Abstract
The textbook view that a primary sequence determines the unique fold of a given protein has been challenged by identification of proteins with variant structures, such as prions. Our recent studies revealed that the transcription factor RfaH simultaneously changes its topology and function. RfaH is a two-domain protein whose N-terminal domain binds to transcribing RNA polymerase, stimulating its processivity. The α-helical C-terminal domain masks the RNA polymerase-binding site of the N-terminal domain, preventing unwarranted recruitment to genes lacking a specific DNA sequence. Upon binding to its DNA target, RfaH domains dissociate, and the C-terminal domain refolds into a β-barrel. This dramatic transformation allows binding to the ribosomal protein S10 and subsequent recruitment of a ribosome, coupling transcription and translation. We define RfaH as first example of “transformer proteins”, in which two alternative structural states have distinct cellular functions and hypothesize that transformer proteins may be widespread in nature.
Keywords: RfaH, NusG, transcription factor, transformer protein, moonlighting protein, metamorphic protein, ops DNA, multifunctionality, protein folding, refolding
NusG: A Processivity Clamp and a Nexus for Gene Regulation
RNA synthesis is the first step in gene expression and is elaborately controlled in all living cells by an astonishingly large number of accessory factors. Proteins that modulate RNA polymerase (RNAP) in response to cellular signals are not conserved across the different kingdoms of life, reflecting very different regulatory requirements and system complexity. By contrast, consistent with their common evolutionary origin, all multi-subunit RNAPs share the catalytic core (composed of the β and β' subunits in bacterial RNAP) and the basic mechanism of nucleotide addition.1-3 These enzymes also face a common challenge - each RNAP must complete synthesis of the entire RNA chain (up to a million nucleotides in length) in one try, overcoming many obstacles, such as unfavorable nucleic acid signals or DNA-bound proteins, along the way.4 RNAP cannot rebind a prematurely released transcript, underscoring the paramount importance of processivity control. Indeed, regulators from the NusG family, which act as RNAP processivity clamps, constitute the only universally conserved family of regulators.5
The family of NusG-like proteins comprises NusG in Bacteria, Spt5 in Archaea and lower eukaryotes, and DSIF in humans,5 as well as gene-specific paralogs present in many bacterial species.6 Ubiquity of these proteins suggested similarities in their functions and structures, a view that has been supported by studies of bacterial, archaeal, and yeast NusGs. Structural analyses of bacterial NusGs7,8 revealed their two-domain architecture (Fig. 1A), and subsequent studies assigned specific functions to each domain. The N-terminal domain (NTD) exhibits a mixed α/β topology with a central antiparallel β-sheet surrounded by α-helices. It binds RNAP and is sufficient to reduce transcriptional pausing in vitro.9,10 The C-terminal domain (CTD), an SH3 β-barrel connected to the NTD by a long flexible linker, interacts with the termination factor Rho9 to silence the expression of foreign DNA11 or with ribosomal protein S1012 to couple transcription to translation.13 Archaeal and eukaryotic NusG homologs have similar structures (whereas several repeats of the CTD are present in eukaryotes) and the domains play analogous roles.5 The NTD bridges the gap between the β' clamp helices (β’CH) and the β gate loop (βGL), the two pincers of the crab-claw RNAP, to form a clamp around the nucleic acid chains.14-17 The closed clamp is thought to resist RNAP isomerization into off-pathway states, favoring uninterrupted RNA synthesis. The CTD acts as a protein-interaction platform that links the elongating RNAP to other macromolecular complexes acting co-transcriptionally.5,18
