin-2 (Servat et al., 2012). On the other hand, a number of these agents have been associated with serious systemic toxicities, including potentially fatal infections and progressive multifocal leukoencephalopathy. Locally administered corticosteroid therapy can avoid the complications of systemically administered drugs. Local administration methods include periocular injection (Ferrante et al., 2004), intravitreal injection (Kok et al., 2005) and, for extended delivery, implantation of sustained release devices (Jaffe et al., 2006; Lowder et al., 2011). However, long-term delivery of corticosteroids to the eye has its own hazards, including the potential to cause visually significant cataract and elevated intraocular pressure requiring medical or surgical interventions (Kempen et al., 2011; Pavesio et al., 2010).
Antimicrobial therapy is the primary treatment for infectious posterior uveitis, but a good outcome with presently available agents is not guaranteed. For ocular toxoplasmosis in particular, the literature lacks strong evidence for effectiveness of treatment for the acute infection and none of the available drugs have proved capable of eradicating encysted parasites from the human retina (Stanford et al., 2003). The rate of clinical recurrence is estimated at 1 episode per 5 years (Holland et al., 2008). In addition, toxicity and teratogenicity are important concerns for the standard antimicrobial treatments (Rothova, 1993).
2.2 Retinal ischemic vasculopathies
Diabetic retinopathy is an ischemic retinal vasculopathy that affects individuals who suffer from diabetes mellitus. Retinopathy is routinely classified by clinical severity (Cheung et al., 2010) as non-proliferative—or background—or proliferative. Both forms involve microvascular lesions, but proliferative disease is distinguished by the presence of retinal neovascularization. Clinically significant macular edema may complicate either form. Diabetic retinopathy is the most common cause of blindness in young adults in Western countries (Congdon et al., 2003), and prevalence is expected to increase in developing countries (Cheung et al., 2010). Statistics from the United States estimate that approximately one-third of diabetic patients have eye involvement and one-third of these people have vision-threatening retinopathy (i.e., preproliferative or proliferative retinopathy and/or macular edema) (Kempen et al., 2004a).
Retinopathy of prematurity is an ischemic retinal vasculopathy that occurs as a complication of premature birth. Retinal vascularization is incomplete prior to term, and therefore the premature infant is born with retinas that are only partially vascularized. The disease is staged according to vascular abnormalities that occur at the junction of vascularized and avascularized retina (International Committee for the Classification of Retinopathy of Prematurity, 2005). Retinopathy of prematurity causes up to 20% of childhood blindness in countries with relatively high incomes and low infant mortality rates (i.e., less than 10 per 1000 live births) (Wheatley et al., 2002). However, rates as high as 33% have been recorded in middle-income countries with intermediate infant mortality rates (i.e., 10–60 per 1000 live births) (Gilbert et al., 1997). Less common retinal ischemic vasculopathies include: retinal vein occlusion, as may complicate glaucoma or systemic hypertension; sickle cell retinopathy, which usually occurs in persons of African descent; and radiation retinopathy, which may follow radiotherapy for ocular tumors.
For more than 20 years, the mainstay of treatment for retinal ischemic vasculopathy has been the destruction of retina, including retinal pigment epithelium, by cryotherapy or photocoagulation (Diabetic Retinopathy Study Research Group, 1981; Cryotherapy for Retinopathy of Prematurity Cooperative Group, 1990; Early Treatment For Retinopathy Of Prematurity Cooperative, 2003). While this therapy is effective, it may contribute to structural complications, particularly if used to treat retinopathy of prematurity (Hovakimyan and Cunningham, 2002; McLoone et al., 2006). Reductions in visual field, color vision, and contrast sensitivity are also well documented in treated diabetic patients (Fong et al., 2007). These disadvantages have provided incentives for investigators to develop biological medical approaches to retinal ischemic vasculopathy.
Following recognition of the key role that VEGF plays in disease pathogenesis (Cheung et al., 2010; Sapieha et al., 2010) and the success of VEGF antibody blockade in treating neovascular age-related macular degeneration (Coleman et al., 2008), the potential effectiveness of targeting VEGF in diabetic retinopathy and retinopathy of prematurity has been explored. Clinical trials and case series indicate the approach might be of benefit (Cheung et al., 2010; Micieli et al., 2009). However, there is potential for toxicity to retinal neurons and glia, for which VEGF is a trophic factor (van Wijngaarden et al., 2005). Even more concerning are reports of thromboembolism and extraocular hemorrhage related to the effects of VEGF on non-ocular vascular beds following the exit of locally delivered anti-VEGF antibody from the eye (Gillies and Wong, 2007; Ueta et al., 2009).
3. Anatomical and physiological considerations
3.1 Anatomy of the retinal microvasculature
The microvasculature that supplies and drains the inner retina is well described in the classic anatomical text, Gray’s Anatomy (Standring, 2008). The central retinal artery derives from the ophthalmic branch of the internal carotid artery, entering the optic nerve within the orbit approximately 12 mm behind the globe and subsequently coursing through the lamina cribrosa to access the retina. On the inner surface of the retina, superior and inferior branches immediately give rise to temporal and nasal arcades, which supply the 4 quadrants of the retina. Corresponding retinal veins drain these quadrants and meet at the optic nerve head as the central retinal vein, which drains into the cavernous sinus both directly and via the superior ophthalmic vein. The other intraocular circulations of the iris and choroid also derive from the ophthalmic artery, but via ciliary arteries, which branch off the main trunk within the orbit subsequent to the central retinal artery.
Applying scanning electron microscopy to methacrylic methyl ester-injection/corrosion ocular vascular casts of 80 human eyes has allowed detailed observations of the 3-dimensional architecture of the retinal vascular network (Zhang, 1994). The retinal arteries and veins lie in the nerve fiber and ganglion cell layers. Arteriolar branches give rise to capillary networks, which exist in trilaminar form at the posterior pole. The layers include: radial peripapillary capillaries in the inner nerve fiber layer, mostly in a “long chain” pattern; an inner capillary plexus in the nerve fiber and ganglion cell layers; and a deep capillary plexus in the inner plexiform layer and inner nuclear layer. These layers reduce to 2 at the equator and only 1 in the macula and far retinal periphery. The capillary networks communicate via vertical “vascular bridges.” The macula contains a ring of terminal capillaries surrounding a central zone 450 to 500 μm in diameter, which appears avascular. Vessels are also absent within 1 disc area of the ora serrata where another terminal anastomosis exists. There are differences between the anatomy of the human retinal microvasculature and that of other species (Zhang, 1994). A new microperfusion fixation and immunostaining technique for processing retinal whole mounts, which are subsequently imaged by confocal microscopy, results in impressive resolution and has permitted novel observations relating to the human retinal microvasculature (Yu et al., 2010a; Yu et al., 2010b). Most notably, in almost 1 in 5 normal human eyes, retinal capillaries are seen to cross the fovea. This observation “may require a change in the concept of a completely avascular fovea and may be relevant to many macular diseases” (Yu et al., 2010b).
3.2 Embryology of the retinal microvasculature
The development of the human retinal circulation in utero remains a subject of much discussion, as exemplified in recent reviews by Fruttiger (Fruttiger, 2007) and Gariano (Gariano, 2010). Studies using human fetal whole mounts and immunohistochemistry for endothelial precursor markers suggest that in the human (Chan-Ling et al., 2004; Hasegawa et al., 2008; Hughes et al., 2000; McLeod et al., 2006), as opposed to other species such as the mouse (Fruttiger, 2002), retinal blood vessel formation begins at the level of the inner capillary plexus and in the region of the optic nerve head. Growth is centripetal by a process of vasculogenesis, which involves the development of rudimentary channels from differentiation of vascular endothelial precursor cells within the tissue. This is followed by expansion of the inner capillary plexus and the appearance of the deep capillary plexus and peripapillary radial plexus, as well as the foveal region and temporal raphe. These latter events occur by the process of angiogenesis, which refers to sprouting from existing endothelial buds. The avascularity of the fovea remains an enigma, but recent findings by the Provis laboratory are potentially highly relevant. These investigators find relatively high expression of potent anti-angiogenic regulator, pigment epithelium-derived factor, and repellent axonal guidance factors (which are likely to also affect endothelial cells) in the macula (Kozulin et al., 2009b). These factors have been localized to the ganglion cell layer in separate studies using macaque retinas (Kozulin et al., 2010; Kozulin et al., 2009a). Another group (Gariano, 2010) presents indirect evidence of a role for lutein and other macular pigments in foveal avascularity.
3.3 Physiology of the retinal circulation
There is disagreement in published literature regarding the presence of autonomic innervation of the retinal vessels within the eye of humans and other species (Collin, 1966; Hogan and Feeney, 1963; Lanigan et al., 1990; Menage et al., 1994). Whether such innervation is present or absent, it is well accepted that blood flow within the retinal circulation relies heavily on autoregulation. Recently the subject was reviewed comprehensively in this journal (Pournaras et al., 2008). Simplistically presented, perfusion pressure and metabolic reactions act to influence the tone of retinal arterioles and capillaries, to regulate retinal blood flow. Although not specifically studied, retinal endothelial cells are likely to assist mural myocytes and pericytes in sensing and transducing mechanical forces. The retinal endothelium is coated with a glycocalyx (Lawrenson et al., 2000), which has been shown to function as a mechanical sensor and transducer in extra-ocular endothelia (Tarbell and Ebong, 2008). Retinal vascular endothelium also has the ability to detect chemical perturbations, including hypoxia and hypercapnia, and the metabolite, lactose (Pournaras et al., 2008). The endothelium communicates a need for retinal vasodilation or vasoconstriction, by production of molecular mediators that include nitric oxide, arachidonic acid metabolites, and endothelin-1 (Pournaras et al., 2008).
3.4 Microanatomy of the retinal endothelial cell
As a general rule, endothelial cells have flattened cytoplasm (except in the area where the nucleus bulges), abundant mitochondria and ribosomes, and pinocytotic vesicles that are more prominent in arterial forms (Rhodin, 1967, 1968). Distinguishing features of endothelial cells of the retinal circulation in particular are a lack of fenestrations and the presence of specialized “zonula occludens” intercellular junctions, which form stable and extremely tight unions with neighboring cells (Hogan et al., 1971). These characteristics contribute in large part to the blood-retinal barrier, which in health excludes circulating solutes from the retina (Cunha-Vaz, 1979). In contrast, choroidal endothelial cells have fenestrations with bridging diaphragms (Hogan et al., 1971). Within the retina, endothelial form varies markedly with vessel order. Studies of the retinal vasculature in human and porcine eyes reveal elongation of arteriolar endothelial cells that is most marked at the start of the tree in comparison to the more polygonal shape of venular cells (Yu et al., 2010b; Yu et al., 1997). This difference may relate to changes in blood flow and resulting local shear stress along the vascular tree, and is reflected in a differential abundance and structure of F-actin microfilaments—or stress fibers—in different retinal endothelial subpopulations. Interestingly, however, these differences do not apply in the macular region, suggesting “special features of macular hemodynamics” (Yu et al., 2010a).
3.5 Microenvironment of the retinal endothelial cell
Although the focus of our research and this review is the retinal endothelial cell, the cell does not function in isolation. On the contrary, interactions with vascular mural cells, neurons, and glial cells are critical for normal retinal endothelial cell functioning, and also contribute to the development of retinal vascular diseases. The intimate relationship between retinal endothelial cells and pericytes was recognized in the 1970s (Matsusaka, 1975), with pericytes embedded within the endothelial basement membrane seen to be making formal adhesive junctions with the endothelial cells. In this position, pericytes regulate multiple aspects of retinal endothelial behavior, including survival and proliferation (Benjamin et al., 1998; Darland et al., 2003). Loss of pericytes adversely impacts retinal endothelial cell function early in the development of diabetic retinopathy, as reviewed (Motiejunaite and Kazlauskas, 2008). Other work has drawn attention to the close relationships between retinal vessels, and astrocytes and Müller cells, as well as retinal neurons (Yu et al., 2010b). The functional importance of the interactions of retinal endothelial cells with these neighboring cells is well exemplified by the essential role of astrocytes in endothelial cell guidance in vascular patterning during retinal development (Dorrell and Friedlander, 2006) and the ability of retinal ganglion cells, reacting to local levels of the metabolite succinate by production of VEGF, to control retinal angiogenesis in health and disease (Sapieha et al., 2008).
3.6 Angiogenesis in the retina
Retinal angiogenesis is part of normal vascular development and a key component of retinal neovascularization in ischemic vasculopathy. In both situations, retinal hypoxia stimulates the synthesis of endothelial growth factors by various retinal cells. Vascular endothelial growth factor (VEGF) was the first angiogenic factor identified in retinal ischemic vasculopathy (Aiello et al., 1994) and almost simultaneously in retinal vascular development (Stone et al., 1995), and while other hypoxia-induced and hypoxia-independent factors have since been identified, it is clear that VEGF—or VEGFA—plays a critical role in normal and pathological angiogenesis (Cheung et al., 2010; Sapieha et al., 2010).
Angiogenesis involves specialization of endothelial cells into “tip cells” and “stalk cells” (Ferrara, 2004; Gerhardt, 2008). The migrating tip cells extend filipodia in association with astrocyte processes. Behind each tip cell, stalk cells proliferate to grow the endothelial sprout that initiates vessel formation. VEGF acts via at least 2 receptors, VEGFR-1 and VEGFR-2, to direct endothelial tip cell migration and stalk cell proliferation. Remarkable work from the Gerhardt group (Jakobsson et al., 2010), using computational modeling and in vitro and in vivo genetic mosaic sprouting assays, shows that endothelial cells compete to become tip cells and that competition is controlled by the balance of VEGFR-1 and VEGFR-2 between a cell and its neighbors. Lower relative VEGFR-1 increases, and lower VEGFR-2 decreases, the likelihood of being a tip cell, as signaled through the Notch system by variation in the level of delta-like ligand 4. Recently, 2 publications have separately implicated retinal microglia in endothelial tip cell anastomosis, which promotes the arborization of vascular networks (Fantin et al., 2010; Rymo et al., 2011). Interestingly, this effect appears to be independent of VEGF.
4. Molecular Phenotype of the Retinal Endothelial Cell
4.1 Molecular heterogeneity of vascular endothelial cells
Vascular endothelial cell heterogeneity refers to the variations in structure and function that differentiate endothelial cell subtypes across the body (Aird, 2006). Of particular interest in translational medical research are the molecular distinctions between these populations, as differences provide insights into disease pathogenesis and are potential targets for specific therapies. Profiling of the vascular endothelium by gene expression microarray, in particular, confirms the existence of heterogeneity between endothelial cells from different tissues, for large vessel versus microvascular endothelial cells, and for arterial versus venous endothelial cells (Chi et al., 2003b). Our research has been directed at defining the unique molecular phenotype of the human retinal endothelial cells, by transcriptomic and proteomic profiling. When we commenced this work, earlier studies on the responses of human retinal versus umbilical vein endothelial cells, and bovine retinal versus brain endothelial cells, to highly concentrated glucose had already suggested specific molecular features of the retinal endothelial cell that might have implications for retinal ischemic vasculopathy in diabetes mellitus (Grammas and Riden, 2003; Rymaszewski et al., 1992). Other pertinent studies have showed differential expression of angiogenic proteins and receptors by bovine retinal and choroidal endothelial cells in response to hypoxia (Brylla et al., 2003), and differential impact of nerve growth factor on the angiogenic properties of human retinal and choroidal endothelial cells (Steinle and Granger, 2003).
4.2 Isolation and culture of human ocular vascular endothelial cells
We have investigated the profile of the retinal endothelial cell at transcript and protein levels, with the goal of increasing understanding of this cell’s involvement in retinal vascular pathology. Since our interest is human disease, we have preferred to study retinal endothelial cells from human eyes, as opposed to eyes of experimental animals. Although the molecular phenotypes of vascular endothelial cells from humans and other species have not been systematically compared, groups working in various areas of vascular endothelial cell pathobiology have observed differences that are likely to impact disease mechanisms (Autar et al., 2011; Choo et al., 1997; Kalsi et al., 1999; Pan et al., 1998; Smolenski et al., 2006). Gene expression may vary considerably between individuals. To address this concern, we took retinal endothelial cells and the comparison cell population from the same human eyes. We selected the choroidal endothelial cell as the control cell for several reasons. Like the retinal endothelial cell, it is microvascular. Since the choroid lies immediately adjacent to the retina, these cells’ microenvironments are as similar as possible, and the choroidal and retinal circulations derive from the same artery. Importantly, the choroidal vasculature is not primarily involved ischemic retinal vasculopathy or the most common forms of posterior uveitis, as reported in large clinical series (Jakob et al., 2009; Rodriguez et al., 1996).
Human cadaver globes provide the source of primary ocular endothelial cells. In our experience, optimal yields are obtained if donors are younger than 50 at the time of death, have no history of vascular disease, and have been deceased for less than 24 hours at the start of the isolation procedure. We use paired globes to prepare each endothelial cell isolate. Following careful dissection of retina and choroid from the globe, and manual removal of the retinal pigment epithelium and pigmented choroidal cells from the choroid, the tissues are digested with graded solutions of type II collagenase (beginning as high as 3 mg/ml) and dispase (beginning as high as 0.3 mg/ml). Concentrations depend on the density of the tissue, which is affected by factors including donor age, presence of vascular disease, and time since death. Digestion of the tissue is facilitated by initial trituration, centrifugation to separate cells and debris after enzymatic treatment, and final passage through a 40-μm filter.
Primary ocular endothelial cell isolates are cultured in MCDB-131 medium with endothelial growth factors (EGM-2 SingleQuots supplement, omitting gentamicin, hydrocortisone, and serum; Lonza Clonetics, St. Louis, MO) and up to 10% fetal bovine serum (with pH strictly maintained at 7.2) until approximately 1 million endothelial cells are present, which may take more than a week for retinal endothelial cells. At this point, Dynabeads (Invitrogen Dynal AS, Oslo, Norway), pre-coated with murine anti-human CD31 antibody (BD Biosciences Pharmingen, San Diego, CA), are used to purify the endothelial cells. Separations may be repeated multiple times to ensure maximum yield. Several rounds of magnetic bead purification and subsequent culture may be needed to obtain endothelial cultures that are no less than 99% pure. Choroidal cultures, in particular, are initially heavily contaminated with stromal cells, which must be removed. While flow cytometric sorting is another potential approach to purification, in our hands this results in lower numbers of cells and inferior purity of the cultures.
4.3 Transcriptome of the human retinal endothelial cell
We initiated studies of the molecular phenotype of the human retinal vascular endothelial cell with gene expression profiling, using oligonucleotide arrays that included probes designed to detect 8746 well-characterized human transcripts (Smith et al., 2007). Retinal and control choroidal samples from 6 human donors were studied and replicates were included for majority. In addition to non-stimulated cells, cells exposed to Toxoplasma gondii and lipopolysaccharide—a commonly utilized inflammatory stimulus that is capable of inducing posterior uveitis in rodents (Ruiz-Moreno et al., 1992)—were also studied. Statistical assessment included normalization procedures developed for oligonucleotide expression arrays (Irizarry et al., 2003; Li and Wong, 2001; Tusher et al., 2001), significance analysis of microarrays (SAM) with the false-discovery rate set at 5% (Tusher et al., 2001), and gene ontology annotation using the United States National Institutes of Health Database for Annotation, Visualization and Integrated Discovery (DAVID).
One notable observation from this work was the demonstration that although gene expression differed between samples from different donors, and between stimulated and non-stimulated cells from the same donor, the most obvious difference in gene expression was between retinal and choroidal endothelial cells. This finding was clear evidence of the existence of vascular endothelial diversity even within the eye, and of a unique molecular phenotype of the retinal endothelial cell. By SAM, 779 transcripts (8.9%) were differentially expressed by retinal endothelial cells compared to choroidal endothelial cells, including 330 transcripts (3.8%) that were relatively highly expressed. Another important finding came from the gene ontology annotation, which showed that retinal endothelial cells expressed relatively high levels of transcripts involved in the immune response, including cell adhesion molecules, cytokines, chemokines, receptors, and enzymes involved in synthesizing inflammatory proteins. This finding was consistent with the known role of retinal blood vessels in leukocyte trafficking and regulation of inflammation in uveitis. Retinal endothelial cells also expressed relatively high levels of certain transcripts involved in response to stress, cell proliferation and adhesion, suggesting the possibility of a unique reaction to ischemia and specific regulation of neovascularization.
In considering the results of this work, we speculated that differential gene expression reflected differences in the interactions of transcription factors and respective cis-regulatory motifs(s) in human retinal and control choroidal endothelial cells. Taking an in silico approach, we used TRANSFAC Professional v11.4 (BIOBASE, Wolfenbuettel, Germany) and CisModule (Zhou and Wong, 2004) to identify cis-regulatory motifs in promoter sequences of genes that were differentially expressed by the 2 endothelial subpopulations (Choi et al., 2008). Motifs corresponding to 5 transcription factors were significantly more abundant in genes that were relatively highly expressed in retinal endothelial cells (i.e., glucocorticoid receptor, GCCR; high mobility group AT-hook 1, HMGA1; heat shock transcription factor 1, HSF1; p53, vitamin D receptor, VDR). As discussed in our publication (Choi et al., 2008), there is ample evidence that all 5 transcription factors regulate cellular processes involved in the growth of new vessels, including cell proliferation and migration and endothelial monolayer integrity, as well as effects on apoptotic pathways.
These same transcription factors have also been implicated in inflammatory disease. GCCR levels drop in the retina in endotoxin-induced uveitis, an effect which is reversed by exogenous corticosteroid, suggesting an immunomodulatory function (Zhao et al., 2011). Calcitriol (1,25-dihydroxyvitamin D3), which acts via VDR, can prevent or limit experimental autoimmune uveoretinitis by preventing Th17 responses in particular (Tang et al., 2009). Although not studied in relation to uveitis, HMGA1 and HSF1 have both been implicated in systemic inflammatory responses. Inhibition of HMGA1 binding to the promoters of inducible nitric oxide synthase and P-selectin increases survival and reduces lung and liver inflammation in murine endotoxemia (Baron et al., 2010; Grant et al., 2009). Genetic HSF1 deficiency promotes inflammation in a murine model of inflammatory bowel disease, a systemic disease that is associated with uveitis (Tanaka et al., 2007). p53 is a master tumor suppressor transcription factor, but also acts to reciprocally down-regulate activity of NF-κb, which is a central transcription factor in inflammation (Gudkov et al., 2011).
Our gene expression microarray study reinforced an important consideration for designing similar experiments (Smith et al., 2007). Global gene expression patterns of retinal endothelial cell isolates were examined graphically by multi-dimensional scaling (MDS). The MDS plot simplifies a data set such that differences between samples can be viewed as 2- or 3-dimensional distances; points representing samples with similar gene expression are clustered, and those representing divergent profiles are far apart. From the MDS plot presented in Figure 1A, it is obvious that retinal endothelial cells from different donors have distinct gene expression profiles. We separately examined the expression levels of E-selectin, intercellular adhesion molecule (ICAM)-1, vascular cell adhesion molecule (VCAM)-1, CD44, and CX3CL1, and found that different retinal endothelial cell isolates expressed different levels of these adhesion molecules (Figure 1B). Research is often based on cells derived from a single human donor, but these data show the importance of studying multiple donors when investigating the molecular profile of an endothelial cell population.
Figure 1.
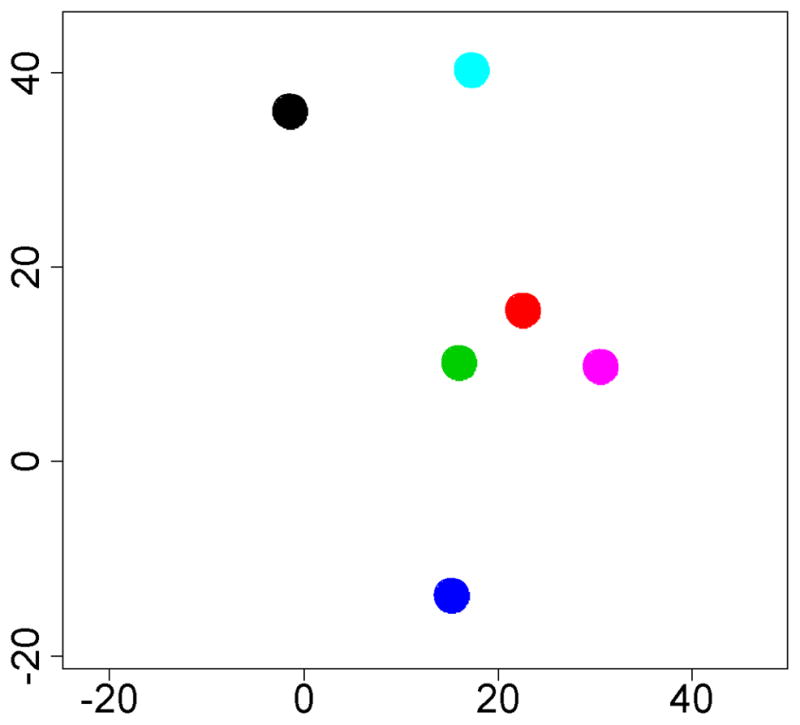
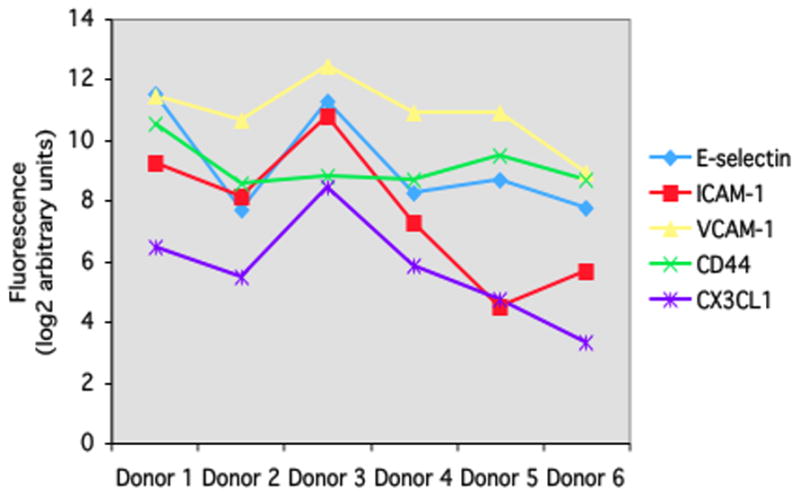
(A) Multi-dimensional scaling plot shows global gene expression by retinal endothelial cells from 6 human donors. Circles designate individual donors. (B) Relative gene expression of selected adhesion molecules in retinal endothelial cells from the same human donors. Normalized fluorescence intensity, which reflects hybridization to the relevant array probes, was averaged for each donor and expressed in log2 scale. Generated using previously published data (Smith et al., 2007).
Recently an independent group published a transcriptomic profiling study, also comparing cultured primary human retinal and choroidal endothelial cells (Browning et al., 2012), but having some methodological differences from our study. Gene expression of ocular endothelial cells, isolated from retina, choroid, and iris of 3 human donors, was profiled using oligonucleotide arrays. Human umbilical vein endothelial cells from 3 additional donors were also studied. A similar percentage of transcripts differed significantly between the retinal and choroidal endothelial cells in both this study (i.e., 8.4%) and our study (i.e., 8.9%). RNA was extracted when endothelial cells were 80% confluent and therefore actively proliferating, which has obvious relevance to angiogenesis. Differential expression of genes related to cell cycle, DNA replication, cell morphology, cell-to-cell interactions, cell movement, and gene expression were highlighted. Interestingly, transcriptomes of iris and choroidal endothelial cells differed less than 1%; although the authors did not speculate on this reason for this finding, it might reflect the shared uveal location of the respective vascular beds. As expected, differences were noted in the transcriptomes of retinal and umbilical vein endothelial cells, leading the authors to conclude that the latter “are probably not a suitable surrogate for the study of ocular … disorders”.
4.4 Proteome of the human retinal endothelial cell
After we identified significant differences in the transcriptome of human vascular endothelial cells of retinal versus choroidal origin, the logical next step was to compare the proteomes of these 2 endothelial cell subsets. We combined 2-dimensional difference gel electrophoresis (2D-DIGE) and tandem mass spectrometry to do this. Cultured retinal and choroidal endothelial cells from 5 human donors were separately lysed by treatment with 40 mM Tris-2% SDS buffer extraction, followed by sonication. The samples were then acetone-precipitated and labeled with Cy5 or Cy3 dyes. Isoelectrofocusing of the pooled protein samples was done with pH 4–7 gradient strips and a Protean IEF Cell (Bio-Rad Laboratories, Hercules, CA). Second-dimension separation employed 12% SDS-polyacrylamide gels. Gels were scanned using the DIGE-enabled Typhoon 9400 imager (GE Healthcare, Piscataway, NJ). Fluorescence intensities were analyzed by Phoretix 2D Evolution v2005 (Nonlinear Dynamics, Durham, NC). Log2 signal intensities were normalized across the gels by matching medians to remove dye biases. Proteins that were differentially abundant in at least 4 of 5 donors were identified by SAM with the FDR set at 5%, as also employed in our gene expression microarray study.
Of 2514 protein spots detected on 2D-DIGE, 123 spots qualified for analysis; 20 spots were more abundant in retinal endothelial cell samples and 11 were more abundant in choroidal endothelial cell samples. These 31 protein spots were excised from gels and digested with trypsin. Peptides were separated by reverse-phase chromatography, and 17 proteins (including 11 more abundant in retina cells and 6 more abundant in choroid cells) were identified by tandem mass spectrometry with a LTQ linear ion trap (Thermo Finnigan, San Jose, CA). Eleven proteins more abundant in retinal endothelial cells included proteins implicated in inflammation (i.e., calreticulin, peroxiredoxin-4, protein disulfide isomerase, serpinB9, coactosin-like protein, vimentin, cathepsin B, annexin A3) and angiogenesis (i.e., calreticulin, peroxiredoxin-4, protein disulfide isomerase, vimentin, cathepsin B, annexin A3).
The data indicated that differences existed in the protein composition of human retinal and choroidal vascular endothelium. However, only a small number of differentially expressed proteins were identified. Several obstacles limit the yield by this method (Corthals et al., 2000; Santoni et al., 2000). Proteins with extreme isoelectric points or molecular weights go undetected in a single gel. Low abundance proteins may be missed due to the limited range of available stains. Membrane proteins are poorly solubilized in the aqueous buffers required for iso-electric separation and thus are also largely undetected; this is a particular concern for our investigation of the retinal endothelial proteome because cell surface proteins are an important aspect of an endothelial cell’s signature. We recently turned to shotgun proteomics to establish a comprehensive list of retinal endothelial proteins.
Shotgun proteomics—“the direct and rapid analysis of the entire protein complement within a complex protein mixture” (Wu and MacCoss, 2002)—combines a gel-free approach to protein separation termed multidimensional protein identification technology (MudPIT) with tandem mass spectrometry (MS/MS) and sophisticated software for rapid spectrum matching. In MudPIT, digested protein is separated by 2-dimensional liquid chromatography (2D-LC) and fed directly into the mass spectrometer (Link et al., 1999). Various methods exist for measuring relative protein abundance in 2D-LC-MS/MS. The most straightforward method is spectral counting. Within a complex sample, higher abundance proteins produce more peptides, and consequently, a larger number of mass spectra. The number of mass spectra assigned to a protein is directly related to abundance in the sample (Liu et al., 2004).
We conducted a pilot study to evaluate the value of MudPIT in profiling the human retinal endothelial cell. Following standard isolation from 1 donor, retinal endothelial cells were lysed by sonication. Whole protein extracts were solubilized in 8M urea, reduced/alkylated, and digested with trypsin following dilution of urea to a 2M concentration. The digest was analyzed by 2D-LC-MS/MS, using the LTQ linear ion trap. This yielded approximately 400,000 MS/MS spectra. The data set was analyzed by Sequest software v27 rev12 (Thermo Finnigan) to identity peptides, and the numbers of spectra matching individual proteins were tabulated using the program, Scaffold (Proteome Software, Portland, OR). Only high (i.e., greater than 0.99) confidence proteins that could be identified by 2 or more unique peptides were included, resulting in the identification of 2457 proteins (Table 1). This result illustrates the superiority of the shotgun approach for protein identification in human retinal endothelial cells. A study comparing the retinal and choroidal endothelial proteomes of multiple donors by this method is ongoing in our laboratory.
Table 1.
Human retinal endothelial proteins identified by 2 or more peptides using multidimensional protein identification technology. Protein descriptions and accessions are from a human Sprot database search. If proteins could not be distinguished on the basis of observed peptides, more than one accession will be listed. Molecular weight is calculated and given in Da. Total SpC is the total spectral count and proteins are ranked in decreasing order of Total SpC. Sequence-reversed decoy protein matches are denoted by REVERSED.
| Protein Description | Accession Number | Molecular Weight | Total SpC |
|---|---|---|---|
| Vimentin | P08670|VIME_HUMAN | 53,635 | 1955 |
| Myosin-9 (Myosin heavy chain 9) (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A) | P35579|MYH9_HUMAN | 226,520 | 1386 |
| Actin, cytoplasmic 1 (Beta-actin) | P60709|ACTB_HUMAN, P63261|ACTG_HUMAN | 41,720 | 1239 |
| Filamin-B (FLN-B) (Beta-filamin) (Actin-binding-like protein) (Thyroid autoantigen) (Truncated actin-binding protein) (Truncated ABP) (ABP-280 homolog) (ABP-278) (Filamin 3) (Filamin homolog 1) (Fh1) | O75369|FLNB_HUMAN | 278,172 | 711 |
| Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin) | P21333|FLNA_HUMAN | 280,711 | 678 |
| Tubulin beta chain (Tubulin beta-5 chain) | P07437|TBB5_HUMAN | 49,653 | 629 |
| Annexin A2 (Annexin II) (Lipocortin II) (Calpactin I heavy chain) (Chromobindin-8) (p36) (Protein I) (Placental anticoagulant protein IV) (PAP-IV) | P07355|ANXA2_HUMAN | 38,588 | 585 |
| Alpha-enolase (EC 4.2.1.11) (2-phospho-D-glycerate hydro-lyase) (Non-neural enolase) (NNE) (Enolase 1) (Phosphopyruvate hydratase) (C-myc promoter-binding protein) (MBP-1) (MPB-1) (Plasminogen-binding protein) | P06733|ENOA_HUMAN | 47,152 | 567 |
| Glyceraldehyde-3-phosphate dehydrogenase (EC 1.2.1.12) (GAPDH) | P04406|G3P_HUMAN | 36,035 | 521 |
| Pyruvate kinase isozymes M1/M2 (EC 2.7.1.40) (Pyruvate kinase muscle isozyme) (Pyruvate kinase 2/3) (Cytosolic thyroid hormone-binding protein) (CTHBP) (THBP1) | P14618|KPYM_HUMAN | 57,920 | 512 |
| Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC) | P98160|PGBM_HUMAN | 468,788 | 485 |
| Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1) (Plectin-11) | Q15149|PLEC1_HUMAN | 531,708 | 477 |
| Heat shock protein HSP 90-beta (HSP 84) (HSP 90) | P08238|HS90B_HUMAN | 83,249 | 456 |
| Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8) | P11142|HSP7C_HUMAN | 70,882 | 430 |
| Neuroblast differentiation-associated protein AHNAK (Desmoyokin) (Fragments) | Q09666|AHNK_HUMAN | 312,479 | 395 |
| 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP) (Endoplasmic reticulum lumenal Ca(2+)-binding protein grp78) | P11021|GRP78_HUMAN | 72,317 | 392 |
| Talin-1 | Q9Y490|TLN1_HUMAN | 269,747 | 346 |
| Peptidyl-prolyl cis-trans isomerase A (EC 5.2.1.8) (PPIase A) (Rotamase A) (Cyclophilin A) (Cyclosporin A-binding protein) | P62937|PPIA_HUMAN | 17,995 | 343 |
| Alpha-actinin-4 (Non-muscle alpha-actinin 4) (F-actin cross-linking protein) | O43707|ACTN4_HUMAN | 104,839 | 331 |
| Tubulin alpha-6 chain (Alpha-tubulin 6) | Q9BQE3|TBA6_HUMAN | 49,877 | 321 |
| Tubulin beta-6 chain | Q9BUF5|TBB6_HUMAN | 49,839 | 319 |
| Elongation factor 2 (EF-2) | P13639|EF2_HUMAN | 95,322 | 316 |
| Elongation factor 1-alpha 1 (EF-1-alpha-1) (Elongation factor 1 A-1) (eEF1A-1) (Elongation factor Tu) (EF-Tu) (Leukocyte receptor cluster member 7) | P68104|EF1A1_HUMAN | 50,123 | 315 |
| Lamin-A/C (70 kDa lamin) (Renal carcinoma antigen NY-REN-32) | P02545|LMNA_HUMAN | 74,123 | 301 |
| Moesin (Membrane-organizing extension spike protein) | P26038|MOES_HUMAN | 67,804 | 297 |
| Endoplasmin precursor (Heat shock protein 90 kDa beta member 1) (94 kDa glucose-regulated protein) (GRP94) (gp96 homolog) (Tumor rejection antigen 1) | P14625|ENPL_HUMAN | 92,454 | 278 |
| Galectin-1 (Lectin galactoside-binding soluble 1) (Beta-galactoside-binding lectin L-14-I) (Lactose-binding lectin 1) (S-Lac lectin 1) (Galaptin) (14 kDa lectin) (HPL) (HBL) (Putative MAPK-activating protein MP12) | P09382|LEG1_HUMAN | 14,698 | 271 |
| von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)] | P04275|VWF_HUMAN | 309,268 | 270 |
| Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain) | Q13813|SPTA2_HUMAN | 284,525 | 269 |
| Protein disulfide-isomerase precursor (EC 5.3.4.1) (PDI) (Prolyl 4-hydroxylase subunit beta) (Cellular thyroid hormone-binding protein) (p55) | P07237|PDIA1_HUMAN | 57,100 | 262 |
| Protein disulfide-isomerase A3 precursor (EC 5.3.4.1) (Disulfide isomerase ER-60) (ERp60) (58 kDa microsomal protein) (p58) (ERp57) (58 kDa glucose-regulated protein) | P30101|PDIA3_HUMAN | 56,767 | 260 |
| Cofilin-1 (Cofilin, non-muscle isoform) (18 kDa phosphoprotein) (p18) | P23528|COF1_HUMAN | 18,485 | 246 |
| Fibronectin precursor (FN) (Cold-insoluble globulin) (CIG) | P02751|FINC_HUMAN | 262,581 | 243 |
| Tropomyosin alpha-4 chain (Tropomyosin-4) (TM30p1) | P67936|TPM4_HUMAN | 28,504 | 241 |
| Histone H4 | P62805|H4_HUMAN | 11,350 | 241 |
| Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain) | Q01082|SPTB2_HUMAN | 274,595 | 235 |
| Annexin A1 (Annexin I) (Lipocortin I) (Calpactin II) (Chromobindin-9) (p35) (Phospholipase A2 inhibitory protein) | P04083|ANXA1_HUMAN | 38,698 | 232 |
| Clathrin heavy chain 1 (CLH-17) | Q00610|CLH1_HUMAN | 191,601 | 223 |
| 60 kDa heat shock protein, mitochondrial precursor (Hsp60) (60 kDa chaperonin) (CPN60) (Heat shock protein 60) (HSP-60) (Mitochondrial matrix protein P1) (P60 lymphocyte protein) (HuCHA60) | P10809|CH60_HUMAN | 61,038 | 223 |
| Profilin-1 (Profilin I) | P07737|PROF1_HUMAN | 15,036 | 220 |
| Heat shock protein HSP 90-alpha (HSP 86) (Renal carcinoma antigen NY-REN-38) | P07900|HS90A_HUMAN | 84,645 | 216 |
| Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)-ATPase p97 subunit) (Valosin-containing protein) (VCP) | P55072|TERA_HUMAN | 89,307 | 209 |
| Fructose-bisphosphate aldolase A (EC 4.1.2.13) (Muscle-type aldolase) (Lung cancer antigen NY-LU-1) | P04075|ALDOA_HUMAN | 39,403 | 209 |
| Vinculin (Metavinculin) | P18206|VINC_HUMAN | 123,783 | 204 |
| Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1) | Q14204|DYHC_HUMAN | 532,388 | 201 |
| Phosphoglycerate kinase 1 (EC 2.7.2.3) (Primer recognition protein 2) (PRP 2) (Cell migration-inducing gene 10 protein) | P00558|PGK1_HUMAN | 44,597 | 201 |
| Triosephosphate isomerase (EC 5.3.1.1) (TIM) (Triose-phosphate isomerase) | P60174|TPIS_HUMAN | 26,651 | 190 |
| Alpha-actinin-1 (Alpha-actinin cytoskeletal isoform) (Non-muscle alpha-actinin-1) (F-actin cross-linking protein) | P12814|ACTN1_HUMAN | 103,043 | 183 |
| Calreticulin precursor (CRP55) (Calregulin) (HACBP) (ERp60) (grp60) | P27797|CALR_HUMAN | 48,125 | 182 |
| Thrombospondin-1 precursor | P07996|TSP1_HUMAN | 129,364 | 181 |
| Nestin | P48681|NEST_HUMAN | 176,687 | 178 |
| WD repeat protein 1 (Actin-interacting protein 1) (AIP1) (NORI-1) | O75083|WDR1_HUMAN | 66,175 | 169 |
| Nucleolin (Protein C23) | P19338|NUCL_HUMAN | 76,598 | 162 |
| Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17) | P60660|MYL6_HUMAN | 16,912 | 159 |
| Heterogeneous nuclear ribonucleoprotein U (hnRNP U) (Scaffold attachment factor A) (SAF-A) (p120) (pp120) | Q00839|HNRPU_HUMAN | 90,496 | 158 |
| Fascin (Singed-like protein) (55 kDa actin-bundling protein) (p55) | Q16658|FSCN1_HUMAN | 54,512 | 158 |
| TRYPSIN PRECURSOR. | CONT|sp|P00761|TRYP_PI G | 24,391 | 157 |
| Heterogeneous nuclear ribonucleoproteins A2/B1 (hnRNP A2/hnRNP B1) | P22626|ROA2_HUMAN | 37,412 | 152 |
| Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein) | Q9P2E9|RRBP1_HUMAN | 152,453 | 147 |
| Heterogeneous nuclear ribonucleoprotein K (hnRNP K) (Transformation up-regulated nuclear protein) (TUNP) | P61978|HNRPK_HUMAN | 50,961 | 143 |
| Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I) | P60842|IF4A1_HUMAN | 46,137 | 143 |
| Ras GTPase-activating-like protein IQGAP1 (p195) | P46940|IQGA1_HUMAN | 189,241 | 139 |
| Thioredoxin domain-containing protein 5 precursor (Thioredoxin-like protein p46) (Endoplasmic reticulum protein ERp46) | Q8NBS9|TXND5_HUMAN | 47,611 | 138 |
| Serpin H1 precursor (Collagen-binding protein) (Colligin) (47 kDa heat shock protein) (Rheumatoid arthritis-related antigen RA-A47) (Arsenic-transactivated protein 3) (AsTP3) (Proliferation-inducing gene 14 protein) | P50454|SERPH_HUMAN | 46,424 | 138 |
| Transgelin-2 (SM22-alpha homolog) | P37802|TAGL2_HUMAN | 22,374 | 138 |
| Tubulin alpha-ubiquitous chain (Alpha-tubulin ubiquitous) (Tubulin K-alpha-1) | P68363|TBAK_HUMAN | 50,134 | 138 |
| ATP synthase subunit beta, mitochondrial precursor (EC 3.6.3.14) | P06576|ATPB_HUMAN | 56,543 | 134 |
| Heterogeneous nuclear ribonucleoprotein A1 (Helix-destabilizing protein) (Single-strand RNA-binding protein) (hnRNP core protein A1) | P09651|ROA1_HUMAN | 38,828 | 131 |
| ATP synthase subunit alpha, mitochondrial precursor (EC 3.6.3.14) | P25705|ATPA_HUMAN | 59,734 | 128 |
| Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S-acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S-malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)] | P49327|FAS_HUMAN | 273,382 | 124 |
| Major vault protein (MVP) (Lung resistance-related protein) | Q14764|MVP_HUMAN | 99,308 | 124 |
| 60S ribosomal protein L3 (HIV-1 TAR RNA-binding protein B) (TARBP-B) | P39023|RL3_HUMAN | 46,092 | 124 |
| Histone H2A type 1-B | P04908|H2A1B_HUMAN, P28001|H2A1E_HUMAN, Q7L7L0|H2A3_HUMAN, Q93077|H2A1C_HUMAN | 14,118 | 121 |
| Platelet endothelial cell adhesion molecule precursor (PECAM-1) (EndoCAM) (GPIIA’) (CD31 antigen) | P16284|PECA1_HUMAN | 82,518 | 118 |
| Protein disulfide-isomerase A4 precursor (EC 5.3.4.1) (Protein ERp-72) (ERp72) | P13667|PDIA4_HUMAN | 72,916 | 118 |
| Chloride intracellular channel protein 1 (Nuclear chloride ion channel 27) (NCC27) (Chloride channel ABP) (Regulatory nuclear chloride ion channel protein) (hRNCC) | O00299|CLIC1_HUMAN | 26,905 | 117 |
| Annexin A5 (Annexin V) (Lipocortin V) (Endonexin II) (Calphobindin I) (CBP-I) (Placental anticoagulant protein I) (PAP-I) (PP4) (Thromboplastin inhibitor) (Vascular anticoagulant-alpha) (VAC-alpha) (Anchorin CII) | P08758|ANXA5_HUMAN | 35,921 | 116 |
| Cytosol aminopeptidase (EC 3.4.11.1) (Leucine aminopeptidase) (LAP) (Leucyl aminopeptidase) (Leucine aminopeptidase 3) (Proline aminopeptidase) (EC 3.4.11.5) (Prolyl aminopeptidase) (Peptidase S) | P28838|AMPL_HUMAN | 56,150 | 115 |
| Peroxiredoxin-1 (EC 1.11.1.15) (Thioredoxin peroxidase 2) (Thioredoxin-dependent peroxide reductase 2) (Proliferation-associated gene protein) (PAG) (Natural killer cell-enhancing factor A) (NKEF-A) | Q06830|PRDX1_HUMAN | 22,093 | 114 |
| Heat-shock protein beta-1 (HspB1) (Heat shock 27 kDa protein) (HSP 27) (Stress-responsive protein 27) (SRP27) (Estrogen-regulated 24 kDa protein) (28 kDa heat shock protein) | P04792|HSPB1_HUMAN | 22,765 | 112 |
| ATP-citrate synthase (EC 2.3.3.8) (ATP-citrate (pro-S-)-lyase) (Citrate cleavage enzyme) | P53396|ACLY_HUMAN | 120,825 | 111 |
| Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63) | Q07065|CKAP4_HUMAN | 66,004 | 111 |
| Staphylococcal nuclease domain-containing protein 1 (p100 co-activator) (100 kDa coactivator) (EBNA2 coactivator p100) (Tudor domain-containing protein 11) | Q7KZF4|SND1_HUMAN | 101,981 | 110 |
| Protein-glutamine gamma-glutamyltransferase 2 (EC 2.3.2.13) (Tissue transglutaminase) (TGase C) (TGC) (TG(C)) (Transglutaminase-2) (TGase-H) | P21980|TGM2_HUMAN | 77,311 | 110 |
| SERUM ALBUMIN PRECURSOR. | CONT|sp|P02769|ALBU_BO VIN | 69,253 | 110 |
| 60S ribosomal protein L4 (L1) | P36578|RL4_HUMAN | 47,681 | 109 |
| Malate dehydrogenase, mitochondrial precursor (EC 1.1.1.37) | P40926|MDHM_HUMAN | 35,514 | 108 |
| Transketolase (EC 2.2.1.1) (TK) | P29401|TKT_HUMAN | 67,861 | 107 |
| Heterogeneous nuclear ribonucleoprotein M (hnRNP M) | P52272|HNRPM_HUMAN | 77,499 | 106 |
| 40S ribosomal protein S3a | P61247|RS3A_HUMAN | 29,927 | 106 |
| PDZ and LIM domain protein 1 (Elfin) (LIM domain protein CLP-36) (C-terminal LIM domain protein 1) | O00151|PDLI1_HUMAN | 36,053 | 106 |
| Rab GDP dissociation inhibitor beta (Rab GDI beta) (Guanosine diphosphate dissociation inhibitor 2) (GDI-2) | P50395|GDIB_HUMAN | 50,648 | 104 |
| Heterogeneous nuclear ribonucleoprotein H (hnRNP H) | P31943|HNRH1_HUMAN | 49,212 | 104 |
| Actin, alpha skeletal muscle (Alpha-actin-1) | P68133|ACTS_HUMAN | 42,034 | 103 |
| Protein disulfide-isomerase A6 precursor (EC 5.3.4.1) (Protein disulfide isomerase P5) (Thioredoxin domain-containing protein 7) | Q15084|PDIA6_HUMAN | 48,104 | 102 |
| 6-phosphofructokinase type C (EC 2.7.1.11) (Phosphofructokinase 1) (Phosphohexokinase) (Phosphofructo-1-kinase isozyme C) (PFK-C) (6-phosphofructokinase, platelet type) | Q01813|K6PP_HUMAN | 85,579 | 101 |
| Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)] | P07814|SYEP_HUMAN | 163,011 | 101 |
| 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up-regulated 1) | Q9Y4L1|OXRP_HUMAN | 111,319 | 101 |
| Polyadenylate-binding protein 1 (Poly(A)-binding protein 1) (PABP 1) | P11940|PABP1_HUMAN | 70,653 | 101 |
| 14-3-3 protein epsilon (14-3-3E) | P62258|1433E_HUMAN | 29,157 | 101 |
| Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (Transformation-sensitive protein IEF SSP 3521) (NY-REN-11 antigen) | P31948|STIP1_HUMAN | 62,624 | 98 |
| Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (Mortalin) (MOT) | P38646|GRP75_HUMAN | 73,663 | 98 |
| ATP-dependent DNA helicase 2 subunit 1 (ATP-dependent DNA helicase II 70 kDa subunit) (Lupus Ku autoantigen protein p70) (Ku70) (70 kDa subunit of Ku antigen) (Thyroid-lupus autoantigen) (TLAA) (CTC box-binding factor 75 kDa subunit) (CTCBF) (CTC75) (DNA-repair protein XRCC6) | P12956|KU70_HUMAN | 69,828 | 98 |
| 14-3-3 protein zeta/delta (Protein kinase C inhibitor protein 1) (KCIP-1) | P63104|1433Z_HUMAN | 27,728 | 98 |
| Microtubule-associated protein 4 (MAP 4) | P27816|MAP4_HUMAN | 121,003 | 97 |
| Cell surface glycoprotein MUC18 precursor (Melanoma-associated antigen MUC18) (Melanoma cell adhesion molecule) (Melanoma-associated antigen A32) (S-endo 1 endothelial-associated antigen) (Cell surface glycoprotein P1H12) (CD146 antigen) | P43121|MUC18_HUMAN | 71,589 | 97 |
| Heterogeneous nuclear ribonucleoprotein Q (hnRNP Q) (hnRNP-Q) (Synaptotagmin-binding, cytoplasmic RNA-interacting protein) (Glycine- and tyrosine-rich RNA-binding protein) (GRY-RBP) (NS1-associated protein 1) | O60506|HNRPQ_HUMAN | 69,586 | 96 |
| Neutral alpha-glucosidase AB precursor (EC 3.2.1.84) (Glucosidase II subunit alpha) | Q14697|GANAB_HUMAN | 106,858 | 92 |
| Glucose-6-phosphate isomerase (EC 5.3.1.9) (GPI) (Phosphoglucose isomerase) (PGI) (Phosphohexose isomerase) (PHI) (Neuroleukin) (NLK) (Sperm antigen 36) (SA-36) | P06744|G6PI_HUMAN | 63,131 | 92 |
| Ubiquitin-activating enzyme E1 (A1S9 protein) | P22314|UBE1_HUMAN | 117,832 | 91 |
| DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460) | P78527|PRKDC_HUMAN | 469,078 | 90 |
| Catenin alpha-1 (Cadherin-associated protein) (Alpha E-catenin) (NY-REN-13 antigen) | P35221|CTNA1_HUMAN | 100,055 | 90 |
| 6-phosphogluconate dehydrogenase, decarboxylating (EC 1.1.1.44) | P52209|6PGD_HUMAN | 53,124 | 90 |
| Heterogeneous nuclear ribonucleoproteins C1/C2 (hnRNP C1/hnRNP C2) | P07910|HNRPC_HUMAN | 33,653 | 90 |
| Plastin-3 (T-plastin) | P13797|PLST_HUMAN | 70,421 | 89 |
| Peptidyl-prolyl cis-trans isomerase B precursor (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin B) (S-cyclophilin) (SCYLP) (CYP-S1) | P23284|PPIB_HUMAN | 22,725 | 89 |
| Interleukin enhancer-binding factor 3 (Nuclear factor of activated T-cells 90 kDa) (NF-AT-90) (Double-stranded RNA-binding protein 76) (DRBP76) (Translational control protein 80) (TCP80) (Nuclear factor associated with dsRNA) (NFAR) (M-phase phosphoprotein 4) (MPP4) | Q12906|ILF3_HUMAN | 95,321 | 88 |
| T-complex protein 1 subunit beta (TCP-1-beta) (CCT-beta) | P78371|TCPB_HUMAN | 57,472 | 87 |
| HLA class I histocompatibility antigen, A-2 alpha chain precursor (MHC class I antigen A*2) | P01892|1A02_HUMAN | 40,903 | 87 |
| Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1] | P46821|MAP1B_HUMAN | 270,602 | 86 |
| Heat shock 70 kDa protein 1 (HSP70.1) (HSP70-1/HSP70-2) | P08107|HSP71_HUMAN | 70,022 | 85 |
| Polymerase I and transcript release factor (PTRF protein) | Q6NZI2|PTRF_HUMAN | 43,459 | 85 |
| 40S ribosomal protein S3 | P23396|RS3_HUMAN | 26,671 | 85 |
| 40S ribosomal protein S4, X isoform (Single copy abundant mRNA protein) (SCR10) | P62701|RS4X_HUMAN | 29,581 | 85 |
| Heterogeneous nuclear ribonucleoprotein D0 (hnRNP D0) (AU-rich element RNA-binding protein 1) | Q14103|HNRPD_HUMAN | 38,417 | 85 |
| T-complex protein 1 subunit epsilon (TCP-1-epsilon) (CCT-epsilon) | P48643|TCPE_HUMAN | 59,654 | 84 |
| Fatty acid-binding protein, epidermal (E-FABP) (Psoriasis-associated fatty acid-binding protein homolog) (PA-FABP) | Q01469|FABPE_HUMAN | 15,146 | 83 |
| Nucleophosmin (NPM) (Nucleolar phosphoprotein B23) (Numatrin) (Nucleolar protein NO38) | P06748|NPM_HUMAN | 32,557 | 83 |
| Nucleoside diphosphate kinase B (EC 2.7.4.6) (NDK B) (NDP kinase B) (nm23-H2) (C-myc purine-binding transcription factor PUF) | P22392|NDKB_HUMAN | 17,280 | 82 |
| Tryptophanyl-tRNA synthetase, cytoplasmic (EC 6.1.1.2) (Tryptophan--tRNA ligase) (TrpRS) (IFP53) (hWRS) | P23381|SYWC_HUMAN | 53,150 | 81 |
| Dihydropyrimidinase-related protein 2 (DRP-2) (Collapsin response mediator protein 2) (CRMP-2) (N2A3) | Q16555|DPYL2_HUMAN | 62,276 | 81 |
| EH domain-containing protein 2 | Q9NZN4|EHD2_HUMAN | 61,145 | 80 |
| Vigilin (High density lipoprotein-binding protein) (HDL-binding protein) | Q00341|VIGLN_HUMAN | 141,424 | 80 |
| T-complex protein 1 subunit delta (TCP-1-delta) (CCT-delta) (Stimulator of TAR RNA-binding) | P50991|TCPD_HUMAN | 57,908 | 80 |
| Adenylyl cyclase-associated protein 1 (CAP 1) | Q01518|CAP1_HUMAN | 51,838 | 80 |
| L-lactate dehydrogenase B chain (EC 1.1.1.27) (LDH-B) (LDH heart subunit) (LDH-H) (Renal carcinoma antigen NY-REN-46) | P07195|LDHB_HUMAN | 36,621 | 80 |
| 40S ribosomal protein S6 (Phosphoprotein NP33) | P62753|RS6_HUMAN | 28,664 | 80 |
| 60S ribosomal protein L10 (QM protein) (Tumor suppressor QM) (Laminin receptor homolog) | P27635|RL10_HUMAN | 24,560 | 79 |
| Phosphoglycerate mutase 1 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13) (Phosphoglycerate mutase isozyme B) (PGAM-B) (BPG-dependent PGAM 1) | P18669|PGAM1_HUMAN | 28,787 | 78 |
| 60S ribosomal protein L8 | P62917|RL8_HUMAN | 28,007 | 78 |
| Dihydropyrimidinase-related protein 3 (DRP-3) (Unc-33-like phosphoprotein) (ULIP protein) (Collapsin response mediator protein 4) (CRMP-4) | Q14195|DPYL3_HUMAN | 61,946 | 78 |
| Filamin-C (Gamma-filamin) (Filamin-2) (Protein FLNc) (Actin-binding-like protein) (ABP-L) (ABP-280-like protein) | Q14315|FLNC_HUMAN | 290,934 | 76 |
| Actin-like protein 3 (Actin-related protein 3) | P61158|ARP3_HUMAN | 47,354 | 76 |
| 60S ribosomal protein L5 | P46777|RL5_HUMAN | 34,346 | 76 |
| ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9) | Q08211|DHX9_HUMAN | 140,944 | 75 |
| Eukaryotic translation initiation factor 5A-1 (eIF-5A-1) (eIF-5A1) (Eukaryotic initiation factor 5A isoform 1) (eIF-5A) (eIF-4D) (Rev-binding factor) | P63241|IF5A1_HUMAN | 16,815 | 75 |
| Thioredoxin reductase 1, cytoplasmic precursor (EC 1.8.1.9) (TR) (TR1) | Q16881|TRXR1_HUMAN | 54,689 | 74 |
| Glutathione S-transferase P (EC 2.5.1.18) (GST class-pi) (GSTP1-1) | P09211|GSTP1_HUMAN | 23,339 | 74 |
| Myosin regulatory light chain 2, nonsarcomeric (Myosin RLC) | P19105|MLRM_HUMAN | 19,777 | 74 |
| ATP-dependent DNA helicase 2 subunit 2 (EC 3.6.1.-) (ATP-dependent DNA helicase II 80 kDa subunit) (Lupus Ku autoantigen protein p86) (Ku86) (Ku80) (86 kDa subunit of Ku antigen) (Thyroid-lupus autoantigen) (TLAA) (CTC box-binding factor 85 kDa subunit) (CTCBF) (CTC85) (Nuclear factor IV) (DNA-repair protein XRCC5) | P13010|KU86_HUMAN | 82,689 | 73 |
| L-lactate dehydrogenase A chain (EC 1.1.1.27) (LDH-A) (LDH muscle subunit) (LDH-M) (Proliferation-inducing gene 19 protein) (Renal carcinoma antigen NY-REN-59) | P00338|LDHA_HUMAN | 36,671 | 73 |
| T-complex protein 1 subunit alpha (TCP-1-alpha) (CCT-alpha) | P17987|TCPA_HUMAN | 60,327 | 72 |
| Heterogeneous nuclear ribonucleoprotein A3 (hnRNP A3) | P51991|ROA3_HUMAN | 39,577 | 72 |
| Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1) (RAI) (Placental ribonuclease inhibitor) (RNase inhibitor) (RI) | P13489|RINI_HUMAN | 49,956 | 72 |
| Chloride intracellular channel protein 4 (Intracellular chloride ion channel protein p64H1) | Q9Y696|CLIC4_HUMAN | 28,756 | 72 |
| Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma) | P26641|EF1G_HUMAN | 50,101 | 72 |
| GTP-binding nuclear protein Ran (GTPase Ran) (Ras-like protein TC4) (Androgen receptor-associated protein 24) | P62826|RAN_HUMAN | 24,405 | 72 |
| T-complex protein 1 subunit theta (TCP-1-theta) (CCT-theta) (Renal carcinoma antigen NY-REN-15) | P50990|TCPQ_HUMAN | 59,603 | 71 |
| Guanine nucleotide-binding protein subunit beta 2-like 1 (Guanine nucleotide-binding protein subunit beta-like protein 12.3) (Receptor of activated protein kinase C 1) (RACK1) (Receptor for activated C kinase) | P63244|GBLP_HUMAN | 35,059 | 71 |
| 40S ribosomal protein S2 (S4) (LLRep3 protein) | P15880|RS2_HUMAN | 31,307 | 71 |
| Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2) (HSP70RY) | P34932|HSP74_HUMAN | 94,283 | 70 |
| 40S ribosomal protein S8 | P62241|RS8_HUMAN | 24,188 | 70 |
| Coronin-1C (Coronin-3) (hCRNN4) | Q9ULV4|COR1C_HUMAN | 53,232 | 69 |
| Non-POU domain-containing octamer-binding protein (NonO protein) (54 kDa nuclear RNA-and DNA-binding protein) (p54(nrb)) (p54nrb) (55 kDa nuclear protein) (NMT55) (DNA-binding p52/p100 complex, 52 kDa subunit) | Q15233|NONO_HUMAN | 54,214 | 69 |
| Proliferation-associated protein 2G4 (Cell cycle protein p38-2G4 homolog) (hG4-1) (ErbB3-binding protein 1) | Q9UQ80|PA2G4_HUMAN | 43,769 | 68 |
| Actin-like protein 2 (Actin-related protein 2) | P61160|ARP2_HUMAN | 44,744 | 68 |
| 10 kDa heat shock protein, mitochondrial (Hsp10) (10 kDa chaperonin) (CPN10) (Early-pregnancy factor) (EPF) | P61604|CH10_HUMAN | 10,914 | 67 |
| Keratin, type II cytoskeletal 1 (Cytokeratin-1) (CK-1) (Keratin-1) (K1) (67 kDa cytokeratin) (Hair alpha protein) | P04264|K2C1_HUMAN | 66,001 | 66 |
| Interferon-induced GTP-binding protein Mx1 (Interferon-regulated resistance GTP-binding protein MxA) (Interferon-induced protein p78) (IFI-78K) | P20591|MX1_HUMAN | 75,519 | 66 |
| Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 67 kDa subunit precursor (EC 2.4.1.119) (Ribophorin I) (RPN-I) | P04843|RIB1_HUMAN | 68,553 | 65 |
| Voltage-dependent anion-selective channel protein 2 (VDAC-2) (hVDAC2) (Outer mitochondrial membrane protein porin 2) | P45880|VDAC2_HUMAN | 38,076 | 65 |
| Myoferlin (Fer-1-like protein 3) | Q9NZM1|MYOF_HUMAN | 234,698 | 64 |
| Laminin subunit beta-1 precursor (Laminin B1 chain) | P07942|LAMB1_HUMAN | 198,045 | 64 |
| Coatomer subunit alpha (Alpha-coat protein) (Alpha-COP) (HEPCOP) (HEP-COP) [Contains: Xenin (Xenopsin-related peptide); Proxenin] | P53621|COPA_HUMAN | 138,317 | 64 |
| Spliceosome RNA helicase BAT1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (ATP-dependent RNA helicase p47) (HLA-B-associated transcript-1) | Q13838|UAP56_HUMAN | 48,974 | 64 |
| Trifunctional enzyme subunit alpha, mitochondrial precursor (TP-alpha) (78 kDa gastrin-binding protein) [Includes: Long-chain enoyl-CoA hydratase (EC 4.2.1.17); Long chain 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.211)] | P40939|ECHA_HUMAN | 82,984 | 63 |
| EH domain-containing protein 1 (Testilin) (hPAST1) | Q9H4M9|EHD1_HUMAN | 60,611 | 63 |
| Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1) (Nucleic acid-binding protein SUB2.3) | Q15365|PCBP1_HUMAN | 37,480 | 63 |
| Zyxin (Zyxin-2) | Q15942|ZYX_HUMAN | 61,258 | 63 |
| Nucleosome assembly protein 1-like 1 (NAP-1-related protein) (hNRP) | P55209|NP1L1_HUMAN | 45,357 | 63 |
| Histone H1.4 (Histone H1b) | P10412|H14_HUMAN, P16402|H13_HUMAN | 21,849 | 63 |
| Calnexin precursor (Major histocompatibility complex class I antigen-binding protein p88) (p90) (IP90) | P27824|CALX_HUMAN | 67,552 | 63 |
| Tubulin beta-2C chain (Tubulin beta-2 chain) | P68371|TBB2C_HUMAN | 49,813 | 63 |
| Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2) | Q9UDY2|ZO2_HUMAN | 133,957 | 62 |
| 60S acidic ribosomal protein P0 (L10E) | P05388|RLA0_HUMAN | 34,256 | 62 |
| Programmed cell death 6-interacting protein (PDCD6-interacting protein) (ALG-2-interacting protein 1) (Hp95) | Q8WUM4|PDC6I_HUMAN | 96,007 | 61 |
| 60S acidic ribosomal protein P2 (Renal carcinoma antigen NY-REN-44) | P05387|RLA2_HUMAN | 11,648 | 61 |
| Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC) | P33176|KINH_HUMAN | 109,668 | 60 |
| A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin) | Q02952|AKA12_HUMAN | 191,414 | 60 |
| Splicing factor, proline- and glutamine-rich (Polypyrimidine tract-binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100) | P23246|SFPQ_HUMAN | 76,132 | 60 |
| Keratin, type II cytoskeletal 7 (Cytokeratin-7) (CK-7) (Keratin-7) (K7) (Sarcolectin) | P08729|K2C7_HUMAN | 51,401 | 60 |
| 5′-nucleotidase precursor (EC 3.1.3.5) (Ecto-5′-nucleotidase) (5′-NT) (CD73 antigen) | P21589|5NTD_HUMAN | 63,351 | 60 |
| Cytosolic nonspecific dipeptidase (Glutamate carboxypeptidase-like protein 1) (CNDP dipeptidase 2) | Q96KP4|CNDP2_HUMAN | 52,862 | 59 |
| EH domain-containing protein 4 (Hepatocellular carcinoma-associated protein 10/11) | Q9H223|EHD4_HUMAN | 61,160 | 59 |
| Microtubule-actin cross-linking factor 1, isoform 4 | Q96PK2|MACF4_HUMAN | 670,132 | 59 |
| 40S ribosomal protein S7 | P62081|RS7_HUMAN | 22,110 | 58 |
| High mobility group protein B1 (High mobility group protein 1) (HMG-1) | P09429|HMGB1_HUMAN | 24,877 | 58 |
| Inosine-5′-monophosphate dehydrogenase 2 (EC 1.1.1.205) (IMP dehydrogenase 2) (IMPDH-II) (IMPD 2) | P12268|IMDH2_HUMAN | 55,788 | 57 |
| T-complex protein 1 subunit zeta (TCP-1-zeta) (CCT-zeta) (CCT-zeta-1) (Tcp20) (HTR3) (Acute morphine dependence-related protein 2) | P40227|TCPZ_HUMAN | 58,007 | 57 |
| C-1-tetrahydrofolate synthase, cytoplasmic (C1-THF synthase) [Includes: Methylenetetrahydrofolate dehydrogenase (EC 1.5.1.5); Methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9); Formyltetrahydrofolate synthetase (EC 6.3.4.3)] | P11586|C1TC_HUMAN | 101,544 | 56 |
| Far upstream element-binding protein 2 (FUSE-binding protein 2) (KH type-splicing regulatory protein) (KSRP) (p75) | Q92945|FUBP2_HUMAN | 72,691 | 56 |
| Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (Nuclear corepressor KAP-1) (KRAB-associated protein 1) (KAP-1) (KRAB-interacting protein 1) (KRIP-1) (RING finger protein 96) | Q13263|TIF1B_HUMAN | 88,531 | 56 |
| Dysferlin (Dystrophy-associated fer-1-like protein) (Fer-1-like protein 1) | O75923|DYSF_HUMAN | 237,284 | 55 |
| Rho GDP-dissociation inhibitor 2 (Rho GDI 2) (Rho-GDI beta) (Ly-GDI) | P52566|GDIS_HUMAN | 22,970 | 55 |
| 60S ribosomal protein L6 (TAX-responsive enhancer element-binding protein 107) (TAXREB107) (Neoplasm-related protein C140) | Q02878|RL6_HUMAN | 32,711 | 55 |
| Heterogeneous nuclear ribonucleoprotein L (hnRNP L) | P14866|HNRPL_HUMAN | 60,169 | 54 |
| Cathepsin B precursor (EC 3.4.22.1) (Cathepsin B1) (APP secretase) (APPS) [Contains: Cathepsin B light chain; Cathepsin B heavy chain] | P07858|CATB_HUMAN | 37,803 | 54 |
| RING finger protein 213 | Q63HN8|RN213_HUMAN | 373,963 | 54 |
| Peroxiredoxin-6 (EC 1.11.1.15) (Antioxidant protein 2) (1-Cys peroxiredoxin) (1-Cys PRX) (Acidic calcium-independent phospholipase A2) (EC 3.1.1.-) (aiPLA2) (Non-selenium glutathione peroxidase) (EC 1.11.1.7) (NSGPx) (24 kDa protein) (Liver 2D page spot 40) (Red blood cells page spot 12) | P30041|PRDX6_HUMAN | 25,018 | 54 |
| Protein CYR61 precursor (Cysteine-rich, angiogenic inducer, 61) (Insulin-like growth factor-binding protein 10) (Protein GIG1) | O00622|CYR61_HUMAN | 42,008 | 54 |
| Annexin A6 (Annexin VI) (Lipocortin VI) (P68) (P70) (Protein III) (Chromobindin-20) (67 kDa calelectrin) (Calphobindin-II) (CPB-II) | P08133|ANXA6_HUMAN | 75,860 | 53 |
| T-complex protein 1 subunit eta (TCP-1-eta) (CCT-eta) (HIV-1 Nef-interacting protein) | Q99832|TCPH_HUMAN | 59,350 | 53 |
| Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17) (RNA-dependent helicase p72) (DEAD box protein p72) | Q92841|DDX17_HUMAN | 72,355 | 53 |
| A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2) (AKAP-KL) | Q9Y2D5|AKAP2_HUMAN | 96,084 | 53 |
| Transaldolase (EC 2.2.1.2) | P37837|TALDO_HUMAN | 37,524 | 53 |
| Proteasome subunit alpha type 1 (EC 3.4.25.1) (Proteasome component C2) (Macropain subunit C2) (Multicatalytic endopeptidase complex subunit C2) (Proteasome nu chain) (30 kDa prosomal protein) (PROS-30) | P25786|PSA1_HUMAN | 29,538 | 53 |
| Nuclease sensitive element-binding protein 1 (Y-box-binding protein 1) (Y-box transcription factor) (YB-1) (CCAAT-binding transcription factor I subunit A) (CBF-A) (Enhancer factor I subunit A) (EFI-A) (DNA-binding protein B) (DBPB) | P67809|YBOX1_HUMAN | 35,906 | 53 |
| 60S ribosomal protein L17 (L23) | P18621|RL17_HUMAN | 21,379 | 53 |
| 26S protease regulatory subunit S10B (Proteasome 26S subunit ATPase 6) (Proteasome subunit p42) | P62333|PRS10_HUMAN | 44,157 | 52 |
| 60S ribosomal protein L7a (Surfeit locus protein 3) (PLA-X polypeptide) | P62424|RL7A_HUMAN | 29,978 | 52 |
| Procollagen-lysine,2-oxoglutarate 5-dioxygenase 1 precursor (EC 1.14.11.4) (Lysyl hydroxylase 1) (LH1) | Q02809|PLOD1_HUMAN | 83,535 | 52 |
| 40S ribosomal protein S18 (Ke-3) (Ke3) | P62269|RS18_HUMAN | 17,701 | 52 |
| T-complex protein 1 subunit gamma (TCP-1-gamma) (CCT-gamma) (hTRiC5) | P49368|TCPG_HUMAN | 60,517 | 51 |
| PDZ and LIM domain protein 5 (Enigma homolog) (Enigma-like PDZ and LIM domains protein) | Q96HC4|PDLI5_HUMAN | 63,953 | 51 |
| BTB/POZ domain-containing protein KCTD12 (Pfetin) (Predominantly fetal expressed T1 domain) | Q96CX2|KCD12_HUMAN | 35,684 | 51 |
| Laminin subunit gamma-1 precursor (Laminin B2 chain) | P11047|LAMC1_HUMAN | 177,587 | 50 |
| Hexokinase-1 (EC 2.7.1.1) (Hexokinase type I) (HK I) (Brain form hexokinase) | P19367|HXK1_HUMAN | 102,470 | 50 |
| Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)) | O60716|CTND1_HUMAN | 108,154 | 50 |
| 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1) | P62191|PRS4_HUMAN | 49,168 | 50 |
| 40S ribosomal protein S17 | P08708|RS17_HUMAN | 15,533 | 50 |
| Translationally-controlled tumor protein (TCTP) (p23) (Histamine-releasing factor) (HRF) (Fortilin) | P13693|TCTP_HUMAN | 19,578 | 50 |
| 60S ribosomal protein L7 | P18124|RL7_HUMAN | 29,210 | 50 |
| Reticulocalbin-1 precursor | Q15293|RCN1_HUMAN | 38,873 | 50 |
| Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP) (DNA helicase V) (HDH V) | Q96AE4|FUBP1_HUMAN | 67,543 | 49 |
| Glucose-6-phosphate 1-dehydrogenase (EC 1.1.1.49) (G6PD) | P11413|G6PD_HUMAN | 59,240 | 49 |
| 40S ribosomal protein SA (p40) (34/67 kDa laminin receptor) (Colon carcinoma laminin-binding protein) (NEM/1CHD4) (Multidrug resistance-associated protein MGr1-Ag) | P08865|RSSA_HUMAN | 32,836 | 49 |
| 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (Thyroid hormone receptor-interacting protein 1) (TRIP1) | P62195|PRS8_HUMAN | 45,609 | 49 |
| Collagen alpha-1(IV) chain precursor (Arresten) | P02462|CO4A1_HUMAN | 160,600 | 49 |
| Protein FAM62A (Membrane-bound C2 domain-containing protein) | Q9BSJ8|FA62A_HUMAN | 122,841 | 48 |
| Proteasome subunit alpha type 6 (EC 3.4.25.1) (Proteasome iota chain) (Macropain iota chain) (Multicatalytic endopeptidase complex iota chain) (27 kDa prosomal protein) (PROS-27) (p27K) | P60900|PSA6_HUMAN | 27,382 | 48 |
| Heterogeneous nuclear ribonucleoprotein F (hnRNP F) (Nucleolin-like protein mcs94-1) | P52597|HNRPF_HUMAN | 45,654 | 48 |
| Heterogeneous nuclear ribonucleoprotein G (hnRNP G) (RNA-binding motif protein, X chromosome) (Glycoprotein p43) | P38159|HNRPG_HUMAN | 42,316 | 48 |
| 60S ribosomal protein L23 (Ribosomal protein L17) | P62829|RL23_HUMAN | 14,848 | 48 |
| Serpin B6 (Placental thrombin inhibitor) (Cytoplasmic antiproteinase) (CAP) (Protease inhibitor 6) (PI-6) | P35237|SPB6_HUMAN | 42,573 | 48 |
| Thymosin beta-4 (T beta 4) (Fx) [Contains: Hematopoietic system regulatory peptide (Seraspenide)] | P62328|TYB4_HUMAN | 5,035 | 48 |
| Tubulin beta-3 chain (Tubulin beta-III) (Tubulin beta-4) | Q13509|TBB3_HUMAN | 50,415 | 47 |
| Keratin, type I cytoskeletal 18 (Cytokeratin-18) (CK-18) (Keratin-18) (K18) | P05783|K1C18_HUMAN | 48,041 | 47 |
| Septin-2 (Protein NEDD5) | Q15019|SEPT2_HUMAN | 41,470 | 47 |
| Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin] | P39060|COIA1_HUMAN | 178,142 | 47 |
| Transforming protein RhoA precursor (H12) | P61586|RHOA_HUMAN | 21,750 | 47 |
| Thymosin beta-10 | P63313|TYB10_HUMAN | 5,008 | 47 |
| Ubiquitin carboxyl-terminal hydrolase isozyme L1 (EC 3.4.19.12) (EC 6.-.-.-) (UCH-L1) (Ubiquitin thioesterase L1) (Neuron cytoplasmic protein 9.5) (PGP 9.5) (PGP9.5) | P09936|UCHL1_HUMAN | 24,806 | 46 |
| Guanine nucleotide-binding protein G(i), alpha-2 subunit (Adenylate cyclase-inhibiting G alpha protein) | P04899|GNAI2_HUMAN | 40,434 | 46 |
| Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37) | Q16543|CDC37_HUMAN | 44,450 | 46 |
| Protein DJ-1 (Oncogene DJ1) (Parkinson disease protein 7) | Q99497|PARK7_HUMAN | 19,873 | 46 |
| Aspartate aminotransferase, mitochondrial precursor (EC 2.6.1.1) (Transaminase A) (Glutamate oxaloacetate transaminase 2) | P00505|AATM_HUMAN | 47,459 | 46 |
| 60S ribosomal protein L12 | P30050|RL12_HUMAN | 17,801 | 46 |
| Histone H2B type 1-K (H2B K) (HIRA-interacting protein 1) | O60814|H2B1K_HUMAN, P57053|H2BFS_HUMAN, P58876|H2B1D_HUMAN, P62807|H2B1C_HUMAN, Q5QNW6|H2B2F_HUMAN, Q93079|H2B1H_HUMAN, Q99877|H2B1N_HUMAN, Q99879|H2B1M_HUMAN, Q99880|H2B1L_HUMAN | 13,873 | 46 |
| CAD protein [Includes: Glutamine-dependent carbamoyl-phosphate synthase (EC 6.3.5.5); Aspartate carbamoyltransferase (EC 2.1.3.2); Dihydroorotase (EC 3.5.2.3)] | P27708|PYR1_HUMAN | 242,965 | 45 |
| Integrin beta-1 precursor (Fibronectin receptor subunit beta) (Integrin VLA-4 subunit beta) (CD29 antigen) | P05556|ITB1_HUMAN | 88,447 | 45 |
| Peroxisomal multifunctional enzyme type 2 (MFE-2) (D-bifunctional protein) (DBP) (17-beta-hydroxysteroid dehydrogenase 4) (17-beta-HSD 4) (D-3-hydroxyacyl-CoA dehydratase) (EC 4.2.1.107) (3-alpha,7-alpha,12-alpha-trihydroxy-5-beta-cholest-24-enoyl-CoA hydratase) (3-hydroxyacyl-CoA dehydrogenase) (EC 1.1.1.35) | P51659|DHB4_HUMAN | 79,670 | 45 |
| Importin beta-1 subunit (Karyopherin beta-1 subunit) (Nuclear factor P97) (Importin 90) | Q14974|IMB1_HUMAN | 97,153 | 45 |
| Polypyrimidine tract-binding protein 1 (PTB) (Heterogeneous nuclear ribonucleoprotein I) (hnRNP I) (57 kDa RNA-binding protein PPTB-1) | P26599|PTBP1_HUMAN | 57,205 | 45 |
| ADP/ATP translocase 2 (Adenine nucleotide translocator 2) (ANT 2) (ADP,ATP carrier protein 2) (Solute carrier family 25 member 5) (ADP,ATP carrier protein, fibroblast isoform) | P05141|ADT2_HUMAN | 32,879 | 45 |
| 40S ribosomal protein S14 | P62263|RS14_HUMAN | 16,255 | 45 |
| Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein) | P14314|GLU2B_HUMAN | 59,408 | 45 |
| Cysteine and glycine-rich protein 1 (Cysteine-rich protein 1) (CRP1) (CRP) | P21291|CSRP1_HUMAN | 20,549 | 45 |
| Proactivator polypeptide precursor [Contains: Saposin A (Protein A); Saposin B-Val; Saposin B (Sphingolipid activator protein 1) (SAP-1) (Cerebroside sulfate activator) (CSAct) (Dispersin) (Sulfatide/GM1 activator); Saposin C (Co-beta-glucosidase) (A1 activator) (Glucosylceramidase activator) (Sphingolipid activator protein 2) (SAP-2); Saposin D (Protein C) (Component C)] | P07602|SAP_HUMAN | 58,094 | 45 |
| Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma) (hTM5) | P06753|TPM3_HUMAN | 32,802 | 45 |
| 40S ribosomal protein S5 | P46782|RS5_HUMAN | 22,859 | 45 |
| CD59 glycoprotein precursor (Membrane attack complex inhibition factor) (MACIF) (MAC-inhibitory protein) (MAC-IP) (Protectin) (MEM43 antigen) (Membrane inhibitor of reactive lysis) (MIRL) (20 kDa homologous restriction factor) (HRF-20) (HRF20) (1F5 antigen) | P13987|CD59_HUMAN | 14,159 | 45 |
| Rho-related GTP-binding protein RhoC precursor (H9) | P08134|RHOC_HUMAN | 21,989 | 45 |
| Sodium/potassium-transporting ATPase alpha-1 chain precursor (EC 3.6.3.9) (Sodium pump 1) (Na(+)/K(+) ATPase 1) | P05023|AT1A1_HUMAN | 112,882 | 44 |
| Elongation factor Tu, mitochondrial precursor (EF-Tu) (P43) | P49411|EFTU_HUMAN | 49,524 | 44 |
| GCN1-like protein 1 (HsGCN1) | Q92616|GCN1L_HUMAN | 292,732 | 44 |
