Abstract
The dynamic regulation of transcriptional events is fundamental to many aspects of neuronal cell functions. However, proteomics methods have not been routinely used in global neuroproteomics analyses of transcriptional regulators because they are much less abundant than the “house-keeping” proteins in cells and tissues. Recent improvements in both biochemical preparations of nuclear proteins and detection sensitivities of proteomics technologies have made the global analysis of nuclear transcriptional regulators possible. We report here an optimized neuroproteomic method for the analysis of transcriptional regulators in the nuclear extracts of SHSY-5Y neuroblastoma cells by combining an improved nuclear protein extraction procedure with multidimensional peptide separation approaches. We found that rigorous removal of cytoplasmic proteins and solubilization of DNA-associated proteins improved the number of nuclear proteins identified. Furthermore, we discovered that multidimensional peptide separations by either strong cation exchange (SCX) chromatography or electrostatic repulsion-hydrophilic interaction chromatography (ERLIC) analysis detected more than 1,800 nuclear proteins through the application of our technique, which constitute one of the largest datasets of nuclear proteins reported for a neuronal cell. Thus, in-depth analyses of transcriptional regulators for studying neurological diseases are increasingly feasible.
Keywords: Neuroproteomics, Neuroblastoma, Transcription factor, Strong Cation Exchange (SCX), Electrostatic Repulsion-Hydrophilic Interaction Chromatographic (ERLIC), SHSY-5Y cell
1. Introduction
Proteomics approaches have been successfully used for large-scale analyses of protein expression patterns, post-translational modifications and protein–protein interactions (Cahill, 2001; Pandey and Mann, 2000). The rapid evolution of quantitative proteomics technologies has enabled routine analyses of global proteomic changes among diverse tissues and cells (Gauss et al., 1999; Shevchenko et al., 1996; Yan et al., 2001). More recently, specialized proteomic studies such as those of neuroproteomics are becoming increasingly useful for understanding the dynamic regulatory protein networks underlying neuronal development and neurological diseases (Lin et al., 2009; Liu et al., 2006; Tyler et al., 2011). In addition, neuroproteomics has branched into more in-depth studies of the sub-proteomes, including synaptoproteomics and neural plasma membrane proteomics (Zhang, 2010). However, compared with other high-throughput tools for system-wide analyses of genes and proteins, the sensitivities of proteomics technologies for the characterization of less abundant signaling molecules and transcriptional regulators have remained low for routine biochemical studies.
The eukaryotic nucleus is an important organelle for regulating gene expression and other diverse functions (Trinkle-Mulcahy and Lamond, 2008). Within the nuclear proteomes, many cellular signals, including the ones for stress response, growth and differentiation, ultimately target specific gene promoters to induce alterations in gene expression or DNA replication. The ability to comprehensively identify and quantify transcriptional regulators is important for understanding their functions under different physiological and diseased conditions. Unfortunately, transcription factors are often underrepresented in global proteomic studies due to their relatively low abundance in comparison with the “house-keeping” proteins, such as metabolic enzymes, cytoskeletal proteins and heat shock proteins. To address this limitation, subcellular fractionation approaches for organelle-specific proteomic analyses have been attempted for more sensitive examinations of low-abundance proteins (Andersen et al., 2002; Boisvert et al., 2010; Dreger et al., 2001; Trinkle-Mulcahy and Lamond, 2008). For example, several groups have investigated the nuclear or chromatin proteomes, using a variety of biochemical approaches for the enrichment of nuclear proteins from diverse cell lines and primary cells (reviewed by Albrethsen, J. et al.) (Albrethsen et al., 2009). In one study, a 2D gel electrophoresis (2DE) reference map of total nuclear proteins isolated from human liver was established (Jung et al., 2000); however, both heat shock proteins and cytoskeletal proteins were still abundantly represented. Additional subnuclear fractionation can further improve the depth of the proteome coverage. For instance, the nuclear proteome of human HeLa cells was extensively analyzed by Andersen et al., leading to the identification of 271 nucleolar proteins (Andersen et al., 2002). Similarly, Tchapyjnikov et al. used a nanospray LC/MS/MS-based approach to analyze cell nuclei extracted with commercially available nuclear extraction kit and identified 154 transcription factors and numerous other transcriptional co-regulators, kinases and phosphatases (Tchapyjnikov et al., 2010). Shakib et al. analyzed the nuclear proteins from NRK49F rat kidney fibroblasts after prolonged hypoxia by 2DE. Among the 791 proteins identified, 17 transcription factors or cofactors were found to be possibly regulated by hypoxia (Shakib et al., 2005). In addition to 2DE, LC-based shotgun proteomics methods have also been used effectively for the analysis of nuclear proteomes. Shiio and Eisenman used the isotope-coded affinity tag (ICAT) approach to identify Myc-induced changes in the nuclear proteome (Shiio et al., 2003). After chromatin enrichment, they applied ICAT in combination with LC/MS/MS and identified 282 proteins, including 64 known nuclear proteins. Among the 18 transcription factors identified, ATF-3 reduction and NIFK induction were found to be Myc-modulated. Recently, several advanced mass spectrometry-based studies have made notable progress in characterizing human chromatin. Garcia’s group used three different chromatin extraction methods and identified over 1,900 proteins, 40% of which were classified as nuclear proteins by independent bioinformatics analyses (Torrente et al., 2011). Overall, it appears that a balance needs to be reached between nuclear protein specificity and the depths of the nuclear proteome coverage.