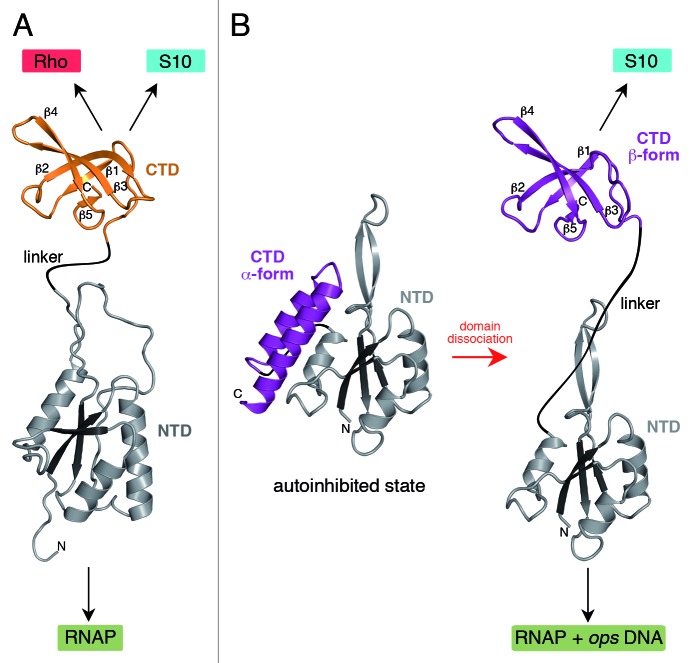
Figure 1. Structures of NusG (A) and RfaH (B). All structures are shown in ribbon representation whereas the central β-sheet of the NTD is highlighted in dark colors. The flexible linker is depicted as black line. Black arrows indicate interaction partners of the respective domain. In (B) the transformation of RfaH from an autoinhibited into an active state is shown as its CTD refolds from an all-α into an all-β state. The figure was created using PyMOL.36 PDB IDs: 2K06, NusG NTD; 2JVV, NusG CTD; 2OUG, RfaH; 2LCL, RfaH CTD
Given its universally important role in maintaining high RNAP processivity, the NTD has justifiably been the focus of attention in the recent years. However, it is important to note that the CTD plays equally important and diverse roles in the regulation of gene expression. Escherichia coli NusG CTD has two opposite functions: on the one hand, it acts in synergy with Rho to terminate transcription of horizontally transferred genes;11 on the other hand, the CTD interaction with S1012 would block Rho access to the nascent RNA, inhibiting termination. Eukaryotic Spt factors mediate the assembly of protein complexes with diverse regulatory functions; examples of yeast Spt5 interaction partners include the capping enzyme,19 BRCA1,20 FACT,21 cleavage factor I22 and PAF1 complex.23
Unprecedented Structural Transitions of RfaH CTD
E. coli RfaH, a gene-specific paralog of NusG, is only recruited to operons that contain an operon polarity suppressor (ops) element in the leader region. The crystal structure of RfaH revealed that the NTD fold was very similar to that observed in NusG, whereas the CTD formed an α-helical hairpin, in stark contrast to the β-barrel NusG CTD (Fig. 1B).10 In RfaH the two domains interact tightly whereby the CTD masks the RNAP-binding surface of the NTD. Thus, in contrast to NusG in which the NTD is fully exposed, RfaH was captured in a silent, autoinhibited state. Interaction with the ops DNA element presented in the elongation complex would be necessary to trigger RfaH domain separation and to allow RfaH recruitment to the elongating RNAP. Functional analysis supported the existence and the key role of these interdomain contacts.10 Interestingly, homology modeling indicated that the primary sequence of the RfaH CTD could be easily integrated into the NusG β-barrel. Studies on RfaH suggested that the surprisingly broad repertoire of its CTD partners may be in part explained by the ability of the CTD to adopt two alternative folds. This let to our proposal that RfaH CTD refolds when released from the NTD.6
Using nuclear magnetic resonance (NMR) spectroscopy, we demonstrated that the RfaH structure in solution was essentially identical to that captured in the crystal.24 Furthermore, the two domains moved with identical correlation times, confirming tight domain interaction. Surprisingly, the isolated, separately expressed RfaH CTD (Fig. 2B) did not contain any α-helical elements, but consisted of five β-strands organized in an antiparallel β-sheet forming a β-barrel virtually identical to that of E. coli NusG CTD in solution (backbone root mean square deviation: 0.76 Å). On the one hand, this analysis revealed that the RfaH CTD exists in an all-α-helical state when stabilized by interactions with the NTD. On the other hand, these experiments suggest that the CTD may completely refold into an all-β form when the domain interface is disrupted after NTD recruitment to ops-paused RNAP.