| Rho GDP-dissociation inhibitor 1 (Rho GDI 1) (Rho-GDI alpha) | P52565|GDIR_HUMAN | 23,190 | 44 |
| Voltage-dependent anion-selective channel protein 1 (VDAC-1) (hVDAC1) (Outer mitochondrial membrane protein porin 1) (Plasmalemmal porin) (Porin 31HL) (Porin 31HM) | P21796|VDAC1_HUMAN | 30,756 | 44 |
| Phosphatidylethanolamine-binding protein 1 (PEBP-1) (Prostatic-binding protein) (HCNPpp) (Neuropolypeptide h3) (Raf kinase inhibitor protein) (RKIP) [Contains: Hippocampal cholinergic neurostimulating peptide (HCNP)] | P30086|PEBP1_HUMAN | 21,039 | 44 |
| 14-3-3 protein beta/alpha (Protein kinase C inhibitor protein 1) (KCIP-1) (Protein 1054) | P31946|1433B_HUMAN | 28,065 | 44 |
| Coronin-1B (Coronin-2) | Q9BR76|COR1B_HUMAN | 54,217 | 44 |
| 60S ribosomal protein L10a (CSA-19) | P62906|RL10A_HUMAN | 24,814 | 44 |
| 40S ribosomal protein S10 | P46783|RS10_HUMAN | 18,880 | 44 |
| HLA class I histocompatibility antigen, B-49 alpha chain precursor (MHC class I antigen B*49) (B-21) | P30487|1B49_HUMAN, P30488|1B50_HUMAN | 40,563 | 44 |
| Vasodilator-stimulated phosphoprotein (VASP) | P50552|VASP_HUMAN | 39,811 | 43 |
| Valyl-tRNA synthetase (EC 6.1.1.9) (Valine--tRNA ligase) (ValRS) (Protein G7a) | P26640|SYV_HUMAN | 140,460 | 43 |
| F-actin capping protein subunit beta (CapZ beta) | P47756|CAPZB_HUMAN | 31,334 | 43 |
| Actin-related protein 2/3 complex subunit 2 (ARP2/3 complex 34 kDa subunit) (p34-ARC) | O15144|ARPC2_HUMAN | 34,316 | 43 |
| Myosin-10 (Myosin heavy chain 10) (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B) | P35580|MYH10_HUMAN | 228,927 | 42 |
| Lamin-B1 | P20700|LMNB1_HUMAN | 66,392 | 42 |
| Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform (PP2A, subunit A, PR65-alpha isoform) (PP2A, subunit A, R1-alpha isoform) (Medium tumor antigen-associated 61 kDa protein) | P30153|2AAA_HUMAN | 65,207 | 42 |
| Heat-shock protein 105 kDa (Heat shock 110 kDa protein) (Antigen NY-CO-25) | Q92598|HS105_HUMAN | 96,848 | 42 |
| Annexin A3 (Annexin III) (Lipocortin III) (Placental anticoagulant protein III) (PAP-III) (35-alpha calcimedin) (Inositol 1,2-cyclic phosphate 2-phosphohydrolase) | P12429| ANXA3_HU MAN | 36,359 | 42 |
| Prohibitin | P35232|PHB_HUMAN | 29,787 | 42 |
| Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta 1 (Transducin beta chain 1) | P62873|GBB1_HUMAN | 37,360 | 42 |
| Glycyl-tRNA synthetase (EC 6.1.1.14) (Glycine--tRNA ligase) (GlyRS) | P41250|SYG_HUMAN | 83,124 | 42 |
| Caldesmon (CDM) | Q05682|CALD1_HUMAN | 93,233 | 42 |
| 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7) | P43686|PRS6B_HUMAN | 47,350 | 42 |
| 40S ribosomal protein S11 | P62280|RS11_HUMAN | 18,413 | 42 |
| Nicotinamide phosphoribosyltransferase (EC 2.4.2.12) (NAmPRTase) (Nampt) (Pre-B cell-enhancing factor) (Pre-B-cell colony-enhancing factor 1) (Visfatin) | P43490|NAMPT_HUMAN | 55,505 | 42 |
| Histone H2A type 1 (H2A.1) | P0C0S8|H2A1_HUMAN, P20671|H2A1D_HUMAN, Q16777|H2A2C_HUMAN, Q6FI13|H2A2A_HUMAN, Q96KK5|H2A1H_HUMAN, Q99878|H2A1J_HUMAN | 14,074 | 42 |
| Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135) | Q14203|DYNA_HUMAN | 141,680 | 41 |
| Myosin-Ic (Myosin I beta) (MMI-beta) (MMIb) | O00159|MYO1C_HUMAN | 118,024 | 41 |
| Coatomer subunit delta (Delta-coat protein) (Delta-COP) (Archain) | P48444|COPD_HUMAN | 57,193 | 41 |
| Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M-type) (M-calpain) (Millimolar-calpain) (Calpain large polypeptide L2) | P17655|CAN2_HUMAN | 79,995 | 41 |
| Adenosylhomocysteinase (EC 3.3.1.1) (S-adenosyl-L-homocysteine hydrolase) (AdoHcyase) | P23526|SAHH_HUMAN | 47,699 | 41 |
| Drebrin (Developmentally-regulated brain protein) | Q16643|DREB_HUMAN | 71,407 | 41 |
| Eukaryotic translation initiation factor 3 subunit 8 (eIF3 p110) (eIF3c) | Q99613|IF38_HUMAN | 105,329 | 41 |
| Coatomer subunit beta′ (Beta′-coat protein) (Beta′-COP) (p102) | P35606|COPB2_HUMAN | 102,471 | 41 |
| Proteasome subunit alpha type 7 (EC 3.4.25.1) (Proteasome subunit RC6-1) (Proteasome subunit XAPC7) | O14818|PSA7_HUMAN | 27,869 | 41 |
| 40S ribosomal protein S12 | P25398|RS12_HUMAN | 14,508 | 41 |
| 60S ribosomal protein L38 | P63173|RL38_HUMAN | 8,201 | 41 |
| Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein) | P80723|BASP_HUMAN | 22,675 | 41 |
| Histone H3.1 (H3/a) (H3/b) (H3/c) (H3/d) (H3/f) (H3/h) (H3/i) (H3/j) (H3/k) (H3/l) | P68431|H31_HUMAN, P84243|H33_HUMAN, Q71DI3|H32_HUMAN | 15,387 | 41 |
| HLA class I histocompatibility antigen, B-8 alpha chain precursor (MHC class I antigen B*8) | P30460|1B08_HUMAN | 40,313 | 41 |
| High mobility group protein B2 (High mobility group protein 2) (HMG-2) | P26583|HMGB2_HUMAN | 24,017 | 40 |
| Calponin-2 (Calponin H2, smooth muscle) (Neutral calponin) | Q99439|CNN2_HUMAN | 33,680 | 40 |
| 40S ribosomal protein S13 | P62277|RS13_HUMAN | 17,205 | 40 |
| Elongation factor 1-delta (EF-1-delta) (Antigen NY-CO-4) | P29692|EF1D_HUMAN | 31,104 | 40 |
| Coatomer subunit gamma (Gamma-coat protein) (Gamma-COP) | Q9Y678|COPG_HUMAN | 97,701 | 39 |
| Ras-interacting protein 1 (Rain) | Q5U651|RAIN_HUMAN | 103,442 | 39 |
| Histone H1.5 (Histone H1a) | P16401|H15_HUMAN | 22,564 | 39 |
| Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2) | Q15075|EEA1_HUMAN | 162,450 | 39 |
| Trifunctional enzyme subunit beta, mitochondrial precursor (TP-beta) [Includes: 3-ketoacyl-CoA thiolase (EC 2.3.1.16) (Acetyl-CoA acyltransferase) (Beta-ketothiolase)] | P55084|ECHB_HUMAN | 51,278 | 39 |
| F-actin capping protein subunit alpha-1 (CapZ alpha-1) | P52907|CAZA1_HUMAN | 32,905 | 39 |
| Leucine-rich repeat-containing protein 59 | Q96AG4|LRC59_HUMAN | 34,913 | 39 |
| Isocitrate dehydrogenase [NADP], mitochondrial precursor (EC 1.1.1.42) (Oxalosuccinate decarboxylase) (IDH) (NADP(+)-specific ICDH) (IDP) (ICD-M) | P48735|IDHP_HUMAN | 50,892 | 39 |
| Synaptic vesicle membrane protein VAT-1 homolog (EC 1.-.-.-) | Q99536|VAT1_HUMAN | 41,902 | 39 |
| Calumenin precursor (Crocalbin) (IEF SSP 9302) | O43852|CALU_HUMAN | 37,090 | 39 |
| Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220) | Q04637|IF4G1_HUMAN | 175,520 | 38 |
| UPF0027 protein C22orf28 | Q9Y3I0|CV028_HUMAN | 55,192 | 38 |
| Src substrate cortactin (Amplaxin) (Oncogene EMS1) | Q14247|SRC8_HUMAN | 61,618 | 38 |
| Acidic leucine-rich nuclear phosphoprotein 32 family member A (Potent heat-stable protein phosphatase 2A inhibitor I1PP2A) (Acidic nuclear phosphoprotein pp32) (Leucine-rich acidic nuclear protein) (Lanp) (Putative HLA-DR-associated protein I) (PHAPI) (Mapmodulin) | P39687|AN32A_HUMAN | 28,568 | 38 |
| Endoglin precursor (CD105 antigen) | P17813|EGLN_HUMAN | 70,561 | 38 |
| Hematopoietic lineage cell-specific protein (Hematopoietic cell-specific LYN substrate 1) (LckBP1) (p75) | P14317|HCLS1_HUMAN | 53,979 | 38 |
| Alpha-centractin (Centractin) (Centrosome-associated actin homolog) (Actin-RPV) (ARP1) | P61163|ACTZ_HUMAN | 42,597 | 38 |
| 60S ribosomal protein L27a | P46776|RL27A_HUMAN | 16,543 | 38 |
| Catenin beta-1 (Beta-catenin) | P35222|CTNB1_HUMAN | 85,479 | 38 |
| 60S ribosomal protein L22 (Epstein-Barr virus small RNA-associated protein) (EBER-associated protein) (EAP) (Heparin-binding protein HBp15) | P35268|RL22_HUMAN | 14,769 | 38 |
| Small nuclear ribonucleoprotein Sm D2 (snRNP core protein D2) (Sm-D2) | P62316|SMD2_HUMAN | 13,509 | 38 |
| SAM domain and HD domain-containing protein 1 (Dendritic cell-derived IFNG-induced protein) (DCIP) (Monocyte protein 5) (MOP-5) | Q9Y3Z3|SAMH1_HUMAN | 72,185 | 38 |
| Beta crystallin B1 | P53674|CRBB1_HUMAN | 28,006 | 38 |
| Elongation factor 1-beta (EF-1-beta) | P24534|EF1B_HUMAN | 24,746 | 38 |
| 130 kDa leucine-rich protein (LRP 130) (GP130) (Leucine-rich PPR motif-containing protein) | P42704|LPPRC_HUMAN | 145,190 | 37 |
| Arginyl-tRNA synthetase, cytoplasmic (EC 6.1.1.19) (Arginine--tRNA ligase) (ArgRS) | P54136|SYRC_HUMAN | 75,364 | 37 |
| Heterogeneous nuclear ribonucleoprotein R (hnRNP R) | O43390|HNRPR_HUMAN | 70,926 | 37 |
| ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1) (DEAD box protein retinoblastoma) (DBP-RB) | Q92499|DDX1_HUMAN | 82,415 | 37 |
| 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (Tat-binding protein 1) (TBP-1) (Proteasome subunit P50) | P17980|PRS6A_HUMAN | 49,187 | 37 |
| Splicing factor 3B subunit 2 (Spliceosome-associated protein 145) (SAP 145) (SF3b150) (Pre-mRNA-splicing factor SF3b 145 kDa subunit) | Q13435|SF3B2_HUMAN | 97,641 | 37 |
| Eukaryotic translation initiation factor 2 subunit 1 (Eukaryotic translation initiation factor 2 subunit alpha) (eIF-2-alpha) (EIF-2alpha) (EIF-2A) | P05198|IF2A_HUMAN | 36,095 | 37 |
| Ras GTPase-activating protein-binding protein 1 (EC 3.6.1.-) (G3BP-1) (ATP-dependent DNA helicase VIII) (HDH-VIII) (GAP SH3 domain-binding protein 1) | Q13283|G3BP1_HUMAN | 52,145 | 37 |
| Tropomyosin-1 alpha chain (Alpha-tropomyosin) | P09493|TPM1_HUMAN | 32,692 | 37 |
| Transmembrane emp24 domain-containing protein 10 precursor (Transmembrane protein Tmp21) (21 kDa transmembrane-trafficking protein) (p24delta) (S31III125) (S31I125) (Tmp-21-I) | P49755|TMEDA_HUMAN | 24,960 | 37 |
| 60S ribosomal protein L11 (CLL-associated antigen KW-12) | P62913|RL11_HUMAN | 20,235 | 37 |
| FK506-binding protein 1A (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (12 kDa FKBP) (FKBP-12) (Immunophilin FKBP12) | P62942|FKB1A_HUMAN | 11,933 | 37 |
| Phosphoribosylformylglycinamidine synthase (EC 6.3.5.3) (FGAM synthase) (FGAMS) (Formylglycinamide ribotide amidotransferase) (FGARAT) (Formylglycinamide ribotide synthetase) | O15067|PUR4_HUMAN | 144,646 | 36 |
| Matrin-3 | P43243|MATR3_HUMAN | 94,609 | 36 |
| Trifunctional purine biosynthetic protein adenosine-3 [Includes: Phosphoribosylamine--glycine ligase (EC 6.3.4.13) (GARS) (Glycinamide ribonucleotide synthetase) (Phosphoribosylglycinamide synthetase); Phosphoribosylformylglycinamidine cyclo-ligase (EC 6.3.3.1) (AIRS) (Phosphoribosyl-aminoimidazole synthetase) (AIR synthase); Phosphoribosylglycinamide formyltransferase (EC 2.1.2.2) (GART) (GAR transformylase) (5′-phosphoribosylglycinamide transformylase)] | P22102|PUR2_HUMAN | 107,750 | 36 |
| Very-long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.-) (VLCAD) | P49748|ACADV_HUMAN | 70,374 | 36 |
| 3-ketoacyl-CoA thiolase, mitochondrial (EC 2.3.1.16) (Beta-ketothiolase) (Acetyl-CoA acyltransferase) (Mitochondrial 3-oxoacyl-CoA thiolase) (T1) | P42765|THIM_HUMAN | 41,906 | 36 |
| Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) | Q14152|IF3A_HUMAN | 166,557 | 36 |
| 60S ribosomal protein L13 (Breast basic conserved protein 1) | P26373|RL13_HUMAN | 24,244 | 36 |
| Multimerin-2 precursor (EMILIN-3) (Elastin microfibril interface located protein 3) (Elastin microfibril interfacer 3) (EndoGlyx-1 p125/p140 subunit) | Q9H8L6|MMRN2_HUMAN | 104,398 | 36 |
| Integrin alpha-5 precursor (Fibronectin receptor subunit alpha) (Integrin alpha-F) (VLA-5) (CD49e antigen) [Contains: Integrin alpha-5 heavy chain; Integrin alpha-5 light chain] | P08648|ITA5_HUMAN | 114,521 | 36 |
| 26S proteasome non-ATPase regulatory subunit 6 (26S proteasome regulatory subunit S10) (p42A) (Proteasome regulatory particle subunit p44S10) (Phosphonoformate immuno-associated protein 4) (Breast cancer-associated protein SGA-113M) | Q15008|PSMD6_HUMAN | 45,515 | 36 |
| Procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 precursor (EC 1.14.11.4) (Lysyl hydroxylase 2) (LH2) | O00469|PLOD2_HUMAN | 84,669 | 36 |
| Lupus La protein (Sjoegren syndrome type B antigen) (SS-B) (La ribonucleoprotein) (La autoantigen) | P05455|LA_HUMAN | 46,821 | 36 |
| UMP-CMP kinase (EC 2.7.4.14) (Cytidylate kinase) (Deoxycytidylate kinase) (Cytidine monophosphate kinase) (Uridine monophosphate/cytidine monophosphate kinase) (UMP/CMP kinase) (UMP/CMPK) (Uridine monophosphate kinase) | P30085|KCY_HUMAN | 22,205 | 36 |
| 60S ribosomal protein L26-like 1 | Q9UNX3|RL26L_HUMAN | 17,238 | 36 |
| 14-3-3 protein theta (14-3-3 protein tau) (14-3-3 protein T-cell) (HS1 protein) | P27348|1433T_HUMAN | 27,747 | 36 |
| Ras-related protein Ral-A precursor | P11233|RALA_HUMAN | 23,549 | 36 |
| Heterogeneous nuclear ribonucleoprotein U-like protein 2 (Scaffold-attachment factor A2) (SAF-A2) | Q1KMD3|HNRL2_HUMAN | 85,087 | 35 |
| ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X-chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform) | O00571|DDX3X_HUMAN | 73,228 | 35 |
| Inorganic pyrophosphatase (EC 3.6.1.1) (Pyrophosphate phospho-hydrolase) (PPase) | Q15181|IPYR_HUMAN | 32,643 | 35 |
| Cullin-associated NEDD8-dissociated protein 1 (Cullin-associated and neddylation-dissociated protein 1) (p120 CAND1) (TBP-interacting protein TIP120A) (TBP-interacting protein of 120 kDa A) | Q86VP6|CAND1_HUMAN | 136,363 | 35 |
| Putative GTP-binding protein 9 | Q9NTK5|GTPB9_HUMAN | 44,727 | 35 |
| 60S ribosomal protein L13a (23 kDa highly basic protein) | P40429|RL13A_HUMAN | 23,560 | 35 |
| Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (EC 3.6.3.8) (Calcium pump 2) (SERCA2) (SR Ca(2+)-ATPase 2) (Calcium-transporting ATPase sarcoplasmic reticulum type, slow twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase) | P16615|AT2A2_HUMAN | 114,741 | 34 |
| Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T) | P45974|UBP5_HUMAN | 95,770 | 34 |
| 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1) | P35998|PRS7_HUMAN | 48,618 | 34 |
| UDP-glucose:glycoprotein glucosyltransferase 1 precursor (EC 2.4.1.-) (UDP-glucose ceramide glucosyltransferase-like 1) (UDP--Glc:glycoprotein glucosyltransferase) (HUGT1) | Q9NYU2|UGGG1_HUMAN | 174,965 | 34 |
| 26S proteasome non-ATPase regulatory subunit 2 (26S proteasome regulatory subunit RPN1) (26S proteasome regulatory subunit S2) (26S proteasome subunit p97) (Tumor necrosis factor type 1 receptor-associated protein 2) (55.11 protein) | Q13200|PSMD2_HUMAN | 100,184 | 34 |
| UDP-glucose 6-dehydrogenase (EC 1.1.1.22) (UDP-Glc dehydrogenase) (UDP-GlcDH) (UDPGDH) | O60701|UGDH_HUMAN | 55,007 | 34 |
| Peroxiredoxin-2 (EC 1.11.1.15) (Thioredoxin peroxidase 1) (Thioredoxin-dependent peroxide reductase 1) (Thiol-specific antioxidant protein) (TSA) (PRP) (Natural killer cell-enhancing factor B) (NKEF-B) | P32119|PRDX2_HUMAN | 21,874 | 34 |
| Endoplasmic reticulum protein ERp29 precursor (ERp31) (ERp28) | P30040|ERP29_HUMAN | 28,977 | 34 |
| Asparaginyl-tRNA synthetase, cytoplasmic (EC 6.1.1.22) (Asparagine--tRNA ligase) (AsnRS) | O43776|SYNC_HUMAN | 62,926 | 34 |
| 26S proteasome non-ATPase regulatory subunit 11 (26S proteasome regulatory subunit S9) (26S proteasome regulatory subunit p44.5) | O00231|PSD11_HUMAN | 47,448 | 34 |
| 60S ribosomal protein L23a | P62750|RL23A_HUMAN | 17,678 | 34 |
| Ubiquitin thioesterase protein OTUB1 (EC 3.4.-.-) (Otubain-1) (OTU domain-containing ubiquitin aldehyde-binding protein 1) (Ubiquitin-specific-processing protease OTUB1) (Deubiquitinating enzyme OTUB1) | Q96FW1|OTUB1_HUMAN | 31,267 | 34 |
| Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein) (Foocen) (Neuroendocrine-specific protein) (NSP) (Neuroendocrine-specific protein C homolog) (RTN-x) (Reticulon-5) | Q9NQC3|RTN4_HUMAN | 129,917 | 34 |
| Plexin-D1 precursor | Q9Y4D7|PLXD1_HUMAN | 212,078 | 33 |
| 116 kDa U5 small nuclear ribonucleoprotein component (U5 snRNP-specific protein, 116 kDa) (U5-116 kDa) (Elongation factor Tu GTP-binding domain protein 2) (hSNU114) | Q15029|U5S1_HUMAN | 109,420 | 33 |
| Golgi apparatus protein 1 precursor (Golgi sialoglycoprotein MG-160) (E-selectin ligand 1) (ESL-1) (Cysteine-rich fibroblast growth factor receptor) (CFR-1) | Q92896|GSLG1_HUMAN | 134,577 | 33 |
| Carbonyl reductase [NADPH] 1 (EC 1.1.1.184) (NADPH-dependent carbonyl reductase 1) (Prostaglandin-E(2) 9-reductase) (EC 1.1.1.189) (Prostaglandin 9-ketoreductase) (15-hydroxyprostaglandin dehydrogenase [NADP+]) (EC 1.1.1.197) | P16152|DHCA_HUMAN | 30,357 | 33 |
| Prohibitin-2 (B-cell receptor-associated protein BAP37) (Repressor of estrogen receptor activity) (D-prohibitin) | Q99623|PHB2_HUMAN | 33,280 | 33 |
| Cathepsin D precursor (EC 3.4.23.5) [Contains: Cathepsin D light chain; Cathepsin D heavy chain] | P07339|CATD_HUMAN | 44,535 | 33 |
| Glutamate dehydrogenase 1, mitochondrial precursor (EC 1.4.1.3) (GDH) | P00367|DHE3_HUMAN | 61,382 | 33 |
| Microtubule-associated protein RP/EB family member 1 (APC-binding protein EB1) (End-binding protein 1) (EB1) | Q15691|MARE1_HUMAN | 29,982 | 33 |
| Calpain-1 catalytic subunit (EC 3.4.22.52) (Calpain-1 large subunit) (Calcium-activated neutral proteinase 1) (CANP 1) (Calpain mu-type) (muCANP) (Micromolar-calpain) | P07384|CAN1_HUMAN | 81,875 | 33 |
| Septin-11 | Q9NVA2|SEP11_HUMAN | 49,381 | 33 |
| Coatomer subunit beta (Beta-coat protein) (Beta-COP) | P53618|COPB_HUMAN | 107,128 | 33 |
| Proteasome activator complex subunit 1 (Proteasome activator 28-alpha subunit) (PA28alpha) (PA28a) (Activator of multicatalytic protease subunit 1) (11S regulator complex subunit alpha) (REG-alpha) (Interferon gamma up-regulated I-5111 protein) (IGUP I-5111) | Q06323|PSME1_HUMAN | 28,706 | 33 |
| Splicing factor, arginine/serine-rich 1 (pre-mRNA-splicing factor SF2, P33 subunit) (Alternative-splicing factor 1) (ASF-1) | Q07955|SFRS1_HUMAN | 27,727 | 33 |
| 60S ribosomal protein L32 | P62910|RL32_HUMAN | 15,842 | 33 |
| 60S ribosomal protein L21 | P46778|RL21_HUMAN | 18,547 | 33 |
| 40S ribosomal protein S19 | P39019|RS19_HUMAN | 16,043 | 33 |
| Protein transport protein Sec23A (SEC23-related protein A) | Q15436|SC23A_HUMAN | 86,145 | 32 |
| AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha-1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit) | O95782|AP2A1_HUMAN | 107,540 | 32 |
| Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116) (eIF3 p110) (eIF3b) (Prt1 homolog) (hPrt1) | P55884|IF39_HUMAN | 92,475 | 32 |
| Signal transducer and activator of transcription 1-alpha/beta (Transcription factor ISGF-3 components p91/p84) | P42224|STAT1_HUMAN | 87,319 | 32 |
| 40S ribosomal protein S9 | P46781|RS9_HUMAN | 22,575 | 32 |
| Proteasome subunit alpha type 2 (EC 3.4.25.1) (Proteasome component C3) (Macropain subunit C3) (Multicatalytic endopeptidase complex subunit C3) | P25787|PSA2_HUMAN | 25,881 | 32 |
| Multifunctional protein ADE2 [Includes: Phosphoribosylaminoimidazole-succinocarboxamide synthase (EC 6.3.2.6) (SAICAR synthetase); Phosphoribosylaminoimidazole carboxylase (EC 4.1.1.21) (AIR carboxylase) (AIRC)] | P22234|PUR6_HUMAN | 47,062 | 32 |
| Malate dehydrogenase, cytoplasmic (EC 1.1.1.37) (Cytosolic malate dehydrogenase) | P40925|MDHC_HUMAN | 36,409 | 32 |
| Nuclear autoantigenic sperm protein (NASP) | P49321|NASP_HUMAN | 85,218 | 31 |
| Ran GTPase-activating protein 1 | P46060|RGP1_HUMAN | 63,525 | 31 |
| Heterogeneous nuclear ribonucleoprotein H3 (hnRNP H3) (hnRNP 2H9) | P31942|HNRH3_HUMAN | 36,910 | 31 |
| Cysteine-rich protein 2 (CRP2) (Protein ESP1) | P52943|CRIP2_HUMAN | 22,475 | 31 |
| Serpin B9 (Cytoplasmic antiproteinase 3) (CAP-3) (CAP3) (Protease inhibitor 9) | P50453|SPB9_HUMAN | 42,386 | 31 |
| Agrin precursor | O00468|AGRIN_HUMAN | 214,820 | 31 |
| Thioredoxin-dependent peroxide reductase, mitochondrial precursor (EC 1.11.1.15) (Peroxiredoxin-3) (PRX III) (Antioxidant protein 1) (AOP-1) (Protein MER5 homolog) (HBC189) | P30048|PRDX3_HUMAN | 27,675 | 31 |
| Interferon-induced guanylate-binding protein 1 (GTP-binding protein 1) (Guanine nucleotide-binding protein 1) (GBP-1) (HuGBP-1) | P32455|GBP1_HUMAN | 67,886 | 31 |
| Mannose-6-phosphate receptor-binding protein 1 (Cargo selection protein TIP47) (47 kDa mannose 6-phosphate receptor-binding protein) (47 kDa MPR-binding protein) (Placental protein 17) (PP17) | O60664|M6PBP_HUMAN | 47,028 | 30 |
| Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit) | O75533|SF3B1_HUMAN | 145,802 | 30 |
| Probable transcription factor PML (Tripartite motif-containing protein 19) (RING finger protein 71) | P29590|PML_HUMAN | 97,499 | 30 |
| RuvB-like 2 (EC 3.6.1.-) (48 kDa TATA box-binding protein-interacting protein) (48 kDa TBP-interacting protein) (TIP49b) (Repressing pontin 52) (Reptin 52) (51 kDa erythrocyte cytosolic protein) (ECP-51) (TIP60-associated protein 54-beta) (TAP54-beta) | Q9Y230|RUVB2_HUMAN | 51,140 | 30 |
| Integrin-linked protein kinase (EC 2.7.11.1) (ILK-1) (ILK-2) (59 kDa serine/threonine-protein kinase) (p59ILK) | Q13418|ILK_HUMAN | 51,402 | 30 |
| Aconitate hydratase, mitochondrial precursor (EC 4.2.1.3) (Citrate hydro-lyase) (Aconitase) | Q99798|ACON_HUMAN | 85,410 | 30 |
| Tyrosyl-tRNA synthetase, cytoplasmic (EC 6.1.1.1) (Tyrosyl--tRNA ligase) (TyrRS) | P54577|SYYC_HUMAN | 59,127 | 30 |
| Glutaminyl-tRNA synthetase (EC 6.1.1.18) (Glutamine--tRNA ligase) (GlnRS) | P47897|SYQ_HUMAN | 87,782 | 30 |
| Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2) | O60841|IF2P_HUMAN | 138,786 | 30 |
| 26S proteasome non-ATPase regulatory subunit 3 (26S proteasome regulatory subunit S3) (Proteasome subunit p58) | O43242|PSMD3_HUMAN | 60,962 | 30 |
| Serine-threonine kinase receptor-associated protein (UNR-interacting protein) (WD-40 repeat protein PT-WD) (MAP activator with WD repeats) | Q9Y3F4|STRAP_HUMAN | 38,421 | 30 |
| 1,4-alpha-glucan branching enzyme (EC 2.4.1.18) (Glycogen branching enzyme) (Brancher enzyme) | Q04446|GLGB_HUMAN | 80,445 | 30 |
| Aspartyl-tRNA synthetase, cytoplasmic (EC 6.1.1.12) (Aspartate--tRNA ligase) (AspRS) (Cell proliferation-inducing gene 40 protein) | P14868|SYDC_HUMAN | 57,119 | 30 |
| 60S ribosomal protein L18 | Q07020|RL18_HUMAN | 21,617 | 30 |
| 40S ribosomal protein S16 | P62249|RS16_HUMAN | 16,428 | 30 |
| 40S ribosomal protein S25 | P62851|RS25_HUMAN | 13,725 | 30 |
| 60S ribosomal protein L14 (CAG-ISL 7) | P50914|RL14_HUMAN | 23,272 | 30 |
| Ubiquinol-cytochrome-c reductase complex core protein 1, mitochondrial precursor (EC 1.10.2.2) (Core I protein) | P31930|UQCR1_HUMAN | 52,628 | 29 |
| LIM and SH3 domain protein 1 (LASP-1) (MLN 50) | Q14847|LASP1_HUMAN | 29,699 | 29 |
| Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up-frameshift suppressor 1 homolog) (hUpf1) | Q92900|RENT1_HUMAN | 124,329 | 29 |
| Integrin alpha-2 precursor (Platelet membrane glycoprotein Ia) (GPIa) (Collagen receptor) (VLA-2 alpha chain) (CD49b antigen) | P17301|ITA2_HUMAN | 129,280 | 29 |
| DNA-(apurinic or apyrimidinic site) lyase (EC 4.2.99.18) (AP endonuclease 1) (APEX nuclease) (APEN) (REF-1 protein) | P27695|APEX1_HUMAN | 35,538 | 29 |
| 14-3-3 protein eta (Protein AS1) | Q04917|1433F_HUMAN | 28,202 | 29 |
| Splicing factor 3B subunit 3 (Spliceosome-associated protein 130) (SAP 130) (SF3b130) (Pre-mRNA-splicing factor SF3b 130 kDa subunit) (STAF130) | Q15393|SF3B3_HUMAN | 135,561 | 29 |
| FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59) | Q02790|FKBP4_HUMAN | 51,788 | 29 |
| Small nuclear ribonucleoprotein-associated proteins B and B′ (snRNP-B) (Sm protein B/B′) (Sm-B/Sm-B′) (SmB/SmB′) | P14678|RSMB_HUMAN, P63162|RSMN_HUMAN | 24,593 | 29 |
| Superoxide dismutase [Cu-Zn] (EC 1.15.1.1) | P00441|SODC_HUMAN | 15,917 | 29 |
| 60S ribosomal protein L24 (Ribosomal protein L30) | P83731|RL24_HUMAN | 17,762 | 29 |
| Serine/threonine-protein phosphatase PP1-beta catalytic subunit (EC 3.1.3.16) (PP-1B) | P62140|PP1B_HUMAN | 37,170 | 29 |
| Beta-2-microglobulin precursor [Contains: Beta-2-microglobulin variant pI 5.3] | P61769|B2MG_HUMAN | 13,697 | 29 |
| CD44 antigen precursor (Phagocytic glycoprotein I) (PGP-1) (HUTCH-I) (Extracellular matrix receptor-III) (ECMR-III) (GP90 lymphocyte homing/adhesion receptor) (Hermes antigen) (Hyaluronate receptor) (Heparan sulfate proteoglycan) (Epican) (CDw44) | P16070|CD44_HUMAN | 81,535 | 29 |
| Nucleoprotein TPR | P12270|TPR_HUMAN | 265,580 | 28 |
| Dynactin subunit 2 (Dynactin complex 50 kDa subunit) (50 kDa dynein-associated polypeptide) (p50 dynamitin) (DCTN-50) | Q13561|DCTN2_HUMAN | 44,214 | 28 |
| Acetyl-CoA acetyltransferase, mitochondrial precursor (EC 2.3.1.9) (Acetoacetyl-CoA thiolase) (T2) | P24752|THIL_HUMAN | 45,182 | 28 |
| Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21) | Q9NR30|DDX21_HUMAN | 87,328 | 28 |
| Serine hydroxymethyltransferase, mitochondrial precursor (EC 2.1.2.1) (Serine methylase) (Glycine hydroxymethyltransferase) (SHMT) | P34897|GLYM_HUMAN | 55,977 | 28 |
| Threonyl-tRNA synthetase, cytoplasmic (EC 6.1.1.3) (Threonine--tRNA ligase) (ThrRS) | P26639|SYTC_HUMAN | 83,420 | 28 |
| Eukaryotic translation initiation factor 3 subunit 7 (eIF-3 zeta) (eIF3 p66) (eIF3d) | O15371|IF37_HUMAN | 63,956 | 28 |
| Protein-L-isoaspartate(D-aspartate) O-methyltransferase (EC 2.1.1.77) (Protein-beta-aspartate methyltransferase) (PIMT) (Protein L-isoaspartyl/D-aspartyl methyltransferase) (L-isoaspartyl protein carboxyl methyltransferase) | P22061|PIMT_HUMAN | 24,633 | 28 |
| ATP-binding cassette sub-family E member 1 (RNase L inhibitor) (Ribonuclease 4 inhibitor) (RNS4I) (2′-5′ oligoadenylate-binding protein) (HuHP68) | P61221|ABCE1_HUMAN | 67,298 | 28 |
| 60S ribosomal protein L9 | P32969|RL9_HUMAN | 21,846 | 28 |
| Core histone macro-H2A.1 (Histone macroH2A1) (mH2A1) (H2A.y) (H2A/y) (Medulloblastoma antigen MU-MB-50.205) | O75367|H2AY_HUMAN | 39,601 | 28 |
| 26S proteasome non-ATPase regulatory subunit 1 (26S proteasome regulatory subunit RPN2) (26S proteasome regulatory subunit S1) (26S proteasome subunit p112) | Q99460|PSMD1_HUMAN | 105,821 | 28 |
| Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase) (DBI-related protein 1) (DRS-1) (Hepatocellular carcinoma-associated antigen 88) (Renal carcinoma antigen NY-REN-1) | O75521|PECI_HUMAN | 39,592 | 28 |
| DNA damage-binding protein 1 (Damage-specific DNA-binding protein 1) (UV-damaged DNA-binding factor) (DDB p127 subunit) (DDBa) (UV-damaged DNA-binding protein 1) (UV-DDB 1) (Xeroderma pigmentosum group E-complementing protein) (XPCe) (XPE-binding factor) (XPE-BF) (X-associated protein 1) (XAP-1) | Q16531|DDB1_HUMAN | 126,952 | 28 |
| Purine nucleoside phosphorylase (EC 2.4.2.1) (Inosine phosphorylase) (PNP) | P00491|PNPH_HUMAN | 32,100 | 28 |
| Thioredoxin (Trx) (ATL-derived factor) (ADF) (Surface-associated sulphydryl protein) (SASP) | P10599|THIO_HUMAN | 11,719 | 28 |
| Stabilin-1 precursor (Fasciclin, EGF-like, laminin-type EGF-like and link domain-containing scavenger receptor 1) (FEEL-1) (MS-1 antigen) | Q9NY15|STAB1_HUMAN | 275,449 | 28 |
| Putative RNA-binding protein 3 (RNA-binding motif protein 3) (RNPL) | P98179|RBM3_HUMAN | 17,153 | 28 |
| GTPase IMAP family member 4 (Immunity-associated protein 4) (Immunity-associated nucleotide 1 protein) (hIAN1) | Q9NUV9|GIMA4_HUMAN | 37,517 | 28 |
| Aldo-keto reductase family 1 member C3 (EC 1.-.-.-) (Trans-1,2-dihydrobenzene-1,2-diol dehydrogenase) (EC 1.3.1.20) (3-alpha-hydroxysteroid dehydrogenase type 2) (EC 1.1.1.213) (3-alpha-HSD type 2) (3-alpha-HSD type II, brain) (Prostaglandin F synthase) (EC 1.1.1.188) (PGFS) (Estradiol 17-beta-dehydrogenase) (EC 1.1.1.62) (17-beta-hydroxysteroid dehydrogenase type 5) (17-beta-HSD 5) (Chlordecone reductase homolog HAKRb) (HA1753) (Dihydrodiol dehydrogenase type I) (Dihydrodiol dehydrogenase 3) (DD3) (DD-3) | P42330|AK1C3_HUMAN | 36,827 | 28 |
| Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22) | P16949|STMN1_HUMAN | 17,285 | 28 |
| Cystatin-B (Stefin-B) (Liver thiol proteinase inhibitor) (CPI-B) | P04080|CYTB_HUMAN | 11,121 | 28 |
| U5 small nuclear ribonucleoprotein 200 kDa helicase (EC 3.6.1.-) (U5 snRNP-specific 200 kDa protein) (U5-200KD) (Activating signal cointegrator 1 complex subunit 3-like 1) (BRR2 homolog) | O75643|U520_HUMAN | 244,496 | 27 |
| Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) | Q9P0K7|RAI14_HUMAN | 110,025 | 27 |
| Paxillin | P49023|PAXI_HUMAN | 64,515 | 27 |
| Nuclear migration protein nudC (Nuclear distribution protein C homolog) | Q9Y266|NUDC_HUMAN | 38,226 | 27 |
| Splicing factor 3 subunit 1 (Spliceosome-associated protein 114) (SAP 114) (SF3a120) | Q15459|SF3A1_HUMAN | 88,868 | 27 |
| 60S ribosomal protein L18a | Q02543|RL18A_HUMAN | 20,745 | 27 |
| Sequestosome-1 (Phosphotyrosine-independent ligand for the Lck SH2 domain of 62 kDa) (Ubiquitin-binding protein p62) (EBI3-associated protein of 60 kDa) (p60) (EBIAP) | Q13501|SQSTM_HUMAN | 47,669 | 27 |
| Presequence protease, mitochondrial precursor (EC 3.4.24.-) (hPreP) (Pitrilysin metalloproteinase 1) (Metalloprotease 1) (hMP1) | Q5JRX3|PREP_HUMAN | 117,439 | 27 |
| Ca(2+)/calmodulin-dependent protein kinase phosphatase (EC 3.1.3.16) (CaM-kinase phosphatase) (CaMKPase) (Partner of PIX 2) (hFEM-2) (Protein phosphatase 1F) | P49593|PPM1F_HUMAN | 49,812 | 27 |
| Protein SET (Phosphatase 2A inhibitor I2PP2A) (I-2PP2A) (Template-activating factor I) (TAF-I) (HLA-DR-associated protein II) (PHAPII) (Inhibitor of granzyme A-activated DNase) (IGAAD) | Q01105|SET_HUMAN | 33,471 | 27 |
| Proteasome activator complex subunit 2 (Proteasome activator 28-subunit beta) (PA28beta) (PA28b) (Activator of multicatalytic protease subunit 2) (11S regulator complex subunit beta) (REG-beta) | Q9UL46|PSME2_HUMAN | 27,344 | 27 |
| EGF-containing fibulin-like extracellular matrix protein 1 precursor (Fibulin-3) (FIBL-3) (Fibrillin-like protein) (Extracellular protein S1-5) | Q12805|FBLN3_HUMAN | 54,621 | 27 |
| Prostaglandin E synthase 3 (EC 5.3.99.3) (Cytosolic prostaglandin E2 synthase) (cPGES) (Telomerase-binding protein p23) (Hsp90 co-chaperone) (Progesterone receptor complex p23) | Q15185|TEBP_HUMAN | 18,680 | 27 |
| 60S ribosomal protein L27 | P61353|RL27_HUMAN | 15,780 | 27 |
| Cathepsin Z precursor (EC 3.4.22.-) (Cathepsin X) (Cathepsin P) | Q9UBR2|CATZ_HUMAN | 33,850 | 27 |
| Laminin subunit alpha-4 precursor | Q16363|LAMA4_HUMAN | 202,512 | 26 |
| Tubulointerstitial nephritis antigen-like precursor (Tubulointerstitial nephritis antigen-related protein) (TIN Ag-related protein) (TIN-Ag-RP) (Glucocorticoid-inducible protein 5) (Oxidized LDL-responsive gene 2 protein) (OLRG-2) | Q9GZM7|TINAL_HUMAN | 52,369 | 26 |
| Kinectin (Kinesin receptor) (CG-1 antigen) | Q86UP2|KTN1_HUMAN | 156,258 | 26 |
| Septin-7 (CDC10 protein homolog) | Q16181|SEPT7_HUMAN | 50,662 | 26 |
| Proliferating cell nuclear antigen (PCNA) (Cyclin) | P12004|PCNA_HUMAN | 28,751 | 26 |
| Eukaryotic translation initiation factor 3 subunit 2 (eIF-3 beta) (eIF3 p36) (eIF3i) (TGF-beta receptor-interacting protein 1) (TRIP-1) | Q13347|IF32_HUMAN | 36,484 | 26 |
| Serine/threonine-protein phosphatase PP1-alpha catalytic subunit (EC 3.1.3.16) (PP-1A) | P62136|PP1A_HUMAN | 37,496 | 26 |
| Ubiquitin-associated protein 2-like (Protein NICE-4) | Q14157|UBP2L_HUMAN | 114,516 | 26 |
| Electron transfer flavoprotein subunit alpha, mitochondrial precursor (Alpha-ETF) | P13804|ETFA_HUMAN | 35,062 | 26 |
| Dynamin-2 (EC 3.6.5.5) | P50570|DYN2_HUMAN | 98,050 | 26 |
| Heterogeneous nuclear ribonucleoprotein U-like protein 1 (Adenovirus early region 1B-associated protein 5) (E1B-55 kDa-associated protein 5) (E1B-AP5) | Q9BUJ2|HNRL1_HUMAN | 95,722 | 26 |
| Cytoplasmic dynein 1 intermediate chain 2 (Dynein intermediate chain 2, cytosolic) (DH IC-2) (Cytoplasmic dynein intermediate chain 2) | Q13409|DC1I2_HUMAN | 71,438 | 26 |
| AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B) | P63010|AP2B1_HUMAN | 104,537 | 26 |
| ELAV-like protein 1 (Hu-antigen R) (HuR) | Q15717|ELAV1_HUMAN | 36,075 | 26 |
| Methionyl-tRNA synthetase, cytoplasmic (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS) | P56192|SYMC_HUMAN | 101,100 | 26 |
| Fumarate hydratase, mitochondrial precursor (EC 4.2.1.2) (Fumarase) | P07954|FUMH_HUMAN | 54,620 | 26 |
| Phenylalanyl-tRNA synthetase beta chain (EC 6.1.1.20) (Phenylalanine--tRNA ligase beta chain) (PheRS) | Q9NSD9|SYFB_HUMAN | 66,115 | 26 |
| Catechol O-methyltransferase (EC 2.1.1.6) | P21964|COMT_HUMAN | 30,020 | 26 |
| Switch-associated protein 70 (SWAP-70) | Q9UH65|SWP70_HUMAN | 68,981 | 26 |
| Leucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS) | Q9P2J5|SYLC_HUMAN | 134,453 | 26 |
| Glutathione peroxidase 1 (EC 1.11.1.9) (GSHPx-1) (GPx-1) (Cellular glutathione peroxidase) | P07203|GPX1_HUMAN | 21,882 | 26 |
| Proteasome subunit beta type 1 precursor (EC 3.4.25.1) (Proteasome component C5) (Macropain subunit C5) (Multicatalytic endopeptidase complex subunit C5) (Proteasome gamma chain) | P20618|PSB1_HUMAN | 26,473 | 26 |
| ATP synthase D chain, mitochondrial (EC 3.6.3.14) | O75947|ATP5H_HUMAN | 18,474 | 26 |
| Twinfilin-2 (Twinfilin-1-like protein) (A6-related protein) (hA6RP) (Protein tyrosine kinase 9-like) | Q6IBS0|TWF2_HUMAN | 39,531 | 26 |
| Histone H2B type 1-J (H2B.r) (H2B/r) (H2B.1) | P06899|H2B1J_HUMAN, P23527|H2B1O_HUMAN, P33778|H2B1B_HUMAN, Q16778|H2B2E_HUMAN | 13,887 | 26 |
| Transcription factor BTF3 (RNA polymerase B transcription factor 3) | P20290|BTF3_HUMAN | 22,150 | 26 |
| Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15) (ATP-dependent RNA helicase #46) | O43143|DHX15_HUMAN | 90,917 | 26 |
| Aminopeptidase N (EC 3.4.11.2) (hAPN) (Alanyl aminopeptidase) (Microsomal aminopeptidase) (Aminopeptidase M) (gp150) (Myeloid plasma membrane glycoprotein CD13) (CD13 antigen) | P15144|AMPN_HUMAN | 109,496 | 25 |
| Isocitrate dehydrogenase [NADP] cytoplasmic (EC 1.1.1.42) (Cytosolic NADP-isocitrate dehydrogenase) (Oxalosuccinate decarboxylase) (IDH) (NADP(+)-specific ICDH) (IDP) | O75874|IDHC_HUMAN | 46,643 | 25 |
| Gamma-interferon-inducible protein Ifi-16 (Interferon-inducible myeloid differentiation transcriptional activator) (IFI 16) | Q16666|IF16_HUMAN | 88,240 | 25 |
| G1 to S phase transition protein 1 homolog (GTP-binding protein GST1-HS) | P15170|GSPT1_HUMAN | 55,739 | 25 |
| Puromycin-sensitive aminopeptidase (EC 3.4.11.-) (PSA) | P55786|PSA_HUMAN | 103,261 | 25 |
| 6-phosphofructokinase, liver type (EC 2.7.1.11) (Phosphofructokinase 1) (Phosphohexokinase) (Phosphofructo-1-kinase isozyme B) (PFK-B) | P17858|K6PL_HUMAN | 85,001 | 25 |
| Pre-mRNA-processing factor 19 (EC 6.3.2.-) (PRP19/PSO4 homolog) (hPso4) (Nuclear matrix protein 200) (Senescence evasion factor) | Q9UMS4|PRP19_HUMAN | 55,163 | 25 |
| Gelsolin precursor (Actin-depolymerizing factor) (ADF) (Brevin) (AGEL) | P06396|GELS_HUMAN | 85,680 | 25 |
| Transportin-1 (Importin beta-2) (Karyopherin beta-2) (M9 region interaction protein) (MIP) | Q92973|TNPO1_HUMAN | 101,296 | 25 |
| Adenylate kinase isoenzyme 2, mitochondrial (EC 2.7.4.3) (ATP-AMP transphosphorylase) | P54819|KAD2_HUMAN | 26,461 | 25 |
| Dynein light chain 1, cytoplasmic (Dynein light chain LC8-type 1) (8 kDa dynein light chain) (DLC8) (Protein inhibitor of neuronal nitric oxide synthase) (PIN) | P63167|DYL1_HUMAN | 10,348 | 25 |
| Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C) | O15355|PP2CG_HUMAN | 59,254 | 25 |
| Collagen alpha-2(IV) chain precursor | P08572|CO4A2_HUMAN | 167,522 | 25 |
| Destrin (Actin-depolymerizing factor) (ADF) | P60981|DEST_HUMAN | 18,488 | 25 |
| Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14) | P54578|UBP14_HUMAN | 56,052 | 25 |
| Bifunctional 3′-phosphoadenosine 5′-phosphosulfate synthetase 2 (PAPS synthetase 2) (PAPSS 2) (Sulfurylase kinase 2) (SK2) (SK 2) [Includes: Sulfate adenylyltransferase (EC 2.7.7.4) (Sulfate adenylate transferase) (SAT) (ATP-sulfurylase); Adenylyl-sulfate kinase (EC 2.7.1.25) (Adenylylsulfate 3′-phosphotransferase) (APS kinase) (Adenosine-5′-phosphosulfate 3′-phosphotransferase) (3′-phosphoadenosine-5′-phosphosulfate synthetase)] | O95340|PAPS2_HUMAN | 69,484 | 25 |
| 40S ribosomal protein S20 | P60866|RS20_HUMAN | 13,355 | 25 |
| Acidic leucine-rich nuclear phosphoprotein 32 family member B (PHAPI2 protein) (Silver-stainable protein SSP29) (Acidic protein rich in leucines) | Q92688|AN32B_HUMAN | 28,771 | 25 |
| Transgelin (Smooth muscle protein 22-alpha) (SM22-alpha) (WS3-10) (22 kDa actin-binding protein) | Q01995|TAGL_HUMAN | 22,593 | 25 |
| Actin-related protein 2/3 complex subunit 1B (ARP2/3 complex 41 kDa subunit) (p41-ARC) | O15143|ARC1B_HUMAN | 40,932 | 24 |
| Signal transducer and activator of transcription 3 (Acute-phase response factor) | P40763|STAT3_HUMAN | 88,052 | 24 |
| Hepatoma-derived growth factor (HDGF) (High-mobility group protein 1-like 2) (HMG-1L2) | P51858|HDGF_HUMAN | 26,771 | 24 |
| Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial precursor (EC 1.3.5.1) (Fp) (Flavoprotein subunit of complex II) | P31040|DHSA_HUMAN | 72,674 | 24 |
| Poly(rC)-binding protein 2 (Alpha-CP2) (hnRNP-E2) | Q15366|PCBP2_HUMAN | 38,563 | 24 |
| NAD(P)H dehydrogenase [quinone] 1 (EC 1.6.5.2) (Quinone reductase 1) (NAD(P)H:quinone oxidoreductase 1) (QR1) (DT-diaphorase) (DTD) (Azoreductase) (Phylloquinone reductase) (Menadione reductase) | P15559|NQO1_HUMAN | 30,851 | 24 |
| Protein flightless-1 homolog | Q13045|FLII_HUMAN | 144,737 | 24 |
| Filamin-binding LIM protein 1 (FBLP-1) (Mitogen-inducible 2-interacting protein) (MIG2-interacting protein) (Migfilin) | Q8WUP2|FBLI1_HUMAN | 40,651 | 24 |
| Zinc finger CCCH type antiviral protein 1 (Zinc finger CCCH domain-containing protein 2) | Q7Z2W4|ZCC2_HUMAN | 101,428 | 24 |
| Nascent polypeptide-associated complex subunit alpha (NAC-alpha) (Alpha-NAC) (Hom s 2.02) | Q13765|NACA_HUMAN | 23,365 | 24 |
| Glycogen phosphorylase, brain form (EC 2.4.1.1) | P11216|PYGB_HUMAN | 96,680 | 24 |
| 14-3-3 protein gamma (Protein kinase C inhibitor protein 1) (KCIP-1) | P61981|1433G_HUMAN | 28,285 | 24 |
| 40S ribosomal protein S28 | P62857|RS28_HUMAN | 7,823 | 24 |
| 40S ribosomal protein S15a | P62244|RS15A_HUMAN | 14,822 | 24 |
| 60S ribosomal protein L19 | P84098|RL19_HUMAN | 23,449 | 24 |
| SH3 domain-binding glutamic acid-rich-like protein 3 (SH3 domain-binding protein 1) (SH3BP-1) | Q9H299|SH3L3_HUMAN | 10,420 | 24 |
| Alpha crystallin A chain (Heat-shock protein beta-4) (HspB4) [Contains: Alpha crystallin A chain, short form] | P02489|CRYAA_HUMAN | 19,892 | 24 |
| Collagen alpha-1(XII) chain precursor | Q99715|COCA1_HUMAN | 333,174 | 24 |
| D-dopachrome decarboxylase (EC 4.1.1.84) (D-dopachrome tautomerase) (Phenylpyruvate tautomerase II) | P30046|DOPD_HUMAN | 12,694 | 24 |
| S-formylglutathione hydrolase (EC 3.1.2.12) (FGH) (Esterase D) | P10768|ESTD_HUMAN | 31,446 | 23 |
| HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1) | Q7Z6Z7|HUWE1_HUMAN | 481,874 | 23 |
| Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1) | P09874|PARP1_HUMAN | 113,070 | 23 |
| 2-oxoglutarate dehydrogenase E1 component, mitochondrial precursor (EC 1.2.4.2) (Alpha-ketoglutarate dehydrogenase) | Q02218|ODO1_HUMAN | 113,459 | 23 |
| Ras-related C3 botulinum toxin substrate 1 precursor (p21-Rac1) (Ras-like protein TC25) (Cell migration-inducing gene 5 protein) | P63000|RAC1_HUMAN | 21,433 | 23 |
| Adenylate kinase isoenzyme 1 (EC 2.7.4.3) (ATP-AMP transphosphorylase) (AK1) (Myokinase) | P00568|KAD1_HUMAN | 21,617 | 23 |
| Ras-related protein Rab-1B | Q9H0U4|RAB1B_HUMAN | 22,154 | 23 |
| FK506-binding protein 3 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (25 kDa FKBP) (FKBP-25) (Rapamycin-selective 25 kDa immunophilin) | Q00688|FKBP3_HUMAN | 25,159 | 23 |
| Nonspecific lipid-transfer protein (EC 2.3.1.176) (Propanoyl-CoA C-acyltransferase) (NSL-TP) (Sterol carrier protein 2) (SCP-2) (Sterol carrier protein X) (SCP-X) (SCP-chi) (SCPX) | P22307|NLTP_HUMAN | 58,977 | 23 |
| Regulator of chromosome condensation (Chromosome condensation protein 1) (Cell cycle regulatory protein) | P18754|RCC1_HUMAN | 44,950 | 23 |
| Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Core protein II) (Complex III subunit II) | P22695|UQCR2_HUMAN | 48,425 | 23 |
| Myosin-VI (Unconventional myosin VI) | Q9UM54|MYO6_HUMAN | 149,679 | 23 |
| Ran-specific GTPase-activating protein (Ran-binding protein 1) (RanBP1) | P43487|RANG_HUMAN | 23,293 | 23 |
| U1 small nuclear ribonucleoprotein 70 kDa (U1 snRNP 70 kDa) (snRNP70) (U1-70K) | P08621|RU17_HUMAN | 51,540 | 23 |
| 60S ribosomal protein L29 (Cell surface heparin-binding protein HIP) | P47914|RL29_HUMAN | 17,735 | 23 |
| Alanyl-tRNA synthetase, cytoplasmic (EC 6.1.1.7) (Alanine--tRNA ligase) (AlaRS) (Renal carcinoma antigen NY-REN-42) | P49588|SYAC_HUMAN | 106,795 | 22 |
| Serine/threonine-protein kinase PAK 2 (EC 2.7.11.1) (p21-activated kinase 2) (PAK-2) (PAK65) (Gamma-PAK) (S6/H4 kinase) | Q13177|PAK2_HUMAN | 57,988 | 22 |
| E3 SUMO-protein ligase RanBP2 (Ran-binding protein 2) (Nuclear pore complex protein Nup358) (Nucleoporin Nup358) (358 kDa nucleoporin) (p270) | P49792|RBP2_HUMAN | 358,180 | 22 |
| Niban-like protein (Meg-3) | Q96TA1|NIBL_HUMAN | 82,666 | 22 |
| RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (Nuclear matrix protein 238) (NMP 238) (54 kDa erythrocyte cytosolic protein) (ECP-54) (TIP60-associated protein 54-alpha) (TAP54-alpha) | Q9Y265|RUVB1_HUMAN | 50,211 | 22 |
| Mitochondrial inner membrane protein (Mitofilin) (p87/89) (Proliferation-inducing gene 4 protein) | Q16891|IMMT_HUMAN | 83,661 | 22 |
| Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68) | P17844|DDX5_HUMAN | 69,132 | 22 |
| NSFL1 cofactor p47 (p97 cofactor p47) | Q9UNZ2|NSF1C_HUMAN | 40,555 | 22 |
| Tripartite motif-containing protein 25 (Zinc finger protein 147) (Estrogen-responsive finger protein) (Efp) (RING finger protein 147) | Q14258|TRI25_HUMAN | 70,971 | 22 |
| Kinesin light chain 1 (KLC 1) | Q07866|KLC1_HUMAN | 64,769 | 22 |
| Protein RCC2 (Telophase disk protein of 60 kDa) (RCC1-like protein TD-60) | Q9P258|RCC2_HUMAN | 56,067 | 22 |
| Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (HECH) | Q13185|CBX3_HUMAN | 20,794 | 22 |
| ARMET protein precursor (Arginine-rich protein) | P55145|ARMET_HUMAN | 20,240 | 22 |
| Aldehyde dehydrogenase, mitochondrial precursor (EC 1.2.1.3) (ALDH class 2) (ALDHI) (ALDH-E2) | P05091|ALDH2_HUMAN | 56,363 | 22 |
| Vacuolar protein sorting-associated protein 35 (Vesicle protein sorting 35) (hVPS35) (Maternal-embryonic 3) | Q96QK1|VPS35_HUMAN | 91,692 | 22 |
| Eukaryotic translation initiation factor 3 subunit 3 (eIF-3 gamma) (eIF3 p40 subunit) (eIF3h) | O15372|IF33_HUMAN | 39,913 | 22 |
| Cytochrome c oxidase subunit VIb isoform 1 (EC 1.9.3.1) (COX VIb-1) | P14854|CX6B1_HUMAN | 10,174 | 22 |
| Caveolin-1 | Q03135|CAV1_HUMAN | 20,454 | 22 |
| Proteasome subunit alpha type 5 (EC 3.4.25.1) (Proteasome zeta chain) (Macropain zeta chain) (Multicatalytic endopeptidase complex zeta chain) | P28066|PSA5_HUMAN | 26,393 | 22 |
| Ubiquitin-conjugating enzyme E2 L3 (EC 6.3.2.19) (Ubiquitin-protein ligase L3) (Ubiquitin carrier protein L3) (UbcH7) (E2-F1) (L-UBC) | P68036|UB2L3_HUMAN | 17,844 | 22 |
| Plasminogen activator inhibitor 1 precursor (PAI-1) (Endothelial plasminogen activator inhibitor) (PAI) | P05121|PAI1_HUMAN | 45,042 | 22 |
| Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen) | Q14980|NUMA1_HUMAN | 238,257 | 21 |
| Lamin-B2 | Q03252|LMNB2_HUMAN | 67,672 | 21 |
| Methionine aminopeptidase 2 (EC 3.4.11.18) (MetAP 2) (Peptidase M 2) (Initiation factor 2-associated 67 kDa glycoprotein) (p67) (p67eIF2) | P50579|AMPM2_HUMAN | 52,874 | 21 |
| Ezrin (p81) (Cytovillin) (Villin-2) | P15311|EZRI_HUMAN | 69,397 | 21 |
| Sialic acid synthase (N-acetylneuraminate synthase) (EC 2.5.1.56) (N-acetylneuraminic acid synthase) (N-acetylneuraminate-9-phosphate synthase) (EC 2.5.1.57) (N-acetylneuraminic acid phosphate synthase) | Q9NR45|SIAS_HUMAN | 40,290 | 21 |
| Heme oxygenase 2 (EC 1.14.99.3) (HO-2) | P30519|HMOX2_HUMAN | 36,016 | 21 |
| Hsc70-interacting protein (Hip) (Suppression of tumorigenicity protein 13) (Putative tumor suppressor ST13) (Protein FAM10A1) (Progesterone receptor-associated p48 protein) (Renal carcinoma antigen NY-REN-33) | P50502|F10A1_HUMAN | 41,314 | 21 |
| PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma) | Q9NR12|PDLI7_HUMAN | 49,826 | 21 |
| cAMP-dependent protein kinase type I-alpha regulatory subunit (Tissue-specific extinguisher 1) (TSE1) | P10644|KAP0_HUMAN | 42,964 | 21 |
| Calponin-3 (Calponin, acidic isoform) | Q15417|CNN3_HUMAN | 36,397 | 21 |
| ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DDX19-like protein) | Q9NUU7|DD19A_HUMAN | 53,958 | 21 |
| Procollagen-lysine,2-oxoglutarate 5-dioxygenase 3 precursor (EC 1.14.11.4) (Lysyl hydroxylase 3) (LH3) | O60568|PLOD3_HUMAN | 84,769 | 21 |
| Calpastatin (Calpain inhibitor) (Sperm BS-17 component) | P20810|ICAL_HUMAN | 76,557 | 21 |
| Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1) | Q07157|ZO1_HUMAN | 195,442 | 21 |
| Synaptopodin | Q8N3V7|SYNPO_HUMAN | 99,446 | 21 |
| Iron-responsive element-binding protein 1 (IRE-BP 1) (Iron regulatory protein 1) (IRP1) (Ferritin repressor protein) (Aconitate hydratase) (EC 4.2.1.3) (Citrate hydro-lyase) (Aconitase) | P21399|IREB1_HUMAN | 98,383 | 21 |
| F-actin capping protein subunit alpha-2 (CapZ alpha-2) | P47755|CAZA2_HUMAN | 32,931 | 21 |
| Astrocytic phosphoprotein PEA-15 (Phosphoprotein enriched in diabetes) (PED) | Q15121|PEA15_HUMAN | 15,023 | 21 |
| Poly [ADP-ribose] polymerase 4 (EC 2.4.2.30) (PARP-4) (Vault poly(ADP-ribose) polymerase) (VPARP) (193 kDa vault protein) (PARP-related/IalphaI-related H5/proline-rich) (PH5P) | Q9UKK3|PARP4_HUMAN | 192,574 | 21 |
| Lysyl-tRNA synthetase (EC 6.1.1.6) (Lysine--tRNA ligase) (LysRS) | Q15046|SYK_HUMAN | 68,032 | 21 |
| Junctional adhesion molecule A precursor (JAM-A) (Junctional adhesion molecule 1) (JAM-1) (Platelet adhesion molecule 1) (PAM-1) (Platelet F11 receptor) (CD321 antigen) | Q9Y624|JAM1_HUMAN | 32,565 | 21 |
| ADP-sugar pyrophosphatase (EC 3.6.1.13) (EC 3.6.1.-) (Nucleoside diphosphate-linked moiety X motif 5) (Nudix motif 5) (YSA1H) | Q9UKK9|NUDT5_HUMAN | 24,310 | 21 |
| Vesicular integral-membrane protein VIP36 precursor (GP36b glycoprotein) (Lectin, mannose-binding 2) | Q12907|LMAN2_HUMAN | 40,212 | 21 |
| Endothelin-converting enzyme 1 (EC 3.4.24.71) (ECE-1) | P42892|ECE1_HUMAN | 87,147 | 21 |
| Leukocyte elastase inhibitor (LEI) (Serpin B1) (Monocyte/neutrophil elastase inhibitor) (M/NEI) (EI) | P30740|ILEU_HUMAN | 42,726 | 21 |
| Platelet-activating factor acetylhydrolase IB subunit beta (EC 3.1.1.47) (PAF acetylhydrolase 30 kDa subunit) (PAF-AH 30 kDa subunit) (PAF-AH subunit beta) (PAFAH subunit beta) | P68402|PA1B2_HUMAN | 25,552 | 21 |
| Drebrin-like protein (SH3 domain-containing protein 7) (Drebrin-F) (Cervical SH3P7) (HPK1-interacting protein of 55 kDa) (HIP-55) (Cervical mucin-associated protein) | Q9UJU6|DBNL_HUMAN | 48,188 | 20 |
| Adenylosuccinate synthetase isozyme 2 (EC 6.3.4.4) (Adenylosuccinate synthetase, acidic isozyme) (IMP--aspartate ligase 2) (AdSS 2) (AMPSase 2) | P30520|PURA2_HUMAN | 50,080 | 20 |
| Protein KIAA1967 (Deleted in breast cancer gene 1 protein) (DBC.1) (DBC-1) (p30 DBC) | Q8N163|K1967_HUMAN | 102,885 | 20 |
| Delta 1-pyrroline-5-carboxylate synthetase (P5CS) (Aldehyde dehydrogenase 18 family member A1) [Includes: Glutamate 5-kinase (EC 2.7.2.11) (Gamma-glutamyl kinase) (GK); Gamma-glutamyl phosphate reductase (GPR) (EC 1.2.1.41) (Glutamate-5-semialdehyde dehydrogenase) (Glutamyl-gamma-semialdehyde dehydrogenase)] | P54886|P5CS_HUMAN | 87,285 | 20 |
| Leukotriene A-4 hydrolase (EC 3.3.2.6) (LTA-4 hydrolase) (Leukotriene A(4) hydrolase) | P09960|LKHA4_HUMAN | 69,269 | 20 |
| Cadherin-5 precursor (Vascular endothelial-cadherin) (VE-cadherin) (7B4 antigen) (CD144 antigen) | P33151|CADH5_HUMAN | 87,499 | 20 |
| Thioredoxin-like protein 1 (32 kDa thioredoxin-related protein) | O43396|TXNL1_HUMAN | 32,233 | 20 |
| Importin beta-3 (Karyopherin beta-3) (Ran-binding protein 5) (RanBP5) | O00410|IMB3_HUMAN | 123,614 | 20 |
| Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (136 kDa double-stranded RNA-binding protein) (P136) (K88DSRBP) (Interferon-inducible protein 4) (IFI-4 protein) | P55265|DSRAD_HUMAN | 135,981 | 20 |
| Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1) | Q9UHD8|SEPT9_HUMAN | 65,384 | 20 |
| Coatomer subunit epsilon (Epsilon-coat protein) (Epsilon-COP) | O14579|COPE_HUMAN | 34,465 | 20 |
| Heat shock protein 75 kDa, mitochondrial precursor (HSP 75) (Tumor necrosis factor type 1 receptor-associated protein) (TRAP-1) (TNFR-associated protein 1) | Q12931|TRAP1_HUMAN | 80,095 | 20 |
| Eukaryotic translation initiation factor 2 subunit 2 (Eukaryotic translation initiation factor 2 subunit beta) (eIF-2-beta) | P20042|IF2B_HUMAN | 38,372 | 20 |
| 3-hydroxyacyl-CoA dehydrogenase type-2 (EC 1.1.1.35) (3-hydroxyacyl-CoA dehydrogenase type II) (Type II HADH) (3-hydroxy-2-methylbutyryl-CoA dehydrogenase) (EC 1.1.1.178) (Endoplasmic reticulum-associated amyloid beta-peptide-binding protein) (Short-chain type dehydrogenase/reductase XH98G2) | Q99714|HCD2_HUMAN | 26,905 | 20 |
| RNA-binding protein FUS (Oncogene FUS) (Oncogene TLS) (Translocated in liposarcoma protein) (POMp75) (75 kDa DNA-pairing protein) | P35637|FUS_HUMAN | 53,408 | 20 |
| Ras-related protein Rab-14 | P61106|RAB14_HUMAN | 23,880 | 20 |
| 26S proteasome non-ATPase regulatory subunit 4 (26S proteasome regulatory subunit S5A) (Rpn10) (Multiubiquitin chain-binding protein) (Antisecretory factor 1) (AF) (ASF) | P55036|PSMD4_HUMAN | 40,719 | 20 |
| NADH-cytochrome b5 reductase (EC 1.6.2.2) (B5R) (Diaphorase-1) (Cytochrome b5 reductase 3) [Contains: NADH-cytochrome b5 reductase membrane-bound form; NADH-cytochrome b5 reductase soluble form] | P00387|NCB5R_HUMAN | 34,218 | 20 |
| 60S ribosomal protein L30 | P62888|RL30_HUMAN | 12,767 | 20 |
| Sulfide:quinone oxidoreductase, mitochondrial precursor (EC 1.-.-.-) | Q9Y6N5|SQRD_HUMAN | 49,944 | 20 |
| NG,NG-dimethylarginine dimethylaminohydrolase 2 (EC 3.5.3.18) (Dimethylargininase-2) (Dimethylarginine dimethylaminohydrolase 2) (DDAHII) (DDAH-2) (S-phase protein) (Protein G6a) | O95865|DDAH2_HUMAN | 29,626 | 20 |
| 40S ribosomal protein S24 | P62847|RS24_HUMAN | 15,406 | 20 |
| 40S ribosomal protein S21 | P63220|RS21_HUMAN | 9,094 | 20 |
| 60S ribosomal protein L36 | Q9Y3U8|RL36_HUMAN | 12,236 | 20 |
| Eukaryotic translation initiation factor 3 subunit 4 (eIF-3 delta) (eIF3 p44) (eIF-3 RNA-binding subunit) (eIF3 p42) (eIF3g) | O75821|IF34_HUMAN | 35,594 | 20 |
| Eukaryotic translation initiation factor 6 (eIF-6) (B4 integrin interactor) (CAB) (p27(BBP)) (B(2)GCN homolog) | P56537|IF6_HUMAN | 26,580 | 20 |
| Protein S100-A11 (S100 calcium-binding protein A11) (Protein S100C) (Calgizzarin) (MLN 70) | P31949|S10AB_HUMAN | 11,723 | 20 |
| Proteasome subunit beta type 2 (EC 3.4.25.1) (Proteasome component C7-I) (Macropain subunit C7-I) (Multicatalytic endopeptidase complex subunit C7-I) | P49721|PSB2_HUMAN | 22,820 | 20 |
| Hemoglobin subunit alpha (Hemoglobin alpha chain) (Alpha-globin) | P69905|HBA_HUMAN | 15,240 | 20 |
| Lamina-associated polypeptide 2, isoforms beta/gamma (Thymopoietin, isoforms beta/gamma) (TP beta/gamma) (Thymopoietin-related peptide isoforms beta/gamma) (TPRP isoforms beta/gamma) [Contains: Thymopoietin (TP) (Splenin); Thymopentin (TP5)] | P42167|LAP2B_HUMAN | 50,653 | 20 |
| Complement component 1 Q subcomponent-binding protein, mitochondrial precursor (Glycoprotein gC1qBP) (C1qBP) (GC1q-R protein) (Hyaluronan-binding protein 1) (Mitochondrial matrix protein p32) (p33) | Q07021|C1QBP_HUMAN | 31,345 | 20 |
| Integrin alpha-6 precursor (VLA-6) (CD49f antigen) [Contains: Integrin alpha-6 heavy chain; Integrin alpha-6 light chain] | P23229|ITA6_HUMAN | 126,604 | 19 |
| Alpha-taxilin | P40222|TXLNA_HUMAN | 61,873 | 19 |
| 6-phosphogluconolactonase (EC 3.1.1.31) (6PGL) | O95336|6PGL_HUMAN | 27,530 | 19 |
| Structural maintenance of chromosomes protein 1A (SMC1alpha protein) (Sb1.8) | Q14683|SMC1A_HUMAN | 143,220 | 19 |
| NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (Complex I-75kD) (CI-75kD) | P28331|NDUS1_HUMAN | 79,450 | 19 |
| Bifunctional purine biosynthesis protein PURH [Includes: Phosphoribosylaminoimidazolecarboxamide formyltransferase (EC 2.1.2.3) (5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase) (AICAR transformylase); IMP cyclohydrolase (EC 3.5.4.10) (Inosinicase) (IMP synthetase) (ATIC)] | P31939|PUR9_HUMAN | 64,599 | 19 |
| Cold shock domain-containing protein E1 (UNR protein) (N-ras upstream gene protein) | O75534|CSDE1_HUMAN | 88,867 | 19 |
| Xaa-Pro aminopeptidase 1 (EC 3.4.11.9) (X-Pro aminopeptidase 1) (X-prolyl aminopeptidase 1, soluble) (Cytosolic aminopeptidase P) (Soluble aminopeptidase P) (sAmp) (Aminoacylproline aminopeptidase) | Q9NQW7|XPP1_HUMAN | 69,901 | 19 |
| Ras-related protein Rab-7 | P51149|RAB7_HUMAN | 23,472 | 19 |
| Vacuolar ATP synthase catalytic subunit A, ubiquitous isoform (EC 3.6.3.14) (V-ATPase subunit A 1) (Vacuolar proton pump alpha subunit 1) (V-ATPase 69 kDa subunit 1) (Isoform VA68) | P38606|VATA1_HUMAN | 68,287 | 19 |
| Seryl-tRNA synthetase, cytoplasmic (EC 6.1.1.11) (Serine--tRNA ligase) (SerRS) | P49591|SYSC_HUMAN | 58,761 | 19 |
| Protein kinase C and casein kinase substrate in neurons protein 2 | Q9UNF0|PACN2_HUMAN | 55,721 | 19 |
| Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1) | P30084|ECHM_HUMAN | 31,370 | 19 |
| 26S proteasome non-ATPase regulatory subunit 13 (26S proteasome regulatory subunit S11) (26S proteasome regulatory subunit p40.5) | Q9UNM6|PSD13_HUMAN | 42,901 | 19 |
| Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase) (Glycine cleavage system L protein) | P09622|DLDH_HUMAN | 54,132 | 19 |
| 40S ribosomal protein S26 | P62854|RS26_HUMAN | 12,998 | 19 |
| Dynamin-1-like protein (EC 3.6.5.5) (Dynamin-like protein) (Dnm1p/Vps1p-like protein) (DVLP) (Dynamin family member proline-rich carboxyl-terminal domain less) (Dymple) (Dynamin-related protein 1) (Dynamin-like protein 4) (Dynamin-like protein IV) (HdynIV) | O00429|DNM1L_HUMAN | 81,861 | 19 |
| Multisynthetase complex auxiliary component p43 [Contains: Endothelial monocyte-activating polypeptide 2 (EMAP-II) (Small inducible cytokine subfamily E member 1)] | Q12904|MCA1_HUMAN | 34,335 | 19 |
| Leucine-rich repeat flightless-interacting protein 1 (LRR FLII-interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2) | Q32MZ4|LRRF1_HUMAN | 89,235 | 19 |
| Actin-related protein 2/3 complex subunit 5 (ARP2/3 complex 16 kDa subunit) (p16-ARC) | O15511|ARPC5_HUMAN | 16,303 | 19 |
| 4F2 cell-surface antigen heavy chain (4F2hc) (Lymphocyte activation antigen 4F2 large subunit) (4F2 heavy chain antigen) (CD98 antigen) | P08195|4F2_HUMAN | 57,929 | 19 |
| Alcohol dehydrogenase [NADP+] (EC 1.1.1.2) (Aldehyde reductase) (Aldo-keto reductase family 1 member A1) | P14550|AK1A1_HUMAN | 36,556 | 19 |
| CTP synthase 1 (EC 6.3.4.2) (UTP--ammonia ligase 1) (CTP synthetase 1) | P17812|PYRG1_HUMAN | 66,673 | 19 |
| Phenylalanyl-tRNA synthetase alpha chain (EC 6.1.1.20) (Phenylalanine--tRNA ligase alpha chain) (PheRS) (CML33) | Q9Y285|SYFA_HUMAN | 57,547 | 19 |
| DNA-binding protein A (Cold shock domain-containing protein A) (Single-strand DNA-binding protein NF-GMB) | P16989|DBPA_HUMAN | 40,071 | 19 |
| Glucosamine--fructose-6-phosphate aminotransferase [isomerizing] 1 (EC 2.6.1.16) (Hexosephosphate aminotransferase 1) (D-fructose-6-phosphate amidotransferase 1) (GFAT 1) (GFAT1) | Q06210|GFPT1_HUMAN | 78,790 | 19 |
| 26S proteasome non-ATPase regulatory subunit 7 (26S proteasome regulatory subunit rpn8) (26S proteasome regulatory subunit S12) (Proteasome subunit p40) (Mov34 protein homolog) | P51665|PSD7_HUMAN | 37,008 | 19 |
| Proteasome subunit alpha type 3 (EC 3.4.25.1) (Proteasome component C8) (Macropain subunit C8) (Multicatalytic endopeptidase complex subunit C8) | P25788|PSA3_HUMAN | 28,416 | 19 |
| Protein C14orf166 | Q9Y224|CN166_HUMAN | 28,051 | 19 |
| Myotrophin (Protein V-1) | P58546|MTPN_HUMAN | 12,877 | 19 |
| Peroxiredoxin-4 (EC 1.11.1.15) (Prx-IV) (Thioredoxin peroxidase AO372) (Thioredoxin-dependent peroxide reductase A0372) (Antioxidant enzyme AOE372) (AOE37-2) | Q13162|PRDX4_HUMAN | 30,523 | 19 |
| 40S ribosomal protein S27 (Metallopan-stimulin 1) (MPS-1) | P42677|RS27_HUMAN | 9,443 | 19 |
| Mannosyl-oligosaccharide glucosidase (EC 3.2.1.106) (Processing A-glucosidase I) | Q13724|GCS1_HUMAN | 91,901 | 19 |
| Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein) | P29966|MARCS_HUMAN | 31,536 | 19 |
| Platelet-activating factor acetylhydrolase IB subunit alpha (PAF acetylhydrolase 45 kDa subunit) (PAF-AH 45 kDa subunit) (PAF-AH alpha) (PAFAH alpha) (Lissencephaly-1 protein) (LIS-1) | P43034|LIS1_HUMAN | 46,619 | 19 |
| Acyl-CoA-binding protein (ACBP) (Diazepam-binding inhibitor) (DBI) (Endozepine) (EP) | P07108|ACBP_HUMAN | 10,027 | 19 |
| Reticulon-1 (Neuroendocrine-specific protein) | Q16799|RTN1_HUMAN | 83,602 | 19 |
| General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein) | O60763|VDP_HUMAN | 107,880 | 18 |
| Connective tissue growth factor precursor (Hypertrophic chondrocyte-specific protein 24) | P29279|CTGF_HUMAN | 38,073 | 18 |
| Isoleucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS) (IRS) | P41252|SYIC_HUMAN | 144,944 | 18 |
| Tubulin-specific chaperone A (Tubulin-folding cofactor A) (CFA) (TCP1-chaperonin cofactor A) | O75347|TBCA_HUMAN | 12,837 | 18 |
| Protein LYRIC (Lysine-rich CEACAM1 co-isolated protein) (3D3/lyric) (Metastasis adhesion protein) (Metadherin) (Astrocyte elevated gene-1 protein) (AEG-1) | Q86UE4|LYRIC_HUMAN | 63,820 | 18 |
| Nucleobindin-1 precursor (CALNUC) | Q02818|NUCB1_HUMAN | 53,862 | 18 |
| Pleckstrin homology domain-containing family C member 1 (Kindlin-2) (Mitogen-inducible gene 2 protein) (Mig-2) | Q96AC1|PKHC1_HUMAN | 77,846 | 18 |
| Glyoxylate reductase/hydroxypyruvate reductase (EC 1.1.1.79) | Q9UBQ7|GRHPR_HUMAN | 35,651 | 18 |
| Tripeptidyl-peptidase 2 (EC 3.4.14.10) (Tripeptidyl-peptidase II) (TPP-II) (Tripeptidyl aminopeptidase) | P29144|TPP2_HUMAN | 138,335 | 18 |
| Tropomodulin-3 (Ubiquitous tropomodulin) (U-Tmod) | Q9NYL9|TMOD3_HUMAN | 39,578 | 18 |
| Nuclear protein Hcc-1 (Proliferation-associated cytokine-inducible protein CIP29) (Cytokine-induced protein of 29 kDa) | P82979|HCC1_HUMAN | 23,653 | 18 |
| Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (Mitochondrial ATP-dependent protease Lon) (LONHs) (Serine protease 15) | P36776|LONM_HUMAN | 106,473 | 18 |
| DNA-directed RNA polymerase II 140 kDa polypeptide (EC 2.7.7.6) (RNA polymerase II subunit 2) (RPB2) | P30876|RPB2_HUMAN | 133,883 | 18 |
| DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2) (HSDJ) | P31689|DNJA1_HUMAN | 44,851 | 18 |
| Apoptosis regulator BAX, membrane isoform alpha | Q07812|BAXA_HUMAN, Q07814|BAXB_HUMAN | 21,167 | 18 |
| Proteasome subunit beta type 8 precursor (EC 3.4.25.1) (Proteasome component C13) (Macropain subunit C13) (Multicatalytic endopeptidase complex subunit C13) | P28062|PSB8_HUMAN | 30,337 | 18 |
| S-adenosylmethionine synthetase isoform type-2 (EC 2.5.1.6) (Methionine adenosyltransferase 2) (AdoMet synthetase 2) (Methionine adenosyltransferase II) (MAT-II) | P31153|METK2_HUMAN | 43,643 | 18 |
| Cytosolic acyl coenzyme A thioester hydrolase (EC 3.1.2.2) (Long chain acyl-CoA thioester hydrolase) (CTE-II) (CTE-IIa) (Brain acyl-CoA hydrolase) (Acyl-CoA thioesterase 7) | O00154|BACH_HUMAN | 41,777 | 18 |
| Adipocyte-derived leucine aminopeptidase precursor (EC 3.4.11.-) (A-LAP) (ARTS-1) (Aminopeptidase PILS) (Puromycin-insensitive leucyl-specific aminopeptidase) (PILS-AP) (Type 1 tumor necrosis factor receptor shedding aminopeptidase regulator) | Q9NZ08|ARTS1_HUMAN | 105,832 | 18 |
| Protein S100-A10 (S100 calcium-binding protein A10) (Calpactin-1 light chain) (Calpactin I light chain) (p10 protein) (p11) (Cellular ligand of annexin II) | P60903|S10AA_HUMAN | 11,186 | 18 |
| Sorting nexin-3 (Protein SDP3) | O60493|SNX3_HUMAN | 18,745 | 18 |
| Fibulin-1 precursor | P23142|FBLN1_HUMAN | 77,241 | 18 |
| Arginine/serine-rich-splicing factor 10 (Transformer-2-beta) (HTRA2-beta) (Transformer 2 protein homolog) | P62995|TRA2B_HUMAN | 33,649 | 18 |
| Phosphate carrier protein, mitochondrial precursor (PTP) (Solute carrier family 25 member 3) | Q00325|MPCP_HUMAN | 40,078 | 18 |
| Macrophage migration inhibitory factor (MIF) (Phenylpyruvate tautomerase) (EC 5.3.2.1) (Glycosylation-inhibiting factor) (GIF) | P14174|MIF_HUMAN | 12,459 | 18 |
| Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma) | Q93052|LPP_HUMAN | 65,728 | 17 |
| Vacuolar ATP synthase subunit B, brain isoform (EC 3.6.3.14) (V-ATPase B2 subunit) (Vacuolar proton pump B isoform 2) (Endomembrane proton pump 58 kDa subunit) (HO57) | P21281|VATB2_HUMAN | 56,484 | 17 |
| AP-2 complex subunit mu-1 (Adaptin mu-1) (AP-2 mu-2 chain) (Clathrin coat assembly protein AP50) (Clathrin coat-associated protein AP50) (Plasma membrane adaptor AP-2 50 kDa protein) (HA2 50 kDa subunit) (Clathrin assembly protein complex 2 medium chain) | Q96CW1|AP2M1_HUMAN | 49,638 | 17 |
| Phosphoglucomutase-2 (EC 5.4.2.2) (Glucose phosphomutase 2) (PGM 2) | Q96G03|PGM2_HUMAN | 68,268 | 17 |
| Stomatin-like protein 2 (SLP-2) (EPB72-like 2) | Q9UJZ1|STML2_HUMAN | 38,517 | 17 |
| Exportin-2 (Exp2) (Importin-alpha re-exporter) (Chromosome segregation 1-like protein) (Cellular apoptosis susceptibility protein) | P55060|XPO2_HUMAN | 110,404 | 17 |
| ATPase family AAA domain-containing protein 3A | Q9NVI7|ATD3A_HUMAN | 71,352 | 17 |
| Phospholipase A-2-activating protein (PLAP) (PLA2P) | Q9Y263|PLAP_HUMAN | 87,141 | 17 |
| Nucleolar protein NOP5 (Nucleolar protein 5) (NOP58) | Q9Y2X3|NOP5_HUMAN | 59,562 | 17 |
| Ubiquitin-conjugating enzyme E2 N (EC 6.3.2.19) (Ubiquitin-protein ligase N) (Ubiquitin carrier protein N) (Ubc13) (Bendless-like ubiquitin-conjugating enzyme) | P61088|UBE2N_HUMAN | 17,121 | 17 |
| Junction plakoglobin (Desmoplakin-3) (Desmoplakin III) | P14923|PLAK_HUMAN | 81,613 | 17 |
| Long-chain-fatty-acid--CoA ligase 3 (EC 6.2.1.3) (Long-chain acyl-CoA synthetase 3) (LACS 3) | O95573|ACSL3_HUMAN | 80,405 | 17 |
| Myosin-Ib (Myosin I alpha) (MMI-alpha) (MMIa) (MYH-1c) | O43795|MYO1B_HUMAN | 131,973 | 17 |
| Vesicle-trafficking protein SEC22b (SEC22 vesicle-trafficking protein homolog B) (SEC22 vesicle-trafficking protein-like 1) (ERS24) (ERS-24) | O75396|SC22B_HUMAN | 24,723 | 17 |
| SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40) | P09486|SPRC_HUMAN | 34,614 | 17 |
| 60S ribosomal protein L35 | P42766|RL35_HUMAN | 14,535 | 17 |
| DNA topoisomerase 1 (EC 5.99.1.2) (DNA topoisomerase I) | P11387|TOP1_HUMAN | 90,712 | 17 |
| Nuclear protein localization protein 4 homolog (Protein NPL4) | Q8TAT6|NPL4_HUMAN | 68,103 | 17 |
| Histidyl-tRNA synthetase, cytoplasmic (EC 6.1.1.21) (Histidine--tRNA ligase) (HisRS) | P12081|SYHC_HUMAN | 57,395 | 17 |
| Splicing factor, arginine/serine-rich 3 (Pre-mRNA-splicing factor SRP20) | P84103|SFRS3_HUMAN | 19,312 | 17 |
| CD109 antigen precursor (p180) (150 KDa TGF-beta-1-binding protein) (r150) (Platelet-specific Gov antigen) | Q6YHK3|CD109_HUMAN | 161,674 | 17 |
| Four and a half LIM domains protein 1 (FHL-1) (Skeletal muscle LIM-protein 1) (SLIM 1) (SLIM) | Q13642|FHL1_HUMAN | 36,244 | 17 |
| Coactosin-like protein | Q14019|COTL1_HUMAN | 15,927 | 17 |
| UV excision repair protein RAD23 homolog B (hHR23B) (XP-C repair-complementing complex 58 kDa protein) (p58) | P54727|RD23B_HUMAN | 43,153 | 17 |
| ATP synthase O subunit, mitochondrial precursor (EC 3.6.3.14) (Oligomycin sensitivity conferral protein) (OSCP) | P48047|ATPO_HUMAN | 23,259 | 17 |
| Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit precursor (EC 2.4.1.119) (Oligosaccharyl transferase 48 kDa subunit) (DDOST 48 kDa subunit) | P39656|OST48_HUMAN | 48,793 | 17 |
| B-cell receptor-associated protein 31 (BCR-associated protein Bap31) (p28 Bap31) (Protein CDM) (6C6-AG tumor-associated antigen) | P51572|BAP31_HUMAN | 27,975 | 17 |
| Putative quinone oxidoreductase (EC 1.-.-.-) (Tumor protein p53-inducible protein 3) (p53-induced protein 3) | Q53FA7|QORX_HUMAN | 35,519 | 17 |
| Granulins precursor (Proepithelin) (PEPI) [Contains: Acrogranin; Paragranulin; Granulin-1 (Granulin G); Granulin-2 (Granulin F); Granulin-3 (Granulin B); Granulin-4 (Granulin A); Granulin-5 (Granulin C); Granulin-6 (Granulin D); Granulin-7 (Granulin E)] | P28799|GRN_HUMAN | 63,522 | 17 |
| Aspartyl/asparaginyl beta-hydroxylase (EC 1.14.11.16) (Aspartate beta-hydroxylase) (ASP beta-hydroxylase) (Peptide-aspartate beta-dioxygenase) | Q12797|ASPH_HUMAN | 85,873 | 17 |
| 40S ribosomal protein S23 | P62266|RS23_HUMAN | 15,790 | 17 |
| Flavin reductase (EC 1.5.1.30) (FR) (NADPH-dependent diaphorase) (NADPH-flavin reductase) (FLR) (Biliverdin reductase B) (EC 1.3.1.24) (BVR-B) (Biliverdin-IX beta-reductase) (Green heme-binding protein) (GHBP) | P30043|BLVRB_HUMAN | 22,101 | 17 |
| Eukaryotic translation initiation factor 1A, X-chromosomal (eIF-1A X isoform) (eIF-4C) | P47813|IF1AX_HUMAN | 16,443 | 17 |
| Ras-related protein Rap-1b precursor (GTP-binding protein smg p21B) | P61224|RAP1B_HUMAN | 20,807 | 17 |
| Acetyl-CoA acetyltransferase, cytosolic (EC 2.3.1.9) (Cytosolic acetoacetyl-CoA thiolase) (Acetyl CoA transferase-like protein) | Q9BWD1|THIC_HUMAN | 41,332 | 16 |
| Dipeptidyl-peptidase 3 (EC 3.4.14.4) (Dipeptidyl-peptidase III) (DPP III) (Dipeptidyl aminopeptidase III) (Dipeptidyl arylamidase III) | Q9NY33|DPP3_HUMAN | 82,574 | 16 |
| Zinc finger protein 185 (LIM domain protein ZNF185) (P1-A) | O15231|ZN185_HUMAN | 49,169 | 16 |
| Annexin A4 (Annexin IV) (Lipocortin IV) (Endonexin I) (Chromobindin-4) (Protein II) (P32.5) (Placental anticoagulant protein II) (PAP-II) (PP4-X) (35-beta calcimedin) (Carbohydrate-binding protein P33/P41) (P33/41) | P09525|ANXA4_HUMAN | 35,866 | 16 |
| Tumor protein D54 (hD54) (Tumor protein D52-like 2) | O43399|TPD54_HUMAN | 22,220 | 16 |
| Uncharacterized protein C17orf25 | Q9HC38|CQ025_HUMAN | 34,776 | 16 |
| Eukaryotic translation initiation factor 2 subunit 3 (Eukaryotic translation initiation factor 2 subunit gamma) (eIF-2-gamma) | P41091|IF2G_HUMAN | 51,092 | 16 |
| Interleukin enhancer-binding factor 2 (Nuclear factor of activated T-cells 45 kDa) | Q12905|ILF2_HUMAN | 43,045 | 16 |
| Calpain small subunit 1 (CSS1) (Calcium-dependent protease small subunit 1) (Calcium-dependent protease small subunit) (CDPS) (Calpain regulatory subunit) (Calcium-activated neutral proteinase small subunit) (CANP small subunit) | P04632|CPNS1_HUMAN | 28,299 | 16 |
| Putative RNA-binding protein Luc7-like 2 | Q9Y383|LC7L2_HUMAN | 46,497 | 16 |
| Histidine triad nucleotide-binding protein 1 (Adenosine 5′-monophosphoramidase) (Protein kinase C inhibitor 1) (Protein kinase C-interacting protein 1) (PKCI-1) | P49773|HINT1_HUMAN | 13,784 | 16 |
| Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) | O60610|DIAP1_HUMAN | 138,966 | 16 |
| CD2-associated protein (Cas ligand with multiple SH3 domains) (Adapter protein CMS) | Q9Y5K6|CD2AP_HUMAN | 71,436 | 16 |
| Testin (TESS) | Q9UGI8|TES_HUMAN | 47,978 | 16 |
| Selenide, water dikinase 1 (EC 2.7.9.3) (Selenophosphate synthetase 1) (Selenium donor protein 1) | P49903|SPS1_HUMAN | 42,893 | 16 |
| Cell division cycle 5-like protein (Cdc5-like protein) (Pombe cdc5-related protein) | Q99459|CDC5L_HUMAN | 92,236 | 16 |
| Phosphatidylinositol transfer protein beta isoform (PtdIns transfer protein beta) (PtdInsTP) (PI-TP-beta) | P48739|PIPNB_HUMAN | 31,522 | 16 |
| Peroxiredoxin-5, mitochondrial precursor (EC 1.11.1.15) (Prx-V) (Peroxisomal antioxidant enzyme) (PLP) (Thioredoxin reductase) (Thioredoxin peroxidase PMP20) (Antioxidant enzyme B166) (AOEB166) (TPx type VI) (Liver tissue 2D-page spot 71B) (Alu corepressor 1) | P30044|PRDX5_HUMAN | 22,008 | 16 |
| Heterogeneous nuclear ribonucleoprotein A0 (hnRNP A0) | Q13151|ROA0_HUMAN | 30,823 | 16 |
| Nuclear pore complex protein Nup93 (Nucleoporin Nup93) (93 kDa nucleoporin) | Q8N1F7|NUP93_HUMAN | 93,473 | 16 |
| Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled-coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2) | O75116|ROCK2_HUMAN | 160,901 | 16 |
| Ribonucleoside-diphosphate reductase large subunit (EC 1.17.4.1) (Ribonucleoside-diphosphate reductase M1 subunit) (Ribonucleotide reductase large chain) | P23921|RIR1_HUMAN | 90,056 | 16 |
| Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase) | O94804|STK10_HUMAN | 112,120 | 16 |
| Thioredoxin-like protein 2 (PKC-interacting cousin of thioredoxin) (PKC-theta-interacting protein) (PKCq-interacting protein) | O76003|TXNL2_HU MAN | 37,415 | 16 |
| Lactoylglutathione lyase (EC 4.4.1.5) (Methylglyoxalase) (Aldoketomutase) (Glyoxalase I) (Glx I) (Ketone-aldehyde mutase) (S-D-lactoylglutathione methylglyoxal lyase) | Q04760|LGUL_HUMAN | 20,761 | 16 |
| MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS) | P49006|MRP_HUMAN | 19,511 | 16 |
| Sec1 family domain-containing protein 1 (Syntaxin-binding protein 1-like 2) (Sly1p) | Q8WVM8|SCFD1_HUMAN | 72,364 | 16 |
| 40S ribosomal protein S4, Y isoform 1 | P22090|RS4Y1_HUMAN | 29,438 | 16 |
| NMDA receptor-regulated protein 1 (N-terminal acetyltransferase) (Protein tubedown-1) (Tbdn100) (Gastric cancer antigen Ga19) | Q9BXJ9|NARG1_HUMAN | 101,260 | 16 |
| Polyadenylate-binding protein 4 (Poly(A)-binding protein 4) (PABP 4) (Inducible poly(A)-binding protein) (iPABP) (Activated-platelet protein 1) (APP-1) | Q13310|PABP4_HUMAN | 70,766 | 16 |
| Interferon-induced guanylate-binding protein 2 (GTP-binding protein 2) (Guanine nucleotide-binding protein 2) (GBP-2) (HuGBP-2) | P32456|GBP2_HUMAN | 67,167 | 16 |
| Ras-related protein Rab-11B (GTP-binding protein YPT3) | P62491|RB11A_HUMAN, Q15907|RB11B_HUMAN | 24,471 | 16 |
| Alpha-soluble NSF attachment protein (SNAP-alpha) (N-ethylmaleimide-sensitive factor attachment protein, alpha) | P54920|SNAA_HUMAN | 33,216 | 16 |
| Integrin alpha-V precursor (Vitronectin receptor subunit alpha) (CD51 antigen) [Contains: Integrin alpha-V heavy chain; Integrin alpha-V light chain] | P06756|ITAV_HUMAN | 116,037 | 16 |
| N-acetylglucosamine-6-sulfatase precursor (EC 3.1.6.14) (G6S) (Glucosamine-6-sulfatase) | P15586|GNS_HUMAN | 62,066 | 16 |
| Cytochrome c oxidase subunit 5A, mitochondrial precursor (EC 1.9.3.1) (Cytochrome c oxidase polypeptide Va) | P20674|COX5A_HUMAN | 16,757 | 16 |
| Heterogeneous nuclear ribonucleoprotein A/B (hnRNP A/B) (APOBEC-1-binding protein 1) (ABBP-1) | Q99729|ROAA_HUMAN | 36,595 | 16 |
| Basigin precursor (Leukocyte activation antigen M6) (Collagenase stimulatory factor) (Extracellular matrix metalloproteinase inducer) (EMMPRIN) (5F7) (Tumor cell-derived collagenase stimulatory factor) (TCSF) (OK blood group antigen) (CD147 antigen) | P35613|BASI_HUMAN | 42,182 | 16 |
| Coiled-coil domain-containing protein 50 (Protein Ymer) | Q8IVM0|CCD50_HUMAN | 35,804 | 16 |
| Protein S100-A6 (S100 calcium-binding protein A6) (Calcyclin) (Prolactin receptor-associated protein) (PRA) (Growth factor-inducible protein 2A9) (MLN 4) | P06703|S10A6_HUMAN | 10,162 | 16 |
| 40S ribosomal protein S29 | P62273|RS29_HUMAN | 6,659 | 16 |
| Nucleoside diphosphate kinase A (EC 2.7.4.6) (NDK A) (NDP kinase A) (Tumor metastatic process-associated protein) (Metastasis inhibition factor nm23) (nm23-H1) (Granzyme A-activated DNase) (GAAD) | P15531|NDKA_HUMAN | 17,131 | 16 |
| Nucleosome assembly protein 1-like 4 (Nucleosome assembly protein 2) (NAP2) | Q99733|NP1L4_HUMAN | 42,806 | 15 |
| Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50) (56 kDa autoantigen) | P50995|ANX11_HUMAN | 54,374 | 15 |
| RNA-binding protein Raly (hnRNP associated with lethal yellow homolog) (Autoantigen p542) | Q9UKM9|RALY_HUMAN | 32,446 | 15 |
| Eukaryotic translation initiation factor 3 subunit 6-interacting protein | Q9Y262|IF3I_HUMAN | 66,711 | 15 |
| Acyl-coenzyme A thioesterase 9 (EC 3.1.2.-) (Acyl-CoA thioesterase 9) (Acyl-CoA thioester hydrolase 9) | Q9Y305|ACOT9_HUMAN | 46,337 | 15 |
| SUMO-activating enzyme subunit 2 (EC 6.3.2.-) (Ubiquitin-like 1-activating enzyme E1B) (Anthracycline-associated resistance ARX) | Q9UBT2|SAE2_HUMAN | 71,207 | 15 |
| Glycogen phosphorylase, liver form (EC 2.4.1.1) | P06737|PYGL_HUMAN | 97,134 | 15 |
| Splicing factor, arginine/serine-rich 7 (Splicing factor 9G8) | Q16629|SFRS7_HUMAN | 27,350 | 15 |