Nuclear proteomics analyses of neuronal cells have not been widely reported, in part due to the difficulties associated with the unusual morphologies and processes of cells of the central and peripheral nervous systems. In this study, we have developed a comprehensive approach for the characterization of the nuclear proteome from a SHSY-5Y neuroblastoma cell line. We found that by both rigorous removal of cytoplasmic proteins and extensive extraction of chromatin-associated proteins, we can dramatically improve the nuclear proteome coverage in this cell line. Furthermore, by adopting multidimensional chromatographic approaches including ERLIC and SCX fractionations to further expand the nuclear proteome coverage, we were able to achieve one of the most in-depth identifications of transcription factors and regulators in SHSY-5Y cells.
2. Materials and Methods
Materials
HPLC-grade solvents and water were purchased from Mallinckrodt Baker, Inc. (Phillipsburg, NJ). Triethylammonium bicarbonate (TEAB), protease inhibitors cocktail and phosphatase inhibitors cocktail were purchased from Sigma (St. Louis, MO). Sequencing-grade modified trypsin was purchased from Promega Corp. (Madison, WI). PepClean C18 spin columns were purchased from Pierce (Rockford, IL). Western blot reagents were obtained from BioRad (Redmond, WA). The antibody against actin was purchased from Santa Cruz Biotechnology (Santa Cruz, CA), and the antibody against histone H1 (Clone AE-4) was purchased from Millipore (Billerica, MA).
Cell lines and cell culture
Human neuroblastoma cell line SHSY-5Y was obtained from ATCC (Manassas, VA). Cells were propagated as monolayers in a 1:1 mixture of Dulbecco’s Modified Eagle Medium (DMEM) and F12 medium supplemented with 0.1 mMol/L nonessential amino acids, 1% penicillin/streptomycin and 10% fetal bovine serum at 37°C in 5% CO2. Exponentially growing and nearly confluent (90%) cells were harvested after several passages and washed twice with PBS.
2.1 Nuclear protein extraction and analysis
2.1.1 Basic extraction method
Nuclear extracts were prepared from the SHSY-5Y cells using a cell lysis and salt extraction procedure described by Dignam et al. (Dignam et al., 1983). Briefly, the PBS-washed cell pellets were gently resuspended in a hypotonic lysis buffer consisting of 10 mM HEPES (pH 7.9), 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, protease inhibitors and phosphatase inhibitors. After incubation of the re-suspended cells on ice for 15 min, 0.5% NP-40 was added, and the extracts were vigorously vortexed for 10 sec to disrupt the cell membranes. The cellular extracts were then centrifuged at 800 × g for 10 min at 4 °C to separate the cytoplasmic components (supernatants) from the nuclei-enriched fractions (pellets). The cytoplasmic fractions (supernatants) were stored at −80 °C until subsequent analyses. The nuclear pellets were resuspended in a hypertonic buffer (20 mM HEPES, pH 7.9, 1.5 mM MgCl2, 420 mM NaCl, 25% v/v glycerol, 0.5 mM DTT, 0.2 mM EDTA and cocktails of protease and phosphatase inhibitors). Nuclear proteins were extracted via vigorous agitation for 15 min on ice. The solutions were further sonicated 3 times at 10 sec intervals on ice. The resulting solutions were centrifuged at 16,000 × g and 4 °C for 15 min. The supernatants containing solubilized nuclear proteins were stored at −80 °C until further analyses. Protein extracts were further concentrated by the addition of 5 volumes of ethanol for precipitation. After centrifugation at 16,000 × g for 15 min, the protein pellets were re-suspended in 500 μl of a buffer containing 8 M urea, 50 mM TEAB (pH 8.0) and protease and phosphatase inhibitor cocktails. Protein concentrations were determined using the Bradford assay according to the manufacturer’s instructions (Bio-Rad). The proteins were reduced with 10 mM DTT at RT for 1 h and alkylated with 50 mM iodoacetamide for 30 min in the dark. Afterwards, for trypsin diegestion, urea concentration was diluted to 1 M with the addition of 50 mM TEAB. For in-solution proteolytic digestion, trypsin was added into the protein solutions at a ratio of 1:25 (trypsin/protein by weight), and the solutions were incubated overnight at 37 °C. The resulting tryptic peptides were desalted using C18 spin columns (Pierce) and stored at −80°C until LC/MS/MS analyses.