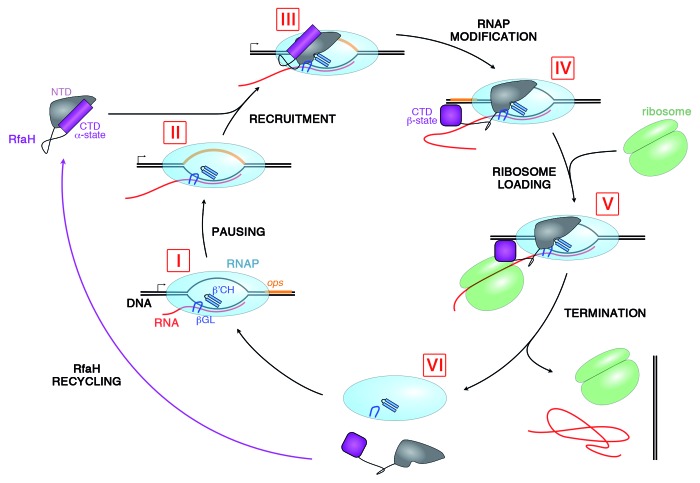
Figure 2. Lifecycle of RfaH. During transcription of RfaH-controlled operons RNAP pauses at the ops element in the leader region (I and II). RfaH, in its autoinhibited form, interacts with the ops DNA via its NTD; the domains then separate, allowing RfaH NTD to bind RNAP (III). Together with β’CH and βGL, RfaH NTD forms a clamp around the nucleic acids chains to increase the processivity of RNAP. The freed RfaH CTD completely refolds from its α-helical form into a β-barrel (IV). The transformed CTD binds S10, recruiting a ribosome to the mRNA, and translation commences (V). After termination of translation, the transcription elongation complex dissociates (VI). The CTD of released RfaH folds back into its α-form and RfaH is recycled into its autoinhibited state, ready for the next round of recruitment.
To confirm that such a dramatic structural rearrangement is possible within full-length RfaH we used two approaches to weaken NTD-CTD interactions.24 First, we introduced a recognition site for TEV protease into the flexible linker and ensured that this insertion did not alter RfaH’s function or structure. Then [1H,15N]-heteronuclear single quantum coherence experiments were used to monitor changes in this RfaH variant upon incubation with catalytic amounts of TEV protease. We detected the appearance of signals of the β-barrel CTD while simultaneously signals of the α-helical CTD as well as those of the NTD disappeared. The latter can be explained by NTD precipitation on domain release as this domain is nearly insoluble unless stabilized by the α-helical CTD or RNAP.10
Second, we constructed an RfaH variant with a Glu48Ser substitution in the NTD that breaks the salt bridge to Arg138 in the CTD, without altering the CTD itself. Remarkably, this full-length RfaH variant existed in two roughly equally populated states, one with the CTD in the α-helical form and one with the CTD in the β-barrel conformation, as shown by NMR spectroscopy.
Our results demonstrate that RfaH CTD is able to adopt two strikingly different structures with completely different internal amino acid contacts. In the domain-closed form of RfaH, the large hydrophobic interdomain surface may foster denser packing of the α-helical CTD than would be possible were it in the β-fold.10 Domain separation thus does not only allow establishment of interactions between the hydrophobic surface of the NTD and RNAP, but also leads to refolding of RfaH CTD into a β-barrel. Small structural rearrangements of proteins are not uncommon, especially as all molecules are subject to dynamic changes. Larger conformational changes have also been described, but they often involve transitions between structured and unstructured regions.25-28 However, the complete conformational switch from one state with stable secondary structure elements (all-α-helical fold) to another state with completely different stable secondary structure elements (all-β structure) surpasses the extent of all structural transitions that have been described so far, singling RfaH out as a thus far unique system.