| Nucleolar protein Nop56 (Nucleolar protein 5A) | O00567|NOP56_HUMAN | 66,034 | 15 |
| FK506-binding protein 10 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (65 kDa FK506-binding protein) (FKBP65) (Immunophilin FKBP65) | Q96AY3|FKB10_HUMAN | 64,228 | 15 |
| Pyruvate dehydrogenase E1 component alpha subunit, somatic form, mitochondrial precursor (EC 1.2.4.1) (PDHE1-A type I) | P08559|ODPA_HUMAN | 43,279 | 15 |
| Adenylosuccinate lyase (EC 4.3.2.2) (Adenylosuccinase) (ASL) (ASASE) | P30566|PUR8_HUMAN | 54,873 | 15 |
| Serum deprivation-response protein (Phosphatidylserine-binding protein) (PS-p68) | O95810|SDPR_HUMAN | 47,155 | 15 |
| Suppressor of G2 allele of SKP1 homolog (Sgt1) (Putative 40-6-3 protein) | Q9Y2Z0|SUGT1_HUMAN | 41,007 | 15 |
| H/ACA ribonucleoprotein complex subunit 4 (EC 5.4.99.-) (Dyskerin) (Nucleolar protein family A member 4) (snoRNP protein DKC1) (Nopp140-associated protein of 57 kDa) (Nucleolar protein NAP57) (CBF5 homolog) | O60832|DKC1_HUMAN | 57,657 | 15 |
| Uroporphyrinogen decarboxylase (EC 4.1.1.37) (URO-D) (UPD) | P06132|DCUP_HUMAN | 40,769 | 15 |
| Deoxyuridine 5′-triphosphate nucleotidohydrolase, mitochondrial precursor (EC 3.6.1.23) (dUTPase) (dUTP pyrophosphatase) | P33316|DUT_HUMAN | 26,689 | 15 |
| Small nuclear ribonucleoprotein Sm D3 (snRNP core protein D3) (Sm-D3) | P62318|SMD3_HUMAN | 13,899 | 15 |
| Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta) | Q9Y5S2|MRCKB_HUMAN | 194,300 | 15 |
| Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70) | O94826|TOM70_HUMAN | 67,439 | 15 |
| Shwachman-Bodian-Diamond syndrome protein | Q9Y3A5|SBDS_HUMAN | 28,746 | 15 |
| Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2-oxoglutarate dehydrogenase complex) (E2) (E2K) | P36957|ODO2_HUMAN | 48,622 | 15 |
| Glutathione transferase omega-1 (EC 2.5.1.18) (GSTO 1-1) | P78417|GSTO1_HUMAN | 27,549 | 15 |
| MIR-interacting saposin-like protein precursor (Transmembrane protein 4) (Putative secreted protein ZSIG9) | Q9Y2B0|MSAP_HUMAN | 20,635 | 15 |
| RRP5 protein homolog (Programmed cell death protein 11) | Q14690|RRP5_HUMAN | 208,719 | 15 |
| Succinyl-CoA ligase [GDP-forming] beta-chain, mitochondrial precursor (EC 6.2.1.4) (Succinyl-CoA synthetase, betaG chain) (SCS-betaG) (GTP-specific succinyl-CoA synthetase subunit beta) | Q96I99|SUCB2_HUMAN | 46,494 | 15 |
| Eukaryotic translation initiation factor 4H (eIF-4H) (Williams-Beuren syndrome chromosome region 1 protein) | Q15056|IF4H_HUMAN | 27,368 | 15 |
| Spermine synthase (EC 2.5.1.22) (Spermidine aminopropyltransferase) (SPMSY) | P52788|SPSY_HUMAN | 41,252 | 15 |
| Proteasome subunit beta type 5 precursor (EC 3.4.25.1) (Proteasome epsilon chain) (Macropain epsilon chain) (Multicatalytic endopeptidase complex epsilon chain) (Proteasome subunit X) (Proteasome chain 6) (Proteasome subunit MB1) | P28074|PSB5_HUMAN | 22,879 | 15 |
| S-phase kinase-associated protein 1A (Cyclin A/CDK2-associated protein p19) (p19A) (p19skp1) (RNA polymerase II elongation factor-like protein) (Organ of Corti protein 2) (OCP-II protein) (OCP-2) (Transcription elongation factor B) (SIII) | P63208|SKP1_HUMAN | 18,640 | 15 |
| KH domain-containing, RNA-binding, signal transduction-associated protein 1 (p21 Ras GTPase-activating protein-associated p62) (GAP-associated tyrosine phosphoprotein p62) (Src-associated in mitosis 68 kDa protein) (Sam68) (p68) | Q07666|SAM68_HUMAN | 48,210 | 15 |
| Ras-related protein Rab-1A (YPT1-related protein) | P62820|RAB1A_HUMAN | 22,661 | 15 |
| Thioredoxin-like protein 5 (14 kDa thioredoxin-related protein) (TRP14) (Protein 42-9-9) | Q9BRA2|TXNL5_HUMAN | 13,922 | 15 |
| Proline synthetase co-transcribed bacterial homolog protein | O94903|PROSC_HUMAN | 30,326 | 15 |
| Vesicle-associated membrane protein-associated protein A (VAMP-associated protein A) (VAMP-A) (VAP-A) (33 kDa Vamp-associated protein) (VAP-33) | Q9P0L0|VAPA_HUMAN | 27,876 | 15 |
| Calcyclin-binding protein (CacyBP) (hCacyBP) (Siah-interacting protein) (S100A6-binding protein) | Q9HB71|CYBP_HUMAN | 26,192 | 15 |
| Microtubule-associated protein RP/EB family member 2 (APC-binding protein EB2) (End-binding protein 2) (EB2) | Q15555|MARE2_HUMAN | 37,014 | 15 |
| Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 63 kDa subunit precursor (EC 2.4.1.119) (Ribophorin II) (RPN-II) (RIBIIR) | P04844|RIB2_HUMAN | 69,267 | 15 |
| 60S ribosomal protein L36a (60S ribosomal protein L44) (Cell migration-inducing gene 6 protein) | P83881|RL36A_HUMAN | 12,423 | 15 |
| ADAMTS-1 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 1) (ADAM-TS 1) (ADAM-TS1) (METH-1) | Q9UHI8|ATS1_HUMAN | 105,340 | 15 |
| Fibrillin-1 precursor | P35555|FBN1_HUMAN | 312,283 | 15 |
| Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I) | O95786|DDX58_HUMAN | 106,586 | 15 |
| ADP-ribosylation factor 4 | P18085|ARF4_HUMAN, P61204|ARF3_HUMAN, P84077|ARF1_HUMAN | 20,494 | 15 |
| Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease) | P14735|IDE_HUMAN | 118,009 | 15 |
| NADP-dependent leukotriene B4 12-hydroxydehydrogenase (EC 1.3.1.74) (15-oxoprostaglandin 13-reductase) (EC 1.3.1.48) | Q14914|LTB4D_HUMAN | 35,853 | 14 |
| DNA replication licensing factor MCM4 (CDC21 homolog) (P1-CDC21) | P33991|MCM4_HUMAN | 96,543 | 14 |
| Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N-ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein) | P46459|NSF_HUMAN | 82,545 | 14 |
| Eukaryotic translation initiation factor 4 gamma 2 (eIF-4-gamma 2) (eIF-4G 2) (eIF4G 2) (p97) (Death-associated protein 5) (DAP-5) | P78344|IF4G2_HUMAN | 102,349 | 14 |
| NADPH:adrenodoxin oxidoreductase, mitochondrial precursor (EC 1.18.1.2) (Adrenodoxin reductase) (AR) (Ferredoxin reductase) (Ferredoxin--NADP(+) reductase) | P22570|ADRO_HUMAN | 53,819 | 14 |
| TAR DNA-binding protein 43 (TDP-43) | Q13148|TADBP_HUMAN | 44,722 | 14 |
| GMP synthase [glutamine-hydrolyzing] (EC 6.3.5.2) (Glutamine amidotransferase) (GMP synthetase) | P49915|GUAA_HUMAN | 76,699 | 14 |
| 4-trimethylaminobutyraldehyde dehydrogenase (EC 1.2.1.47) (TMABADH) (Aldehyde dehydrogenase 9A1) (EC 1.2.1.3) (Aldehyde dehydrogenase E3 isozyme) (Gamma-aminobutyraldehyde dehydrogenase) (EC 1.2.1.19) (R-aminobutyraldehyde dehydrogenase) | P49189|AL9A1_HUMAN | 53,784 | 14 |
| Alcohol dehydrogenase class 3 chi chain (EC 1.1.1.1) (Alcohol dehydrogenase class III chi chain) (S-(hydroxymethyl)glutathione dehydrogenase) (EC 1.1.1.284) (Glutathione-dependent formaldehyde dehydrogenase) (FDH) | P11766|ADHX_HUMAN | 39,706 | 14 |
| Tubulin beta-2A chain | Q13885|TBB2A_HUMAN | 49,889 | 14 |
| Pre-mRNA-processing-splicing factor 8 (Splicing factor Prp8) (PRP8 homolog) (220 kDa U5 snRNP-specific protein) (p220) | Q6P2Q9|PRP8_HUMAN | 273,591 | 14 |
| Serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform (EC 3.1.3.16) (PP2A-alpha) (Replication protein C) (RP-C) | P67775|PP2AA_HUMAN | 35,577 | 14 |
| Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150) | Q9Y2W1|TR150_HUMAN | 108,651 | 14 |
| Apoptotic chromatin condensation inducer in the nucleus (Acinus) | Q9UKV3|ACINU_HUMAN | 151,870 | 14 |
| COP9 signalosome complex subunit 2 (Signalosome subunit 2) (SGN2) (JAB1-containing signalosome subunit 2) (Thyroid receptor-interacting protein 15) (TRIP-15) (Alien homolog) | P61201|CSN2_HUMAN | 51,582 | 14 |
| GPI-anchored membrane protein 1 (GPI-anchored protein p137) (p137GPI) (Membrane component chromosome 11 surface marker 1) | Q14444|GPIA1_HUMAN | 72,732 | 14 |
| Cytoplasmic FMR1-interacting protein 1 (Specifically Rac1-associated protein 1) (Sra-1) (p140sra-1) | Q7L576|CYFP1_HUMAN | 145,169 | 14 |
| Sorcin (22 kDa protein) (CP-22) (V19) | P30626|SORCN_HUMAN | 21,659 | 14 |
| 14 kDa phosphohistidine phosphatase (EC 3.1.3.-) (Phosphohistidine phosphatase 1) (Protein janus-A homolog) | Q9NRX4|PHP14_HUMAN | 13,815 | 14 |
| Aminopeptidase B (EC 3.4.11.6) (Ap-B) (Arginyl aminopeptidase) (Arginine aminopeptidase) | Q9H4A4|AMPB_HUMAN | 72,579 | 14 |
| COP9 signalosome complex subunit 4 (Signalosome subunit 4) (SGN4) (JAB1-containing signalosome subunit 4) | Q9BT78|CSN4_HUMAN | 46,252 | 14 |
| Twinfilin-1 (Protein A6) (Protein tyrosine kinase 9) | Q12792|TWF1_HUMAN | 42,192 | 14 |
| Clathrin light chain A (Lca) | P09496|CLCA_HUMAN | 27,059 | 14 |
| Mitogen-activated protein kinase 1 (EC 2.7.11.24) (Extracellular signal-regulated kinase 2) (ERK-2) (Mitogen-activated protein kinase 2) (MAP kinase 2) (MAPK 2) (p42-MAPK) (ERT1) | P28482|MK01_HUMAN | 41,374 | 14 |
| DnaJ homolog subfamily C member 13 (Required for receptor-mediated endocytosis 8) | O75165|DNJCD_HUMAN | 254,410 | 14 |
| N-terminal acetyltransferase complex ARD1 subunit homolog A (EC 2.3.1.88) (EC 2.3.1.-) | P41227|ARD1H_HUMAN | 26,441 | 14 |
| NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin-activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin-activating enzyme E1C) | Q8TBC4|UBA3_HUMAN | 51,835 | 14 |
| Pirin | O00625|PIR_HUMAN | 32,096 | 14 |
| Low molecular weight phosphotyrosine protein phosphatase (EC 3.1.3.48) (LMW-PTPase) (LMW-PTP) (Low molecular weight cytosolic acid phosphatase) (EC 3.1.3.2) (Red cell acid phosphatase 1) (Adipocyte acid phosphatase) | P24666|PPAC_HUMAN | 18,025 | 14 |
| Copine-3 (Copine III) | O75131|CPNE3_HUMAN | 60,114 | 14 |
| Programmed cell death protein 5 (Protein TFAR19) (TF-1 cell apoptosis-related gene 19 protein) | O14737|PDCD5_HUMAN | 14,267 | 14 |
| 26S proteasome non-ATPase regulatory subunit 14 (26S proteasome regulatory subunit rpn11) (26S proteasome-associated PAD1 homolog 1) | O00487|PSDE_HUMAN | 34,559 | 14 |
| Plasminogen activator inhibitor 1 RNA-binding protein (PAI1 RNA-binding protein 1) (PAI-RBP1) (SERPINE1 mRNA-binding protein 1) | Q8NC51|PAIRB_HUMAN | 44,948 | 14 |
| Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10) | P13645|K1C10_HUMAN | 59,502 | 14 |
| Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP-7) (IGF-binding protein 7) (MAC25 protein) (Prostacyclin-stimulating factor) (PGI2-stimulating factor) (IGFBP-rP1) | Q16270|IBP7_HUMAN | 29,112 | 14 |
| Interferon-induced, double-stranded RNA-activated protein kinase (EC 2.7.11.1) (Interferon-inducible RNA-dependent protein kinase) (Eukaryotic translation initiation factor 2-alpha kinase 2) (eIF-2A protein kinase 2) (Protein kinase RNA-activated) (PKR) (p68 kinase) (P1/eIF-2A protein kinase) | P19525|E2AK2_HUMAN | 62,079 | 14 |
| Signal recognition particle 72 kDa protein (SRP72) | O76094|SRP72_HUMAN | 74,590 | 14 |
| Uncharacterized protein C19orf10 precursor (Stromal cell-derived growth factor SF20) (Interleukin-25) (IL-25) | Q969H8|CS010_HUMAN | 18,777 | 14 |
| Tripartite motif-containing protein 16 (Estrogen-responsive B box protein) | O95361|TRI16_HUMAN | 63,979 | 14 |
| Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B alpha isoform (PP2A, subunit B, B-alpha isoform) (PP2A, subunit B, B55-alpha isoform) (PP2A, subunit B, PR55-alpha isoform) (PP2A, subunit B, R2-alpha isoform) | P63151|2ABA_HUMAN | 51,675 | 14 |
| Crk-like protein | P46109|CRKL_HUMAN | 33,759 | 14 |
| Hematological and neurological expressed 1 protein (Androgen-regulated protein 2) | Q9UK76|HN1_HUMAN | 15,997 | 14 |
| Spermidine synthase (EC 2.5.1.16) (Putrescine aminopropyltransferase) (SPDSY) | P19623|SPEE_HUMAN | 33,806 | 14 |
| Small nuclear ribonucleoprotein Sm D1 (snRNP core protein D1) (Sm-D1) (Sm-D autoantigen) | P62314|SMD1_HUMAN | 13,264 | 14 |
| Podocalyxin-like protein 1 precursor | O00592|PODXL_HUMAN | 55,578 | 14 |
| Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (LC64P) | P13796|PLSL_HUMAN | 70,274 | 14 |
| 182 kDa tankyrase 1-binding protein | Q9C0C2|TB182_HUMAN | 181,763 | 13 |
| Utrophin (Dystrophin-related protein 1) (DRP1) (DRP) | P46939|UTRO_HUMAN | 394,477 | 13 |
| Plasma membrane calcium-transporting ATPase 4 (EC 3.6.3.8) (PMCA4) (Plasma membrane calcium pump isoform 4) (Plasma membrane calcium ATPase isoform 4) (Matrix-remodelling-associated protein 1) | P23634|AT2B4_HUMAN | 137,906 | 13 |
| Alkyldihydroxyacetonephosphate synthase, peroxisomal precursor (EC 2.5.1.26) (Alkyl-DHAP synthase) (Alkylglycerone-phosphate synthase) (Aging-associated gene 5 protein) | O00116|ADAS_HUMAN | 72,895 | 13 |
| Aldehyde dehydrogenase family 7 member A1 (EC 1.2.1.3) (Antiquitin-1) | P49419|AL7A1_HUMAN | 55,349 | 13 |
| Heme oxygenase 1 (EC 1.14.99.3) (HO-1) | P09601|HMOX1_HUMAN | 32,801 | 13 |
| Signal recognition particle 68 kDa protein (SRP68) | Q9UHB9|SRP68_HUMAN | 70,714 | 13 |
| Bcl-2-associated transcription factor 1 (Btf) | Q9NYF8|BCLF1_HUMAN | 106,107 | 13 |
| Complement component C1q receptor precursor (Complement component 1 q subcomponent receptor 1) (C1qR) (C1qRp) (C1qR(p)) (C1q/MBL/SPA receptor) (Matrix-remodelling-associated protein 4) (CD93 antigen) (CDw93) | Q9NPY3|C1QR1_HUMAN | 68,541 | 13 |
| Leucine-rich repeat-containing protein 47 | Q8N1G4|LRC47_HUMAN | 63,457 | 13 |
| N-acetylglucosamine kinase (EC 2.7.1.59) (GlcNAc kinase) | Q9UJ70|NAGK_HUMAN | 37,359 | 13 |
| Ribosomal L1 domain-containing protein 1 (Cellular senescence-inhibited gene protein) (Protein PBK1) (CATX-11) | O76021|RL1D1_HUMAN | 54,957 | 13 |
| Histone H2A.Z (H2A/z) | P0C0S5|H2AZ_HUMAN, Q71UI9|H2AV_HUMAN | 13,535 | 13 |
| Voltage-dependent anion-selective channel protein 3 (VDAC-3) (hVDAC3) (Outer mitochondrial membrane protein porin 3) | Q9Y277|VDAC3_HUMAN | 30,642 | 13 |
| Disabled homolog 2 (Differentially expressed protein 2) (DOC-2) | P98082|DAB2_HUMAN | 82,490 | 13 |
| cAMP-dependent protein kinase type II-alpha regulatory subunit | P13861|KAP2_HUMAN | 45,501 | 13 |
| UPF0318 protein FAM120A | Q9NZB2|F120A_HUMAN | 116,684 | 13 |
| LIM and senescent cell antigen-like-containing domain protein 1 (Particularly interesting new Cys-His protein 1) (PINCH-1) (Renal carcinoma antigen NY-REN-48) | P48059|LIMS1_HUMAN | 37,233 | 13 |
| PRKC apoptosis WT1 regulator protein (Prostate apoptosis response 4 protein) (Par-4) | Q96IZ0|PAWR_HUMAN | 36,550 | 13 |
| Uncharacterized protein KIAA1949 | Q6NYC8|K1949_HUMAN | 67,925 | 13 |
| Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase-targeting subunit 1) (Myosin phosphatase target subunit 1) (Protein phosphatase myosin-binding subunit) | O14974|MYPT1_HUMAN | 115,265 | 13 |
| Mitotic checkpoint protein BUB3 | O43684|BUB3_HUMAN | 37,137 | 13 |
| Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen) | Q9UQ35|SRRM2_HUMAN | 299,604 | 13 |
| Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1) | P52292|IMA2_HUMAN | 57,845 | 13 |
| 60S ribosomal protein L28 | P46779|RL28_HUMAN | 15,730 | 13 |
| Rho-related GTP-binding protein RhoG precursor | P84095|RHOG_HUMAN | 21,290 | 13 |
| Adenine phosphoribosyltransferase (EC 2.4.2.7) (APRT) | P07741|APT_HUMAN | 19,591 | 13 |
| Proteasome activator complex subunit 3 (Proteasome activator 28-gamma subunit) (PA28gamma) (PA28g) (Activator of multicatalytic protease subunit 3) (11S regulator complex subunit gamma) (REG-gamma) (Ki nuclear autoantigen) | P61289|PSME3_HUMAN | 29,489 | 13 |
| Ras-related protein Rab-2A | P61019|RAB2A_HUMAN | 23,528 | 13 |
| Ornithine aminotransferase, mitochondrial precursor (EC 2.6.1.13) (Ornithine--oxo-acid aminotransferase) [Contains: Ornithine aminotransferase, hepatic form; Ornithine aminotransferase, renal form] | P04181|OAT_HUMAN | 48,518 | 13 |
| Farnesyl pyrophosphate synthetase (FPP synthetase) (FPS) (Farnesyl diphosphate synthetase) [Includes: Dimethylallyltranstransferase (EC 2.5.1.1); Geranyltranstransferase (EC 2.5.1.10)] | P14324|FPPS_HUMAN | 40,516 | 13 |
| Thioredoxin domain-containing protein 4 precursor (Endoplasmic reticulum resident protein ERp44) | Q9BS26|TXND4_HUMAN | 46,955 | 13 |
| Translin | Q15631|TSN_HUMAN | 26,165 | 13 |
| Signal recognition particle 14 kDa protein (SRP14) (18 kDa Alu RNA-binding protein) | P37108|SRP14_HUMAN | 14,553 | 13 |
| Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1) | Q13526|PIN1_HUMAN | 18,226 | 13 |
| Signal recognition particle 9 kDa protein (SRP9) | P49458|SRP09_HUMAN | 10,094 | 13 |
| Glutaminase kidney isoform, mitochondrial precursor (EC 3.5.1.2) (GLS) (L-glutamine amidohydrolase) (K-glutaminase) | O94925|GLSK_HUMAN | 73,444 | 13 |
| Sad1/unc-84-like protein 2 (Rab5-interacting protein) (Rab5IP) | Q9UH99|UN84B_HUMAN | 80,294 | 13 |
| Eukaryotic translation initiation factor 5 (eIF-5) | P55010|IF5_HUMAN | 49,205 | 13 |
| Caspase-3 precursor (EC 3.4.22.56) (CASP-3) (Apopain) (Cysteine protease CPP32) (Yama protein) (CPP-32) (SREBP cleavage activity 1) (SCA-1) [Contains: Caspase-3 p17 subunit; Caspase-3 p12 subunit] | P42574|CASP3_HUMAN | 31,591 | 13 |
| Aldose reductase (EC 1.1.1.21) (AR) (Aldehyde reductase) | P15121|ALDR_HUMAN | 35,836 | 13 |
| Tripeptidyl-peptidase 1 precursor (EC 3.4.14.9) (Tripeptidyl-peptidase I) (TPP-1) (TPP-I) (Tripeptidyl aminopeptidase) (Lysosomal pepstatin insensitive protease) (LPIC) (Cell growth-inhibiting gene 1 protein) | O14773|TPP1_HUMAN | 61,230 | 13 |
| Nodal modulator 3 precursor (pM5 protein 3) | P69849|NOMO3_HUMAN, Q15155|NOMO1_HUMAN, Q5JPE7|NOMO2_HUMAN | 134,165 | 13 |
| ATP synthase gamma chain, mitochondrial precursor (EC 3.6.3.14) | P36542|ATPG_HUMAN | 32,980 | 13 |
| Activated RNA polymerase II transcriptional coactivator p15 (SUB1 homolog) (Positive cofactor 4) (PC4) (p14) | P53999|TCP4_HUMAN | 14,378 | 13 |
| Anamorsin (Cytokine-induced apoptosis inhibitor 1) (CUA001) | Q6FI81|CPIN1_HUMAN | 33,565 | 13 |
| Activator of 90 kDa heat shock protein ATPase homolog 1 (AHA1) (p38) | O95433|AHSA1_HUMAN | 38,256 | 13 |
| Histone-binding protein RBBP7 (Retinoblastoma-binding protein 7) (RBBP-7) (Retinoblastoma-binding protein p46) (Histone acetyltransferase type B subunit 2) (Nucleosome remodeling factor subunit RBAP46) | Q16576|RBBP7_HUMAN | 47,802 | 13 |
| Phosphoacetylglucosamine mutase (EC 5.4.2.3) (PAGM) (Acetylglucosamine phosphomutase) (N-acetylglucosamine-phosphate mutase) (Phosphoglucomutase 3) | O95394|AGM1_HUMAN | 59,834 | 13 |
| Reticulocalbin-2 precursor (Calcium-binding protein ERC-55) (E6-binding protein) (E6BP) | Q14257|RCN2_HUMAN | 36,860 | 13 |
| 60S ribosomal protein L35a | P18077|RL35A_HUMAN | 12,520 | 13 |
| Arylacetamide deacetylase-like 1 (EC 3.1.1.-) | Q6PIU2|ADCL1_HUMAN | 45,791 | 13 |
| 60S acidic ribosomal protein P1 | P05386|RLA1_HUMAN | 11,496 | 13 |
| High mobility group protein HMG-I/HMG-Y (HMG-I(Y)) (High mobility group AT-hook protein 1) (High mobility group protein A1) (High mobility group protein-R) | P17096|HMGA1_HUMAN | 11,658 | 13 |
| Calmodulin (CaM) | P62158|CALM_HUMAN | 16,820 | 13 |
| 40S ribosomal protein S30 | P62861|RS30_HUMAN | 6,630 | 13 |
| CD9 antigen (p24) (Leukocyte antigen MIC3) (Motility-related protein) (MRP-1) (Tetraspanin-29) (Tspan-29) | P21926|CD9_HUMAN | 25,399 | 13 |
| Serine/threonine-protein phosphatase 2A catalytic subunit beta isoform (EC 3.1.3.16) (PP2A-beta) | P62714|PP2AB_HUMAN | 35,557 | 13 |
| Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) | Q9Y2J2|E41L3_HUMAN | 120,662 | 13 |
| GTPase NRas precursor (Transforming protein N-Ras) | P01111|RASN_HUMAN | 21,211 | 13 |
| DNA replication licensing factor MCM3 (DNA polymerase alpha holoenzyme-associated protein P1) (RLF subunit beta) (P102 protein) (P1-MCM3) | P25205|MCM3_HUMAN | 90,965 | 12 |
| Dedicator of cytokinesis protein 6 | Q96HP0|DOCK6_HUMAN | 229,643 | 12 |
| Structural maintenance of chromosomes protein 2 (Chromosome-associated protein E) (hCAP-E) (XCAP-E homolog) | O95347|SMC2_HUMAN | 135,767 | 12 |
| Cysteinyl-tRNA synthetase, cytoplasmic (EC 6.1.1.16) (Cysteine--tRNA ligase) (CysRS) | P49589|SYCC_HUMAN | 85,458 | 12 |
| Cytoplasmic dynein 1 light intermediate chain 1 (Dynein light intermediate chain 1, cytosolic) (Dynein light chain A) (DLC-A) | Q9Y6G9|DC1L1_HUMAN | 56,562 | 12 |
| ATP-binding cassette sub-family F member 1 (ATP-binding cassette 50) (TNF-alpha-stimulated ABC protein) | Q8NE71|ABCF1_HUMAN | 95,910 | 12 |
| High mobility group protein B3 (High mobility group protein 4) (HMG-4) (High mobility group protein 2a) (HMG-2a) | O15347|HMGB3_HUMAN | 22,963 | 12 |
| Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1) (hSGT2) | Q86W92|LIPB1_HUMAN | 114,010 | 12 |
| Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit) (hTom34) | Q15785|OM34_HUMAN | 34,542 | 12 |
| Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1) (MTA1-L1 protein) (p53 target protein in deacetylase complex) | O94776|MTA2_HUMAN | 75,007 | 12 |
| Far upstream element-binding protein 3 (FUSE-binding protein 3) | Q96I24|FUBP3_HUMAN | 61,622 | 12 |
| NHP2-like protein 1 (High mobility group-like nuclear protein 2 homolog 1) (U4/U6.U5 tri-snRNP 15.5 kDa protein) (OTK27) (hSNU13) | P55769|NH2L1_HUMAN | 14,156 | 12 |
| PDZ and LIM domain protein 4 (LIM protein RIL) (Reversion-induced LIM protein) | P50479|PDLI4_HUMAN | 35,380 | 12 |
| Large proline-rich protein BAT3 (HLA-B-associated transcript 3) (Protein G3) | P46379|BAT3_HUMAN | 119,389 | 12 |
| Rab GDP dissociation inhibitor alpha (Rab GDI alpha) (Guanosine diphosphate dissociation inhibitor 1) (GDI-1) (XAP-4) (Oligophrenin-2) | P31150|GDIA_HUMAN | 50,566 | 12 |
| GrpE protein homolog 1, mitochondrial precursor (Mt-GrpE#1) (HMGE) | Q9HAV7|GRPE1_HUMAN | 24,261 | 12 |
| UNC45 homolog A (UNC-45A) (Smooth muscle cell-associated protein 1) (SMAP-1) | Q9H3U1|UN45A_HUMAN | 103,061 | 12 |
| La-related protein 1 (La ribonucleoprotein domain family member 1) | Q6PKG0|LARP1_HUMAN | 123,495 | 12 |
| Barrier-to-autointegration factor (Breakpoint cluster region protein 1) | O75531|BAF_HUMAN | 10,041 | 12 |
| Emerin | P50402|EMD_HUMAN | 28,977 | 12 |
| Ubiquitin carboxyl-terminal hydrolase isozyme L3 (EC 3.4.19.12) (UCH-L3) (Ubiquitin thioesterase L3) | P15374|UCHL3_HUMAN | 26,165 | 12 |
| Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.12) (Pyruvate dehydrogenase complex E2 subunit) (PDCE2) (E2) (Dihydrolipoamide S-acetyltransferase component of pyruvate dehydrogenase complex) (PDC-E2) (70 kDa mitochondrial autoantigen of primary biliary cirrhosis) (PBC) (M2 antigen complex 70 kDa subunit) | P10515|ODP2_HUMAN | 65,764 | 12 |
| Methionine aminopeptidase 1 (EC 3.4.11.18) (MetAP 1) (MAP 1) (Peptidase M 1) | P53582|AMPM1_HUMAN | 43,197 | 12 |
| Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15) (Unph-2) (Unph4) | Q9Y4E8|UBP15_HUMAN | 112,405 | 12 |
| 40S ribosomal protein S15 (RIG protein) | P62841|RS15_HUMAN | 17,023 | 12 |
| Copper transport protein ATOX1 (Metal transport protein ATX1) | O00244|ATOX1_HUMAN | 7,384 | 12 |
| Nucleobindin-2 precursor (DNA-binding protein NEFA) (Gastric cancer antigen Zg4) | P80303|NUCB2_HUMAN | 50,206 | 12 |
| Galactokinase (EC 2.7.1.6) (Galactose kinase) | P51570|GALK1_HUMAN | 42,254 | 12 |
| Squamous cell carcinoma antigen recognized by T-cells 3 (SART-3) (hSART-3) (Tat-interacting protein of 110 kDa) (Tip110) | Q15020|SART3_HUMAN | 109,918 | 12 |
| Electron transfer flavoprotein subunit beta (Beta-ETF) | P38117|ETFB_HUMAN | 27,826 | 12 |
| Importin-4 (Importin 4b) (Imp4b) (Ran-binding protein 4) (RanBP4) | Q8TEX9|IPO4_HUMAN | 118,701 | 12 |
| SH3 domain-binding glutamic acid-rich-like protein | O75368|SH3L1_HUMAN | 12,757 | 12 |
| 26S proteasome non-ATPase regulatory subunit 5 (26S proteasome subunit S5B) (26S protease subunit S5 basic) | Q16401|PSMD5_HUMAN | 56,179 | 12 |
| Eukaryotic translation initiation factor 3 subunit 1 (eIF-3 alpha) (eIF3 p35) (eIF3j) | O75822|IF31_HUMAN | 29,045 | 12 |
| Mitogen-activated protein kinase 3 (EC 2.7.11.24) (Extracellular signal-regulated kinase 1) (ERK-1) (Insulin-stimulated MAP2 kinase) (MAP kinase 1) (MAPK 1) (p44-ERK1) (ERT2) (p44-MAPK) (Microtubule-associated protein 2 kinase) | P27361|MK03_HUMAN | 43,119 | 12 |
| Cellular nucleic acid-binding protein (CNBP) (Zinc finger protein 9) | P62633|CNBP_HUMAN | 19,444 | 12 |
| Proteasome-associated protein ECM29 homolog (Ecm29) | Q5VYK3|ECM29_HUMAN | 204,278 | 12 |
| Actin-related protein 2/3 complex subunit 4 (ARP2/3 complex 20 kDa subunit) (p20-ARC) | P59998|ARPC4_HUMAN | 19,649 | 12 |
| Casein kinase II subunit alpha′ (EC 2.7.11.1) (CK II) | P19784|CSK22_HUMAN | 41,197 | 12 |
| Carbonyl reductase [NADPH] 3 (EC 1.1.1.184) (NADPH-dependent carbonyl reductase 3) | O75828|DHC3_HUMAN | 30,832 | 12 |
| DnaJ homolog subfamily C member 9 (DnaJ protein SB73) | Q8WXX5|DNJC9_HUMAN | 29,892 | 12 |
| HLA class I histocompatibility antigen, A-1 alpha chain precursor (MHC class I antigen A*1) | P30443|1A01_HUMAN | 40,828 | 12 |
| Adipocyte plasma membrane-associated protein (BSCv protein) | Q9HDC9|APMAP_HUMAN | 46,464 | 12 |
| ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1) | P46063|RECQ1_HUMAN | 73,440 | 12 |
| PEST proteolytic signal-containing nuclear protein (PEST-containing nuclear protein) (PCNP) | Q8WW12|PCNP_HUMAN | 18,907 | 12 |
| Eukaryotic translation initiation factor 4B (eIF-4B) | P23588|IF4B_HUMAN | 69,209 | 12 |
| Epsin-1 (EPS-15-interacting protein 1) (EH domain-binding mitotic phosphoprotein) | Q9Y6I3|EPN1_HUMAN | 57,558 | 12 |
| DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1) | Q96EP5|DAZP1_HUMAN | 43,365 | 12 |
| 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40) (Cyclophilin-related protein) | Q08752|PPID_HUMAN | 40,747 | 12 |
| Proto-oncogene tyrosine-protein kinase Yes (EC 2.7.10.2) (p61-Yes) (c-Yes) | P07947|YES_HUMAN | 60,785 | 12 |
| Uncharacterized protein KIAA0090 precursor | Q8N766|K0090_HUMAN | 111,743 | 12 |
| Protein transport protein Sec24D (SEC24-related protein D) | O94855|SC24D_HUMAN | 112,984 | 12 |
| FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (40 kDa SR-repressor protein) (SRrp40) (Splicing factor SRp38) | O75494|FUSIP_HUMAN | 31,284 | 12 |
| U1 small nuclear ribonucleoprotein A (U1 snRNP protein A) (U1A protein) (U1-A) | P09012|SNRPA_HUMAN | 31,262 | 12 |
| PRA1 family protein 3 (ARL-6-interacting protein 5) (ADP-ribosylation-like factor 6-interacting protein 5) (Aip-5) (Glutamate transporter EAAC1-interacting protein) (GTRAP3-18) (Prenylated Rab acceptor protein 2) (Protein JWa) (Dermal papilla-derived protein 11) (JM5) (Putative MAPK-activating protein PM27) (Cytoskeleton-related vitamin A-responsive protein) | O75915|PRAF3_HUMAN | 21,598 | 12 |
| Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f) | O00303|IF35_HUMAN | 37,546 | 12 |
| Diphosphoinositol polyphosphate phosphohydrolase 2 (EC 3.6.1.52) (DIPP-2) (Diadenosine 5′,5‴-P1,P6-hexaphosphate hydrolase 2) (EC 3.6.1.-) (Nucleoside diphosphate-linked moiety X motif 4) (Nudix motif 4) | Q9NZJ9|NUDT4_HUMAN | 20,288 | 12 |
| GTPase IMAP family member 1 (Immunity-associated protein 1) (hIMAP1) | Q8WWP7|GIMA1_HUMAN | 34,351 | 12 |
| NEDD8 precursor (Ubiquitin-like protein Nedd8) (Neddylin) | Q15843|NEDD8_HUMAN | 9,054 | 12 |
| Actin-related protein 2/3 complex subunit 3 (ARP2/3 complex 21 kDa subunit) (p21-ARC) | O15145|ARPC3_HUMAN | 20,530 | 12 |
| Antigen peptide transporter 1 (APT1) (Peptide transporter TAP1) (ATP-binding cassette sub-family B member 2) (Peptide transporter PSF1) (Peptide supply factor 1) (PSF-1) (Peptide transporter involved in antigen processing 1) | Q03518|TAP1_HUMAN | 80,948 | 12 |
| 60S ribosomal protein L15 | P61313|RL15_HUMAN | 24,129 | 12 |
| Protein S100-A16 (S100 calcium-binding protein A16) (Protein S100F) (Aging-associated protein 13) | Q96FQ6|S10AG_HUMAN | 11,784 | 12 |
| Cytochrome c oxidase subunit 5B, mitochondrial precursor (EC 1.9.3.1) (Cytochrome c oxidase polypeptide Vb) | P10606|COX5B_HUMAN | 13,678 | 12 |
| LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm) | Q9UHB6|LIMA1_HUMAN | 85,208 | 11 |
| DNA replication licensing factor MCM7 (CDC47 homolog) (P1.1-MCM3) | P33993|MCM7_HUMAN | 81,291 | 11 |
| Leucine zipper protein 1 | Q86V48|LUZP1_HUMAN | 120,259 | 11 |
| S-methyl-5-thioadenosine phosphorylase (EC 2.4.2.28) (5′-methylthioadenosine phosphorylase) (MTA phosphorylase) (MTAPase) | Q13126|MTAP_HUMAN | 31,232 | 11 |
| Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta 2 (Transducin beta chain 2) (G protein beta 2 subunit) | P62879|GBB2_HUMAN | 37,314 | 11 |
| Rho GTPase-activating protein 1 (GTPase-activating protein rhoOGAP) (Rho-related small GTPase protein activator) (CDC42 GTPase-activating protein) (p50-RhoGAP) | Q07960|RHG01_HUMAN | 50,420 | 11 |
| Prolyl endopeptidase (EC 3.4.21.26) (Post-proline cleaving enzyme) (PE) | P48147|PPCE_HUMAN | 80,748 | 11 |
| Macrophage capping protein (Actin-regulatory protein CAP-G) | P40121|CAPG_HUMAN | 38,500 | 11 |
| NAD(P) transhydrogenase, mitochondrial precursor (EC 1.6.1.2) (Pyridine nucleotide transhydrogenase) (Nicotinamide nucleotide transhydrogenase) | Q13423|NNTM_HUMAN | 113,881 | 11 |
| rRNA 2′-O-methyltransferase fibrillarin (EC 2.1.1.-) (34 kDa nucleolar scleroderma antigen) | P22087|FBRL_HUMAN | 33,766 | 11 |
| Citrate synthase, mitochondrial precursor (EC 2.3.3.1) | O75390|CISY_HUMAN | 51,696 | 11 |
| Neurolysin, mitochondrial precursor (EC 3.4.24.16) (Neurotensin endopeptidase) (Mitochondrial oligopeptidase M) (Microsomal endopeptidase) (MEP) | Q9BYT8|NEUL_HUMAN | 80,636 | 11 |
| Arsenite-resistance protein 2 | Q9BXP5|ARS2_HUMAN | 100,652 | 11 |
| Prolyl 4-hydroxylase subunit alpha-1 precursor (EC 1.14.11.2) (4-PH alpha-1) (Procollagen-proline,2-oxoglutarate-4-dioxygenase alpha-1 subunit) | P13674|P4HA1_HUMAN | 61,034 | 11 |
| Quaking protein (Hqk) | Q96PU8|QKI_HUMAN | 37,654 | 11 |
| 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-1) (Phospholipase C-gamma-1) (PLC-II) (PLC-148) | P19174|PLCG1_HUMAN | 148,518 | 11 |
| Rho guanine nucleotide exchange factor 1 (p115-RhoGEF) (p115RhoGEF) (115 kDa guanine nucleotide exchange factor) (Sub1.5) | Q92888|ARHG1_HUMAN | 102,420 | 11 |
| Alpha-synuclein (Non-A beta component of AD amyloid) (Non-A4 component of amyloid precursor) (NACP) | P37840|SYUA_HUMAN | 14,441 | 11 |
| BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene-3) (BAG-3) (Bcl-2-binding protein Bis) (Docking protein CAIR-1) | O95817|BAG3_HUMAN | 61,575 | 11 |
| 60S ribosomal protein L31 | P62899|RL31_HUMAN | 14,445 | 11 |
| Single-stranded DNA-binding protein, mitochondrial precursor (Mt-SSB) (MtSSB) (PWP1-interacting protein 17) | Q04837|SSB_HUMAN | 17,242 | 11 |
| Translocon-associated protein subunit delta precursor (TRAP-delta) (Signal sequence receptor subunit delta) (SSR-delta) | P51571|SSRD_HUMAN | 18,981 | 11 |
| Intercellular adhesion molecule 1 precursor (ICAM-1) (Major group rhinovirus receptor) (CD54 antigen) | P05362|ICAM1_HUMAN | 57,807 | 11 |
| Lanosterol synthase (EC 5.4.99.7) (Oxidosqualene--lanosterol cyclase) (2,3-epoxysqualene--lanosterol cyclase) (OSC) | P48449|ERG7_HUMAN | 83,292 | 11 |
| 3-mercaptopyruvate sulfurtransferase (EC 2.8.1.2) (MST) | P25325|THTM_HUMAN | 33,161 | 11 |
| Vacuolar protein sorting-associating protein 4B (Suppressor of K(+) transport growth defect 1) (Protein SKD1) | O75351|VPS4B_HUMAN | 49,286 | 11 |
| Transforming acidic coiled-coil-containing protein 1 (Taxin 1) (Gastric cancer antigen Ga55) | O75410|TACC1_HUMAN | 87,778 | 11 |
| Prolyl 3-hydroxylase 1 precursor (EC 1.14.11.7) (Leucine- and proline-enriched proteoglycan 1) (Leprecan-1) (Growth suppressor 1) | Q32P28|P3H1_HUMAN | 83,377 | 11 |
| Pyruvate dehydrogenase E1 component subunit beta, mitochondrial precursor (EC 1.2.4.1) (PDHE1-B) | P11177|ODPB_HUMAN | 39,215 | 11 |
| 26S proteasome non-ATPase regulatory subunit 10 (26S proteasome regulatory subunit p28) (Gankyrin) | O75832|PSD10_HUMAN | 24,410 | 11 |
| Glutathione S-transferase kappa 1 (EC 2.5.1.18) (GST 13-13) (Glutathione S-transferase subunit 13) (GST class-kappa) (GSTK1-1) (hGSTK1) | Q9Y2Q3|GSTK1_HUMAN | 25,480 | 11 |
| Proteasome subunit beta type 4 precursor (EC 3.4.25.1) (Proteasome beta chain) (Macropain beta chain) (Multicatalytic endopeptidase complex beta chain) (Proteasome chain 3) (HSN3) (HsBPROS26) | P28070|PSB4_HUMAN | 29,187 | 11 |
| Nitric-oxide synthase, endothelial (EC 1.14.13.39) (EC-NOS) (NOS type III) (NOSIII) (Endothelial NOS) (eNOS) (Constitutive NOS) (cNOS) | P29474|NOS3_HUMAN | 133,272 | 11 |
| Ubiquitin-activating enzyme E1-like protein 2 (Monocyte protein 4) (MOP-4) | A0AVT1|UB1L2_HUMAN | 117,955 | 11 |
| Hypoxanthine-guanine phosphoribosyltransferase (EC 2.4.2.8) (HGPRT) (HGPRTase) | P00492|HPRT_HUMAN | 24,562 | 11 |
| Basic leucine zipper and W2 domain-containing protein 2 | Q9Y6E2|BZW2_HUMAN | 48,146 | 11 |
| Protein arginine N-methyltransferase 1 (EC 2.1.1.-) (Interferon receptor 1-bound protein 4) | Q99873|ANM1_HUMAN | 41,468 | 11 |
| Casein kinase II subunit beta (CK II beta) (Phosvitin) (G5a) | P67870|CSK2B_HUMAN | 24,925 | 11 |
| Synaptosomal-associated protein 23 (SNAP-23) (Vesicle-membrane fusion protein SNAP-23) | O00161|SNP23_HUMAN | 23,337 | 11 |
| Protein S100-A13 (S100 calcium-binding protein A13) | Q99584|S10AD_HUMAN | 11,454 | 11 |
| Splicing factor 3A subunit 3 (Spliceosome-associated protein 61) (SAP 61) (SF3a60) | Q12874|SF3A3_HUMAN | 58,833 | 11 |
| Vesicle-associated membrane protein-associated protein B/C (VAMP-associated protein B/C) (VAMP-B/VAMP-C) (VAP-B/VAP-C) | O95292|VAPB_HUMAN | 27,211 | 11 |
| EF-hand domain-containing protein 2 (Swiprosin-1) | Q96C19|EFHD2_HUMAN | 26,680 | 11 |
| AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor-interacting protein) (Immunophilin homolog ARA9) (HBV-X-associated protein 2) | O00170|AIP_HUMAN | 37,647 | 11 |
| UDP-N-acetylhexosamine pyrophosphorylase (Antigen X) (AGX) (Sperm-associated antigen 2) [Includes: UDP-N-acetylgalactosamine pyrophosphorylase (EC 2.7.7.-) (AGX-1); UDP-N-acetylglucosamine pyrophosphorylase (EC 2.7.7.23) (AGX-2)] | Q16222|UAP1_HUMAN | 58,752 | 11 |
| Guanylate-binding protein 4 (GTP-binding protein 4) (Guanine nucleotide-binding protein 4) (GBP-4) | Q96PP9|GBP4_HUMAN | 73,148 | 11 |
| TATA-binding protein-associated factor 2N (RNA-binding protein 56) (TAFII68) (TAF(II)68) | Q92804|RBP56_HUMAN | 61,813 | 11 |
| ADP/ATP translocase 3 (Adenine nucleotide translocator 2) (ANT 3) (ADP,ATP carrier protein 3) (Solute carrier family 25 member 6) (ADP,ATP carrier protein, isoform T2) | P12236|ADT3_HUMAN | 32,849 | 11 |
| Protein TFG (TRK-fused gene protein) | Q92734|TFG_HUMAN | 43,416 | 11 |
| 60S ribosomal protein L37a | P61513|RL37A_HUMAN | 10,257 | 11 |
| Acidic leucine-rich nuclear phosphoprotein 32 family member E (LANP-like protein) (LANP-L) | Q9BTT0|AN32E_HUMAN | 30,675 | 11 |
| CD99 antigen precursor (T-cell surface glycoprotein E2) (E2 antigen) (Protein MIC2) (12E7) | P14209|CD99_HUMAN | 18,830 | 11 |
| N6-adenosine-methyltransferase 70 kDa subunit (EC 2.1.1.62) (MT-A70) (Methyltransferase-like protein 3) | Q86U44|MTA70_HUMAN | 64,457 | 11 |
| Plasminogen activator inhibitor 2 precursor (PAI-2) (Placental plasminogen activator inhibitor) (Monocyte Arg-serpin) (Urokinase inhibitor) | P05120|PAI2_HUMAN | 46,580 | 11 |
| Radixin | P35241|RADI_HUMAN | 68,548 | 10 |
| Rho GTPase-activating protein 18 (MacGAP) | Q8N392|RHG18_HUMAN | 74,931 | 10 |
| Nidogen-1 precursor (Entactin) | P14543|NID1_HUMAN | 136,434 | 10 |
| Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PP-T) (PPT) | P53041|PPP5_HUMAN | 56,862 | 10 |
| Ubiquinol-cytochrome c reductase complex 14 kDa protein (EC 1.10.2.2) (Complex III subunit VI) (QP-C) | P14927|UCR6_HUMAN | 13,513 | 10 |
| ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) | Q8IUD2|RB6I2_HUMAN | 128,073 | 10 |
| Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip) | Q6WCQ1|MRIP_HUMAN | 116,430 | 10 |
| Secernin-1 | Q12765|SCRN1_HUMAN | 46,364 | 10 |
| DnaJ homolog subfamily C member 8 (Splicing protein spf31) | O75937|DNJC8_HUMAN | 29,824 | 10 |
| Acyl-protein thioesterase 2 (EC 3.1.2.-) (Lysophospholipase II) (LPL-I) | O95372|LYPA2_HUMAN | 24,719 | 10 |
| Heme-binding protein 1 (p22HBP) | Q9NRV9|HEBP1_HUMAN | 21,079 | 10 |
| Heme-binding protein 2 (Protein SOUL) (Placental protein 23) (PP23) | Q9Y5Z4|HEBP2_HUMAN | 22,858 | 10 |
| Leucine zipper-EF-hand-containing transmembrane protein 1, mitochondrial precursor | O95202|LETM1_HUMAN | 83,338 | 10 |
| Protein NOXP20 (Nervous system over-expressed protein 20) (Protein FAM114A1) | Q8IWE2|NXP20_HUMAN | 60,807 | 10 |
| Dipeptidyl-peptidase 1 precursor (EC 3.4.14.1) (Dipeptidyl-peptidase I) (DPP-I) (DPPI) (Cathepsin C) (Cathepsin J) (Dipeptidyl transferase) [Contains: Dipeptidyl-peptidase 1 exclusion domain chain (Dipeptidyl-peptidase I exclusion domain chain); Dipeptidyl-peptidase 1 heavy chain (Dipeptidyl-peptidase I heavy chain); Dipeptidyl-peptidase 1 light chain (Dipeptidyl-peptidase I light chain)] | P53634|CATC_HUMAN | 51,824 | 10 |
| Programmed cell death protein 6 (Probable calcium-binding protein ALG-2) | O75340|PDCD6_HUMAN | 21,851 | 10 |
| Uncharacterized protein KIAA0310 | O15027|K0310_HUMAN | 233,497 | 10 |
| Casein kinase II subunit alpha (EC 2.7.11.1) (CK II) | P68400|CSK21_HUMAN | 45,127 | 10 |
| Structural maintenance of chromosomes protein 4 (Chromosome-associated polypeptide C) (hCAP-C) (XCAP-C homolog) | Q9NTJ3|SMC4_HUMAN | 147,170 | 10 |
| Apoptosis-inducing factor 1, mitochondrial precursor (EC 1.-.-.-) (Programmed cell death protein 8) | O95831|AIFM1_HUMAN | 66,884 | 10 |
| DNA fragmentation factor subunit alpha (DNA fragmentation factor 45 kDa subunit) (DFF-45) (Inhibitor of CAD) (ICAD) | O00273|DFFA_HUMAN | 36,505 | 10 |
| Vacuolar ATP synthase subunit E (EC 3.6.3.14) (V-ATPase E subunit) (Vacuolar proton pump E subunit) (V-ATPase 31 kDa subunit) (P31) | P36543|VATE_HUMAN | 26,128 | 10 |
| Latexin (Endogenous carboxypeptidase inhibitor) (ECI) (Tissue carboxypeptidase inhibitor) (TCI) (MUM) | Q9BS40|LXN_HUMAN | 25,751 | 10 |
| Coiled-coil-helix-coiled-coil-helix domain-containing protein 3 | Q9NX63|CHCH3_HUMAN | 26,135 | 10 |
| Splicing factor U2AF 65 kDa subunit (U2 auxiliary factor 65 kDa subunit) (U2 snRNP auxiliary factor large subunit) (hU2AF(65)) | P26368|U2AF2_HUMAN | 53,483 | 10 |
| 26S proteasome non-ATPase regulatory subunit 12 (26S proteasome regulatory subunit p55) | O00232|PSD12_HUMAN | 52,888 | 10 |
| Epididymal secretory protein E1 precursor (Niemann-Pick disease type C2 protein) (hE1) | P61916|NPC2_HUMAN | 16,552 | 10 |
| Coiled-coil domain-containing protein 47 precursor | Q96A33|CCD47_HUMAN | 55,857 | 10 |
| Signal recognition particle receptor subunit beta (SR-beta) (Protein APMCF1) | Q9Y5M8|SRPRB_HUMAN | 29,685 | 10 |
| Protein arginine N-methyltransferase 5 (EC 2.1.1.125) (EC 2.1.1.-) (Shk1 kinase-binding protein 1 homolog) (SKB1Hs) (Jak-binding protein 1) (72 kDa ICln-binding protein) | O14744|ANM5_HUMAN | 72,667 | 10 |
| Importin-7 (Imp7) (Ran-binding protein 7) (RanBP7) | O95373|IPO7_HUMAN | 119,502 | 10 |
| Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase) | O95819|M4K4_HUMAN | 142,083 | 10 |
| Palmdelphin (Paralemnin-like protein) | Q9NP74|PALMD_HUMAN | 62,741 | 10 |
| Histone deacetylase 1 (HD1) | Q13547|HDAC1_HUMAN | 55,086 | 10 |
| Vinexin (Sorbin and SH3 domain-containing protein 3) (SH3-containing adapter molecule 1) (SCAM-1) | O60504|VINEX_HUMAN | 75,312 | 10 |
| Reticulocalbin-3 precursor (EF-hand calcium-binding protein RLP49) | Q96D15|RCN3_HUMAN | 37,475 | 10 |
| C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8-interacting protein 4) (Human lung cancer protein 6) (HLC-6) (Proliferation-inducing protein 6) (Sperm-specific protein) (Sperm surface protein) (Protein highly expressed in testis) (PHET) (Sunday driver 1) | O60271|JIP4_HUMAN | 146,187 | 10 |
| Histone acetyltransferase type B catalytic subunit (EC 2.3.1.48) | O14929|HAT1_HUMAN | 49,496 | 10 |
| Nuclear factor NF-kappa-B p105 subunit (DNA-binding factor KBF1) (EBP-1) [Contains: Nuclear factor NF-kappa-B p50 subunit] | P19838|NFKB1_HUMAN | 105,341 | 10 |
| Basic leucine zipper and W2 domain-containing protein 1 (Protein Orf) | Q7L1Q6|BZW1_HUMAN | 48,027 | 10 |
| Inosine triphosphate pyrophosphatase (EC 3.6.1.19) (ITPase) (Inosine triphosphatase) (Putative oncogene protein hlc14-06-p) | Q9BY32|ITPA_HUMAN | 21,428 | 10 |
| SEC13-related protein (SEC13-like protein 1) | P55735|SEC13_HUMAN | 35,522 | 10 |
| SUMO-conjugating enzyme UBC9 (EC 6.3.2.-) (SUMO-protein ligase) (Ubiquitin-conjugating enzyme E2 I) (Ubiquitin-protein ligase I) (Ubiquitin carrier protein I) (Ubiquitin carrier protein 9) (p18) | P63279|UBC9_HUMAN | 17,990 | 10 |
| U6 snRNA-associated Sm-like protein LSm4 (Glycine-rich protein) (GRP) | Q9Y4Z0|LSM4_HUMAN | 15,332 | 10 |
| RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2) | Q15287|RNPS1_HUMAN | 34,192 | 10 |
| DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa) | Q13217|DNJC3_HUMAN | 57,564 | 10 |
| Pre-mRNA-processing factor 6 (PRP6 homolog) (U5 snRNP-associated 102 kDa protein) (U5-102 kDa protein) | O94906|PRP6_HUMAN | 106,910 | 10 |
| Endothelial cell-selective adhesion molecule precursor | Q96AP7|ESAM_HUMAN | 41,158 | 10 |
| Cytochrome c oxidase subunit 2 (EC 1.9.3.1) (Cytochrome c oxidase polypeptide II) | P00403|COX2_HUMAN | 25,548 | 10 |
| Replication protein A 70 kDa DNA-binding subunit (RP-A) (RF-A) (Replication factor-A protein 1) (Single-stranded DNA-binding protein) (p70) | P27694|RFA1_HUMAN | 68,121 | 10 |
| Peptidyl-tRNA hydrolase 2, mitochondrial precursor (EC 3.1.1.29) (PTH 2) (Bcl-2 inhibitor of transcription 1) | Q9Y3E5|PTH2_HUMAN | 19,176 | 10 |
| Transmembrane protein 173 | Q86WV6|TM173_HUMAN | 42,176 | 10 |
| Keratin, type I cytoskeletal 9 (Cytokeratin-9) (CK-9) (Keratin-9) (K9) | P35527|K1C9_HUMAN | 62,113 | 10 |
| Splicing factor U2AF 35 kDa subunit (U2 auxiliary factor 35 kDa subunit) (U2 snRNP auxiliary factor small subunit) | Q01081|U2AF1_HUMAN | 27,854 | 10 |
| Charged multivesicular body protein 5 (Chromatin-modifying protein 5) (Vacuolar protein sorting 60) (Vps60) (hVps60) (SNF7 domain-containing protein 2) | Q9NZZ3|CHMP5_HUMAN | 24,554 | 10 |
| Interferon-induced 17 kDa protein precursor [Contains: Ubiquitin cross-reactive protein (hUCRP) (Interferon-induced 15 kDa protein)] | P05161|UCRP_HUMAN | 17,869 | 10 |
| Inosine-5′-monophosphate dehydrogenase 1 (EC 1.1.1.205) (IMP dehydrogenase 1) (IMPDH-I) (IMPD 1) | P20839|IMDH1_HUMAN | 55,389 | 10 |
| ERO1-like protein alpha precursor (EC 1.8.4.-) (ERO1-Lalpha) (Oxidoreductin-1-Lalpha) (Endoplasmic oxidoreductin-1-like protein) (ERO1-L) | Q96HE7|ERO1A_HUMAN | 54,377 | 10 |
| Keratin, type I cytoskeletal 16 (Cytokeratin-16) (CK-16) (Keratin-16) (K16) | P08779|K1C16_HUMAN | 51,251 | 10 |
| Thymidine phosphorylase precursor (EC 2.4.2.4) (TdRPase) (TP) (Platelet-derived endothelial cell growth factor) (PD-ECGF) (Gliostatin) | P19971|TYPH_HUMAN | 49,938 | 10 |
| Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620) | Q9UPN3|MACF1_HUMAN | 620,397 | 9 |
| STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9) | Q9H2G2|SLK_HUMAN | 142,680 | 9 |
| Structural maintenance of chromosomes protein 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) | Q9UQE7|SMC3_HUMAN | 141,529 | 9 |
| Myosin-Id | O94832|MYO1D_HUMAN | 116,188 | 9 |
| Coronin-7 (70 kDa WD repeat tumor rejection antigen homolog) | P57737|CORO7_HUMAN | 100,558 | 9 |
| Signal recognition particle receptor subunit alpha (SR-alpha) (Docking protein alpha) (DP-alpha) | P08240|SRPR_HUMAN | 69,795 | 9 |
| Biliverdin reductase A precursor (EC 1.3.1.24) (Biliverdin-IX alpha-reductase) (BVR A) | P53004|BIEA_HUMAN | 33,411 | 9 |
| SEC23-interacting protein (p125) | Q9Y6Y8|S23IP_HUMAN | 111,060 | 9 |
| Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (AICD-59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (AICD-57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)) (AICD-50); C31] | P05067|A4_HUMAN | 86,923 | 9 |
| Fragile X mental retardation syndrome-related protein 1 (hFXR1p) | P51114|FXR1_HUMAN | 69,674 | 9 |
| Flap endonuclease 1 (EC 3.1.-.-) (Flap structure-specific endonuclease 1) (FEN-1) (Maturation factor 1) (MF1) (hFEN-1) (DNase IV) | P39748|FEN1_HUMAN | 42,576 | 9 |
| Protein transport protein Sec24C (SEC24-related protein C) | P53992|SC24C_HUMAN | 118,297 | 9 |
| 6-phosphofructokinase, muscle type (EC 2.7.1.11) (Phosphofructokinase 1) (Phosphohexokinase) (Phosphofructo-1-kinase isozyme A) (PFK-A) (Phosphofructokinase-M) | P08237|K6PF_HUMAN | 85,166 | 9 |
| Transcription elongation factor A protein 1 (Transcription elongation factor S-II protein 1) (Transcription elongation factor TFIIS.o) | P23193|TCEA1_HUMAN | 33,953 | 9 |
| Ras-related C3 botulinum toxin substrate 2 precursor (p21-Rac2) (Small G protein) (GX) | P15153|RAC2_HUMAN | 21,411 | 9 |
| SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170) | Q8TAQ2|SMRC2_HUMAN | 132,862 | 9 |
| DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase HsaI) (DNA MTase HsaI) (MCMT) (M.HsaI) | P26358|DNMT1_HUMAN | 183,151 | 9 |
| Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52) | O75439|MPPB_HUMAN | 54,349 | 9 |
| RNA-binding protein EWS (EWS oncogene) (Ewing sarcoma breakpoint region 1 protein) | Q01844|EWS_HUMAN | 68,460 | 9 |
| Ras-related protein R-Ras precursor (p23) | P10301|RRAS_HUMAN | 23,463 | 9 |
| GDP-fucose protein O-fucosyltransferase 1 precursor (EC 2.4.1.221) (Peptide-O-fucosyltransferase 1) (O-FucT-1) | Q9H488|OFUT1_HUMAN | 43,938 | 9 |
| 40S ribosomal protein S27a | P62979|RS27A_HUMAN | 9,400 | 9 |
| WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP) | Q9Y2W2|WBP11_HUMAN | 69,982 | 9 |
| FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (FACT 80 kDa subunit) (FACTp80) (Chromatin-specific transcription elongation factor 80 kDa subunit) (Structure-specific recognition protein 1) (hSSRP1) (Recombination signal sequence recognition protein 1) (T160) | Q08945|SSRP1_HUMAN | 81,060 | 9 |
| DNA replication licensing factor MCM6 (p105MCM) | Q14566|MCM6_HUMAN | 92,873 | 9 |
| Bifunctional 3′-phosphoadenosine 5′-phosphosulfate synthetase 1 (PAPS synthetase 1) (PAPSS 1) (Sulfurylase kinase 1) (SK1) (SK 1) [Includes: Sulfate adenylyltransferase (EC 2.7.7.4) (Sulfate adenylate transferase) (SAT) (ATP-sulfurylase); Adenylyl-sulfate kinase (EC 2.7.1.25) (Adenylylsulfate 3′-phosphotransferase) (APS kinase) (Adenosine-5′-phosphosulfate 3′-phosphotransferase) (3′-phosphoadenosine-5′-phosphosulfate synthetase)] | O43252|PAPS1_HUMAN | 70,815 | 9 |
| Syntaxin-12 | Q86Y82|STX12_HUMAN | 31,625 | 9 |
| Peptidyl-prolyl cis-trans isomerase, mitochondrial precursor (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin F) | P30405|PPIF_HUMAN | 22,022 | 9 |
| 39S ribosomal protein L12, mitochondrial precursor (L12mt) (MRP-L12) (5c5-2) | P52815|RM12_HUMAN | 21,330 | 9 |
| Ubiquitin | P62988|UBIQ_HUMAN | 8,547 | 9 |
| THO complex subunit 4 (Tho4) (Ally of AML-1 and LEF-1) (Transcriptional coactivator Aly/REF) (bZIP-enhancing factor BEF) | Q86V81|THOC4_HUMAN | 26,871 | 9 |
| Proteasome subunit beta type 6 precursor (EC 3.4.25.1) (Proteasome delta chain) (Macropain delta chain) (Multicatalytic endopeptidase complex delta chain) (Proteasome subunit Y) | P28072|PSB6_HUMAN | 25,340 | 9 |
| Protein phosphatase 1 regulatory subunit 14B (Phospholipase C beta 3 neighbouring gene protein) | Q96C90|PP14B_HUMAN | 15,894 | 9 |
| Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25) | P45973|CBX5_HUMAN | 22,208 | 9 |
| Supervillin (Archvillin) (p205/p250) | O95425|SVIL_HUMAN | 247,689 | 9 |
| NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 51 kDa subunit) (Complex I-51kD) (CI-51kD) (NADH dehydrogenase flavoprotein 1) | P49821|NDUV1_HUMAN | 50,800 | 9 |
| ETHE1 protein, mitochondrial precursor (EC 3.-.-.-) (Ethylmalonic encephalopathy protein 1) (Hepatoma subtracted clone one protein) | O95571|ETHE1_HUMAN | 27,855 | 9 |
| Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-) | Q13011|ECH1_HUMAN | 35,798 | 9 |
| DnaJ homolog subfamily B member 4 (Heat shock 40 kDa protein 1 homolog) (Heat shock protein 40 homolog) (HSP40 homolog) | Q9UDY4|DNJB4_HUMAN | 37,791 | 9 |
| Cytochrome c oxidase polypeptide VIc precursor (EC 1.9.3.1) | P09669|COX6C_HUMAN | 8,764 | 9 |
| Ras suppressor protein 1 (Rsu-1) (RSP-1) | Q15404|RSU1_HUMAN | 31,524 | 9 |
| Developmentally-regulated GTP-binding protein 1 (DRG 1) | Q9Y295|DRG1_HUMAN | 40,526 | 9 |
| Prefoldin subunit 2 | Q9UHV9|PFD2_HUMAN | 16,630 | 9 |
| AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha-2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit) (Huntingtin-interacting protein HYPJ) | O94973|AP2A2_HUMAN | 103,945 | 9 |
| 2′,3′-cyclic-nucleotide 3′-phosphodiesterase (EC 3.1.4.37) (CNP) (CNPase) | P09543|CN37_HUMAN | 47,563 | 9 |
| Golgi-specific brefeldin A-resistance guanine nucleotide exchange factor 1 (BFA-resistant GEF 1) | Q92538|GBF1_HUMAN | 206,433 | 9 |
| Ubiquitin-conjugating enzyme E2-25 kDa (EC 6.3.2.19) (Ubiquitin-protein ligase) (Ubiquitin carrier protein) (E2(25K)) (Huntingtin-interacting protein 2) (HIP-2) | P61086|UBC1_HUMAN | 22,389 | 9 |
| Cytospin-A (SPECC1-like protein) (Renal carcinoma antigen NY-REN-22) | Q69YQ0|CYTSA_HUMAN | 124,578 | 9 |
| Transferrin receptor protein 1 (TfR1) (TR) (TfR) (Trfr) (CD71 antigen) (T9) (p90) | P02786|TFR1_HUMAN | 84,856 | 9 |
| Succinyl-CoA ligase [GDP-forming] subunit alpha, mitochondrial precursor (EC 6.2.1.4) (Succinyl-CoA synthetase subunit alpha) (SCS-alpha) | P53597|SUCA_HUMAN | 35,030 | 9 |
| Ubiquinol-cytochrome c reductase complex 11 kDa protein, mitochondrial precursor (EC 1.10.2.2) (Mitochondrial hinge protein) (Cytochrome c1 nonheme 11 kDa protein) (Complex III subunit VIII) | P07919|UCRH_HUMAN | 10,721 | 9 |
| GDP-L-fucose synthetase (EC 1.1.1.271) (Protein FX) (Red cell NADP(H)-binding protein) (GDP-4-keto-6-deoxy-D-mannose-3,5-epimerase-4-reductase) | Q13630|FCL_HUMAN | 35,875 | 9 |
| Protein disulfide-isomerase A5 precursor (EC 5.3.4.1) (Protein disulfide isomerase-related protein) | Q14554|PDIA5_HUMAN | 59,577 | 9 |
| Adhesion-regulating molecule 1 precursor (110 kDa cell membrane glycoprotein) (Gp110) | Q16186|ADRM1_HUMAN | 42,136 | 9 |
| Proto-oncogene C-crk (p38) (Adapter molecule crk) | P46108|CRK_HUMAN | 33,854 | 9 |
| Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7–60 protein) | Q9NZT2|OGFR_HUMAN | 73,307 | 9 |
| Eukaryotic translation initiation factor 3 subunit 12 (eIF-3 p25) (eIF-3 p28) (eIF3k) (Muscle-specific gene M9 protein) (PLAC-24) | Q9UBQ5|IF3C_HUMAN | 25,042 | 9 |
| Arsenical pump-driving ATPase (EC 3.6.3.16) (Arsenite-translocating ATPase) (Arsenical resistance ATPase) (Arsenite-transporting ATPase) (ARSA) (ASNA-I) | O43681|ARSA1_HUMAN | 38,776 | 9 |
| Glutamate-rich WD repeat-containing protein 1 | Q9BQ67|GRWD1_HUMAN | 49,400 | 9 |
| Poly [ADP-ribose] polymerase 14 (EC 2.4.2.30) (PARP-14) (B aggressive lymphoma protein 2) | Q460N5|PAR14_HUMAN | 193,737 | 9 |
| Small nuclear ribonucleoprotein E (snRNP-E) (Sm protein E) (Sm-E) (SmE) | P62304|RUXE_HUMAN | 10,786 | 9 |
| Tyrosine-protein phosphatase non-receptor type 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1B) (PTP-1B) | P18031|PTN1_HUMAN | 49,950 | 9 |
| 52 kDa Ro protein (Sjoegren syndrome type A antigen) (SS-A) (Ro(SS-A)) (52 kDa ribonucleoprotein autoantigen Ro/SS-A) (Tripartite motif-containing protein 21) (RING finger protein 81) | P19474|RO52_HUMAN | 54,152 | 9 |
| Parathymosin | P20962|PTMS_HUMAN | 11,512 | 9 |
| Nucleolysin TIAR (TIA-1-related protein) | Q01085|TIAR_HUMAN | 41,572 | 9 |
| NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 30 kDa subunit) (Complex I-30kD) (CI-30kD) | O75489|NDUS3_HUMAN | 30,224 | 9 |
| Protein FAM49A | Q9H0Q0|FA49A_HUMAN | 37,296 | 9 |
| WD repeat protein 74 (NOP seven-associated protein 1) | Q6RFH5|WDR74_HUMAN | 42,423 | 9 |
| BolA-like protein 2 | Q9H3K6|BOLA2_HUMAN | 10,098 | 9 |
| Ubiquitin-conjugating enzyme E2 variant 1 (UEV-1) (CROC-1) (Ubiquitin-conjugating enzyme variant Kua) (TRAF6-regulated IKK activator 1 beta Uev1A) | Q13404|UB2V1_HUMAN | 25,779 | 9 |
| Cordon-bleu protein-like 1 | Q53SF7|CBLL1_HUMAN | 131,771 | 9 |
| Cytochrome c1 heme protein, mitochondrial precursor (Cytochrome c-1) | P08574|CY1_HUMAN | 35,373 | 9 |
| Cytochrome b5 type B precursor (Cytochrome b5 outer mitochondrial membrane isoform) | O43169|CYB5B_HUMAN | 16,314 | 9 |
| Sorting nexin-6 (TRAF4-associated factor 2) | Q9UNH7|SNX6_HUMAN | 46,632 | 8 |
| RNA-binding protein 14 (RNA-binding motif protein 14) (RRM-containing coactivator activator/modulator) (Synaptotagmin-interacting protein) (SYT-interacting protein) | Q96PK6|RBM14_HUMAN | 69,474 | 8 |
| Tubulin-specific chaperone B (Tubulin folding cofactor B) (Cytoskeleton-associated protein 1) (Cytoskeleton-associated protein CKAPI) | Q99426|TBCB_HUMAN | 27,308 | 8 |
| Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10) | Q14694|UBP10_HUMAN | 87,117 | 8 |
| SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 (EC 3.6.1.-) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin A5) (Sucrose nonfermenting protein 2 homolog) (hSNF2H) | O60264|SMCA5_HUMAN | 121,893 | 8 |
| Succinate dehydrogenase [ubiquinone] iron-sulfur subunit, mitochondrial precursor (EC 1.3.5.1) (Ip) (Iron-sulfur subunit of complex II) | P21912|DHSB_HUMAN | 31,613 | 8 |
| Protein FAM21C | Q9Y4E1|FA21C_HUMAN | 144,897 | 8 |
| Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP-dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta) | Q14839|CHD4_HUMAN | 217,977 | 8 |
| Putative RNA methyltransferase NOL1 (EC 2.1.1.-) (Proliferating-cell nucleolar antigen p120) (Proliferation-associated nucleolar protein p120) | P46087|NOL1_HUMAN | 89,286 | 8 |
| Dynactin subunit 4 (Dynactin subunit p62) | Q9UJW0|DCTN4_HUMAN | 52,320 | 8 |
| U4/U6 small nuclear ribonucleoprotein Prp4 (U4/U6 snRNP 60 kDa protein) (WD splicing factor Prp4) (hPrp4) (PRP4 homolog) | O43172|PRP4_HUMAN | 58,432 | 8 |
| Elongation factor G 1, mitochondrial precursor (mEF-G 1) (Elongation factor G1) | Q96RP9|EFG1_HUMAN | 83,456 | 8 |
| Protocadherin-1 precursor (Protocadherin-42) (PC42) (Cadherin-like protein 1) | Q08174|PCDH1_HUMAN | 111,253 | 8 |
| Catalase (EC 1.11.1.6) | P04040|CATA_HUMAN | 59,739 | 8 |
| Actin-binding LIM protein 1 (Actin-binding LIM protein family member 1) (abLIM-1) (Actin-binding double-zinc-finger protein) (LIMAB1) (Limatin) | O14639|ABLM1_HUMAN | 87,628 | 8 |
| Splicing factor, arginine/serine-rich 9 (Pre-mRNA-splicing factor SRp30C) | Q13242|SFRS9_HUMAN | 25,525 | 8 |
| Thymidylate kinase (EC 2.7.4.9) (dTMP kinase) | P23919|DTYMK_HUMAN | 23,802 | 8 |
| 28 kDa heat- and acid-stable phosphoprotein (PDGF-associated protein) (PAP) (PDGFA-associated protein 1) (PAP1) | Q13442|HAP28_HUMAN | 20,613 | 8 |
| Thimet oligopeptidase (EC 3.4.24.15) (Endopeptidase 24.15) (MP78) | P52888|MEPD_HUMAN | 78,823 | 8 |
| Sorting nexin-2 (Transformation-related gene 9 protein) (TRG-9) | O60749|SNX2_HUMAN | 58,454 | 8 |
| Aspartate aminotransferase, cytoplasmic (EC 2.6.1.1) (Transaminase A) (Glutamate oxaloacetate transaminase 1) | P17174|AATC_HUMAN | 46,230 | 8 |
| Proteasome subunit alpha type 4 (EC 3.4.25.1) (Proteasome component C9) (Macropain subunit C9) (Multicatalytic endopeptidase complex subunit C9) (Proteasome subunit L) | P25789|PSA4_HUMAN | 29,467 | 8 |
| NADP-dependent malic enzyme (EC 1.1.1.40) (NADP-ME) (Malic enzyme 1) | P48163|MAOX_HUMAN | 64,133 | 8 |
| Cleavage and polyadenylation specificity factor 5 (Cleavage and polyadenylation specificity factor 25 kDa subunit) (CPSF 25 kDa subunit) (Pre-mRNA cleavage factor Im 25 kDa subunit) (Nucleoside diphosphate-linked moiety X motif 21) (Nudix motif 21) | O43809|CPSF5_HUMAN | 26,210 | 8 |
| Host cell factor (HCF) (HCF-1) (C1 factor) (VP16 accessory protein) (VCAF) (CFF) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N-terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6] | P51610|HCFC1_HUMAN | 208,816 | 8 |
| NG,NG-dimethylarginine dimethylaminohydrolase 1 (EC 3.5.3.18) (Dimethylargininase-1) (Dimethylarginine dimethylaminohydrolase 1) (DDAHI) (DDAH-1) | O94760|DDAH1_HUMAN | 31,104 | 8 |
| SH3 domain GRB2-like protein B1 (EC 2.3.1.-) (Endophilin-B1) (Bax-interacting factor 1) (Bif-1) | Q9Y371|SHLB1_HUMAN | 40,780 | 8 |
| Glutathione reductase, mitochondrial precursor (EC 1.8.1.7) (GR) (GRase) | P00390|GSHR_HUMAN | 56,239 | 8 |
| Tripartite motif-containing protein 47 (Gene overexpressed in astrocytoma protein) (RING finger protein 100) | Q96LD4|TRI47_HUMAN | 69,513 | 8 |
| SNW domain-containing protein 1 (Nuclear protein SkiP) (Ski-interacting protein) (Nuclear receptor coactivator NCoA-62) | Q13573|SNW1_HUMAN | 61,478 | 8 |
| Acylamino-acid-releasing enzyme (EC 3.4.19.1) (AARE) (Acyl-peptide hydrolase) (APH) (Acylaminoacyl-peptidase) (Oxidized protein hydrolase) (OPH) (DNF15S2 protein) | P13798|ACPH_HUMAN | 81,206 | 8 |
| Guanine nucleotide-binding protein G(s) subunit alpha isoforms short (Adenylate cyclase-stimulating G alpha protein) | P63092|GNAS2_HUMAN, Q5JWF2|GNAS1_HUMAN | 45,647 | 8 |
| Vacuolar protein sorting-associated protein 26A (Vesicle protein sorting 26A) (hVPS26) | O75436|VP26A_HUMAN | 38,153 | 8 |
| 26S proteasome non-ATPase regulatory subunit 9 (26S proteasome regulatory subunit p27) | O00233|PSMD9_HUMAN | 24,635 | 8 |
| BH3-interacting domain death agonist (BID) (p22 BID) [Contains: BH3-interacting domain death agonist p15 (p15 BID); BH3-interacting domain death agonist p13 (p13 BID); BH3-interacting domain death agonist p11 (p11 BID)] | P55957|BID_HUMAN | 21,977 | 8 |
| Coatomer subunit zeta-1 (Zeta-1 coat protein) (Zeta-1 COP) | P61923|COPZ1_HUMAN | 20,181 | 8 |
| Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (NPI-1) | P52294|IMA1_HUMAN | 60,232 | 8 |
| Isoleucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS) | Q9NSE4|SYIM_HUMAN | 113,776 | 8 |
| Cytoplasmic dynein 1 light intermediate chain 2 (Dynein light intermediate chain 2, cytosolic) (LIC53/55) (LIC-2) | O43237|DC1L2_HUMAN | 54,082 | 8 |
| Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP-1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2) | Q15599|NHRF2_HUMAN | 37,395 | 8 |
| COP9 signalosome complex subunit 3 (Signalosome subunit 3) (SGN3) (JAB1-containing signalosome subunit 3) | Q9UNS2|CSN3_HUMAN | 47,857 | 8 |
| Prefoldin subunit 3 (Von Hippel-Lindau-binding protein 1) (VHL-binding protein 1) (VBP-1) (HIBBJ46) | P61758|PFD3_HUMAN | 22,641 | 8 |
| CD166 antigen precursor (Activated leukocyte-cell adhesion molecule) (ALCAM) | Q13740|CD166_HUMAN | 65,086 | 8 |
| Ubiquitin-fold modifier 1 precursor | P61960|UFM1_HUMAN | 9,100 | 8 |
| Eukaryotic translation initiation factor 3 subunit 6 (eIF-3 p48) (eIF3e) (Viral integration site protein INT-6 homolog) | P60228|IF36_HUMAN | 52,205 | 8 |
| Small glutamine-rich tetratricopeptide repeat-containing protein A (Vpu-binding protein) (UBP) | O43765|SGTA_HUMAN | 34,046 | 8 |
| UPF0317 protein C14orf159, mitochondrial precursor | Q7Z3D6|CN159_HUMAN | 66,419 | 8 |
| Actin-related protein 2/3 complex subunit 5-like protein (ARP2/3 complex 16 kDa subunit 2) (ARC16-2) | Q9BPX5|ARP5L_HUMAN | 16,923 | 8 |
| 2,4-dienoyl-CoA reductase, mitochondrial precursor (EC 1.3.1.34) (2,4-dienoyl-CoA reductase [NADPH]) (4-enoyl-CoA reductase [NADPH]) | Q16698|DECR_HUMAN | 36,051 | 8 |
| Eukaryotic translation initiation factor 1 (eIF1) (Protein translation factor SUI1 homolog) (Sui1iso1) (A121) | P41567|EIF1_HUMAN | 12,715 | 8 |
| Nicalin precursor (Nicastrin-like protein) | Q969V3|NCLN_HUMAN | 62,957 | 8 |
| Ras GTPase-activating protein-binding protein 2 (G3BP-2) (GAP SH3 domain-binding protein 2) | Q9UN86|G3BP2_HUMAN | 54,102 | 8 |
| Growth factor receptor-bound protein 2 (Adapter protein GRB2) (SH2/SH3 adapter GRB2) (Protein Ash) | P62993|GRB2_HUMAN | 25,189 | 8 |
| Ras-related protein Rab-18 | Q9NP72|RAB18_HUMAN | 22,960 | 8 |
| Complement decay-accelerating factor precursor (CD55 antigen) | P08174|DAF_HUMAN | 41,382 | 8 |
| TRM112-like protein | Q9UI30|TR112_HUMAN | 14,181 | 8 |
| Cytochrome c oxidase subunit 4 isoform 1, mitochondrial precursor (EC 1.9.3.1) (Cytochrome c oxidase subunit IV isoform 1) (COX IV-1) (Cytochrome c oxidase polypeptide IV) | P13073|COX41_HUMAN | 19,559 | 8 |
| Transmembrane emp24 domain-containing protein 9 precursor (Glycoprotein 25L2) | Q9BVK6|TMED9_HUMAN | 25,087 | 8 |
| Nuclear transport factor 2 (NTF-2) (Placental protein 15) (PP15) | P61970|NTF2_HUMAN | 14,461 | 8 |
| 3′(2′),5′-bisphosphate nucleotidase 1 (EC 3.1.3.7) (Bisphosphate 3′-nucleotidase 1) (PAP-inositol-1,4-phosphatase) (PIP) | O95861|BPNT1_HUMAN | 33,375 | 8 |
| Tyrosine-protein kinase CSK (EC 2.7.10.2) (C-SRC kinase) (Protein-tyrosine kinase CYL) | P41240|CSK_HUMAN | 50,687 | 8 |
| Guanylate kinase (EC 2.7.4.8) (GMP kinase) | Q16774|KGUA_HUMAN | 21,708 | 8 |
| Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII-4/7) | Q16630|CPSF6_HUMAN | 59,193 | 8 |
| Prolyl 4-hydroxylase subunit alpha-2 precursor (EC 1.14.11.2) (4-PH alpha-2) (Procollagen-proline,2-oxoglutarate-4-dioxygenase alpha-2 subunit) | O15460|P4HA2_HUMAN | 60,885 | 8 |
| Vesicle-associated membrane protein 3 (VAMP-3) (Synaptobrevin-3) (Cellubrevin) (CEB) | Q15836|VAMP3_HUMAN | 11,291 | 8 |
| Rho GTPase-activating protein 17 (Rho-type GTPase-activating protein 17) (RhoGAP interacting with CIP4 homologs protein 1) (RICH-1) | Q68EM7|RHG17_HUMAN | 95,419 | 8 |
| Cleavage stimulation factor 50 kDa subunit (CSTF 50 kDa subunit) (CF-1 50 kDa subunit) (CstF-50) | Q05048|CSTF1_HUMAN | 48,341 | 8 |
| ATP synthase B chain, mitochondrial precursor (EC 3.6.3.14) | P24539|AT5F1_HUMAN | 28,891 | 8 |
| Putative adenosylhomocysteinase 2 (EC 3.3.1.1) (S-adenosyl-L-homocysteine hydrolase 2) (AdoHcyase 2) (S-adenosylhomocysteine hydrolase-like 1) (DC-expressed AHCY-like molecule) | O43865|SAHH2_HUMAN | 58,934 | 8 |
| C-type lectin domain family 14 member A precursor (Epidermal growth factor receptor 5) (EGFR-5) | Q86T13|CLC14_HUMAN | 51,618 | 8 |
| UDP-glucose 4-epimerase (EC 5.1.3.2) (Galactowaldenase) (UDP-galactose 4-epimerase) | Q14376|GALE_HUMAN | 38,264 | 8 |
| Proteasome subunit beta type 7 precursor (EC 3.4.25.1) (Proteasome subunit Z) (Macropain chain Z) (Multicatalytic endopeptidase complex chain Z) | Q99436|PSB7_HUMAN | 29,948 | 8 |
| Importin-9 (Imp9) (Ran-binding protein 9) (RanbP9) | Q96P70|IPO9_HUMAN | 115,946 | 8 |
| Protein deltex 3-like protein (B-lymphoma- and BAL-associated protein) (Rhysin-2) (Rhysin2) | Q8TDB6|DTX3L_HUMAN | 83,538 | 8 |
| DnaJ homolog subfamily B member 11 precursor (ER-associated dnaJ protein 3) (ErJ3) (ER-associated Hsp40 co-chaperone) (hDj9) (PWP1-interacting protein 4) | Q9UBS4|DJB11_HUMAN | 40,497 | 8 |
| Phosphoglucomutase-1 (EC 5.4.2.2) (Glucose phosphomutase 1) (PGM 1) | P36871|PGM1_HUMAN | 61,433 | 8 |
| Elongation factor Ts, mitochondrial precursor (EF-Ts) (EF-TsMt) | P43897|EFTS_HUMAN | 35,373 | 8 |
| Membrane-associated progesterone receptor component 1 (mPR) | O00264|PGRC1_HUMAN | 21,654 | 8 |
| Ubiquitin carboxyl-terminal hydrolase 7 (EC 3.1.2.15) (Ubiquitin thioesterase 7) (Ubiquitin-specific-processing protease 7) (Deubiquitinating enzyme 7) (Herpesvirus-associated ubiquitin-specific protease) | Q93009|UBP7_HUMAN | 128,257 | 8 |
| Huntingtin-interacting protein 1 (HIP-I) | O00291|HIP1_HUMAN | 115,426 | 8 |
| Signal peptidase complex subunit 2 (EC 3.4.-.-) (Microsomal signal peptidase 25 kDa subunit) (SPase 25 kDa subunit) | Q15005|SPCS2_HUMAN | 24,986 | 8 |
| Solute carrier family 2, facilitated glucose transporter member 1 (Glucose transporter type 1, erythrocyte/brain) (HepG2 glucose transporter) | P11166|GTR1_HUMAN | 54,067 | 8 |
| Intercellular adhesion molecule 2 precursor (ICAM-2) (CD102 antigen) | P13598|ICAM2_HUMAN | 30,636 | 8 |
| Small nuclear ribonucleoprotein G (snRNP-G) (Sm protein G) (Sm-G) (SmG) | P62308|RUXG_HUMAN | 8,478 | 8 |
| FK506-binding protein 2 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (13 kDa FKBP) (FKBP-13) | P26885|FKBP2_HUMAN | 15,632 | 8 |
| Transcription elongation factor B polypeptide 2 (RNA polymerase II transcription factor SIII subunit B) (SIII p18) (Elongin B) (EloB) (Elongin 18 kDa subunit) | Q15370|ELOB_HUMAN | 13,115 | 8 |
| ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (Gp58) (Intracellular mannose-specific lectin MR60) | P49257|LMAN1_HUMAN | 57,531 | 8 |
| Protein FAM107B | Q9H098|F107B_HUMAN | 15,540 | 8 |
| Splicing factor, arginine/serine-rich 2 (Splicing factor SC35) (SC-35) (Splicing component, 35 kDa) (Protein PR264) | Q01130|SFRS2_HUMAN | 25,459 | 8 |
| WD repeat protein 61 (Meiotic recombination REC14 protein homolog) | Q9GZS3|WDR61_HUMAN | 33,563 | 8 |
| Scavenger mRNA decapping enzyme DcpS (EC 3.-.-.-) (DCS-1) (Hint-related 7meGMP-directed hydrolase) (Histidine triad protein member 5) (HINT-5) | Q96C86|DCPS_HUMAN | 38,592 | 8 |
| Thyroid receptor-interacting protein 6 (TRIP-6) (OPA-interacting protein 1) (Zyxin-related protein 1) (ZRP-1) | Q15654|TRIP6_HUMAN | 50,269 | 8 |
| Transcription factor BTF3 homolog 4 (Basic transcription factor 3-like 4) | Q96K17|BT3L4_HUMAN | 17,253 | 8 |
| 39S ribosomal protein L47, mitochondrial precursor (L47mt) (MRP-L47) | Q9HD33|RM47_HUMAN | 29,561 | 8 |
| Replication protein A 14 kDa subunit (RP-A) (RF-A) (Replication factor-A protein 3) (p14) | P35244|RFA3_HUMAN | 13,551 | 8 |
| AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain) | O00203|AP3B1_HUMAN | 121,336 | 8 |
| ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39) (Nuclear RNA helicase URH49) | O00148|DDX39_HUMAN | 49,112 | 7 |
| Flotillin-1 | O75955|FLOT1_HUMAN | 47,337 | 7 |
| Uncharacterized protein C10orf58 precursor | Q9BRX8|CJ058_HUMAN | 25,748 | 7 |
| Flotillin-2 (Epidermal surface antigen) (ESA) | Q14254|FLOT2_HUMAN | 41,667 | 7 |
| Calcium/calmodulin-dependent protein kinase type II delta chain (EC 2.7.11.17) (CaM-kinase II delta chain) (CaM kinase II subunit delta) (CaMK-II subunit delta) | Q13557|KCC2D_HUMAN | 56,353 | 7 |
| Thiosulfate sulfurtransferase (EC 2.8.1.1) (Rhodanese) | Q16762|THTR_HUMAN | 33,411 | 7 |
| Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1) | Q12888|TP53B_HUMAN | 213,554 | 7 |
| DNA replication licensing factor MCM2 (Minichromosome maintenance protein 2 homolog) (Nuclear protein BM28) | P49736|MCM2_HUMAN | 101,880 | 7 |
| Ras-related protein Rab-5C (RAB5L) (L1880) | P51148|RAB5C_HUMAN | 23,465 | 7 |
| Signal recognition particle 54 kDa protein (SRP54) | P61011|SRP54_HUMAN | 55,688 | 7 |
| Mothers against decapentaplegic homolog 3 (SMAD 3) (Mothers against DPP homolog 3) (Mad3) (hMAD-3) (JV15-2) (hSMAD3) | P84022|SMAD3_HUMAN | 48,063 | 7 |
| Uncharacterized protein C12orf5 | Q9NQ88|CL005_HUMAN | 30,045 | 7 |
| Nck-associated protein 1 (NAP 1) (p125Nap1) (Membrane-associated protein HEM-2) | Q9Y2A7|NCKP1_HUMAN | 128,777 | 7 |
| Inositol monophosphatase (EC 3.1.3.25) (IMPase) (IMP) (Inositol-1(or 4)-monophosphatase) (Lithium-sensitive myo-inositol monophosphatase A1) | P29218|IMPA1_HUMAN | 30,171 | 7 |
| Sideroflexin-3 | Q9BWM7|SFXN3_HUMAN | 35,486 | 7 |
| Serine palmitoyltransferase 2 (EC 2.3.1.50) (Long chain base biosynthesis protein 2) (LCB 2) (Serine-palmitoyl-CoA transferase 2) (SPT 2) | O15270|LCB2_HUMAN | 62,908 | 7 |
| mRNA-associated protein mrnp 41 (Rae1 protein homolog) | P78406|RAE1L_HUMAN | 40,951 | 7 |
| Protein NDRG1 (N-myc downstream-regulated gene 1 protein) (Differentiation-related gene 1 protein) (DRG-1) (Reducing agents and tunicamycin-responsive protein) (RTP) (Nickel-specific induction protein Cap43) (Rit42) | Q92597|NDRG1_HUMAN | 42,817 | 7 |
| UTP--glucose-1-phosphate uridylyltransferase 2 (EC 2.7.7.9) (UDP-glucose pyrophosphorylase 2) (UDPGP 2) (UGPase 2) | Q07131|UGPA1_HUMAN, Q16851|UGPA2_HUMAN | 56,924 | 7 |
| Glutathione synthetase (EC 6.3.2.3) (Glutathione synthase) (GSH synthetase) (GSH-S) | P48637|GSHB_HUMAN | 52,368 | 7 |
| Superkiller viralicidic activity 2-like 2 (EC 3.6.1.-) (ATP-dependent helicase SKIV2L2) | P42285|SK2L2_HUMAN | 117,790 | 7 |
| ATP-dependent RNA helicase DDX42 (EC 3.6.1.-) (DEAD box protein 42) (Splicing factor 3B-associated 125 kDa protein) (SF3b125) (RNA helicase-related protein) (RNAHP) (RNA helicase-like protein) (RHELP) (SF3b DEAD-box protein) | Q86XP3|DDX42_HUMAN | 102,959 | 7 |
| Adenosine kinase (EC 2.7.1.20) (AK) (Adenosine 5′-phosphotransferase) | P55263|ADK_HUMAN | 40,529 | 7 |
| ATP synthase coupling factor 6, mitochondrial precursor (EC 3.6.3.14) (ATPase subunit F6) | P18859|ATP5J_HUMAN | 12,570 | 7 |
| U6 snRNA-associated Sm-like protein LSm8 | O95777|LSM8_HUMAN | 10,385 | 7 |
| NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 24 kDa subunit) | P19404|NDUV2_HUMAN | 27,374 | 7 |
| Laminin subunit alpha-5 precursor | O15230|LAMA5_HUMAN | 399,725 | 7 |
| Nucleoporin 50 kDa (Nuclear pore-associated protein 60 kDa-like) | Q9UKX7|NUP50_HUMAN | 50,127 | 7 |
| Mitochondrial carrier homolog 2 (Met-induced mitochondrial protein) | Q9Y6C9|MTCH2_HUMAN | 33,314 | 7 |
| Sulfatase-modifying factor 2 precursor (C-alpha-formyglycine-generating enzyme 2) | Q8NBJ7|SUMF2_HUMAN | 33,839 | 7 |
| Programmed cell death protein 4 (Nuclear antigen H731-like) (Neoplastic transformation inhibitor protein) (Protein 197/15a) | Q53EL6|PDCD4_HUMAN | 51,704 | 7 |
| RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (hLark) | Q9BWF3|RBM4_HUMAN | 40,296 | 7 |
| Exocyst complex component 4 (Exocyst complex component Sec8) | Q96A65|EXOC4_HUMAN | 110,485 | 7 |
| Ubiquilin-1 (Protein linking IAP with cytoskeleton 1) (PLIC-1) (hPLIC-1) | Q9UMX0|UBQL1_HUMAN | 62,502 | 7 |
| NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 13 kDa-A subunit) (Complex I-13kD-A) (CI-13kD-A) | O75380|NDUS6_HUMAN | 13,693 | 7 |
| Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55) | Q10713|MPPA_HUMAN | 58,236 | 7 |
| Enhancer of mRNA decapping protein 4 (Human enhancer of decapping large subunit) (Hedls) (Autoantigen Ge-1) (Autoantigen RCD-8) | Q6P2E9|EDC4_HUMAN | 151,644 | 7 |
| ARF GTPase-activating protein GIT2 (G protein-coupled receptor kinase-interactor 2) (GRK-interacting protein 2) (Cool-interacting tyrosine-phosphorylated protein 2) (CAT2) (CAT-2) | Q14161|GIT2_HUMAN | 84,526 | 7 |
| Protein transport protein Sec24B (SEC24-related protein B) | O95487|SC24B_HUMAN | 137,773 | 7 |
| Diphosphomevalonate decarboxylase (EC 4.1.1.33) (Mevalonate pyrophosphate decarboxylase) (Mevalonate (diphospho)decarboxylase) | P53602|ERG19_HUMAN | 43,387 | 7 |
| Sorbitol dehydrogenase (EC 1.1.1.14) (L-iditol 2-dehydrogenase) | Q00796|DHSO_HUMAN | 38,279 | 7 |
| Sodium/potassium-transporting ATPase subunit beta-3 (Sodium/potassium-dependent ATPase beta-3 subunit) (ATPB-3) (CD298 antigen) | P54709|AT1B3_HUMAN | 31,496 | 7 |
| Splicing factor 3B subunit 5 (SF3b5) (Pre-mRNA-splicing factor SF3b 10 kDa subunit) | Q9BWJ5|SF3B5_HUMAN | 10,118 | 7 |
| Angiopoietin-2 precursor (ANG-2) | O15123|ANGP2_HUMAN | 56,902 | 7 |
| Dynein light chain roadblock-type 1 (Dynein light chain 2A, cytoplasmic) (Dynein-associated protein Km23) (Bithoraxoid-like protein) (BLP) | Q9NP97|DLRB1_HUMAN | 10,904 | 7 |
| Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein) | P35658|NU214_HUMAN | 213,748 | 7 |
| Glycylpeptide N-tetradecanoyltransferase 1 (EC 2.3.1.97) (Peptide N-myristoyltransferase 1) (Myristoyl-CoA:protein N-myristoyltransferase 1) (NMT 1) (Type I N-myristoyltransferase) | P30419|NMT1_HUMAN | 56,789 | 7 |
| 39S ribosomal protein L11, mitochondrial precursor (L11mt) (MRP-L11) | Q9Y3B7|RM11_HUMAN | 20,666 | 7 |
| SPFH domain-containing protein 1 precursor (Protein KE04) | O75477|SPFH1_HUMAN | 38,909 | 7 |
| Glycogen debranching enzyme (Glycogen debrancher) [Includes: 4-alpha-glucanotransferase (EC 2.4.1.25) (Oligo-1,4-1,4-glucantransferase); Amylo-alpha-1,6-glucosidase (EC 3.2.1.33) (Amylo-1,6-glucosidase) (Dextrin 6-alpha-D-glucosidase)] | P35573|GDE_HUMAN | 174,750 | 7 |
| Integrin beta-3 precursor (Platelet membrane glycoprotein IIIa) (GPIIIa) (CD61 antigen) | P05106|ITB3_HUMAN | 87,040 | 7 |
| ATP-binding cassette sub-family F member 2 (Iron-inhibited ABC transporter 2) | Q9UG63|ABCF2_HUMAN | 71,275 | 7 |
| Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog) | Q9BUQ8|DDX23_HUMAN | 95,633 | 7 |
| Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48) (Eukaryotic initiation factor 4A-like NUK-34) (Nuclear matrix protein 265) (hNMP 265) (Eukaryotic translation initiation factor 4A isoform 3) | P38919|DDX48_HUMAN | 46,854 | 7 |
| Synaptic glycoprotein SC2 | Q9NZ01|GPSN2_HUMAN | 36,018 | 7 |
| ATPase family AAA domain-containing protein 3B | Q5T9A4|ATD3B_HUMAN | 72,556 | 7 |
| FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP-51) (54 kDa progesterone receptor-associated immunophilin) (FKBP54) (P54) (FF1 antigen) (HSP90-binding immunophilin) (Androgen-regulated protein 6) | Q13451|FKBP5_HUMAN | 51,196 | 7 |
| 39S ribosomal protein L15, mitochondrial precursor (L15mt) (MRP-L15) | Q9P015|RM15_HUMAN | 33,404 | 7 |
| Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK) | P26196|DDX6_HUMAN | 54,401 | 7 |
| Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA-binding region-containing protein 7) (Protein S164) | P49756|RBM25_HUMAN | 94,108 | 7 |
| 3-ketoacyl-CoA thiolase, peroxisomal precursor (EC 2.3.1.16) (Beta-ketothiolase) (Acetyl-CoA acyltransferase) (Peroxisomal 3-oxoacyl-CoA thiolase) | P09110|THIK_HUMAN | 44,274 | 7 |
| FK506-binding protein 8 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (38 kDa FK506-binding protein) (FKBPR38) (hFKBP38) | Q14318|FKBP8_HUMAN | 38,391 | 7 |
| AP-1 complex subunit beta-1 (Adapter-related protein complex 1 beta-1 subunit) (Beta-adaptin 1) (Adaptor protein complex AP-1 beta-1 subunit) (Golgi adaptor HA1/AP1 adaptin beta subunit) (Clathrin assembly protein complex 1 beta large chain) | Q10567|AP1B1_HUMAN | 104,591 | 7 |
| Transcription elongation factor B polypeptide 1 (RNA polymerase II transcription factor SIII subunit C) (SIII p15) (Elongin-C) (EloC) (Elongin 15 kDa subunit) | Q15369|ELOC_HUMAN | 12,455 | 7 |
| Tricarboxylate transport protein, mitochondrial precursor (Citrate transport protein) (CTP) (Tricarboxylate carrier protein) (Solute carrier family 25 member 1) | P53007|TXTP_HUMAN | 33,995 | 7 |
| Caspase-7 precursor (EC 3.4.22.60) (CASP-7) (ICE-like apoptotic protease 3) (ICE-LAP3) (Apoptotic protease Mch-3) (CMH-1) [Contains: Caspase-7 subunit p20; Caspase-7 subunit p11] | P55210|CASP7_HUMAN | 34,260 | 7 |
| 40S ribosomal protein S27-like protein | Q71UM5|RS27L_HUMAN | 9,459 | 7 |
| E3 ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20) | Q5VTR2|BRE1A_HUMAN | 113,648 | 7 |
| Acetyl-CoA carboxylase 1 (EC 6.4.1.2) (ACC-alpha) [Includes: Biotin carboxylase (EC 6.3.4.14)] | Q13085|COA1_HUMAN | 265,538 | 7 |
| Matrix metalloproteinase-14 precursor (EC 3.4.24.80) (MMP-14) (Membrane-type matrix metalloproteinase 1) (MT-MMP 1) (MTMMP1) (Membrane-type-1 matrix metalloproteinase) (MT1-MMP) (MT1MMP) (MMP-X1) | P50281|MMP14_HUMAN | 65,868 | 7 |
| Cell division control protein 2 homolog (EC 2.7.11.22) (EC 2.7.11.23) (p34 protein kinase) (Cyclin-dependent kinase 1) (CDK1) | P06493|CDC2_HUMAN | 34,079 | 7 |
| Proteasome subunit beta type 9 precursor (EC 3.4.25.1) (Proteasome chain 7) (Macropain chain 7) (Multicatalytic endopeptidase complex chain 7) (RING12 protein) (Low molecular mass protein 2) | P28065|PSB9_HUMAN | 23,246 | 7 |
| Salivary alpha-amylase precursor (EC 3.2.1.1) (1,4-alpha-D-glucan glucanohydrolase) | P04745|AMYS_HUMAN | 57,750 | 7 |
| Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase G1) (PTPG1) | Q05209|PTN12_HUMAN | 88,104 | 7 |
| Uncharacterized protein KIAA0179 | Q14684|K0179_HUMAN | 84,396 | 7 |
| Glutamate--cysteine ligase regulatory subunit (EC 6.3.2.2) (Gamma-glutamylcysteine synthetase) (Gamma-ECS) (GCS light chain) (Glutamate--cysteine ligase modifier subunit) | P48507|GSH0_HUMAN | 30,709 | 7 |
| Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP) | Q15637|SF01_HUMAN | 68,313 | 7 |
| Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3) | Q9H307|PININ_HUMAN | 81,595 | 7 |
| Histone-binding protein RBBP4 (Retinoblastoma-binding protein 4) (RBBP-4) (Retinoblastoma-binding protein p48) (Chromatin assembly factor 1 subunit C) (CAF-1 subunit C) (Chromatin assembly factor I p48 subunit) (CAF-I 48 kDa subunit) (CAF-I p48) (Nucleosome remodeling factor subunit RBAP48) | Q09028|RBBP4_HUMAN | 47,638 | 7 |
| Phosphoserine aminotransferase (EC 2.6.1.52) (PSAT) | Q9Y617|SERC_HUMAN | 40,405 | 7 |
| SAPK substrate protein 1 (UBA/UBX 33.3 kDa protein) | Q04323|SAKS1_HUMAN | 33,307 | 7 |
| 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase (EC 1.13.-.-) (Aci-reductone dioxygenase) (ARD) (Membrane-type 1 matrix metalloproteinase cytoplasmic tail-binding protein 1) (MTCBP-1) (Submergence-induced protein 2 homolog) (SIPL) | Q9BV57|MTND_HUMAN | 21,481 | 7 |
| Mitochondrial 28S ribosomal protein S36 (S36mt) (MRP-S36) | P82909|RT36_HUMAN | 11,448 | 7 |
| Nuclear inhibitor of protein phosphatase 1 (NIPP-1) (Protein phosphatase 1 regulatory inhibitor subunit 8) [Includes: Activator of RNA decay (EC 3.1.4.-) (ARD-1)] | Q12972|PP1R8_HUMAN | 38,462 | 7 |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2 (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase B8 subunit) (Complex I-B8) (CI-B8) | O43678|NDUA2_HUMAN | 10,904 | 7 |
| Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA-IntP) | Q96CV9|OPTN_HUMAN | 65,905 | 7 |
| Dual specificity mitogen-activated protein kinase kinase 1 (EC 2.7.12.2) (MAP kinase kinase 1) (MAPKK 1) (ERK activator kinase 1) (MAPK/ERK kinase 1) (MEK1) | Q02750|MP2K1_HUMAN | 43,422 | 7 |
| Monoamine-sulfating phenol sulfotransferase (EC 2.8.2.1) (Aryl sulfotransferase 1A3) (Sulfotransferase, monoamine-preferring) (M-PST) (Thermolabile phenol sulfotransferase) (TL-PST) (Placental estrogen sulfotransferase) (Catecholamine-sulfating phenol sulfotransferase) (HAST3) | P50224|ST1A3_HUMAN | 34,179 | 7 |
| GMP reductase 2 (EC 1.7.1.7) (Guanosine 5′-monophosphate oxidoreductase 2) (Guanosine monophosphate reductase 2) | Q9P2T1|GMPR2_HUMAN | 37,857 | 7 |
| Protein transport protein Sec23B (SEC23-related protein B) | Q15437|SC23B_HUMAN | 86,463 | 7 |
| Interferon-induced 35 kDa protein (IFP 35) | P80217|IN35_HUMAN | 31,496 | 7 |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 42 kDa subunit) (Complex I-42kD) (CI-42kD) | O95299|NDUAA_HUMAN | 40,734 | 7 |
| Mitochondrial import receptor subunit TOM22 homolog (Translocase of outer membrane 22 kDa subunit homolog) (hTom22) (1C9-2) | Q9NS69|TOM22_HUMAN | 15,504 | 7 |
| Uncharacterized protein C1orf123 | Q9NWV4|CA123_HUMAN | 18,031 | 7 |
| Cathepsin L precursor (EC 3.4.22.15) (Major excreted protein) (MEP) [Contains: Cathepsin L heavy chain; Cathepsin L light chain] | P07711|CATL_HUMAN | 37,546 | 7 |
| Small acidic protein | O00193|SMAP_HUMAN | 20,315 | 7 |
| Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17) | Q9HCF4|ALO17_HUMAN | 174,882 | 7 |
| Ras-related protein Ral-B precursor | P11234|RALB_HUMAN | 23,391 | 7 |
| Exportin-5 (Exp5) (Ran-binding protein 21) | Q9HAV4|XPO5_HUMAN | 136,297 | 7 |
| Erythrocyte band 7 integral membrane protein (Stomatin) (Protein 7.2b) | P27105|STOM_HUMAN | 31,714 | 7 |
| Interleukin-6 receptor subunit beta precursor (IL-6R-beta) (Interleukin-6 signal transducer) (Membrane glycoprotein 130) (gp130) (Oncostatin-M receptor alpha subunit) (CD130 antigen) (CDw130) | P40189|IL6RB_HUMAN | 103,505 | 7 |
| Retinol dehydrogenase 11 (EC 1.1.1.-) (Retinal reductase 1) (RalR1) (Prostate short-chain dehydrogenase/reductase 1) (Androgen-regulated short-chain dehydrogenase/reductase 1) (HCV core-binding protein HCBP12) | Q8TC12|RDH11_HUMAN | 35,369 | 7 |
| Dedicator of cytokinesis protein 9 (Cdc42 guanine nucleotide exchange factor zizimin-1) | Q9BZ29|DOCK9_HUMAN | 236,433 | 7 |
| Cisplatin resistance-associated overexpressed protein (cAMP regulatory element-associated protein 1) (CRE-associated protein 1) (CREAP-1) (Luc7A) (Okadaic acid-inducible phosphoprotein OA48-18) | O95232|CROP_HUMAN | 51,449 | 7 |
| Abhydrolase domain-containing protein 14B (CCG1-interacting factor B) | Q96IU4|AB14B_HUMAN | 22,328 | 7 |
| Alpha crystallin B chain (Alpha(B)-crystallin) (Rosenthal fiber component) (Heat-shock protein beta-5) (HspB5) (Renal carcinoma antigen NY-REN-27) | P02511|CRYAB_HUMAN | 20,141 | 7 |
| Netrin-4 precursor (Beta-netrin) (Hepar-derived netrin-like protein) | Q9HB63|NET4_HUMAN | 70,052 | 7 |
| Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Qip1 protein) | O00629|IMA4_HUMAN | 57,869 | 7 |
| Unc-112-related protein 2 (Kindlin-3) (MIG2-like) | Q86UX7|URP2_HUMAN | 75,937 | 7 |
| Thioredoxin domain-containing protein 1 precursor (Transmembrane Trx-related protein) (Thioredoxin-related transmembrane protein) | Q9H3N1|TXND1_HUMAN | 31,774 | 7 |
| Mannose-6-phosphate isomerase (EC 5.3.1.8) (Phosphomannose isomerase) (PMI) (Phosphohexomutase) | P34949|MANA_HUMAN | 46,639 | 7 |
| C-terminal-binding protein 1 (EC 1.1.1.-) (CtBP1) | Q13363|CTBP1_HUMAN | 47,517 | 7 |
| Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1) | Q9H1E3|NUCKS_HUMAN | 27,279 | 7 |
| C-Myc-binding protein (Associate of Myc 1) (AMY-1) | Q99417|MYCBP_HUMAN | 11,949 | 7 |
| Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE) | Q8WXH0|SYNE2_HUMAN | 796,436 | 7 |
| Translin-associated protein X (Translin-associated factor X) | Q99598|TSNAX_HUMAN | 33,095 | 7 |
| Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1) | P22064|LTB1S_HUMAN | 152,767 | 7 |
| Protein enabled homolog | Q8N8S7|ENAH_HUMAN | 66,493 | 7 |
| Argininosuccinate lyase (EC 4.3.2.1) (Arginosuccinase) (ASAL) | P04424|ARLY_HUMAN | 51,641 | 7 |
| REVERSED | REV|Q9HCU4|CELR2 | 317,431 | 7 |
| Cadherin-6 precursor (Kidney-cadherin) (K-cadherin) | P55285|CADH6_HUMAN | 88,293 | 7 |
| Pancreatic alpha-amylase precursor (EC 3.2.1.1) (PA) (1,4-alpha-D-glucan glucanohydrolase) | P04746|AMYP_HUMAN | 57,689 | 7 |
| Bone marrow stromal antigen 2 precursor (BST-2) (CD317 antigen) (HM1.24 antigen) | Q10589|BST2_HUMAN | 19,751 | 7 |
| Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas) | P56945|BCAR1_HUMAN | 93,343 | 6 |
| Exportin-1 (Exp1) (Chromosome region maintenance 1 protein homolog) | O14980|XPO1_HUMAN | 123,371 | 6 |
| DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1) | P24928|RPB1_HUMAN | 217,193 | 6 |
| RNA-binding protein Nova-2 (Neuro-oncological ventral antigen 2) (Astrocytic NOVA1-like RNA-binding protein) | Q9UNW9|NOVA2_HUMAN | 48,991 | 6 |
| Afadin (Protein AF-6) | P55196|AFAD_HUMAN | 205,592 | 6 |
| Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog) | Q7L014|DDX46_HUMAN | 117,348 | 6 |
| SON protein (SON3) (Negative regulatory element-binding protein) (NRE-binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1) | P18583|SON_HUMAN | 263,828 | 6 |
| General transcription factor II-I (GTFII-I) (TFII-I) (Bruton tyrosine kinase-associated protein 135) (BTK-associated protein 135) (BAP-135) (SRF-Phox1-interacting protein) (SPIN) (Williams-Beuren syndrome chromosome region 6 protein) | P78347|GTF2I_HUMAN | 112,400 | 6 |
| Tyrosine-protein kinase JAK1 (EC 2.7.10.2) (Janus kinase 1) (JAK-1) | P23458|JAK1_HUMAN | 131,943 | 6 |
| Coiled-coil domain-containing protein 124 | Q96CT7|CC124_HUMAN | 25,818 | 6 |
| Osteoclast-stimulating factor 1 | Q92882|OSTF1_HUMAN | 23,770 | 6 |
| Sorting nexin-1 | Q13596|SNX1_HUMAN | 59,053 | 6 |
| Charged multivesicular body protein 4b (Chromatin-modifying protein 4b) (CHMP4b) (Vacuolar protein sorting 7-2) (SNF7-2) (hSnf7-2) (SNF7 homolog associated with Alix 1) (hVps32) | Q9H444|CHM4B_HUMAN | 24,933 | 6 |
| Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170) | Q08378|GOGA3_HUMAN | 167,338 | 6 |
| Transcription elongation factor SPT6 (hSPT6) (Tat-cotransactivator 2 protein) (Tat-CT2 protein) | Q7KZ85|SPT6H_HUMAN | 199,059 | 6 |
| Cytoskeleton-associated protein 5 (Colonic and hepatic tumor over-expressed protein) (Ch-TOG protein) | Q14008|CKAP5_HUMAN | 225,498 | 6 |
| Aflatoxin B1 aldehyde reductase member 2 (EC 1.-.-.-) (AFB1-AR 1) (Aldoketoreductase 7) | O43488|ARK72_HUMAN | 39,571 | 6 |
| Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His-domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14) | Q9H3S7|PTN23_HUMAN | 178,957 | 6 |
| DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50) | Q92878|RAD50_HUMAN | 153,880 | 6 |
| Alpha-parvin (Calponin-like integrin-linked kinase-binding protein) (CH-ILKBP) (Matrix-remodelling-associated protein 2) | Q9NVD7|PARVA_HUMAN | 42,227 | 6 |
| RNA-binding protein 39 (RNA-binding motif protein 39) (RNA-binding region-containing protein 2) (Hepatocellular carcinoma protein 1) (Splicing factor HCC1) | Q14498|RBM39_HUMAN | 59,363 | 6 |
| Dual specificity mitogen-activated protein kinase kinase 2 (EC 2.7.12.2) (MAP kinase kinase 2) (MAPKK 2) (ERK activator kinase 2) (MAPK/ERK kinase 2) (MEK2) | P36507|MP2K2_HUMAN | 44,407 | 6 |
| Hematological and neurological expressed 1-like protein (HN1-like protein) | Q9H910|HN1L_HUMAN | 20,046 | 6 |
| U1 small nuclear ribonucleoprotein C (U1 snRNP protein C) (U1C protein) (U1-C) | P09234|RU1C_HUMAN | 17,376 | 6 |
| Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372) | Q14789|GOGB1_HUMAN | 376,058 | 6 |
| Double-stranded RNA-binding protein Staufen homolog 1 | O95793|STAU1_HUMAN | 63,165 | 6 |
| U2 small nuclear ribonucleoprotein A′ (U2 snRNP-A′) | P09661|RU2A_HUMAN | 28,399 | 6 |
| COP9 signalosome complex subunit 1 (Signalosome subunit 1) (SGN1) (JAB1-containing signalosome subunit 1) (G protein pathway suppressor 1) (Protein GPS1) (Protein MFH) | Q13098|CSN1_HUMAN | 53,356 | 6 |
| Cofilin-2 (Cofilin, muscle isoform) | Q9Y281|COF2_HUMAN | 18,719 | 6 |
| DnaJ homolog subfamily B member 1 (Heat shock 40 kDa protein 1) (Heat shock protein 40) (HSP40) (DnaJ protein homolog 1) (HDJ-1) | P25685|DNJB1_HUMAN | 38,028 | 6 |
| Polyadenylate-binding protein 2 (Poly(A)-binding protein 2) (Poly(A)-binding protein II) (PABII) (Polyadenylate-binding nuclear protein 1) (Nuclear poly(A)-binding protein 1) | Q86U42|PABP2_HUMAN | 32,732 | 6 |
| ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor | O76031|CLPX_HUMAN | 69,207 | 6 |
| Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C) | O43847|NRDC_HUMAN | 131,558 | 6 |
| Nuclear receptor coactivator 5 (NCoA-5) (Coactivator independent of AF-2) (CIA) | Q9HCD5|NCOA5_HUMAN | 65,520 | 6 |
| Gem-associated protein 5 (Gemin5) | Q8TEQ6|GEMI5_HUMAN | 168,545 | 6 |
| Coiled-coil domain-containing protein 6 (H4 protein) (Papillary thyroid carcinoma-encoded protein) | Q16204|CCDC6_HUMAN | 65,901 | 6 |
| 5′-3′ exoribonuclease 2 (EC 3.1.13.-) (DHM1-like protein) (DHP protein) | Q9H0D6|XRN2_HUMAN | 108,568 | 6 |
| Melanoma-associated antigen D2 (MAGE-D2 antigen) (MAGE-D) (Breast cancer-associated gene 1 protein) (BCG-1) (11B6) (Hepatocellular carcinoma-associated protein JCL-1) | Q9UNF1|MAGD2_HUMAN | 64,938 | 6 |
| Transcription elongation regulator 1 (TATA box-binding protein-associated factor 2S) (Transcription factor CA150) | O14776|TCRG1_HUMAN | 123,944 | 6 |
| Vacuolar ATP synthase subunit G 1 (EC 3.6.3.14) (V-ATPase G subunit 1) (Vacuolar proton pump G subunit 1) (V-ATPase 13 kDa subunit 1) (Vacuolar ATP synthase subunit M16) | O75348|VATG1_HUMAN | 13,740 | 6 |
| Tyrosine-protein phosphatase non-receptor type 11 (EC 3.1.3.48) (Protein-tyrosine phosphatase 2C) (PTP-2C) (PTP-1D) (SH-PTP3) (SH-PTP2) (SHP-2) (Shp2) | Q06124|PTN11_HUMAN | 68,418 | 6 |
| Cell division control protein 42 homolog precursor (G25K GTP-binding protein) | P60953|CDC42_HUMAN | 21,293 | 6 |
| Exocyst complex component 7 (Exocyst complex component Exo70) | Q9UPT5|EXOC7_HUMAN | 83,367 | 6 |
| Beta-catenin-like protein 1 (Nuclear-associated protein) (NAP) (Testis development protein NYD-SP19) | Q8WYA6|CTBL1_HUMAN | 65,157 | 6 |
| H/ACA ribonucleoprotein complex subunit 2 (Nucleolar protein family A member 2) (snoRNP protein NHP2) | Q9NX24|NOLA2_HUMAN | 17,183 | 6 |
| Putative helicase MOV-10 (EC 3.6.1.-) (Moloney leukemia virus 10 protein) | Q9HCE1|MOV10_HUMAN | 113,658 | 6 |
| Syntenin-1 (Syndecan-binding protein 1) (Melanoma differentiation-associated protein 9) (MDA-9) (Scaffold protein Pbp1) (Pro-TGF-alpha cytoplasmic domain-interacting protein 18) (TACIP18) | O00560|SDCB1_HUMAN | 32,427 | 6 |
| Serine palmitoyltransferase 1 (EC 2.3.1.50) (Long chain base biosynthesis protein 1) (LCB 1) (Serine-palmitoyl-CoA transferase 1) (SPT 1) (SPT1) | O15269|LCB1_HUMAN | 52,728 | 6 |
| Vascular endothelial growth factor receptor 2 precursor (EC 2.7.10.1) (VEGFR-2) (Kinase insert domain receptor) (Protein-tyrosine kinase receptor Flk-1) (CD309 antigen) | P35968|VGFR2_HUMAN | 151,511 | 6 |
| Calcium-binding mitochondrial carrier protein Aralar2 (Mitochondrial aspartate glutamate carrier 2) (Solute carrier family 25 member 13) (Citrin) | Q9UJS0|CMC2_HUMAN | 74,159 | 6 |
| Periodic tryptophan protein 1 homolog (Keratinocyte protein IEF SSP 9502) | Q13610|PWP1_HUMAN | 55,811 | 6 |
| Ras-related protein Rab-21 | Q9UL25|RAB21_HUMAN | 24,330 | 6 |
| Dual specificity mitogen-activated protein kinase kinase 3 (EC 2.7.12.2) (MAP kinase kinase 3) (MAPKK 3) (MAPK/ERK kinase 3) | P46734|MP2K3_HUMAN | 39,301 | 6 |
| Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II) | Q14240|IF4A2_HUMAN | 46,386 | 6 |
| Glia maturation factor gamma (GMF-gamma) | O60234|GMFG_HUMAN | 16,783 | 6 |
| NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10 (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase PDSW subunit) (Complex I-PDSW) (CI-PDSW) | O96000|NDUBA_HUMAN | 20,759 | 6 |
| Lysosomal alpha-glucosidase precursor (EC 3.2.1.20) (Acid maltase) (Aglucosidase alfa) [Contains: 76 kDa lysosomal alpha-glucosidase; 70 kDa lysosomal alpha-glucosidase] | P10253| LYAG_HU MAN | 105,321 | 6 |
| Heterogeneous nuclear ribonucleoprotein H′ (hnRNP H′) (FTP-3) | P55795|HNRH2_HUMAN | 49,246 | 6 |
| Cullin-3 (CUL-3) | Q13618|CUL3_HUMAN | 88,914 | 6 |
| Tyrosine-protein kinase receptor Tie-1 precursor (EC 2.7.10.1) | P35590|TIE1_HUMAN | 125,073 | 6 |
| DnaJ homolog subfamily A member 2 (HIRA-interacting protein 4) (Cell cycle progression restoration gene 3 protein) (Dnj3) (Renal carcinoma antigen NY-REN-14) | O60884|DNJA2_HUMAN | 45,728 | 6 |
| Serum paraoxonase/arylesterase 2 (EC 3.1.1.2) (EC 3.1.8.1) (PON 2) (Serum aryldialkylphosphatase 2) (A-esterase 2) (Aromatic esterase 2) | Q15165|PON2_HUMAN | 39,381 | 6 |
| Cadherin-2 precursor (Neural-cadherin) (N-cadherin) (CD325 antigen) (CDw325) | P19022|CADH2_HUMAN | 99,794 | 6 |
| FK506-binding protein 9 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) | O95302|FKBP9_HUMAN | 63,067 | 6 |
| A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95) | O43823|AKAP8_HUMAN | 76,091 | 6 |
| Serine/threonine-protein kinase 24 (EC 2.7.11.1) (STE20-like kinase MST3) (MST-3) (Mammalian STE20-like protein kinase 3) | Q9Y6E0|STK24_HUMAN | 49,293 | 6 |
| Isocitrate dehydrogenase [NAD] subunit beta, mitochondrial precursor (EC 1.1.1.41) (Isocitric dehydrogenase) (NAD(+)-specific ICDH) | O43837|IDH3B_HUMAN | 42,194 | 6 |
| Striatin | O43815|STRN_HUMAN | 86,116 | 6 |
| Calcium-binding protein p22 (Calcium-binding protein CHP) (Calcineurin homologous protein) (Calcineurin B homolog) | Q99653|CHP1_HUMAN | 22,439 | 6 |
| Torsin-1A-interacting protein 1 | Q5JTV8|TOIP1_HUMAN | 66,231 | 6 |
| Adaptin ear-binding coat-associated protein 2 (NECAP-2) | Q9NVZ3|NECP2_HUMAN | 28,321 | 6 |
| Endothelial differentiation-related factor 1 (EDF-1) (Multiprotein-bridging factor 1) (MBF1) | O60869|EDF1_HUMAN | 16,351 | 6 |
| Gamma-synuclein (Persyn) (Breast cancer-specific gene 1 protein) (Synoretin) (SR) | O76070|SYUG_HUMAN | 13,312 | 6 |
| Dedicator of cytokinesis protein 4 | Q8N1I0|DOCK4_HUMAN | 225,137 | 6 |
| Serine/threonine-protein phosphatase 4 regulatory subunit 1 | Q8TF05|PP4R1_HUMAN | 106,988 | 6 |
| Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (Prolyl hydroxylase domain-containing protein 2) (PHD2) (SM-20) | Q9GZT9|EGLN1_HUMAN | 46,004 | 6 |
| Transcriptional activator protein Pur-beta (Purine-rich element-binding protein B) | Q96QR8|PURB_HUMAN | 33,224 | 6 |
| Dual specificity protein phosphatase 3 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase VHR) | P51452|DUS3_HUMAN | 20,461 | 6 |
| Semaphorin-6B precursor (Semaphorin Z) (Sema Z) | Q9H3T3|SEM6B_HUMAN | 95,267 | 6 |
| NADPH--cytochrome P450 reductase (EC 1.6.2.4) (CPR) (P450R) | P16435|NCPR_HUMAN | 76,673 | 6 |
| N-acylneuraminate cytidylyltransferase (EC 2.7.7.43) (CMP-N-acetylneuraminic acid synthetase) (CMP-NeuNAc synthetase) | Q8NFW8|NEUA_HUMAN | 48,362 | 6 |
| Isocitrate dehydrogenase [NAD] subunit gamma, mitochondrial precursor (EC 1.1.1.41) (Isocitric dehydrogenase) (NAD(+)-specific ICDH) | P51553|IDH3G_HUMAN | 42,776 | 6 |
| DNA replication licensing factor MCM5 (CDC46 homolog) (P1-CDC46) | P33992|MCM5_HUMAN | 82,270 | 6 |
| Coiled-coil-helix-coiled-coil-helix domain-containing protein 2 (HCV NS2 trans-regulated protein) (NS2TP) (Aging-associated gene 10 protein) | Q9Y6H1|CHCH2_HUMAN | 15,494 | 6 |
| SRA stem-loop-interacting RNA-binding protein, mitochondrial precursor | Q9GZT3|SLIRP_HUMAN | 12,331 | 6 |
| Histone H1.0 (Histone H1(0)) (Histone H1′) | P07305|H10_HUMAN | 20,846 | 6 |
| Nuclear factor NF-kappa-B p100 subunit (DNA-binding factor KBF2) (H2TF1) (Lymphocyte translocation chromosome 10) (Oncogene Lyt-10) (Lyt10) [Contains: Nuclear factor NF-kappa-B p52 subunit] | Q00653|NFKB2_HUMAN | 96,733 | 6 |
| 2′-5′-oligoadenylate synthetase 3 (EC 2.7.7.-) ((2–5′)oligo(A) synthetase 3) (2–5A synthetase 3) (p100 OAS) (p100OAS) | Q9Y6K5|OAS3_HUMAN | 121,149 | 6 |
| ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18) (Myc-regulated DEAD box protein) (MrDb) | Q9NVP1|DDX18_HUMAN | 75,392 | 6 |
| Sperm-specific antigen 2 (Cleavage signal-1 protein) (CS-1) (Ki-ras-induced actin-interacting protein) | P28290|SSFA2_HUMAN | 138,368 | 6 |
| Estradiol 17-beta-dehydrogenase 12 (EC 1.1.1.62) (17-beta-HSD 12) (17-beta-hydroxysteroid dehydrogenase 12) (3-ketoacyl-CoA reductase) (EC 1.3.1.-) (KAR) | Q53GQ0|DHB12_HUMAN | 34,334 | 6 |
| Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase pez) | Q15678|PTN14_HUMAN | 135,221 | 6 |
| ATP synthase e chain, mitochondrial (EC 3.6.3.14) | P56385|ATP5I_HUMAN | 7,916 | 6 |
| Torsin-1A-interacting protein 2 (Lumenal domain-like LAP1) | Q8NFQ8|TOIP2_HUMAN | 51,245 | 6 |
| Multimerin-1 precursor (Endothelial cell multimerin 1) (EMILIN-4) (Elastin microfibril interface located protein 4) (Elastin microfibril interfacer 4) | Q13201|MMRN1_HUMAN | 138,150 | 6 |
| 60S ribosomal protein L34 | P49207|RL34_HUMAN | 13,275 | 6 |
| NAD-dependent malic enzyme, mitochondrial precursor (EC 1.1.1.38) (NAD-ME) (Malic enzyme 2) | P23368|MAOM_HUMAN | 65,428 | 6 |
| Aspartyl aminopeptidase (EC 3.4.11.21) | Q9ULA0|DNPEP_HUMAN | 52,411 | 6 |
| Acyl-coenzyme A oxidase 1, peroxisomal (EC 1.3.3.6) (Palmitoyl-CoA oxidase) (AOX) (Straight-chain acyl-CoA oxidase) (SCOX) | Q15067|ACOX1_HUMAN | 74,407 | 6 |
| Eukaryotic peptide chain release factor subunit 1 (eRF1) (Eukaryotic release factor 1) (TB3-1) (Protein Cl1) | P62495|ERF1_HUMAN | 49,015 | 6 |
| Small ubiquitin-related modifier 2 precursor (SUMO-2) (Ubiquitin-like protein SMT3B) (SMT3 homolog 2) (Sentrin-2) (HSMT3) (SUMO-3) | P55854|SUMO3_HUMAN, P61956|SUMO2_HUMAN, Q6EEV6|SUMO4_HUMAN | 10,853 | 6 |
| Myb-binding protein 1A | Q9BQG0|MBB1A_HUMAN | 148,840 | 6 |
| Cation-dependent mannose-6-phosphate receptor precursor (CD Man-6-P receptor) (CD-MPR) (46 kDa mannose 6-phosphate receptor) (MPR 46) | P20645|MPRD_HUMAN | 30,975 | 6 |
| Nucleolar GTP-binding protein 1 (Chronic renal failure gene protein) (GTP-binding protein NGB) | Q9BZE4|NOG1_HUMAN | 73,949 | 6 |
| Tetratricopeptide repeat protein 1 (TPR repeat protein 1) | Q99614|TTC1_HUMAN | 33,509 | 6 |
| Autophagy-related protein 7 (APG7-like) (Ubiquitin-activating enzyme E1-like protein) (hAGP7) | O95352|ATG7_HUMAN | 77,944 | 6 |
| Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon) | P23469|PTPRE_HUMAN | 80,625 | 6 |
| Ubiquitin/ISG15-conjugating enzyme E2 L6 (EC 6.3.2.19) (Ubiquitin-protein ligase L6) (Ubiquitin carrier protein L6) (UbcH8) (Retinoic acid-induced gene B protein) (RIG-B) | O14933|UB2L6_HUMAN | 17,620 | 6 |
| Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (USP on chromosome 21) | Q9UHP3|UBP25_HUMAN | 125,736 | 6 |
| Receptor-interacting serine/threonine-protein kinase 2 (EC 2.7.11.1) (RIP-like-interacting CLARP kinase) (Receptor-interacting protein 2) (RIP-2) (CARD-containing interleukin-1 beta-converting enzyme-associated kinase) (CARD-containing IL-1 beta ICE-kinase) | O43353|RIPK2_HUMAN | 61,179 | 6 |
| Protein FAM49B (L1) | Q9NUQ9|FA49B_HUMAN | 36,731 | 6 |
| Ribosomal protein L22-like 1 | Q6P5R6|RL22L_HUMAN | 14,589 | 6 |
| Glutaredoxin-1 (Thioltransferase-1) (TTase-1) | P35754|GLRX1_HUMAN | 11,758 | 6 |
| Integrin-linked kinase-associated serine/threonine phosphatase 2C (EC 3.1.3.16) (ILKAP) | Q9H0C8|ILKAP_HUMAN | 42,889 | 6 |
| Receptor-type tyrosine-protein phosphatase beta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase beta) (R-PTP-beta) | P23467|PTPRB_HUMAN | 224,250 | 6 |
| Interferon-induced protein with tetratricopeptide repeats 1 (IFIT-1) (Interferon-induced 56 kDa protein) (IFI-56K) | P09914|IFIT1_HUMAN | 55,344 | 6 |
| Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre-mRNA-processing factor 4 homolog) (PRP4 kinase) | Q13523|PRP4B_HUMAN | 116,961 | 6 |
| Membrane-associated progesterone receptor component 2 (Progesterone membrane-binding protein) (Steroid receptor protein DG6) | O15173|PGRC2_HUMAN | 23,801 | 6 |
| Signal transducer and activator of transcription 2 (p113) | P52630|STAT2_HUMAN | 97,901 | 6 |
| TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin) | Q9H2D6|TARA_HUMAN | 261,356 | 5 |
| SH3-containing GRB2-like protein 1 (Endophilin-2) (Endophilin-A2) (SH3 domain protein 2B) (Extra eleven-nineteen leukemia fusion gene) (EEN) (EEN fusion partner of MLL) | Q99961|SH3G1_HUMAN | 41,473 | 5 |
| Serine protease HTRA1 precursor (EC 3.4.21.-) (L56) (Serine protease 11) | Q92743|HTRA1_HUMAN | 51,269 | 5 |
| Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein) | Q8WWM7|ATX2L_HUMAN | 113,355 | 5 |
| Copine-1 (Copine I) | Q99829|CPNE1_HUMAN | 59,041 | 5 |
| Palmitoyl-protein thioesterase 1 precursor (EC 3.1.2.22) (PPT-1) (Palmitoyl-protein hydrolase 1) | P50897|PPT1_HUMAN | 34,176 | 5 |
| Protein DEK | P35659|DEK_HUMAN | 42,658 | 5 |
| Phospholipid hydroperoxide glutathione peroxidase, mitochondrial precursor (EC 1.11.1.12) (PHGPx) (GPX-4) | P36969|GPX4_HUMAN | 22,110 | 5 |
| Exosome component 10 (Polymyositis/scleroderma autoantigen 2) (Autoantigen PM/Scl 2) (Polymyositis/scleroderma autoantigen 100 kDa) (PM/Scl-100) (P100 polymyositis-scleroderma overlap syndrome-associated autoantigen) | Q01780|EXOSX_HUMAN | 100,816 | 5 |
| Beta-arrestin-1 (Arrestin beta 1) | P49407|ARRB1_HUMAN | 47,048 | 5 |
| DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2) | Q99615|DNJC7_HUMAN | 56,425 | 5 |
| CAP-Gly domain-containing linker protein 1 (Restin) (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed-Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1) | P30622|CLIP1_HUMAN | 160,975 | 5 |
| Nucleotide-binding protein 2 (NBP 2) | Q9Y5Y2|NUBP2_HUMAN | 28,807 | 5 |
| Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54) | Q05519|SFR11_HUMAN | 53,526 | 5 |
| Peptidyl-prolyl cis-trans isomerase-like 1 (EC 5.2.1.8) (PPIase) (Rotamase) | Q9Y3C6|PPIL1_HUMAN | 18,219 | 5 |
| Quinone oxidoreductase (EC 1.6.5.5) (NADPH:quinone reductase) (Zeta-crystallin) | Q08257|QOR_HUMAN | 35,189 | 5 |
| LIM and senescent cell antigen-like-containing domain protein 3 (Particularly interesting new Cys-His protein 3) (PINCH-3) | Q9HB10|LIMS3_HUMAN | 13,233 | 5 |
| UPF0404 protein C11orf59 | Q6IAA8|CK059_HUMAN | 17,727 | 5 |
| Succinyl-CoA ligase [ADP-forming] beta-chain, mitochondrial precursor (EC 6.2.1.5) (Succinyl-CoA synthetase, betaA chain) (SCS-betaA) (ATP-specific succinyl-CoA synthetase subunit beta) (Renal carcinoma antigen NY-REN-39) | Q9P2R7|SUCB1_HUMAN | 50,300 | 5 |
| Polyribonucleotide nucleotidyltransferase 1, mitochondrial precursor (EC 2.7.7.8) (PNPase 1) (Polynucleotide phosphorylase-like protein) (PNPase old-35) (3′-5′ RNA exonuclease OLD35) | Q8TCS8|PNPT1_HUMAN | 85,934 | 5 |
| Protein NDRG3 | Q9UGV2|NDRG3_HUMAN | 41,391 | 5 |
| Glycylpeptide N-tetradecanoyltransferase 2 (EC 2.3.1.97) (Peptide N-myristoyltransferase 2) (Myristoyl-CoA:protein N-myristoyltransferase 2) (NMT 2) (Type II N-myristoyltransferase) | O60551|NMT2_HUMAN | 56,964 | 5 |
| UPF0368 protein Cxorf26 | Q9BVG4|CX026_HUMAN | 26,040 | 5 |
| Pyrroline-5-carboxylate reductase 1 (EC 1.5.1.2) (P5CR 1) (P5C reductase 1) | P32322|P5CR1_HUMAN | 33,343 | 5 |
| ATP-binding cassette sub-family F member 3 | Q9NUQ8|ABCF3_HUMAN | 79,729 | 5 |
| COP9 signalosome complex subunit 8 (Signalosome subunit 8) (SGN8) (JAB1-containing signalosome subunit 8) (COP9 homolog) (hCOP9) | Q99627|CSN8_HUMAN | 23,208 | 5 |
| Negative elongation factor E (NELF-E) (RD protein) | P18615|NELFE_HUMAN | 43,223 | 5 |
| Carnitine O-palmitoyltransferase I, liver isoform (EC 2.3.1.21) (CPT I) (CPTI-L) (Carnitine palmitoyltransferase 1A) | P50416|CPT1A_HUMAN | 88,352 | 5 |
| Glucosamine 6-phosphate N-acetyltransferase (EC 2.3.1.4) (Phosphoglucosamine transacetylase) (Phosphoglucosamine acetylase) | Q96EK6|GNA1_HUMAN | 20,731 | 5 |
| Vacuolar protein sorting-associated protein 29 (Vesicle protein sorting 29) (hVPS29) (PEP11) | Q9UBQ0|VPS29_HUMAN | 20,488 | 5 |
| Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90) | P17480|UBF1_HUMAN | 89,392 | 5 |
| Alanyl-tRNA synthetase domain-containing protein 1 | Q9BTE6|AASD1_HUMAN | 45,462 | 5 |
| WD repeat protein 57 (Prp8-binding protein) (hPRP8BP) (U5 snRNP-specific 40 kDa protein) (38 kDa-splicing factor) | Q96DI7|WDR57_HUMAN | 39,293 | 5 |
| NudC domain-containing protein 2 | Q8WVJ2|NUDC2_HUMAN | 17,658 | 5 |
| Mitochondrial antiviral-signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced-signaling adapter) (CARD adapter inducing interferon-beta) (Cardif) (Putative NF-kappa-B-activating protein 031N) | Q7Z434|MAVS_HUMAN | 56,510 | 5 |
| PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (Transcriptional coactivator p75/p52) (Dense fine speckles 70 kDa protein) (DFS 70) (CLL-associated antigen KW-7) | O75475|PSIP1_HUMAN | 60,086 | 5 |
| Thyroid receptor-interacting protein 12 (TRIP-12) | Q14669|TRIPC_HUMAN | 220,420 | 5 |
| Rab3 GTPase-activating protein catalytic subunit (RAB3 GTPase-activating protein 130 kDa subunit) (Rab3-GAP p130) (Rab3-GAP) | Q15042|RB3GP_HUMAN | 110,508 | 5 |
| Coproporphyrinogen III oxidase, mitochondrial precursor (EC 1.3.3.3) (Coproporphyrinogenase) (Coprogen oxidase) (COX) | P36551|HEM6_HUMAN | 50,134 | 5 |
| Ubiquitin-activating enzyme E1 domain-containing protein 1 (UFM1-activating enzyme) (Ubiquitin-activating enzyme 5) (ThiFP1) | Q9GZZ9|UE1D1_HUMAN | 44,845 | 5 |
| ADP-dependent glucokinase (EC 2.7.1.147) (ADPGK) (ADP-GK) (RbBP-35) | Q9BRR6|ADPGK_HUMAN | 54,071 | 5 |
| Mitochondrial 28S ribosomal protein S22 (S22mt) (MRP-S22) | P82650|RT22_HUMAN | 41,264 | 5 |
| Mitochondrial Rho GTPase 2 (EC 3.6.5.-) (MIRO-2) (hMiro-2) (Ras homolog gene family member T2) | Q8IXI1|MIRO2_HUMAN | 68,100 | 5 |
| Polypeptide N-acetylgalactosaminyltransferase 2 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 2) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase 2) (Polypeptide GalNAc transferase 2) (GalNAc-T2) (pp-GaNTase 2) [Contains: Polypeptide N-acetylgalactosaminyltransferase 2 soluble form] | Q10471|GALT2_HUMAN | 64,715 | 5 |
| GRIP1-associated protein 1 (GRASP-1) | Q4V328|GRAP1_HUMAN | 95,973 | 5 |
| Septin-8 | Q92599|SEPT8_HUMAN | 55,738 | 5 |
| Tubulin gamma-1 chain (Gamma-1 tubulin) (Gamma-tubulin complex component 1) (GCP-1) | P23258|TBG1_HUMAN | 51,153 | 5 |
| Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M) (mtGPD) | P43304|GPDM_HUMAN | 80,818 | 5 |
| ADP-ribosylation factor GTPase-activating protein 1 (ADP-ribosylation factor 1 GTPase-activating protein) (ARF1 GAP) (ARF1-directed GTPase-activating protein) (GAP protein) | Q8N6T3|ARFG1_HUMAN | 44,649 | 5 |
| Scaffold attachment factor B (Scaffold attachment factor B1) (SAF-B) (HSP27 estrogen response element-TATA box-binding protein) (HSP27 ERE-TATA-binding protein) | Q15424|SAFB1_HUMAN | 102,625 | 5 |
| Nucleoporin-like protein RIP (HIV-1 Rev-binding protein) (Rev-interacting protein) (Rev/Rex activation domain-binding protein) | P52594|NUPL_HUMAN | 58,242 | 5 |
| Putative RNA-binding protein Luc7-like 1 (SR+89) (Putative SR protein LUC7B1) | Q9NQ29|LUC7L_HUMAN | 43,711 | 5 |
| RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN) | Q9NTZ6|RBM12_HUMAN | 97,379 | 5 |
| Transformer-2 protein homolog (TRA-2 alpha) | Q13595|TRA2A_HUMAN | 32,671 | 5 |
| AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) | O14617|AP3D1_HUMAN | 130,144 | 5 |
| Protein memo (Mediator of ErbB2-driven cell motility) (C21orf19-like protein) (Hepatitis C virus NS5A-transactivated protein 7) (HCV NS5A-transactivated protein 7) | Q9Y316|MEMO_HUMAN | 33,716 | 5 |
| AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1) | Q2M2I8|AAK1_HUMAN | 93,560 | 5 |
| Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1) | O14745|NHERF_HUMAN | 38,850 | 5 |
| Multisynthetase complex auxiliary component p38 (Protein JTV-1) | Q13155|MCA2_HUMAN | 35,331 | 5 |
| Proteasome subunit beta type 3 (EC 3.4.25.1) (Proteasome theta chain) (Proteasome chain 13) (Proteasome component C10-II) | P49720|PSB3_HUMAN | 22,932 | 5 |
| Amyloid-like protein 2 precursor (Amyloid protein homolog) (APPH) (CDEI box-binding protein) (CDEBP) | Q06481|APLP2_HUMAN | 86,937 | 5 |
| Muscleblind-like protein (Triplet-expansion RNA-binding protein) | Q9NR56|MBNL_HUMAN | 41,799 | 5 |
| DNA topoisomerase 2-beta (EC 5.99.1.3) (DNA topoisomerase II, beta isozyme) | Q02880|TOP2B_HUMAN | 183,255 | 5 |
| Glycogenin-1 (EC 2.4.1.186) | P46976|GLYG_HUMAN | 39,366 | 5 |
| ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24) | Q9GZR7|DDX24_HUMAN | 96,317 | 5 |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8 (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 19 kDa subunit) (Complex I-19kD) (CI-19kD) (Complex I-PGIV) (CI-PGIV) | P51970|NDUA8_HUMAN | 20,087 | 5 |
| Probable G-protein coupled receptor 116 precursor | Q8IZF2|GP116_HUMAN | 149,441 | 5 |
| Prefoldin subunit 4 (Protein C-1) | Q9NQP4|PFD4_HUMAN | 15,297 | 5 |
| Smu-1 suppressor of mec-8 and unc-52 protein homolog | Q2TAY7|SMU1_HUMAN | 57,527 | 5 |
| Peptidyl-prolyl cis-trans isomerase H (EC 5.2.1.8) (PPIase H) (Rotamase H) (U-snRNP-associated cyclophilin SnuCyp-20) (USA-CYP) (Small nuclear ribonucleoprotein particle-specific cyclophilin H) (CypH) | O43447|PPIH_HUMAN | 19,190 | 5 |
| Exocyst complex component 5 (Exocyst complex component Sec10) (hSec10) | O00471|EXOC5_HUMAN | 81,837 | 5 |
| Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2) (Lysyl oxidase-related protein 2) (Lysyl oxidase-related protein WS9-14) | Q9Y4K0|LOXL2_HUMAN | 86,705 | 5 |
| N-acetyltransferase 10 (EC 2.3.1.-) | Q9H0A0|NAT10_HUMAN | 115,690 | 5 |
| G patch domain and KOW motifs-containing protein (G patch domain-containing protein 5) (Protein MOS2 homolog) (Protein T54) | Q92917|GPKOW_HUMAN | 52,211 | 5 |
| Putative ATP-dependent Clp protease proteolytic subunit, mitochondrial precursor (EC 3.4.21.92) (Endopeptidase Clp) | Q16740|CLPP_HUMAN | 30,163 | 5 |
| Ufm1-conjugating enzyme 1 (Ubiquitin-fold modifier-conjugating enzyme 1) | Q9Y3C8|UFC1_HUMAN | 19,441 | 5 |
| Nuclear pore complex protein Nup205 (Nucleoporin Nup205) (205 kDa nucleoporin) | Q92621|NU205_HUMAN | 227,909 | 5 |
| Beta-hexosaminidase alpha chain precursor (EC 3.2.1.52) (N-acetyl-beta-glucosaminidase) (Beta-N-acetylhexosaminidase) (Hexosaminidase A) | P06865|HEXA_HUMAN | 60,672 | 5 |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5 (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 13 kDa-B subunit) (Complex I-13kD-B) (CI-13kD-B) (Complex I subunit B13) | Q16718|NDUA5_HUMAN | 13,441 | 5 |
| Large proline-rich protein BAT2 (HLA-B-associated transcript 2) | P48634|BAT2_HUMAN | 228,845 | 5 |
| Acyl-CoA dehydrogenase family member 9, mitochondrial precursor (EC 1.3.99.-) (ACAD-9) | Q9H845|ACAD9_HUMAN | 68,745 | 5 |
| Xaa-Pro dipeptidase (EC 3.4.13.9) (X-Pro dipeptidase) (Proline dipeptidase) (Prolidase) (Imidodipeptidase) | P12955|PEPD_HUMAN | 54,530 | 5 |
| Thymidylate synthase (EC 2.1.1.45) (TS) (TSase) | P04818|TYSY_HUMAN | 35,699 | 5 |
| Oligoribonuclease, mitochondrial precursor (EC 3.1.-.-) (Small fragment nuclease) (RNA exonuclease 2 homolog) | Q9Y3B8|ORN_HUMAN | 26,816 | 5 |
| Huntingtin-interacting protein HYPK (Huntingtin yeast partner K) | Q9NX55|HYPK_HUMAN | 19,314 | 5 |
| Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3) | Q8IZL8|PELP1_HUMAN | 119,683 | 5 |
| U6 snRNA-associated Sm-like protein LSm6 (Sm protein F) | P62312|LSM6_HUMAN | 9,110 | 5 |
| Dpy-30-like protein | Q9C005|DPY30_HUMAN | 11,232 | 5 |
| H/ACA ribonucleoprotein complex subunit 1 (Nucleolar protein family A member 1) (snoRNP protein GAR1) | Q9NY12|NOLA1_HUMAN | 22,331 | 5 |
| Isovaleryl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.10) (IVD) | P26440|IVD_HUMAN | 46,303 | 5 |
| CD81 antigen (26 kDa cell surface protein TAPA-1) (Target of the antiproliferative antibody 1) (Tetraspanin-28) (Tspan-28) | P60033|CD81_HUMAN | 25,792 | 5 |
| Putative rRNA methyltransferase 3 (EC 2.1.1.-) (rRNA (uridine-2′-O-)-methyltransferase 3) | Q8IY81|RRMJ3_HUMAN | 96,560 | 5 |
| Regulator of G-protein signaling 10 (RGS10) | O43665|RGS10_HUMAN | 20,219 | 5 |
| Long-chain fatty acid transport protein 4 (EC 6.2.1.-) (Fatty acid transport protein 4) (FATP-4) (Solute carrier family 27 member 4) | Q6P1 M0|S27A4_HUMAN | 72,048 | 5 |
| Probable cation-transporting ATPase 13A1 (EC 3.6.3.-) | Q9HD20|AT131_HUMAN | 132,940 | 5 |
| Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat-cotransactivator 1 protein) (Tat-CT1 protein) | O00267|SPT5H_HUMAN | 120,982 | 5 |
| Glycogen [starch] synthase, muscle (EC 2.4.1.11) | P13807|GYS1_HUMAN | 83,769 | 5 |
| Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain] | P46531|NOTC1_HUMAN | 272,471 | 5 |
| BRCA2 and CDKN1A-interacting protein (Protein TOK-1) | Q9P287|BCCIP_HUMAN | 35,962 | 5 |
| Eukaryotic translation initiation factor 4E (eIF4E) (eIF-4E) (mRNA cap-binding protein) (eIF-4F 25 kDa subunit) | P06730|IF4E_HUMAN | 25,080 | 5 |
| AP-3 complex subunit mu-1 (Adapter-related protein complex 3 mu-1 subunit) (Mu-adaptin 3A) (AP-3 adapter complex mu3A subunit) | Q9Y2T2|AP3M1_HUMAN | 46,922 | 5 |
| Import inner membrane translocase subunit TIM50, mitochondrial precursor | Q3ZCQ8|TIM50_HUMAN | 39,630 | 5 |
| Translocation protein SEC63 homolog | Q9UGP8|SEC63_HUMAN | 87,981 | 5 |
| ATP synthase delta chain, mitochondrial precursor (EC 3.6.3.14) | P30049|ATPD_HUMAN | 17,472 | 5 |
| Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52) | Q9NZV1|CRIM1_HUMAN | 113,717 | 5 |
| RNA-binding protein 9 (RNA-binding motif protein 9) (Hexaribonucleotide-binding protein 2) (Repressor of tamoxifen transcriptional activity) | O43251|RBM9_HUMAN | 41,356 | 5 |
| Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2) | Q9Y6W5|WASF2_HU MAN | 54,267 | 5 |
| MAK16-like protein RBM13 (RNA-binding motif protein 13) (NNP78) | Q9BXY0|RBM13_HUMAN | 35,352 | 5 |
| Pre-mRNA 3′-end-processing factor FIP1 (FIP1-like 1) (Factor interacting with PAP) (hFip1) (Rearranged in hypereosinophilia) | Q6UN15|FIP1_HUMAN | 66,509 | 5 |
| Polypeptide N-acetylgalactosaminyltransferase 1 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 1) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase 1) (Polypeptide GalNAc transferase 1) (GalNAc-T1) (pp-GaNTase 1) [Contains: Polypeptide N-acetylgalactosaminyltransferase 1 soluble form] | Q10472|GALT1_HUMAN | 64,202 | 5 |
| Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1) (Death inducer with SAP domain) | Q8IX12|CCAR1_HUMAN | 132,806 | 5 |
| Receptor expression-enhancing protein 5 (Polyposis locus protein 1) (TB2 protein) | Q00765|REEP5_HUMAN | 21,477 | 5 |
| Profilin-2 (Profilin II) | P35080|PROF2_HUMAN | 15,028 | 5 |
| Septin-10 | Q9P0V9|SEP10_HUMAN | 59,964 | 5 |
| Serine/threonine-protein kinase Nek9 (EC 2.7.11.1) (NimA-related protein kinase 9) (Never in mitosis A-related kinase 9) (Nercc1 kinase) (NIMA-related kinase 8) (Nek8) | Q8TD19|NEK9_HUMAN | 107,152 | 5 |
| Syntaxin-binding protein 2 (Unc-18 homolog 2) (Unc-18B) (Unc18-2) | Q15833|STXB2_HUMAN | 66,423 | 5 |
| YTH domain family protein 2 (High-glucose-regulated protein 8) (CLL-associated antigen KW-14) (Renal carcinoma antigen NY-REN-2) | Q9Y5A9|YTHD2_HUMAN | 62,316 | 5 |
| Ras-related protein Rab-6A (Rab-6) | P20340|RAB6A_HUMAN | 23,575 | 5 |
| Thioredoxin domain-containing protein 12 precursor (EC 1.8.4.2) (Thioredoxin-like protein p19) (Endoplasmic reticulum protein ERp19) (ERp18) (hTLP19) | O95881|TXD12_HUMAN | 19,188 | 5 |
| Protein phosphatase inhibitor 2 (IPP-2) | P41236|IPP2_HUMAN | 22,998 | 5 |
| E3 ubiquitin-protein ligase RNF25 (EC 6.3.2.-) (RING finger protein 25) | Q96BH1|RNF25_HUMAN | 51,200 | 5 |
| ADP-ribosylation factor-like protein 8A (ADP-ribosylation factor-like protein 10B) (Novel small G protein indispensable for equal chromosome segregation 2) | Q96BM9|ARL8A_HUMAN | 21,399 | 5 |
| Amidophosphoribosyltransferase precursor (EC 2.4.2.14) (Glutamine phosphoribosylpyrophosphate amidotransferase) (ATASE) (GPAT) | Q06203|PUR1_HUMAN | 57,381 | 5 |
| Poliovirus receptor-related protein 2 precursor (Herpes virus entry mediator B) (HveB) (Nectin-2) (CD112 antigen) | Q92692|PVRL2_HUMAN | 57,724 | 5 |
| N-acetylgalactosaminyltransferase 7 (EC 2.4.1.-) (Protein-UDP acetylgalactosaminyltransferase 7) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase 7) (Polypeptide GalNAc transferase 7) (GalNAc-T7) (pp-GaNTase 7) | Q86SF2|GALT7_HUMAN | 75,373 | 5 |
| Platelet-activating factor acetylhydrolase IB subunit gamma (EC 3.1.1.47) (PAF acetylhydrolase 29 kDa subunit) (PAF-AH 29 kDa subunit) (PAF-AH subunit gamma) (PAFAH subunit gamma) | Q15102|PA1B3_HUMAN | 25,716 | 5 |
| Nuclear pore glycoprotein p62 (62 kDa nucleoporin) | P37198|NUP62_HUMAN | 53,238 | 5 |
| Caspase-4 precursor (EC 3.4.22.57) (CASP-4) (ICH-2 protease) (TX protease) (ICE(rel)-II) [Contains: Caspase-4 subunit 1; Caspase-4 subunit 2] | P49662| CASP4_HU MAN | 43,245 | 5 |
| Galectin-9 (HOM-HD-21) (Ecalectin) | O00182|LEG9_HUMAN | 39,500 | 5 |
| Isopentenyl-diphosphate Delta-isomerase 1 (EC 5.3.3.2) (IPP isomerase 1) (Isopentenyl pyrophosphate isomerase 1) (IPPI1) | Q13907|IDI1_HUMAN | 26,302 | 5 |
| TGF-beta receptor type-2 precursor (EC 2.7.11.30) (TGF-beta receptor type II) (TGFR-2) (TGF-beta type II receptor) (Transforming growth factor-beta receptor type II) (TbetaR-II) | P37173|TGFR2_HUMAN | 64,551 | 5 |
| Mps one binder kinase activator-like 1A (Mob1 homolog 1A) (Mob1A) (Mob1B) (Protein Mob4A) | Q7L9L4|MOL1A_HUMAN, Q9H8S9|MOL1B_HUMAN | 25,074 | 5 |
| Ribosome biogenesis protein BOP1 (Block of proliferation 1 protein) | Q14137|BOP1_HUMAN | 83,611 | 5 |
| Protein BUD31 homolog (Protein G10 homolog) (EDG-2) | P41223|BUD31_HUMAN | 16,982 | 5 |
| Keratin, type II cytoskeletal 2 epidermal (Cytokeratin-2e) (K2e) (CK 2e) (Keratin 2) | P35908|K22E_HUMAN | 65,848 | 5 |
| Import inner membrane translocase subunit TIM44, mitochondrial precursor | O43615|TIM44_HUMAN | 51,339 | 5 |
| Lysosome membrane protein 2 (Lysosome membrane protein II) (LIMP II) (Scavenger receptor class B member 2) (85 kDa lysosomal membrane sialoglycoprotein) (LGP85) (CD36 antigen-like 2) | Q14108|SCRB2_HUMAN | 54,274 | 5 |
| Focal adhesion kinase 1 (EC 2.7.10.2) (FADK 1) (pp125FAK) (Protein-tyrosine kinase 2) | Q05397|FAK1_HUMAN | 119,218 | 5 |
| MICAL-like protein 1 (Molecule interacting with Rab13) (MIRab13) | Q8N3F8|MILK1_HUMAN | 93,424 | 5 |
| Protein FAM98A | Q8NCA5|FA98A_HUMAN | 55,383 | 5 |
| Apolipoprotein-L2 (Apolipoprotein L-II) (ApoL-II) | Q9BQE5|APOL2_HUMAN | 37,075 | 5 |
| Splicing factor, arginine/serine-rich 5 (Pre-mRNA-splicing factor SRP40) (Delayed-early protein HRS) | Q13243|SFRS5_HUMAN | 31,247 | 5 |
| Sulfotransferase family cytosolic 1B member 1 (EC 2.8.2.-) (Sulfotransferase 1B2) (Thyroid hormone sulfotransferase) | O43704|ST1B1_HUMAN | 34,883 | 5 |
| Transmembrane protein 165 (Transmembrane protein TPARL) (Transmembrane protein PT27) | Q9HC07|TM165_HUMAN | 34,888 | 5 |
| Ras-related protein R-Ras2 precursor (Ras-like protein TC21) (Teratocarcinoma oncogene) | P62070|RRAS2_HUMAN | 23,382 | 5 |
| ATP synthase f chain, mitochondrial (EC 3.6.3.14) | P56134|ATPK_HUMAN | 10,900 | 5 |
| Probable rRNA-processing protein EBP2 (EBNA1-binding protein 2) (Nucleolar protein p40) | Q99848|EBP2_HUMAN | 34,835 | 5 |
| Activity-dependent neuroprotector (Activity-dependent neuroprotective protein) | Q9H2P0|ADNP_HUMAN | 123,546 | 5 |
| Keratin, type II cytoskeletal 6A (Cytokeratin-6A) (CK 6A) (K6a keratin) | P02538|K2C6A_HUMAN, P48666|K2C6C_HUMAN | 60,028 | 5 |
| Dihydroxyacetone kinase (EC 2.7.1.29) (Glycerone kinase) (DHA kinase) | Q3LXA3|DAK_HUMAN | 58,960 | 5 |
| Cullin-1 (CUL-1) | Q13616|CUL1_HUMAN | 89,663 | 5 |
| Interferon-induced protein with tetratricopeptide repeats 5 (IFIT-5) (Retinoic acid- and interferon-inducible 58 kDa protein) | Q13325|IFIT5_HUMAN | 55,831 | 5 |
| Urokinase plasminogen activator surface receptor precursor (uPAR) (U-PAR) (Monocyte activation antigen Mo3) (CD87 antigen) | Q03405|UPAR_HUMAN | 36,959 | 5 |
| Sjoegren syndrome/scleroderma autoantigen 1 (Autoantigen p27) | O60232|SSA27_HUMAN | 21,457 | 5 |
| Ras-related protein Rab-8A (Oncogene c-mel) | P61006|RAB8A_HUMAN | 23,652 | 5 |
| Multidrug resistance-associated protein 1 (ATP-binding cassette sub-family C member 1) (Leukotriene C(4) transporter) (LTC4 transporter) | P33527|MRP1_HUMAN | 171,547 | 5 |
| Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55) (p59) (Golgi phosphoprotein 6) (GOLPH6) | Q9H8Y8|GORS2_HUMAN | 47,128 | 4 |
| Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA-binding protein 2) (IMP-2) (Hepatocellular carcinoma autoantigen p62) | Q9Y6M1|IF2B2_HUMAN | 61,825 | 4 |
| Gamma-soluble NSF attachment protein (SNAP-gamma) (N-ethylmaleimide-sensitive factor attachment protein, gamma) | Q99747|SNAG_HUMAN | 34,729 | 4 |
| Rho guanine nucleotide exchange factor 7 (PAK-interacting exchange factor beta) (Beta-Pix) (COOL-1) (p85) | Q14155|ARHG7_HUMAN | 89,996 | 4 |
| Cation-independent mannose-6-phosphate receptor precursor (CI Man-6-P receptor) (CI-MPR) (M6PR) (Insulin-like growth factor 2 receptor) (Insulin-like growth factor II receptor) (IGF-II receptor) (M6P/IGF2 receptor) (M6P/IGF2R) (300 kDa mannose 6-phosphate receptor) (MPR 300) (MPR300) (CD222 antigen) | P11717|MPRI_HUMAN | 274,256 | 4 |
| Ephrin type-A receptor 2 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor ECK) (Epithelial cell kinase) | P29317|EPHA2_HUMAN | 108,237 | 4 |
| Tubulin--tyrosine ligase-like protein 12 | Q14166|TTL12_HUMAN | 74,386 | 4 |
| YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113) | P49750|YLPM1_HUMAN | 219,970 | 4 |
| Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial precursor (EC 1.1.1.35) (Short chain 3-hydroxyacyl-CoA dehydrogenase) (HCDH) (Medium and short chain L-3-hydroxyacyl-coenzyme A dehydrogenase) | Q16836|HCDH_HUMAN | 34,260 | 4 |
| Uncharacterized protein C1orf77 | Q9Y3Y2|CA077_HUMAN | 26,380 | 4 |
| Density-regulated protein (DRP) (Protein DRP1) (Smooth muscle cell-associated protein 3) (SMAP-3) | O43583|DENR_HUMAN | 22,074 | 4 |
| Prefoldin subunit 5 (C-myc-binding protein Mm-1) (Myc modulator 1) | Q99471|PFD5_HUMAN | 17,310 | 4 |
| 5′-AMP-activated protein kinase catalytic subunit alpha-1 (EC 2.7.11.1) (AMPK alpha-1 chain) | Q13131|AAPK1_HUMAN | 62,791 | 4 |
| Neuropilin-1 precursor (Vascular endothelial cell growth factor 165 receptor) (CD304 antigen) | O14786|NRP1_HUMAN | 103,105 | 4 |
| Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G) | O43491|E41L2_HUMAN | 112,570 | 4 |
| Annexin A7 (Annexin VII) (Synexin) | P20073|ANXA7_HUMAN | 52,723 | 4 |
| NEDD8-conjugating enzyme Ubc12 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme E2 M) (NEDD8 protein ligase) (NEDD8 carrier protein) | P61081|UBC12_HUMAN | 20,883 | 4 |
| Hydroxymethylglutaryl-CoA synthase, cytoplasmic (EC 2.3.3.10) (HMG-CoA synthase) (3-hydroxy-3-methylglutaryl coenzyme A synthase) | Q01581|HMCS1_HUMAN | 57,277 | 4 |
| Cleavage stimulation factor 64 kDa subunit (CSTF 64 kDa subunit) (CF-1 64 kDa subunit) (CstF-64) | P33240|CSTF2_HUMAN | 60,941 | 4 |
| Translation initiation factor eIF-2B subunit delta (eIF-2B GDP-GTP exchange factor subunit delta) | Q9UI10|EI2BD_HUMAN | 57,539 | 4 |
| Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs) | O14964|HGS_HUMAN | 86,174 | 4 |
| Ubiquitin-conjugating enzyme E2 O (EC 6.3.2.19) (Ubiquitin-protein ligase O) (Ubiquitin carrier protein O) (Ubiquitin-conjugating enzyme E2 of 230 kDa) (E2-230K) | Q9C0C9|UBE2O_HUMAN | 141,336 | 4 |
| Glycogen synthase kinase-3 beta (EC 2.7.11.26) (GSK-3 beta) | P49841|GSK3B_HUMAN | 46,727 | 4 |
| Mitochondrial 39S ribosomal protein L4 (L4mt) (MRP-L4) | Q9BYD3|RM04_HUMAN | 34,902 | 4 |
| Ribose-phosphate pyrophosphokinase I (EC 2.7.6.1) (Phosphoribosyl pyrophosphate synthetase I) (PRS-I) (PPRibP) | P60891|PRPS1_HUMAN | 34,817 | 4 |
| 3-hydroxyisobutyrate dehydrogenase, mitochondrial precursor (EC 1.1.1.31) (HIBADH) | P31937|3HIDH_HUMAN | 35,312 | 4 |
| Medium-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.3) (MCAD) | P11310|ACADM_HUMAN | 46,572 | 4 |
| RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein) (CAST) | O15446|PAF49_HUMAN | 54,968 | 4 |
| Guanine nucleotide-binding protein alpha-13 subunit (G alpha-13) | Q14344|GNA13_HUMAN | 44,033 | 4 |
| Serine/threonine-protein kinase N1 (EC 2.7.11.13) (Protein kinase C-like 1) (Protein-kinase C-related kinase 1) (Protein kinase C-like PKN) (Serine-threonine protein kinase N) (Protein kinase PKN-alpha) | Q16512|PKN1_HUMAN | 103,975 | 4 |
| SPFH domain-containing protein 2 precursor | O94905|SPFH2_HUMAN | 37,822 | 4 |
| Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-12 subunit precursor | Q9UBI6|GBG12_HUMAN | 7,989 | 4 |
| Nuclear pore complex protein Nup98-Nup96 precursor [Contains: Nuclear pore complex protein Nup98 (Nucleoporin Nup98) (98 kDa nucleoporin); Nuclear pore complex protein Nup96 (Nucleoporin Nup96) (96 kDa nucleoporin)] | P52948|NUP98_HUMAN | 187,776 | 4 |
| Actin-like protein 6A (53 kDa BRG1-associated factor A) (Actin-related protein Baf53a) (ArpNbeta) | O96019|ACL6A_HUMAN | 47,443 | 4 |
| Protein LAP4 (Protein scribble homolog) (hScrib) | Q14160|LAP4_HUMAN | 174,915 | 4 |
| Tetratricopeptide repeat protein 37 (TPR repeat protein 37) | Q6PGP7|TTC37_HUMAN | 175,474 | 4 |
| ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta) | Q9BQ39|DDX50_HUMAN | 82,549 | 4 |
| Prostaglandin E synthase 2 (EC 5.3.99.3) (Microsomal prostaglandin E synthase 2) (mPGES-2) [Contains: Prostaglandin E synthase 2 truncated form] | Q9H7Z7|PGES2_HUMAN | 41,926 | 4 |
| Peroxisomal biogenesis factor 19 (Peroxin-19) (Peroxisomal farnesylated protein) (33 kDa housekeeping protein) | P40855|PEX19_HUMAN | 32,789 | 4 |
| Selenocysteine-specific elongation factor (Elongation factor sec) (Eukaryotic elongation factor, selenocysteine-tRNA-specific) | P57772|SELB_HUMAN | 65,322 | 4 |
| GTPase-activating protein ZNF289 | Q8N6H7|ZN289_HUMAN | 56,703 | 4 |
| CUG triplet repeat RNA-binding protein 1 (CUG-BP1) (RNA-binding protein BRUNOL-2) (Deadenylation factor CUG-BP) (50 kDa Nuclear polyadenylated RNA-binding protein) (EDEN-BP) | Q92879|CUGB1_HUMAN | 52,046 | 4 |
| FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23) | Q6ZNL6|FGD5_HUMAN | 159,890 | 4 |
| Succinyl-CoA:3-ketoacid-coenzyme A transferase 1, mitochondrial precursor (EC 2.8.3.5) (Somatic-type succinyl CoA:3-oxoacid CoA-transferase) (Scot-S) | P55809|SCOT_HUMAN | 56,141 | 4 |
| Zinc phosphodiesterase ELAC protein 2 (EC 3.1.26.11) (Ribonuclease Z 2) (RNase Z 2) (tRNase Z 2) (tRNA 3 endonuclease 2) (ElaC homolog protein 2) (Heredity prostate cancer protein 2) | Q9BQ52|RNZ2_HUMAN | 92,202 | 4 |
| Alpha-N-acetylgalactosaminidase precursor (EC 3.2.1.49) (Alpha-galactosidase B) | P17050|NAGAB_HUMAN | 46,548 | 4 |
| Activating signal cointegrator 1 (ASC-1) (Thyroid receptor-interacting protein 4) (TRIP-4) | Q15650|TRIP4_HUMAN | 66,130 | 4 |
| Uncharacterized potential DNA-binding protein C17orf49 | Q8IXM2|CQ049_HUMAN | 17,883 | 4 |
| Translation initiation factor eIF-2B subunit beta (eIF-2B GDP-GTP exchange factor subunit beta) (S20I15) (S20III15) | P49770|EI2BB_HUMAN | 38,972 | 4 |
| Dehydrogenase/reductase SDR family member 4 (EC 1.1.1.184) (NADPH-dependent carbonyl reductase/NADP-retinol dehydrogenase) (CR) (PHCR) (Peroxisomal short-chain alcohol dehydrogenase) (NADPH-dependent retinol dehydrogenase/reductase) (NDRD) (SCAD-SRL) (humNRDR) (PSCD) | Q9BTZ2|DHRS4_HUMAN | 27,554 | 4 |
| Cadherin-13 precursor (Truncated-cadherin) (T-cadherin) (T-cad) (Heart-cadherin) (H-cadherin) (P105) | P55290|CAD13_HUMAN | 78,270 | 4 |
| Transcriptional activator protein Pur-alpha (Purine-rich single-stranded DNA-binding protein alpha) | Q00577|PURA_HUMAN | 34,893 | 4 |
| Guanine nucleotide-binding protein subunit beta 4 (Transducin beta chain 4) | Q9HAV0|GBB4_HUMAN | 37,550 | 4 |
| Peptidyl-prolyl cis-trans isomerase-like 3 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL3) (Cyclophilin J) (CyPJ) | Q9H2H8|PPIL3_HUMAN | 18,137 | 4 |
| Abl interactor 1 (Abelson interactor 1) (Abi-1) (Spectrin SH3 domain-binding protein 1) (Eps8 SH3 domain-binding protein) (Eps8-binding protein) (e3B1) (Nap1-binding protein) (Nap1BP) (Abl-binding protein 4) (AblBP4) | Q8IZP0|ABI1_HUMAN | 55,064 | 4 |
| Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1) | Q14978|NOLC1_HUMAN | 73,703 | 4 |
| FK506-binding protein 11 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (19 kDa FK506-binding protein) (FKBP-19) | Q9NYL4|FKB11_HUMAN | 22,163 | 4 |
| cAMP-dependent protein kinase, alpha-catalytic subunit (EC 2.7.11.11) (PKA C-alpha) | P17612|KAPCA_HUMAN | 40,573 | 4 |
| Promega trypsin artifact 5 K to R mods (2239.1, 2914)(1987, 2003) | CONT|Trypa5|PromTArt5 | 5,543 | 4 |
| Serine/threonine-protein kinase OSR1 (EC 2.7.11.1) (Oxidative stress-responsive 1 protein) | O95747|OXSR1_HUMAN | 58,005 | 4 |
| GTPase IMAP family member 7 (Immunity-associated nucleotide 7 protein) | Q8NHV1|GIMA7_HUMAN | 34,492 | 4 |
| Armadillo repeat protein deleted in velo-cardio-facial syndrome | O00192|ARVC_HUMAN | 104,624 | 4 |
| NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1) | Q8TDZ2|MICA1_HUMAN | 117,858 | 4 |
| COP9 signalosome complex subunit 6 (Signalosome subunit 6) (SGN6) (JAB1-containing signalosome subunit 6) (Vpr-interacting protein) (hVIP) (MOV34 homolog) | Q7L5N1|CSN6_HUMAN | 36,145 | 4 |
| Protein phosphatase methylesterase 1 (EC 3.1.1.-) (PME-1) | Q9Y570|PPME1_HUMAN | 42,298 | 4 |
| Putative eukaryotic translation initiation factor 3 subunit (eIF-3) | O75153|IF3X_HUMAN | 146,654 | 4 |
| Histone-arginine methyltransferase CARM1 (EC 2.1.1.125) (EC 2.1.1.-) (Protein arginine N-methyltransferase 4) (Coactivator-associated arginine methyltransferase 1) | Q86X55|CARM1_HUMAN | 63,442 | 4 |
| Transcriptional repressor p66 alpha (Hp66alpha) (GATA zinc finger domain-containing protein 2A) | Q86YP4|P66A_HUMAN | 68,045 | 4 |
| Interferon-inducible double stranded RNA-dependent protein kinase activator A (Protein kinase, interferon-inducible double stranded RNA-dependent activator) (Protein activator of the interferon-induced protein kinase) (PKR-associated protein X) (PKR-associating protein X) | O75569|PRKRA_HUMAN | 34,387 | 4 |
| Pseudouridylate synthase 7 homolog (EC 5.4.99.-) | Q96PZ0|PUS7_HUMAN | 75,020 | 4 |
| Synembryn-A (Protein Ric-8A) | Q9NPQ8|RIC8A_HUMAN | 59,595 | 4 |
| Apoptosis inhibitor 5 (API-5) (Fibroblast growth factor 2-interacting factor) (FIF) (Protein XAGL) (Antiapoptosis clone 11 protein) (AAC-11) | Q9BZZ5|API5_HUMAN | 57,545 | 4 |
| UBX domain-containing protein 2 | Q92575|UBXD2_HUMAN | 56,760 | 4 |
| Methylosome subunit pICln (Chloride conductance regulatory protein ICln) (I(Cln)) (Chloride channel, nucleotide sensitive 1A) (Chloride ion current inducer protein) (ClCI) (Reticulocyte pICln) | P54105|ICLN_HUMAN | 26,197 | 4 |
| Nitric oxide synthase-interacting protein (eNOS-interacting protein) | Q9Y314|NOSIP_HUMAN | 33,154 | 4 |
| RNA 3′-terminal phosphate cyclase (EC 6.5.1.4) (RNA-3′-phosphate cyclase) (RNA cyclase) | O00442|RTC1_HUMAN | 39,320 | 4 |
| RAC-alpha serine/threonine-protein kinase (EC 2.7.11.1) (RAC-PK-alpha) (Protein kinase B) (PKB) (C-AKT) | P31749|AKT1_HUMAN | 55,670 | 4 |
| Nuclear receptor-binding protein | Q9UHY1|NRBP_HUMAN | 59,827 | 4 |
| Ubiquitin-associated protein 2 | Q5T6F2|UBAP2_HUMAN | 117,097 | 4 |
| Symplekin | Q92797|SYMPK_HUMAN | 141,136 | 4 |
| Stromal cell-derived factor 2-like protein 1 precursor (SDF2-like protein 1) (PWP1-interacting protein 8) | Q9HCN8|SDF2L_HUMAN | 23,580 | 4 |
| Eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1) (eIF4E-binding protein 1) (Phosphorylated heat- and acid-stable protein regulated by insulin 1) (PHAS-I) | Q13541|4EBP1_HUMAN | 12,562 | 4 |
| Deoxyhypusine hydroxylase (EC 1.14.99.29) (Deoxyhypusine monooxygenase) (hDOHH) (HEAT-like repeat-containing protein 1) | Q9BU89|DOHH_HUMAN | 32,886 | 4 |
| Protein FAM96B | Q9Y3D0|FA96B_HUMAN | 17,645 | 4 |
| Acyl-protein thioesterase 1 (EC 3.1.2.-) (Lysophospholipase 1) (Lysophospholipase I) | O75608|LYPA1_HUMAN | 24,652 | 4 |
| Cytoplasmic protein NCK1 (NCK adaptor protein 1) (SH2/SH3 adaptor protein NCK-alpha) | P16333|NCK1_HUMAN | 42,846 | 4 |
| DnaJ homolog subfamily C member 11 | Q9NVH1|DCJ11_HUMAN | 63,261 | 4 |
| Queuine tRNA-ribosyltransferase (EC 2.4.2.29) (tRNA-guanine transglycosylase) (Guanine insertion enzyme) | Q9BXR0|TGT_HUMAN | 42,474 | 4 |
| WD repeat protein 36 (T-cell activation WD repeat-containing protein) (TA-WDRP) | Q8NI36|WDR36_HUMAN | 105,308 | 4 |
| U6 snRNA-associated Sm-like protein LSm2 (snRNP core Sm-like protein Sm-x5) (Small nuclear ribonuclear protein D homolog) (Protein G7b) | Q9Y333|LSM2_HUMAN | 10,817 | 4 |
| Dynamin-like 120 kDa protein, mitochondrial precursor (Optic atrophy protein 1) [Contains: Dynamin-like 120 kDa protein, form S1] | O60313|OPA1_HUMAN | 111,643 | 4 |
| Monocarboxylate transporter 1 (MCT 1) (Solute carrier family 16 member 1) | P53985|MOT1_HUMAN | 53,942 | 4 |
| Golgi resident protein GCP60 (Acyl-CoA-binding domain-containing protein 3) (Golgi phosphoprotein 1) (GOLPH1) (Golgi complex-associated protein 1) (GOCAP1) (PBR- and PKA-associated protein 7) (Peripheral benzodiazepine receptor-associated protein PAP7) | Q9H3P7|GCP60_HUMAN | 60,575 | 4 |
| SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 (BRG1-associated factor 57) | Q969G3|SMCE1_HUMAN | 46,632 | 4 |
| Ubiquitin fusion degradation protein 1 homolog (UB fusion protein 1) | Q92890|UFD1_HUMAN | 38,708 | 4 |
| EGF, latrophilin and seven transmembrane domain-containing protein 1 precursor (EGF-TM7-latrophilin-related protein) (ETL protein) | Q9HBW9|ELTD1_HUMAN | 77,809 | 4 |
| Phospholipid scramblase 1 (PL scramblase 1) (Ca(2+)-dependent phospholipid scramblase 1) (Erythrocyte phospholipid scramblase) (MmTRA1b) | O15162|PLS1_HUMAN | 35,031 | 4 |
| U4/U6 small nuclear ribonucleoprotein Prp31 (Pre-mRNA-processing factor 31) (U4/U6 snRNP 61 kDa protein) (hPrp31) (Protein 61K) (Serologically defined breast cancer antigen NY-BR-99) | Q8WWY3|PRP31_HUMAN | 55,439 | 4 |
| Helicase SKI2W (EC 3.6.1.-) (Helicase-like protein) (HLP) | Q15477|SKIV2_HUMAN | 137,784 | 4 |
| Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X-linked) (Ubiquitin-specific protease 9, X chromosome) | Q93008|USP9X_HUMAN | 289,527 | 4 |
| FACT complex subunit SPT16 (Facilitates chromatin transcription complex subunit SPT16) (hSPT16) (FACT 140 kDa subunit) (FACTp140) (Chromatin-specific transcription elongation factor 140 kDa subunit) | Q9Y5B9|SPT16_HUMAN | 119,899 | 4 |
| Superoxide dismutase [Mn], mitochondrial precursor (EC 1.15.1.1) | P04179|SODM_HUMAN | 24,705 | 4 |
| Lamin-B receptor (Integral nuclear envelope inner membrane protein) (LMN2R) | Q14739|LBR_HUMAN | 70,688 | 4 |
| H/ACA ribonucleoprotein complex subunit 3 (Nucleolar protein family A member 3) (snoRNP protein NOP10) | Q9NPE3|NOLA3_HUMAN | 7,688 | 4 |
| Hook homolog 3 (hHK3) | Q86VS8|HOOK3_HUMAN | 83,110 | 4 |
| Nucleolar complex protein 3 homolog (NOC3 protein homolog) (NOC3-like protein) (Nucleolar complex-associated protein 3-like protein) (Factor for adipocyte differentiation 24) | Q8WTT2|NOC3L_HUMAN | 92,534 | 4 |
| Tumor susceptibility gene 101 protein | Q99816|TS101_HUMAN | 43,928 | 4 |
| Protein disulfide-isomerase TXNDC10 precursor (EC 5.3.4.1) (Thioredoxin domain-containing protein 10) (Thioredoxin-related transmembrane protein 3) | Q96JJ7|TXD10_HUMAN | 51,855 | 4 |
| Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform (EC 3.1.3.16) (Calmodulin-dependent calcineurin A subunit alpha isoform) (CAM-PRP catalytic subunit) | Q08209|PP2BA_HUMAN | 58,672 | 4 |
| cAMP-dependent protein kinase inhibitor gamma (PKI-gamma) | Q9Y2B9|IPKG_HUMAN | 7,892 | 4 |
| Polyglutamine-binding protein 1 (Polyglutamine tract-binding protein 1) (PQBP-1) (38 kDa nuclear protein containing a WW domain) (Npw38) | O60828|PQBP1_HUMAN | 30,455 | 4 |
| Ubiquitin-conjugating enzyme E2 Z (EC 6.3.2.19) (Ubiquitin-protein ligase Z) (Ubiquitin carrier protein Z) | Q9H832|UBE2Z_HUMAN | 38,193 | 4 |
| cAMP-dependent protein kinase type II-beta regulatory subunit | P31323|KAP3_HUMAN | 46,329 | 4 |
| Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit) | Q04206|TF65_HUMAN | 60,202 | 4 |
| Neutral amino acid transporter B(0) (ATB(0)) (Sodium-dependent neutral amino acid transporter type 2) (RD114/simian type D retrovirus receptor) (Baboon M7 virus receptor) | Q15758|AAAT_HUMAN | 56,582 | 4 |
| HBS1-like protein (ERFS) | Q9Y450|HBS1L_HUMAN | 75,456 | 4 |
| ATP synthase subunit g, mitochondrial (EC 3.6.3.14) (ATPase subunit g) | O75964|ATP5L_HUMAN | 11,411 | 4 |
| Sorting and assembly machinery component 50 homolog | Q9Y512|SAM50_HUMAN | 51,960 | 4 |
| Gasdermin domain-containing protein 1 | P57764|GSDC1_HUMAN | 52,783 | 4 |
| DCN1-like protein 1 (Defective in cullin neddylation protein 1-like protein 1) (DCUN1 domain-containing protein 1) (Squamous cell carcinoma-related oncogene) | Q96GG9|DCNL1_HUMAN | 30,108 | 4 |
| Small nuclear ribonucleoprotein F (snRNP-F) (Sm protein F) (Sm-F) (SmF) | P62306|RUXF_HUMAN | 9,707 | 4 |
| Developmentally-regulated GTP-binding protein 2 (DRG 2) | P55039|DRG2_HUMAN | 40,730 | 4 |
| Nuclear fragile X mental retardation-interacting protein 2 (FMRP-interacting protein 2) (82 kDa FMRP-interacting protein) (82-FIP) (Proliferation-inducing gene 1 protein) | Q7Z417|NUFP2_HUMAN | 76,103 | 4 |
| Phosphomannomutase 2 (EC 5.4.2.8) (PMM 2) | O15305|PMM2_HUMAN | 28,065 | 4 |
| Uridine phosphorylase 1 (EC 2.4.2.3) (UrdPase 1) (UPase 1) | Q16831|UPP1_HUMAN | 33,917 | 4 |
| Hepatitis B virus X-interacting protein (HBX-interacting protein) (HBV X-interacting protein) | O43504|XIP_HUMAN | 9,596 | 4 |
| Sorting nexin-4 | O95219|SNX4_HUMAN | 51,892 | 4 |
| Oxidoreductase HTATIP2 (EC 1.1.1.-) (HIV-1 TAT-interactive protein 2) (30 kDa HIV-1 TAT-interacting protein) | Q9BUP3|TIP30_HUMAN | 27,100 | 4 |
| Rho-related GTP-binding protein RhoB precursor (H6) | P62745|RHOB_HUMAN | 22,105 | 4 |
| Sorting nexin-12 | Q9UMY4|SNX12_HUMAN | 19,713 | 4 |
| Caspase-8 precursor (EC 3.4.22.61) (CASP-8) (ICE-like apoptotic protease 5) (MORT1-associated CED-3 homolog) (MACH) (FADD-homologous ICE/CED-3-like protease) (FADD-like ICE) (FLICE) (Apoptotic cysteine protease) (Apoptotic protease Mch-5) (CAP4) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10] | Q14790|CASP8_HUMAN | 55,376 | 4 |
| Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin) | O94833|BPAEA_HUMAN | 590,974 | 4 |
| Serpin B8 (Cytoplasmic antiproteinase 2) (CAP-2) (CAP2) (Protease inhibitor 8) | P50452|SPB8_HUMAN | 42,769 | 4 |
| Leucine-rich repeat flightless-interacting protein 2 (LRR FLII-interacting protein 2) | Q9Y608|LRRF2_HUMAN | 82,157 | 4 |
| UPF0384 protein CGI-117 (HBV pre-S2 trans-regulated protein 3) | Q9Y3C1|U384_HUMAN | 21,171 | 4 |
| Bifunctional coenzyme A synthase (CoA synthase) (NBP) (POV-2) [Includes: Phosphopantetheine adenylyltransferase (EC 2.7.7.3) (Pantetheine-phosphate adenylyltransferase) (PPAT) (Dephospho-CoA pyrophosphorylase); Dephospho-CoA kinase (EC 2.7.1.24) (DPCK) (Dephosphocoenzyme A kinase) (DPCOAK)] | Q13057|COASY_HUMAN | 62,312 | 4 |
| Protein ariadne-1 homolog (ARI-1) (Ubiquitin-conjugating enzyme E2-binding protein 1) (UbcH7-binding protein) (UbcM4-interacting protein) (HHARI) (H7-AP2) (Monocyte protein 6) (MOP-6) | Q9Y4X5|ARI1_HUMAN | 64,099 | 4 |
| Angiotensin-converting enzyme, somatic isoform precursor (EC 3.4.15.1) (Dipeptidyl carboxypeptidase I) (Kininase II) (CD143 antigen) [Contains: Angiotensin-converting enzyme, somatic isoform, soluble form] | P12821|ACE_HUMAN | 149,701 | 4 |
| GTP-binding protein SAR1a (COPII-associated small GTPase) | Q9NR31|SAR1A_HUMAN | 22,350 | 4 |
| E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR) | Q9ULT8|HECD1_HUMAN | 289,580 | 4 |
| BET1 homolog (Golgi vesicular membrane-trafficking protein p18) (hBET1) | O15155|BET1_HUMAN | 13,272 | 4 |
| Transmembrane protein 109 precursor (Mitsugumin-23) (Mg23) | Q9BVC6|TM109_HUMAN | 26,193 | 4 |
| Alpha-endosulfine (ARPP-19e) | O43768|ENSA_HUMAN, P56211|ARP19_HUMAN | 13,371 | 4 |
| mRNA turnover protein 4 homolog | Q9UKD2|MRT4_HUMAN | 27,543 | 4 |
| CAAX prenyl protease 1 homolog (EC 3.4.24.84) (Prenyl protein-specific endoprotease 1) (Farnesylated proteins-converting enzyme 1) (FACE-1) (Zinc metalloproteinase Ste24 homolog) | O75844|FACE1_HUMAN | 54,798 | 4 |
| 26S proteasome non-ATPase regulatory subunit 8 (26S proteasome regulatory subunit S14) (p31) | P48556|PSMD8_HUMAN | 29,989 | 4 |
| Ubiquitin-activating enzyme E1 homolog (D8) | P41226|UBE1L_HUMAN | 111,703 | 4 |
| Metaxin-1 | Q13505|MTX1_HUMAN | 35,760 | 4 |
| Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating (EC 1.1.1.170) (H105e3 protein) | Q15738|NSDHL_HUMAN | 41,883 | 4 |
| Protein FAM98B | Q52LJ0|FA98B_HUMAN | 37,175 | 4 |
| RNA-binding protein 34 (RNA-binding motif protein 34) | P42696|RBM34_HUMAN | 48,548 | 4 |
| Dolichyl-phosphate beta-glucosyltransferase (EC 2.4.1.117) (DolP-glucosyltransferase) | Q9Y673|ALG5_HUMAN | 36,930 | 4 |
| Glutamine-dependent NAD(+) synthetase (EC 6.3.5.1) (NAD(+) synthase [glutamine-hydrolyzing]) (NAD(+) synthetase 1) | Q6IA69|NADE1_HUMAN | 79,277 | 4 |
| Brix domain-containing protein 1 | Q9H7B2|BXDC1_HUMAN | 35,568 | 4 |
| Actin-related protein 10 (hARP11) | Q9NZ32|ARP10_HUMAN | 46,290 | 4 |
| Cysteine-rich with EGF-like domain protein 2 precursor | Q6UXH1|CREL2_HUMAN | 38,173 | 4 |
| Cysteine-rich protein 1 (Cysteine-rich intestinal protein) (CRIP) (Cysteine-rich heart protein) (hCRHP) | P50238|CRIP1_HUMAN | 8,515 | 4 |
| MMS19-like protein (hMMS19) (MET18 homolog) | Q96T76|MMS19_HUMAN | 113,259 | 4 |
| Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Calcium-binding protein BDR-1) (HLP2) | P37235|HPCL1_HUMAN, P84074|HPCA_HUMAN | 22,296 | 4 |
| Echinoderm microtubule-associated protein-like 1 (EMAP-1) (HuEMAP-1) | O00423|EMAL1_HUMAN | 79,009 | 4 |
| Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30) | Q7L2E3|DHX30_HUMAN | 133,924 | 4 |
| Glutathione peroxidase 7 precursor (EC 1.11.1.9) (CL683) | Q96SL4|GPX7_HUMAN | 20,978 | 4 |
| Dedicator of cytokinesis protein 1 (180 kDa protein downstream of CRK) (DOCK180) | Q14185|DOCK1_HUMAN | 215,365 | 4 |
| Striatin-3 (Cell-cycle autoantigen SG2NA) (S/G2 antigen) | Q13033|STRN3_HUMAN | 87,117 | 4 |
| Lipopolysaccharide-responsive and beige-like anchor protein (CDC4-like protein) (Beige-like protein) | P50851|LRBA_HUMAN | 319,145 | 4 |
| 2′-5′-oligoadenylate synthetase 2 (EC 2.7.7.-) ((2–5′)oligo(A) synthetase 2) (2–5A synthetase 2) (p69 OAS/p71 OAS) (p69OAS/p71OAS) | P29728|OAS2_HUMAN | 82,415 | 4 |
| mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (Smad4-interacting transcriptional co-activator) | Q9NPI6|DCP1A_HUMAN | 63,261 | 4 |
| Exocyst complex component 6 (Exocyst complex component Sec15A) (Sec15-like 1) | Q8TAG9|EXOC6_HUMAN | 93,620 | 4 |
| Cullin-4A (CUL-4A) | Q13619|CUL4A_HUMAN | 87,666 | 4 |
| Ephrin-B2 precursor (EPH-related receptor tyrosine kinase ligand 5) (LERK-5) (HTK ligand) (HTK-L) | P52799|EFNB2_HUMAN | 36,906 | 4 |
| Testis-expressed sequence 10 protein | Q9NXF1|TEX10_HUMAN | 105,661 | 4 |
| Protein mago nashi homolog | P61326|MGN_HUMAN, Q96A72|MGN2_HUMAN | 17,146 | 4 |
| Serine/threonine-protein phosphatase 2A regulatory subunit B′ (PP2A, subunit B′, PR53 isoform) (Phosphotyrosyl phosphatase activator) (PTPA) | Q15257|PTPA_HUMAN | 40,650 | 4 |
| Dedicator of cytokinesis protein 10 (Zizimin-3) | Q96BY6|DOC10_HUMAN | 249,300 | 4 |
| Fatty aldehyde dehydrogenase (EC 1.2.1.3) (Aldehyde dehydrogenase, microsomal) (Aldehyde dehydrogenase family 3 member A2) (Aldehyde dehydrogenase 10) | P51648|AL3A2_HUMAN | 54,832 | 4 |
| Multidrug resistance protein 1 (EC 3.6.3.44) (ATP-binding cassette sub-family B member 1) (P-glycoprotein 1) (CD243 antigen) | P08183|MDR1_HUMAN | 141,450 | 4 |
| Transmembrane emp24 domain-containing protein 7 precursor | Q9Y3B3|TMED7_HUMAN | 25,154 | 4 |
| Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B (EC 2.4.1.119) (Oligosaccharyl transferase subunit STT3B) (STT3-B) (Source of immunodominant MHC-associated peptides homolog) | Q8TCJ2|STT3B_HUMAN | 93,660 | 4 |
| Mitochondrial 2-oxoglutarate/malate carrier protein (OGCP) (Solute carrier family 25 member 11) | Q02978|M2OM_HUMAN | 34,045 | 4 |
| Forkhead box protein K1 (Myocyte nuclear factor) (MNF) | P85037|FOXK1_HUMAN | 75,439 | 4 |
| Lysosomal acid phosphatase precursor (EC 3.1.3.2) (LAP) | P11117|PPAL_HUMAN | 48,327 | 4 |
| Uveal autoantigen with coiled-coil domains and ankyrin repeats protein | Q9BZF9|UACA_HUMAN | 162,492 | 3 |
| Ubiquitin-conjugating enzyme E2 variant 2 (MMS2) (Enterocyte differentiation-associated factor EDAF-1) (Enterocyte differentiation-promoting factor) (EDPF-1) (Vitamin D3-inducible protein) (DDVit 1) | Q15819|UB2V2_HUMAN | 16,345 | 3 |
| UPF0160 protein MYG1 | Q9HB07|MYG1_HUMAN | 42,428 | 3 |
| U4/U6 small nuclear ribonucleoprotein Prp3 (Pre-mRNA-splicing factor 3) (U4/U6 snRNP 90 kDa protein) (hPrp3) | O43395|PRPF3_HUMAN | 77,513 | 3 |
| Uncharacterized protein C2orf32 | Q96F85|CB032_HUMAN | 18,630 | 3 |
| Four and a half LIM domains protein 2 (FHL-2) (Skeletal muscle LIM-protein 3) (SLIM 3) (LIM domain protein DRAL) | Q14192|FHL2_HUMAN | 32,174 | 3 |
| Myosin-Va (Dilute myosin heavy chain, non-muscle) (Myosin-12) (Myosin heavy chain 12) (Myoxin) | Q9Y4I1|MYO5A_HUMAN | 215,411 | 3 |
| Fumarylacetoacetate hydrolase domain-containing protein 1 (EC 3.-.-.-) (YISK-like) | Q6P587|FAHD1_HUMAN | 24,825 | 3 |
| Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (SRP1-gamma) | O00505|IMA3_HUMAN | 57,793 | 3 |
| SHC-transforming protein 1 (SH2 domain protein C1) (Src homology 2 domain-containing-transforming protein C1) | P29353|SHC1_HUMAN | 62,835 | 3 |
| Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66) | Q15428|SF3A2_HUMAN | 49,237 | 3 |
| Target of Myb protein 1 | O60784|TOM1_HUMAN | 53,801 | 3 |
| STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70-interacting protein) (E3 ubiquitin protein ligase CHIP) (CLL-associated antigen KW-8) (Antigen NY-CO-7) | Q9UNE7|STUB1_HUMAN | 34,839 | 3 |
| ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1) (Meg-4) (Presenilin-associated metalloprotease) (PAMP) | Q96TA2|YMEL1_HUMAN | 86,440 | 3 |
| Rab11 family-interacting protein 5 (Rab11-FIP5) (Rab11-interacting protein Rip11) (Gamma-SNAP-associated factor 1) (Gaf-1) (Phosphoprotein pp75) | Q9BXF6|RFIP5_HUMAN | 70,398 | 3 |