2.1.2 Rigorous nuclear protein enrichment method
Similar to the basic method, cytoplasmic extracts and nuclear pellets were first separated by low-speed centrifugation after treating the cells with a hypotonic buffer and NP-40. For more rigorous enrichment of the nuclear proteins, we washed the nuclear pellets additional 2–3 times with the fresh hypotonic buffer and NP-40 to more thoroughly remove the cytoplasmic proteins. The proteins in the resulting nuclear pellets were extracted with the hypertonic buffer, followed by the sonication and centrifugation steps as described above. The nuclear proteins in the supernatant were concentrated by ethanol precipitation and resuspended in the urea buffer. Tryptic digestion and LC/MS/MS analysis were performed as described in Method 1.
2.1.3 Solubilization of chromatin-associated proteins with nuclease treatment
After hypertonic buffer extraction of proteins from the nuclear pellets as described in Method 2, the remaining nuclear pellets after centrifugation may still contain proteins that were trapped within the DNA and chromatin. These proteins were extracted from the pellets by the addition of ≥250 units of Benzonase® nuclease (Sigma, St. Louis, MO) into the nuclease buffer (20 mM Tris HCl, pH 8.0, 2 mM MgCl2, and 20 mM NaCl) and incubation at 37 °C for 1 h. The resulting proteins were resuspended in the buffer with 8 M urea, digested with trypsin and analyzed by LC/MS/MS as described for Method 1.
2.2 Peptide fractionation and LC/MS/MS
2.2.1 Strong cation exchange chromatography (SCX)
For each SCX separation of the tryptic digests, peptides derived from 300 μg of nuclear proteins were separated on a BioCAD™ Perfusion Chromatography System (ABI) equipped with a polysulfoethyl A column (4.6 × 200 mm, 5 μm, 300 Å, Poly LC, Columbia, MD) plus an upstream guard column (4 × 10 mm). The column was first washed isocratically with the mobile phase A (10 mM KH2PO4, 20% acetonitrile (ACN), pH 2.7) for 10 min to remove the unbound materials. Retained peptides were then eluted with a 30 min linear gradient from 0 to 15% mobile phase B (600 mM KCl, 10 mM KH2PO4 and 20% ACN, pH 2.7), then another 20 min gradient from 15 to 50% mobile phase B, followed by a final 10 min linear gradient from 50 to 100% mobile phase B, at a flow rate of 1 ml/min. The peptides in each 2 min fraction were desalted via PepClean C18 spin columns, dried in a speedvac and then combined into 12 fractions with comparable complexities according to MS signals observed by MALDI-TOF/TOF MS on 4800 Protein Analyzer (AB Sciex).
2.2.2 Electrostatic repulsion hydrophilic interaction chromatography (ERLIC)
For each ERLIC analysis of the tryptic digests, peptides derived from 300 μg of nuclear proteins were fractionated in ERLIC mode, using a Poly-WAX LP column (4.6 × 200 mm, 5 μm, 300 Å, PolyLC, Columbia, MD), on the BioCAD HPLC. A gradient consisting of mobile phase A (10 mM ammonium acetate in 85% ACN/1% formic acid (FA)) and mobile phase B (30% ACN/0.1% FA) was used for the separation, which was conducted at an initial gradient from 0 to 15% B for 10 min, then from 15 to 30% B for 25 min, followed by a gradient from 30 to 100% B for 5 min and finally 100% B for 10 min, at a flow rate of 1 ml/min. After 2-min fractions were collected, C18 spin columns were used to desalt the peptides; certain neighboring fractions were combined into 12 fractions based on the MALDI MS signals.