The Refolded CTD Interacts with S10
The α-helical RfaH CTD plays a key role in the regulatory function of the protein: it restricts RfaH action to only those operons that have an ops element in their leader region because domain dissociation, and consequently RNAP binding, requires ops DNA. This autoinhibitory mechanism is critical to ensure that RfaH is not recruited to operons normally controlled by NusG, particularly because RfaH and NusG play opposite roles in the cell: RfaH enhances expression of a small set of horizontally-transferred genes whereas one of the essential functions of NusG is thought to silence foreign DNA.11 The in vivo analysis demonstrates that RfaH and NusG are indeed restricted to their regulatory niches.6
After RfaH recruitment, the NTD remains bound to RNAP for the duration of transcription, whereas the CTD becomes free and spontaneously refolds. One could ask whether the dramatic CTD refolding merely serves to hide an exposed hydrophobic surface or plays additional roles. Several lines of evidence led us to favor the second possibility. First, a large discrepancy exists between RfaH effects in vitro and in vivo. Second, even though the antipausing activity of RfaH is universally conserved, our in vivo analysis showed that this activity plays only a minor role in the overall activation of gene expression.17 Third, all RfaH-controlled genes are horizontally transferred and have many properties suggesting that their translation may be rate-limiting. RfaH effects on translation could explain its dramatic effects on gene expression in the cell,17 a scenario supported by our recent report of direct contacts between NusG and S10 that link the transcription and translation machineries.12 A similar activity of RfaH would explain its documented ability to block the action of Rho,17 and contacts with the translation apparatus have been suggested by an early report.29
Given that refolded RfaH CTD is structurally nearly identical to NusG CTD, the possibility of S10 binding by RfaH CTD suggested itself. Indeed, we observed direct interactions between RfaH CTD and S10 (bound to NusB) by gel filtration as well as NMR spectroscopy, and chemical shift mapping indicated that RfaH CTD and NusG CTD use similar sets of interactions to bind S10. Targeted ChIP-chip analysis and mass spectroscopy identification of E. coli proteins interacting with RfaH showed that S10 is associated with the RfaH-controlled rfb operon and with RfaH, respectively, lending support to physiological relevance of these contacts.
RfaH Substitutes for a Missing Translation Initiation Signal
RfaH-controlled operons have numerous rare codons and, most strikingly, lack Shine-Dalgarno (SD) elements; strong polarity in the rfb operon is consistent with its poor translation and is nearly abolished by RfaH. Activation of translation mediated by RfaH interactions with S10 would explain this effect, but the underlying mechanism remains to be elucidated.
Three mechanisms of translation initiation exist in E. coli.30 In the first and best understood pathway, interactions between an SD element located 7–10 nucleotides upstream of the AUG start codon with an anti-SD sequence in the 16S rRNA recruit the 30S subunit of the ribosome (bound to fMet-tRNAfMet and initiation factors IF1, IF2, and IF3) to mRNA. Following complex conformational rearrangements that align fMet-tRNAfMet with the start codon, the 50S subunit joins to form the 70S initiation complex, while IFs are released. The second pathway comprises leaderless mRNAs. Here, a nearly complete 70S ribosome can initiate translation at a 5′-terminal AUG codon. The third type is related to genes that have an internal AUG or, more likely, GUG start codon but lack SD elements in their unstructured, AU-rich untranslated leader region. Practically nothing is known about initiation of these SD-lacking mRNAs. All known RfaH targets belong to the latter class: they have 150+ nucleotide leader regions, with the ops element located within 100 nucleotides of the annotated translation start (commonly GUG).