| Mesoderm development candidate 2 (Renal carcinoma antigen NY-REN-61) | Q14696|MESD2_HUMAN | 26,060 | 3 |
| Sad1/unc-84 protein-like 1 (Unc-84 homolog A) | O94901|UN84A_HUMAN | 90,046 | 3 |
| Serine beta-lactamase-like protein LACTB, mitochondrial precursor (EC 3.4.-.-) | P83111|LACTB_HUMAN | 60,677 | 3 |
| Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin) (Clathrin-interacting protein localized in the trans-Golgi region) (Clint) | Q14677|EPN4_HUMAN | 68,243 | 3 |
| Cleavage stimulation factor 77 kDa subunit (CSTF 77 kDa subunit) (CF-1 77 kDa subunit) (CstF-77) | Q12996|CSTF3_HUMAN | 82,906 | 3 |
| Protein FAM3C precursor (Protein GS3786) | Q92520|FAM3C_HUMAN | 24,663 | 3 |
| Protein bicaudal D homolog 2 (Bic-D 2) | Q8TD16|BICD2_HUMAN | 93,518 | 3 |
| Centromere/kinetochore protein zw10 homolog | O43264|ZW10_HUMAN | 88,815 | 3 |
| ES1 protein homolog, mitochondrial precursor (Protein KNP-I) (Protein GT335) | P30042|ES1_HUMAN | 28,152 | 3 |
| Probable saccharopine dehydrogenase (EC 1.5.1.9) | Q8NBX0|SCPDH_HUMAN | 47,135 | 3 |
| Nuclear pore complex protein Nup133 (Nucleoporin Nup133) (133 kDa nucleoporin) | Q8WUM0|NU133_HUMAN | 129,000 | 3 |
| Serine protease 23 precursor (EC 3.4.21.-) (Putative secreted protein ZSIG13) | O95084|PRS23_HUMAN | 42,984 | 3 |
| AP-1 complex subunit mu-1 (Adaptor-related protein complex 1 mu-1 subunit) (Mu-adaptin 1) (Adaptor protein complex AP-1 mu-1 subunit) (Golgi adaptor HA1/AP1 adaptin mu-1 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 1) (AP-mu chain family member mu1A) (Clathrin coat assembly protein AP47) (Clathrin coat-associated protein AP47) | Q9BXS5|AP1 M1_HUMAN | 48,570 | 3 |
| Phosphatidylinositol-4-phosphate 5-kinase type-2 alpha (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type II alpha) (1-phosphatidylinositol-4-phosphate 5-kinase 2-alpha) (PtdIns(4)P-5-kinase isoform 2-alpha) (PIP5KII-alpha) (Diphosphoinositide kinase 2-alpha) (PtdIns(4)P-5-kinase B isoform) (PIP5KIII) (PtdIns(4)P-5-kinase C isoform) | P48426|PI52A_HUMAN | 46,208 | 3 |
| Phosphoenolpyruvate carboxykinase [GTP], mitochondrial precursor (EC 4.1.1.32) (Phosphoenolpyruvate carboxylase) (PEPCK-M) | Q16822|PPCKM_HUMAN | 70,620 | 3 |
| Autophagy-related protein 3 (APG3-like) (hApg3) (Protein PC3-96) | Q9NT62|ATG3_HUMAN | 35,846 | 3 |
| Dihydropteridine reductase (EC 1.5.1.34) (HDHPR) (Quinoid dihydropteridine reductase) | P09417|DHPR_HUMAN | 25,772 | 3 |
| Long-chain-fatty-acid--CoA ligase 4 (EC 6.2.1.3) (Long-chain acyl-CoA synthetase 4) (LACS 4) | O60488|ACSL4_HUMAN | 79,174 | 3 |
| 39S ribosomal protein L1, mitochondrial precursor (L1mt) (MRP-L1) | Q9BYD6|RM01_HUMAN | 34,436 | 3 |
| Histone-lysine N-methyltransferase, H3 lysine-4 specific SET7 (EC 2.1.1.43) (Histone H3-K4 methyltransferase) (H3-K4-HMTase) (SET domain-containing protein 7) (Set9) (SET7/9) | Q8WTS6|SETD7_HUMAN | 40,703 | 3 |
| Bifunctional protein NCOAT (Nuclear cytoplasmic O-GlcNAcase and acetyltransferase) (Meningioma-expressed antigen 5) [Includes: Beta-hexosaminidase (EC 3.2.1.52) (N-acetyl-beta-glucosaminidase) (Beta-N-acetylhexosaminidase) (Hexosaminidase C) (N-acetyl-beta-D-glucosaminidase) (O-GlcNAcase); Histone acetyltransferase (EC 2.3.1.48) (HAT)] | O60502|NCOAT_HUMAN | 102,900 | 3 |
| Tubulin alpha-1 chain (Alpha-tubulin 1) (Testis-specific alpha-tubulin) (Tubulin H2-alpha) | P68366|TBA1_HUMAN | 49,907 | 3 |
| Condensin complex subunit 1 (Non-SMC condensin I complex subunit D2) (Chromosome condensation-related SMC-associated protein 1) (Chromosome-associated protein D2) (hCAP-D2) (XCAP-D2 homolog) | Q15021|CND1_HUMAN | 157,155 | 3 |
| SH2 domain-containing protein 3C (Novel SH2-containing protein 3) | Q8N5H7|SH2D3_HUMAN | 94,394 | 3 |
| NADH dehydrogenase [ubiquinone] iron-sulfur protein 8, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 23 kDa subunit) (Complex I-23kD) (CI-23kD) (TYKY subunit) | O00217|NDUS8_HUMAN | 23,688 | 3 |
| Translation initiation factor eIF-2B subunit gamma (eIF-2B GDP-GTP exchange factor subunit gamma) | Q9NR50|EI2BG_HUMAN | 50,223 | 3 |
| Peptidyl-prolyl cis-trans isomerase NIMA-interacting 4 (EC 5.2.1.8) (Rotamase Pin4) (PPIase Pin4) (Parvulin 14) (Par14) (Peptidyl-prolyl cis/trans isomerase EPVH) (hPar14) | Q9Y237|PIN4_HUMAN | 13,792 | 3 |
| Eukaryotic translation initiation factor 2C 2 (eIF2C 2) (eIF-2C 2) (Argonaute-2) (Slicer protein) (PAZ Piwi domain protein) (PPD) | Q9UKV8|I2C2_HUMAN | 96,780 | 3 |
| Dihydrofolate reductase (EC 1.5.1.3) | P00374|DYR_HUMAN | 21,435 | 3 |
| Ferritin light chain (Ferritin L subunit) | P02792|FRIL_HUMAN | 20,003 | 3 |
| Programmed cell death protein 10 (TF-1 cell apoptosis-related protein 15) (Cerebral cavernous malformations 3 protein) | Q9BUL8|PDC10_HUMAN | 24,685 | 3 |
| Transmembrane protein 111 | Q9P0I2|TM111_HUMAN | 29,935 | 3 |
| Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2) | O60879|DIAP2_HUMAN | 125,558 | 3 |
| Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif) | Q9P2R3|ANFY1_HUMAN | 128,384 | 3 |
| Cdc42-interacting protein 4 (Thyroid receptor-interacting protein 10) (TRIP-10) (Protein Felic) (Salt-tolerant protein) (hSTP) | Q15642|CIP4_HUMAN | 68,335 | 3 |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 39 kDa subunit) (Complex I-39kD) (CI-39kD) | Q16795|NDUA9_HUMAN | 42,492 | 3 |
| Replication protein A 32 kDa subunit (RP-A) (RF-A) (Replication factor-A protein 2) (p32) (p34) | P15927|RFA2_HUMAN | 29,229 | 3 |
| Rho/Rac guanine nucleotide exchange factor 2 (GEF-H1 protein) (Proliferating cell nucleolar antigen p40) | Q92974|ARHG2_HUMAN | 101,159 | 3 |
| Ferritin heavy chain (EC 1.16.3.1) (Ferritin H subunit) (Proliferation-inducing gene 15 protein) | P02794|FRIH_HUMAN | 21,208 | 3 |
| Translation initiation factor eIF-2B subunit alpha (eIF-2B GDP-GTP exchange factor subunit alpha) | Q14232|EI2BA_HUMAN | 33,695 | 3 |
| 39S ribosomal protein L37, mitochondrial precursor (L37mt) (MRP-L37) | Q9BZE1|RM37_HUMAN | 48,085 | 3 |
| Poly(ADP-ribose) glycohydrolase ARH3 (EC 3.2.1.143) ([Protein ADP-ribosylarginine] hydrolase-like protein 2) (ADP-ribosylhydrolase 3) | Q9NX46|ARHL2_HUMAN | 38,929 | 3 |
| Isochorismatase domain-containing protein 2, mitochondrial precursor | Q96AB3|ISOC2_HUMAN | 22,319 | 3 |
| Probable mitochondrial import receptor subunit TOM40 homolog (Translocase of outer membrane 40 kDa subunit homolog) (Haymaker protein) (p38.5) | O96008|TOM40_HUMAN | 37,875 | 3 |
| Mitochondrial import inner membrane translocase subunit Tim8 A (Deafness dystonia protein 1) (X-linked deafness dystonia protein) | O60220|TIM8A_HUMAN | 10,980 | 3 |
| Glutaredoxin-related protein 5 | Q86SX6|GLRX5_HUMAN | 16,610 | 3 |
| Secretory carrier-associated membrane protein 3 (Secretory carrier membrane protein 3) | O14828|SCAM3_HUMAN | 38,302 | 3 |
| 60 kDa SS-A/Ro ribonucleoprotein (60 kDa Ro protein) (60 kDa ribonucleoprotein Ro) (RoRNP) (Ro 60 kDa autoantigen) (TROVE domain family member 2) (Sjoegren syndrome type A antigen) (SS-A) (Sjoegren syndrome antigen A2) | P10155|RO60_HUMAN | 60,654 | 3 |
| COP9 signalosome complex subunit 5 (EC 3.4.-.-) (Signalosome subunit 5) (SGN5) (Jun activation domain-binding protein 1) | Q92905|CSN5_HUMAN | 37,562 | 3 |
| 28S ribosomal protein S7, mitochondrial precursor (S7mt) (MRP-S7) (bMRP27a) (bMRP-27a) | Q9Y2R9|RT07_HUMAN | 28,145 | 3 |
| NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (FIP-3) | Q9Y6K9|NEMO_HUMAN | 48,179 | 3 |
| Kinesin-like protein KIF1B (Klp) | O60333|KIF1B_HUMAN | 204,463 | 3 |
| APAF1-interacting protein | Q96GX9|APIP_HUMAN | 27,107 | 3 |
| Oxysterol-binding protein 1 | P22059|OSBP1_HUMAN | 89,404 | 3 |
| Syntaxin-4 (Renal carcinoma antigen NY-REN-31) | Q12846|STX4_HUMAN | 34,162 | 3 |
| Beta-hexosaminidase beta chain precursor (EC 3.2.1.52) (N-acetyl-beta-glucosaminidase) (Beta-N-acetylhexosaminidase) (Hexosaminidase B) (Cervical cancer proto-oncogene 7) (HCC-7) [Contains: Beta-hexosaminidase beta-B chain; Beta-hexosaminidase beta-A chain] | P07686|HEXB_HUMAN | 63,095 | 3 |
| Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein) | Q99700|ATX2_HUMAN | 140,120 | 3 |
| Kinesin light chain 2 (KLC 2) | Q9H0B6|KLC2_HUMAN | 68,918 | 3 |
| Translocation protein SEC62 (Translocation protein 1) (TP-1) (hTP-1) | Q99442|SEC62_HUMAN | 45,845 | 3 |
| ADP-ribose pyrophosphatase, mitochondrial precursor (EC 3.6.1.13) (ADP-ribose diphosphatase) (Adenosine diphosphoribose pyrophosphatase) (ADPR-PPase) (ADP-ribose phosphohydrolase) (Nucleoside diphosphate-linked moiety X motif 9) (Nudix motif 9) | Q9BW91|NUDT9_HUMAN | 39,108 | 3 |
| Charged multivesicular body protein 6 (Chromatin-modifying protein 6) (Vacuolar protein sorting-associated protein 20) (hVps20) | Q96FZ7|CHMP6_HUMAN | 23,467 | 3 |
| Bifunctional polynucleotide phosphatase/kinase (Polynucleotide kinase-3′-phosphatase) (DNA 5′-kinase/3′-phosphatase) [Includes: Polynucleotide 3′-phosphatase (EC 3.1.3.32) (2′(3′)-polynucleotidase); Polynucleotide 5′-hydroxyl-kinase (EC 2.7.1.78)] | Q96T60|PNKP_HUMAN | 57,059 | 3 |
| Ras-related protein Rap-2b precursor | P61225|RAP2B_HUMAN | 20,486 | 3 |
| UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit) | O15294|OGT1_HUMAN | 116,910 | 3 |
| NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase ASHI subunit) (Complex I-ASHI) (CI-ASHI) | O95169|NDUB8_HUMAN | 21,748 | 3 |
| Valacyclovir hydrolase precursor (EC 3.1.-.-) (VACVase) (Biphenyl hydrolase-like protein) (Biphenyl hydrolase-related protein) (Bph-rp) (Breast epithelial mucin-associated antigen) (MCNAA) | Q86WA6|BPHL_HUMAN | 32,525 | 3 |
| Thioredoxin domain-containing protein 9 (Protein 1-4) (ATP-binding protein associated with cell differentiation) | O14530|TXND9_HUMAN | 26,517 | 3 |
| Ubiquitin-conjugating enzyme E2 Q1 (EC 6.3.2.19) (Ubiquitin-protein ligase Q1) (Ubiquitin carrier protein Q1) (Protein NICE-5) | Q7Z7E8|UB2Q1_HUMAN | 46,110 | 3 |
| AFG3-like protein 2 (EC 3.4.24.-) (Paraplegin-like protein) | Q9Y4W6|AFG32_HUMAN | 88,569 | 3 |
| Huntingtin-interacting protein 1-related protein (Hip1-related) (Hip 12) | O75146|HIP1R_HUMAN | 119,372 | 3 |
| 45 kDa calcium-binding protein precursor (Cab45) (Stromal cell-derived factor 4) (SDF-4) | Q9BRK5|CAB45_HUMAN | 41,789 | 3 |
| Uncharacterized protein C20orf116 precursor | Q96HY6|CT116_HUMAN | 35,593 | 3 |
| ADP-ribosylation factor-like protein 3 | P36405|ARL3_HUMAN | 20,438 | 3 |
| Protein phosphatase 2C isoform alpha (EC 3.1.3.16) (PP2C-alpha) (IA) (Protein phosphatase 1A) | P35813|PP2CA_HUMAN | 42,429 | 3 |
| Mitochondrial 39S ribosomal protein L39 (L39mt) (MRP-L39) (MRP-L5) | Q9NYK5|RM39_HUMAN | 38,704 | 3 |
| Tyrosine-protein kinase Lyn (EC 2.7.10.2) | P07948|LYN_HUMAN | 58,558 | 3 |
| PERQ amino acid-rich with GYF domain-containing protein 2 (Grb10-interacting GYF protein 2) (Trinucleotide repeat-containing protein 15) | Q6Y7W6|PERQ2_HUMAN | 150,051 | 3 |
| Exosome complex exonuclease RRP43 (EC 3.1.13.-) (Ribosomal RNA-processing protein 43) (Exosome component 8) (p9) (Opa-interacting protein 2) | Q96B26|EXOS8_HUMAN | 30,022 | 3 |
| 28S ribosomal protein S35, mitochondrial precursor (S35mt) (MRP-S35) (Mitochondrial ribosomal protein S28) (MRP-S28) | P82673|RT35_HUMAN | 36,827 | 3 |
| Zinc finger protein 313 (RING finger protein 114) | Q9Y508|ZN313_HUMAN | 25,676 | 3 |
| Diablo homolog, mitochondrial precursor (Second mitochondria-derived activator of caspase) (Smac protein) (Direct IAP-binding protein with low pI) | Q9NR28|DBLOH_HUMAN | 27,113 | 3 |
| Enhancer of rudimentary homolog | P84090|ERH_HUMAN | 12,241 | 3 |
| 39S ribosomal protein L28, mitochondrial precursor (L28mt) (MRP-L28) (Melanoma antigen p15) (Melanoma-associated antigen recognized by T lymphocytes) | Q13084|RM28_HUMAN | 30,140 | 3 |
| Nucleotide-binding protein 1 (NBP 1) | P53384|NUBP1_HUMAN | 34,571 | 3 |
| RRP12-like protein | Q5JTH9|RRP12_HUMAN | 143,687 | 3 |
| Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2) | Q92785|REQU_HUMAN | 44,138 | 3 |
| Cold-inducible RNA-binding protein (Glycine-rich RNA-binding protein CIRP) (A18 hnRNP) | Q14011|CIRBP_HUMAN | 18,630 | 3 |
| Receptor-type tyrosine-protein phosphatase F precursor (EC 3.1.3.48) (LAR protein) (Leukocyte antigen related) | P10586|PTPRF_HUMAN | 211,826 | 3 |
| Breast carcinoma amplified sequence 2 (DNA amplified in mammary carcinoma 1 protein) (Spliceosome-associated protein SPF 27) | O75934|BCAS2_HUMAN | 26,114 | 3 |
| 28S ribosomal protein S18b, mitochondrial precursor (MRP-S18-b) (Mrps18b) (MRP-S18-2) | Q9Y676|RT18B_HUMAN | 29,378 | 3 |
| 28S ribosomal protein S31, mitochondrial precursor (S31mt) (MRP-S31) (Imogen 38) | Q92665|RT31_HUMAN | 45,302 | 3 |
| Ribosome biogenesis protein BMS1 homolog | Q14692|BMS1_HUMAN | 145,794 | 3 |
| FKBP12-rapamycin complex-associated protein (FK506-binding protein 12-rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR) | P42345|FRAP_HUMAN | 288,878 | 3 |
| Peroxisomal proliferator-activated receptor A-interacting complex 285 kDa protein (EC 3.6.1.-) (ATP-dependent helicase PRIC285) (PPAR-alpha-interacting complex protein 285) (PPAR-gamma DBD-interacting protein 1) (PDIP1) | Q9BYK8|PR285_HUMAN | 294,512 | 3 |
| DNA-directed RNA polymerase I 40 kDa polypeptide (EC 2.7.7.6) (RPA40) (RPA39) | O15160|RPA5_HUMAN | 39,232 | 3 |
| Vacuolar protein sorting-associating protein 4A (Protein SKD2) (hVPS4) (VPS4-1) | Q9UN37|VPS4A_HUMAN | 48,881 | 3 |
| Gephyrin | Q9NQX3|GEPH_HUMAN | 79,732 | 3 |
| Core-binding factor subunit beta (CBF-beta) (Polyomavirus enhancer-binding protein 2 beta subunit) (PEBP2-beta) (PEA2-beta) (SL3-3 enhancer factor 1 beta subunit) (SL3/AKV core-binding factor beta subunit) | Q13951|PEBB_HUMAN | 21,490 | 3 |
| Zinc finger CCCH domain-containing protein 7B (Rotavirus ‘X’-associated non-structural protein) (RoXaN) | Q9UGR2|Z3H7B_HUMAN | 111,563 | 3 |
| Nuclear cap-binding protein subunit 2 (20 kDa nuclear cap-binding protein) (NCBP 20 kDa subunit) (CBP20) (NCBP-interacting protein 1) (NIP1) (Cell proliferation-inducing gene 55 protein) | P52298|NCBP2_HUMAN | 17,984 | 3 |
| Dynamin-binding protein (Scaffold protein Tuba) | Q6XZF7|DNMBP_HUMAN | 177,332 | 3 |
| Putative C10 protein | Q99622|C10_HUMAN | 13,160 | 3 |
| Double-strand-break repair protein rad21 homolog (hHR21) (Nuclear matrix protein 1) (NXP-1) (SCC1 homolog) | O60216|RAD21_HUMAN | 71,674 | 3 |
| Intron-binding protein aquarius (Intron-binding protein of 160 kDa) (IBP160) | O60306|AQR_HUMAN | 171,282 | 3 |
| Dehydrogenase/reductase SDR family member 1 (EC 1.1.-.-) | Q96LJ7|DHRS1_HUMAN | 33,890 | 3 |
| Raftlin (Raft-linking protein) (Cell migration-inducing gene 2 protein) | Q14699|RFTN1_HUMAN | 63,127 | 3 |
| 15 kDa selenoprotein precursor | O60613|SEP15_HUMAN | 17,726 | 3 |
| 39S ribosomal protein L45, mitochondrial precursor (L45mt) (MRP-L45) | Q9BRJ2|RM45_HUMAN | 35,333 | 3 |
| WD repeat protein 82 | Q6UXN9|WDR82_HUMAN | 35,062 | 3 |
| Placenta growth factor precursor (PlGF) | P49763|PLGF_HUMAN | 24,770 | 3 |
| Nucleoplasmin-3 | O75607|NPM3_HUMAN | 19,325 | 3 |
| Pescadillo homolog 1 | O00541|PESC_HUMAN | 67,987 | 3 |
| Probable peptidyl-tRNA hydrolase (EC 3.1.1.29) (PTH) | Q86Y79|PTH_HUMAN | 22,919 | 3 |
| Poly [ADP-ribose] polymerase 10 (EC 2.4.2.30) (PARP-10) | Q53GL7|PAR10_HUMAN | 109,979 | 3 |
| Nuclear RNA export factor 1 (Tip-associating protein) (Tip-associated protein) (mRNA export factor TAP) | Q9UBU9|NXF1_HUMAN | 70,167 | 3 |
| Legumain precursor (EC 3.4.22.34) (Asparaginyl endopeptidase) (Protease, cysteine 1) | Q99538|LGMN_HUMAN | 49,393 | 3 |
| Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (Box-dependent myc-interacting protein 1) | O00499|BIN1_HUMAN | 64,681 | 3 |
| Centrin-2 (Caltractin isoform 1) | P41208|CETN2_HUMAN | 19,722 | 3 |
| Methionine synthase (EC 2.1.1.13) (5-methyltetrahydrofolate--homocysteine methyltransferase) (Methionine synthase, vitamin-B12 dependent) (MS) | Q99707|METH_HUMAN | 140,514 | 3 |
| Geranylgeranyl transferase type-2 alpha subunit (EC 2.5.1.60) (Geranylgeranyl transferase type II alpha subunit) (Rab geranylgeranyltransferase alpha subunit) (Rab geranyl-geranyltransferase alpha subunit) (Rab GG transferase alpha) (Rab GGTase alpha) | Q92696|PGTA_HUMAN | 65,055 | 3 |
| SET domain-containing protein 3 | Q86TU7|SETD3_HUMAN | 67,241 | 3 |
| Dolichol-phosphate mannosyltransferase (EC 2.4.1.83) (Dolichol-phosphate mannose synthase) (Dolichyl-phosphate beta-D-mannosyltransferase) (Mannose-P-dolichol synthase) (MPD synthase) (DPM synthase) | O60762|DPM1_HUMAN | 29,618 | 3 |
| Signal transducing adapter molecule 1 (STAM-1) | Q92783|STAM1_HUMAN | 59,162 | 3 |
| Nuclear autoantigen Sp-100 (Speckled 100 kDa) (Nuclear dot-associated Sp100 protein) (Lysp100b) | P23497|SP100_HUMAN | 100,401 | 3 |
| Phosphatidylinositol-binding clathrin assembly protein (Clathrin assembly lymphoid myeloid leukemia protein) | Q13492|PICAL_HUMAN | 70,738 | 3 |
| Cartilage-associated protein precursor | O75718|CRTAP_HUMAN | 46,546 | 3 |
| Condensin complex subunit 2 (Non-SMC condensin I complex subunit H) (Barren homolog protein 1) (Chromosome-associated protein H) (hCAP-H) (XCAP-H homolog) | Q15003|CND2_HUMAN | 82,547 | 3 |
| BTB/POZ domain-containing protein 14B (Nucleus accumbens-1) (NAC-1) | Q96RE7|BTB14_HUMAN | 57,239 | 3 |
| Lysosome-associated membrane glycoprotein 1 precursor (LAMP-1) (CD107a antigen) | P11279|LAMP1_HUMAN | 44,756 | 3 |
| Cell death regulator Aven | Q9NQS1|AVEN_HUMAN | 38,488 | 3 |
| Uncharacterized protein C6orf55 (Dopamine-responsive protein DRG-1) | Q9NP79|CF055_HUMAN | 33,862 | 3 |
| WD repeat protein 6 | Q9NNW5|WDR6_HUMAN | 121,706 | 3 |
| WD repeat and FYVE domain-containing protein 1 (WD40- and FYVE domain-containing protein 1) (Phosphoinositide-binding protein 1) (FENS-1) (Zinc finger FYVE domain-containing protein 17) | Q8IWB7|WDFY1_HUMAN | 46,306 | 3 |
| Vacuolar ATP synthase subunit H (EC 3.6.3.14) (V-ATPase H subunit) (Vacuolar proton pump subunit H) (V-ATPase 50/57 kDa subunits) (Vacuolar proton pump subunit SFD) (VMA13) (Nef-binding protein 1) (NBP1) | Q9UI12|VATH_HUMAN | 55,865 | 3 |
| Polymerase delta-interacting protein 3 (46 kDa DNA polymerase delta interaction protein) (p46) | Q9BY77|PDIP3_HUMAN | 46,072 | 3 |
| Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) | Q8WWN8|CEND3_HUMAN | 169,830 | 3 |
| ANK repeat and LEM domain-containing KIAA0692 | Q86XL3|K0692_HUMAN | 104,106 | 3 |
| Sulfite oxidase, mitochondrial precursor (EC 1.8.3.1) | P51687|SUOX_HUMAN | 53,865 | 3 |
| Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin) | P49790|NU153_HUMAN | 153,872 | 3 |
| Pleiotropic regulator 1 | O43660|PLRG1_HUMAN | 57,175 | 3 |
| Coiled-coil domain-containing protein 44 | Q9BSH4|CCD44_HUMAN | 32,459 | 3 |
| Brix domain-containing protein 2 (Ribosome biogenesis protein Brix) | Q8TDN6|BXDC2_HUMAN | 41,385 | 3 |
| SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 1 (SWI/SNF complex 60 kDa subunit) (60 kDa BRG-1/Brm-associated factor subunit A) (BRG1-associated factor 60A) | Q96GM5|SMRD1_HUMAN | 54,928 | 3 |
| Coronin-1A (Coronin-like protein p57) (Coronin-like protein A) (Clipin-A) (Tryptophan aspartate-containing coat protein) (TACO) | P31146|COR1A_HUMAN | 51,008 | 3 |
| Cleavage and polyadenylation specificity factor subunit 1 (Cleavage and polyadenylation specificity factor 160 kDa subunit) (CPSF 160 kDa subunit) | Q10570|CPSF1_HUMAN | 160,868 | 3 |
| Kinesin-like protein KIF2A (Kinesin-2) (HK2) | O00139|KIF2A_HUMAN | 76,938 | 3 |
| Uncharacterized protein C10orf119 | Q9BTE3|CJ119_HUMAN | 72,963 | 3 |
| Tripartite motif-containing protein 22 (RING finger protein 94) (50 kDa-stimulated trans-acting factor) (Staf-50) | Q8IYM9|TRI22_HUMAN | 56,929 | 3 |
| Ribosome recycling factor, mitochondrial precursor | Q96E11|RRFM_HUMAN | 29,260 | 3 |
| Glutathione S-transferase theta-2 (EC 2.5.1.18) (GST class-theta-2) | P30712|GSTT2_HUMAN | 27,490 | 3 |
| 39S ribosomal protein L24, mitochondrial precursor (L24mt) (MRP-L24) | Q96A35|RM24_HUMAN | 24,897 | 3 |
| Signal transducer and activator of transcription 5B | P42229|STA5A_HUMAN, P51692|STA5B_HUMAN | 89,849 | 3 |
| Tensin-1 | Q9HBL0|TENS1_HUMAN | 185,660 | 3 |
| REVERSED | REV|Q9HCF4|ALO17_HUM AN | 174,882 | 3 |
| Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit delta isoform (PP2A, B subunit, B′ delta isoform) (PP2A, B subunit, B56 delta isoform) (PP2A, B subunit, PR61 delta isoform) (PP2A, B subunit, R5 delta isoform) | Q14738|2A5D_HUMAN | 69,976 | 3 |
| Transcriptional repressor CTCF (CCCTC-binding factor) (CTCFL paralog) (11-zinc finger protein) | P49711|CTCF_HUMAN | 82,766 | 3 |
| Mitogen-activated protein kinase kinase kinase kinase 5 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 5) (MEK kinase kinase 5) (MEKKK 5) (Kinase homologous to SPS1/STE20) (KHS) | Q9Y4K4|M4K5_HUMAN | 95,025 | 3 |
| Acylglycerol kinase, mitochondrial precursor (EC 2.7.1.94) (EC 2.7.1.107) (hAGK) (Multiple substrate lipid kinase) (Multi-substrate lipid kinase) (MuLK) (HsMuLK) | Q53H12|AGK_HUMAN | 47,120 | 3 |
| Arylsulfatase A precursor (EC 3.1.6.8) (ASA) (Cerebroside-sulfatase) [Contains: Arylsulfatase A component B; Arylsulfatase A component C] | P15289|ARSA_HUMAN | 53,571 | 3 |
| Zinc finger CCCH domain-containing protein 11A | O75152|ZC11A_HUMAN | 89,113 | 3 |
| c-Myc-responsive protein Rcl | O43598|RCL_HUMAN | 19,090 | 3 |
| Tyrosyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.1) (Tyrosine--tRNA ligase) (TyrRS) | Q9 Y2Z4|SYYM_HU M AN | 53,183 | 3 |
| E3 ubiquitin-protein ligase LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL) | Q6UWE0|LRSM1_HUMAN | 83,578 | 3 |
| CAP-Gly domain-containing linker protein 2 (Cytoplasmic linker protein 2) (Cytoplasmic linker protein 115) (CLIP-115) (Williams-Beuren syndrome chromosome region 4 protein) | Q9UDT6|CLIP2_HUMAN | 115,821 | 3 |
| Gamma-enolase (EC 4.2.1.11) (2-phospho-D-glycerate hydro-lyase) (Neural enolase) (Neuron-specific enolase) (NSE) (Enolase 2) | P09104|ENOG_HUMAN | 47,252 | 3 |
| 60S ribosomal protein L10-like | Q96L21|RL10L_HUMAN | 24,501 | 3 |
| Thioredoxin reductase 2, mitochondrial precursor (EC 1.8.1.9) (TR3) (TR-beta) (Selenoprotein Z) (SelZ) | Q9NNW7|TRXR2_HUMAN | 56,441 | 3 |
| Mitochondrial 28S ribosomal protein S27 (S27mt) (MRP-S27) | Q92552|RT27_HUMAN | 47,653 | 3 |
| SAPS domain family member 1 | Q9UPN7|SAPS1_HUMAN | 96,706 | 3 |
| PHD finger-like domain-containing protein 5A (PHD finger-like domain protein 5A) (Splicing factor 3B-associated 14 kDa protein) (SF3b14b) | Q7RTV0|PHF5A_HUMAN | 12,387 | 3 |
| Nuclear cap-binding protein subunit 1 (80 kDa nuclear cap-binding protein) (NCBP 80 kDa subunit) (CBP80) | Q09161|NCBP1_HUMAN | 91,823 | 3 |
| Transcriptional regulator ISGF3 subunit gamma (Interferon regulatory factor 9) (IRF-9) (IFN-alpha-responsive transcription factor subunit) (Interferon-stimulated gene factor 3 gamma) (ISGF3 p48 subunit) (ISGF-3 gamma) | Q00978|IRTF_HUMAN | 43,678 | 3 |
| Sodium/hydrogen exchanger 1 (Na(+)/H(+) exchanger 1) (NHE-1) (Solute carrier family 9 member 1) (Na(+)/H(+) antiporter, amiloride-sensitive) (APNH) | P19634|SL9A1_HUMAN | 90,748 | 3 |
| Epoxide hydrolase 1 (EC 3.3.2.9) (Microsomal epoxide hydrolase) (Epoxide hydratase) | P07099|HYEP_HUMAN | 52,933 | 3 |
| Serine/threonine-protein kinase receptor R3 precursor (EC 2.7.11.30) (SKR3) (Activin receptor-like kinase 1) (ALK-1) (TGF-B superfamily receptor type I) (TSR-I) | P37023|ACVL1_HUMAN | 56,106 | 3 |
| Small ubiquitin-related modifier 1 precursor (SUMO-1) (Sentrin) (Ubiquitin-like protein SMT3C) (SMT3 homolog 3) (Ubiquitin-homology domain protein PIC1) (Ubiquitin-like protein UBL1) (GAP-modifying protein 1) (GMP1) | P63165|SUMO1_HUMAN | 11,539 | 3 |
| Phosphomevalonate kinase (EC 2.7.4.2) (PMKase) | Q15126|PMVK_HUMAN | 21,977 | 3 |
| Actin-related protein 2/3 complex subunit 1A (SOP2-like protein) | Q92747|ARC1A_HUMAN | 41,551 | 3 |
| Exosome complex exonuclease RRP45 (EC 3.1.13.-) (Exosome component 9) (Polymyositis/scleroderma autoantigen 1) (Autoantigen PM/Scl 1) (Polymyositis/scleroderma autoantigen 75 kDa) (PM/Scl-75) (P75 polymyositis-scleroderma overlap syndrome-associated autoantigen) | Q06265|EXOS9_HUMAN | 46,961 | 3 |
| N-myc-interactor (Nmi) (N-myc and STAT interactor) | Q13287|NMI_HUMAN | 35,039 | 3 |
| Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17) | Q15811|ITSN1_HUMAN | 195,407 | 3 |
| Lysosome-associated membrane glycoprotein 2 precursor (LAMP-2) (CD107b antigen) | P13473|LAMP2_HUMAN | 44,943 | 3 |
| Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22) (Zinc finger CCCH domain-containing protein 16) | Q9NW64|RBM22_HUMAN | 46,879 | 3 |
| CCR4-NOT transcription complex subunit 2 (CCR4-associated factor 2) | Q9NZN8|CNOT2_HUMAN | 59,721 | 3 |
| Coiled-coil-helix-coiled-coil-helix domain-containing protein 6 | Q9BRQ6|CHCH6_HUMAN | 26,439 | 3 |
| Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3-methylcrotonyl-CoA carboxylase biotin-containing subunit) | Q96RQ3|MCCA_HUMAN | 80,456 | 3 |
| THO complex subunit 2 (Tho2) | Q8NI27|THOC2_HUMAN | 169,569 | 3 |
| Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1) | Q14162|SREC_HUMAN | 87,408 | 3 |
| Proteolipid protein 2 (Intestinal membrane A4 protein) (Differentiation-dependent protein A4) | Q04941|PLP2_HUMAN | 16,673 | 3 |
| Glucosamine-6-phosphate isomerase (EC 3.5.99.6) (Glucosamine-6-phosphate deaminase) (GNPDA) (GlcN6P deaminase) (Oscillin) | P46926|GNPI_HUMAN | 32,651 | 3 |
| Heat shock factor-binding protein 1 (Nasopharyngeal carcinoma-associated antigen 13) (NPC-A-13) | O75506|HSBP1_HUMAN | 8,526 | 3 |
| U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10) | O00566|MPP10_HUMAN | 78,849 | 3 |
| Implantation-associated protein precursor (IAP) (Magnesium transporter protein 1) (MagT1) | Q9H0U3|IAG2_HUMAN | 38,019 | 3 |
| Acyl-coenzyme A oxidase 3, peroxisomal (EC 1.3.3.6) (Pristanoyl-CoA oxidase) (Branched-chain acyl-CoA oxidase) (BRCACox) | O15254|ACOX3_HUMAN | 77,613 | 3 |
| Poliovirus receptor precursor (Nectin-like protein 5) (Necl-5) (CD155 antigen) | P15151|PVR_HUMAN | 45,284 | 3 |
| ARL-6-interacting protein 1 (ADP-ribosylation-like factor 6-interacting protein 1) (Aip-1) | Q15041|AR6P1_HUMAN | 23,346 | 3 |
| Alpha-2-macroglobulin receptor-associated protein precursor (Alpha-2-MRAP) (Low density lipoprotein receptor-related protein-associated protein 1) (RAP) | P30533|AMRP_HUMAN | 41,450 | 3 |
| Antigen KI-67 | P46013|KI67_HUMAN | 358,678 | 3 |
| Ubiquitin-conjugating enzyme E2 D3 (EC 6.3.2.19) (Ubiquitin-protein ligase D3) (Ubiquitin carrier protein D3) (Ubiquitin-conjugating enzyme E2-17 kDa 3) (E2(17)KB 3) | P61077|UB2D3_HUMAN, P62837|UB2D2_HUMAN, Q9NTT1|U2D3L_HUMAN | 16,670 | 3 |
| Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3A (EC 2.4.1.119) (Oligosaccharyl transferase subunit STT3A) (STT3-A) (B5) (Integral membrane protein 1) (TMC) | P46977|STT3A_HUMAN | 80,457 | 3 |
| WD repeat protein 44 (Rabphilin-11) | Q5JSH3|WDR44_HUMAN | 101,351 | 3 |
| Uncharacterized calcium-binding protein KIAA0494 | O75071|K0494_HUMAN | 55,015 | 3 |
| 39S ribosomal protein L16, mitochondrial precursor (L16mt) (MRP-L16) | Q9NX20|RM16_HUMAN | 28,432 | 3 |
| Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3) | Q5T447|HECD3_HUMAN | 97,096 | 3 |
| Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2] | P78559|MAP1A_HUMAN | 306,456 | 3 |
| Nicotinamide N-methyltransferase (EC 2.1.1.1) | P40261|NNMT_HUMAN | 29,557 | 3 |
| Transcription initiation factor IIE subunit alpha (TFIIE-alpha) (General transcription factor IIE subunit 1) (General transcription factor IIE 56 kDa subunit) | P29083|T2EA_HUMAN | 49,435 | 3 |
| Triple functional domain protein (EC 2.7.11.1) (PTPRF-interacting protein) | O75962|TRIO_HUMAN | 341,600 | 3 |
| AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma-1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain) | O43747|AP1G1_HUMAN | 91,376 | 3 |
| HLA class I histocompatibility antigen, Cw-7 alpha chain precursor (MHC class I antigen Cw*7) | P10321|1C07_HUMAN | 40,630 | 3 |
| Syntaxin-binding protein 1 (Unc-18 homolog) (Unc-18A) (Unc-18-1) (N-Sec1) (p67) | P61764|STXB1_HUMAN | 67,554 | 3 |
| Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin) | O15020|SPTN2_HUMAN | 271,278 | 3 |
| Vacuolar protein sorting-associated protein 36 (ELL-associated protein of 45 kDa) | Q86VN1|VPS36_HUMAN | 43,800 | 3 |
| Histone H2A.x (H2a/x) | P16104|H2AX_HUMAN, Q96QV6|H2A1A_HUMAN | 15,127 | 3 |
| Pre-mRNA-splicing factor ISY1 homolog | Q9ULR0|ISY1_HUMAN | 37,549 | 3 |
| Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L) | Q9UBV2|SEL1L_HUMAN | 88,739 | 3 |
| Toll-interacting protein | Q9H0E2|TOLIP_HUMAN | 30,263 | 3 |
| Sorting nexin-18 (Sorting nexin-associated Golgi protein 1) (SH3 and PX domain-containing protein 3B) | Q96RF0|SNX18_HUMAN | 68,863 | 3 |
| Secernin-2 | Q96FV2|SCRN2_HUMAN | 46,546 | 3 |
| Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit) | Q15427|SF3B4_HUMAN | 44,369 | 3 |
| Serine/threonine-protein phosphatase 2B catalytic subunit beta isoform (EC 3.1.3.16) (Calmodulin-dependent calcineurin A subunit beta isoform) (CAM-PRP catalytic subunit) | P16298|PP2BB_HUMAN | 59,007 | 3 |
| HD domain-containing protein 3 | Q8N4P3|HDDC3_HUMAN | 20,311 | 3 |
| Protein AATF (Apoptosis-antagonizing transcription factor) (Rb-binding protein Che-1) | Q9NY61|AATF_HUMAN | 63,117 | 3 |
| Metaxin-2 | O75431|MTX2_HUMAN | 29,745 | 3 |
| Choline transporter-like protein 2 (Solute carrier family 44 member 2) | Q8IWA5|CTL2_HUMAN | 80,138 | 3 |
| UPF0364 protein C6orf211 | Q9H993|CF211_HUMAN | 51,156 | 3 |
| Splicing factor, arginine/serine-rich 6 (Pre-mRNA-splicing factor SRP55) | Q13247|SFRS6_HUMAN | 39,570 | 3 |
| Cathepsin S precursor (EC 3.4.22.27) | P25774|CATS_HUMAN | 37,478 | 3 |
| Inhibitor of Bruton tyrosine kinase (IBtk) | Q9P2D0|IBTK_HUMAN | 150,512 | 3 |
| REVERSED | REV|O94909|K0819_HUMA N | 126,510 | 3 |
| Neuroplastin precursor (Stromal cell-derived receptor 1) (SDR-1) | Q9Y639|NPTN_HUMAN | 31,274 | 3 |
| Mitochondrial fission 1 protein (Fis1 homolog) (hFis1) (Tetratricopeptide repeat protein 11) (TPR repeat protein 11) | Q9Y3D6|FIS1_HUMAN | 16,920 | 3 |
| 5′-nucleotidase domain-containing protein 2 | Q9H857|NT5D2_HUMAN | 60,703 | 3 |
| Phosphoglucomutase-2-like 1 (EC 5.4.2.2) (PMMLP) | Q6PCE3|PGM2L_HUMAN | 70,439 | 3 |
| Bleomycin hydrolase (EC 3.4.22.40) (BLM hydrolase) (BMH) (BH) | Q13867|BLMH_HUMAN | 52,545 | 3 |
| Synaptojanin-2-binding protein (Mitochondrial outer membrane protein 25) | P57105|SYJ2B_HUMAN | 15,910 | 3 |
| Cytoplasmic tyrosine-protein kinase BMX (EC 2.7.10.2) (Bone marrow tyrosine kinase gene in chromosome X protein) (Epithelial and endothelial tyrosine kinase) (ETK) (NTK38) | P51813|BMX_HUMAN | 77,994 | 3 |
| Huntingtin (Huntington disease protein) (HD protein) | P42858|HD_HUMAN | 347,841 | 3 |
| Phosphoribosyl pyrophosphate synthetase-associated protein 2 (PRPP synthetase-associated protein 2) (41 kDa phosphoribosypyrophosphate synthetase-associated protein) (PAP41) | O60256|KPRB_HUMAN | 40,909 | 3 |
| Eukaryotic translation elongation factor 1 epsilon-1 (Multisynthetase complex auxiliary component p18) (Elongation factor p18) | O43324|MCA3_HUMAN | 19,793 | 3 |
| Collagen alpha-2(VI) chain precursor | P12110|CO6A2_HUMAN | 108,563 | 3 |
| Leupaxin | O60711|LPXN_HUMAN | 43,314 | 3 |
| Casein kinase I isoform alpha (EC 2.7.11.1) (CKI-alpha) (CK1) | P48729|KC1A_HUMAN | 38,899 | 3 |
| REVERSED | REV|Q05469|LIPS_HUMAN | 116,552 | 3 |
| CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150) | O60244|CRSP2_HUMAN | 160,649 | 3 |
| ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97) | Q8TDD1|DDX54_HUMAN | 98,580 | 3 |
| REVERSED | REV|O14497|ARI1A_HUMA N | 242,026 | 3 |
| Sulfhydryl oxidase 2 precursor (EC 1.8.3.2) (Quiescin Q6-like protein 1) (Neuroblastoma-derived sulfhydryl oxidase) | Q6ZRP7|QSC6L_HUMAN | 77,526 | 3 |
| SAPS domain family member 3 (Sporulation-induced transcript 4-associated protein SAPL) (Protein phosphatase 6 regulatory subunit 3) | Q5H9R7|SAPS3_HUMAN | 97,653 | 3 |
| Transmembrane emp24 domain-containing protein 2 precursor (Membrane protein p24A) | Q15363|TMED2_HUMAN | 22,743 | 3 |
| Tapasin precursor (TPSN) (TPN) (TAP-binding protein) (TAP-associated protein) (NGS-17) | O15533|TPSN_HUMAN | 47,609 | 3 |
| Beta crystallin B2 (Beta-crystallin Bp) | P43320|CRBB2_HUMAN | 23,362 | 3 |
| REVERSED | REV|O14980|XPO1_HUMA N | 123,371 | 3 |
| Lamina-associated polypeptide 2 isoform alpha (Thymopoietin isoform alpha) (TP alpha) (Thymopoietin-related peptide isoform alpha) (TPRP isoform alpha) [Contains: Thymopoietin (TP) (Splenin); Thymopentin (TP5)] | P42166|LAP2A_HUMAN | 75,476 | 2 |
| Ras-related protein Rap-1A precursor (GTP-binding protein smg-p21A) (Ras-related protein Krev-1) (C21KG) (G-22K) | P62834|RAP1A_HUMAN | 20,969 | 2 |
| Prefoldin subunit 6 (Protein Ke2) | O15212|PFD6_HUMAN | 14,565 | 2 |
| Branched-chain-amino-acid aminotransferase, cytosolic (EC 2.6.1.42) (BCAT(c)) (ECA39 protein) | P54687|BCAT1_HUMAN | 42,935 | 2 |
| CD2 antigen cytoplasmic tail-binding protein 2 (CD2 cytoplasmic domain-binding protein) (CD2 tail-binding protein) | O95400|CD2B2_HUMAN | 37,629 | 2 |
| Homer protein homolog 3 (Homer-3) | Q9NSC5|HOME3_HUMAN | 39,818 | 2 |
| Absent in melanoma 1 protein | Q9Y4K1|AIM1_HUMAN | 188,659 | 2 |
| PDZ and LIM domain protein 2 (PDZ-LIM protein mystique) (PDZ-LIM protein) | Q96JY6|PDLI2_HUMAN | 37,442 | 2 |
| Phosphoribosyl pyrophosphate synthetase-associated protein 1 (PRPP synthetase-associated protein 1) (39 kDa phosphoribosypyrophosphate synthetase-associated protein) (PAP39) | Q14558|KPRA_HUMAN | 39,377 | 2 |
| Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) | Q9UBC2|EP15R_HUMAN | 94,240 | 2 |
| FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1) | Q9Y613|FHOD1_HUMAN | 126,537 | 2 |
| Chloride intracellular channel protein 2 (XAP121) | O15247|CLIC2_HUMAN | 28,340 | 2 |
| Adenylyl cyclase-associated protein 2 (CAP 2) | P40123|CAP2_HUMAN | 52,806 | 2 |
| Ribulose-phosphate 3-epimerase (EC 5.1.3.1) (Ribulose-5-phosphate-3-epimerase) | Q96AT9|RPE_HUMAN | 24,910 | 2 |
| MKI67 FHA domain-interacting nucleolar phosphoprotein (Nucleolar protein interacting with the FHA domain of pKI-67) (hNIFK) (Nucleolar phosphoprotein Nopp34) | Q9BYG3|MK67I_HUMAN | 34,205 | 2 |
| U4/U6.U5 tri-snRNP-associated protein 1 (U4/U6.U5 tri-snRNP-associated 110 kDa protein) (Squamous cell carcinoma antigen recognized by T cells 1) (SART-1) (hSART-1) (hSnu66) | O43290|SNUT1_HUMAN | 90,239 | 2 |
| Rap1 GTPase-GDP dissociation stimulator 1 (SMG P21 stimulatory GDP/GTP exchange protein) (SMG GDS protein) (Exchange factor smgGDS) | P52306|GDS1_HUMAN | 66,386 | 2 |
| TBC1 domain family member 13 | Q9NVG8|TBC13_HUMAN | 32,143 | 2 |
| Syntaxin-7 | O15400|STX7_HUMAN | 29,798 | 2 |
| Wolframin | O76024|WFS1_HUMAN | 100,290 | 2 |
| RUN and FYVE domain-containing protein 1 (FYVE-finger protein EIP1) (Zinc finger FYVE domain-containing protein 12) (La-binding protein 1) (Rab4-interacting protein) | Q96T51|RUFY1_HUMAN | 79,801 | 2 |
| Hydroxyacylglutathione hydrolase (EC 3.1.2.6) (Glyoxalase II) (GLX II) | Q16775|GLO2_HUMAN | 28,842 | 2 |
| Solute carrier family 12 member 2 (Bumetanide-sensitive sodium-(potassium)-chloride cotransporter 1) (Basolateral Na-K-Cl symporter) | P55011|S12A2_HUMAN | 131,434 | 2 |
| RNA-binding motif, single-stranded-interacting protein 2 (Suppressor of CDC2 with RNA-binding motif 3) | Q15434|RBMS2_HUMAN | 43,941 | 2 |
| Pituitary tumor-transforming gene 1 protein-interacting protein precursor (Pituitary tumor-transforming gene protein-binding factor) (PTTG-binding factor) (PBF) | P53801|PTTG_HUMAN | 20,306 | 2 |
| Cell division protein kinase 2 (EC 2.7.11.22) (p33 protein kinase) | P24941|CDK2_HUMAN | 33,913 | 2 |
| Breakpoint cluster region protein (EC 2.7.11.1) (Renal carcinoma antigen NY-REN-26) | P11274|BCR_HUMAN | 142,792 | 2 |
| DNA damage-binding protein 2 (Damage-specific DNA-binding protein 2) (DDB p48 subunit) (DDBb) (UV-damaged DNA-binding protein 2) (UV-DDB 2) | Q92466|DDB2_HUMAN | 47,847 | 2 |
| Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha) | Q86UU1|PHLB1_HUMAN | 151,147 | 2 |
| Double-strand break repair protein MRE11A (MRE11 homolog 1) (MRE11 meiotic recombination 11 homolog A) | P49959|MRE11_HUMAN | 80,577 | 2 |
| NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7 (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase B18 subunit) (Complex I-B18) (CI-B18) (Cell adhesion protein SQM1) | P17568|NDUB7_HUMAN | 16,384 | 2 |
| Signal recognition particle 19 kDa protein (SRP19) | P09132|SRP19_HUMAN | 16,138 | 2 |
| Uncharacterized protein C12orf23 | Q8WUH6|CL023_HUMAN | 11,730 | 2 |
| ARF GTPase-activating protein GIT1 (G protein-coupled receptor kinase-interactor 1) (GRK-interacting protein 1) (Cool-associated and tyrosine-phosphorylated protein 1) (Cat-1) | Q9Y2X7|GIT1_HUMAN | 84,324 | 2 |
| Cullin-2 (CUL-2) | Q13617|CUL2_HUMAN | 86,967 | 2 |
| Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa) | Q15262|PTPRK_HUMAN | 162,071 | 2 |
| Mitochondrial 39S ribosomal protein L50 (L50mt) (MRP-L50) | Q8N5N7|RM50_HUMAN | 18,307 | 2 |
| Sorting nexin-9 (SH3 and PX domain-containing protein 1) (Protein SDP1) (SH3 and PX domain-containing protein 3A) | Q9Y5X1|SNX9_HUMAN | 66,575 | 2 |
| Protein FAM50A (Protein XAP-5) (Protein HXC-26) | Q14320|FA50A_HUMAN | 40,225 | 2 |
| La-related protein 7 (La ribonucleoprotein domain family member 7) | Q4G0J3|LARP7_HUMAN | 66,883 | 2 |
| Nucleoporin NUP53 (Nuclear pore complex protein Nup53) (Nucleoporin Nup35) (35 kDa nucleoporin) (Mitotic phosphoprotein 44) (MP-44) | Q8NFH5|NUP53_HUMAN | 34,757 | 2 |
| Zinc-finger protein ZPR1 (Zinc finger protein 259) | O75312|ZPR1_HUMAN | 50,908 | 2 |
| Uncharacterized protein KIAA0152 precursor | Q14165|K0152_HUMAN | 32,216 | 2 |
| Cleavage and polyadenylation specificity factor subunit 3 (Cleavage and polyadenylation specificity factor 73 kDa subunit) (CPSF 73 kDa subunit) | Q9UKF6|CPSF3_HUMAN | 77,470 | 2 |
| Cleavage and polyadenylation specificity factor subunit 2 (Cleavage and polyadenylation specificity factor 100 kDa subunit) (CPSF 100 kDa subunit) | Q9P2I0|CPSF2_HUMAN | 88,472 | 2 |
| Treacle protein (Treacher Collins syndrome protein) | Q13428|TCOF_HUMAN | 152,085 | 2 |
| Protein FAM125A (CIN85/CD2AP family-binding protein) | Q96EY5|F125A_HUMAN | 28,766 | 2 |
| CTTNBP2 N-terminal-like protein | Q9P2B4|CT2NL_HUMAN | 70,141 | 2 |
| AP-1 complex subunit sigma-1A (Adapter-related protein complex 1 sigma-1A subunit) (Sigma-adaptin 1A) (Adaptor protein complex AP-1 sigma-1A subunit) (Golgi adaptor HA1/AP1 adaptin sigma-1A subunit) (Clathrin assembly protein complex 1 sigma-1A small chain) (Clathrin coat assembly protein AP19) (HA1 19 kDa subunit) (Sigma 1a subunit of AP-1 clathrin) | P61966|AP1S1_HUMAN | 18,716 | 2 |
| 28S ribosomal protein S9, mitochondrial precursor (S9mt) (MRP-S9) | P82933|RT09_HUMAN | 45,806 | 2 |
| Probable ribosome biogenesis protein RLP24 (Ribosomal protein L24-like) | Q9UHA3|RLP24_HUMAN | 19,604 | 2 |
| Transcription factor 25 (Nuclear localized protein 1) | Q9BQ70|TCF25_HUMAN | 76,651 | 2 |
| Exosome complex exonuclease RRP42 (EC 3.1.13.-) (Ribosomal RNA-processing protein 42) (Exosome component 7) (p8) | Q15024|EXOS7_HUMAN | 31,817 | 2 |
| Down syndrome critical region protein 3 (Down syndrome critical region protein A) | O14972|DSCR3_HUMAN | 32,992 | 2 |
| Stromal interaction molecule 1 precursor | Q13586|STIM1_HUMAN | 77,475 | 2 |
| Angio-associated migratory cell protein | Q13685|AAMP_HUMAN | 46,732 | 2 |
| Ubiquitin-conjugating enzyme E2 G2 (EC 6.3.2.19) (Ubiquitin-protein ligase G2) (Ubiquitin carrier protein G2) | P60604|UB2G2_HUMAN | 18,549 | 2 |
| Guanine nucleotide-binding protein-like 3 (Nucleolar GTP-binding protein 3) (Nucleostemin) (E2-induced gene 3-protein) (Novel nucleolar protein 47) (NNP47) | Q9BVP2|GNL3_HUMAN | 61,981 | 2 |
| Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X-associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex) | Q96T23|RSF1_HUMAN | 162,992 | 2 |
| TBC1 domain family member 15 | Q8TC07|TBC15_HUMAN | 79,445 | 2 |
| Ras and Rab interactor 2 (Ras interaction/interference protein 2) (Ras inhibitor JC265) (Ras association domain family 4) | Q8WYP3|RIN2_HUMAN | 100,148 | 2 |
| Ubiquinol-cytochrome c reductase complex ubiquinone-binding protein QP-C (EC 1.10.2.2) (Ubiquinol-cytochrome c reductase complex 9.5 kDa protein) (Complex III subunit VII) | O14949|UCRQ_HUMAN | 9,889 | 2 |
| AT-rich interactive domain-containing protein 1A (ARID domain-containing protein 1A) (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A) | O14497|ARI1A_HUMAN | 242,026 | 2 |
| Neural Wiskott-Aldrich syndrome protein (N-WASP) | O00401|WASL_HUMAN | 54,810 | 2 |
| Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4) | O95104|SFR15_HUMAN | 125,850 | 2 |
| C-terminal-binding protein 2 (CtBP2) | P56545|CTBP2_HUMAN | 48,927 | 2 |
| Cytosolic purine 5′-nucleotidase (EC 3.1.3.5) (5′-nucleotidase cytosolic II) | P49902|5NTC_HUMAN | 64,955 | 2 |
| Uncharacterized protein KIAA0776 | O94874|K0776_HUMAN | 89,580 | 2 |
| Protein numb homolog (h-Numb) (Protein S171) | P49757|NUMB_HUMAN | 70,784 | 2 |
| Exosome complex exonuclease RRP44 (EC 3.1.13.-) (Ribosomal RNA-processing protein 44) (DIS3 protein homolog) | Q9Y2L1|RRP44_HUMAN | 108,988 | 2 |
| Mitochondrial 28S ribosomal protein S14 (S14mt) (MRP-S14) | O60783|RT14_HUMAN | 15,121 | 2 |
| Proteasome subunit beta type 10 precursor (EC 3.4.25.1) (Proteasome MECl-1) (Macropain subunit MECl-1) (Multicatalytic endopeptidase complex subunit MECl-1) | P40306|PSB10_HUMAN | 28,918 | 2 |
| Conserved oligomeric Golgi complex component 7 | P83436|COG7_HUMAN | 86,329 | 2 |
| Uncharacterized protein C17orf28 (Down-regulated in multiple cancers-1) | Q8IV36|CQ028_HUMAN | 88,729 | 2 |
| Zinc finger protein 207 | O43670|ZN207_HUMAN | 50,733 | 2 |
| Rab GTPase-binding effector protein 2 (Rabaptin-5beta) | Q9H5N1|RABE2_HUMAN | 63,524 | 2 |
| Zinc finger protein 326 | Q5BKZ1|ZN326_HUMAN | 65,636 | 2 |
| Beta-galactosidase precursor (EC 3.2.1.23) (Lactase) (Acid beta-galactosidase) | P16278|BGAL_HUMAN, P16279|BGAM_HUMAN | 76,076 | 2 |
| Alpha-adducin (Erythrocyte adducin subunit alpha) | P35611|ADDA_HUMAN | 80,938 | 2 |
| Exocyst complex component 1 (Exocyst complex component Sec3) | Q9NV70|EXOC1_HUMAN | 101,966 | 2 |
| Prefoldin subunit 1 | O60925|PFD1_HUMAN | 14,193 | 2 |
| Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit epsilon isoform (PP2A, B subunit, B′ epsilon isoform) (PP2A, B subunit, B56 epsilon isoform) (PP2A, B subunit, PR61 epsilon isoform) (PP2A, B subunit, R5 epsilon isoform) | Q16537|2A5E_HUMAN | 54,684 | 2 |
| Myosin regulatory light chain 2, smooth muscle isoform (Myosin RLC) (Myosin regulatory light chain 9) (LC20) | P24844|MLRN_HUMAN | 19,810 | 2 |
| Heat shock 70 kDa protein 12B | Q96MM6|HS12B_HUMAN | 75,670 | 2 |
| RAC-gamma serine/threonine-protein kinase (EC 2.7.11.1) (RAC-PK-gamma) (Protein kinase Akt-3) (Protein kinase B, gamma) (PKB gamma) (STK-2) | Q9Y243|AKT3_HUMAN | 55,758 | 2 |
| Kanadaptin (Kidney anion exchanger adapter protein) (Solute carrier family 4 anion exchanger member 1 adapter protein) (Lung cancer oncogene 3 protein) | Q9BWU0|NADAP_HUMAN | 88,798 | 2 |
| Protein NipSnap1 | Q9BPW8|NIPS1_HUMAN | 33,293 | 2 |
| Glycine cleavage system H protein, mitochondrial precursor | P23434|GCSH_HUMAN | 18,893 | 2 |
| GPI-anchor transamidase precursor (EC 3.-.-.-) (GPI transamidase) (Phosphatidylinositol-glycan biosynthesis class K protein) (PIG-K) (hGPI8) | Q92643|GPI8_HUMAN | 45,235 | 2 |
| Mitochondrial 39S ribosomal protein L23 (L23mt) (MRP-L23) (L23 mitochondrial-related protein) (Ribosomal protein L23-like) | Q16540|RM23_HUMAN | 17,763 | 2 |
| RNA-binding protein 10 (RNA-binding motif protein 10) (G patch domain-containing protein 9) | P98175|RBM10_HUMAN | 103,444 | 2 |
| Serine/threonine-protein kinase TBK1 (EC 2.7.11.1) (TANK-binding kinase 1) (T2K) (NF-kappa-B-activating kinase) | Q9UHD2|TBK1_HUMAN | 83,627 | 2 |
| Sideroflexin-1 (Tricarboxylate carrier protein) (TCC) | Q9H9B4|SFXN1_HUMAN | 35,602 | 2 |
| Leucine-rich repeat-containing protein 8A | Q8IWT6|LRC8A_HUMAN | 94,186 | 2 |
| KDEL motif-containing protein 2 precursor | Q7Z4H8|KDEL2_HUMAN | 58,556 | 2 |
| Serine/threonine-protein kinase N2 (EC 2.7.11.13) (Protein kinase C-like 2) (Protein-kinase C-related kinase 2) | Q16513|PKN2_HUMAN | 112,020 | 2 |
| Phakinin (Beaded filament structural protein 2) (Lens fiber cell beaded filament protein CP 49) (CP49) (49 kDa cytoskeletal protein) (CP 47) (CP47) (Lens intermediate filament-like light) (LIFL-L) | Q13515|BFSP2_HUMAN | 45,861 | 2 |
| Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10) | Q13427|PPIG_HUMAN | 88,602 | 2 |
| Glucosylceramidase precursor (EC 3.2.1.45) (Beta-glucocerebrosidase) (Acid beta-glucosidase) (D-glucosyl-N-acylsphingosine glucohydrolase) (Alglucerase) (Imiglucerase) | P04062|GLCM_HUMAN | 59,700 | 2 |
| Trafficking protein particle complex subunit 3 (BET3 homolog) | O43617|TPPC3_HUMAN | 20,257 | 2 |
| Kinetochore protein Hec1 (HsHec1) (Kinetochore-associated protein 2) (Highly expressed in cancer protein) (Retinoblastoma-associated protein HEC) | O14777|KNTC2_HUMAN | 73,897 | 2 |
| Mitochondrial import inner membrane translocase subunit Tim9 | Q9Y5J7|TIM9_HUMAN | 10,360 | 2 |
| Mitochondrial ribosomal protein S23 (S23mt) (MRP-S23) | Q9Y3D9|RT23_HUMAN | 21,753 | 2 |
| XPA-binding protein 1 | Q9HCN4|XAB1_HUMAN | 41,722 | 2 |
| Cat eye syndrome critical region protein 5 precursor | Q9BXW7|CECR5_HUMAN | 46,304 | 2 |
| Mitogen-activated protein kinase kinase kinase 7-interacting protein 1 (TGF-beta-activated kinase 1-binding protein 1) (TAK1-binding protein 1) | Q15750|TAB1_HUMAN | 54,626 | 2 |
| Cdc42 effector protein 1 (Binder of Rho GTPases 5) (Serum protein MSE55) | Q00587|BORG5_HUMAN | 40,277 | 2 |
| Alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase (EC 2.4.1.101) (N-glycosyl-oligosaccharide-glycoprotein N-acetylglucosaminyltransferase I) (GNT-I) (GlcNAc-T I) | P26572|MGAT1_HUMAN | 50,845 | 2 |
| SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2) | O75044|FNBP2_HUMAN | 120,865 | 2 |
| HECT domain and RCC1-like domain-containing protein 4 (EC 6.3.2.-) | Q5GLZ8|HERC4_HUMAN | 118,549 | 2 |
| Kinesin-like protein KIF13B (Kinesin-like protein GAKIN) | Q9NQT8|KI13B_HUMAN | 202,649 | 2 |
| Heterogeneous nuclear ribonucleoprotein L-like (Stromal RNA-regulating factor) (BLOCK24 protein) | Q8WVV9|HNRLL_HUMAN | 60,065 | 2 |
| Acyl-coenzyme A thioesterase 2 (EC 3.1.2.2) (Acyl-CoA thioesterase 2) (Peroxisomal acyl-coenzyme A thioester hydrolase 2a) (Peroxisomal long-chain acyl-coA thioesterase 2) (ZAP128) (CTE-Ia) | P49753|ACOT2_HUMAN, Q86TX2|ACOT1_HUMAN | 53,240 | 2 |
| IkappaB kinase complex-associated protein (IKK complex-associated protein) (p150) | O95163|IKAP_HUMAN | 150,174 | 2 |
| Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B) | Q96SB3|NEB2_HUMAN | 89,174 | 2 |
| Serine/threonine-protein kinase VRK1 (EC 2.7.11.1) (Vaccinia-related kinase 1) | Q99986|VRK1_HUMAN | 45,460 | 2 |
| RNA polymerase-associated protein RTF1 homolog | Q92541|RTF1_HUMAN | 76,565 | 2 |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13 (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase B16.6 subunit) (Complex I-B16.6) (CI-B16.6) (Gene associated with retinoic-interferon-induced mortality 19 protein) (GRIM-19) (Cell death-regulatory protein GRIM-19) | Q9P0J0|NDUAD_HUMAN | 16,681 | 2 |
| Mitogen-activated protein-binding protein-interacting protein (Late endosomal/lysosomal Mp1-interacting protein) (p14) | Q9Y2Q5| MAPIP_HU MAN | 13,490 | 2 |
| Zinc finger Ran-binding domain-containing protein 2 (Zinc finger protein 265) (Zinc finger, splicing) | O95218|ZRAB2_HUMAN | 38,206 | 2 |
| RNA-binding motif, single-stranded-interacting protein 1 (Single-stranded DNA-binding protein MSSP-1) (Suppressor of CDC2 with RNA-binding motif 2) | P29558|RBMS1_HUMAN | 44,488 | 2 |
| Caseinolytic peptidase B protein homolog (Suppressor of potassium transport defect 3) | Q9H078|CLPB_HUMAN | 78,713 | 2 |
| Tubulin-specific chaperone C (Tubulin-folding cofactor C) (CFC) | Q15814|TBCC_HUMAN | 39,202 | 2 |
| Dipeptidyl peptidase 9 (EC 3.4.14.5) (Dipeptidyl peptidase IX) (DP9) (Dipeptidyl peptidase-like protein 9) (DPLP9) (Dipeptidyl peptidase IV-related protein 2) (DPRP-2) | Q86TI2|DPP9_HUMAN | 98,246 | 2 |
| Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP-dependent helicase CHD1) (CHD-1) | O14646|CHD1_HUMAN | 196,507 | 2 |
| Inositol polyphosphate 5-phosphatase OCRL-1 (EC 3.1.3.36) (Lowe oculocerebrorenal syndrome protein) | Q01968|OCRL_HUMAN | 104,190 | 2 |
| Microsomal signal peptidase 18 kDa subunit (EC 3.4.-.-) (SPase 18 kDa subunit) (SPC18) (Endopeptidase SP18) (SEC11-like 1) | P67812|SPC18_HUMAN | 20,608 | 2 |
| Ubiquitin carboxyl-terminal hydrolase 19 (EC 3.1.2.15) (Ubiquitin thioesterase 19) (Ubiquitin-specific-processing protease 19) (Deubiquitinating enzyme 19) (Zinc finger MYND domain-containing protein 9) (Fragment) | O94966|UBP19_HUMAN | 151,323 | 2 |
| NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9 (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase B22 subunit) (Complex I-B22) (CI-B22) | Q9Y6M9|NDUB9_HUMAN | 21,813 | 2 |
| Synaptojanin-2 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5-phosphatase 2) | O15056|SYNJ2_HUMAN | 165,523 | 2 |
| Cysteine-rich with EGF-like domain protein 1 precursor | Q96HD1|CREL1_HUMAN | 45,389 | 2 |
| Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial precursor (EC 1.1.1.41) (Isocitric dehydrogenase) (NAD(+)-specific ICDH) | P50213|IDH3A_HUMAN | 39,575 | 2 |
| Steroid receptor RNA activator 1 (Steroid receptor RNA activator protein) (SRAP) | Q9HD15|SRA1_HUMAN | 25,655 | 2 |
| Pentraxin-related protein PTX3 precursor (Pentaxin-related protein PTX3) (Tumor necrosis factor-inducible protein TSG-14) | P26022|PTX3_HUMAN | 42,002 | 2 |
| N-acetylserotonin O-methyltransferase-like protein (ASMTL) | O95671|ASML_HUMAN | 68,840 | 2 |
| Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform (PP2A, subunit A, PR65-beta isoform) (PP2A, subunit A, R1-beta isoform) | P30154|2AAB_HUMAN | 66,197 | 2 |
| Development and differentiation-enhancing factor 2 (Pyk2 C-terminus-associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3) | O43150|DDEF2_HUMAN | 111,635 | 2 |
| GTP-binding protein 1 (G-protein 1) (GP-1) (GP1) | O00178|GTPB1_HUMAN | 71,446 | 2 |
| IWS1 homolog (IWS1-like protein) | Q96ST2|IWS1_HUMAN | 91,938 | 2 |
| SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) | Q92922|SMRC1_HUMAN | 122,735 | 2 |
| tRNA (guanine-N(7)-)-methyltransferase (EC 2.1.1.33) (tRNA(m7G46)-methyltransferase) (Methyltransferase-like protein 1) | Q9UBP6|TRMB_HUMAN | 31,454 | 2 |
| Mitogen-activated protein kinase kinase 1-interacting protein 1 (MEK-binding partner 1) (Mp1) | Q9UHA4|MK1I1_HUMAN | 13,605 | 2 |
| Serine/threonine-protein kinase D2 (EC 2.7.11.13) (nPKC-D2) | Q9BZL6|KPCD2_HUMAN | 96,706 | 2 |
| Myeloid differentiation primary response protein MyD88 | Q99836|MYD88_HUMAN | 33,216 | 2 |
| Histone H1x | Q92522|H1X_HUMAN | 22,470 | 2 |
| Probable aminopeptidase NPEPL1 (EC 3.4.11.-) (Aminopeptidase-like 1) | Q8NDH3|PEPL1_HUMAN | 55,843 | 2 |
| E3 ubiquitin-protein ligase RING1 (EC 6.3.2.-) (Polycomb complex protein RING1) (RING finger protein 1) | Q06587|RING1_HUMAN, Q99496|RING2_HUMAN | 42,412 | 2 |
| BAG family molecular chaperone regulator 2 (BCL2-associated athanogene 2) (BAG-2) | O95816|BAG2_HUMAN | 23,755 | 2 |
| PDZ domain-containing protein GIPC1 (RGS19-interacting protein 1) (GAIP C-terminus-interacting protein) (RGS-GAIP-interacting protein) (Tax interaction protein 2) (TIP-2) | O14908|GIPC1_HUMAN | 36,033 | 2 |
| Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6) | Q9Y3D8|KAD6_HUMAN | 20,044 | 2 |
| Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase alpha) | P51003|PAPOA_HUMAN | 82,826 | 2 |
| Endothelial protein C receptor precursor (Endothelial cell protein C receptor) (Activated protein C receptor) (APC receptor) (CD201 antigen) | Q9UNN8|EPCR_HUMAN | 26,653 | 2 |
| NNP-1 protein (Novel nuclear protein 1) (Nucleolar protein Nop52) (D21S2056E) | P56182|NNP1_HUMAN | 52,823 | 2 |
| Clathrin light chain B (Lcb) | P09497|CLCB_HUMAN | 25,173 | 2 |
| Transcription factor SOX-18 | P35713|SOX18_HUMAN | 40,875 | 2 |
| Proteasome maturation protein (Proteassemblin) (Protein UMP1 homolog) (hUMP1) (Voltage-gated K channel beta subunit 4.1) | Q9Y244|POMP_HUMAN | 15,772 | 2 |
| Caskin-2 | Q8WXE0|CSKI2_HUMAN | 126,696 | 2 |
| ADAM 9 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 9) (Metalloprotease/disintegrin/cysteine-rich protein 9) (Myeloma cell metalloproteinase) (Meltrin gamma) (Cellular disintegrin-related protein) | Q13443|ADAM9_HUMAN | 90,538 | 2 |
| Splicing factor 4 (RNA-binding protein RBP) | Q8IWZ8|SF04_HUMAN | 72,455 | 2 |
| Anaphase-promoting complex subunit 7 (APC7) (Cyclosome subunit 7) | Q9UJX3|APC7_HUMAN | 63,117 | 2 |
| Ras-related protein Rab-10 | P61026|RAB10_HUMAN | 22,524 | 2 |
| Putative GTP-binding protein Parf (Partner of ARF) | Q3YEC7|PARF_HUMAN | 79,532 | 2 |
| Pyridoxine-5′-phosphate oxidase (EC 1.4.3.5) (Pyridoxamine-phosphate oxidase) | Q9NVS9|PNPO_HUMAN | 29,970 | 2 |
| Mitochondrial import inner membrane translocase subunit Tim10 | P62072|TIM10_HUMAN | 10,315 | 2 |
| Translocon-associated protein subunit gamma (TRAP-gamma) (Signal sequence receptor subunit gamma) (SSR-gamma) | Q9UNL2|SSRG_HUMAN | 21,063 | 2 |
| Methyl-CpG-binding domain protein 2 (Methyl-CpG-binding protein MBD2) (Demethylase) (DMTase) | Q9UBB5|MBD2_HUMAN | 43,237 | 2 |
| Exocyst complex component 8 (Exocyst complex 84 kDa subunit) | Q8IYI6|EXOC8_HUMAN | 81,782 | 2 |
| Adipose most abundant gene transcript 2 | Q15847|APM2_HUMAN | 7,837 | 2 |
| Small EDRK-rich factor 2 (4F5rel) (h4F5rel) (Gastric cancer-related protein VRG107) | P84101|SERF2_HUMAN | 6,882 | 2 |
| Fumarylacetoacetase (EC 3.7.1.2) (Fumarylacetoacetate hydrolase) (Beta-diketonase) (FAA) | P16930|FAAA_HUMAN | 46,358 | 2 |
| Cytochrome c oxidase subunit 7C, mitochondrial precursor (EC 1.9.3.1) (Cytochrome c oxidase polypeptide VIIc) | P15954|COX7C_HUMAN | 7,228 | 2 |
| Pyruvate carboxylase, mitochondrial precursor (EC 6.4.1.1) (Pyruvic carboxylase) (PCB) | P11498|PYC_HUMAN | 129,617 | 2 |
| Hexokinase-2 (EC 2.7.1.1) (Hexokinase type II) (HK II) (Muscle form hexokinase) | P52789|HXK2_HUMAN | 102,363 | 2 |
| Regulator of differentiation 1 (Rod1) | O95758|ROD1_HUMAN | 56,486 | 2 |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12 (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase subunit B17.2) (Complex I-B17.2) (CI-B17.2) (CIB17.2) (13 kDa differentiation-associated protein) | Q9UI09|NDUAC_HUMAN | 17,097 | 2 |
| Carbohydrate kinase-like protein (EC 2.7.1.-) | Q9UHJ6|CARKL_HUMAN | 51,485 | 2 |
| ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3) | Q9NP61|ARFG3_HUMAN | 56,911 | 2 |
| Enhancer of mRNA decapping protein 3 (YjeF domain-containing protein 1) | Q96F86|EDC3_HUMAN | 56,059 | 2 |
| Minor histocompatibility antigen H13 (EC 3.4.99.-) (Signal peptide peptidase) (Presenilin-like protein 3) (hIMP1 protein) | Q8TCT9|HM13_HUMAN | 41,473 | 2 |
| Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3) (Phosphatidylinositol 3-kinase p100 subunit) | Q8NEB9|PK3C3_HUMAN | 101,535 | 2 |
| Mixed lineage kinase domain-like protein | Q8NB16|MLKL_HUMAN | 54,463 | 2 |
| Prolyl 3-hydroxylase 3 precursor (EC 1.14.11.7) (Leprecan-like protein 2) (Protein B) | Q8IVL6|P3H3_HUMAN | 81,820 | 2 |
| SAM and SH3 domain-containing protein 1 (Proline-glutamate repeat-containing protein) | O94885|SASH1_HUMAN | 136,666 | 2 |
| NADH dehydrogenase [ubiquinone] iron-sulfur protein 2, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 49 kDa subunit) (Complex I-49kD) (CI-49kD) | O75306|NDUS2_HUMAN | 52,529 | 2 |
| GDP-mannose 4,6 dehydratase (EC 4.2.1.47) (GDP-D-mannose dehydratase) (GMD) | O60547|GMDS_HUMAN | 41,932 | 2 |
| Branched-chain-amino-acid aminotransferase, mitochondrial precursor (EC 2.6.1.42) (BCAT(m)) (Placental protein 18) (PP18) | O15382|BCAT2_HUMAN | 44,270 | 2 |
| Multiple inositol polyphosphate phosphatase 1 precursor (EC 3.1.3.62) (Inositol (1,3,4,5)-tetrakisphosphate 3-phosphatase) (Ins(1,3,4,5)P(4) 3-phosphatase) | Q9UNW1|MINP1_HUMAN | 55,035 | 2 |
| Double-stranded RNA-binding protein Staufen homolog 2 | Q9NUL3|STAU2_HUMAN | 62,624 | 2 |
| Pantothenate kinase 2, mitochondrial precursor (EC 2.7.1.33) (Pantothenic acid kinase 2) (hPANK2) | Q9BZ23|PANK2_HUMAN | 62,665 | 2 |
| Apolipoprotein O precursor (Protein FAM121B) | Q9BUR5|APOO_HUMAN | 22,267 | 2 |
| Stromal cell-derived factor 2 precursor (SDF-2) | Q99470|SDF2_HUMAN | 23,007 | 2 |
| THO complex subunit 1 (Tho1) (Nuclear matrix protein p84) | Q96FV9|THOC1_HUMAN | 75,651 | 2 |
| Histone deacetylase 7a (HD7a) | Q8WUI4|HDAC7_HUMAN | 102,910 | 2 |
| HEAT repeat-containing protein 3 | Q7Z4Q2|HEAT3_HUMAN | 74,567 | 2 |
| COMM domain-containing protein 6 | Q7Z4G1|COMD6_HUMAN | 9,620 | 2 |
| GTP-binding protein Rheb precursor (Ras homolog enriched in brain) | Q15382|RHEB_HUMAN | 20,480 | 2 |
| Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5-trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3) | Q14573|ITPR3_HUMAN | 304,024 | 2 |
| Eukaryotic translation initiation factor 4E-binding protein 2 (4E-BP2) (eIF4E-binding protein 2) | Q13542|4EBP2_HUMAN | 12,922 | 2 |
| Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1) | Q13439|GOGA4_HUMAN | 261,126 | 2 |
| Protein OS-9 precursor (Amplified in osteosarcoma 9) | Q13438|OS9_HUMAN | 75,546 | 2 |
| Ubiquitin-protein ligase E3A (EC 6.3.2.-) (E6AP ubiquitin-protein ligase) (Oncogenic protein-associated protein E6-AP) (Human papillomavirus E6-associated protein) (Renal carcinoma antigen NY-REN-54) | Q05086|UBE3A_HUMAN | 100,632 | 2 |
| Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (p190) | P78357|CNTP1_HUMAN | 156,250 | 2 |
| Acylphosphatase-1 (EC 3.6.1.7) (Acylphosphate phosphohydrolase 1) (Acylphosphatase, organ-common type isozyme) (Acylphosphatase, erythrocyte isozyme) | P07311|ACYP1_HUMAN | 11,243 | 2 |
| Protein phosphatase 1 regulatory subunit 11 (Protein phosphatase inhibitor 3) (Hemochromatosis candidate protein V) (HCG V) (HCG-V) | O60927|PP1RB_HUMAN | 13,934 | 2 |
| Cdc42 effector protein 2 (Binder of Rho GTPases 1) | O14613|BORG1_HUMAN | 22,467 | 2 |
| Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp) | Q13469|NFAC2_HUMAN | 100,128 | 2 |
| L-aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase (EC 2.7.8.-) (4′-phosphopantetheinyl transferase) (Alpha-aminoadipic semialdehyde dehydrogenase-phosphopantetheinyl transferase) (AASD-PPT) (LYS5 ortholog) | Q9NRN7|ADPPT_HUMAN | 35,759 | 2 |
| Transcriptional repressor p66 beta (p66/p68) (GATA zinc finger domain-containing protein 2B) | Q8WXI9|P66B_HUMAN | 65,244 | 2 |
| Chitinase domain-containing protein 1 precursor (Stabilin-1-interacting chitinase-like protein) (SI-CLP) | Q9BWS9|CHID1_HUMAN | 44,923 | 2 |
| Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial precursor [Includes: NAD-dependent methylenetetrahydrofolate dehydrogenase (EC 1.5.1.15); Methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9)] | P13995|MTDC_HUMAN | 37,877 | 2 |
| Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17) | Q96I25|SPF45_HUMAN | 44,944 | 2 |
| OTU domain-containing protein 6B | Q8N6M0|OTU6B_HUMAN | 33,795 | 2 |
| 39S ribosomal protein L34, mitochondrial precursor (L34mt) (MRP-L34) | Q9BQ48|RM34_HUMAN | 10,147 | 2 |
| Gamma-aminobutyric acid receptor-associated protein (GABA(A) receptor-associated protein) (MM46) | O95166|GBRAP_HUMAN | 13,901 | 2 |
| Pantothenate kinase 4 (EC 2.7.1.33) (Pantothenic acid kinase 4) (hPanK4) | Q9NVE7|PANK4_HUMAN | 85,975 | 2 |
| Integrator complex subunit 1 (Int1) | Q8N201|INT1_HUMAN | 244,285 | 2 |
| Alpha-1,3-mannosyltransferase ALG2 (EC 2.4.1.-) (GDP-Man:Man(1)GlcNAc(2)-PP-dolichol mannosyltransferase) | Q9H553|ALG2_HUMAN | 47,074 | 2 |
| Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP-dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM) | P51531|SMCA2_HUMAN | 180,749 | 2 |
| Calcium-transporting ATPase type 2C member 1 (EC 3.6.3.8) (ATPase 2C1) (ATP-dependent Ca(2+) pump PMR1) | P98194|AT2C1_HUMAN | 100,561 | 2 |
| RNA-binding protein 26 (RNA-binding motif protein 26) (CTCL tumor antigen se70-2) | Q5T8P6|RBM26_HUMAN | 113,582 | 2 |
| Follistatin-related protein 1 precursor (Follistatin-like 1) | Q12841|FSTL1_HUMAN | 34,967 | 2 |
| 3′-5′ exoribonuclease CSL4 homolog (EC 3.1.13.-) (Exosome component 1) | Q9Y3B2|EXOS1_HUMAN | 21,434 | 2 |
| Spartin (Trans-activated by hepatitis C virus core protein 1) | Q8N0X7|SPG20_HUMAN | 72,815 | 2 |
| Hedgehog-interacting protein precursor (HHIP) (HIP) | Q96QV1|HHIP_HUMAN | 78,835 | 2 |
| Transcription initiation factor IIF subunit alpha (EC 2.7.11.1) (TFIIF-alpha) (General transcription factor IIF subunit 1) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein) | P35269|T2FA_HUMAN | 58,224 | 2 |
| 39S ribosomal protein L19, mitochondrial precursor (L19mt) (MRP-L19) (MRP-L15) | P49406|RM19_HUMAN | 33,518 | 2 |
| Tumor necrosis factor receptor type 1-associated DEATH domain protein (TNFR1-associated DEATH domain protein) (TNFRSF1A-associated via death domain) | Q15628|TRADD_HUMAN | 34,230 | 2 |
| Disabled homolog 2-interacting protein (DAB2-interacting protein) (DAB2 interaction protein) (ASK-interacting protein 1) | Q5 VWQ8| DAB2P_HUMAN | 131,612 | 2 |
| TSC22 domain family protein 1 (Transforming growth factor beta-1-induced transcript 4 protein) (Regulatory protein TSC-22) (TGFB-stimulated clone 22 homolog) (Cerebral protein 2) | Q15714|T22D1_HUMAN | 15,662 | 2 |
| Integrin beta-5 precursor | P18084|ITB5_HUMAN | 88,037 | 2 |
| Cytochrome c oxidase polypeptide VIIa-liver/heart, mitochondrial precursor (EC 1.9.3.1) (Cytochrome c oxidase subunit VIIa-L) (VIIaL) | P14406|CX7A2_HUMAN | 9,379 | 2 |
| Ribonucleases P/MRP protein subunit POP1 (EC 3.1.26.5) (hPOP1) | Q99575|POP1_HUMAN | 114,692 | 2 |
| E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-) | P46934|NEDD4_HUMAN | 114,922 | 2 |
| Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein homolog) | Q13107|UBP4_HUMAN | 108,549 | 2 |
| Asparagine synthetase [glutamine-hydrolyzing] (EC 6.3.5.4) (Glutamine-dependent asparagine synthetase) (Cell cycle control protein TS11) | P08243|ASNS_HUMAN | 64,354 | 2 |
| DnaJ homolog subfamily C member 1 (DnaJ protein homolog MTJ1) | Q96KC8|DNJC1_HUMAN | 63,867 | 2 |
| F-box-like/WD repeat protein TBL1XR1 (Transducin beta-like 1X-related protein 1) (Nuclear receptor corepressor/HDAC3 complex subunit TBLR1) (TBL1-related protein 1) | O60907|TBL1X_HUMAN, Q9BQ87|TBL1Y_HUMAN, Q9BZK7|TBL1R_HUMAN | 55,577 | 2 |
| Tripartite motif-containing protein 5 (EC 6.3.2.-) (RING finger protein 88) | Q9C035|TRIM5_HUMAN | 56,320 | 2 |
| Gamma-glutamyl hydrolase precursor (EC 3.4.19.9) (Gamma-Glu-X carboxypeptidase) (Conjugase) (GH) | Q92820|GGH_HUMAN | 35,948 | 2 |
| Histidine triad nucleotide-binding protein 2 (EC 3.-.-.-) (HINT-2) (HINT-3) (HIT-17kDa) (PKCI-1-related HIT protein) | Q9BX68|HINT2_HUMAN | 17,144 | 2 |
| Activating signal cointegrator 1 complex subunit 3 (EC 3.6.1.-) (ASC-1 complex subunit p200) (Trip4 complex subunit p200) (Helicase, ATP binding 1) | Q8N3C0|HELC1_HUMAN | 251,479 | 2 |
| Mitochondrial 28S ribosomal protein S34 (S34mt) (MRP-S34) | P82930|RT34_HUMAN | 25,633 | 2 |
| LanC-like protein 1 (40 kDa erythrocyte membrane protein) (p40) | O43813|LANC1_HUMAN | 45,267 | 2 |
| Baculoviral IAP repeat-containing protein 4 (Inhibitor of apoptosis protein 3) (X-linked inhibitor of apoptosis protein) (X-linked IAP) (IAP-like protein) (HILP) | P98170|BIRC4_HUMAN | 56,667 | 2 |
| Collagen alpha-1(VI) chain precursor | P12109|CO6A1_HUMAN | 108,513 | 2 |
| Death-associated protein kinase 3 (EC 2.7.11.1) (DAP kinase 3) (DAP-like kinase) (Dlk) (ZIP-kinase) | O43293|DAPK3_HUMAN | 52,520 | 2 |
| Caskin-1 (CASK-interacting protein 1) | Q8WXD9|CSKI1_HUMAN | 149,797 | 2 |
| Immunoglobulin-binding protein 1 (CD79a-binding protein 1) (B cell signal transduction molecule alpha 4) (Alpha 4 protein) (Renal carcinoma antigen NY-REN-16) | P78318|IGBP1_HUMAN | 39,205 | 2 |
| Protein furry homolog-like (ALL1-fused gene from chromosome 4p12) | O94915|FRYL_HUMAN | 339,585 | 2 |
| Mitochondrial carrier homolog 1 (Presenilin-associated protein) | Q9NZJ7|MTCH1_HUMAN | 41,527 | 2 |
| Transmembrane 9 superfamily protein member 4 | Q92544|TM9S4_HUMAN | 72,526 | 2 |
| Ubiquitin-conjugating enzyme E2 R1 (EC 6.3.2.19) (Ubiquitin-protein ligase R1) (Ubiquitin-conjugating enzyme E2-32 kDa complementing) (E2-CDC34) | P49427|UB2R1_HUMAN | 26,719 | 2 |
| Pre-mRNA branch site protein p14 (SF3B 14 kDa subunit) | Q9Y3B4|PM14_HUMAN | 14,568 | 2 |
| UV excision repair protein RAD23 homolog A (hHR23A) | P54725|RD23A_HUMAN | 39,591 | 2 |
| Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin) | Q96C24|SYTL4_HUMAN | 75,993 | 2 |
| Pyridoxal kinase (EC 2.7.1.35) (Pyridoxine kinase) | O00764|PDXK_HUMAN | 35,084 | 2 |
| Target of EGR1 protein 1 | Q96GM8|TOE1_HUMAN | 56,530 | 2 |
| PCI domain-containing protein 2 (CSN12-like protein) | Q5JVF3|PCID2_HUMAN | 46,013 | 2 |
| Inorganic pyrophosphatase 2, mitochondrial precursor (EC 3.6.1.1) (PPase 2) (Pyrophosphatase SID6-306) | Q9H2U2|IPYR2_HUMAN | 37,903 | 2 |
| Transportin-3 (Transportin-SR) (TRN-SR) (Importin-12) | Q9Y5L0|TNPO3_HUMAN | 109,798 | 2 |
| Cdc42 effector protein 3 (Binder of Rho GTPases 2) (MSE55-related Cdc42-binding protein) | Q9UKI2|BORG2_HUMAN | 27,661 | 2 |
| THUMP domain-containing protein 1 | Q9NXG2|THUM1_HUMAN | 39,298 | 2 |
| Cell growth-regulating nucleolar protein | Q9NX58|LYAR_HUMAN | 43,597 | 2 |
| Transmembrane protein C1orf78 | Q9NVM1|CA078_HUMAN | 18,356 | 2 |
| WD repeat protein 55 | Q9H6Y2|WDR55_HUMAN | 42,106 | 2 |
| Phosducin-like protein 3 (Viral IAP-associated factor 1) (VIAF-1) (HTPHLP) | Q9H2J4|PDCL3_HUMAN | 27,598 | 2 |
| Mediator of RNA polymerase II transcription subunit 28 (Mediator complex subunit 28) (Tumor angiogenesis marker EG-1) (Endothelial-derived protein 1) (Merlin and Grb2-interacting cytoskeletal protein) (Magicin) | Q9H204|MED28_HUMAN | 19,502 | 2 |
| Nucleolar protein 10 | Q9BSC4|NOL10_HUMAN | 80,287 | 2 |
| Exocyst complex component 2 (Exocyst complex component Sec5) | Q96KP1|EXOC2_HUMAN | 104,052 | 2 |
| Uncharacterized protein KIAA1279 | Q96EK5|K1279_HUMAN | 71,798 | 2 |
| Glutaryl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.7) (GCD) | Q92947|GCDH_HUMAN | 48,111 | 2 |
| Protein PTDSR (Phosphatidylserine receptor) | Q6NYC1|PTDSR_HUMAN | 46,445 | 2 |
| Sterile alpha motif domain-containing protein 9 | Q5K651|SAMD9_HUMAN | 184,272 | 2 |
| Glutamine-rich protein 1 | Q2TAL8|QRIC1_HUMAN | 86,415 | 2 |
| Active breakpoint cluster region-related protein | Q12979|ABR_HUMAN | 97,682 | 2 |
| Filensin (Beaded filament structural protein 1) (Lens fiber cell beaded-filament structural protein CP 115) (CP115) (Lens intermediate filament-like heavy) (LIFL-H) | Q12934|BFSP1_HUMAN | 74,527 | 2 |
| Forkhead box protein O1A (Forkhead in rhabdomyosarcoma) | Q12778|FOXO1_HUMAN | 69,644 | 2 |
| Guanine nucleotide-binding protein-like 1 (GTP-binding protein HSR1) | P36915|GNL1_HUMAN | 47,447 | 2 |
| Colorectal mutant cancer protein (Protein MCC) | P23508|CRCM_HUMAN | 93,039 | 2 |
| Lysosomal protective protein precursor (EC 3.4.16.5) (Cathepsin A) (Carboxypeptidase C) (Protective protein for beta-galactosidase) [Contains: Lysosomal protective protein 32 kDa chain; Lysosomal protective protein 20 kDa chain] | P10619|PPGB_HUMAN | 54,450 | 2 |
| Alpha-2-macroglobulin precursor (Alpha-2-M) | P01023|A2MG_HUMAN | 163,259 | 2 |
| Prothrombin precursor (EC 3.4.21.5) (Coagulation factor II) [Contains: Activation peptide fragment 1; Activation peptide fragment 2; Thrombin light chain; Thrombin heavy chain] | P00734|THRB_HUMAN | 70,019 | 2 |
| Carboxypeptidase D precursor (EC 3.4.17.22) (Metallocarboxypeptidase D) (gp180) | O75976|CBPD_HUMAN | 152,915 | 2 |
| Glycosylphosphatidylinositol anchor attachment 1 protein (GPI anchor attachment protein 1) (GAA1 protein homolog) (hGAA1) | O43292|GPAA1_HUMAN | 67,607 | 2 |
| Tumor necrosis factor receptor superfamily member 10B precursor (Death receptor 5) (TNF-related apoptosis-inducing ligand receptor 2) (TRAIL receptor 2) (TRAIL-R2) (CD262 antigen) | O14763|TR10B_HUMAN | 47,832 | 2 |
| Copper chaperone for superoxide dismutase (Superoxide dismutase copper chaperone) | O14618|CCS_HUMAN | 29,022 | 2 |
| Nuclear factor 1 B-type (Nuclear factor 1/B) (NF1-B) (NFI-B) (NF-I/B) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein) | O00712|NFIB_HUMAN | 47,425 | 2 |
| DNA-directed RNA polymerase III subunit 127.6 kDa polypeptide (EC 2.7.7.6) (RNA polymerase III subunit 2) (RPC2) | Q9NW08|RPC2_HUMAN | 127,771 | 2 |
| Transmembrane 9 superfamily protein member 2 precursor (p76) | Q99805|TM9S2_HUMAN | 75,761 | 2 |
| Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein) | Q92870|APBB2_HUMAN | 83,328 | 2 |
| 39S ribosomal protein L38, mitochondrial precursor (L38mt) (MRP-L38) | Q96DV4|RM38_HUMAN | 44,579 | 2 |
| Nucleoporin SEH1-like (SEC13-like protein) | Q96EE3|SEH1L_HUMAN | 46,560 | 2 |
| Diacylglycerol kinase alpha (EC 2.7.1.107) (Diglyceride kinase alpha) (DGK-alpha) (DAG kinase alpha) (80 kDa diacylglycerol kinase) | P23743|DGKA_HUMAN | 82,656 | 2 |
| Nicastrin precursor | Q92542|NICA_HUMAN | 78,394 | 2 |
| RWD domain-containing protein 1 | Q9H446|RWDD1_HUMAN | 27,922 | 2 |
| Bystin | Q13895|BYST_HUMAN | 49,585 | 2 |
| Transforming growth factor beta-1 precursor (TGF-beta-1) [Contains: Latency-associated peptide (LAP)] | P01137|TGFB1_HUMAN | 44,324 | 2 |
| REVERSED | REV|Q14573|ITPR3 | 304,024 | 2 |
| REVERSED | REV|P14618|KPYM | 57,920 | 2 |
| NFX1-type zinc finger-containing protein 1 | Q9P2E3|ZNFX1_HUMAN | 220,207 | 2 |
| Diphosphoinositol polyphosphate phosphohydrolase 1 (EC 3.6.1.52) (DIPP-1) (Diadenosine 5′,5‴-P1,P6-hexaphosphate hydrolase 1) (EC 3.6.1.-) (Nucleoside diphosphate-linked moiety X motif 3) (Nudix motif 3) | O95989|NUDT3_HUMAN | 19,453 | 2 |
| NAD-dependent deacetylase sirtuin-5 (EC 3.5.1.-) (SIR2-like protein 5) | Q9NXA8|SIRT5_HUMAN | 33,863 | 2 |
| Gamma-aminobutyric acid receptor-associated protein-like 2 (GABA(A) receptor-associated protein-like 2) (Ganglioside expression factor 2) (GEF-2) (General protein transport factor p16) (MAP1 light chain 3-related protein) | P60520|GBRL2_HUMAN | 13,649 | 2 |
| Squalene synthetase (EC 2.5.1.21) (SQS) (SS) (Farnesyl-diphosphate farnesyltransferase) (FPP:FPP farnesyltransferase) | P37268|FDFT_HUMAN | 48,098 | 2 |
| REVERSED | REV|Q9Y623|MYH4 | 223,001 | 2 |
| Activating signal cointegrator 1 complex subunit 1 (ASC-1 complex subunit p50) (Trip4 complex subunit p50) | Q8N9N2|ASCC1_HUMAN | 45,492 | 2 |
| cAMP-dependent protein kinase, beta-catalytic subunit (EC 2.7.11.11) (PKA C-beta) | P22694|KAPCB_HUMAN | 40,606 | 2 |
| DNA polymerase delta catalytic subunit (EC 2.7.7.7) (DNA polymerase subunit delta p125) | P28340|DPOD1_HUMAN | 123,616 | 2 |
| Myosin-2 (Myosin heavy chain 2) (Myosin heavy chain 2a) (MyHC-2a) (Myosin heavy chain, skeletal muscle, adult 2) (Myosin heavy chain IIa) (MyHC-IIa) | Q9UKX2|MYH2_HUMAN | 223,032 | 2 |
| 1-acyl-sn-glycerol-3-phosphate acyltransferase gamma (EC 2.3.1.51) (1-AGP acyltransferase 3) (1-AGPAT 3) (Lysophosphatidic acid acyltransferase gamma) (LPAAT-gamma) (1-acylglycerol-3-phosphate O-acyltransferase 3) | Q9NRZ7|PLCC_HUMAN | 43,364 | 2 |
| REVERSED | REV|P35609|ACTN2 | 103,840 | 2 |
| Transcription factor MafK (Erythroid transcription factor NF-E2 p18 subunit) | O60675|MAFK_HUMAN | 17,505 | 2 |
| REVERSED | REV|Q92673|SORL | 248,424 | 2 |
| Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein) | O14686|MLL2_HUMAN | 564,164 | 2 |