2.2.3 Reversed-phase liquid chromatography and tandem mass spectrometry (RPLC/MS/MS)
Tryptic peptides obtained from total nuclear extracts were either analyzed by LC/MS/MS directly or subjected to further SCX or ERLIC fractionations prior to LC/MS/MS. RPLC/MS/MS was performed on an Ultimate™ 3000 Chromatography System that was equipped with an Ultimate™ 3000 autosampler (Dionex, Sunnyvale, CA) and coupled with an LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific). Desalted peptides were first trapped on a cartridge (Pepmap C18, 0.5 cm × 300 μm, Dionex) at 2% mobile phase B (mobile phase A: 2% ACN and 0.1% FA; mobile phase B: 85% ACN and 0.1% FA) with a flow rate of 30 μl/min, then transferred onto a 75 μm × 150 mm capillary Acclaim® PepMap100 column (C18, 3 μm, 100 Å, Dionex) and fractionated using a 75-min gradient (3–45% B) at a flow rate of 250 nl/min. After each gradient cycle, the column was washed isocratically at 95% mobile phase B to clean the column and minimize carry-overs, followed by a re-equilibration step with mobile phase A. The eluted peptides were ionized at 2.0 kV via a Proxeon nano electrospray ion source and introduced into an LTQ-Orbitrap Velos mass spectrometer. The capillary temperature was 275 °C. The MS was operated in a data-dependent mode. Full scan MS spectra (from m/z 300–2000) were acquired in the Orbitrap analyzer operating at a resolution of 60,000 at m/z of 400. The lock mass option was enabled to achieve high mass accuracy. The 10 most intense peptide ions with charge states ≥2 were sequentially isolated to a target value of 10,000 and fragmented in the linear ion trap by low-energy collision-induced dissociation (CID), with a normalized collision energy of 35%. The ion selection threshold was set at 5,000 counts for MS/MS. The maximum allowed ion accumulation times were 500 ms for full scans in the Orbitrap and 100 ms for CID measurements in the LTQ. Ions with single or unassigned charge states were excluded from fragmentation.
2.3 Protein database search
All the raw LC/MS/MS spectra were analyzed by the Proteome Discoverer (version 1.3.0.339) software suite, using both SEQUEST (Thermo, CA, U.S.) and Mascot (version 2.3, Matrix Science, London, U.K.) search engines. Carbamidomethyl cysteine (+57 Da) was set as a fixed modification, and methionine oxidation (+16 Da) was set as an optional modification. Up to two missed internal tryptic cleavage sites were allowed. Parameters common to all database searches included the use of monoisotopic masses and a mass error tolerance of 10 ppm for the precursor ions and 0.5 Da for the CID fragment ions. The database search was performed against all human proteins (20,233 sequence entries) annotated in the SwissProt protein database (released in October, 2012).
The database search results (.msf files) were further filtered and compiled into a list of nonredundant proteins with Scaffold (version Scaffold_3_3_1; Proteome Software, Inc., Portland, OR). Peptide identifications were accepted if they could be established at ≥95% probability as specified by the Peptide Prophet algorithm (Keller et al., 2002). Protein identifications were accepted if they could be established at ≥95.0% probability with a false discovery rate (FDR) no more than 1% based on the forward and reverse database search approach, and if they contained at least one uniquely identified peptide. Protein probabilities were assigned by the Protein Prophet algorithm (Nesvizhskii et al., 2003). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped together to satisfy the principles of parsimony. The accession numbers of the identified proteins were uploaded into the Ingenuity Pathway Analysis (IPA) software (http://www.ingenuity.com) to retrieve the putative cellular localization and functional information for respective protein.
3. Results and Discussion
3.1 Effective nuclear proteome analysis with the basic method
SHSY-5Y is a well-characterized human neuroblastoma cell line that has been widely used as a model for investigating neurodegenerative disorders, such as Alzheimer’s and Parkinson’s diseases (Molina-Holgado et al., 2008). In this study, we aimed to optimize a neuroproteomics method for the identification of low-abundance transcriptional regulators in SHSY-5Y cells. We isolated cell nuclei based on a procedure described by Dignam et al. (Dignam et al., 1983) that utilizes a hypotonic procedure for cell lysis and a high-salt extraction to isolate proteins from nuclear pellets (Method 1, Materials and Methods).