The available evidence indicates that RfaH can substitute for an SD element during initiation. We showed that simply deleting the SD element in front of a reporter luxCDABE operon makes luciferase expression dependent on RfaH (> 1,000 fold activation). RfaH CTD is required for this dramatic stimulation, and a single substitution of Ile146 at the interface with S10 reduces lux expression in the absence, but not in the presence, of the SD sequence. The simplest mechanism suggested by these observations is that RfaH interactions with S10 increase the local 30S concentration, thereby recruiting a ribosome to the nascent mRNA that is being made by RfaH-bound RNAP. We hypothesize that NusG may play a similar role in the majority of SD-less operons.
RfaH is a Transformer Protein
Traditionally it is believed that proteins adopt a unique fold determined by their amino acid sequences under defined environmental conditions to accomplish a specific function.31 This rule had to be stretched when moonlighting proteins32 (e.g., NusE/S1012,33) or metamorphic proteins28 (e.g., Aquifex aeolicus ribosomal protein L2027) were discovered. Moonlighting proteins have one three-dimensional fold but are able to carry out two or more different functions. Metamorphic proteins can adopt different folds in identical environments, but typically this involves transitions between structured and unstructured regions. Proteins that combine features of the two classes have also been described; for example, the chemokine lymphotactin has two alternative functions which are regulated by transition of an N-terminal loop into a β-strand while the C-terminal α-helix becomes unstructured.25 RfaH goes beyond the scope of these changes and is hence the first member of the class of transformer proteins,34 in which a dramatic and complete transition between two conformations underlies an equally dramatic change in function. Never before has a protein been described that simultaneously changes both its complete stable topology of conservative secondary structure elements and its function. This includes prion proteins26 and the β-amyloid A4 peptide,35 which both undergo large conformational changes but do not coexist in two forms in identical solution; rather, a soluble (functional) form is nearly irreversibly converted into an insoluble (pathogenic) form. In contrast, both stable forms of RfaH play very different key roles in the regulation of transcription and translation, two separate processes which are yet tightly coordinated through RfaH and NusG.12,24
Summary and Future Questions
Our studies characterize RfaH as a transformer protein that uses two alternative folds to carry out two separate functions: autoinhibition of RNAP binding and recruitment of a ribosome. Most strikingly, we were able to construct an RfaH variant with a single substitution in the NTD which exists as a mixture of the α-helical and the β-barrel CTD states under physiological conditions. In wild-type RfaH, this equilibrium is shifted basically completely toward the α-state by interaction with the NTD, a situation which is reversed when the NTD docks to RNAP, releasing the CTD which then refolds and binds S10. The structural switch thus allows the transformation of RfaH from a transcription into a translation factor.
Our observations open a myriad of interesting questions, some of which we are currently pursuing. First, what is the pathway of RfaH domain dissociation and how fast does the CTD refold? Second, what is the molecular mechanism of the α→β transition? Third, what happens when RNAP runs to the end of the operon and releases nucleic acids and RfaH? Does RfaH refold back into the closed, silent state or is it degraded to maintain the observed regulatory specificity? Fourth, is the α↔β transition a unique feature of RfaH CTD, or is it widespread among the NusG family of regulators? Perhaps not all interactions of NusG family proteins are mediated by the β-fold, as commonly assumed. Finally, we propose that the genuine concept of transformer proteins is not a unique feature of at least some NusG-like proteins, but is a much more general principle of nature.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (Ro617/18–1) and the National Institutes of Health (GM67153).