| REVERSED | REV|P20929|NEBU | 773,209 | 2 |
| Uncharacterized protein C6orf115 | Q9P1F3|CF115_HUMAN | 9,039 | 2 |
| REVERSED | REV|Q9UJ83|HACL1 | 63,711 | 2 |
| REVERSED | REV|Q9HCE9|TM16H | 136,019 | 2 |
| Ryanodine receptor 3 (Brain-type ryanodine receptor) (RyR3) (RYR-3) (Brain ryanodine receptor-calcium release channel) | Q15413|RYR3_HUMAN | 551,916 | 2 |
| REVERSED | REV|P25391|LAMA1 | 337,133 | 2 |
| Aldo-keto reductase family 1 member C1 (EC 1.1.1.-) (20-alpha-hydroxysteroid dehydrogenase) (EC 1.1.1.149) (20-alpha-HSD) (Trans-1,2-dihydrobenzene-1,2-diol dehydrogenase) (EC 1.3.1.20) (High-affinity hepatic bile acid-binding protein) (HBAB) (Chlordecone reductase homolog HAKRC) (Dihydrodiol dehydrogenase 1/2) (DD1/DD2) | Q04828|AK1C1_HUMAN | 36,771 | 2 |
| Nucleolysin TIA-1 isoform p40 (RNA-binding protein TIA-1) (p40-TIA-1) [Contains: Nucleolysin TIA-1 isoform p15 (p15-TIA-1)] | P31483|TIA1_HUMAN | 42,942 | 2 |
| REVERSED | REV|Q9NYC9|DYH9 | 511,915 | 2 |
| REVERSED | REV|Q9NRM7|LATS2 | 120,177 | 2 |
| Casein kinase I isoform gamma-2 (EC 2.7.11.1) (CKI-gamma 2) | P78368|KC1G2_HUMAN, Q9Y6M4|KC1G3_HUMAN | 47,441 | 2 |
| Protein TRS85 homolog | Q9Y2L5|TRS85_HUMAN | 160,926 | 2 |
| ATP-binding cassette sub-family G member 2 (Placenta-specific ATP-binding cassette transporter) (Breast cancer resistance protein) (Mitoxantrone resistance-associated protein) (CD338 antigen) (CDw338) | Q9UNQ0|ABCG2_HUMAN | 72,299 | 2 |
| Uncharacterized protein C1orf149 (Sarcoma antigen NY-SAR-91) | Q9HAF1|CA149_HUMAN | 21,617 | 2 |
| Vacuolar protein sorting-associated protein 16 homolog (hVPS16) | Q9H269|VPS16_HUMAN | 94,678 | 2 |
| Sialidase-1 precursor (EC 3.2.1.18) (Lysosomal sialidase) (N-acetyl-alpha-neuraminidase 1) (Acetylneuraminyl hydrolase) (G9 sialidase) | Q99519|NEUR1_HUMAN | 45,449 | 2 |
| Putative RNA-binding protein 15 (RNA-binding motif protein 15) (One-twenty two protein) | Q96T37|RBM15_HUMAN | 107,175 | 2 |
| Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a) | Q96RN5|PCQAP_HUMAN | 86,733 | 2 |
| COMM domain-containing protein 1 (Protein Murr1) | Q8N668|COMD1_HUMAN | 21,161 | 2 |
| Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160) | Q8IYB3|SRRM1_HUMAN | 102,319 | 2 |
| 39S ribosomal protein L21, mitochondrial precursor (L21mt) (MRP-L21) | Q7Z2W9|RM21_HUMAN | 23,142 | 2 |
| SSXT protein (Synovial sarcoma, translocated to X chromosome) (SYT protein) | Q15532|SSXT_HUMAN | 45,910 | 2 |
| Transmembrane protein 33 (DB83 protein) | P57088|TMM33_HUMAN | 27,962 | 2 |
| Cyclin-H (MO15-associated protein) (p37) (p34) | P51946|CCNH_HUMAN | 37,627 | 2 |
| E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto-oncogene) (RING finger protein 55) | P22681|CBL_HUMAN | 99,631 | 2 |
| Reticulon-3 (Neuroendocrine-specific protein-like 2) (NSP-like protein II) (NSPLII) | O95197|RTN3_HUMAN | 112,596 | 2 |
| A-kinase anchor protein 3 (Protein kinase A-anchoring protein 3) (PRKA3) (A-kinase anchor protein 110 kDa) (AKAP 110) (Sperm oocyte-binding protein) (Fibrousheathin-1) (Fibrousheathin I) (Fibrous sheath protein of 95 kDa) (FSP95) | O75969|AKAP3_HUMAN | 94,720 | 2 |
| U2-associated protein SR140 (140 kDa Ser/Arg-rich domain protein) | O15042|SR140_HUMAN | 118,278 | 2 |
| Ferrochelatase, mitochondrial precursor (EC 4.99.1.1) (Protoheme ferro-lyase) (Heme synthetase) | P22830|HEMH_HUMAN | 47,845 | 2 |
| Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (HMFG) (Breast epithelial antigen BA46) (MFGM) [Contains: Lactadherin short form; Medin] | Q08431|MFGM_HUMAN | 43,105 | 2 |
| REVERSED | REV|Q14678|ANR15 | 147,270 | 2 |
| Probable histone-lysine N-methyltransferase ASH1L (EC 2.1.1.43) (ASH1-like protein) (Absent small and homeotic disks protein 1 homolog) (huASH1) | Q9NR48|ASH1L_HUMAN | 332,778 | 2 |
| Transcription initiation factor IIE subunit beta (TFIIE-beta) (General transcription factor IIE subunit 2) | P29084|T2EB_HUMAN | 33,027 | 2 |
| REVERSED | REV|Q8TDM6|DLG5 | 202,043 | 2 |
| Ubiquitin carboxyl-terminal hydrolase isozyme L5 (EC 3.4.19.12) (UCH-L5) (Ubiquitin thioesterase L5) (Ubiquitin C-terminal hydrolase UCH37) | Q9Y5K5|UCHL5_HUMAN | 37,589 | 2 |
| REVERSED | REV|Q96CN5|LRC45 | 75,935 | 2 |
4.5 Immortalization of human retinal endothelial cells
While the human retinal endothelial cell is the ideal cell for investigating its role in retinal vascular diseases, only limited numbers of cells can be isolated from paired human globes. In our hands, the maximum number is approximately 12 million cells, which is insufficient for follow-up studies of interesting molecules identified by molecular profiling. For this reason, we investigated the possibility of obtaining large numbers of retinal endothelial cells, without compromising the cell phenotype, by immortalizing the cells using LXSN16E6E7 (generously provided by Denise A. Galloway, PhD, Fred Hutchinson Cancer Institute, Seattle, WA). This murine amphotropic retroviral construct encodes the E6 and E7 oncogenes of human papilloma virus 16 and a gene conferring G418 antibiotic resistance (Halbert et al., 1991).
Actively proliferating retinal endothelial cells were exposed for 24 hours to LXSN16E6E7 harvested from PA317 packaging cells, with 5 mg/ml hexadimethrine bromide in some cases, and subsequently cultured in the presence of G418 antibiotic for a minimum of 4 days. Attempted immortalization of retinal endothelial cells from 11 donors was successful in 4 (36%), defined on the basis of growth to greater than 6 passages with 1:3 or greater split at passage. Reasons for failure included low growth rate and overgrowth of non-endothelial cells. Phenotype of the endothelial cells was retained following immortalization, as demonstrated by cobblestone morphology, expression of CD31 and von Willebrand factor (VWF), and capillary-like tube formation on Matrigel (BD Biosciences Discovery Labware, Franklin Lakes, NJ) (Figure 2). Although the success rate of this procedure, per our definition, was less than 50%, when successful, the procedure provided substantial numbers of endothelial cells for use in multiple studies. Human choroidal endothelial cells may be similarly immortalized.
Figure 2.
Photomicrographs of human retinal endothelial cells immortalized by transduction with LXSN16E6E7. Cell retain an endothelial phenotype as indicated by: (A) Cobblestone morphology. Original magnification: 200X; (B) Capillary-like tube formation after 24-hour incubation in 5% CO2 and at 37 °C on Matrigel (BD Biosciences Discovery Labware, Bedford, MA). Original Magnification: 100X. (C–D) Expression of (C) CD31, as detected by mouse monoclonal anti-human CD31 antibody (concentration: 10 μg/ml; clone: JC70A: isotype IgG1κ; BD Pharmingen Biosciences, San Diego, CA) and Alexa Fluor 488-conjugated goat anti-mouse IgG antibody (concentration: 5 μg/ml; Life Technologies, Molecular Probes, Eugene, OR) and (D) VWF, as detected by Alexa Fluor 488-conjugated rabbit polyclonal anti-human VWF antibody (concentration: 16 μg/ml; fraction: IgG; DAKO, Glostrup, Denmark) and Alexa Fluor 488-conjugated anti-rabbit IgG antibody (concentration: 5 μg/ml; Life Technologies, Molecular Probes). Propidium iodide nuclear counterstain (Life Technologies, Molecular Probes). Original magnification: 400X. Paired cell cultures stained with antibody directed against an irrelevant antigen showed no positive staining.
4.6 Supervillin: a retinal endothelial protein identified by molecular profiling
Molecular profiling has the potential to identify many novel proteins with potential relevance to retinal vascular diseases. We illustrate this by presenting work on supervillin, which was detected with relatively high expression in retinal endothelial cells by transcriptomic profiling (Smith et al., 2007). Although originally described in 1997 (Pestonjamasp et al., 1997), at the time of writing, there are only 21 research publications that relate to supervillin. The presence and role(s) of supervillin in endothelium have never been investigated.
Supervillin is a 205 kDa member of the gelsolin superfamily of actin-binding proteins (Silacci et al., 2004). A muscle-specific isoform, termed archvillin, is a closely related 250 KDa protein (Oh et al., 2003). Supervillin contains 6 gelsolin-related repeating units, plus an actin-binding domain/nuclear localization signal (Archer et al., 2004). This protein is expressed in human and mouse, and the structure is well conserved across species (Oh et al., 2003; Pope et al., 1998). Expression is widespread, but varies considerably between tissues; supervillin is expressed at very low levels in neural tissue (Pope et al., 1998). As well as actin, supervillin binds myosin and filamin (Chen et al., 2003), and has been implicated in cytoarchitecture at the plasma membrane and within the nucleus (Pestonjamasp et al., 1997; Wulfkuhle et al., 1999). Supervillin promotes disassembly of cell-substrate focal adhesions, inhibits cell spreading, and promotes invadopodial function (Crowley et al., 2009; Liu et al., 2011; Takizawa et al., 2007; Takizawa et al., 2006). In addition, studies in HeLa cells indicate that supervillin promotes cell motility (Fang et al., 2010). Although the gelsolin family proteins control actin organization, various members have other unrelated functions in cell processes, from controlling apoptosis to regulating phagocytosis (Silacci et al., 2004). Nonetheless, a role for supervillin in endothelial cell function has not yet been investigated, although CapG, another gelsolin family protein, is known to promote endothelial cell motility (Pellieux et al., 2003).
We independently confirmed expression of supervillin in primary human retinal endothelial cells by RT-PCR and western blot of total RNA and protein lysate, respectively, for 5 of 6 donors (Fig. 3A and 3B). One donor either did not produce supervillin or expressed it at a level undetectable by western blot. Reports of supervillin involvement in cell motility suggested a potential role in retinal vessel formation, which we studied using the immortalized human retinal endothelial cells. We observed upregulation of supervillin when endothelial cells were stimulated with human VEGF for up to 24 hours (Fig. 3C). We used the CyQuant NF Cell Proliferation Assay (Life Technologies, Molecular Probes, Eugene, OR), in which cellular DNA is tagged for quantification by fluorescent microplate reader, to examine supervillin’s involvement in cell proliferation, which is a key component of vessel growth. In separate experiments using endothelial isolates derived from 2 human donors, supervillin-targeted siRNA knockdown resulted in significantly less proliferation than non-targeted siRNA (Fig. 3D). In both experiments and shown for 1 donor (Fig. 3E), western blot of protein extracts from siRNA-treated endothelial cells confirmed knockdown by the targeted siRNA by 48 hours after transfection. These findings, taken with our previous observation of high relative expression of supervillin by human retinal endothelial cells, suggest the hypothesis that supervillin may specifically regulate growth of retinal blood vessels and make it an excellent candidate for further study in relation to the pathogenesis of neovascularization in the retinal ischemic vasculopathies.
Figure 3.
Gel images of supervillin (A) RT-PCR product (165 bp) and (B) protein (205 kDa, β-actin at 42 kDa) from primary retinal endothelial cells of 6 human donors (D1–D6). L = ladder./ = no cDNA. (C) Gel image of supervillin protein in human retinal endothelial cells stimulated with VEGF (20 ng/ml, Millipore, Temecula, CA) for 0, 4 and 24 hours. (D) Graph showing proliferation of immortalized retinal endothelial cells from 2 human donors initially plated at 3000 cells/wells in a 96-well plate, 96 hours after transfection with supervillin (SV) or non-targeted (NT) siRNA using Targefect-siRNA transfection kit (Targeting Systems, El Cajon, CA). SV siRNA: sense = 5′-GGCGGUCCCUCAUCAAGAAGC-3′, anti-sense = 5′-UUCUUGAUGAGGGACCGCCCU-3′ (designed using siDirect (Naito et al., 2004)). NT siRNA: sense = 5′-CGCCGACGUUUAACGGAAGCC-3′, anti-sense = 5′-CUUCCGUUAAACGUCGGCGCA-3′. n = 4–8 wells/condition. D = donor. * = p ≤ 0.003. (E) Gel image of supervillin protein in the siRNA-treated human retinal endothelial cells that were used in the proliferation experiment shown in (D – donor 1), 48 hours following transfection.
5. Autoimmune posterior uveitis
5.1 In vivo models
Experimental autoimmune uveoretinitis (EAU) is the standard in vivo model of human autoimmune uveitis. The model was first described in the 1960s as “experimental allergic uveitis” in guinea pigs immunized with homologous retina emulsified in complete Freund’s adjuvant (Wacker and Lipton, 1965). Work conducted mainly at the National Eye Institute in the 1980s (e.g. (Caspi et al., 1988; Gery et al., 1986)) led to the development of models in genetically susceptible inbred mice and rats, and today these species are almost invariably used for studies of EAU. Inflammation is induced by a variety of different protocols, but frequently common is the use of bovine interphotoreceptor retinoid binding protein as the uveitogenic antigen. While uveitis affects both anterior and posterior segments of the eye in mouse EAU, the inflammation is based posteriorly and therefore this is appropriately viewed as a model of autoimmune posterior uveitis. Inflamed eyes demonstrate findings that include retinal vasculitis, retinitis and serous retinal detachments, subretinal neovascularization, choroiditis and vitritis (Caspi et al., 1988). Late in the course of the inflammation, retinal neovascular membranes develop (Chen et al., 2012).
The ocular leukocytic infiltrate is heterogeneous in EAU, including lymphocyte subsets, macrophages and neutrophils; resident cells, such as perivascular macrophages, microglia and retinal pigment epithelium also participate in the inflammation (Kerr et al., 2008). Different subpopulations of CD4+ helper T cell may initiate EAU, including Th1 cells and Th17 cells (Luger and Caspi, 2008; Nussenblatt, 1991). Th1 cells differentiate from naïve CD4+ T cells when interleukin (IL)-12 activates STAT-4 and T-bet, and their signature cytokine is interferon-gamma (IFN-γ). Th17 cells differentiate under the control of RORγt and are characterized by synthesis of IL-17—or IL-17A—as well as other inflammatory cytokines (i.e., IL-17F, IL-6, IL-21, IL-22 and tumor necrosis factor [TNF]-α). A relatively small, but persistent, subset of helper T cells produce IL-17 and IFN-γ, and have been designated Th17/Th1 cells (Shi et al., 2008). A landmark study from the Caspi group (Luger et al., 2008) shows that the effector response in EAU—Th1 or Th17—depends on the mode and the environment of antigen presentation. Once EAU is initiated, macrophages play a major role in the ensuing retinal damage, as convincingly demonstrated by the Dick laboratory (Copland et al., 2007).
To uncover pathogenic mechanisms, the effect of relevant manipulations of the immune system on the severity of EAU is determined. Traditionally, severity of EAU is measured by standard histopathology on tissue sections, usually scoring both cellular infiltration and structural damage to the retina (Dick et al., 1994). More recently, dissection of retinal whole mounts has been combined with intravital or immunohistochemical staining of retinal cells and molecules, which is often imaged by confocal microscopy (Xu et al., 2003a). Various imaging systems provide a means for studying disease severity in vivo. Until recently, fundus imaging in the mouse was not commonly performed due to limited availability and cost of equipment. Five years ago, topical endoscopic imaging (TEFI) was first described. This simple, inexpensive system involves applanation to the mouse cornea of a human tele-otoscope, which is attached to a digital single-lens reflex camera and illuminated with a xenon lamp (Paques et al., 2007). Several groups have used TEFI successfully to image the posterior eye EAU (Copland et al., 2008; Xu et al., 2008), and a severity grading has been developed that describes disease according to retinal infiltrates, optic disc inflammation, retinal vasculitis and structural damage (Xu et al., 2008). TEFI may be adapted for fluorescein angiographic imaging of retinal vessels. Optical coherence tomography has been applied to imaging retinal microstructure in mice with EAU (Oh et al., 2011). Scanning laser confocal microscopy allows imaging of individual leukocyte interactions with the retinal vessel wall following administration of appropriate intravital dyes (Parnaby-Price et al., 1998a; Xu et al., 2002).
5.2 In vitro models
While most studies of retinal endothelial involvement in autoimmune posterior uveitis have been conducted with animal models, some standard immunological methods are readily adapted for the investigation of retinal endothelial properties during inflammation. Simple experiments for evaluation of leukocyte-retinal endothelial cell interactions and retinal endothelial responses to inflammatory molecules may be conducted using confluent cultured cells. Various flow chambers may be used to simulate blood flow across cell monolayers. The Boyden chamber, which is divided into upper and lower chambers by a filter (Boyden, 1962), may be used to study retinal endothelial transmigration of leukocytes when the filter is coated with basement membrane substitute and seeded with endothelial cells.
The Woodruff-Stamper binding assay is helpful to address the concern of phenotypic drift by cultured endothelial cells. Originally this assay was developed to examine the interaction between lymphocytes and the endothelium of high endothelial venules in peripheral lymph nodes (Stamper and Woodruff, 1977). In that setting, a suspension of lymphocytes is overlaid onto cryostat-cut sections of unfixed lymph node, with agitation designed to simulate blood flow. The assay has been modified by replacing lymph node with retina, for observation of leukocyte binding to retinal vascular endothelium (Hill et al., 1997). As well as demonstrating the affinity of a particular vessel for any given cell population, this assay makes it possible to study molecules that potentially mediate an interaction, using specific blocking antibodies.
5.3 Role of retinal endothelium in autoimmune posterior uveitis
Early work on EAU in the Lewis rat suggested important roles for the retinal endothelium in the development of autoimmune posterior uveitis. Immunohistochemical examination of eyes enucleated at the onset of EAU revealed expression of activation markers, class II antigen and fibronectin, on retinal endothelium, which stressed “the importance of the local vasculature in the development of the immune response” (Fujikawa et al., 1987a). By electron microscopy, endothelial cells in retinal venules showed morphologic changes coinciding with maximum tissue inflammation (McMenamin et al., 1992). The changes, which were described as “high endothelial venule-ness”, included increased numbers of cytoplasmic organelles, and increased thickness and irregularity, with deep intercellular clefts that contained lymphocytes and monocytes. These findings implied an essential participation of the retinal endothelium in leukocyte trafficking into the retina in posterior uveitis.
5.4 Leukocyte trafficking across retinal endothelium
Experiments applying scanning laser ophthalmoscopy and confocal microscopy to rat EAU have demonstrated that leukocytes migrate into the posterior segment of the eye via the retinal microvasculature (Parnaby-Price et al., 1998b). T cells also traffic across these vessels in small numbers to conduct immune surveillance of this region (Xu et al., 2003b). The experimental findings in EAU are supported by clinical observations in patients suffering from autoimmune posterior uveitis, who show frequent involvement of the retinal vessels (Sanders and Graham, 1988). One notable difference between human uveitis and EAU is that patients with different forms of posterior uveitis may have involvement of retinal arteries and/or retinal veins, whereas in EAU, leukocyte migration occurs primarily at the level of the post-capillary venule (Xu et al., 2003a). A series of intravital and postmortem observations made in mouse EAU indicate that trafficking of leukocytes across the retinal endothelium requires 3 sets of conditions: up-regulation of adhesive proteins by the endothelium; priming of circulating leukocytes; and conducive hydrodynamic force within the retinal vasculature (Xu et al., 2003a; Xu et al., 2004a).
Leukocytes cross the endothelium by moving either between (paracellular) or through (transcellular) endothelial cells (Engelhardt and Wolburg, 2004). Transcellular migration is considered the rule for organs with extremely tight endothelial junctions, such as the brain (Garrido-Urbani et al., 2008). During transcellular migration, lymphocytic projections or ‘podosomes’ extend into endothelial ‘podoprints’, ultimately creating a transcellular pore (Carman et al., 2007). Electron microscopic studies show that in retina, where endothelial cells are connected by very tight junctions, leukocytes move transcellularly (Greenwood et al., 1994; McMenamin et al., 1992). However, a thorough immunohistochemical examination of retinal whole mounts from mice with EAU (Xu et al., 2005) reveals that leukocyte adhesion to and transmigration across the endothelium triggers disruption and loss of the junctional protein, occludin-1, and astrocyte disensheathment of the affected vessel. The changes occur only in retinal venules and spare other vessels despite close anatomical proximity. In light of these observations, one cannot discount the possibility of paracellular movement of leukocytes through the retinal endothelium in posterior uveitis.
Regardless of the route of transmigration, circulating leukocytes access any tissue as a result of complex molecular interactions with the local vascular endothelium. Chemokines control leukocyte movement through the endothelium, and adhesion molecules tether leukocytes to the endothelium (Ley et al., 2007). Although different leukocyte populations interact similarly with the vascular endothelium, the relative importance of specific adhesion molecules and chemokines appears to vary between the subsets. This is an area of active research in relation to both uveitis and extra-ocular inflammatory diseases.
5.4.1 Adhesion molecules
While the field of leukocyte extravasation continues to progress at fast pace, the literature contains excellent reviews that summarize the molecular events involved in leukocyte migration in general (Chavakis et al., 2009; Ley et al., 2007; Nourshargh et al., 2010) and across the retinal endothelium (Crane and Liversidge, 2008). Families of adhesion molecules that have been studied in relation to posterior uveitis in particular, include the selectins and members of the immunoglobulin superfamily. Selectins on endothelial cells (i.e., P-selectin; CD62P, E-selectin; CD62E) and leukocytes (i.e., L-selectin; CD62L) tether leukocytes to the endothelium via carbohydrate ligands (e.g., P-selectin glycoprotein ligand (PSGL)-1; CD162). This binding readily dissociates, and leukocytes begin to roll along the endothelial surface at low velocity. Chemokines on the endothelial surface activate integrins, resulting in arrest and firm adhesion of leukocytes. Well-described adhesive interactions occur between leukocyte function associated antigen-1 (LFA-1; CD11a/CD18) and very late antigen 4 (VLA-4; CD49d/CD29) on leukocytes, and ICAM-1 (CD54) and VCAM-1 (CD106) on endothelial cells, respectively. Gene expression profiling shows that human retinal endothelial cells constitutively express relatively high levels of ICAM-1, VCAM-1 and E-selectin (Smith et al., 2007), which might predispose the retina to inflammation if leukocyte activation status and shear stress were conducive.
Increased expression of P- and E-selectin on retinal venules is observed in tissue whole-mounts one day prior to leukocyte extravasation in mouse EAU (Xu et al., 2003a). Using the same model, but with adoptive transfer of CD4+ T cells polarized in vitro, a role for these selectins in EAU was confirmed (Xu et al., 2004b). Exposure to antibody directed against P-selectin glycoprotein ligand 1 inhibited rolling and infiltration of Th1-polarized, but not Th2-polarized, cells. Endotoxin-induced uveitis is a rodent model that is widely employed to study anterior uveitis, but retinal involvement is also reported (Ruiz-Moreno et al., 1992). Consistent with the observations made in EAU, in rats injected systemically with lipopolysaccharide, cellular infiltration of the posterior segment was reduced by treatment with the antibodies directed against E- and P-selectin (Suzuma et al., 1998a). Participation of CD44 in posterior uveitis is also described. T cells and endothelium express this transmembrane glycoprotein, which binds L-selectin and E-selectin, and may bind itself via a hyaluronan bridge (Bonder et al., 2006; Dimitroff et al., 2001a; Dimitroff et al., 2001b). Retinal venules demonstrate increased CD44 expression during initiation of EAU in mice (Xu et al., 2004b). The importance of this finding was revealed with the adoptive transfer model (Xu et al., 2004b). Anti-CD44 antibody inhibited rolling and infiltration of CD4+ T cells, and the effect was most apparent for Th1-polarized cells.
Paralleling the changes in expression of E- and P-selectin during mouse EAU, ICAM-1 is detected early and on retinal venules, where leukocytes extravasate (Xu et al., 2003a). In contrast, VCAM-1 is expressed after the onset of leukocyte extravasation and occurs mainly in the retinal arterioles. The timing of ICAM-1 versus VCAM-1 expression is consistent with in vitro observations from an independent laboratory working in the rat (Greenwood et al., 1995). Migration of rat CD4+ T cells across a non-activated retinal endothelial monolayer in vitro was blocked by anti-ICAM-1, but not anti-VCAM-1, antibody. Yet, after endothelial activation by IL-1β, anti-VCAM-1 antibody inhibited migration. Several groups have demonstrated the inhibitory effect of targeting the ICAM-1/LFA-1 interaction in murine EAU (Uchio et al., 1994; Whitcup et al., 1993; Xu et al., 2003a). One study showed that this blockade targeted Th1-polarized cells in preference to Th2-polarized cells (Xu et al., 2004b). We observed that anti-ICAM-1 antibody did not inhibit rat experimental melanin-induced uveitis, a T cell-mediated inflammation affecting primarily the anterior uvea (Smith et al., 2000). In other words, ICAM-1 appears to play a more important role in posterior uveitis. A peptide inhibitor of VLA-4, termed α4-api, has been used to ameliorate mouse EAU, implicating VCAM-1/VLA-4 interactions in leukocyte extravasation (Martin et al., 2005).
Translational research using human cells or tissues supports a role for adhesion molecules in the development of autoimmune posterior uveitis. By immunohistochemistry, E-selectin and ICAM-1 were detected on retinal endothelium of cadaver eyes with no history of disease (Duguid et al., 1992). In an independent report, ICAM-1 was detected on endothelial cells in the posterior segment of eyes from 6 patients with uveitis, but not 7 normal eyes (Whitcup et al., 1992). Another paper described expression of E-selectin, ICAM-1, VCAM-1, and CD44 to be significantly increased in an eye with acute sympathetic ophthalmia, in comparison to an eye in the late, fibrotic stage and several normal eyes (Kuppner et al., 1993). Elevated serum levels of soluble adhesion molecules (i.e., P-selectin, E-selectin and ICAM-1) have been measured in patients with different forms of posterior uveitis, including primary retinal vasculitis, Behcet’s disease, sarcoidosis and idiopathic disease (Arocker-Mettinger et al., 1992; Aydintug et al., 1995; Haznedaroglu et al., 2000; Lee et al., 2007a; Sari et al., 2005; Zaman et al., 1994). Serum levels of ICAM-1 and VCAM-1 drop in patients with Behcet’s disease after immunosuppressive treatment (Verity et al., 1998). In one functional study, antibodies directed against the LFA-1 subunit, CD18, or VLA-4 subunit, CD29, significantly inhibited binding of peripheral blood lymphocytes to retinal endothelium in a modified Woodruff-Stamper assay (Hill et al., 1997). In other work, anti-ICAM-1 antibody reduced adhesion of human CD4+ T cells to a human retinal endothelial cell line by up to 50% (Liversidge et al., 1990).
Gene expression profiling in our laboratory using oligonucleotide arrays has showed that expression of E-selectin, ICAM-1, and VCAM-1 increases significantly in retinal endothelial cells after a 4-hour incubation with the general inflammatory stimulus, lipopolysaccharide (Smith et al., 2007). To follow-up on this result, we used real-time quantitative RT-PCR to investigate the effect of several pro-inflammatory cytokines on the retinal endothelial cell expression of adhesion molecules implicated in posterior uveitis: tumor necrosis factor (TNF)-α, which is a master cytokine, and interferon (IFN)-γ and interleukin (IL)-17, which are prototype Th1 and Th17 cytokines, respectively (Figure 4). In comparison to unstimulated cells, immortalized human retinal endothelial cells treated for 4 hours with TNF-α showed a significant increase in the expression of ICAM-1, VCAM-1, and E-selectin. A similarly timed exposure to IFN-γ significantly up-regulated ICAM-1 expression and trended to increased VCAM-1 expression (p = 0.055), while exposure to IL-17 significantly up-regulated E-selectin expression. We also examined the expression of P-selectin and CD44 and found that none of the selected cytokines significantly altered transcript levels in human retinal endothelial cells, with the exception of IL-17, which induced a modest increase in P-selectin expression.
Figure 4.
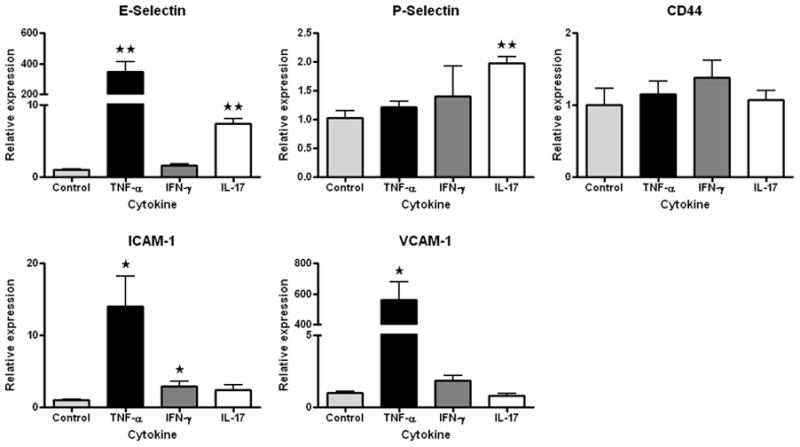
Graphs showing relative expression of E-selectin, P-selectin, ICAM-1, VCAM-1, and CD44 transcript by immortalized human retinal endothelial cells following exposure to one of the following conditions: medium alone; TNF-α (10ng/ml, R&D Systems, Minneapolis, MN); IFN-γ (20ng/ml, R&D Systems); and IL-17 (100ng/ml, R&D Systems). Endothelial cells were cultured to confluence in modified MCDB-131 medium (Sigma-Aldrich, St. Louis, MO) with 2.5% FBS (Hyclone, Logan, UT) and endothelial growth factors (EGM-2 SingleQuots supplement (Clonetics-Lonza, St. Louis, MO), omitting gentamicin, hydrocortisone and serum, at 1:4 dilution), and subsequently incubated with or without cytokine for 4 hours. Total RNA was isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA), and cDNA was synthesized using the iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA). Relative expression of gene products, normalized to GAPDH, was determined by using the Chromo4 Thermocycler and iQ SYBR Green Supermix (both from Bio-Rad Laboratories). Data were analyzed using Chromo4 Opticon Monitor 3 software. Primer sequences appear in Table 2. In all graphs, bars represent mean and error bars represent standard error of mean (n = 3 wells; * = p<0.05, ** = p<0.01, two-tailed Student’s t-test).
5.4.2 Chemokines
Chemokines are a large family of low molecular weight cytokines with multiple chemoattractant activities. These functions include directing chemokine receptor-bearing leukocytes along a concentration gradient towards a site of inflammation, which may involve crossing an endothelium (Rot and von Andrian, 2004). Endothelial cells synthesize an array of chemokines that are expressed on the luminal surface in association with glycosaminoglycans or atypical chemokine receptors; additionally, they trancystose chemokine produced by neighboring non-endothelial cells and similarly express the cytokines (Middleton et al., 2002; Ulvmar et al., 2011). Chemokines also contribute to leukocyte migration by interacting with G protein-coupled chemokine receptors on the endothelial cell surface, activating integrins, and promoting firm adhesion between leukocytes and endothelium, as discussed above. We have reported that human retinal endothelial cells constitutively express relatively high levels of multiple chemokines, including CCL2, CXCL1, CXCL2, CXCL3, CXCL6, CXCL8, CXCL10, CXCL11, and CX3CL1 (Smith et al., 2007). This spectrum of chemokines sets up the endothelium to attract a spectrum of leukocytes, including T cells, B cells, NK cells, monocytes, dendritic cells, and neutrophils. Experiments conducted in mouse EAU indicate the participation of retinal endothelial cell chemokines, including CCL2, CCL3, and CXCL10. Applying immunohistochemistry to ocular cross-sections from mice in the early phase of EAU, one group demonstrated expression of CXCL10 on retinal endothelium (Keino et al., 2003), and a second group independently reported expression of CCL3 and CCL2 on retinal endothelium (Crane et al., 2001). Treatment with anti-CCL3 antibody significantly reduced the inflammatory and structural EAU scores, and leukocytes demonstrated faster velocity, lower rolling efficiency, and less tissue infiltration, in comparison to mice treated with irrelevant antibody (Crane et al., 2001). CCL5 has been detected in inflamed eyes, but in association with T cells, not vascular endothelium (Crane et al., 2001), and consistent with this, CCR5-knock-out mice and mice treated with anti-CCR5 antibody exhibit alterations in T cell emigration into the eye. (Crane et al., 2006; Takeuchi et al., 2005).
Several published studies using cultured human retinal endothelial cells implicate multiple endothelial chemokines in the development of retinal inflammation, covering a broad range of leukocyte specificities and including CCL2, CCL3, and CXCL10. In one report, stimulation of human retinal endothelial cells with the combination of master cytokines, TNF-α and IL-1β, increased levels of all tested chemokines (i.e., CCL2, CCL3, CCL4, CCL5, CXCL1, CXCL5, and CXCL8), according to ELISA (Crane et al., 2000). Another publication focused on CX3CL1—also known as fractalkine—which is a unique protein that functions as an adhesion molecule in its membrane-anchored form and a chemokine when cleaved (Silverman et al., 2003). Immunostaining of human ocular tissue confirmed constitutive expression of CX3CL1 on retinal endothelium. Per ELICA, cultured human retinal endothelial cells up-regulated the chemokine on exposure to TNF-α, IFN-γ, and CD40 ligand, but not IL-13 or IL-17, and down-regulated it in response to IL-4. Involvement of this chemokine in uveitis may depend on species, since mice without functional CX3CR1 develop EAU of equivalent severity to that observed in wild-type animals (Kezic and McMenamin, 2010). In our microarray profiling study, stimulating human retinal endothelial cells with lipopolysaccharide increased gene transcripts of CCL2, CCL8, CCL20, CXCL1, CXCL2, CXCL6, CXCL18, CXCL10, and CX3CL1 (Smith et al., 2007). We were interested to know the effects of TNF-α, IFN-γ, and IL-17 on expression of CXCL10 or CCL20, which specifically attract Th1 or Th17 cells, respectively (Sallusto et al., 1998; Singh et al., 2008), and approached this with real-time quantitative RT-PCR. When immortalized human retinal endothelial cells were stimulated for 4 hours with TNF-α, levels of both CXCL10 and CCL20 transcript increased significantly. As expected, the Th1 cytokine, IFN-γ, significantly increased expression of CXCL10 alone, but surprisingly, stimulation with IL-17 did not impact expression of CCL20 significantly (Figure 5).
Figure 5.
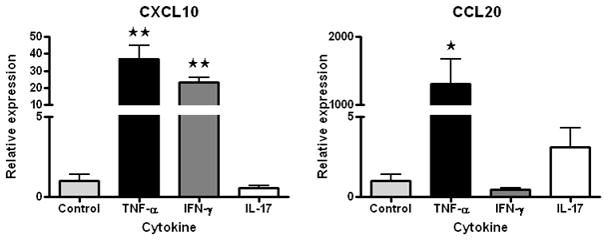
Graphs showing relative expression of CXCL10 and CCL20 transcript by immortalized human retinal endothelial cells following exposure to one of the following conditions: medium alone; TNF-α (10ng/ml); IFN-γ (20ng/ml); and IL-17 (100ng/ml). Experimental conditions and real-time quantitative RT-PCR are described in the Figure 4 legend. Primer sequences appear in Table 2. In both graphs, bars represent mean and error bars represent standard error of mean (n = 3 wells; * = p<0.05, ** = p<0.01, two-tailed Student’s t-test).
5.5 Effects of retinal endothelium on the inflammatory response
In posterior uveitis, retinal endothelial cells respond to a variety of molecular signals, but they also produce molecules that may influence the course of the inflammation, including certain membrane-bound proteins, enzymes and cytokines. Cytokines have long been recognized as important molecular mediators of uveitis (Wakefield and Lloyd, 1992). Interleukin-1β and TNF-α are master cytokines with multiple pro-inflammatory activities that are clearly involved in autoimmune posterior uveitis and its animal model. Patients with posterior uveitis may exhibit elevated intraocular levels of TNF-α and IL-β (Ahn et al., 2006; Franks et al., 1992; Kuiper et al., 2011), and polymorphisms in the genes encoding these cytokines predispose to Behcet’s disease, which commonly manifests as posterior uveitis (Du et al., 2009). Therapies that inhibit the activity of TNF-α have become standard in the management of recalcitrant uveitis involving the posterior segment of the eye (Sfikakis, 2010), and there is recent interest in targeting IL-β for treatment of the disease (Gul et al., 2012). Consistent with the human experience, intravitreal injection of TNF-α or IL-β in rabbits induces retinal perivascular inflammation and breakdown of the blood-retinal barrier (Luna et al., 1997). Both TNF-α and IL-β are expressed at significantly elevated levels in eyes of mice with active EAU (Hashida et al., 2005), and severity of the inflammation is significantly reduced by intervention that lowers the levels of these cytokines (Kitamei et al., 2006).
Human retinal endothelial cells produce TNF-α and IL-β, as well as IL-6, which may be secreted in response to the 2 master cytokines (Smith et al., 2007; Tezel et al., 2001). IL-6 is a pleiotropic cytokine that induces the differentiation and/or activation of various leukocyte subsets (Kishimoto et al., 1995). In our microarray study, human retinal endothelial cells constitutively expressed relatively high levels of IL-6 transcript, and increased this expression almost 20-fold following a brief exposure to lipopolysaccharide (Smith et al., 2007). Expression of pro-inflammatory cytokines by these cells appears to be modulated, since lipopolysaccharide also induced up-regulation of suppressor of cytokine signaling (SOCS)1 (Smith et al., 2007), which down-regulates pro-inflammatory cytokine signaling (Krebs and Hilton, 2001). SOCS1 is highly induced in the retina at the onset of peak of inflammation (Takase et al., 2005), and SOCS1 transgenic rats or mice develop relatively mild EAU by clinical and standard histopathological assessment, with reduced expansion and retinal immigration of CD4+ T cells producing IFN-γ or IL-17 (Yu et al., 2011).
Human retinal endothelial cells also are capable of immunomodulation through the production of IFN-β when the innate immune toll-like receptor 3 is activated, as might occur in retinal vasculitis (Lee et al., 2007b). Following the pioneering work by Koetter and Zierhut, it has become clear that Type I IFNs, including IFN-α and IFN-β, may be highly effective anti-inflammatory agents in various forms of severe uveitis, particularly that due to Behcet’s disease (Becker et al., 2005; Bodaghi et al., 2007; Deuter et al., 2010). Elevated levels of Type 1 interferons have been detected in the serum of patients with different forms of retinal vasculitis (Kotter et al., 2005; Lee et al., 2007b). Studies in rodents suggest these cytokines effect inhibition of uveitis though the reduced production of pro-inflammatory cytokines by cell populations that include CD4+ T cells (Okada et al., 1998a; Okada et al., 1998b; Sun et al., 2011).
Enzymes synthesized by retinal endothelial cells include members of the matrix metalloproteinase (MMP) family (Smith et al., 2007). These zinc-dependent endopeptidases have multiple activities in health and disease, the most studied of which is degradation of various components of the extracellular matrix (McCawley and Matrisian, 2001). As one example, leukocytes use MMPs to enable them to transmigrate the endothelial basement membrane and access target tissue during inflammation. The activity of MMPs is regulated by a family of promiscuous tissue inhibitor of metalloproteinases (TIMPs) that also are synthesized by endothelial cells (Smith et al., 2007).
Early evidence that MMPs were involved in the development of posterior uveitis came from an interventional study in rat EAU with BB-1101, which is a broad-spectrum MMP inhibitor (Wallace et al., 1999). Incidence and clinical severity of inflammation were significantly reduced, and retina was protected per histological examination. Subsequent work revealed similar anti-inflammatory activity of a selective MMP-2/MMP-9 small peptide inhibitor in mouse EAU (El-Shabrawi et al., 2004). High levels of MMP-2 and MMP-9 have been measured in ocular fluids taken from patients with different forms of uveitis (El-Shabrawi et al., 2000), and both MMPs have been detected in human and bovine retinal endothelial cells by RT-PCR (Behzadian et al., 2001; Li et al., 2010). Studies using bovine cells show MMP-2 and MMP-9 probably contribute to breakdown of the blood-retinal barrier in posterior uveitis; purified MMP-2 or MMP-9 increases the permeability of simulated bovine retinal endothelium in a transwell system and effects degradation of junctional occludin (Behzadian et al., 2001; Giebel et al., 2005). Work from our group (Smith et al., 2007) and others (Li et al., 2010) with human retinal endothelial cells suggests roles for additional MMPs and TIMPs in posterior uveitis, including MMP-3, MMP-10, MMP-12, MMP-14, and TIMP1, but additional investigation is required before conclusions can be drawn.
From multiple studies conducted in rodent EAU, backed by research using diseased human eyes, an important role for oxidative stress in posterior uveitis has been recognized (Nguyen and Rao, 2011; Rao, 1990). A key player is nitric oxide, which is produced from arginine and oxygen through the action of inducible nitric oxide synthase (NOS2) or constitutively expressed neuronal (NOS1) and endothelial (NOS3) forms (Lowenstein et al., 1994). Although NOS is not essential for EAU to be manifest, inhibition of the enzyme reduces inflammation in and protects the structure of the retina (Liversidge et al., 2002; Silver et al., 1999; Thillaye-Goldenberg et al., 2000). Immunohistochemistry was used to show NOS2 expression in the retina of eyes from 8 patients with sympathetic ophthalmia, which is a form of uveitis that follows sensitization to retinal antigens (Parikh et al., 2008). Ahead of leukocyte infiltration in rodent EAU, photoreceptors demonstrate NOS2 activity (Rajendram et al., 2007), and subsequently, infiltrating macrophages and resident microglia are major producers (Broderick et al., 2002; Zhang et al., 1999). Retinal endothelium may be a supplementary source of NOS in posterior uveitis. Our shotgun proteomic profiling confirms human retinal endothelial cell expression of NOS3, as well as eNOS interacting protein, which regulates enzymatic activity. Research conducted with bovine cells shows that endothelial cells express NOS2 when stimulated with TNF-α and/or IFN-γ (Chakravarthy et al., 1995). Other enzymes provide protection against oxidative stress, such as superoxide dismutase (SOD), which converts superoxide to hydrogen peroxide, and is up-regulated early in mouse EAU (Saraswathy and Rao, 2009). Our gene expression array profiling detected high levels of SOD2 in human retinal endothelial cells, with significant increase following stimulation with lipopolysaccharide (Smith et al., 2007).
Vascular endothelium, including that in retina, does not constitutively express Class II antigen and thus cannot be considered a professional antigen-presenting cell. However, expression of Class II antigen by Lewis rat retinal endothelial cells is induced during EAU and in culture, leading to speculation of a potential capacity to present antigen to T cells non-professionally (Fujikawa et al., 1987b; Fujikawa et al., 1989). While it is unclear what might occur in vivo, subconfluent rat retinal endothelial cells support retinal antigen-induced T cell proliferation that is potentiated by IFN-γ (Wang et al., 1995). Retinal endothelium also may impact the activation of the infiltrating monocytes that are responsible for much of the damage to local tissue during posterior uveitis. Involvement of CD200 and receptor, CD200R, in this process has been thoroughly investigated in the Dick laboratory (Dick et al., 2003). CD200 is a member of the immunoglobulin superfamily that is expressed on vascular endothelium and neurons within human and rodent retina (Dick et al., 2001). CD200R is expressed on cells of myeloid derivation. Agonist CD200R antibody inhibited IFN-γ-induced production of nitric oxide and IL-6 by mouse bone marrow-derived macrophages (Copland et al., 2007). Consistent with this observation, retinas of CD200 gene-deficient mice exhibited relatively high NOS2 activity early in EAU, and experienced earlier onset and greater severity of inflammation, with prominent apoptosis of ganglion cells and photoreceptors (Broderick et al., 2002). Parallel studies in rats with EAU treated with anti-CD200R antibody yielded similar results (Banerjee and Dick, 2004).
6. Infectious posterior uveitis
6.1 Interaction of the retinal endothelium with infectious pathogens
As highlighted in several editorials and dedicated issues of Thrombosis and Haemostasis (Herwald, 2007; Schnittler and Preissner, 2009; Schnittler and Preissner, 2005), it is only recently that the critical involvement of the vascular endothelium in limiting the access of infectious pathogens to body sites, and the mechanisms by which disease microbes target the endothelium, have been recognized. Mechanisms involve invading or transmigrating host endothelial cells, as well as impacting host cell machinery that may support or inhibit replication of the microbe, or modulate the immune response. Some pathogens utilize a leukocyte taxi to move across an endothelial barrier; this mechanism is considered particularly important in infections of the central nervous system (CNS) because of the presence of the blood-brain barrier. Most endogenous ocular infections that present as posterior uveitis are acquired following systemic infection, and therefore require that the retinal endothelium be breached by the responsible micro-organism. The most common infectious posterior uveitis is caused by Toxoplasma gondii.
6.2 Ocular toxoplasmosis
T. gondii is an obligate intracellular protozoan parasite with an “apical complex,” placing it in phylum Apicomplexan (Black and Boothroyd, 2000). The complex, which defines the anterior end of the parasite, contains specialized secretory granules known as micronemes and rhoptries that are critical for host cell invasion. T. gondii exists in 3 forms: (1) an oocyst, which is produced by sexual reproduction and, after maturation, contains a small number of highly infectious sporozoites; (2) a tachyzoite, the form that characterizes an active infection; and (3) a tissue cyst, which contains multiple relatively dormant bradyzoites (Dubey et al., 1998). Although sexual reproduction takes place only in the intestine of the feline primary host, all other mammals and birds may act as secondary hosts for T. gondii. Most humans are infected following oral ingestion of cysts in pork and lamb, or oocysts released into the environment in the feces of infected cats (Montoya and Liesenfeld, 2004). In a minority of cases, tachyzoites cross the placenta from a newly infected pregnant woman to her unborn child.
After ingestion by a human host and conversion to the tachyzoite form within the intestinal epithelium, T. gondii disseminates throughout the body primarily via the blood stream and lymphatics (Roberts and McLeod, 1999). Tachyzoites proliferate to high numbers within cells of target organs, and lyse cells on egress (Black and Boothroyd, 2000). Although the parasite infects all nucleated cells, in the human host, the infection persists in retina, brain, and muscle. The reasons for the CNS localization of T. gondii are poorly understood, but it is hypothesized that either: (1) the mechanisms responsible for immune privilege promote persistence of the parasite; or (2) tachyzoite entry into the CNS is facilitated (Montoya and Remington, 1997). Certainly immunomodulatory cytokines expressed in the retina promote tachyzoite proliferation (Nagineni et al., 2002). Studies from our group have explored the interaction between T. gondii and the human retinal endothelial cells.
It seems very possible that retinal endothelium is exposed to free T. gondii tachyzoites following infection, since circulating tachyzoites have been detected in the blood of immunocompetent patients (Silveira et al., 2011). We have addressed the possibility that tachyzoites might exhibit specific tropism for retinal endothelial cells by comparing infection in different subtypes of endothelial cells, including retinal endothelial cells, in simple infectivity assays (Smith et al., 2004). Intracellular growth of RH strain T. gondii tachyzoites was measured by uptake of 3H-uracil in cell cultures, since parasites, but not mammalian endothelial cells, incorporate uracil directly through pyrimidine salvage. Results of these studies showed that tachyzoites proliferated more rapidly within human retinal endothelial cells than aortic, human umbilical, and dermal endothelial cells. New work from our group suggests that, in addition to the possibility of entering the retina by infecting endothelial cells, T. gondii tachyzoites may also gain access to the tissue by a different interaction with the retinal vascular endothelium. Using the Boyden chamber assay, we have demonstrated that tachyzoites are capable of transmigrating simulated human retinal endothelium, without significant disruption of the endothelium (Furtado et al., 2012).
Infection or transmigration are initiated by molecular attachment of the T. gondii tachyzoite to the host cell. Tachyzoites bind vascular endothelium in intact human retina under flow conditions (Chipps et al., 2006), and the binding-invasion step occurs relatively quickly for human retinal endothelial cells (Zamora et al., 2008). Sulfated proteoglycans have been implicated as host receptors on non-endothelial cells (Carruthers et al., 2000; Monteiro et al., 1998; Ortega-Barria and Boothroyd, 1999). In this respect, results of our gene expression microarray study might be relevant, showing that human retinal endothelial cells express relatively high levels of multiple sulfotransferases. These include carbohydrate sulfotransferases 1, 2, 11, and 12, N-deacetylase/N-sulfotransferases 1 and 2, sulfotransferase 1B1, and tyrosylprotein sulfotransferase 1 (Smith et al., 2007). High levels of these enzymes may confer high retinal endothelial surface expression of sulfated proteoglycans, which would promote tachyzoite adhesion. Other potential host receptors for T. gondii tachyzoites that are expressed at high levels by human retinal endothelial cells, per our transcriptomic profiling study, include integrin αv and ICAM-1. Tachyzoites bind laminin (Furtado et al., 1992), as does endothelial integrin αv when complexed with integrin β3 (Kramer et al., 1990); a laminin bridge is used by the tachyzoites to attach to human fibroblasts (Furtado et al., 1992). Our work using the Boyden chamber assay reveals that ICAM-1, but not VCAM-1, blockade significantly inhibits T. gondii tachyzoite migration across simulated human retinal vascular endothelium (Furtado et al., 2012). This is consistent with work conducted previously in the Sibley laboratory, showing ICAM-1 binds tachyzoite micronemal protein, MIC2, and facilitates migration across simulated human intestinal epithelium (Barragan et al., 2005).
Infection of retinal endothelial cells with T. gondii results in expression of various molecules that participate in the immune response. We used gene expression microarray profiling to study the response of human retinal endothelial cells to infection with T. gondii tachyzoites. The 62 transcripts that were up-regulated 4 hours following exposure included ICAM-1 and VCAM-1. These cell adhesion molecules are used by lymphocytes, dendritic cells, monocytes, and innate immune cells that are expected to react to a retinal infection. It is possible that this up-regulation also promotes migration of infected leukocytes across retinal endothelium; these cells have been shown to traffic tachyzoites across the blood-brain barrier (Courret et al., 2006; Lambert et al., 2006). Also upregulated were CCL2, CXCL2, and CCL8, which could attract a wide variety of innate and adaptive immune cells to a site of infection. Finally, we noted up-regulation of IL-6, which has the ability to proliferate and activate T cells and differentiate B cells. Retinal endothelial cells might also regulate the reactive inflammation in ocular toxoplasmosis, since the nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (IKBA), was also found to be up-regulated. Results from an independent group’s investigation of the response of rat retinal endothelial cells to infection are very consistent with our own observations (Knight et al., 2005). At 2 hours post-infection, these cells up-regulate chemokines, CCL2, CCL4, CCL5, CXCL1, and CX3CL1, and adhesion molecule, ICAM-1.
6.3 Endogenous endophthalmitis
Our observations of T. gondii tachyzoite interactions with the retinal endothelium leads to questions about how other systematically contracted infectious agents might move into the retina. Endogenous bacterial endophthalmitis is an uncommon, but eye-threatening infection characterized by diffuse intraocular inflammation that is acquired via the retinal circulation (Jackson et al., 2003). Listeria monocytogenes is one of the most common Gram-positive organisms recovered from eyes with endogenous bacterial endophthalmitis, and Escherichia coli is a common Gram-negative pathogen in this disease (Jackson et al., 2003). Endogenous endophthalmitis may also result from fungal infection, most commonly with the yeast Candida albicans (Chhablani, 2011). Although the route of bacterial or fungal migration from the bloodstream to the retina has not been studied, interactions between L. monocytogenes, E. coli, and C. albicans with non-ocular endothelial subpopulations have been investigated.
L. monocytogenes crosses the blood-brain barrier to cause meningitis. Infectivity studies in human brain vascular endothelial cells show bacterial invasion, intracellular replication, and spread to adjacent cells, leaving the endothelium intact (Greiffenberg et al., 1998). At least 2 bacterial proteins mediate host cell surface recognition, invasion, or both: internalin A, recognizing host epithelial (E)-cadherin; and internalin B, recognizing hepatocyte growth factor receptor, Met (Bonazzi et al., 2009). However, the studies implicating E-cadherin have been conducted in non-endothelial cell populations (Bonazzi et al., 2009), and whether Met-internalin B interactions account for brain tropism is undetermined. One group (Greiffenberg et al., 1998) observed no difference in invasion of human brain endothelial cells when internalin A was deleted from L. monocytogenes, but significantly reduced invasion when internalin B was deleted. On the other hand, an independent group (Wilson and Drevets, 1998) observed no difference in binding or invasion of these cells by internalin A-deleted, internalin B-deleted and doubled deleted L. monocytogenes mutants. While the non-observation of a protein in a proteomics experiment does not discount its presence on a cell in vivo, our pilot MudPIT analysis of cultured human retinal endothelial cells did not identify either E-cadherin or Met in these cells.
Similar to our finding of T. gondii tachyzoite trophism for human retinal endothelial cells, human vascular endothelial invasion by E. coli is specific to brain endothelial cells and is not observed for other endothelial subpopulations, including umbilical vein and aortic endothelial cells (Kim, 2000). E. coli trafficking is a complicated process involving multiple host-bacterial interactions (Kim, 2003). Some identified bacterial-human brain endothelial ligands that specifically mediate binding and invasion include OmpA with gp96 homolog (Prasadarao et al., 2003), IbeA with vimentin (Zou et al., 2006), and cytotoxic necotizing factor 1 with 67 kDa laminin receptor (Kim et al., 2005a). According to our shotgun proteomics analysis, human retinal endothelial cells express each of these ligands. Although tested in human umbilical vein cells, an interesting twist on this observation is that FadA adhesion of Fusobacterium nucleatum, which is an oral commensal, binds vascular endothelial (VE)-cadherin (Fardini et al., 2011). As expected for an endothelial junctional protein, we find VE-cadherin in human retinal endothelial cells, although not differentially expressed. Binding translocates VE-cadherin from junctional complexes, and this is associated with increased permeability of the endothelium and increased transmigration of E. coli as evaluated by transwell migration assay. In other words, F. nucleatum, which could readily be introduced into the blood stream during tooth brushing, may enable E. coli passage across endothelium.
In a flow chamber, using immortalized microvascular endothelial monolayers to simulate the vascular wall, different forms of C. albicans bind readily when shear stress is matched to that in capillaries and postcapillary venules (Grubb et al., 2009). Using a transwell system populated with human brain endothelial cells on collagen, transendothelial migration occurs without loss of monolayer integrity, and by transmission electron microscopy, it is possible to observe the fungus in intracellular vacuoles without disruption to the cell (Jong et al., 2001). Studies of human umbilical vein cells indicate that endothelial N (neural)-cadherin may coordinate endocytosis of C. albicans (Phan et al., 2005). By affinity purification, with subsequent protein sequencing and confirmatory immunblotting and immunostaining, neuronal (N)-cadherin was identified as a C. albicans hyphal binding protein. However, when blockade by siRNA almost completely silenced N-cadherin, endocytosis was only reduced by approximately 30%, suggesting the involvement of other ligands. Indirect evidence suggests the possible involvement of αv integrin in fungal adhesion. Human umbilical vein endothelial αvβ3 binds vitronectin when presented in clustered form (Zanetti et al., 1994), and C. albicans germ tubes bind vitronectin (Santoni et al., 2001). Thus it is possible that clusters of vitronectin on the fungus provide a molecular connection to the vascular endothelium. These observations of a role for αv integrin and N-cadherin in C. albicans-endothelial binding are particularly interesting in light of the relatively high levels of expression of these 2 molecules by human retinal endothelial cells, per our gene expression microarray analysis (Smith et al., 2007).
6.4 Viral infections of the retina
A number of systemically acquired viruses infect the human retina, and these presumably reach this tissue via the hematogenous route. On the other hand, studies of viral infection of the retinal vascular endothelium are scant. Cytomegalovirus (CMV) is a herpesvirus that infects most persons across the globe, and causes clinically significant retinal infections in the context of immune compromise. CMV retinitis develops in 20% to 25% of untreated persons infected with human immunodeficiency virus (Holland, 1992), and the condition continues to be a common infection in patients with acquired immune deficiency syndrome (AIDS) despite the introduction of highly active anti-retroviral therapy (Jabs, 2011). In the context of AIDS, CMV has a wide cellular tropism, with generalized organ involvement. On the other hand, in a healthy individual, the virus infects a limited number of cells to establish latency, and endothelial cells are one such cell (Jarvis and Nelson, 2007). The virus demonstrates differential infectivity of different endothelial subpopulations (Jarvis and Nelson, 2007), but although CMV is known to infect human retinal endothelial cells (Rao et al., 1998), there have been no studies that have specifically addressed susceptibility of retinal endothelial cells to infection. Following initial contact with heparin sulfate proteoglycans, interactions with epidermal growth factor receptor (EGFR) (Chan et al., 2009), platelet-derived growth factor receptor (Soroceanu et al., 2008), and various integrins (Feire et al., 2004) mediate virus entry into the cell. Our shotgun proteomics study shows that human retinal endothelial cells express EGFR and multiple integrins.
7. Retinal ischemic vasculopathies
7.1 In vivo models
The classic model of retinal ischemic vasculopathy was developed by in the early 1990s (Smith et al., 1994). In summary, when newborn mouse pups are placed in a hyperoxic environment (i.e., 75% oxygen) from postnatal day (P)7 through P12, and returned to atmospheric air at P12, all the pups develop retinal neovascularization. This peaks at P17 through P21, with subsequent regression of the new vessels and neovascular tufts and development of normal vascular morphology by P24. With the development of standardized methods of quantifying retinal neovascularization and the availability of genetically modified mice, the model of oxygen-induced retinopathy (OIR) has become routine in the investigation of processes of retinal neovascularization and neovascularization in general (Aguilar et al., 2008). Variants of the model in different species have distinct advantages and disadvantages. Having a larger eye than the mouse, the rat is suited for therapeutic experiments that involve intraocular injection (Hartnett, 2010; Penn et al., 1993). However, the method of inducing retinopathy is relatively labor-intensive, requiring alternating high and low oxygen levels every 12 to 24 hours for 7 to 14 days after birth, followed by room air for up to 7 days. Kittens and dog pups have eyes sized closer to those of humans, but these species are expensive to maintain and practically difficult to study (Kremer et al., 1987; McLeod et al., 1998). Retinal detachment, which may be observed in advanced retinal ischemic vasculopathy is a clinically-relevant feature of the canine model. A zebrafish model of hypoxia-induced retinopathy has been developed, which allows for rapid screening of orally administered antiangiogenic agents (Cao et al., 2008).
Standard methods for retinal neovascularization measurement in murine OIR are: (1) fluorescein-dextran angiography to determine the extent of retinal neovascularization by fluorescence microscopy on flat mounted retinas; (2) lectin staining of retinal vasculature to demarcate neovascular tufts on flat mounted retinas; and (3) staining and counting of neovascular endothelial nuclei in whole eye cross sections to determine the number of endothelial cells that have penetrated the vitreal side of the internal limiting membrane (Connor et al., 2009). Digital imaging and analysis software allow the investigator to rapidly and accurately quantify vaso-obliteration, neovascularization, and regression of new vessels in the retina (Connor et al., 2009; Stahl et al., 2009).
Identification of molecules involved in the development of retinal ischemic vasculopathy has provided the basis for other animal models. Given that VEGF is a key stimulus for retinal neovascularization, it is not surprising that intravitreal injections of VEGF or other methods to increase intraocular VEGF have been used in various species to mimic the neovascular phase of the disease in particular (Lebherz et al., 2005; Ohno-Matsui et al., 2002; Rakoczy et al., 2003; Shen et al., 2006; Tolentino et al., 1996). Another example, that also highlights the value of an unbiased profiling approach, is the rat carbonic anhydrase (CA)-1 retinal vascular permeability model. When a proteomics screen of vitreous samples from patients with diabetes mellitus identified elevated CA-1 in the presence of retinopathy, investigators developed a rat model in which intraocular injection of CA-1 resulted in increased retinal vascular permeability and intraretinal edema (Gao et al., 2007).
Although widely used to study ischemic retinal neovascularization, OIR most closely represents retinopathy of prematurity. Additional animal models have been developed to specifically investigate other forms of retinal ischemic vasculopathy. The streptozotocin rat model of diabetic retinopathy is typically induced in rats by intravenous or intraperitoneal injection of streptozotocin (50 – 100 mg/kg body weight) (Papachristodoulou et al., 1976; Sosula et al., 1972). Multiple spontaneous rodent and primate models of human diabetic retinopathy are also reported (e.g. (Johnson et al., 2005; Kim et al., 2005b; Shinohara et al., 2000; Sima et al., 1985)). Although the proliferative stage of diabetic retinopathy is typically not observed, these models are very useful for studying background pathology in the retinal vasculature, including changes in capillary form and capillary leakage. Diabetic dog and cat models offer the advantage of a larger eye and may exhibit retinal changes that mimic the human condition better than rodent models, but are less easily supported (Gardiner et al., 1994; Linsenmeier et al., 1998). Recently, the Kimba transgenic mouse that overexpresses VEGF within photoreceptors was crossed with the Akita mice with hyperglycemic background (Rakoczy et al., 2010). The resulting “Akimba mouse” exhibits severe retinal pathology, including retinal edema and neovascularization. Retinal vein occlusion may be recapitulated in mice or rats by argon laser photocoagulation of retinal veins after intravenous administration of the photolabile dye, rose bengal (Zhang et al., 2007; Zhang et al., 2008). In the central retinal vein occlusion model, the retina becomes ischemic and edematous within minutes, while occlusion of a branch retinal vein results in less severe disease. Laser-induced retinal vessel occlusion has been performed in rabbits, pigs, and nonhuman primates (Ameri et al., 2008; Mendrinos et al., 2011; Virdi and Hayreh, 1982).
7.2 In vitro models
Assays using cultured retinal endothelial cells or retinal tissue have been instrumental for the dissection of the molecular mechanisms of retinal ischemic vasculopathy. Integrity of the retinal endothelial component of the blood-retinal barrier may be readily studied in a transwell system. Confluent endothelial cells are cultured on the filter barrier to establish tight junctions, and subsequently the rate of diffusion of a tagged high molecular weight compound (e.g., rhodamine-isothiocynate labeled 70-kDa dextran) between chambers is measured (Harhaj et al., 2006).
Capillary-like tubule formation is commonly employed to study retinal endothelial cell participation in the formation of blood vessels. This assay has been developed and modified across many laboratories and over many years, using different endothelial cell subpopulations, including human retinal endothelial cells ((Bishop et al., 1999; Crabtree and Subramanian, 2007; Donovan et al., 2001; Kanzawa et al., 1993; Lawley and Kubota, 1989; Sanz et al., 2002) and Figure 2). Endothelial cells are grown on an extracellular matrix substitute (e.g., Matrigel; BD Biosciences, Franklin Lakes, NJ), which may be supplemented with growth factors or various non-endothelial cell populations. Proliferation and migration of the endothelial cells results in formation of capillary-like tube structures over a period of 24 hours, and these may be quantified using digital images and computer software, for length of tubes and number of branch points. Secondary sprouting may also be measured after the original tubules collapse (Castellon et al., 2002). To recognize the impact of the microenvironment on endothelial cell behavior, and the potential for dedifferentiation of endothelial cells in culture, other groups have described technically challenging assays, in which outgrowth of vessels from retinal explants is measured (Im et al., 2005; Knott et al., 1999)
Blood vessel growth requires several activities from the endothelial cell, which may be interrogated separately in different assays. These include endothelial cell proliferation, endothelial cell migration, and degradation of the local basement membrane. To quantify proliferation simply, cells are counted using a hemocytometer with trypan blue stain, either manually or using an automated system such as Countess (Life Technologies, Carlsbad, CA). There are several commercially available indirect methods, such as the CyQuant NF Cell Proliferation Assay (Life Technologies, Molecular Probes), which measures DNA content, and the XTT Cell Proliferation Assay (ATCC, Manassas, VA), which measures cell metabolism. One of the most common endothelial cell migration assays uses the transwell system (Alessandri et al., 1983). Endothelial cells are plated on the upper surface of the filter, and medium in the lower chamber is supplemented with an angiogenic or anti-angiogenic agent under investigation. After incubation, migrated endothelial cells are identified by Wright’s stain, and counted. To examine degradative activity of endothelial cells, one straightforward assay involves seeding cells below confluence on glass cover slips that are pre-coated with fluorescein isothiocyanate-gelatin (Bowden et al., 2001). At the end of the assay, areas of matrix degradation associated with each cell are measured under epifluorescence.
7.3 Role of the retinal endothelium in retinal ischemic vasculopathy
Different types of retinal ischemic vasculopathy are initiated in different pathological settings, but common angiogenic factors are released in response to ischemia, which leads to breakdown of the blood-retinal barrier and/or retinal neovascularization. Over a decade ago, the hypothesis of “hyperglycemia-induced overproduction of superoxide by the mitochondrial electron-transport chain” was first proposed to connect the different pathogenic mechanisms of diabetic vasculopathy including: overactivity of the polyol pathway; high levels of advanced glycation end-products; protein kinase C family activation; and increased hexosamine pathway flux (Brownlee, 2001). Today this theory still holds (Stitt, 2010), although additional interacting mechanisms have been identified (Cheung et al., 2010). Endothelial dysfunction is accompanied by loss of pericytes (Motiejunaite and Kazlauskas, 2008) and basement membrane thickening (Roy et al., 2010), and retinal ischemia ensues. As recently reviewed (Sapieha et al., 2010), within the retina of a premature infant, which lacks normal autoregulation and antioxidants, high levels of oxygen lead to oxidative stress, initiating nitrative stress and lipid peroxidation of cell membranes, and suppress production of growth factors. These events are particularly toxic to endothelial cells. Retinal vascular development ceases, and there is microvascular degeneration, with resultant retinal ischemia.