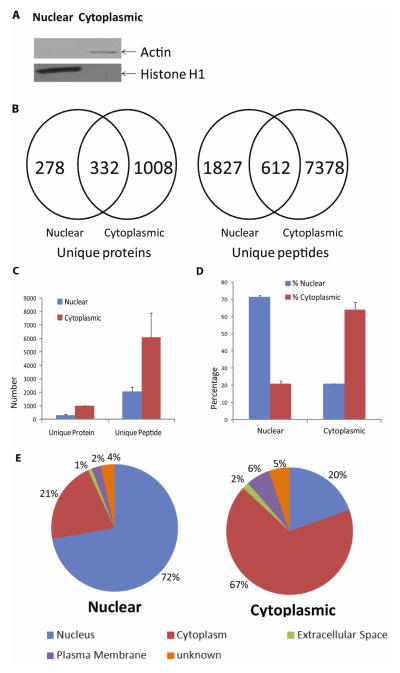
To evaluate the nuclear protein isolation efficiency of the basic method, Western blotting was performed on the nuclear and cytoplasmic extracts (Fig 1A). A histone H1 band (32–33 kDa) was detected specifically in the nuclear fraction but not in the cytoplasmic extracts. On the other hand, actin was only detected in the cytoplasmic fraction but not in the nuclear extracts. These data suggested that the basic nuclear protein enrichment method was effective. To further evaluate the quality of the nuclear preparations and to assess the degree of cytoplasmic protein contamination of the nuclear extracts, we examined both cytoplasmic and nuclear extracts by MS (Fig 1. B to E & Supplemental Tables 1 & 2). Among the approximate 1,400 proteins identified from the cytoplasmic extracts, ~two-third were annotated primarily as cytoplasmic proteins according to the bioinformatics analysis by IPA, whose contents are curated mainly from scientific literature. By comparison, among the >600 proteins found in the nuclear extract, ~72% were annotated as nuclear proteins by IPA, suggesting that the basic method was highly effective and produced a level of nuclear extract purity comparable to that from many published studies on nuclear proteomes (Abdolzade-Bavil et al., 2004; Dreger, 2003; Escobar et al., 2005; Salzano et al., 2006). For example, among the 1,900 proteins identified from the chromatin preparations by Garcia’s group, ~40% were classified as nuclear proteins by DAVID Bioinformatic Resources (http://david.abcc.ncifcrf.gov/) (Torrente et al., 2011). On the other hand, Henrich et al. identified 124 unique proteins from the human Burkitt’s lymphoma B-cell line, using a sucrose density gradient centrifugation followed by 2DE (Henrich et al., 2007). Using PSORT, a protein subcellular localization prediction algorithm, they determined that over 90% of the identified proteins were predicted to be nuclear. Of note, the different approaches to protein localization using PSORT, DAVID, IPA and other methods do not enable a direct comparison of these nuclear protein preparations.
Fig. 1. Comparison of nuclear and cytoplasmic extracts isolated with the basic method.
(A) Western blot detection of cellular compartment-specific proteins in SHSY-5Y cell nuclear and cytoplasmic extracts. Histone H1 (32–33 kD) was found mainly in the nuclear extracts, while actin (42 kD) was enriched mainly in the cytoplasmic extracts. (B) and (C) Comparison of unique proteins and peptides identified from the nuclear and cytoplasmic extracts. Nuclear (Blue bar); Cytoplasmic (Red bar), (B) shows data from one of the representative experiment. Peptide and protein identification criteria are specified in Materials and Methods. Scaffold was used to filter for and compare unique proteins and peptides identified from the analyses of 50 μg of each cytoplasmic or nuclear extract. As expected, more proteins and peptides were discovered in the cytoplasmic extracts. However, 278 unique proteins and 1,827 unique peptides were found only in the nuclear extracts in Fig 1.B (Supplemental Tables 1 & 2). (D) and (E) Cellular localization of the proteins identified from the cytoplasmic and nuclear extracts. (E) shows data from one of the representative experiment. Predictions were made by IPA software. Subcellular localization annotation: Nucleus (Blue); Cytoplasm (Red); Extracellular Space (Green); Plasma Membrane (Purple); Unknown (Orange).
Using the basic approach, we found 332 overlapping proteins common to both nuclear and cytoplasmic extracts in one of the representative experiment (Fig. 1B). The overlap between the two extracts may have been caused by either inefficiencies associated with the extraction methods or intracellular translocations of select proteins. Overall, the basic method was effective at producing enriched nuclear proteins.