Glossary
Abbreviations:
- RNAP
RNA polymerase
- NTD
N-terminal domain
- CTD
C-terminal domain
- β’CH
β’ clamp helices
- βGL
β gate loop
- ops
operon polarity suppressor
- NMR
nuclear magnetic resonance
- SD
Shine Dalgarno
- IF
initiation factor
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/22724
References
- 1.Lane WJ, Darst SA. Molecular evolution of multisubunit RNA polymerases: sequence analysis. J Mol Biol. 2010;395:671–85. doi: 10.1016/j.jmb.2009.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lane WJ, Darst SA. Molecular evolution of multisubunit RNA polymerases: structural analysis. J Mol Biol. 2010;395:686–704. doi: 10.1016/j.jmb.2009.10.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vassylyev DG. Elongation by RNA polymerase: a race through roadblocks. Curr Opin Struct Biol. 2009;19:691–700. doi: 10.1016/j.sbi.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santangelo TJ, Artsimovitch I. Termination and antitermination: RNA polymerase runs a stop sign. Nat Rev Microbiol. 2011;9:319–29. doi: 10.1038/nrmicro2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Werner F. A nexus for gene expression-molecular mechanisms of Spt5 and NusG in the three domains of life. J Mol Biol. 2012;417:13–27. doi: 10.1016/j.jmb.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belogurov GA, Mooney RA, Svetlov V, Landick R, Artsimovitch I. Functional specialization of transcription elongation factors. EMBO J. 2009;28:112–22. doi: 10.1038/emboj.2008.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mooney RA, Davis SE, Peters JM, Rowland JL, Ansari AZ, Landick R. Regulator trafficking on bacterial transcription units in vivo. Mol Cell. 2009;33:97–108. doi: 10.1016/j.molcel.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steiner T, Kaiser JT, Marinkoviç S, Huber R, Wahl MC. Crystal structures of transcription factor NusG in light of its nucleic acid- and protein-binding activities. EMBO J. 2002;21:4641–53. doi: 10.1093/emboj/cdf455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mooney RA, Schweimer K, Rösch P, Gottesman ME, Landick R. Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J Mol Biol. 2009;391:341–58. doi: 10.1016/j.jmb.2009.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belogurov GA, Vassylyeva MN, Svetlov V, Klyuyev S, Grishin NV, Vassylyev DG, et al. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol Cell. 2007;26:117–29. doi: 10.1016/j.molcel.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cardinale CJ, Washburn RS, Tadigotla VR, Brown LM, Gottesman ME, Nudler E. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science. 2008;320:935–8. doi: 10.1126/science.1152763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burmann BM, Schweimer K, Luo X, Wahl MC, Stitt BL, Gottesman ME, et al. A NusE:NusG complex links transcription and translation. Science. 2010;328:501–4. doi: 10.1126/science.1184953. [DOI] [PubMed] [Google Scholar]
- 13.Proshkin S, Rahmouni AR, Mironov A, Nudler E. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science. 2010;328:504–8. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grohmann D, Nagy J, Chakraborty A, Klose D, Fielden D, Ebright RH, et al. The initiation factor TFE and the elongation factor Spt4/5 compete for the RNAP clamp during transcription initiation and elongation. Mol Cell. 2011;43:263–74. doi: 10.1016/j.molcel.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein BJ, Bose D, Baker KJ, Yusoff ZM, Zhang X, Murakami KS. RNA polymerase and transcription elongation factor Spt4/5 complex structure. Proc Natl Acad Sci U S A. 2011;108:546–50. doi: 10.1073/pnas.1013828108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez-Rucobo FW, Sainsbury S, Cheung AC, Cramer P. Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. EMBO J. 2011;30:1302–10. doi: 10.1038/emboj.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sevostyanova A, Belogurov GA, Mooney RA, Landick R, Artsimovitch I. The β subunit gate loop is required for RNA polymerase modification by RfaH and NusG. Mol Cell. 2011;43:253–62. doi: 10.1016/j.molcel.