7.4 Molecular mediators of retinal ischemic vasculopathy
As previously stressed (see section 3.6), VEGF is the key regulator of normal angiogenesis, as well as pathogenic mechanisms involved in retinal ischemic vasculopathy. However, multiple factors have been implicated in the development of, or protection against, this group of diseases. Even an extensive review cannot do justice to the considerable body of literature on the subject. Included in this section is a limited discussion of research relating to VEGF, the angiopoietin-Tie system, and the insulin-like growth factor system, with focus on the participation of the retinal endothelial cell in retinal ischemic vasculopathy.
7.4.1 Vascular endothelial growth factor
More than 2000 reports have been published on subjects relating to VEGF and the retina to date. In all forms of retinal ischemic vasculopathy, leakage from the vasculature and neovascularization occur in large part as the consequence of elevated VEGF levels in the face of hypoxia. Various biological therapies that inhibit these processes act by impeding the action of this potent endothelial growth factor. Per one recent comprehensive review (Otrock et al., 2007), the VEGF family of secreted glycoproteins includes VEGF -A, -B, -C, -D, -E, -F, and placental growth factor. Unqualified VEGF, which has been the primary focus of research and development in relation to retinal ischemic vasculopathy, refers to VEGF-A. The gene encoding VEGF yields alternatively spliced products that are translated into angiogenic isoforms of different lengths, designated by the length of the amino acid chain (i.e., VEGF121, VEGF145, VEGF165, VEGF183, VEGF189, and VEGF206). Of its multiple isoforms, VEGF165 is considered the most relevant to retinal ischemic pathology (Ishida et al., 2003a). Differential splicing also generates anti-angiogenic VEGF isoforms, which are identified by ‘b’ following the amino acid number (i.e., VEGF121b, VEGF165b, and VEGF189b)(Qiu et al., 2009). Vascular endothelial growth factor may be synthesized by multiple populations within human retina, including endothelial cells, pericytes, neurons, and the retinal pigment epithelium (Adamis et al., 1993; Boulton et al., 1998; Lutty et al., 1996; Vidro et al., 2008).
There are at least 5 transmembrane tyrosine kinase receptors for VEGF family members (i.e., VEGFR-1 (Flt-1), VEGFR-2 (KDR), VEGFR-3 (Flt-4), neuropilin (NP)-1, and NP-2) (Otrock et al., 2007). Vascular endothelial growth factor binds VEGFR-1 and -2, and NP-1 and -2; VEGFR-2 activation is primarily responsible for signaling the pro-angiogenic activities of this glycoprotein, but VEGFR-1 binding may influence cell migration (Kanno et al., 2000; Otrock et al., 2007). Immunohistochemical observations of human eyes from 18 cadavers without history of diabetes mellitus and 27 cadavers with such history indicated constitutive expression of VEGFR-1, but not VEGFR-2, in health, and up-regulation of VEGF-R1 and new expression of VEGFR-2 in diabetes for the majority (Witmer et al., 2002).
Consistent with this observation, cultured proliferating human retinal endothelial cells express both VEGFR-1 and -2 (Stewart et al., 2011). Hypoxia induces VEGF receptor expression by retinal endothelial cells. The endothelium of retinal vessels in mouse pups immunostains for VEGFR-2, and when OIR is induced, the number of immunoreactive vessels in avascular areas is significantly increased in comparison to room air-exposed control mice (Suzuma et al., 1998b). Studies from independent groups using bovine retinal endothelial cells show up-regulation of transcript encoding VEFGR-2 (Takagi et al., 1996), or VEGFR-1 and -2 (Brylla et al., 2003) when cells are exposed to hypoxia. The Rhesus macaque retinochoroidal cell line, RF/6A, consistently expresses VEGFR-2 constitutively, and this expression is increased on exposure to low oxygen tension (Ottino et al., 2004).
Hypoxia inducible factor (HIF) is a transcription factor that binds a hypoxia-response element (HRE) within the promoter of the VEGF gene when cellular oxygen is low (Forsythe et al., 1996; Levy et al., 1995). The HIF heterodimer consists of oxygen-sensitive and -insensitive HIF-α and HIF-β subunits. Different HIF-α subunits give rise to different HIFs, although most research to date has focused on HIF-1. In normal oxygen tension, HIF-α is targeted for degradation via proline hydroxylation and von Hippel-Lindau protein-chaperoned ubiquitination; oxygen starvation permits dimerization of HIF-α and HIF-β subunits to generate active HIF (Ivan et al., 2001; Jaakkola et al., 2001). Although HIF is arguably the most important transcriptional regulator of VEGF expression, a microarray gene expression analysis suggests that more than 2% of all genes in human endothelial cells are regulated by HIF-1 (Manalo et al., 2005).
The importance of HIF-1 in the development of retinal ischemic vasculopathy is clear from work with animal models. In OIR, retinal HIF-1α and HIF-2α become markedly elevated within hours of return from hyperoxia to room air in neurons and glial cells, respectively (Mowat et al., 2010; Ozaki et al., 1999). Elegant studies that investigated the association between acute intensive insulin therapy and diabetic retinopathy in the streptozotocin rat, showed such therapy produced breakdown of the blood-retinal barrier, as a consequence of increased VEGF expression, via HIF-1α (Poulaki et al., 2002). Various interventions targeting HIF, including knockout of the relevant VEGF promoter binding site, and administration of digoxin or the small chemical inhibitor, YC-1, stabilize the blood-retinal barrier and/or reduce neovascularization in mouse OIR (DeNiro et al., 2010; Vinores et al., 2006; Yoshida et al., 2010).
While studies using in vivo models have focused on non-endothelial sources of HIF, in vitro research using tissues and cells support a role for retinal endothelial cell HIF in retinal ischemic vasculopathy. Immunostaining showed the presence of HIF-1α in pre-retinal fibrovascular membranes obtained at vitrectomy from 12 patients with proliferative diabetic retinopathy, and this was more obvious in predominantly vascular in comparison to predominantly fibrotic specimens (Lim et al., 2010). Human retinal endothelial cells are capable of producing both HIF-1α and HIF-2α when exposed to hypoxia (DeNiro et al., 2009), and electrophoretic mobility shift assays demonstrate HIF-1 binding to VEGF promoter sequences following hypoxic exposure in bovine retinal endothelial cells (Miyamoto et al., 2002). Under normoxia and hypoxia, YC-1 inhibits proliferation and migration of human retinal endothelial cell, and reduces formation of capillary-like tubules by these cells (DeNiro et al., 2009). The same inhibitor reduces outgrowth of vessels from mouse retinal explants cultured under hypoxic conditions (DeNiro et al., 2009).
Transcription enhancer activator domain family member 4 (TEAD4)—also referred to as transcriptional enhancer factor (TEF)-3 or related transcriptional enhancer factor (RTEF)-1—is a member of the TEA DNA binding family. Interaction of this transcription factor with VEGF was originally described in relation to bovine aortic endothelial cells. In these cells, binding of TEAD4 to a sequence of Sp1 response elements remote from HRE in the VEGF promoter increases the expression of VEGF under hypoxic conditions (Shie et al., 2004). Subsequent publications from the same group have showed a role for TEAD4 in the transcriptional regulation of HIF-1α and VEGF-B in extra-ocular endothelial cell populations (Jin et al., 2011; Xu et al., 2011). One of us (BA) has investigated the expression of TEAD4 in cells that include human retinal endothelial cells, and in mouse OIR and laser-induced central retinal artery occlusion in the Rhesus macaque (Appukuttan et al., 2007; Appukuttan et al., 2012).
Analysis of cultured primary human retinal endothelial cells reveals that TEAD4 message is alternatively spliced to produce multiple isoforms, as well as the full-length TEAD41305, and that certain isoforms are produced only under hypoxic stress. There are differences in the ability of each isoform to regulate the VEGF promoter, and although the majority of the isoforms activate the promoter, TEAD4216 is unique in repressing gene expression whether cells are exposed to normal or low oxygen tension. Different isoforms are detected within the developing mouse retina, and expression similarly varies between mice with OIR and room air-exposed pups. Levels of TEAD4 are also responsive to oxygen tension; 24 hours after occlusion of the central retinal artery, TEAD4 is up-regulated in the Rhesus retina (Appukuttan et al., 2012).
7.4.2 Angiopoietin-Tie system
Recently summarized in relation to diabetic retinopathy (Hammes et al., 2011), the angiopoietin(Ang)-Tie system is essential for development of a normal retinal vasculature and for maintenance of a mature vasculature. On the other hand, perturbations in the system contribute to retinal vascular pathology in a complex manner that depends in part on temporal and environmental factors. Ang1 activates the endothelial cell receptor tyrosine kinase, Tie2. Ang2 also binds Tie2, but with lower affinity and less potency, and may act as an agonist or an antagonist of Tie2 (Maisonpierre et al., 1997; Yuan et al., 2009). As was applied to humans by our shotgun proteomic profiling of human retinal endothelial cells, Tie2 and Ang2, but not Ang1, are expressed by endothelial cells. Retinal pericytes are one source of Ang1 (Wakui et al., 2006). Pericytes also express Tie2 and collaborate with endothelial cells in effecting the vascular changes that are mediated by the Ang-Tie system (Pfister et al., 2008). Ang2 has been implicated in human retinal ischemic vasculopathy. In one study, immunostaining co-localized Ang2 with Tie2 and an endothelial cell marker (i.e., von Willebrand factor) in approximately half of 38 fibrovascular membranes removed from eyes with advanced retinopathy of prematurity (Umeda et al., 2003). Another study showed vitreous levels of Ang2 were significantly elevated in eyes of 30 patients with active proliferative diabetic retinopathy, when compared with levels in eyes of 11 patients with inactive proliferative disease and 18 persons who did not have diabetes mellitus (Watanabe et al., 2005).
Many investigators have addressed the role(s) of the Ang-Tie system in ischemic retinal vasculopathy in in vivo models and in vitro systems. Several studies involving mice OIR show that Ang2 expression increases in the retina during this model, with the level peaking at P17 when retinal neovascularization is active, while Ang1 expression does not vary (Das et al., 2003; Hackett et al., 2000; Oh et al., 1999). In adult rats exposed briefly to hypoxia, up-regulation in the retina of Ang2 protein is also detected (Sivakumar et al., 2008). Despite a difference in kinetics, 2 reports agree that expression of Ang2 message and protein rises in the retina of streptozotocin-injected diabetic rats, but not non-diabetic control rats, while Ang1 levels are not affected (Ohashi et al., 2004; Rangasamy et al., 2011).
Systemic treatment of mouse OIR with a Tie2 antagonist (i.e., muTEK delta Fc) significantly reduces the formation of neovascular capillary fronds in the retina (Das et al., 2003). In the same model, heterozygous or homozygous Ang2 gene-deletion results in less or no retinal neovascularization by histopathological examination at P17, despite a rudimentary retinal vasculature that would be expected to confer marked ischemia (Feng et al., 2009; Hackett et al., 2002). As anticipated from this result, retinal neovascularization is decreased when Ang2 is overexpressed in the retina from P12 to P17 by inducible transgene; however, if Ang2 is overexpressed from P20, when neovascularization begins to regress, this regression is accelerated (Oshima et al., 2005). The findings are consistent with Ang2 having either vasoproliferative or vasoregressive activity depending on the presence in ischemic retina or absence, respectively, of VEGF (Hammes et al., 2011). In mice that transgenically overexpress Ang1 within the retina between P12 and P17, OIR-related neovascularization is reduced (Nambu et al., 2004), although if expression is activated on P20, retinal new vessels regress at normal speed (Nambu et al., 2005). In adult streptozotocin-injected rats, intravitreal injection of Ang1 reduces retinal vascular leakage, while intravitreal injection of Ang2 increases leakage, as demonstrated by extravasation of Evans blue-tagged albumin (Joussen et al., 2002; Rangasamy et al., 2011). Following intravitreal injection of VEGF, retinal Ang1 transgenic mice demonstrate reduced retinal vascular leakage of [3H]-mannitol in comparison to wild-type controls (Nambu et al., 2004).
Research with cultured retinal endothelial cells provides additional insights into the involvement of the Ang-Tie system in retinal ischemic vasculopathy. VEGF stimulates the expression of Ang2, but not Ang1, in cultured bovine retinal endothelial cells (Oh et al., 1999). Hypoxia also induces Ang2 expression in these cells, although by a non-VEGF-directed mechanism since the effect is not neutralized with anti-VEGF antibody. Human retinal endothelial cells cultured in highly concentrated glucose, at a level similar to that in the blood in untreated diabetic mellitus, up-regulate Ang2 expression (Rangasamy et al., 2011). Endogenous Ang2 causes increased permeability of simulated human retinal endothelium in a transwell system (Rangasamy et al., 2011). Coincident with increased permeability, gaps form between endothelial cells, and VE-cadherin staining is lost as the junctional protein is phosphorylated. Studies using porcine retinal endothelial cells and the transwell system show that Ang2 effects on vascular permeability are potentiated by VEGF (Peters et al., 2007). Cultured bovine retinal endothelial cells produce elevated levels of MMP-9 when treated with Ang1 or Ang2 (Das et al., 2003), consistent with the well established role of the MMPs in angiogenesis (Stetler-Stevenson, 1999).
7.4.3 Insulin-like growth factor system
Since insulin-like growth factor (IGF)-1 was first sequenced over 30 years ago (Rinderknecht and Humbel, 1978), a complex IGF system involving ligands, receptors, and binding proteins has been recognized (Firth and Baxter, 2002; Martin and Baxter, 2011). In relation to retinal ischemic vasculopathy, most work in this area has focused on IGF-1. Insulin-like growth factor-1 is produced within the eye and also synthesized extraocularly, primarily by the liver. Thus and although there is debate about the relative importance of the 2 sources, IGF-1 may achieve effects by autocrine, paracrine, and endocrine routes. Soluble IGF-1 binds with highest affinity to cell surface receptor, IGF-1R, to signal activity. The bioavailability of IGF-1 is impacted by the insulin receptors and insulin-like growth factor binding proteins (IGFBPs), which may sequester IGF-1 or concentrate the growth factor near its receptor. Six high affinity IGBPs have been described to date, and intraocular activities independent of IGF-1 are described (Chang et al., 2007; Kielczewski et al., 2011; Lofqvist et al., 2007).
Examination of rodent eyes indicates that IGF-1 may be produced by many retinal cells including photoreceptors and other neuronal populations, glial cells and retinal vascular endothelium (Lofqvist et al., 2009; Sivakumar et al., 2008). Immunostaining of cultured primary human retinal endothelial cells reveals presence of IGF-1, IGF-1R, and IGFBP1 through IGFBP5 (Spoerri et al., 1998), and we detected IGFBP6 transcript in human retinal endothelium in our expression profiling study (Smith et al., 2007). Early work on the involvement of the IGF-1 family in retinal ischemic vasculopathy was nicely summarized in a review from the Grant laboratory (Shaw and Grant, 2004), which has made major contributions on the subject.
Several groups have investigated the expression of IGF-1 and related proteins in human ocular material taken from patients with diabetic retinopathy. Changes in the level of IGF-1 and increase in IGF-1R expression are supportive of a role for the growth factor in retinal ischemic vasculopathy, and apparent contradictions between results of different studies may reflect patient factors such as duration of disease and level of ischemia, as well as methodological differences. Application of immunoassays to vitreous of multiple groups of patients with proliferative diabetic retinopathy—and other forms of retinal ischemia—versus patients without retinal ischemic vasculopathy showed significantly increased concentration of IGF-1 in ischemic eyes (Burgos et al., 2000; Meyer-Schwickerath et al., 1993; Spranger et al., 2000). Epiretinal fibrovascular membranes removed from 5 patients with proliferative diabetic retinopathy at vitrectomy strongly bound radioactive IGF-1, and this was inhibited by addition of non-radioactive IGF-1 to the assay, suggesting expression of IGF-1R in retinal neovascular tissue (Ulbig et al., 1995). By colloidal gold quantitative immunocytochemistry, retinal endothelial cells isolated from eyes of 3 deceased persons with a history of non-proliferative diabetic retinopathy showed significantly reduced levels of IGF-1 in comparison to isolates from 3 donors with no past history of diabetes mellitus (Spoerri et al., 1998). This was associated with increased expression of IGF-1R, and IGFBP1, 2, 3, and 5, and reduced expression of IGFBP4. In a separate study, retina from 6 cadavers with a history of diabetes mellitus under 15 years duration contained significantly less IGF-1 transcript and significantly higher IGF-1R than 6 non-diabetic donors (Gerhardinger et al., 2001).
Despite some differences in results, multiple animal studies have similarly supported an important role for IGF-1 in ischemic retinal vasculopathy. Intravitreal injection of human recombinant IGF-1 in pigs induces retinal microvasculopathy with vascular tortuosity and formation of microaneurysms, accompanied by retinal vascular leakage, as might be seen in non-proliferative diabetic retinopathy (Danis and Bingaman, 1997) Intravitreal injection of IGF-1 in mice induces a breakdown of the blood-retinal barrier, as measured by significant extravasation of [3H]-mannitol into the retina versus kidney and lung (Derevjanik et al., 2002). Mice that transgenically express IGF-1 from retinal neurons develop features of background diabetic retinopathy, including intraretinal microvascular abnormalities, as well as retinal neovascularization, in adulthood (Ruberte et al., 2004). They also experience increased retinal vascular permeability when studied with radioactive and fluorescent trackers, which correlates with altered expression of zonula occludens (ZO)-1 and claudin-1 junctional proteins (Haurigot et al., 2009).
The landmark study from the L.E. Smith group demonstrated that in mouse OIR, an IGF-1R peptide antagonist, JB3, significantly reduced peak retinal neovascularization, in comparison to a control peptide (Smith et al., 1999). Subsequent collaboration between Smith and Hellstrom, involving experimental studies with IGF-1 gene deficient mice and clinical observations made in premature infants, led to an important hypothesis of retinopathy of prematurity (Hellstrom et al., 2001). After premature birth and in the absence of sufficient IGF-1, retinal vascular development initially ceases. Hypoxia results, and VEGF increases to a commiserate degree, which if sufficiently high when the IGF-1 level recovers, promotes neovascularization. Insulin-like growth factor-1 has been measured both at increased and decreased levels in streptozotocin-injected diabetic rats in comparison to non-diabetic controls (Gerhardinger et al., 2001; Lowe et al., 1995; Poulaki et al., 2004). However, systemically administered anti-IGF-1R antibody reduced VEGF levels, and decreased retinal vascular leakage and leukostasis—an early hallmark of diabetic retinopathy discussed in Section 7.5—in these animals(Poulaki et al., 2004).
It is clear that IGF-1 plays an important role in retinal endothelial cell survival. Transcriptomic profiling by both our group (Smith et al., 2007) and another team (Browning et al., 2012) demonstrates relatively high expression of IGF-1R on human retinal endothelial cells. Conditions of highly concentrated glucose and serum starvation induce apoptosis of human retinal endothelial cells, but this effect is partially abrogated when cells are cultured with IGF-1 (Wilson et al., 2001). Mitogen-activated protein kinase signaling is used by VEGF to induce endothelial cell proliferation; following treatment with the IGF-1R peptide antagonist, JB3, bovine retinal endothelial cells do not maximally activate MAP kinase activation in response to VEGF (Smith et al., 1999). Similarly, a minimal level of IGF-1, in conjunction with VEGF, is required to activate the Akt pathway, which is important for cell survival (Hellstrom et al., 2001). Experiments in cultured cells suggest that endothelial cells may be induced to produce IGF-1 when exposed to disease-relevant stimuli. Exposure to low oxygen tension increases IGF-1 production by endothelial cells isolated from bovine or human retina ((Eter et al., 2002) and Table 3).
Table 3.
List of genes for which expression was significantly altered in human retinal endothelial cells following exposure to hypoxia for 48 hours, by PCR array of 84 autophagy-related transcripts. Subtable A shows 21 gene transcripts that were up-regulated in hypoxic retinal endothelial cells. Subtable B shows 11 gene transcripts that were down-regulated in hypoxic retinal endothelial cells. Transcripts are ranked according to fold change in expression as a result of hypoxia, from highest to lowest.
| A. Gene transcripts that were up-regulated in hypoxic retinal endothelial cells | ||||
|---|---|---|---|---|
| Symbol | Description | Role in Autophagy* | P-value | Fold Regulation |
| TNF | Tumor necrosis factor | Regulation: Co-regulator of autophagy and apoptosis | 0.008158 | 2.5747 |
| CTSS | Cathepsin S | Regulation: Autophagy in response to other intracellular signals | 0.000073 | 1.8979 |
| CXCR4 | Chemokine (C-X-C motif) receptor 4 | Regulation: Co-regulator of autophagy and apoptosis | 0.000017 | 1.8666 |
| DRAM1 | DNA-damage regulated autophagy modulator 1 | Gene linking autophagosome to lysosome, Co-regulator of autophagy and apoptosis | 0.000343 | 1.6454 |
| BNIP3 | BCL2/adenovirus E1B 19kDa interacting protein 3 | Regulation: Co-regulator of autophagy and apoptosis | 0.000007 | 1.487 |
| GABARAPL1 | GABA(A) receptor-associated protein like 1 | Gene involved in autophagic vacuole formation | 0.002547 | 1.4544 |
| HGS | Hepatocyte growth factor-regulated tyrosine kinase substrate | Autophagy in response to other Intracellular Signals | 0.007782 | 1.3913 |
| IGF1 | Insulin-like growth factor 1 (somatomedin C) | Co-regulator of autophagy and apoptosis | 0.021975 | 1.3913 |
| TNFSF10 | Tumor necrosis factor (ligand) superfamily, member 10 | Regulation: Co-regulator of autophagy and apoptosis | 0.000046 | 1.3836 |
| TGFB1 | Transforming growth factor, beta 1 | Co-regulator of autophagy and apoptosis, co-regulator of autophagy and the cell cycle | 0.005692 | 1.3797 |
| FAM176A | Family with sequence similarity 176, member A | Gene linking autophagosome to lysosome | 0.00415 | 1.3759 |
| EIF2AK3 | Eukaryotic translation initiation factor 2-alpha kinase 3 | Regulation: Co-regulator of autophagy and apoptosis, autophagy induction by intracellular pathogens | 0.002122 | 1.3721 |
| RAB24 | RAB24, member RAS oncogene family | Gene responsible for protein transport | 0.028074 | 1.3383 |
| TGM2 | Transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) | Regulation: Co-regulator of autophagy and apoptosis | 0.006108 | 1.3089 |
| GAA | Glucosidase, alpha; acid | Autophagy in response to other Intracellular Signals | 0.018178 | 1.2838 |
| ARSA | Arylsulfatase A | Autophagy in response to other Intracellular Signals | 0.008347 | 1.2315 |
| ULK2 | Unc-51-like kinase 2 (C. elegans) | Autophagy in response to other Intracellular Signals | 0.027169 | 1.2179 |
| ATG16L2 | ATG16 autophagy related 16-like 2 (S. cerevisiae) | Gene responsible for protein transport | 0.031784 | 1.1978 |
| APP | Amyloid beta (A4) precursor protein | Regulation: Co-regulator of autophagy and apoptosis | 0.037293 | 1.1895 |
| ATG9A | ATG9 autophagy related 9 homolog A (S. cerevisiae) | Gene involved in autophagic vacuole formation, Gene responsible for protein transport | 0.027697 | 1.178 |
| RB1 | Retinoblastoma 1 | Co-regulator of autophagy and the cell cycle | 0.008745 | 1.1748 |
| B. Gene transcripts that were down-regulated in hypoxic retinal endothelial cells | ||||
|---|---|---|---|---|
| Symbol | Description | Role in autophagy* | P-value | Fold Regulation |
| ATG4A | ATG4 autophagy related 4 homolog A (S. cerevisiae) | Gene involved in autophagic vacuole formation, Gene responsible for targeting to vacuole, protein transport, Protease activity | 0.0001 | −1.6196 |
| ATG9B | ATG9 autophagy related 9 homolog B (S. cerevisiae) | Gene involved in autophagic vacuole formation | 0.002454 | −1.5301 |
| ATG4C | ATG4 autophagy related 4 homolog C (S. cerevisiae) | Gene involved in autophagic vacuole formation, Gene responsible for targeting to vacuole, protein transport, Protease activity | 0.000944 | −1.4475 |
| ATG3 | ATG3 autophagy related 3 homolog (S. cerevisiae) | gene involved in protein ubiquitination | 0.00055 | −1.4375 |
| FAS | Fas (TNF receptor superfamily, member 6) | Regulation: Co-regulator of autophagy and apoptosis | 0.001326 | −1.4177 |
| CTSB | Cathepsin B | Regulation: Co-regulator of autophagy and apoptosis | 0.00375 | −1.3866 |
| HSP90AA1 | Heat shock protein 90kDa alpha (cytosolic), class A member 1 | Chaperone medicated autophagy | 0.003395 | −1.3809 |
| HSPA8 | Heat shock 70kDa protein 8 | Chaperone medicated autophagy | 0.014572 | −1.2938 |
| CDKN2A | Cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | Co-regulator of autophagy and apoptosis, and the cell cycle | 0.000663 | −1.2394 |
| CASP8 | Caspase 8, apoptosis-related cysteine peptidase | Regulation: Co-regulator of autophagy and apoptosis | 0.046233 | −1.2359 |
| HDAC1 | Histone deacetylase 1 | Regulation: Co-regulator of autophagy and apoptosis | 0.039701 | −1.2055 |
Role in autophagy is as listed at www.sabiosciences.com/rt_pcr_product/HTML/PAHS-084A.html
The ability of IGF-1 to promote retinal endothelial cell proliferation has been observed for several species, including the human (Browning et al., 2012; Castellon et al., 2002; Devi et al., 2011). Retinal endothelial cells may be particularly responsive to IGF-1. When the response of proliferating human retinal and choroidal endothelial cells is compared, both cell populations show dose-related growth in the presence of VEGF, but retinal endothelial cells alone grow on exposure to IGF-1 (Browning et al., 2012). Further, bovine retinal endothelial cells proliferate at a higher rate than bovine aortic endothelial cells, when treated with IGF-1 (King et al., 1985). Insulin growth factor-1 is able to induce migration of human retinal endothelial cells through a transwell filter (Grant et al., 1987), and promotes formation of capillary-like tubes and secondary sprouting on basement membrane substitute (Castellon et al., 2002). In experiments using a rat retinal endothelial cell line, IGF-1 reduces transelectrical resistance of retinal endothelial monolayers in transwells, and this change is associated with a fall in the level of VE-cadherin (Devi et al., 2011).
7.5 Endothelial cell death and survival in retinal ischemic vasculopathy
A role for inflammatory mediators is increasingly recognized in the retinal ischemic vasculopathies. Observations of leukocytes accumulating within the retinal vasculature and transmigrating retinal vessels have focused most research in this area on the adhesion molecules expressed by the retinal vascular endothelium (Noda et al., 2012). Groundbreaking work has established a model in which retinal hyperoxia induces increased ICAM-1 expression on retinal endothelial cells (Ishida et al., 2003b). In turn, this promotes the accumulation of leukocytes within the retinal vasculature, triggering retinal endothelial apoptosis. Vascular remodeling or vaso-obliteration follows when this occurs in the context of development or disease, respectively. Histopathological examination of human globes from diabetic patients reveals an increased expression of ICAM-1 on the retinal endothelium (McLeod et al., 1995). Other work has shown the ability of rodent retinal endothelial cells to up-regulate ICAM-1 in vivo following streptozotocin-induced hyperglycemia (Gustavsson et al., 2010) and in response to intravitreal injection of VEGF (Lu et al., 1999), which is abundantly present in ischemic retinal vasculopathy. Accordingly, anti-ICAM-1 antibody significantly limits retinal leukostasis and vascular leakage in rat streptozotocin-induced diabetes (Miyamoto et al., 1999). The finding of similar retinal vascular development after a course of OIR in ICAM-1-gene-deleted mice versus wild-type controls remains to be reconciled (Kociok et al., 2009).
Several other retinal endothelial adhesion molecules (i.e., VCAM-1 and E-selectin) have been investigated in relation to retinal leukostasis, albeit in fewer studies. Blockade of integrin α4, which with integrin β1 forms the VCAM-1 ligand, VLA-4, significantly reduces retinal vascular leakage (Iliaki et al., 2009). The authors were concerned that VCAM-1 was not up-regulated in streptozotocin-induced rat diabetes, and suggested that other endothelial ligands such as fibronectin or MadCAM-1 might be responsible for these effects. However, an independent group demonstrated significant upregulation of VCAM-1 on retinal vascular endothelium in the streptozotocin mouse model, using confocal immunofluorescence (Gustavsson et al., 2010). In bovine retinal endothelial cells, migration was promoted by serum from diabetic patients versus healthy controls, and this effect was blocked by specific antibody targeting VCAM-1 (Olson et al., 1997). In the same study, antiE-selectin antibody also reduced the migration potency of diabetic serum. E-selection was not increased on retinal endothelium in human diabetic eyes (McLeod et al., 1995), but the phenomenon of selectin shedding has been invoked to explain this apparent discrepancy (Noda et al., 2012). Our gene expression profiling data, showing high relative expression of ICAM-1, VCAM-1, and E-selectin by human retinal endothelial cells, points to a predisposition of the human retinal vasculature to leukocyte adhesion, which may increase its susceptibility to vasculopathy (Smith et al., 2007).
Leukostasis in retinal vessels results in retinal endothelial loss and breakdown of the blood-retinal barrier via the Fas/Fas ligand apoptosis pathway. As shown in studies conducted using the rat streptozotocin-induced model (Joussen et al., 2003), a diabetic state induces increased expression of Fas on retinal endothelium. In addition to augmented expression of FasL on leukocytes, this sets the stage for apoptotic programmed cell death. Indeed, TUNEL staining reveals retinal endothelial apoptosis in diabetic rats. Antibody directed against FasL prevents retinal endothelial cell loss and reduces retinal vascular permeability in this model. Increased numbers of pre-retinal endothelial nuclei with reduced TUNEL staining, indicative of increased retinal neovascularization and reduced apoptosis, are consistently observed in OIR in the gld C57BL/6 mouse (Barreiro et al., 2003; Davies et al., 2003). In this mouse, FasL has a Fas binding deficit in comparison to FasL in C57BL/6 wild-type mice
Other pathways that result in activation of caspases and consequent apoptosis of retinal endothelial cells are also described in relation to retinal ischemic vasculopathy. Oxidative stress is a known trigger of apoptosis, via release of cytochrome c from mitochondria. After alloxan injection, rats develop a streptozotocin-like diabetic syndrome and exhibit high levels of lipid peroxides and caspase-3 activity within the retina (Kowluru and Koppolu, 2002). On the other hand, a diet supplemented with anti-oxidants reduces the high lipid peroxide and caspase-3 levels. Changes in cultured bovine retinal endothelial cells exposed to highly concentrated glucose mimic those seen in the diabetic rat retina and are similarly reversed with various antioxidants (Kowluru and Koppolu, 2002). This result has been independently replicated in bovine and human retinal endothelial cells (El-Remessy et al., 2011; Mohr et al., 2002). There is controversy in the literature regarding the ability of hyperglycemia to directly activate apoptosis. Highly pure cultures of human retinal endothelial cells do not produce oxygen reactive species on exposure to high glucose concentration (Busik et al., 2008). However, they do so in response to pro-inflammatory cytokines, TNF-α and Il-1β, and other retinal cells, including Mueller cells and retinal pigment epithelial cells, secrete these cytokines when treated with highly concentrated glucose. When co-cultured with human Muller cells and exposed to high concentration glucose, caspase-3 is activated in human retinal endothelial cells. This finding is consistent with our observation of increased expression of caspase-3 when human retinal endothelial cells are stimulated with lipopolysaccharide, which is a general pro-inflammatory stimulus (Smith et al., 2007).
Work using a rat retinal endothelial cell line strongly suggests that caspase-independent (i.e., mediated by apoptosis-inducing factor) apoptosis may also occur in the context of retinal ischemic vasculopathy (Leal et al., 2009). When the endothelial cells were exposed to hydrogen peroxide, caspase-3 was activated. In contrast, exposure to highly concentrated glucose or the nitric oxide donor, NOC-18, induced changes in nuclear morphology and annexin V binding indicative of apoptosis, but did not activate caspase-3. Instead, translocation of apoptosis inducing factor from mitochondria to nucleus was observed.
The autophagy-lysosomal pathway is a survival strategy that allows a cell to degrade non-essential components and potentially survive exposure to multiple physiological and pathological stresses.(Kroemer et al., 2010) Intracellular vesicles, known as autophagosomes, engulf organelles and proteins, and chaperone them to lysosomes for immediate degradation. Hypoxic stress is one initiator of autophagy that has been studied quite extensively in tumor pathobiology (Schlie et al., 2011). Key mediators of hypoxia-induced autophagy, such as Beclin 1, BCL2/adenovirus E1B 19 kDa protein-interacting protein (BNIP)-3, microtubule-associated protein light chain (MAPLC)-3, and autophagy-related gene (ATG)5, have been identified in cancer cells (Bai et al., 2012; Can et al., 2011; Song et al., 2011; Wu et al., 2011). Both HIF-1α-dependent and -independent mechanisms have been recognized in tumor growth. We investigated the ability of human retinal endothelial cells to develop an autophagy response in the setting of oxygen starvation, since this is a key feature of human retinal ischemic vasculopathy.
To address whether human retinal endothelial cells undergo autophagy under hypoxic conditions, we performed a preliminary screen of genes involved in the lysosomal degradation process. Confluent cultures of immortalized human retinal endothelial cells were incubated in modified MCDB-131 medium with 2% FCS and endothelial growth factors, and exposed to 1% oxygen or room air for 48 hours. Subsequently, total RNA was isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA). Response to hypoxia was determined by RT-PCR for VEGF165. Primer sequences and expected product size appear in Table 2. Quantitative real-time RT-PCR was performed with the Chromo4 Thermocycler and iQ SYBR Green Supermix (both from Bio-Rad Laboratories, Hercules, CA). The RT2 Profiler PCR Array System (SABiosciences, Qiagen, Frederick, MA) was used to profile the expression of 84 autophagy-associated genes in hypoxic versus normoxic human retinal endothelial cells. Total RNA was reverse transcribed using the RT2 First Strand Kit (SA Biosciences, Qiagen), and the autophagy array plate was cycled on the Chromo4 Thermocycler, per the manufacturer’s instructions. Data were analyzed using the RT2 Profiler PCR Array Data Analysis software (SABiosciences, Qiagen).
Table 2.
Primer pairs and product sizes for selected gene transcripts studied in human retinal endothelial cells.
| Gene Transcript | Primer Pair (Product Size)Reference (if applicable) |
|---|---|
| Supervillin | for: 5′-TGGTGTTTGATTTTGGTAGTGAA-3′ rev: 5′-TAAGCGGATTGCATTCTCCA-3′(165 bp) |
| ICAM-1 | for: 5′-TAAGCCAAGAGGAAGGAGCA-3′ rev: 5′-CATATCATCAAGGGTTGGGG-3′ (282 bp)1 |
| VCAM-1 | for: 5′-CGTCTCATTGACTTGCAGCACC-3′ rev: 5′-GTGATCGGCTTCCCAGCCTC-3′ (276 bp) 2 |
| E-selectin | for: 5′-GAGCCTTCAGTGTACCTCATC-3′ rev: 5′-GACAATTCATGTAGCCTCGCTC-3′ (296 bp) 2 |
| P-selectin | for: 5′-GGATTGTTCTGACACTCGTGG-3′ rev: 5′-GAGGTTGGAGCAGTTCATCG-3′ (412 bp) |
| CD44 | for: 5′-ACATCAGTCACAGACCTGCC-3′ rev: 3′-GCAAACTGCAAGAATCAAAGCC-3′ (471 bp)3 |
| CXCL10 | for: 5′-AAGAGATGTCTGAATCCAGAATCGAAGG-3′ rev: 5′-CCTCAGTAGAGCTTACATTATAGTGCCAG-3′ (333 bp) |
| CCL20 | for: 5′-TGCTGTACCAAGAGTTTGCTC-3′ rev: 5′-GATTTGCGCACACAGACAAC-3′ (226 bp) |
| VEGF-165 | for: 5′-ATCTTCAAGCCATCCTGTGTGC-3′ rev: 5′-CAAGGCCCACAGGGATTTTC-3′ (224 bp)4 |
| GAPDH | for: 5′-AGCTGAACGGGAAGCTCACTGG-3′ rev: 5′-GGAGTGGGTGTCGCTGTTGAAGTC-3′ (209 bp)5 |
Lu Y, Fukuda K, Nakamura Y, et al. Inhibitory effect of triptolide on chemokine expression induced by proinflammatory cytokines in human corneal fibroblasts. Invest Ophthalmol Vis Sci 2005; 46: 2346–2352.
Smith JR, Choi D, Chipps TJ, Pan Y, Zamora DO, Davies MH, Babra B, Powers MR, Planck SR, Rosenbaum JT. Unique gene expression profiles of donor-matched human retinal and choroidal vascular endothelial cells. Invest Ophthalmol Vis Sci 2007; 48: 2676–2684.
Liu NP, Roberts WL, Hale LP, et al. Expression of CD44 and variant isoforms in cultured human retinal pigment epithelial cells. Invest Ophthalmol Vis Sci 1997; 38: 2027–2037.
Zygalaki E, Stathopoulou A, Kroupis C, Kaklamanis L, Kyriakides Z, Kremastinos D, Lianidou ES. Real-time reverse transcription-PCR quantification of vascular endothelial growth factor splice variants. Clin Chem 2005; 51: 1518–1520.
Silverman MD, Zamora DO, Pan Y, et al. Constitutive and inflammatory mediator-regulated fractalkine expression in human ocular tissues and cultured cells. Invest Ophthalmol Vis Sci 2003; 44: 1608–1615.
Expression of VEGF165 was significantly increased 6-fold for hypoxic human retinal endothelial cells in comparison to normoxic cells, confirming that the treated cells had experienced hypoxic stress (Figure 6). After normalization to 5 house-keeping genes (i.e., β-2-microglobulin, hypoxanthine phosphoribosyltransferase 1, ribosomal protein L13a, glyceraldehyde-3-phosphate dehydrogenase, and β-actin), analysis of data from quintuple PCR arrays found 21 genes significantly up-regulated and 11 genes significantly down-regulated in human retinal endothelial cells as a result of exposure to hypoxia (Table 3). However, just 4 genes (i.e., CXCR4, Cathepsin S, DNA-damage regulated autophagy modulator 1 and tumor necrosis factor) showed increases that were greater than 1.5-fold, and down-regulated transcripts included ATG3, ATG4A, ATG4C and ATG9B, which are involved in autophagic vacuole formation. While it would be appropriate to continue these studies by evaluating autophagy at the protein level, our findings imply that human retinal endothelial cells have quite limited capacity to activate autophagy, which may translate to a susceptibility to stress. In other words, and consistent with the observation of endothelial cell apoptosis in early diabetic retinopathy, human retinal endothelial cells may be particularly susceptible to the low oxygen tensions that occur in the retinal circulation in diabetes mellitus.
Figure 6.
Graphs showing increased relative expression of VEGF165 transcript by immortalized human retinal endothelial cells following a 48-hour exposure to hypoxia. Endothelial cells were cultured to confluence in modified MCDB-131 medium (Sigma-Aldrich, St. Louis, MO) with 2% FBS (Hyclone, Logan, UT) and endothelial growth factors (EGM-2 SingleQuots supplement (Clonetics-Lonza, St. Louis, MO), omitting gentamicin, hydrocortisone and serum), and subsequently incubated for 48 hours in 1% or room air level oxygen. Total RNA was isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA), and cDNA was synthesized using the iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA). Relative expression of gene products, normalized to GAPDH, was determined by using the Chromo4 Thermocycler and iQ SYBR Green Supermix (both from Bio-Rad Laboratories). Data were analyzed using Chromo4 Opticon Monitor 3 software. Primer sequences appear in Table 2. Bars represent mean and error bars represent standard error of mean (n = 3 reactions; * = p < 0.001, two-tailed Student’s t-test).
8. Future Directions
Further clarification of the molecular phenotype of the human retinal endothelial cell, particularly in the context of disease-relevant stimulations, will increase understanding of the pathogenesis of retinal vascular diseases. As molecular biological technologies, analysis software, and reference databases advance, novel information will continue to flow from this form of research. Profiling by multiple approaches has merit as this provides a more global overview of the molecular phenotype. Concordance in abundance identified by different molecular profiling methods increases confidence in the results. However, discordance may suggest new hypotheses with respect to the cell’s biology. Several systems that have been used to interrogate the molecular phenotype of different extraocular endothelial cells would also provide interesting insights about the retinal endothelial cell. These include: genome-wide methylation profiling to elucidate epigenetic signature (Lagarkova et al., 2010); ligand-receptor screening, which is best approached by phage display (Pasqualini et al., 2010); and metabolomics to characterize the spectrum of small-molecule metabolites (Yuan et al., 2011).
Most relevant to our research is the relatively recent development of RNA-seq—also termed whole transcriptome shotgun sequencing—which is likely to replace gene expression microarray as the preferred form of transcriptome profiling within the next 5 years. While this technique has several applications, arguably the most exciting is the identification and quantitation of transcript isoforms related to alternative splicing or single nucleotide polymorphisms, in addition to known full-length transcripts (Ozsolak and Milos, 2011). The methodology has already been applied to an extraocular endothelial cell population, in a study that compared pulmonary microvascular endothelial cell gene expression in response to thrombin (Zhang et al., 2012). The authors observed significantly increased expression of 150 genes, but also up-regulation of 480 known transcript isoforms and 1,775 previously unknown isoforms, in treated versus control endothelial cells.
Of immediate future interest is the specific molecular profile of retinal endothelial cell subpopulations (i.e., arterial and venous retinal endothelial cells). Vascular endothelial molecular diversity differentiates arterial and venous locations outside the eye. In the largest endothelial profiling study to date, combined data for cells isolated from arteries or veins at multiple sites revealed 817 vein-specific genes and 59 artery-specific genes (Chi et al., 2003a). Evidence from clinical and experimental posterior uveitis indicates that such diversity also exists within the eye. The pattern of arterial versus venous involvement varies according to the specific etiology of the uveitis, a feature often used diagnostically (Sanders and Graham, 1988). Studies in mice with EAU, which is characterized by leukocyte immigration at the post-capillary venule, indicate that retinal arterioles and venules differ in terms of cell adhesion molecule expression at the onset of the disease (Xu et al., 2003a).
Arteries can be distinguished from veins on the basis of expression of ephrin-B2 and EphB4 respectively (Wang et al., 1998). This distinction is present at the fetal onset of angiogenesis, when these molecules are important in determining vascular cell fate(Harvey and Oliver, 2004), and persists into adult life (Shin et al., 2001). Capillaries also express ephrin-B2 and EphB4, establishing arterial versus venous identities within the microcirculation (Shin et al., 2001). We have used immunohistochemistry to show that human retinal vessels exist as subpopulations of ephrin-B2-positive endothelial cells, consistent with arterial phenotype, and ephrin-B2-negative endothelial cells, indicative of venous identity (Figure 7). Thus ephrin-B2 expression could be used as the basis for flow cytometric separation of freshly isolated human retinal endothelial cells prior to culture. Alternatively, laser capture microdissection could be used to isolate arterial and venous endothelium from intact human retina, following immunodetection of ephrin-B2, to provide material for transcriptomic, proteomic, and other analyses.
Figure 7.
Photomicrographs of fresh frozen retinal sections from the same human donor immunostained with rabbit anti-human ephrin-B2 polyclonal antibody (1:50 dilution; Santa Cruz Biotechnology, Santa Cruz, CA) diluted 1:50 or rabbit IgG similarly diluted as negative control. Specific staining was identified by Alexa Fluor 594-labelled secondary antibody (1:400 dilution; Invitrogen). (A) Retinal vascular channel expressing ephrin-B2; (B) Absence of positive staining in negative control. Original Magnification: 400X; (C) Ephrin-B2-positive arterial channel (arrowhead) adjacent to ephrin-B2-negative venous channel (arrow).
The ideal treatment for a retinal vascular disease would be a drug that inhibited a key pathogenic mechanism, but had no impact on physiological processes. While posterior uveitis and retinal ischemic vasculopathy involve multiple cell populations, the retinal endothelial cell is a central player. Thus, targeting the involvement of the retinal endothelial cell would be a logical therapeutic approach in these diseases. Inventive treatment strategies directed against the retinal endothelium have already been described in animal models. A multimerized endothelin enhancer upstream of the human Cdc6 promoter chimera effectively delivers an IGF-1R targeted ribozyme to proliferating endothelial cells in vitro (Luz-Madrigal et al., 2007). When injected intravitreally, the ribozyme reduces retinal neovascularization in mouse OIR and laser-induced retinopathy, without adversely affecting the normal retinal vasculature. A baculoviral construct with GFP reporter expression controlled by VEGF-R1 promoter shows expression limited to the retinal endothelium when injected intravitreally in rats (Shaw et al., 2006). This system has not been used to deliver therapeutic proteins to the endothelium, but could readily be adapted for that purpose. The molecular composition of the human retinal endothelial cell in conditions relevant to health and disease provides a strong basis for the development of effective biologic therapies for posterior uveitis and retinal ischemic vasculopathy.
Acknowledgments
This work was supported by the National Eye Institute/National Institutes of Health (R01 EY019875, R01 EY019042, and P30 EY10572), the International Retina Research Foundation; and Research to Prevent Blindness (unrestricted grant to Casey Eye Institute).
The authors wish to thank Ashley Moses, PhD (Vaccine and Gene Therapy Institute, Oregon Health & Science University, Portland, Oregon) for expert advice regarding endothelial cell immortalization, and Sierra Binek for technical assistance. The LXSN16E6E7 viral construct was the generous gift of Denise A. Galloway, PhD (Fred Hutchinson Cancer Institute, Seattle, WA).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adamis AP, Shima DT, Yeo KT, Yeo TK, Brown LF, Berse B, D’Amore PA, Folkman J. Synthesis and secretion of vascular permeability factor/vascular endothelial growth factor by human retinal pigment epithelial cells. Biochem Biophys Res Commun. 1993;193:631–638. doi: 10.1006/bbrc.1993.1671. [DOI] [PubMed] [Google Scholar]
- Aguilar E, Dorrell MI, Friedlander D, Jacobson RA, Johnson A, Marchetti V, Moreno SK, Ritter MR, Friedlander M. Chapter 6. Ocular models of angiogenesis. Methods Enzymol. 2008;444:115–158. doi: 10.1016/S0076-6879(08)02806-1. [DOI] [PubMed] [Google Scholar]
- Ahn JK, Yu HG, Chung H, Park YG. Intraocular cytokine environment in active Behcet uveitis. American journal of ophthalmology. 2006;142:429–434. doi: 10.1016/j.ajo.2006.04.016. [DOI] [PubMed] [Google Scholar]
- Aiello LP, Avery RL, Arrigg PG, Keyt BA, Jampel HD, Shah ST, Pasquale LR, Thieme H, Iwamoto MA, Park JE, et al. Vascular endothelial growth factor in ocular fluid of patients with diabetic retinopathy and other retinal disorders. The New England journal of medicine. 1994;331:1480–1487. doi: 10.1056/NEJM199412013312203. [DOI] [PubMed] [Google Scholar]
- Aird WC. Mechanisms of endothelial cell heterogeneity in health and disease. Circ Res. 2006;98:159–162. doi: 10.1161/01.RES.0000204553.32549.a7. [DOI] [PubMed] [Google Scholar]
- Alessandri G, Raju K, Gullino PM. Mobilization of capillary endothelium in vitro induced by effectors of angiogenesis in vivo. Cancer research. 1983;43:1790–1797. [PubMed] [Google Scholar]
- Ameri H, Ratanapakorn T, Rao NA, Chader GJ, Humayun MS. Natural course of experimental retinal vein occlusion in rabbit; arterial occlusion following venous photothrombosis. Graefe’s archive for clinical and experimental ophthalmology = Albrecht von Graefes Archiv fur klinische und experimentelle. Ophthalmologie. 2008;246:1429–1439. doi: 10.1007/s00417-008-0878-4. [DOI] [PubMed] [Google Scholar]
- Appukuttan B, McFarland TJ, Davies MH, Atchaneeyasakul LO, Zhang Y, Babra B, Pan Y, Rosenbaum JT, Acott T, Powers MR, Stout JT. Identification of novel alternatively spliced isoforms of RTEF-1 within human ocular vascular endothelial cells and murine retina. Invest Ophthalmol Vis Sci. 2007;48:3775–3782. doi: 10.1167/iovs.06-1172. [DOI] [PubMed] [Google Scholar]
- Appukuttan B, McFarland TJ, Stempel A, Kassem JB, Hartzell M, Zhang Y, Bond D, West K, Wilson R, Stout A, Pan Y, Ilias H, Robertson K, Klein ML, Wilson D, Smith JR, Stout JT. The Related Transcriptional Enhancer Factor-1 Isoform, TEAD4(216), Can Repress Vascular Endothelial Growth Factor Expression in Mammalian Cells. PLoS One. 2012;7:e31260. doi: 10.1371/journal.pone.0031260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer SK, Behm CA, Claudianos C, Campbell HD. The flightless I protein and the gelsolin family in nuclear hormone receptor-mediated signalling. Biochem Soc Trans. 2004;32:940–942. doi: 10.1042/BST0320940. [DOI] [PubMed] [Google Scholar]
- Arocker-Mettinger E, Steurer-Georgiew L, Steurer M, Huber-Spitzy V, Hoelzl E, Grabner G, Kuchar A. Circulating ICAM-1 levels in serum of uveitis patients. Curr Eye Res. 1992;11(Suppl):161–166. doi: 10.3109/02713689208999527. [DOI] [PubMed] [Google Scholar]
- Autar A, Faber N, Krabbendam-Peters I, Van Duin R, GE, Van der Giessen WJ, Van Beusekom HM. Endothelial cell behaviour in response to des drugs shows significant species differences: a comparison of human, swine and rabbit endothelium. Circulation. 2011;124:A12675. [Google Scholar]
- Aydintug AO, Tokgoz G, Ozoran K, Duzgun N, Gurler A, Tutkak H. Elevated levels of soluble intercellular adhesion molecule-1 correlate with disease activity in Behcet’s disease. Rheumatology international. 1995;15:75–78. doi: 10.1007/BF00262712. [DOI] [PubMed] [Google Scholar]
- Bai H, Inoue J, Kawano T, Inazawa J. A transcriptional variant of the LC3A gene is involved in autophagy and frequently inactivated in human cancers. Oncogene. 2012 doi: 10.1038/onc.2011.613. [DOI] [PubMed] [Google Scholar]
- Baker KB, Spurrier NJ, Watkins AS, Smith JR, Rosenbaum JT. Retention time for corticosteroid-sparing systemic immunosuppressive agents in patients with inflammatory eye disease. Br J Ophthalmol. 2006;90:1481–1485. doi: 10.1136/bjo.2006.097998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee D, Dick AD. Blocking CD200-CD200 receptor axis augments NOS-2 expression and aggravates experimental autoimmune uveoretinitis in Lewis rats. Ocular immunology and inflammation. 2004;12:115–125. doi: 10.1080/09273940490895326. [DOI] [PubMed] [Google Scholar]
- Baron RM, Lopez-Guzman S, Riascos DF, Macias AA, Layne MD, Cheng G, Harris C, Chung SW, Reeves R, von Andrian UH, Perrella MA. Distamycin A inhibits HMGA1-binding to the P-selectin promoter and attenuates lung and liver inflammation during murine endotoxemia. PLoS One. 2010;5:e10656. doi: 10.1371/journal.pone.0010656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barragan A, Brossier F, Sibley LD. Transepithelial migration of Toxoplasma gondii involves an interaction of intercellular adhesion molecule 1 (ICAM-1) with the parasite adhesin MIC2. Cell Microbiol. 2005;7:561–568. doi: 10.1111/j.1462-5822.2005.00486.x. [DOI] [PubMed] [Google Scholar]
- Barreiro R, Schadlu R, Herndon J, Kaplan HJ, Ferguson TA. The role of Fas-FasL in the development and treatment of ischemic retinopathy. Invest Ophthalmol Vis Sci. 2003;44:1282–1286. doi: 10.1167/iovs.02-0478. [DOI] [PubMed] [Google Scholar]
- Becker MD, Heiligenhaus A, Hudde T, Storch-Hagenlocher B, Wildemann B, Barisani-Asenbauer T, Thimm C, Stubiger N, Trieschmann M, Fiehn C. Interferon as a treatment for uveitis associated with multiple sclerosis. Br J Ophthalmol. 2005;89:1254–1257. doi: 10.1136/bjo.2004.061119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadian MA, Wang XL, Windsor LJ, Ghaly N, Caldwell RB. TGF-beta increases retinal endothelial cell permeability by increasing MMP-9: possible role of glial cells in endothelial barrier function. Invest Ophthalmol Vis Sci. 2001;42:853–859. [PubMed] [Google Scholar]
- Benjamin LE, Hemo I, Keshet E. A plasticity window for blood vessel remodelling is defined by pericyte coverage of the preformed endothelial network and is regulated by PDGF-B and VEGF. Development. 1998;125:1591–1598. doi: 10.1242/dev.125.9.1591. [DOI] [PubMed] [Google Scholar]
- Bishop ET, Bell GT, Bloor S, Broom IJ, Hendry NF, Wheatley DN. An in vitro model of angiogenesis: basic features. Angiogenesis. 1999;3:335–344. doi: 10.1023/a:1026546219962. [DOI] [PubMed] [Google Scholar]
- Black MW, Boothroyd JC. Lytic cycle of Toxoplasma gondii. Microbiol Mol Biol Rev. 2000;64:607–623. doi: 10.1128/mmbr.64.3.607-623.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch-Michel E, Nussenblatt RB. International Uveitis Study Group recommendations for the evaluation of intraocular inflammatory disease. American journal of ophthalmology. 1987;103:234–235. doi: 10.1016/s0002-9394(14)74235-7. [DOI] [PubMed] [Google Scholar]
- Bodaghi B, Gendron G, Wechsler B, Terrada C, Cassoux N, Huong du LT, Lemaitre C, Fradeau C, LeHoang P, Piette JC. Efficacy of interferon alpha in the treatment of refractory and sight threatening uveitis: a retrospective monocentric study of 45 patients. Br J Ophthalmol. 2007;91:335–339. doi: 10.1136/bjo.2006.101550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonazzi M, Lecuit M, Cossart P. Listeria monocytogenes internalin and E-cadherin: from structure to pathogenesis. Cell Microbiol. 2009;11:693–702. doi: 10.1111/j.1462-5822.2009.01293.x. [DOI] [PubMed] [Google Scholar]
- Bonder CS, Clark SR, Norman MU, Johnson P, Kubes P. Use of CD44 by CD4+ Th1 and Th2 lymphocytes to roll and adhere. Blood. 2006;107:4798–4806. doi: 10.1182/blood-2005-09-3581. [DOI] [PubMed] [Google Scholar]
- Boulton M, Foreman D, Williams G, McLeod D. VEGF localisation in diabetic retinopathy. Br J Ophthalmol. 1998;82:561–568. doi: 10.1136/bjo.82.5.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden ET, Coopman PJ, Mueller SC. Invadopodia: unique methods for measurement of extracellular matrix degradation in vitro. Methods Cell Biol. 2001;63:613–627. doi: 10.1016/s0091-679x(01)63033-4. [DOI] [PubMed] [Google Scholar]
- Boyden S. The chemotactic effect of mixtures of antibody and antigen on polymorphonuclear leucocytes. The Journal of experimental medicine. 1962;115:453–466. doi: 10.1084/jem.115.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick C, Hoek RM, Forrester JV, Liversidge J, Sedgwick JD, Dick AD. Constitutive retinal CD200 expression regulates resident microglia and activation state of inflammatory cells during experimental autoimmune uveoretinitis. Am J Pathol. 2002;161:1669–1677. doi: 10.1016/S0002-9440(10)64444-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning AC, Halligan EP, Stewart EA, Swan DC, Dove R, Samaranayake GJ, Amoaku WM. Comparative gene expression profiling of human umbilical vein endothelial cells and ocular vascular endothelial cells. Br J Ophthalmol. 2012;96:128–132. doi: 10.1136/bjophthalmol-2011-300572. [DOI] [PubMed] [Google Scholar]
- Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature. 2001;414:813–820. doi: 10.1038/414813a. [DOI] [PubMed] [Google Scholar]
- Brylla E, Tscheudschilsuren G, Santos AN, Nieber K, Spanel-Borowski K, Aust G. Differences between retinal and choroidal microvascular endothelial cells (MVECs) under normal and hypoxic conditions. Exp Eye Res. 2003;77:527–535. doi: 10.1016/s0014-4835(03)00219-7. [DOI] [PubMed] [Google Scholar]
- Burgos R, Mateo C, Canton A, Hernandez C, Mesa J, Simo R. Vitreous levels of IGF-I, IGF binding protein 1, and IGF binding protein 3 in proliferative diabetic retinopathy: a case-control study. Diabetes Care. 2000;23:80–83. doi: 10.2337/diacare.23.1.80. [DOI] [PubMed] [Google Scholar]
- Busik JV, Mohr S, Grant MB. Hyperglycemia-induced reactive oxygen species toxicity to endothelial cells is dependent on paracrine mediators. Diabetes. 2008;57:1952–1965. doi: 10.2337/db07-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Can G, Ekiz HA, Baran Y. Imatinib induces autophagy through BECLIN-1 and ATG5 genes in chronic myeloid leukemia cells. Hematology. 2011;16:95–99. doi: 10.1179/102453311X12902908412039. [DOI] [PubMed] [Google Scholar]
- Cao R, Jensen LD, Soll I, Hauptmann G, Cao Y. Hypoxia-induced retinal angiogenesis in zebrafish as a model to study retinopathy. PLoS One. 2008;3:e2748. doi: 10.1371/journal.pone.0002748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman CV, Sage PT, Sciuto TE, de la Fuente MA, Geha RS, Ochs HD, Dvorak HF, Dvorak AM, Springer TA. Transcellular diapedesis is initiated by invasive podosomes. Immunity. 2007;26:784–797. doi: 10.1016/j.immuni.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carruthers VB, Hakansson S, Giddings OK, Sibley LD. Toxoplasma gondii uses sulfated proteoglycans for substrate and host cell attachment. Infect Immun. 2000;68:4005–4011. doi: 10.1128/iai.68.7.4005-4011.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi RR, Roberge FG, Chan CC, Wiggert B, Chader GJ, Rozenszajn LA, Lando Z, Nussenblatt RB. A new model of autoimmune disease. Experimental autoimmune uveoretinitis induced in mice with two different retinal antigens. J Immunol. 1988;140:1490–1495. [PubMed] [Google Scholar]
- Castellon R, Hamdi HK, Sacerio I, Aoki AM, Kenney MC, Ljubimov AV. Effects of angiogenic growth factor combinations on retinal endothelial cells. Exp Eye Res. 2002;74:523–535. doi: 10.1006/exer.2001.1161. [DOI] [PubMed] [Google Scholar]
- Chakravarthy U, Stitt AW, McNally J, Bailie JR, Hoey EM, Duprex P. Nitric oxide synthase activity and expression in retinal capillary endothelial cells and pericytes. Current eye research. 1995;14:285–294. doi: 10.3109/02713689509033528. [DOI] [PubMed] [Google Scholar]
- Chan G, Nogalski MT, Yurochko AD. Activation of EGFR on monocytes is required for human cytomegalovirus entry and mediates cellular motility. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:22369–22374. doi: 10.1073/pnas.0908787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan-Ling T, McLeod DS, Hughes S, Baxter L, Chu Y, Hasegawa T, Lutty GA. Astrocyte-endothelial cell relationships during human retinal vascular development. Invest Ophthalmol Vis Sci. 2004;45:2020–2032. doi: 10.1167/iovs.03-1169. [DOI] [PubMed] [Google Scholar]
- Chang KH, Chan-Ling T, McFarland EL, Afzal A, Pan H, Baxter LC, Shaw LC, Caballero S, Sengupta N, Li Calzi S, Sullivan SM, Grant MB. IGF binding protein-3 regulates hematopoietic stem cell and endothelial precursor cell function during vascular development. Proc Natl Acad Sci U S A. 2007;104:10595–10600. doi: 10.1073/pnas.0702072104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavakis E, Choi EY, Chavakis T. Novel aspects in the regulation of the leukocyte adhesion cascade. Thromb Haemost. 2009;102:191–197. doi: 10.1160/TH08-12-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Copland DA, Zhao J, Liu J, Forrester JV, Dick AD, Xu H. Persistent inflammation subverts thrombospondin-1-induced regulation of retinal angiogenesis and is driven by CCR2 ligation. Am J Pathol. 2012;180:235–245. doi: 10.1016/j.ajpath.2011.09.020. [DOI] [PubMed] [Google Scholar]
- Chen Y, Takizawa N, Crowley JL, Oh SW, Gatto CL, Kambara T, Sato O, Li XD, Ikebe M, Luna EJ. F-actin and myosin II binding domains in supervillin. J Biol Chem. 2003;278:46094–46106. doi: 10.1074/jbc.M305311200. [DOI] [PubMed] [Google Scholar]
- Cheung N, Mitchell P, Wong TY. Diabetic retinopathy. Lancet. 2010;376:124–136. doi: 10.1016/S0140-6736(09)62124-3. [DOI] [PubMed] [Google Scholar]
- Chhablani J. Fungal endophthalmitis. Expert Rev Anti Infect Ther. 2011;9:1191–1201. doi: 10.1586/eri.11.139. [DOI] [PubMed] [Google Scholar]
- Chi JT, Chang HY, Haraldsen G, Jahnsen FL, Troyanskaya OG, Chang DS, Wang Z, Rockson SG, van de Rijn M, Botstein D, Brown PO. Endothelial cell diversity revealed by global expression profiling. Proc Natl Acad Sci U S A. 2003a;100:10623–10628. doi: 10.1073/pnas.1434429100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi JT, Chang HY, Haraldsen G, Jahnsen FL, Troyanskaya OG, Chang DS, Wang Z, Rockson SG, van de Rijn M, Botstein D, Brown PO. Endothelial cell diversity revealed by global expression profiling. Proceedings of the National Academy of Sciences of the United States of America. 2003b;100:10623–10628. doi: 10.1073/pnas.1434429100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipps TJ, Streeter PR, Franc DT, Neumeyer K, Planck SR, Rosenbaum JT, Smith JR. Modification of the Woodruff-Stamper assay demonstrates binding of Toxoplasma gondii tachyzoites to retinal vascular endothelium. J Immunol Methods. 2006;312:209–213. doi: 10.1016/j.jim.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Choi D, Appukuttan B, Binek SJ, Planck SR, Stout JT, Rosenbaum JT, Smith JR. Prediction of Cis-Regulatory Elements Controlling Genes Differentially Expressed by Retinal and Choroidal Vascular Endothelial Cells. J Ocul Biol Dis Infor. 2008;1:37–45. doi: 10.1007/s12177-008-9007-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo JK, Seebach JD, Nickeleit V, Shimizu A, Lei H, Sachs DH, Madsen JC. Species differences in the expression of major histocompatibility complex class II antigens on coronary artery endothelium: implications for cell-mediated xenoreactivity. Transplantation. 1997;64:1315–1322. doi: 10.1097/00007890-199711150-00014. [DOI] [PubMed] [Google Scholar]
- Coleman HR, Chan CC, Ferris FL, 3rd, Chew EY. Age-related macular degeneration. Lancet. 2008;372:1835–1845. doi: 10.1016/S0140-6736(08)61759-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin HB. Endothelial cell lined lymphatics in the vascularized rabbit cornea. Invest Ophthalmol. 1966;5:337–354. [PubMed] [Google Scholar]
- Congdon NG, Friedman DS, Lietman T. Important causes of visual impairment in the world today. Jama. 2003;290:2057–2060. doi: 10.1001/jama.290.15.2057. [DOI] [PubMed] [Google Scholar]
- Connor KM, Krah NM, Dennison RJ, Aderman CM, Chen J, Guerin KI, Sapieha P, Stahl A, Willett KL, Smith LE. Quantification of oxygen-induced retinopathy in the mouse: a model of vessel loss, vessel regrowth and pathological angiogenesis. Nat Protoc. 2009;4:1565–1573. doi: 10.1038/nprot.2009.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copland DA, Calder CJ, Raveney BJ, Nicholson LB, Phillips J, Cherwinski H, Jenmalm M, Sedgwick JD, Dick AD. Monoclonal antibody-mediated CD200 receptor signaling suppresses macrophage activation and tissue damage in experimental autoimmune uveoretinitis. Am J Pathol. 2007;171:580–588. doi: 10.2353/ajpath.2007.070272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copland DA, Wertheim MS, Armitage WJ, Nicholson LB, Raveney BJ, Dick AD. The clinical time-course of experimental autoimmune uveoretinitis using topical endoscopic fundal imaging with histologic and cellular infiltrate correlation. Invest Ophthalmol Vis Sci. 2008;49:5458–5465. doi: 10.1167/iovs.08-2348. [DOI] [PubMed] [Google Scholar]
- Corthals GL, Wasinger VC, Hochstrasser DF, Sanchez JC. The dynamic range of protein expression: a challenge for proteomic research. Electrophoresis. 2000;21:1104–1115. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1104::AID-ELPS1104>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Courret N, Darche S, Sonigo P, Milon G, Buzoni-Gatel D, Tardieux I. CD11c- and CD11b-expressing mouse leukocytes transport single Toxoplasma gondii tachyzoites to the brain. Blood. 2006;107:309–316. doi: 10.1182/blood-2005-02-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree B, Subramanian V. Behavior of endothelial cells on Matrigel and development of a method for a rapid and reproducible in vitro angiogenesis assay. In vitro cellular & developmental biology. Animal. 2007;43:87–94. doi: 10.1007/s11626-007-9012-x. [DOI] [PubMed] [Google Scholar]
- Crane IJ, Liversidge J. Mechanisms of leukocyte migration across the blood-retina barrier. Seminars in immunopathology. 2008;30:165–177. doi: 10.1007/s00281-008-0106-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane IJ, McKillop-Smith S, Wallace CA, Lamont GR, Forrester JV. Expression of the chemokines MIP-1alpha, MCP-1, and RANTES in experimental autoimmune uveitis. Invest Ophthalmol Vis Sci. 2001;42:1547–1552. [PubMed] [Google Scholar]
- Crane IJ, Wallace CA, McKillop-Smith S, Forrester JV. Control of chemokine production at the blood-retina barrier. Immunology. 2000;101:426–433. doi: 10.1046/j.1365-2567.2000.00105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane IJ, Xu H, Wallace C, Manivannan A, Mack M, Liversidge J, Marquez G, Sharp PF, Forrester JV. Involvement of CCR5 in the passage of Th1-type cells across the blood-retina barrier in experimental autoimmune uveitis. J Leukoc Biol. 2006;79:435–443. doi: 10.1189/jlb.0305130. [DOI] [PubMed] [Google Scholar]
- Crowley JL, Smith TC, Fang Z, Takizawa N, Luna EJ. Supervillin reorganizes the actin cytoskeleton and increases invadopodial efficiency. Molecular biology of the cell. 2009;20:948–962. doi: 10.1091/mbc.E08-08-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryotherapy for Retinopathy of Prematurity Cooperative Group. Multicenter trial of cryotherapy for retinopathy of prematurity. One-year outcome--structure and function. Arch Ophthalmol. 1990;108:1408–1416. [PubMed] [Google Scholar]
- Cunha-Vaz J. The blood-ocular barriers. Surv Ophthalmol. 1979;23:279–296. doi: 10.1016/0039-6257(79)90158-9. [DOI] [PubMed] [Google Scholar]
- Danis RP, Bingaman DP. Insulin-like growth factor-1 retinal microangiopathy in the pig eye. Ophthalmology. 1997;104:1661–1669. doi: 10.1016/s0161-6420(97)30081-5. [DOI] [PubMed] [Google Scholar]
- Darland DC, Massingham LJ, Smith SR, Piek E, Saint-Geniez M, D’Amore PA. Pericyte production of cell-associated VEGF is differentiation-dependent and is associated with endothelial survival. Developmental biology. 2003;264:275–288. doi: 10.1016/j.ydbio.2003.08.015. [DOI] [PubMed] [Google Scholar]
- Darrell RW, Wagener HP, Kurland LT. Epidemiology of uveitis. Incidence and prevalence in a small urban community. Arch Ophthalmol. 1962;68:502–514. doi: 10.1001/archopht.1962.00960030506014. [DOI] [PubMed] [Google Scholar]
- Das A, Fanslow W, Cerretti D, Warren E, Talarico N, McGuire P. Angiopoietin/Tek interactions regulate mmp-9 expression and retinal neovascularization. Laboratory investigation; a journal of technical methods and pathology. 2003;83:1637–1645. doi: 10.1097/01.lab.0000097189.79233.d8. [DOI] [PubMed] [Google Scholar]
- Davies MH, Eubanks JP, Powers MR. Increased retinal neovascularization in Fas ligand-deficient mice. Invest Ophthalmol Vis Sci. 2003;44:3202–3210. doi: 10.1167/iovs.03-0050. [DOI] [PubMed] [Google Scholar]
- DeNiro M, Al-Halafi A, Al-Mohanna FH, Alsmadi O, Al-Mohanna FA. Pleiotropic effects of YC-1 selectively inhibit pathological retinal neovascularization and promote physiological revascularization in a mouse model of oxygen-induced retinopathy. Mol Pharmacol. 2010;77:348–367. doi: 10.1124/mol.109.061366. [DOI] [PubMed] [Google Scholar]
- DeNiro M, Alsmadi O, Al-Mohanna F. Modulating the hypoxia-inducible factor signaling pathway as a therapeutic modality to regulate retinal angiogenesis. Exp Eye Res. 2009;89:700–717. doi: 10.1016/j.exer.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Derevjanik NL, Vinores SA, Xiao WH, Mori K, Turon T, Hudish T, Dong S, Campochiaro PA. Quantitative assessment of the integrity of the blood-retinal barrier in mice. Invest Ophthalmol Vis Sci. 2002;43:2462–2467. [PubMed] [Google Scholar]
- Deuter CM, Zierhut M, Mohle A, Vonthein R, Stobiger N, Kotter I. Long-term remission after cessation of interferon-alpha treatment in patients with severe uveitis due to Behcet’s disease. Arthritis Rheum. 2010;62:2796–2805. doi: 10.1002/art.27581. [DOI] [PubMed] [Google Scholar]
- Devi TS, Singh LP, Hosoya K, Terasaki T. GSK-3beta/CREB axis mediates IGF-1-induced ECM/adhesion molecule expression, cell cycle progression and monolayer permeability in retinal capillary endothelial cells: Implications for diabetic retinopathy. Biochim Biophys Acta. 2011;1812:1080–1088. doi: 10.1016/j.bbadis.2011.04.007. [DOI] [PubMed] [Google Scholar]
- Dick AD, Broderick C, Forrester JV, Wright GJ. Distribution of OX2 antigen and OX2 receptor within retina. Invest Ophthalmol Vis Sci. 2001;42:170–176. [PubMed] [Google Scholar]
- Dick AD, Carter D, Robertson M, Broderick C, Hughes E, Forrester JV, Liversidge J. Control of myeloid activity during retinal inflammation. Journal of leukocyte biology. 2003;74:161–166. doi: 10.1189/jlb.1102535. [DOI] [PubMed] [Google Scholar]
- Dick AD, Cheng YF, Liversidge J, Forrester JV. Immunomodulation of experimental autoimmune uveoretinitis: a model of tolerance induction with retinal antigens. Eye (Lond) 1994;8 ( Pt 1):52–59. doi: 10.1038/eye.1994.10. [DOI] [PubMed] [Google Scholar]
- Dimitroff CJ, Lee JY, Rafii S, Fuhlbrigge RC, Sackstein R. CD44 is a major E-selectin ligand on human hematopoietic progenitor cells. The Journal of cell biology. 2001a;153:1277–1286. doi: 10.1083/jcb.153.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitroff CJ, Lee JY, Schor KS, Sandmaier BM, Sackstein R. differential L-selectin binding activities of human hematopoietic cell L-selectin ligands, HCELL and PSGL-1. J Biol Chem. 2001b;276:47623–47631. doi: 10.1074/jbc.M105997200. [DOI] [PubMed] [Google Scholar]
- Donovan D, Brown NJ, Bishop ET, Lewis CE. Comparison of three in vitro human ‘angiogenesis’ assays with capillaries formed in vivo. Angiogenesis. 2001;4:113–121. doi: 10.1023/a:1012218401036. [DOI] [PubMed] [Google Scholar]
- Dorrell MI, Friedlander M. Mechanisms of endothelial cell guidance and vascular patterning in the developing mouse retina. Prog Retin Eye Res. 2006;25:277–295. doi: 10.1016/j.preteyeres.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Du L, Kijlstra A, Yang P. Immune response genes in uveitis. Ocular immunology and inflammation. 2009;17:249–256. doi: 10.1080/09273940902999356. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Lindsay DS, Speer CA. Structures of Toxoplasma gondii tachyzoites, bradyzoites, and sporozoites and biology and development of tissue cysts. Clin Microbiol Rev. 1998;11:267–299. doi: 10.1128/cmr.11.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duguid IG, Boyd AW, Mandel TE. Adhesion molecules are expressed in the human retina and choroid. Curr Eye Res. 1992;11(Suppl):153–159. doi: 10.3109/02713689208999526. [DOI] [PubMed] [Google Scholar]
- Early Treatment For Retinopathy Of Prematurity Cooperative G. Revised indications for the treatment of retinopathy of prematurity: results of the early treatment for retinopathy of prematurity randomized trial. Arch Ophthalmol. 2003;121:1684–1694. doi: 10.1001/archopht.121.12.1684. [DOI] [PubMed] [Google Scholar]
- El-Remessy AB, Rajesh M, Mukhopadhyay P, Horvath B, Patel V, Al-Gayyar MM, Pillai BA, Pacher P. Cannabinoid 1 receptor activation contributes to vascular inflammation and cell death in a mouse model of diabetic retinopathy and a human retinal cell line. Diabetologia. 2011;54:1567–1578. doi: 10.1007/s00125-011-2061-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Shabrawi Y, Walch A, Hermann J, Egger G, Foster CS. Inhibition of MMP-dependent chemotaxis and amelioration of experimental autoimmune uveitis with a selective metalloproteinase-2 and -9 inhibitor. J Neuroimmunol. 2004;155:13–20. doi: 10.1016/j.jneuroim.2004.05.010. [DOI] [PubMed] [Google Scholar]
- El-Shabrawi YG, Christen WG, Foster SC. Correlation of metalloproteinase-2 and -9 with proinflammatory cytokines interleukin-1b, interleukin-12 and the interleukin-1 receptor antagonist in patients with chronic uveitis. Curr Eye Res. 2000;20:211–214. [PubMed] [Google Scholar]
- Engelhardt B, Wolburg H. Mini-review: Transendothelial migration of leukocytes: through the front door or around the side of the house? European journal of immunology. 2004;34:2955–2963. doi: 10.1002/eji.200425327. [DOI] [PubMed] [Google Scholar]
- Eter N, Sahm M, Klingmuller D, Spitznas M. Modulation of insulin-like growth factor-I production of cultured retinal vascular endothelial cells by oxygen, glucose and growth hormone. Japanese journal of ophthalmology. 2002;46:226–229. doi: 10.1016/s0021-5155(01)00499-3. [DOI] [PubMed] [Google Scholar]
- Fang Z, Takizawa N, Wilson KA, Smith TC, Delprato A, Davidson MW, Lambright DG, Luna EJ. The membrane-associated protein, supervillin, accelerates F-actin-dependent rapid integrin recycling and cell motility. Traffic (Copenhagen, Denmark) 2010;11:782–799. doi: 10.1111/j.1600-0854.2010.01062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantin A, Vieira JM, Gestri G, Denti L, Schwarz Q, Prykhozhij S, Peri F, Wilson SW, Ruhrberg C. Tissue macrophages act as cellular chaperones for vascular anastomosis downstream of VEGF-mediated endothelial tip cell induction. Blood. 2010;116:829–840. doi: 10.1182/blood-2009-12-257832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fardini Y, Wang X, Temoin S, Nithianantham S, Lee D, Shoham M, Han YW. Fusobacterium nucleatum adhesin FadA binds vascular endothelial cadherin and alters endothelial integrity. Mol Microbiol. 2011;82:1468–1480. doi: 10.1111/j.1365-2958.2011.07905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feire AL, Koss H, Compton T. Cellular integrins function as entry receptors for human cytomegalovirus via a highly conserved disintegrin-like domain. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15470–15475. doi: 10.1073/pnas.0406821101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Wang Y, Pfister F, Hillebrands JL, Deutsch U, Hammes HP. Decreased hypoxia-induced neovascularization in angiopoietin-2 heterozygous knockout mouse through reduced MMP activity. Cell Physiol Biochem. 2009;23:277–284. doi: 10.1159/000218174. [DOI] [PubMed] [Google Scholar]
- Ferrante P, Ramsey A, Bunce C, Lightman S. Clinical trial to compare efficacy and side-effects of injection of posterior sub-Tenon triamcinolone versus orbital floor methylprednisolone in the management of posterior uveitis. Clin Experiment Ophthalmol. 2004;32:563–568. doi: 10.1111/j.1442-9071.2004.00902.x. [DOI] [PubMed] [Google Scholar]
- Ferrara N. Vascular endothelial growth factor: basic science and clinical progress. Endocrine reviews. 2004;25:581–611. doi: 10.1210/er.2003-0027. [DOI] [PubMed] [Google Scholar]
- Firth SM, Baxter RC. Cellular actions of the insulin-like growth factor binding proteins. Endocrine reviews. 2002;23:824–854. doi: 10.1210/er.2001-0033. [DOI] [PubMed] [Google Scholar]
- Fong DS, Girach A, Boney A. Visual side effects of successful scatter laser photocoagulation surgery for proliferative diabetic retinopathy: a literature review. Retina (Philadelphia, Pa. 2007;27:816–824. doi: 10.1097/IAE.0b013e318042d32c. [DOI] [PubMed] [Google Scholar]
- Forsythe JA, Jiang BH, Iyer NV, Agani F, Leung SW, Koos RD, Semenza GL. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol. 1996;16:4604–4613. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks WA, Limb GA, Stanford MR, Ogilvie J, Wolstencroft RA, Chignell AH, Dumonde DC. Cytokines in human intraocular inflammation. Current eye research. 1992;11(Suppl):187–191. doi: 10.3109/02713689208999531. [DOI] [PubMed] [Google Scholar]
- Fruttiger M. Development of the mouse retinal vasculature: angiogenesis versus vasculogenesis. Invest Ophthalmol Vis Sci. 2002;43:522–527. [PubMed] [Google Scholar]
- Fruttiger M. Development of the retinal vasculature. Angiogenesis. 2007;10:77–88. doi: 10.1007/s10456-007-9065-1. [DOI] [PubMed] [Google Scholar]
- Fujikawa LS, Chan CC, McAllister C, Gery I, Hooks JJ, Detrick B, Nussenblatt RB. Retinal vascular endothelium expresses fibronectin and class II histocompatibility complex antigens in experimental autoimmune uveitis. Cell Immunol. 1987a;106:139–150. doi: 10.1016/0008-8749(87)90157-2. [DOI] [PubMed] [Google Scholar]
- Fujikawa LS, Chan CC, McAllister C, Gery I, Hooks JJ, Detrick B, Nussenblatt RB. Retinal vascular endothelium expresses fibronectin and class II histocompatibility complex antigens in experimental autoimmune uveitis. Cellular immunology. 1987b;106:139–150. doi: 10.1016/0008-8749(87)90157-2. [DOI] [PubMed] [Google Scholar]
- Fujikawa LS, Reay C, Morin ME. Class II antigens on retinal vascular endothelium, pericytes, macrophages, and lymphocytes of the rat. Invest Ophthalmol Vis Sci. 1989;30:66–73. [PubMed] [Google Scholar]
- Furtado GC, Cao Y, Joiner KA. Laminin on Toxoplasma gondii mediates parasite binding to the beta 1 integrin receptor alpha 6 beta 1 on human foreskin fibroblasts and Chinese hamster ovary cells. Infect Immun. 1992;60:4925–4931. doi: 10.1128/iai.60.11.4925-4931.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furtado JM, Bharadwaj AS, Chipps TJ, Pan Y, Ashander LM, Smith JR. Toxoplasma gondii tachyzoites cross retinal endothelium assisted by intercellular adhesion molecule-1 in vitro. Immunol Cell Biol. 2012 doi: 10.1038/icb.2012.21. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao BB, Clermont A, Rook S, Fonda SJ, Srinivasan VJ, Wojtkowski M, Fujimoto JG, Avery RL, Arrigg PG, Bursell SE, Aiello LP, Feener EP. Extracellular carbonic anhydrase mediates hemorrhagic retinal and cerebral vascular permeability through prekallikrein activation. Nat Med. 2007;13:181–188. doi: 10.1038/nm1534. [DOI] [PubMed] [Google Scholar]
- Gardiner TA, Stitt AW, Anderson HR, Archer DB. Selective loss of vascular smooth muscle cells in the retinal microcirculation of diabetic dogs. Br J Ophthalmol. 1994;78:54–60. doi: 10.1136/bjo.78.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gariano RF. Special features of human retinal angiogenesis. Eye (Lond) 2010;24:401–407. doi: 10.1038/eye.2009.324. [DOI] [PubMed] [Google Scholar]
- Garrido-Urbani S, Bradfield PF, Lee BP, Imhof BA. Vascular and epithelial junctions: a barrier for leucocyte migration. Biochemical Society transactions. 2008;36:203–211. doi: 10.1042/BST0360203. [DOI] [PubMed] [Google Scholar]
- Gerhardinger C, McClure KD, Romeo G, Podesta F, Lorenzi M. IGF-I mRNA and signaling in the diabetic retina. Diabetes. 2001;50:175–183. doi: 10.2337/diabetes.50.1.175. [DOI] [PubMed] [Google Scholar]
- Gerhardt H. VEGF and endothelial guidance in angiogenic sprouting. Organogenesis. 2008;4:241–246. doi: 10.4161/org.4.4.7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gery I, Wiggert B, Redmond TM, Kuwabara T, Crawford MA, Vistica BP, Chader GJ. Uveoretinitis and pinealitis induced by immunization with interphotoreceptor retinoid-binding protein. Invest Ophthalmol Vis Sci. 1986;27:1296–1300. [PubMed] [Google Scholar]
- Giebel SJ, Menicucci G, McGuire PG, Das A. Matrix metalloproteinases in early diabetic retinopathy and their role in alteration of the blood-retinal barrier. Lab Invest. 2005;85:597–607. doi: 10.1038/labinvest.3700251. [DOI] [PubMed] [Google Scholar]
- Gilbert C, Rahi J, Eckstein M, O’Sullivan J, Foster A. Retinopathy of prematurity in middle-income countries. Lancet. 1997;350:12–14. doi: 10.1016/S0140-6736(97)01107-0. [DOI] [PubMed] [Google Scholar]
- Gillies MC, Wong TY. Ranibizumab for neovascular age-related macular degeneration. The New England journal of medicine. 2007;356:748–749. author reply 749–750. [PubMed] [Google Scholar]
- Gordon DM. Prednisone and prednisolone in ocular disease. American journal of ophthalmology. 1956;41:593–600. [PubMed] [Google Scholar]
- Grammas P, Riden M. Retinal endothelial cells are more susceptible to oxidative stress and increased permeability than brain-derived endothelial cells. Microvasc Res. 2003;65:18–23. doi: 10.1016/s0026-2862(02)00016-x. [DOI] [PubMed] [Google Scholar]
- Grant M, Jerdan J, Merimee TJ. Insulin-like growth factor-I modulates endothelial cell chemotaxis. J Clin Endocrinol Metab. 1987;65:370–371. doi: 10.1210/jcem-65-2-370. [DOI] [PubMed] [Google Scholar]
- Grant MA, Baron RM, Macias AA, Layne MD, Perrella MA, Rigby AC. Netropsin improves survival from endotoxaemia by disrupting HMGA1 binding to the NOS2 promoter. Biochem J. 2009;418:103–112. doi: 10.1042/BJ20081427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood J, Howes R, Lightman S. The blood-retinal barrier in experimental autoimmune uveoretinitis. Leukocyte interactions and functional damage. Lab Invest. 1994;70:39–52. [PubMed] [Google Scholar]
- Greenwood J, Wang Y, Calder VL. Lymphocyte adhesion and transendothelial migration in the central nervous system: the role of LFA-1, ICAM-1, VLA-4 and VCAM-1. off. Immunology. 1995;86:408–415. [PMC free article] [PubMed] [Google Scholar]
- Greiffenberg L, Goebel W, Kim KS, Weiglein I, Bubert A, Engelbrecht F, Stins M, Kuhn M. Interaction of Listeria monocytogenes with human brain microvascular endothelial cells: InlB-dependent invasion, long-term intracellular growth, and spread from macrophages to endothelial cells. Infect Immun. 1998;66:5260–5267. doi: 10.1128/iai.66.11.5260-5267.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritz DC, Wong IG. Incidence and prevalence of uveitis in Northern California; the Northern California Epidemiology of Uveitis Study. Ophthalmology. 2004;111:491–500. doi: 10.1016/j.ophtha.2003.06.014. discussion 500. [DOI] [PubMed] [Google Scholar]
- Grubb SE, Murdoch C, Sudbery PE, Saville SP, Lopez-Ribot JL, Thornhill MH. Adhesion of Candida albicans to endothelial cells under physiological conditions of flow. Infect Immun. 2009;77:3872–3878. doi: 10.1128/IAI.00518-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudkov AV, Gurova KV, Komarova EA. Inflammation and p53: A Tale of Two Stresses. Genes Cancer. 2011;2:503–516. doi: 10.1177/1947601911409747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gul A, Tugal-Tutkun I, Dinarello CA, Reznikov L, Esen BA, Mirza A, Scannon P, Solinger A. Interleukin-1beta-regulating antibody XOMA 052 (gevokizumab) in the treatment of acute exacerbations of resistant uveitis of Behcet’s disease: an open-label pilot study. Annals of the rheumatic diseases. 2012;71:563–566. doi: 10.1136/annrheumdis-2011-155143. [DOI] [PubMed] [Google Scholar]
- Gustavsson C, Agardh CD, Zetterqvist AV, Nilsson J, Agardh E, Gomez MF. Vascular cellular adhesion molecule-1 (VCAM-1) expression in mice retinal vessels is affected by both hyperglycemia and hyperlipidemia. PLoS One. 2010;5:e12699. doi: 10.1371/journal.pone.0012699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett SF, Ozaki H, Strauss RW, Wahlin K, Suri C, Maisonpierre P, Yancopoulos G, Campochiaro PA. Angiopoietin 2 expression in the retina: upregulation during physiologic and pathologic neovascularization. J Cell Physiol. 2000;184:275–284. doi: 10.1002/1097-4652(200009)184:3<275::AID-JCP1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Hackett SF, Wiegand S, Yancopoulos G, Campochiaro PA. Angiopoietin-2 plays an important role in retinal angiogenesis. J Cell Physiol. 2002;192:182–187. doi: 10.1002/jcp.10128. [DOI] [PubMed] [Google Scholar]
- Halbert CL, Demers GW, Galloway DA. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J Virol. 1991;65:473–478. doi: 10.1128/jvi.65.1.473-478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes HP, Feng Y, Pfister F, Brownlee M. Diabetic retinopathy: targeting vasoregression. Diabetes. 2011;60:9–16. doi: 10.2337/db10-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harhaj NS, Felinski EA, Wolpert EB, Sundstrom JM, Gardner TW, Antonetti DA. VEGF activation of protein kinase C stimulates occludin phosphorylation and contributes to endothelial permeability. Invest Ophthalmol Vis Sci. 2006;47:5106–5115. doi: 10.1167/iovs.06-0322. [DOI] [PubMed] [Google Scholar]
- Hartnett ME. The effects of oxygen stresses on the development of features of severe retinopathy of prematurity: knowledge from the 50/10 OIR model. Documenta ophthalmologica. 2010;120:25–39. doi: 10.1007/s10633-009-9181-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey NL, Oliver G. Choose your fate: artery, vein or lymphatic vessel? Curr Opin Genet Dev. 2004;14:499–505. doi: 10.1016/j.gde.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Hasegawa T, McLeod DS, Prow T, Merges C, Grebe R, Lutty GA. Vascular precursors in developing human retina. Invest Ophthalmol Vis Sci. 2008;49:2178–2192. doi: 10.1167/iovs.07-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashida N, Ohguro N, Nakai K, Kobashi-Hashida M, Hashimoto S, Matsushima K, Tano Y. Microarray analysis of cytokine and chemokine gene expression after prednisolone treatment in murine experimental autoimmune uveoretinitis. Invest Ophthalmol Vis Sci. 2005;46:4224–4234. doi: 10.1167/iovs.05-0346. [DOI] [PubMed] [Google Scholar]
- Haurigot V, Villacampa P, Ribera A, Llombart C, Bosch A, Nacher V, Ramos D, Ayuso E, Segovia JC, Bueren JA, Ruberte J, Bosch F. Increased intraocular insulin-like growth factor-I triggers blood-retinal barrier breakdown. J Biol Chem. 2009;284:22961–22969. doi: 10.1074/jbc.M109.014787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haznedaroglu E, Karaaslan Y, Buyukasik Y, Kosar A, Ozcebe O, Haznedaroglu C, Kirazli E, Dundar SV. Selectin adhesion molecules in Behcet’s disease. Annals of the rheumatic diseases. 2000;59:61–63. doi: 10.1136/ard.59.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellstrom A, Perruzzi C, Ju M, Engstrom E, Hard AL, Liu JL, Albertsson-Wikland K, Carlsson B, Niklasson A, Sjodell L, LeRoith D, Senger DR, Smith LE. Low IGF-I suppresses VEGF-survival signaling in retinal endothelial cells: direct correlation with clinical retinopathy of prematurity. Proc Natl Acad Sci U S A. 2001;98:5804–5808. doi: 10.1073/pnas.101113998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herwald H. Haemostasis, vascular biology, and infectious agents. Thromb Haemost. 2007;98:483–484. [PubMed] [Google Scholar]
- Hill TA, Stanford MR, Graham EM, Dumonde DC, Brown KA. A new method for studying the selective adherence of blood lymphocytes to the microvasculature of human retina. Invest Ophthalmol Vis Sci. 1997;38:2608–2618. [PubMed] [Google Scholar]
- Hogan MJ, Alvarado JA, Weddell JE. Histology of the Human Eye: an Atlas and Textbook. W.B. Saunders Company; Philadelphia: 1971. [Google Scholar]
- Hogan MJ, Feeney L. The Ultrastructure of the Retinal Vessels. Iii. Vascular-Glial Relationships. J Ultrastruct Res. 1963;49:47–64. doi: 10.1016/s0022-5320(63)80035-0. [DOI] [PubMed] [Google Scholar]
- Holland GN. Acquired immunodeficiency syndrome and ophthalmology: the first decade. American journal of ophthalmology. 1992;114:86–95. doi: 10.1016/s0002-9394(14)77417-3. [DOI] [PubMed] [Google Scholar]
- Holland GN. Ocular toxoplasmosis: a global reassessment. Part I: epidemiology and course of disease. American journal of ophthalmology. 2003;136:973–988. doi: 10.1016/j.ajo.2003.09.040. [DOI] [PubMed] [Google Scholar]
- Holland GN. Ocular toxoplasmosis: a global reassessment. Part II: disease manifestations and management. Am J Ophthalmol. 2004;137:1–17. [PubMed] [Google Scholar]
- Holland GN, Crespi CM, ten Dam-van Loon N, Charonis AC, Yu F, Bosch-Driessen LH, Rothova A. Analysis of recurrence patterns associated with toxoplasmic retinochoroiditis. Am J Ophthalmol. 2008;145:1007–1013. doi: 10.1016/j.ajo.2008.01.023. [DOI] [PubMed] [Google Scholar]
- Hovakimyan A, Cunningham ET., Jr Ocular toxoplasmosis. Ophthalmol Clin North Am. 2002;15:327–332. doi: 10.1016/s0896-1549(02)00030-5. [DOI] [PubMed] [Google Scholar]
- Hughes S, Yang H, Chan-Ling T. Vascularization of the human fetal retina: roles of vasculogenesis and angiogenesis. Invest Ophthalmol Vis Sci. 2000;41:1217–1228. [PubMed] [Google Scholar]
- Iliaki E, Poulaki V, Mitsiades N, Mitsiades CS, Miller JW, Gragoudas ES. Role of alpha 4 integrin (CD49d) in the pathogenesis of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2009;50:4898–4904. doi: 10.1167/iovs.08-2013. [DOI] [PubMed] [Google Scholar]
- Im E, Venkatakrishnan A, Kazlauskas A. Cathepsin B regulates the intrinsic angiogenic threshold of endothelial cells. Molecular biology of the cell. 2005;16:3488–3500. doi: 10.1091/mbc.E04-11-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Committee for the Classification of Retinopathy of Prematurity. The International Classification of Retinopathy of Prematurity revisited. Arch Ophthalmol. 2005;123:991–999. doi: 10.1001/archopht.123.7.991. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Ishida S, Usui T, Yamashiro K, Kaji Y, Amano S, Ogura Y, Hida T, Oguchi Y, Ambati J, Miller JW, Gragoudas ES, Ng YS, D’Amore PA, Shima DT, Adamis AP. VEGF164-mediated inflammation is required for pathological, but not physiological, ischemia-induced retinal neovascularization. The Journal of experimental medicine. 2003a;198:483–489. doi: 10.1084/jem.20022027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida S, Yamashiro K, Usui T, Kaji Y, Ogura Y, Hida T, Honda Y, Oguchi Y, Adamis AP. Leukocytes mediate retinal vascular remodeling during development and vaso-obliteration in disease. Nat Med. 2003b;9:781–788. doi: 10.1038/nm877. [DOI] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, Ratcliffe PJ. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Jabs DA. Cytomegalovirus retinitis and the acquired immunodeficiency syndrome--bench to bedside: LXVII Edward Jackson Memorial Lecture. American journal of ophthalmology. 2011;151:198–216. e191. doi: 10.1016/j.ajo.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabs DA, Rosenbaum JT, Foster CS, Holland GN, Jaffe GJ, Louie JS, Nussenblatt RB, Stiehm ER, Tessler H, Van Gelder RN, Whitcup SM, Yocum D. Guidelines for the use of immunosuppressive drugs in patients with ocular inflammatory disorders: recommendations of an expert panel. American journal of ophthalmology. 2000;130:492–513. doi: 10.1016/s0002-9394(00)00659-0. [DOI] [PubMed] [Google Scholar]
- Jackson TL, Eykyn SJ, Graham EM, Stanford MR. Endogenous bacterial endophthalmitis: a 17-year prospective series and review of 267 reported cases. Surv Ophthalmol. 2003;48:403–423. doi: 10.1016/s0039-6257(03)00054-7. [DOI] [PubMed] [Google Scholar]
- Jaffe GJ, Martin D, Callanan D, Pearson PA, Levy B, Comstock T. Fluocinolone acetonide implant (Retisert) for noninfectious posterior uveitis: thirty-four-week results of a multicenter randomized clinical study. Ophthalmology. 2006;113:1020–1027. doi: 10.1016/j.ophtha.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Jakob E, Reuland MS, Mackensen F, Harsch N, Fleckenstein M, Lorenz HM, Max R, Becker MD. Uveitis subtypes in a german interdisciplinary uveitis center--analysis of 1916 patients. J Rheumatol. 2009;36:127–136. doi: 10.3899/jrheum.080102. [DOI] [PubMed] [Google Scholar]
- Jakobsson L, Franco CA, Bentley K, Collins RT, Ponsioen B, Aspalter IM, Rosewell I, Busse M, Thurston G, Medvinsky A, Schulte-Merker S, Gerhardt H. Endothelial cells dynamically compete for the tip cell position during angiogenic sprouting. Nat Cell Biol. 2010;12:943–953. doi: 10.1038/ncb2103. [DOI] [PubMed] [Google Scholar]
- Jarvis MA, Nelson JA. Human cytomegalovirus tropism for endothelial cells: not all endothelial cells are created equal. J Virol. 2007;81:2095–2101. doi: 10.1128/JVI.01422-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Wu J, Song X, Song Q, Cully BL, Messmer-Blust A, Xu M, Foo SY, Rosenzweig A, Li J. RTEF-1, an Upstream Gene of Hypoxia-inducible Factor-1{alpha}, Accelerates Recovery from Ischemia. J Biol Chem. 2011;286:22699–22705. doi: 10.1074/jbc.M111.237024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, Lutty GA, McLeod DS, Otsuji T, Flower RW, Sandagar G, Alexander T, Steidl SM, Hansen BC. Ocular structure and function in an aged monkey with spontaneous diabetes mellitus. Exp Eye Res. 2005;80:37–42. doi: 10.1016/j.exer.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Jong AY, Stins MF, Huang SH, Chen SH, Kim KS. Traversal of Candida albicans across human blood-brain barrier in vitro. Infect Immun. 2001;69:4536–4544. doi: 10.1128/IAI.69.7.4536-4544.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joussen AM, Poulaki V, Mitsiades N, Cai WY, Suzuma I, Pak J, Ju ST, Rook SL, Esser P, Mitsiades CS, Kirchhof B, Adamis AP, Aiello LP. Suppression of Fas-FasL-induced endothelial cell apoptosis prevents diabetic blood-retinal barrier breakdown in a model of streptozotocin-induced diabetes. Faseb J. 2003;17:76–78. doi: 10.1096/fj.02-0157fje. [DOI] [PubMed] [Google Scholar]
- Joussen AM, Poulaki V, Tsujikawa A, Qin W, Qaum T, Xu Q, Moromizato Y, Bursell SE, Wiegand SJ, Rudge J, Ioffe E, Yancopoulos GD, Adamis AP. Suppression of diabetic retinopathy with angiopoietin-1. Am J Pathol. 2002;160:1683–1693. doi: 10.1016/S0002-9440(10)61115-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsi KK, Zych M, Slominska EM, Kochan Z, Yacoub MH, Smolenski RT. Adenine incorporation in human and rat endothelium. Biochim Biophys Acta. 1999;1452:145–150. doi: 10.1016/s0167-4889(99)00122-6. [DOI] [PubMed] [Google Scholar]
- Kanno S, Oda N, Abe M, Terai Y, Ito M, Shitara K, Tabayashi K, Shibuya M, Sato Y. Roles of two VEGF receptors, Flt-1 and KDR, in the signal transduction of VEGF effects in human vascular endothelial cells. Oncogene. 2000;19:2138–2146. doi: 10.1038/sj.onc.1203533. [DOI] [PubMed] [Google Scholar]
- Kanzawa S, Endo H, Shioya N. Improved in vitro angiogenesis model by collagen density reduction and the use of type III collagen. Annals of plastic surgery. 1993;30:244–251. doi: 10.1097/00000637-199303000-00008. [DOI] [PubMed] [Google Scholar]
- Keino H, Takeuchi M, Kezuka T, Yamakawa N, Tsukahara R, Usui M. Chemokine and chemokine receptor expression during experimental autoimmune uveoretinitis in mice. Graefe’s archive for clinical and experimental ophthalmology = Albrecht von Graefes Archiv fur klinische und experimentelle. Ophthalmologie. 2003;241:111–115. doi: 10.1007/s00417-002-0556-x. [DOI] [PubMed] [Google Scholar]
- Kempen JH, Altaweel MM, Holbrook JT, Jabs DA, Louis TA, Sugar EA, Thorne JE. Randomized Comparison of Systemic Anti-inflammatory Therapy Versus Fluocinolone Acetonide Implant for Intermediate, Posterior, and Panuveitis: The Multicenter Uveitis Steroid Treatment Trial. Ophthalmology. 2011 doi: 10.1016/j.ophtha.2011.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempen JH, O’Colmain BJ, Leske MC, Haffner SM, Klein R, Moss SE, Taylor HR, Hamman RF. The prevalence of diabetic retinopathy among adults in the United States. Archives of ophthalmology. 2004a;122:552–563. doi: 10.1001/archopht.122.4.552. [DOI] [PubMed] [Google Scholar]
- Kempen JH, O’Colmain BJ, Leske MC, Haffner SM, Klein R, Moss SE, Taylor HR, Hamman RF. The prevalence of diabetic retinopathy among adults in the United States. Arch Ophthalmol. 2004b;122:552–563. doi: 10.1001/archopht.122.4.552. [DOI] [PubMed] [Google Scholar]
- Kerr EC, Copland DA, Dick AD, Nicholson LB. The dynamics of leukocyte infiltration in experimental autoimmune uveoretinitis. Prog Retin Eye Res. 2008;27:527–535. doi: 10.1016/j.preteyeres.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Kezic J, McMenamin PG. The monocyte chemokine receptor CX3CR1 does not play a significant role in the pathogenesis of experimental autoimmune uveoretinitis. Invest Ophthalmol Vis Sci. 2010;51:5121–5127. doi: 10.1167/iovs.10-5325. [DOI] [PubMed] [Google Scholar]
- Kielczewski JL, Li Calzi S, Shaw LC, Cai J, Qi X, Ruan Q, Wu L, Liu L, Hu P, Chan-Ling T, Mames RN, Firth S, Baxter RC, Turowski P, Busik JV, Boulton ME, Grant MB. Free insulin-like growth factor binding protein-3 (IGFBP-3) reduces retinal vascular permeability in association with a reduction of acid sphingomyelinase (ASMase) Invest Ophthalmol Vis Sci. 2011;52:8278–8286. doi: 10.1167/iovs.11-8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KJ, Chung JW, Kim KS. 67-kDa laminin receptor promotes internalization of cytotoxic necrotizing factor 1-expressing Escherichia coli K1 into human brain microvascular endothelial cells. J Biol Chem. 2005a;280:1360–1368. doi: 10.1074/jbc.M410176200. [DOI] [PubMed] [Google Scholar]
- Kim KS. E. coli invasion of brain microvascular endothelial cells as a pathogenetic basis of meningitis. Subcell Biochem. 2000;33:47–59. doi: 10.1007/978-1-4757-4580-1_3. [DOI] [PubMed] [Google Scholar]
- Kim KS. Pathogenesis of bacterial meningitis: from bacteraemia to neuronal injury. Nat Rev Neurosci. 2003;4:376–385. doi: 10.1038/nrn1103. [DOI] [PubMed] [Google Scholar]
- Kim SY, Johnson MA, McLeod DS, Alexander T, Hansen BC, Lutty GA. Neutrophils are associated with capillary closure in spontaneously diabetic monkey retinas. Diabetes. 2005b;54:1534–1542. doi: 10.2337/diabetes.54.5.1534. [DOI] [PubMed] [Google Scholar]
- King GL, Goodman AD, Buzney S, Moses A, Kahn CR. Receptors and growth-promoting effects of insulin and insulinlike growth factors on cells from bovine retinal capillaries and aorta. The Journal of clinical investigation. 1985;75:1028–1036. doi: 10.1172/JCI111764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King H, Aubert RE, Herman WH. Global burden of diabetes, 1995–2025: prevalence, numerical estimates, and projections. Diabetes Care. 1998;21:1414–1431. doi: 10.2337/diacare.21.9.1414. [DOI] [PubMed] [Google Scholar]
- Kishimoto T, Akira S, Narazaki M, Taga T. Interleukin-6 family of cytokines and gp130. Blood. 1995;86:1243–1254. [PubMed] [Google Scholar]
- Kitamei H, Iwabuchi K, Namba K, Yoshida K, Yanagawa Y, Kitaichi N, Kitamura M, Ohno S, Onoe K. Amelioration of experimental autoimmune uveoretinitis (EAU) with an inhibitor of nuclear factor-kappaB (NF-kappaB), pyrrolidine dithiocarbamate. Journal of leukocyte biology. 2006;79:1193–1201. doi: 10.1189/jlb.0805453. [DOI] [PubMed] [Google Scholar]
- Knight BC, Brunton CL, Modi NC, Wallace GR, Stanford MR. The effect of Toxoplasma gondii infection on expression of chemokines by rat retinal vascular endothelial cells. J Neuroimmunol. 2005;160:41–47. doi: 10.1016/j.jneuroim.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Knott RM, Robertson M, Muckersie E, Folefac VA, Fairhurst FE, Wileman SM, Forrester JV. A model system for the study of human retinal angiogenesis: activation of monocytes and endothelial cells and the association with the expression of the monocarboxylate transporter type 1 (MCT-1) Diabetologia. 1999;42:870–877. doi: 10.1007/s001250051240. [DOI] [PubMed] [Google Scholar]
- Kociok N, Radetzky S, Krohne TU, Gavranic C, Liang Y, Semkova I, Joussen AM. ICAM-1 depletion does not alter retinal vascular development in a model of oxygen-mediated neovascularization. Exp Eye Res. 2009;89:503–510. doi: 10.1016/j.exer.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Kok H, Lau C, Maycock N, McCluskey P, Lightman S. Outcome of intravitreal triamcinolone in uveitis. Ophthalmology. 2005;112:1916, e1911–1917. doi: 10.1016/j.ophtha.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Kotter I, Koch S, Vonthein R, Ruckwaldt U, Amberger M, Gunaydin I, Zierhut M, Stubiger N. Cytokines, cytokine antagonists and soluble adhesion molecules in patients with ocular Behcet’s disease treated with human recombinant interferon-alpha2a. Results of an open study and review of the literature. Clin Exp Rheumatol. 2005;23:S20–26. [PubMed] [Google Scholar]
- Kowluru RA, Koppolu P. Diabetes-induced activation of caspase-3 in retina: effect of antioxidant therapy. Free Radic Res. 2002;36:993–999. doi: 10.1080/1071576021000006572. [DOI] [PubMed] [Google Scholar]
- Kozulin P, Natoli R, Bumsted O’Brien KM, Madigan MC, Provis JM. The cellular expression of antiangiogenic factors in fetal primate macula. Invest Ophthalmol Vis Sci. 2010;51:4298–4306. doi: 10.1167/iovs.09-4905. [DOI] [PubMed] [Google Scholar]
- Kozulin P, Natoli R, Madigan MC, O’Brien KM, Provis JM. Gradients of Eph-A6 expression in primate retina suggest roles in both vascular and axon guidance. Mol Vis. 2009a;15:2649–2662. [PMC free article] [PubMed] [Google Scholar]
- Kozulin P, Natoli R, O’Brien KM, Madigan MC, Provis JM. Differential expression of anti-angiogenic factors and guidance genes in the developing macula. Mol Vis. 2009b;15:45–59. [PMC free article] [PubMed] [Google Scholar]
- Kramer RH, Cheng YF, Clyman R. Human microvascular endothelial cells use beta 1 and beta 3 integrin receptor complexes to attach to laminin. The Journal of cell biology. 1990;111:1233–1243. doi: 10.1083/jcb.111.3.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs DL, Hilton DJ. SOCS proteins: negative regulators of cytokine signaling. Stem Cells. 2001;19:378–387. doi: 10.1634/stemcells.19-5-378. [DOI] [PubMed] [Google Scholar]
- Kremer I, Kissun R, Nissenkorn I, Ben-Sira I, Garner A. Oxygen-induced retinopathy in newborn kittens. A model for ischemic vasoproliferative retinopathy. Invest Ophthalmol Vis Sci. 1987;28:126–130. [PubMed] [Google Scholar]
- Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper JJ, Mutis T, de Jager W, de Groot-Mijnes JD, Rothova A. Intraocular interleukin-17 and proinflammatory cytokines in HLA-A29-associated birdshot chorioretinopathy. American journal of ophthalmology. 2011;152:177–182. e171. doi: 10.1016/j.ajo.2011.01.031. [DOI] [PubMed] [Google Scholar]
- Kuppner MC, Liversidge J, McKillop-Smith S, Lumsden L, Forrester JV. Adhesion molecule expression in acute and fibrotic sympathetic ophthalmia. Curr Eye Res. 1993;12:923–934. doi: 10.3109/02713689309020399. [DOI] [PubMed] [Google Scholar]
- Lagarkova MA, Shutova MV, Bogomazova AN, Vassina EM, Glazov EA, Zhang P, Rizvanov AA, Chestkov IV, Kiselev SL. Induction of pluripotency in human endothelial cells resets epigenetic profile on genome scale. Cell Cycle. 2010;9:937–946. doi: 10.4161/cc.9.5.10869. [DOI] [PubMed] [Google Scholar]
- Lambert H, Hitziger N, Dellacasa I, Svensson M, Barragan A. Induction of dendritic cell migration upon Toxoplasma gondii infection potentiates parasite dissemination. Cell Microbiol. 2006;8:1611–1623. doi: 10.1111/j.1462-5822.2006.00735.x. [DOI] [PubMed] [Google Scholar]
- Lanigan LP, Birche R, Clark CV, Hill DW. The effect of cervical sympathectomy on retinal vessel responses to systemic autonomic stimulation. Eye (Lond) 1990;4 ( Pt 1):181–189. doi: 10.1038/eye.1990.24. [DOI] [PubMed] [Google Scholar]
- Lawley TJ, Kubota Y. Induction of morphologic differentiation of endothelial cells in culture. The Journal of investigative dermatology. 1989;93:59S–61S. doi: 10.1111/1523-1747.ep12581070. [DOI] [PubMed] [Google Scholar]
- Lawrenson JG, Cassella JP, Hayes AJ, Firth JA, Allt G. Endothelial glycoconjugates: a comparative lectin study of the brain, retina and myocardium. J Anat. 2000;196 ( Pt 1):55–60. doi: 10.1046/j.1469-7580.2000.19610055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal EC, Aveleira CA, Castilho AF, Serra AM, Baptista FI, Hosoya K, Forrester JV, Ambrosio AF. High glucose and oxidative/nitrosative stress conditions induce apoptosis in retinal endothelial cells by a caspase-independent pathway. Exp Eye Res. 2009;88:983–991. doi: 10.1016/j.exer.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Lebherz C, Maguire AM, Auricchio A, Tang W, Aleman TS, Wei Z, Grant R, Cideciyan AV, Jacobson SG, Wilson JM, Bennett J. Nonhuman primate models for diabetic ocular neovascularization using AAV2-mediated overexpression of vascular endothelial growth factor. Diabetes. 2005;54:1141–1149. doi: 10.2337/diabetes.54.4.1141. [DOI] [PubMed] [Google Scholar]
- Lee MT, Hooper LC, Kump L, Hayashi K, Nussenblatt R, Hooks JJ, Detrick B. Interferon-beta and adhesion molecules (E-selectin and s-intracellular adhesion molecule-1) are detected in sera from patients with retinal vasculitis and are induced in retinal vascular endothelial cells by Toll-like receptor 3 signalling. Clin Exp Immunol. 2007a;147:71–80. doi: 10.1111/j.1365-2249.2006.03253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MT, Hooper LC, Kump L, Hayashi K, Nussenblatt R, Hooks JJ, Detrick B. Interferon-beta and adhesion molecules (E-selectin and s-intracellular adhesion molecule-1) are detected in sera from patients with retinal vasculitis and are induced in retinal vascular endothelial cells by Toll-like receptor 3 signalling. Clinical and experimental immunology. 2007b;147:71–80. doi: 10.1111/j.1365-2249.2006.03253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy AP, Levy NS, Wegner S, Goldberg MA. Transcriptional regulation of the rat vascular endothelial growth factor gene by hypoxia. J Biol Chem. 1995;270:13333–13340. doi: 10.1074/jbc.270.22.13333. [DOI] [PubMed] [Google Scholar]
- Ley K, Laudanna C, Cybulsky MI, Nourshargh S. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat Rev Immunol. 2007;7:678–689. doi: 10.1038/nri2156. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Yoneda M, Takeyama M, Sugita I, Tsunekawa H, Yamada H, Watanabe D, Mukai T, Yamamura M, Iwaki M, Zako M. Effect of infliximab on tumor necrosis factor-alpha-induced alterations in retinal microvascular endothelial cells and retinal pigment epithelial cells. J Ocul Pharmacol Ther. 2010;26:549–556. doi: 10.1089/jop.2010.0079. [DOI] [PubMed] [Google Scholar]
- Lim JI, Spee C, Hinton DR. A comparison of hypoxia-inducible factor-alpha in surgically excised neovascular membranes of patients with diabetes compared with idiopathic epiretinal membranes in nondiabetic patients. Retina (Philadelphia, Pa. 2010;30:1472–1478. doi: 10.1097/IAE.0b013e3181d6df09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., 3rd Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- Linsenmeier RA, Braun RD, McRipley MA, Padnick LB, Ahmed J, Hatchell DL, McLeod DS, Lutty GA. Retinal hypoxia in long-term diabetic cats. Invest Ophthalmol Vis Sci. 1998;39:1647–1657. [PubMed] [Google Scholar]
- Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- Liu HP, Yu MC, Jiang MH, Chen JX, Yan DP, Liu F, Ge BX. Association of supervillin with KIR2DL1 regulates the inhibitory signaling of natural killer cells. Cell Signal. 2011;23:487–496. doi: 10.1016/j.cellsig.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Liversidge J, Dick A, Gordon S. Nitric oxide mediates apoptosis through formation of peroxynitrite and Fas/Fas-ligand interactions in experimental autoimmune uveitis. Am J Pathol. 2002;160:905–916. doi: 10.1016/S0002-9440(10)64913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liversidge J, Sewell HF, Forrester JV. Interactions between lymphocytes and cells of the blood-retina barrier: mechanisms of T lymphocyte adhesion to human retinal capillary endothelial cells and retinal pigment epithelial cells in vitro. Immunology. 1990;71:390–396. [PMC free article] [PubMed] [Google Scholar]
- Lofqvist C, Chen J, Connor KM, Smith AC, Aderman CM, Liu N, Pintar JE, Ludwig T, Hellstrom A, Smith LE. IGFBP3 suppresses retinopathy through suppression of oxygen-induced vessel loss and promotion of vascular regrowth. Proc Natl Acad Sci U S A. 2007;104:10589–10594. doi: 10.1073/pnas.0702031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofqvist C, Willett KL, Aspegren O, Smith AC, Aderman CM, Connor KM, Chen J, Hellstrom A, Smith LE. Quantification and localization of the IGF/insulin system expression in retinal blood vessels and neurons during oxygen-induced retinopathy in mice. Invest Ophthalmol Vis Sci. 2009;50:1831–1837. doi: 10.1167/iovs.08-2903. [DOI] [PubMed] [Google Scholar]
- London NJ, Rathinam SR, Cunningham ET., Jr The epidemiology of uveitis in developing countries. Int Ophthalmol Clin. 2010;50:1–17. doi: 10.1097/IIO.0b013e3181d2cc6b. [DOI] [PubMed] [Google Scholar]
- Lowder C, Belfort R, Jr, Lightman S, Foster CS, Robinson MR, Schiffman RM, Li XY, Cui H, Whitcup SM. Dexamethasone intravitreal implant for noninfectious intermediate or posterior uveitis. Arch Ophthalmol. 2011;129:545–553. doi: 10.1001/archophthalmol.2010.339. [DOI] [PubMed] [Google Scholar]
- Lowe WL, Jr, Florkiewicz RZ, Yorek MA, Spanheimer RG, Albrecht BN. Regulation of growth factor mRNA levels in the eyes of diabetic rats. Metabolism. 1995;44:1038. doi: 10.1016/0026-0495(95)90102-7. [DOI] [PubMed] [Google Scholar]
- Lowenstein CJ, Dinerman JL, Snyder SH. Nitric oxide: a physiologic messenger. Ann Intern Med. 1994;120:227–237. doi: 10.7326/0003-4819-120-3-199402010-00009. [DOI] [PubMed] [Google Scholar]
- Lu M, Perez VL, Ma N, Miyamoto K, Peng HB, Liao JK, Adamis AP. VEGF increases retinal vascular ICAM-1 expression in vivo. Invest Ophthalmol Vis Sci. 1999;40:1808–1812. [PubMed] [Google Scholar]
- Luger D, Caspi RR. New perspectives on effector mechanisms in uveitis. Seminars in immunopathology. 2008;30:135–143. doi: 10.1007/s00281-008-0108-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger D, Silver PB, Tang J, Cua D, Chen Z, Iwakura Y, Bowman EP, Sgambellone NM, Chan CC, Caspi RR. Either a Th17 or a Th1 effector response can drive autoimmunity: conditions of disease induction affect dominant effector category. The Journal of experimental medicine. 2008;205:799–810. doi: 10.1084/jem.20071258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna JD, Chan CC, Derevjanik NL, Mahlow J, Chiu C, Peng B, Tobe T, Campochiaro PA, Vinores SA. Blood-retinal barrier (BRB) breakdown in experimental autoimmune uveoretinitis: comparison with vascular endothelial growth factor, tumor necrosis factor alpha, and interleukin-1beta-mediated breakdown. J Neurosci Res. 1997;49:268–280. doi: 10.1002/(sici)1097-4547(19970801)49:3<268::aid-jnr2>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Lutty GA, McLeod DS, Merges C, Diggs A, Plouet J. Localization of vascular endothelial growth factor in human retina and choroid. Arch Ophthalmol. 1996;114:971–977. doi: 10.1001/archopht.1996.01100140179011. [DOI] [PubMed] [Google Scholar]
- Luz-Madrigal A, Clapp C, Aranda J, Vaca L. In vivo transcriptional targeting into the retinal vasculature using recombinant baculovirus carrying the human flt-1 promoter. Virol J. 2007;4:88. doi: 10.1186/1743-422X-4-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonpierre PC, Suri C, Jones PF, Bartunkova S, Wiegand SJ, Radziejewski C, Compton D, McClain J, Aldrich TH, Papadopoulos N, Daly TJ, Davis S, Sato TN, Yancopoulos GD. Angiopoietin-2, a natural antagonist for Tie2 that disrupts in vivo angiogenesis. Science. 1997;277:55–60. doi: 10.1126/science.277.5322.55. [DOI] [PubMed] [Google Scholar]
- Manalo DJ, Rowan A, Lavoie T, Natarajan L, Kelly BD, Ye SQ, Garcia JG, Semenza GL. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood. 2005;105:659–669. doi: 10.1182/blood-2004-07-2958. [DOI] [PubMed] [Google Scholar]
- Martin AP, de Moraes LV, Tadokoro CE, Commodaro AG, Urrets-Zavalia E, Rabinovich GA, Urrets-Zavalia J, Rizzo LV, Serra HM. Administration of a peptide inhibitor of alpha4-integrin inhibits the development of experimental autoimmune uveitis. Invest Ophthalmol Vis Sci. 2005;46:2056–2063. doi: 10.1167/iovs.04-0418. [DOI] [PubMed] [Google Scholar]
- Martin JL, Baxter RC. Signalling pathways of insulin-like growth factors (IGFs) and IGF binding protein-3. Growth Factors. 2011;29:235–244. doi: 10.3109/08977194.2011.614237. [DOI] [PubMed] [Google Scholar]
- Matsusaka T. Tridimensional views of the relationship of pericytes to endothelial cells of capillaries in the human choroid and retina. J Electron Microsc (Tokyo) 1975;24:13–18. [PubMed] [Google Scholar]
- McCawley LJ, Matrisian LM. Matrix metalloproteinases: they’re not just for matrix anymore! Curr Opin Cell Biol. 2001;13:534–540. doi: 10.1016/s0955-0674(00)00248-9. [DOI] [PubMed] [Google Scholar]
- McLeod DS, D’Anna SA, Lutty GA. Clinical and histopathologic features of canine oxygen-induced proliferative retinopathy. Invest Ophthalmol Vis Sci. 1998;39:1918–1932. [PubMed] [Google Scholar]
- McLeod DS, Hasegawa T, Prow T, Merges C, Lutty G. The initial fetal human retinal vasculature develops by vasculogenesis. Dev Dyn. 2006;235:3336–3347. doi: 10.1002/dvdy.20988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod DS, Lefer DJ, Merges C, Lutty GA. Enhanced expression of intracellular adhesion molecule-1 and P-selectin in the diabetic human retina and choroid. Am J Pathol. 1995;147:642–653. [PMC free article] [PubMed] [Google Scholar]
- McLoone E, O’Keefe M, McLoone S, Lanigan B. Long term functional and structural outcomes of laser therapy for retinopathy of prematurity. Br J Ophthalmol. 2006;90:754–759. doi: 10.1136/bjo.2005.068304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMenamin PG, Forrester JV, Steptoe RJ, Dua HS. Ultrastructural pathology of experimental autoimmune uveitis. Quantitative evidence of activation and possible high endothelial venule-like changes in retinal vascular endothelium. Lab Invest. 1992;67:42–55. [PubMed] [Google Scholar]
- Menage MJ, Robinson JC, Kaufman PL, Sponsel WE. Retinal blood flow after superior cervical ganglionectomy: a laser Doppler study in the cynomolgus monkey. Br J Ophthalmol. 1994;78:49–53. doi: 10.1136/bjo.78.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendrinos E, Petropoulos IK, Mangioris G, Tsilimbaris MK, Papadopoulou DN, Geka A, Pournaras CJ. Vasomotor effect of intravitreal juxta-arteriolar injection of L-lactate on the retinal arterioles after acute branch retinal vein occlusion in minipigs. Invest Ophthalmol Vis Sci. 2011;52:3215–3220. doi: 10.1167/iovs.10-6888. [DOI] [PubMed] [Google Scholar]
- Meyer-Schwickerath R, Pfeiffer A, Blum WF, Freyberger H, Klein M, Losche C, Rollmann R, Schatz H. Vitreous levels of the insulin-like growth factors I and II, and the insulin-like growth factor binding proteins 2 and 3, increase in neovascular eye disease. Studies in nondiabetic and diabetic subjects. The Journal of clinical investigation. 1993;92:2620–2625. doi: 10.1172/JCI116877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micieli JA, Surkont M, Smith AF. A systematic analysis of the off-label use of bevacizumab for severe retinopathy of prematurity. American journal of ophthalmology. 2009;148:536–543. e532. doi: 10.1016/j.ajo.2009.05.031. [DOI] [PubMed] [Google Scholar]
- Middleton J, Patterson AM, Gardner L, Schmutz C, Ashton BA. Leukocyte extravasation: chemokine transport and presentation by the endothelium. Blood. 2002;100:3853–3860. doi: 10.1182/blood.V100.12.3853. [DOI] [PubMed] [Google Scholar]
- Miyamoto K, Khosrof S, Bursell SE, Rohan R, Murata T, Clermont AC, Aiello LP, Ogura Y, Adamis AP. Prevention of leukostasis and vascular leakage in streptozotocin-induced diabetic retinopathy via intercellular adhesion molecule-1 inhibition. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:10836–10841. doi: 10.1073/pnas.96.19.10836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto N, Mandai M, Takagi H, Suzuma I, Suzuma K, Koyama S, Otani A, Oh H, Honda Y. Contrasting effect of estrogen on VEGF induction under different oxygen status and its role in murine ROP. Invest Ophthalmol Vis Sci. 2002;43:2007–2014. [PubMed] [Google Scholar]
- Mohr S, Xi X, Tang J, Kern TS. Caspase activation in retinas of diabetic and galactosemic mice and diabetic patients. Diabetes. 2002;51:1172–1179. doi: 10.2337/diabetes.51.4.1172. [DOI] [PubMed] [Google Scholar]
- Monteiro VG, Soares CP, de Souza W. Host cell surface sialic acid residues are involved on the process of penetration of Toxoplasma gondii into mammalian cells. FEMS Microbiol Lett. 1998;164:323–327. doi: 10.1111/j.1574-6968.1998.tb13105.x. [DOI] [PubMed] [Google Scholar]
- Montoya JG, Liesenfeld O. Toxoplasmosis. Lancet. 2004;363:1965–1976. doi: 10.1016/S0140-6736(04)16412-X. [DOI] [PubMed] [Google Scholar]
- Montoya JG, Remington JS. Toxoplasmosis of the central nervous system. In: Peterson PK, Remington JS, editors. Defense of the Brain: Current Concepts in the Immunopathogenesis and Clinical Aspects of CNS Infections. Blackwell Scientific; Boston: 1997. [Google Scholar]
- Motiejunaite R, Kazlauskas A. Pericytes and ocular diseases. Exp Eye Res. 2008;86:171–177. doi: 10.1016/j.exer.2007.10.013. [DOI] [PubMed] [Google Scholar]
- Mowat FM, Luhmann UF, Smith AJ, Lange C, Duran Y, Harten S, Shukla D, Maxwell PH, Ali RR, Bainbridge JW. HIF-1alpha and HIF-2alpha are differentially activated in distinct cell populations in retinal ischaemia. PLoS One. 2010;5:e11103. doi: 10.1371/journal.pone.0011103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagineni CN, Detrick B, Hooks JJ. Transforming growth factor-beta expression in human retinal pigment epithelial cells is enhanced by Toxoplasma gondii: a possible role in the immunopathogenesis of retinochoroiditis. Clinical and experimental immunology. 2002;128:372–378. doi: 10.1046/j.1365-2249.2002.01815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y, Yamada T, Ui-Tei K, Morishita S, Saigo K. siDirect: highly effective, target-specific siRNA design software for mammalian RNA interference. Nucleic Acids Res. 2004;32:W124–129. doi: 10.1093/nar/gkh442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambu H, Nambu R, Oshima Y, Hackett SF, Okoye G, Wiegand S, Yancopoulos G, Zack DJ, Campochiaro PA. Angiopoietin 1 inhibits ocular neovascularization and breakdown of the blood-retinal barrier. Gene Ther. 2004;11:865–873. doi: 10.1038/sj.gt.3302230. [DOI] [PubMed] [Google Scholar]
- Nambu H, Umeda N, Kachi S, Oshima Y, Akiyama H, Nambu R, Campochiaro PA. Angiopoietin 1 prevents retinal detachment in an aggressive model of proliferative retinopathy, but has no effect on established neovascularization. J Cell Physiol. 2005;204:227–235. doi: 10.1002/jcp.20292. [DOI] [PubMed] [Google Scholar]
- Nguyen AM, Rao NA. Oxidative photoreceptor cell damage in autoimmune uveitis. J Ophthalmic Inflamm Infect. 2011;1:7–13. doi: 10.1007/s12348-010-0007-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda K, Nakao S, Ishida S, Ishibashi T. Leukocyte adhesion molecules in diabetic retinopathy. J Ophthalmol. 2012;2012:279037. doi: 10.1155/2012/279037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourshargh S, Hordijk PL, Sixt M. Breaching multiple barriers: leukocyte motility through venular walls and the interstitium. Nat Rev Mol Cell Biol. 2010;11:366–378. doi: 10.1038/nrm2889. [DOI] [PubMed] [Google Scholar]
- Nussenblatt RB. The natural history of uveitis. Int Ophthalmol. 1990;14:303–308. doi: 10.1007/BF00163549. [DOI] [PubMed] [Google Scholar]
- Nussenblatt RB. Proctor Lecture. Experimental autoimmune uveitis: mechanisms of disease and clinical therapeutic indications. Invest Ophthalmol Vis Sci. 1991;32:3131–3141. [PubMed] [Google Scholar]
- Oh H, Takagi H, Suzuma K, Otani A, Matsumura M, Honda Y. Hypoxia and vascular endothelial growth factor selectively up-regulate angiopoietin-2 in bovine microvascular endothelial cells. J Biol Chem. 1999;274:15732–15739. doi: 10.1074/jbc.274.22.15732. [DOI] [PubMed] [Google Scholar]
- Oh HM, Yu CR, Lee Y, Chan CC, Maminishkis A, Egwuagu CE. Autoreactive memory CD4+ T lymphocytes that mediate chronic uveitis reside in the bone marrow through STAT3-dependent mechanisms. J Immunol. 2011;187:3338–3346. doi: 10.4049/jimmunol.1004019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SW, Pope RK, Smith KP, Crowley JL, Nebl T, Lawrence JB, Luna EJ. Archvillin, a muscle-specific isoform of supervillin, is an early expressed component of the costameric membrane skeleton. J Cell Sci. 2003;116:2261–2275. doi: 10.1242/jcs.00422. [DOI] [PubMed] [Google Scholar]
- Ohashi H, Takagi H, Koyama S, Oh H, Watanabe D, Antonetti DA, Matsubara T, Nagai K, Arai H, Kita T, Honda Y. Alterations in expression of angiopoietins and the Tie-2 receptor in the retina of streptozotocin induced diabetic rats. Mol Vis. 2004;10:608–617. [PubMed] [Google Scholar]
- Ohno-Matsui K, Hirose A, Yamamoto S, Saikia J, Okamoto N, Gehlbach P, Duh EJ, Hackett S, Chang M, Bok D, Zack DJ, Campochiaro PA. Inducible expression of vascular endothelial growth factor in adult mice causes severe proliferative retinopathy and retinal detachment. Am J Pathol. 2002;160:711–719. doi: 10.1016/S0002-9440(10)64891-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada AA, Keino H, Fukai T, Sakai J, Usui M, Mizuguchi J. Effect of type I interferon on experimental autoimmune uveoretinitis in rats. Ocular immunology and inflammation. 1998a;6:215–226. doi: 10.1076/ocii.6.4.215.4024. [DOI] [PubMed] [Google Scholar]
- Okada AA, Keino H, Suzuki J, Sakai J, Usui M, Mizuguchi J. Kinetics of intraocular cytokines in the suppression of experimental autoimmune uveoretinitis by type I IFN. Int Immunol. 1998b;10:1917–1922. doi: 10.1093/intimm/10.12.1917. [DOI] [PubMed] [Google Scholar]
- Olson JA, Whitelaw CM, McHardy KC, Pearson DW, Forrester JV. Soluble leucocyte adhesion molecules in diabetic retinopathy stimulate retinal capillary endothelial cell migration. Diabetologia. 1997;40:1166–1171. doi: 10.1007/s001250050802. [DOI] [PubMed] [Google Scholar]
- Ortega-Barria E, Boothroyd JC. A Toxoplasma lectin-like activity specific for sulfated polysaccharides is involved in host cell infection. J Biol Chem. 1999;274:1267–1276. doi: 10.1074/jbc.274.3.1267. [DOI] [PubMed] [Google Scholar]
- Oshima Y, Oshima S, Nambu H, Kachi S, Takahashi K, Umeda N, Shen J, Dong A, Apte RS, Duh E, Hackett SF, Okoye G, Ishibashi K, Handa J, Melia M, Wiegand S, Yancopoulos G, Zack DJ, Campochiaro PA. Different effects of angiopoietin-2 in different vascular beds: new vessels are most sensitive. Faseb J. 2005;19:963–965. doi: 10.1096/fj.04-2209fje. [DOI] [PubMed] [Google Scholar]
- Otrock ZK, Makarem JA, Shamseddine AI. Vascular endothelial growth factor family of ligands and receptors: review. Blood Cells Mol Dis. 2007;38:258–268. doi: 10.1016/j.bcmd.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Ottino P, Finley J, Rojo E, Ottlecz A, Lambrou GN, Bazan HE, Bazan NG. Hypoxia activates matrix metalloproteinase expression and the VEGF system in monkey choroid-retinal endothelial cells: Involvement of cytosolic phospholipase A2 activity. Mol Vis. 2004;10:341–350. [PubMed] [Google Scholar]
- Ozaki H, Yu AY, Della N, Ozaki K, Luna JD, Yamada H, Hackett SF, Okamoto N, Zack DJ, Semenza GL, Campochiaro PA. Hypoxia inducible factor-1alpha is increased in ischemic retina: temporal and spatial correlation with VEGF expression. Invest Ophthalmol Vis Sci. 1999;40:182–189. [PubMed] [Google Scholar]
- Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Xia L, McEver RP. Comparison of promoters for the murine and human P-selectin genes suggests species-specific and conserved mechanisms for transcriptional regulation in endothelial cells. J Biol Chem. 1998;273:10058–10067. doi: 10.1074/jbc.273.16.10058. [DOI] [PubMed] [Google Scholar]
- Papachristodoulou D, Heath H, Kang SS. The development of retinopathy in sucrose-fed and streptozotocin-diabetic rats. Diabetologia. 1976;12:367–374. doi: 10.1007/BF00420981. [DOI] [PubMed] [Google Scholar]
- Paques M, Guyomard JL, Simonutti M, Roux MJ, Picaud S, Legargasson JF, Sahel JA. Panretinal, high-resolution color photography of the mouse fundus. Invest Ophthalmol Vis Sci. 2007;48:2769–2774. doi: 10.1167/iovs.06-1099. [DOI] [PubMed] [Google Scholar]
- Parikh JG, Saraswathy S, Rao NA. Photoreceptor oxidative damage in sympathetic ophthalmia. American journal of ophthalmology. 2008;146:866–875. e862. doi: 10.1016/j.ajo.2008.03.026. [DOI] [PubMed] [Google Scholar]
- Parnaby-Price A, Stanford MR, Biggerstaff J, Howe L, Whiston RA, Marshall J, Wallace GR. Leukocyte trafficking in experimental autoimmune uveitis in vivo. J Leukoc Biol. 1998a;64:434–440. doi: 10.1002/jlb.64.4.434. [DOI] [PubMed] [Google Scholar]
- Parnaby-Price A, Stanford MR, Biggerstaff J, Howe L, Whiston RA, Marshall J, Wallace GR. Leukocyte trafficking in experimental autoimmune uveitis in vivo. J Leukoc Biol. 1998b;64:434–440. doi: 10.1002/jlb.64.4.434. [DOI] [PubMed] [Google Scholar]
- Pasqualini R, Moeller BJ, Arap W. Leveraging molecular heterogeneity of the vascular endothelium for targeted drug delivery and imaging. Semin Thromb Hemost. 2010;36:343–351. doi: 10.1055/s-0030-1253456. [DOI] [PubMed] [Google Scholar]
- Pavesio C, Zierhut M, Bairi K, Comstock TL, Usner DW. Evaluation of an intravitreal fluocinolone acetonide implant versus standard systemic therapy in noninfectious posterior uveitis. Ophthalmology. 2010;117:567–575. 575, e561. doi: 10.1016/j.ophtha.2009.11.027. [DOI] [PubMed] [Google Scholar]
- Pellieux C, Desgeorges A, Pigeon CH, Chambaz C, Yin H, Hayoz D, Silacci P. Cap G, a gelsolin family protein modulating protective effects of unidirectional shear stress. J Biol Chem. 2003;278:29136–29144. doi: 10.1074/jbc.M300598200. [DOI] [PubMed] [Google Scholar]
- Penn JS, Tolman BL, Lowery LA. Variable oxygen exposure causes preretinal neovascularization in the newborn rat. Invest Ophthalmol Vis Sci. 1993;34:576–585. [PubMed] [Google Scholar]
- Pestonjamasp KN, Pope RK, Wulfkuhle JD, Luna EJ. Supervillin (p205): A novel membrane-associated, F-actin-binding protein in the villin/gelsolin superfamily. The Journal of cell biology. 1997;139:1255–1269. doi: 10.1083/jcb.139.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters S, Cree IA, Alexander R, Turowski P, Ockrim Z, Patel J, Boyd SR, Joussen AM, Ziemssen F, Hykin PG, Moss SE. Angiopoietin modulation of vascular endothelial growth factor: Effects on retinal endothelial cell permeability. Cytokine. 2007;40:144–150. doi: 10.1016/j.cyto.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Pfister F, Feng Y, vom Hagen F, Hoffmann S, Molema G, Hillebrands JL, Shani M, Deutsch U, Hammes HP. Pericyte migration: a novel mechanism of pericyte loss in experimental diabetic retinopathy. Diabetes. 2008;57:2495–2502. doi: 10.2337/db08-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan QT, Fratti RA, Prasadarao NV, Edwards JE, Jr, Filler SG. N-cadherin mediates endocytosis of Candida albicans by endothelial cells. J Biol Chem. 2005;280:10455–10461. doi: 10.1074/jbc.M412592200. [DOI] [PubMed] [Google Scholar]
- Pope RK, Pestonjamasp KN, Smith KP, Wulfkuhle JD, Strassel CP, Lawrence JB, Luna EJ. Cloning, characterization, and chromosomal localization of human superillin (SVIL) Genomics. 1998;52:342–351. doi: 10.1006/geno.1998.5466. [DOI] [PubMed] [Google Scholar]
- Poulaki V, Joussen AM, Mitsiades N, Mitsiades CS, Iliaki EF, Adamis AP. Insulin-like growth factor-I plays a pathogenetic role in diabetic retinopathy. Am J Pathol. 2004;165:457–469. doi: 10.1016/S0002-9440(10)63311-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulaki V, Qin W, Joussen AM, Hurlbut P, Wiegand SJ, Rudge J, Yancopoulos GD, Adamis AP. Acute intensive insulin therapy exacerbates diabetic blood-retinal barrier breakdown via hypoxia-inducible factor-1alpha and VEGF. The Journal of clinical investigation. 2002;109:805–815. doi: 10.1172/JCI13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pournaras CJ, Rungger-Brandle E, Riva CE, Hardarson SH, Stefansson E. Regulation of retinal blood flow in health and disease. Prog Retin Eye Res. 2008;27:284–330. doi: 10.1016/j.preteyeres.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Prasadarao NV, Srivastava PK, Rudrabhatla RS, Kim KS, Huang SH, Sukumaran SK. Cloning and expression of the Escherichia coli K1 outer membrane protein A receptor, a gp96 homologue. Infect Immun. 2003;71:1680–1688. doi: 10.1128/IAI.71.4.1680-1688.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Hoareau-Aveilla C, Oltean S, Harper SJ, Bates DO. The anti-angiogenic isoforms of VEGF in health and disease. Biochem Soc Trans. 2009;37:1207–1213. doi: 10.1042/BST0371207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillen DA, Davis JB, Gottlieb JL, Blodi BA, Callanan DG, Chang TS, Equi RA. The white dot syndromes. American journal of ophthalmology. 2004;137:538–550. doi: 10.1016/j.ajo.2004.01.053. [DOI] [PubMed] [Google Scholar]
- Rajendram R, Saraswathy S, Rao NA. Photoreceptor mitochondrial oxidative stress in early experimental autoimmune uveoretinitis. Br J Ophthalmol. 2007;91:531–537. doi: 10.1136/bjo.2006.101576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoczy EP, Ali Rahman IS, Binz N, Li CR, Vagaja NN, de Pinho M, Lai CM. Characterization of a mouse model of hyperglycemia and retinal neovascularization. Am J Pathol. 2010;177:2659–2670. doi: 10.2353/ajpath.2010.090883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoczy PE, Brankov M, Fonceca A, Zaknich T, Rae BC, Lai CM. Enhanced recombinant adeno-associated virus-mediated vascular endothelial growth factor expression in the adult mouse retina: a potential model for diabetic retinopathy. Diabetes. 2003;52:857–863. doi: 10.2337/diabetes.52.3.857. [DOI] [PubMed] [Google Scholar]
- Rangasamy S, Srinivasan R, Maestas J, McGuire PG, Das A. A potential role for angiopoietin 2 in the regulation of the blood-retinal barrier in diabetic retinopathy. Invest Ophthalmol Vis Sci. 2011;52:3784–3791. doi: 10.1167/iovs.10-6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao NA. Role of oxygen free radicals in retinal damage associated with experimental uveitis. Trans Am Ophthalmol Soc. 1990;88:797–850. [PMC free article] [PubMed] [Google Scholar]
- Rao NA, Zhang J, Ishimoto S. Role of retinal vascular endothelial cells in development of CMV retinitis. Trans Am Ophthalmol Soc. 1998;96:111–123. discussion 124–116. [PMC free article] [PubMed] [Google Scholar]
- Rhodin JA. The ultrastructure of mammalian arterioles and precapillary sphincters. J Ultrastruct Res. 1967;18:181–223. doi: 10.1016/s0022-5320(67)80239-9. [DOI] [PubMed] [Google Scholar]
- Rhodin JA. Ultrastructure of mammalian venous capillaries, venules, and small collecting veins. J Ultrastruct Res. 1968;25:452–500. doi: 10.1016/s0022-5320(68)80098-x. [DOI] [PubMed] [Google Scholar]
- Rinderknecht E, Humbel RE. The amino acid sequence of human insulin-like growth factor I and its structural homology with proinsulin. J Biol Chem. 1978;253:2769–2776. [PubMed] [Google Scholar]
- Roberts F, McLeod R. Pathogenesis of toxoplasmic retinochoroiditis. Parasitol Today. 1999;15:51–57. doi: 10.1016/s0169-4758(98)01377-5. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Calonge M, Pedroza-Seres M, Akova YA, Messmer EM, D’Amico DJ, Foster CS. Referral patterns of uveitis in a tertiary eye care center. Arch Ophthalmol. 1996;114:593–599. doi: 10.1001/archopht.1996.01100130585016. [DOI] [PubMed] [Google Scholar]
- Rot A, von Andrian UH. Chemokines in innate and adaptive host defense: basic chemokinese grammar for immune cells. Annual review of immunology. 2004;22:891–928. doi: 10.1146/annurev.immunol.22.012703.104543. [DOI] [PubMed] [Google Scholar]
- Rothova A. Ocular involvement in toxoplasmosis. Br J Ophthalmol. 1993;77:371–377. doi: 10.1136/bjo.77.6.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothova A, Suttorp-van Schulten MS, Frits Treffers W, Kijlstra A. Causes and frequency of blindness in patients with intraocular inflammatory disease. Br J Ophthalmol. 1996;80:332–336. doi: 10.1136/bjo.80.4.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Ha J, Trudeau K, Beglova E. Vascular basement membrane thickening in diabetic retinopathy. Curr Eye Res. 2010;35:1045–1056. doi: 10.3109/02713683.2010.514659. [DOI] [PubMed] [Google Scholar]
- Ruberte J, Ayuso E, Navarro M, Carretero A, Nacher V, Haurigot V, George M, Llombart C, Casellas A, Costa C, Bosch A, Bosch F. Increased ocular levels of IGF-1 in transgenic mice lead to diabetes-like eye disease. The Journal of clinical investigation. 2004;113:1149–1157. doi: 10.1172/JCI19478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Moreno JM, Thillaye B, de Kozak Y. Retino-choroidal changes in endotoxin-induced uveitis in the rat. Ophthalmic Res. 1992;24:162–168. doi: 10.1159/000267163. [DOI] [PubMed] [Google Scholar]
- Rymaszewski Z, Szymanski PT, Abplanalp WA, Myatt L, Di Salvo J, Cohen RM. Human retinal vascular cells differ from umbilical cells in synthetic functions and their response to glucose. Proc Soc Exp Biol Med. 1992;199:183–191. doi: 10.3181/00379727-199-43345. [DOI] [PubMed] [Google Scholar]
- Rymo SF, Gerhardt H, Wolfhagen Sand F, Lang R, Uv A, Betsholtz C. A two-way communication between microglial cells and angiogenic sprouts regulates angiogenesis in aortic ring cultures. PLoS One. 2011;6:e15846. doi: 10.1371/journal.pone.0015846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F, Lenig D, Mackay CR, Lanzavecchia A. Flexible programs of chemokine receptor expression on human polarized T helper 1 and 2 lymphocytes. The Journal of experimental medicine. 1998;187:875–883. doi: 10.1084/jem.187.6.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders MD, Graham EM. Retinal vasculitis. Postgraduate medical journal. 1988;64:488–496. doi: 10.1136/pgmj.64.753.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni G, Spreghini E, Lucciarini R, Amantini C, Piccoli M. Involvement of alpha(v)beta3 integrin-like receptor and glycosaminoglycans in Candida albicans germ tube adhesion to vitronectin and to a human endothelial cell line. Microb Pathog. 2001;31:159–172. doi: 10.1006/mpat.2001.0459. [DOI] [PubMed] [Google Scholar]
- Santoni V, Molloy M, Rabilloud T. Membrane proteins and proteomics: un amour impossible? Electrophoresis. 2000;21:1054–1070. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1054::AID-ELPS1054>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Sanz L, Pascual M, Munoz A, Gonzalez MA, Salvador CH, Alvarez-Vallina L. Development of a computer-assisted high-throughput screening platform for anti-angiogenic testing. Microvasc Res. 2002;63:335–339. doi: 10.1006/mvre.2001.2389. [DOI] [PubMed] [Google Scholar]
- Sapieha P, Joyal JS, Rivera JC, Kermorvant-Duchemin E, Sennlaub F, Hardy P, Lachapelle P, Chemtob S. Retinopathy of prematurity: understanding ischemic retinal vasculopathies at an extreme of life. The Journal of clinical investigation. 2010;120:3022–3032. doi: 10.1172/JCI42142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapieha P, Sirinyan M, Hamel D, Zaniolo K, Joyal JS, Cho JH, Honore JC, Kermorvant-Duchemin E, Varma DR, Tremblay S, Leduc M, Rihakova L, Hardy P, Klein WH, Mu X, Mamer O, Lachapelle P, Di Polo A, Beausejour C, Andelfinger G, Mitchell G, Sennlaub F, Chemtob S. The succinate receptor GPR91 in neurons has a major role in retinal angiogenesis. Nat Med. 2008;14:1067–1076. doi: 10.1038/nm.1873. [DOI] [PubMed] [Google Scholar]
- Saraswathy S, Rao NA. Mitochondrial proteomics in experimental autoimmune uveitis oxidative stress. Invest Ophthalmol Vis Sci. 2009;50:5559–5566. doi: 10.1167/iovs.08-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sari RA, Kiziltunc A, Taysi S, Akdemir S, Gundogdu M. Levels of soluble E-selectin in patients with active Behcet’s disease. Clinical rheumatology. 2005;24:55–59. doi: 10.1007/s10067-004-0982-8. [DOI] [PubMed] [Google Scholar]
- Schlie K, Spowart JE, Hughson LR, Townsend KN, Lum JJ. When Cells Suffocate: Autophagy in Cancer and Immune Cells under Low Oxygen. International journal of cell biology. 2011;2011:470597. doi: 10.1155/2011/470597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittler H, Preissner KT. Between microbial attack and defence: the endothelium as a vulnerable player in infectious diseases. Thromb Haemost. 2009;102:1011–1013. doi: 10.1160/TH09-11-0760. [DOI] [PubMed] [Google Scholar]
- Schnittler HJ, Preissner KT. Vascular endothelium and infectious diseases: trick and treat. Thromb Haemost. 2005;94:238–239. doi: 10.1160/TH05-07-0469. [DOI] [PubMed] [Google Scholar]
- Servat JJ, Mears KA, Black EH, Huang JJ. Biological agents for the treatment of uveitis. Expert Opin Biol Ther. 2012 doi: 10.1517/14712598.2012.658366. [DOI] [PubMed] [Google Scholar]
- Sfikakis PP. The first decade of biologic TNF antagonists in clinical practice: lessons learned, unresolved issues and future directions. Curr Dir Autoimmun. 2010;11:180–210. doi: 10.1159/000289205. [DOI] [PubMed] [Google Scholar]
- Shaw LC, Grant MB. Insulin like growth factor-1 and insulin-like growth factor binding proteins: their possible roles in both maintaining normal retinal vascular function and in promoting retinal pathology. Rev Endocr Metab Disord. 2004;5:199–207. doi: 10.1023/B:REMD.0000032408.18015.b1. [DOI] [PubMed] [Google Scholar]
- Shaw LC, Pan H, Afzal A, Calzi SL, Spoerri PE, Sullivan SM, Grant MB. Proliferating endothelial cell-specific expression of IGF-I receptor ribozyme inhibits retinal neovascularization. Gene Ther. 2006;13:752–760. doi: 10.1038/sj.gt.3302718. [DOI] [PubMed] [Google Scholar]
- Shen WY, Lai CM, Graham CE, Binz N, Lai YK, Eade J, Guidolin D, Ribatti D, Dunlop SA, Rakoczy PE. Long-term global retinal microvascular changes in a transgenic vascular endothelial growth factor mouse model. Diabetologia. 2006;49:1690–1701. doi: 10.1007/s00125-006-0274-8. [DOI] [PubMed] [Google Scholar]
- Shi G, Cox CA, Vistica BP, Tan C, Wawrousek EF, Gery I. Phenotype switching by inflammation-inducing polarized Th17 cells, but not by Th1 cells. J Immunol. 2008;181:7205–7213. doi: 10.4049/jimmunol.181.10.7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shie JL, Wu G, Wu J, Liu FF, Laham RJ, Oettgen P, Li J. RTEF-1, a novel transcriptional stimulator of vascular endothelial growth factor in hypoxic endothelial cells. J Biol Chem. 2004;279:25010–25016. doi: 10.1074/jbc.M403103200. [DOI] [PubMed] [Google Scholar]
- Shin D, Garcia-Cardena G, Hayashi S, Gerety S, Asahara T, Stavrakis G, Isner J, Folkman J, Gimbrone MA, Jr, Anderson DJ. Expression of ephrinB2 identifies a stable genetic difference between arterial and venous vascular smooth muscle as well as endothelial cells, and marks subsets of microvessels at sites of adult neovascularization. Dev Biol. 2001;230:139–150. doi: 10.1006/dbio.2000.9957. [DOI] [PubMed] [Google Scholar]
- Shinohara M, Masuyama T, Shoda T, Takahashi T, Katsuda Y, Komeda K, Kuroki M, Kakehashi A, Kanazawa Y. A new spontaneously diabetic non-obese Torii rat strain with severe ocular complications. International journal of experimental diabetes research. 2000;1:89–100. doi: 10.1155/EDR.2000.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silacci P, Mazzolai L, Gauci C, Stergiopulos N, Yin HL, Hayoz D. Gelsolin superfamily proteins: key regulators of cellular functions. Cell Mol Life Sci. 2004;61:2614–2623. doi: 10.1007/s00018-004-4225-6. [DOI] [PubMed] [Google Scholar]
- Silveira C, Vallochi AL, Rodrigues da Silva U, Muccioli C, Holland GN, Nussenblatt RB, Belfort R, Rizzo LV. Toxoplasma gondii in the peripheral blood of patients with acute and chronic toxoplasmosis. Br J Ophthalmol. 2011;95:396–400. doi: 10.1136/bjo.2008.148205. [DOI] [PubMed] [Google Scholar]
- Silver PB, Tarrant TK, Chan CC, Wiggert B, Caspi RR. Mice deficient in inducible nitric oxide synthase are susceptible to experimental autoimmune uveoretinitis. Invest Ophthalmol Vis Sci. 1999;40:1280–1284. [PubMed] [Google Scholar]
- Silverman MD, Zamora DO, Pan Y, Texeira PV, Baek SH, Planck SR, Rosenbaum JT. Constitutive and inflammatory mediator-regulated fractalkine expression in human ocular tissues and cultured cells. Invest Ophthalmol Vis Sci. 2003;44:1608–1615. doi: 10.1167/iovs.02-0233. [DOI] [PubMed] [Google Scholar]
- Sima AA, Chakrabarti S, Garcia-Salinas R, Basu PK. The BB-rat--an authentic model of human diabetic retinopathy. Curr Eye Res. 1985;4:1087–1092. doi: 10.3109/02713688509003353. [DOI] [PubMed] [Google Scholar]
- Singh SP, Zhang HH, Foley JF, Hedrick MN, Farber JM. Human T cells that are able to produce IL-17 express the chemokine receptor CCR6. J Immunol. 2008;180:214–221. doi: 10.4049/jimmunol.180.1.214. [DOI] [PubMed] [Google Scholar]
- Sivakumar V, Zhang Y, Ling EA, Foulds WS, Kaur C. Insulin-like growth factors, angiopoietin-2, and pigment epithelium-derived growth factor in the hypoxic retina. Journal of neuroscience research. 2008;86:702–711. doi: 10.1002/jnr.21519. [DOI] [PubMed] [Google Scholar]
- Smith JR, Choi D, Chipps TJ, Pan Y, Zamora DO, Davies MH, Babra B, Powers MR, Planck SR, Rosenbaum JT. Unique gene expression profiles of donor-matched human retinal and choroidal vascular endothelial cells. Invest Ophthalmol Vis Sci. 2007;48:2676–2684. doi: 10.1167/iovs.06-0598. [DOI] [PubMed] [Google Scholar]
- Smith JR, Franc DT, Carter NS, Zamora D, Planck SR, Rosenbaum JT. Susceptibility of retinal vascular endothelium to infection with Toxoplasma gondii tachyzoites. Invest Ophthalmol Vis Sci. 2004;45:1157–1161. doi: 10.1167/iovs.03-1105. [DOI] [PubMed] [Google Scholar]
- Smith JR, O’Rourke LM, Becker MD, Cao M, Williams KA, Planck SR, Rosenbaum JT. Anti-rat ICAM-1 antibody does not influence the course of experimental melanin-induced uveitis. Curr Eye Res. 2000;21:906–912. doi: 10.1076/ceyr.21.5.906.5537. [DOI] [PubMed] [Google Scholar]
- Smith LE, Shen W, Perruzzi C, Soker S, Kinose F, Xu X, Robinson G, Driver S, Bischoff J, Zhang B, Schaeffer JM, Senger DR. Regulation of vascular endothelial growth factor-dependent retinal neovascularization by insulin-like growth factor-1 receptor. Nat Med. 1999;5:1390–1395. doi: 10.1038/70963. [DOI] [PubMed] [Google Scholar]
- Smith LE, Wesolowski E, McLellan A, Kostyk SK, D’Amato R, Sullivan R, D’Amore PA. Oxygen-induced retinopathy in the mouse. Invest Ophthalmol Vis Sci. 1994;35:101–111. [PubMed] [Google Scholar]
- Smolenski RT, Khalpey Z, Osborne FN, Yuen A, Slominska EM, Lipinski M, Lavitrano M, Rose M, Yacoub MH. Species differences of endothelial extracellular nucleotide metabolism and its implications for xenotransplantation. Pharmacol Rep. 2006;58(Suppl):118–125. [PubMed] [Google Scholar]
- Song J, Guo X, Xie X, Zhao X, Li D, Deng W, Song Y, Shen F, Wu M, Wei L. Autophagy in hypoxia protects cancer cells against apoptosis induced by nutrient deprivation through a Beclin1-dependent way in hepatocellular carcinoma. Journal of cellular biochemistry. 2011;112:3406–3420. doi: 10.1002/jcb.23274. [DOI] [PubMed] [Google Scholar]
- Soroceanu L, Akhavan A, Cobbs CS. Platelet-derived growth factor-alpha receptor activation is required for human cytomegalovirus infection. Nature. 2008;455:391–395. doi: 10.1038/nature07209. [DOI] [PubMed] [Google Scholar]
- Sosula L, Beaumont P, Hollows FC, Jonson KM. Dilatation and endothelial proliferation of retinal capillaries in streptozotocin-diabetic rats: quantitative electron microscopy. Invest Ophthalmol. 1972;11:926–935. [PubMed] [Google Scholar]
- Spoerri PE, Ellis EA, Tarnuzzer RW, Grant MB. Insulin-like growth factor: receptor and binding proteins in human retinal endothelial cell cultures of diabetic and non-diabetic origin. Growth hormone & IGF research : official journal of the Growth Hormone Research Society and the International IGF Research Society. 1998;8:125–132. doi: 10.1016/s1096-6374(98)80102-0. [DOI] [PubMed] [Google Scholar]
- Spranger J, Buhnen J, Jansen V, Krieg M, Meyer-Schwickerath R, Blum WF, Schatz H, Pfeiffer AF. Systemic levels contribute significantly to increased intraocular IGF-I, IGF-II and IGF-BP3 [correction of IFG-BP3] in proliferative diabetic retinopathy. Horm Metab Res. 2000;32:196–200. doi: 10.1055/s-2007-978621. [DOI] [PubMed] [Google Scholar]
- Stahl A, Connor KM, Sapieha P, Willett KL, Krah NM, Dennison RJ, Chen J, Guerin KI, Smith LE. Computer-aided quantification of retinal neovascularization. Angiogenesis. 2009;12:297–301. doi: 10.1007/s10456-009-9155-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamper HB, Jr, Woodruff JJ. An in vitro model of lymphocyte homing. I. Characterization of the interaction between thoracic duct lymphocytes and specialized high-endothelial venules of lymph nodes. J Immunol. 1977;119:772–780. [PubMed] [Google Scholar]
- Stanbury RM, Graham EM. Systemic corticosteroid therapy--side effects and their management. Br J Ophthalmol. 1998;82:704–708. doi: 10.1136/bjo.82.6.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standring S. Gray’s Anatomy: the anatomical basis of clinical practice. 40. Churchill Livingston Elsevier; Philadelphia: 2008. [Google Scholar]
- Stanford MR, See SE, Jones LV, Gilbert RE. Antibiotics for toxoplasmic retinochoroiditis: an evidence-based systematic review. Ophthalmology. 2003;110:926–931. doi: 10.1016/S0161-6420(03)00083-6. quiz 931–922. [DOI] [PubMed] [Google Scholar]
- Steinle JJ, Granger HJ. Nerve growth factor regulates human choroidal, but not retinal, endothelial cell migration and proliferation. Auton Neurosci. 2003;108:57–62. doi: 10.1016/S1566-0702(03)00151-6. [DOI] [PubMed] [Google Scholar]
- Stetler-Stevenson WG. Matrix metalloproteinases in angiogenesis: a moving target for therapeutic intervention. The Journal of clinical investigation. 1999;103:1237–1241. doi: 10.1172/JCI6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart EA, Samaranayake GJ, Browning AC, Hopkinson A, Amoaku WM. Comparison of choroidal and retinal endothelial cells: characteristics and response to VEGF isoforms and anti-VEGF treatments. Exp Eye Res. 2011;93:761–766. doi: 10.1016/j.exer.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Stitt AW. AGEs and diabetic retinopathy. Invest Ophthalmol Vis Sci. 2010;51:4867–4874. doi: 10.1167/iovs.10-5881. [DOI] [PubMed] [Google Scholar]
- Stone J, Itin A, Alon T, Pe’er J, Gnessin H, Chan-Ling T, Keshet E. Development of retinal vasculature is mediated by hypoxia-induced vascular endothelial growth factor (VEGF) expression by neuroglia. J Neurosci. 1995;15:4738–4747. doi: 10.1523/JNEUROSCI.15-07-04738.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhler EB, Lloyd MJ, Choi D, Rosenbaum JT, Austin DF. Incidence and prevalence of uveitis in Veterans Affairs Medical Centers of the Pacific Northwest. American journal of ophthalmology. 2008;146:890–896. e898. doi: 10.1016/j.ajo.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Sun M, Yang Y, Yang P, Lei B, Du L, Kijlstra A. Regulatory effects of IFN-beta on the development of experimental autoimmune uveoretinitis in B10RIII mice. PLoS One. 2011;6:e19870. doi: 10.1371/journal.pone.0019870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttorp-Schulten MS, Rothova A. The possible impact of uveitis in blindness: a literature survey. Br J Ophthalmol. 1996;80:844–848. doi: 10.1136/bjo.80.9.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuma I, Mandai M, Suzuma K, Ishida K, Tojo SJ, Honda Y. Contribution of E-selectin to cellular infiltration during endotoxin-induced uveitis. Invest Ophthalmol Vis Sci. 1998a;39:1620–1630. [PubMed] [Google Scholar]
- Suzuma K, Takagi H, Otani A, Suzuma I, Honda Y. Increased expression of KDR/Flk-1 (VEGFR-2) in murine model of ischemia-induced retinal neovascularization. Microvasc Res. 1998b;56:183–191. doi: 10.1006/mvre.1998.2111. [DOI] [PubMed] [Google Scholar]
- Takagi H, King GL, Ferrara N, Aiello LP. Hypoxia regulates vascular endothelial growth factor receptor KDR/Flk gene expression through adenosine A2 receptors in retinal capillary endothelial cells. Invest Ophthalmol Vis Sci. 1996;37:1311–1321. [PubMed] [Google Scholar]
- Takase H, Yu CR, Liu X, Fujimoto C, Gery I, Egwuagu CE. Induction of suppressors of cytokine signaling (SOCS) in the retina during experimental autoimmune uveitis (EAU): potential neuroprotective role of SOCS proteins. J Neuroimmunol. 2005;168:118–127. doi: 10.1016/j.jneuroim.2005.07.021. [DOI] [PubMed] [Google Scholar]
- Takeuchi A, Usui Y, Takeuchi M, Hattori T, Kezuka T, Suzuki J, Okunuki Y, Iwasaki T, Haino M, Matsushima K, Usui M. CCR5-deficient mice develop experimental autoimmune uveoretinitis in the context of a deviant effector response. Invest Ophthalmol Vis Sci. 2005;46:3753–3760. doi: 10.1167/iovs.04-1429. [DOI] [PubMed] [Google Scholar]
- Takizawa N, Ikebe R, Ikebe M, Luna EJ. Supervillin slows cell spreading by facilitating myosin II activation at the cell periphery. J Cell Sci. 2007;120:3792–3803. doi: 10.1242/jcs.008219. [DOI] [PubMed] [Google Scholar]
- Takizawa N, Smith TC, Nebl T, Crowley JL, Palmieri SJ, Lifshitz LM, Ehrhardt AG, Hoffman LM, Beckerle MC, Luna EJ. Supervillin modulation of focal adhesions involving TRIP6/ZRP-1. The Journal of cell biology. 2006;174:447–458. doi: 10.1083/jcb.200512051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Namba T, Arai Y, Fujimoto M, Adachi H, Sobue G, Takeuchi K, Nakai A, Mizushima T. Genetic evidence for a protective role for heat shock factor 1 and heat shock protein 70 against colitis. J Biol Chem. 2007;282:23240–23252. doi: 10.1074/jbc.M704081200. [DOI] [PubMed] [Google Scholar]
- Tang J, Zhou R, Luger D, Zhu W, Silver PB, Grajewski RS, Su SB, Chan CC, Adorini L, Caspi RR. Calcitriol suppresses antiretinal autoimmunity through inhibitory effects on the Th17 effector response. J Immunol. 2009;182:4624–4632. doi: 10.4049/jimmunol.0801543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarbell JM, Ebong EE. The endothelial glycocalyx: a mechano-sensor and -transducer. Sci Signal. 2008;1:pt8. doi: 10.1126/scisignal.140pt8. [DOI] [PubMed] [Google Scholar]
- Tezel G, Li LY, Patil RV, Wax MB. TNF-alpha and TNF-alpha receptor-1 in the retina of normal and glaucomatous eyes. Invest Ophthalmol Vis Sci. 2001;42:1787–1794. [PubMed] [Google Scholar]
- The Diabetic Retinopathy Study Research Group. Photocoagulation treatment of proliferative diabetic retinopathy. Clinical application of Diabetic Retinopathy Study (DRS) findings, DRS Report Number 8. Ophthalmology. 1981;88:583–600. [PubMed] [Google Scholar]
- Thillaye-Goldenberg B, Goureau O, Naud MC, de Kozak Y. Delayed onset and decreased severity of experimental autoimmune uveoretinitis in mice lacking nitric oxide synthase type 2. J Neuroimmunol. 2000;110:31–44. doi: 10.1016/s0165-5728(00)00313-1. [DOI] [PubMed] [Google Scholar]
- Tolentino MJ, Miller JW, Gragoudas ES, Jakobiec FA, Flynn E, Chatzistefanou K, Ferrara N, Adamis AP. Intravitreous injections of vascular endothelial growth factor produce retinal ischemia and microangiopathy in an adult primate. Ophthalmology. 1996;103:1820–1828. doi: 10.1016/s0161-6420(96)30420-x. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchio E, Kijima M, Tanaka S, Ohno S. Suppression of experimental uveitis with monoclonal antibodies to ICAM-1 and LFA-1. Invest Ophthalmol Vis Sci. 1994;35:2626–2631. [PubMed] [Google Scholar]
- Ueta T, Yanagi Y, Tamaki Y, Yamaguchi T. Cerebrovascular accidents in ranibizumab. Ophthalmology. 2009;116:362. doi: 10.1016/j.ophtha.2008.09.046. [DOI] [PubMed] [Google Scholar]
- Ulbig MW, Wolfensberger TJ, Hiscott P, Ationu A, Carter ND, Gregor ZJ. Insulin-like growth factor I (IGF-I) receptor/binding protein in human diabetic epiretinal membranes. Ger J Ophthalmol. 1995;4:264–268. [PubMed] [Google Scholar]
- Ulvmar MH, Hub E, Rot A. Atypical chemokine receptors. Exp Cell Res. 2011;317:556–568. doi: 10.1016/j.yexcr.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda N, Ozaki H, Hayashi H, Miyajima-Uchida H, Oshima K. Colocalization of Tie2, angiopoietin 2 and vascular endothelial growth factor in fibrovascular membrane from patients with retinopathy of prematurity. Ophthalmic Res. 2003;35:217–223. doi: 10.1159/000071173. [DOI] [PubMed] [Google Scholar]
- van Wijngaarden P, Coster DJ, Williams KA. Inhibitors of ocular neovascularization: promises and potential problems. Jama. 2005;293:1509–1513. doi: 10.1001/jama.293.12.1509. [DOI] [PubMed] [Google Scholar]
- Verity DH, Wallace GR, Seed PT, Kanawati CA, Ayesh I, Holland-Gladwish J, Stanford MR. Soluble adhesion molecules in Behcet’s disease. Ocular immunology and inflammation. 1998;6:81–92. doi: 10.1076/ocii.6.2.81.4054. [DOI] [PubMed] [Google Scholar]
- Vidro EK, Gee S, Unda R, Ma JX, Tsin A. Glucose and TGFbeta2 modulate the viability of cultured human retinal pericytes and their VEGF release. Current eye research. 2008;33:984–993. doi: 10.1080/02713680802450976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinores SA, Xiao WH, Aslam S, Shen J, Oshima Y, Nambu H, Liu H, Carmeliet P, Campochiaro PA. Implication of the hypoxia response element of the Vegf promoter in mouse models of retinal and choroidal neovascularization, but not retinal vascular development. J Cell Physiol. 2006;206:749–758. doi: 10.1002/jcp.20525. [DOI] [PubMed] [Google Scholar]
- Virdi PS, Hayreh SS. Ocular neovascularization with retinal vascular occlusion. I. Association with experimental retinal vein occlusion. Arch Ophthalmol. 1982;100:331–341. doi: 10.1001/archopht.1982.01030030333024. [DOI] [PubMed] [Google Scholar]
- Wacker WB, Lipton MM. Experimental allergic uveitis: homologous retina as uveitogenic antigen. Nature. 1965;206:253–254. doi: 10.1038/206253a0. [DOI] [PubMed] [Google Scholar]
- Wakefield D, Lloyd A. The role of cytokines in the pathogenesis of inflammatory eye disease. Cytokine. 1992;4:1–5. doi: 10.1016/1043-4666(92)90028-p. [DOI] [PubMed] [Google Scholar]
- Wakui S, Yokoo K, Muto T, Suzuki Y, Takahashi H, Furusato M, Hano H, Endou H, Kanai Y. Localization of Ang-1, -2, Tie-2, and VEGF expression at endothelial-pericyte interdigitation in rat angiogenesis. Laboratory investigation; a journal of technical methods and pathology. 2006;86:1172–1184. doi: 10.1038/labinvest.3700476. [DOI] [PubMed] [Google Scholar]
- Wallace GR, Whiston RA, Stanford MR, Wells GM, Gearing AJ, Clements JM. The matrix metalloproteinase inhibitor BB-1101 prevents experimental autoimmune uveoretinitis (EAU) Clinical and experimental immunology. 1999;118:364–370. doi: 10.1046/j.1365-2249.1999.01066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HU, Chen ZF, Anderson DJ. Molecular distinction and angiogenic interaction between embryonic arteries and veins revealed by ephrin-B2 and its receptor Eph-B4. Cell. 1998;93:741–753. doi: 10.1016/s0092-8674(00)81436-1. [DOI] [PubMed] [Google Scholar]
- Wang Y, Calder VL, Lightman SL, Greenwood J. Antigen presentation by rat brain and retinal endothelial cells. J Neuroimmunol. 1995;61:231–239. doi: 10.1016/0165-5728(95)00096-k. [DOI] [PubMed] [Google Scholar]
- Watanabe D, Suzuma K, Suzuma I, Ohashi H, Ojima T, Kurimoto M, Murakami T, Kimura T, Takagi H. Vitreous levels of angiopoietin 2 and vascular endothelial growth factor in patients with proliferative diabetic retinopathy. American journal of ophthalmology. 2005;139:476–481. doi: 10.1016/j.ajo.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Wheatley CM, Dickinson JL, Mackey DA, Craig JE, Sale MM. Retinopathy of prematurity: recent advances in our understanding. Br J Ophthalmol. 2002;86:696–700. doi: 10.1136/bjo.86.6.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitcup SM, Chan CC, Li Q, Nussenblatt RB. Expression of cell adhesion molecules in posterior uveitis. Arch Ophthalmol. 1992;110:662–666. doi: 10.1001/archopht.1992.01080170084029. [DOI] [PubMed] [Google Scholar]
- Whitcup SM, DeBarge LR, Caspi RR, Harning R, Nussenblatt RB, Chan CC. Monoclonal antibodies against ICAM-1 (CD54) and LFA-1 (CD11a/CD18) inhibit experimental autoimmune uveitis. Clin Immunol Immunopathol. 1993;67:143–150. doi: 10.1006/clin.1993.1057. [DOI] [PubMed] [Google Scholar]
- Wilson SH, Davis MI, Caballero S, Grant MB. Modulation of retinal endothelial cell behaviour by insulin-like growth factor I and somatostatin analogues: implications for diabetic retinopathy. Growth hormone & IGF research : official journal of the Growth Hormone Research Society and the International IGF Research Society. 2001;11(Suppl A):S53–59. doi: 10.1016/s1096-6374(01)80009-5. [DOI] [PubMed] [Google Scholar]
- Wilson SL, Drevets DA. Listeria monocytogenes infection and activation of human brain microvascular endothelial cells. J Infect Dis. 1998;178:1658–1666. doi: 10.1086/314490. [DOI] [PubMed] [Google Scholar]
- Witmer AN, Blaauwgeers HG, Weich HA, Alitalo K, Vrensen GF, Schlingemann RO. Altered expression patterns of VEGF receptors in human diabetic retina and in experimental VEGF-induced retinopathy in monkey. Invest Ophthalmol Vis Sci. 2002;43:849–857. [PubMed] [Google Scholar]
- Wu CC, MacCoss MJ. Shotgun proteomics: tools for the analysis of complex biological systems. Current opinion in molecular therapeutics. 2002;4:242–250. [PubMed] [Google Scholar]
- Wu SY, Lan SH, Cheng DE, Chen WK, Shen CH, Lee YR, Zuchini R, Liu HS. Ras-Related Tumorigenesis Is Suppressed by BNIP3-Mediated Autophagy through Inhibition of Cell Proliferation. Neoplasia. 2011;13:1171–1182. doi: 10.1593/neo.11888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulfkuhle JD, Donina IE, Stark NH, Pope RK, Pestonjamasp KN, Niswonger ML, Luna EJ. Domain analysis of supervillin, an F-actin bundling plasma membrane protein with functional nuclear localization signals. J Cell Sci. 1999;112 ( Pt 13):2125–2136. doi: 10.1242/jcs.112.13.2125. [DOI] [PubMed] [Google Scholar]
- Xu H, Dawson R, Crane IJ, Liversidge J. Leukocyte diapedesis in vivo induces transient loss of tight junction protein at the blood-retina barrier. Invest Ophthalmol Vis Sci. 2005;46:2487–2494. doi: 10.1167/iovs.04-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Forrester JV, Liversidge J, Crane IJ. Leukocyte trafficking in experimental autoimmune uveitis: breakdown of blood-retinal barrier and upregulation of cellular adhesion molecules. Invest Ophthalmol Vis Sci. 2003a;44:226–234. doi: 10.1167/iovs.01-1202. [DOI] [PubMed] [Google Scholar]
- Xu H, Koch P, Chen M, Lau A, Reid DM, Forrester JV. A clinical grading system for retinal inflammation in the chronic model of experimental autoimmune uveoretinitis using digital fundus images. Exp Eye Res. 2008;87:319–326. doi: 10.1016/j.exer.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Xu H, Manivannan A, Goatman KA, Jiang HR, Liversidge J, Sharp PF, Forrester JV, Crane IJ. Reduction in shear stress, activation of the endothelium, and leukocyte priming are all required for leukocyte passage across the blood--retina barrier. J Leukoc Biol. 2004a;75:224–232. doi: 10.1189/jlb.1002479. [DOI] [PubMed] [Google Scholar]
- Xu H, Manivannan A, Goatman KA, Liversidge J, Sharp PF, Forrester JV, Crane IJ. Improved leukocyte tracking in mouse retinal and choroidal circulation. Exp Eye Res. 2002;74:403–410. doi: 10.1006/exer.2001.1134. [DOI] [PubMed] [Google Scholar]
- Xu H, Manivannan A, Jiang HR, Liversidge J, Sharp PF, Forrester JV, Crane IJ. Recruitment of IFN-gamma-producing (Th1-like) cells into the inflamed retina in vivo is preferentially regulated by P-selectin glycoprotein ligand 1:P/E-selectin interactions. J Immunol. 2004b;172:3215–3224. doi: 10.4049/jimmunol.172.5.3215. [DOI] [PubMed] [Google Scholar]
- Xu H, Manivannan A, Liversidge J, Sharp PF, Forrester JV, Crane IJ. Requirements for passage of T lymphocytes across non-inflamed retinal microvessels. J Neuroimmunol. 2003b;142:47–57. doi: 10.1016/s0165-5728(03)00258-3. [DOI] [PubMed] [Google Scholar]
- Xu M, Jin Y, Song Q, Wu J, Philbrick MJ, Cully BL, An X, Guo L, Gao F, Li J. The endothelium-dependent effect of RTEF-1 in pressure overload cardiac hypertrophy: role of VEGF-B. Cardiovascular research. 2011;90:325–334. doi: 10.1093/cvr/cvq400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Zhang H, Iwase T, Shen J, Semenza GL, Campochiaro PA. Digoxin inhibits retinal ischemia-induced HIF-1alpha expression and ocular neovascularization. Faseb J. 2010;24:1759–1767. doi: 10.1096/fj.09-145664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CR, Mahdi RR, Oh HM, Amadi-Obi A, Levy-Clarke G, Burton J, Eseonu A, Lee Y, Chan CC, Egwuagu CE. Suppressor of cytokine signaling-1 (SOCS1) inhibits lymphocyte recruitment into the retina and protects SOCS1 transgenic rats and mice from ocular inflammation. Invest Ophthalmol Vis Sci. 2011;52:6978–6986. doi: 10.1167/iovs.11-7688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu PK, Balaratnasingam C, Cringle SJ, McAllister IL, Provis J, Yu DY. Microstructure and network organization of the microvasculature in the human macula. Invest Ophthalmol Vis Sci. 2010a;51:6735–6743. doi: 10.1167/iovs.10-5415. [DOI] [PubMed] [Google Scholar]
- Yu PK, Balaratnasingam C, Morgan WH, Cringle SJ, McAllister IL, Yu DY. The structural relationship between the microvasculature, neurons, and glia in the human retina. Invest Ophthalmol Vis Sci. 2010b;51:447–458. doi: 10.1167/iovs.09-3978. [DOI] [PubMed] [Google Scholar]
- Yu PK, Yu D, Alder VA, Seydel U, Su E, Cringle SJ. Heterogeneous endothelial cell structure along the porcine retinal microvasculature. Exp Eye Res. 1997;65:379–389. doi: 10.1006/exer.1997.0340. [DOI] [PubMed] [Google Scholar]
- Yuan HT, Khankin EV, Karumanchi SA, Parikh SM. Angiopoietin 2 is a partial agonist/antagonist of Tie2 signaling in the endothelium. Molecular and cellular biology. 2009;29:2011–2022. doi: 10.1128/MCB.01472-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Zhang J, Li S, Edwards JL. Amine metabolomics of hyperglycemic endothelial cells using capillary LC-MS with isobaric tagging. J Proteome Res. 2011;10:5242–5250. doi: 10.1021/pr200815c. [DOI] [PubMed] [Google Scholar]
- Zaman AG, Edelsten C, Stanford MR, Graham EM, Ellis BA, Direskeneli H, D’Cruz DP, Hughes GR, Dumonde DC, Wallace GR. Soluble intercellular adhesion molecule-1 (sICAM-1) as a marker of disease relapse in idiopathic uveoretinitis. Clinical and experimental immunology. 1994;95:60–65. doi: 10.1111/j.1365-2249.1994.tb06015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamora DO, Rosenbaum JT, Smith JR. Invasion of human retinal vascular endothelial cells by Toxoplasma gondii tachyzoites. Br J Ophthalmol. 2008;92:852–855. doi: 10.1136/bjo.2007.133314. [DOI] [PubMed] [Google Scholar]
- Zanetti A, Conforti G, Hess S, Martin-Padura I, Ghibaudi E, Preissner KT, Dejana E. Clustering of vitronectin and RGD peptides on microspheres leads to engagement of integrins on the luminal aspect of endothelial cell membrane. Blood. 1994;84:1116–1123. [PubMed] [Google Scholar]
- Zhang H, Sonoda KH, Qiao H, Oshima T, Hisatomi T, Ishibashi T. Development of a new mouse model of branch retinal vein occlusion and retinal neovascularization. Japanese journal of ophthalmology. 2007;51:251–257. doi: 10.1007/s10384-007-0445-2. [DOI] [PubMed] [Google Scholar]
- Zhang HR. Scanning electron-microscopic study of corrosion casts on retinal and choroidal angioarchitecture in man and animals. Prog Ret Eye Res. 1994;13:243–270. [Google Scholar]
- Zhang J, Wu LY, Wu GS, Rao NA. Differential expression of nitric oxide synthase in experimental uveoretinitis. Invest Ophthalmol Vis Sci. 1999;40:1899–1905. [PubMed] [Google Scholar]
- Zhang LQ, Cheranova D, Gibson M, Ding S, Heruth DP, Fang D, Ye SQ. RNA-seq reveals novel transcriptome of genes and their isoforms in human pulmonary microvascular endothelial cells treated with thrombin. PLoS One. 2012;7:e31229. doi: 10.1371/journal.pone.0031229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fortune B, Atchaneeyasakul LO, McFarland T, Mose K, Wallace P, Main J, Wilson D, Appukuttan B, Stout JT. Natural history and histology in a rat model of laser-induced photothrombotic retinal vein occlusion. Curr Eye Res. 2008;33:365–376. doi: 10.1080/02713680801939318. [DOI] [PubMed] [Google Scholar]
- Zhao M, Bousquet E, Valamanesh F, Farman N, Jeanny JC, Jaisser F, Behar-Cohen FF. Differential Regulations of AQP4 and Kir4.1 by Triamcinolone Acetonide and Dexamethasone in the Healthy and Inflamed Retina. Invest Ophthalmol Vis Sci. 2011;52:6340–6347. doi: 10.1167/iovs.11-7675. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Wong WH. CisModule: de novo discovery of cis-regulatory modules by hierarchical mixture modeling. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12114–12119. doi: 10.1073/pnas.0402858101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, He L, Huang SH. Identification of a surface protein on human brain microvascular endothelial cells as vimentin interacting with Escherichia coli invasion protein IbeA. Biochem Biophys Res Commun. 2006;351:625–630. doi: 10.1016/j.bbrc.2006.10.091. [DOI] [PubMed] [Google Scholar]