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sevostyanova A, Artsimovitch I. Functional analysis of Thermus thermophilus transcription factor NusG. Nucleic Acids Res. 2010;38:7432–45. doi: 10.1093/nar/gkq623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wen Y, Shatkin AJ. Transcription elongation factor hSPT5 stimulates mRNA capping. Genes Dev. 1999;13:1774–9. doi: 10.1101/gad.13.14.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bennett CB, Westmoreland TJ, Verrier CS, Blanchette CA, Sabin TL, Phatnani HP, et al. Yeast screens identify the RNA polymerase II CTD and SPT5 as relevant targets of BRCA1 interaction. PLoS One. 2008;3:e1448. doi: 10.1371/journal.pone.0001448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindstrom DL, Squazzo SL, Muster N, Burckin TA, Wachter KC, Emigh CA, et al. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol Cell Biol. 2003;23:1368–78. doi: 10.1128/MCB.23.4.1368-1378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayer A, Schreieck A, Lidschreiber M, Leike K, Martin DE, Cramer P. The Spt5 C-terminal region recruits yeast 3′ RNA cleavage factor I. Mol Cell Biol. 2012;32:1321–31. doi: 10.1128/MCB.06310-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou K, Kuo WHW, Fillingham J, Greenblatt JF. Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc Natl Acad Sci U S A. 2009;106:6956–61. doi: 10.1073/pnas.0806302106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burmann BM, Knauer SH, Sevostyanova A, Schweimer K, Mooney RA, Landick R, et al. An α helix to β barrel domain switch transforms the transcription factor RfaH into a translation factor. Cell. 2012;150:291–303. doi: 10.1016/j.cell.2012.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuinstra RL, Peterson FC, Kutlesa S, Elgin ES, Kron MA, Volkman BF. Interconversion between two unrelated protein folds in the lymphotactin native state. Proc Natl Acad Sci U S A. 2008;105:5057–62. doi: 10.1073/pnas.0709518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Surewicz W, Apostol M. Prion protein and its conformational conversion: A structural perspective. In: Tatzelt J, ed. Prion Proteins. Vol 305. Springer Berlin / Heidelberg; 2011:135-167. [DOI] [PubMed] [Google Scholar]
- 27.Timsit Y, Allemand F, Chiaruttini C, Springer M. Coexistence of two protein folding states in the crystal structure of ribosomal protein L20. EMBO Rep. 2006;7:1013–8. doi: 10.1038/sj.embor.7400803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murzin AG. Biochemistry. Metamorphic proteins. Science. 2008;320:1725–6. doi: 10.1126/science.1158868. [DOI] [PubMed] [Google Scholar]
- 29.Bailey MJ, Hughes C, Koronakis V. In vitro recruitment of the RfaH regulatory protein into a specialised transcription complex, directed by the nucleic acid ops element. Mol Gen Genet. 2000;262:1052–9. doi: 10.1007/PL00008648. [DOI] [PubMed] [Google Scholar]
- 30.Milón P, Rodnina MV. Kinetic control of translation initiation in bacteria. Crit Rev Biochem Mol Biol. 2012;47:334–48. doi: 10.3109/10409238.2012.678284. [DOI] [PubMed] [Google Scholar]
- 31.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–30. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 32.Copley SD. Moonlighting is mainstream: paradigm adjustment required. Bioessays. 2012;34:578–88. doi: 10.1002/bies.201100191. [DOI] [PubMed] [Google Scholar]
- 33.Squires CL, Zaporojets D. Proteins shared by the transcription and translation machines. Annu Rev Microbiol. 2000;54:775–98. doi: 10.1146/annurev.micro.54.1.775. [DOI] [PubMed] [Google Scholar]
- 34.Knauer SH, Artsimovitch I, Rösch P. Transformer proteins. Cell Cycle. 2012;11 doi: 10.4161/cc.22468. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sukhanova A, Poly S, Shemetov A, Bronstein I, Nabiev I. Implications of protein structure instability: from physiological to pathological secondary structure. Biopolymers. 2012;97:577–88. doi: 10.1002/bip.22055. [DOI] [PubMed] [Google Scholar]
- 36.Schrödinger L. The PyMOL molecular graphics system, version 1.3. Schrödinger, LLC, Mannheim, Germany 2010. [Google Scholar]