3.2 Improvement of nuclear protein enrichment efficiencies
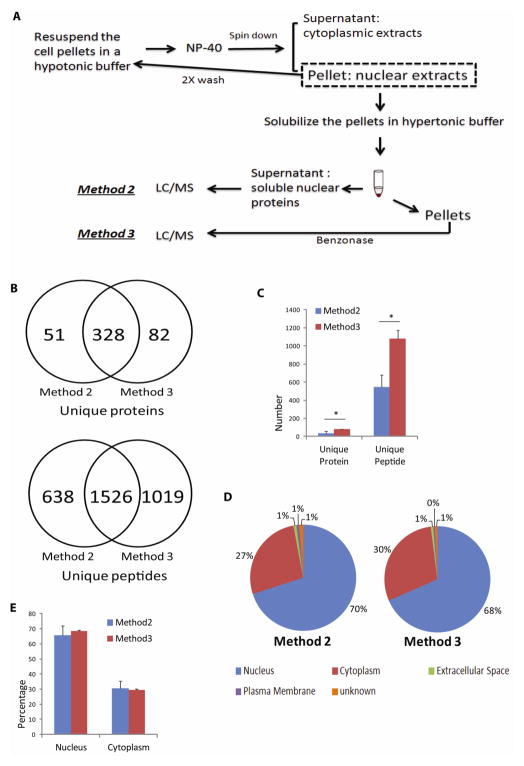
Due to the presence of ~20% cytoplasmic proteins in the nuclear extracts (Fig. 1D), we further evaluated whether repeated washing of the nuclear pellets could improve the removal of cytoplasmic contaminants (Method 2, Materials and Methods, Fig. 2A). With a two-step wash procedure, we found 15.1% ± 0.3% more proteins, with >70% being nuclear proteins. However, there was no further gain with a three-step wash procedure, either with regard to both the protein numbers and percentages of nuclear proteins found (Supplemental Fig 1 & Supplemental Tables 3 & 4), suggesting that the presence of cytoplasmic proteins in the nuclear extracts was unlikely to be the result of casual “contamination”.
Fig. 2. Effect of nuclease treatment on nuclear protein recovery.
(A) After initial isolation of the nuclear pellets from SHSY-5Y cells that included repeated washes to remove loosely associated cytoplasmic proteins, nuclear proteins were extracted with the hypertonic buffer, and the supernatants were digested either immediately (see Materials and Methods: Method 2) or after Benzonase® nuclease digestion of the pellets (see Materials and Methods: Method 3). Free nuclear proteins from Method 2 and DNA-associated proteins recovered from Method 3 were identified by LC/MS/MS analysis. (B) and (C) Comparison of the unique proteins and peptides identified from either method. (B) shows data from one of the representative experiment. In addition to a large degree of protein and peptide overlap found between the two methods, nuclease digestion of DNA resulted in the identification of ~40% more peptides and 20% more proteins in Method 3, when compared with Method 2 alone (* P≤ 0.05). Method 2 (Blue bar); Method 3 (Red bar). (D) and (E) Comparison of IPA-annotated nuclear proteins between the two methods. (D) shows data from one of the representative experiment. Relative percentages of nuclear proteins between the two methods were comparable. Subcellular localization annotation: Nucleus (Blue); Cytoplasm (Red); Extracellular Space (Green); Plasma Membrane (Purple); Unknown (Orange).
Cell nuclei contain large amounts of DNAs and RNAs, which may be associated with transcription factors and other regulators of gene expressions. To release the proteins that might be trapped within the nucleic acids and chromatin, we added Benzonase, a commercially available nuclease, to the pellets after hypertonic extraction to digest the nucleic acids and possibly release more chromatin-associated proteins (Method 3, Materials and Methods, Fig. 2A). The addition of Benzonase markedly reduced cell lysate viscosity during sample processing (data not shown). Indeed, around 82 additional unique proteins and an average 1,082 additional peptides were identified after Benzonase treatment, compared with the yield from Method 2 alone (Fig. 2B and 2C & Supplemental Tables 5 & 6). Methods 2 and 3 enabled the identifications of similar numbers of “transcriptional regulators” classified according to IPA (highlighted in Supplemental Tables 5 & 6), with 18 more unique transcriptional regulators identified only after the Benzonase treatment. Surprisingly, the IPA bioinformatics predictions of protein identified following Benzonase treatment did not find a higher percentage of nuclear proteins; the nuclear protein purity obtained from this method was ~68%, comparable to ~66% from Method 2 (Fig. 2D and 2E), suggesting that some “cytoplasmic proteins” may indeed be associated with nucleic acids or chromatin on occasions and possibly carry out ‘moonlighting’ functions in the nucleus.
3.3 Expanding the nuclear proteome coverage by multidimensional ractionations
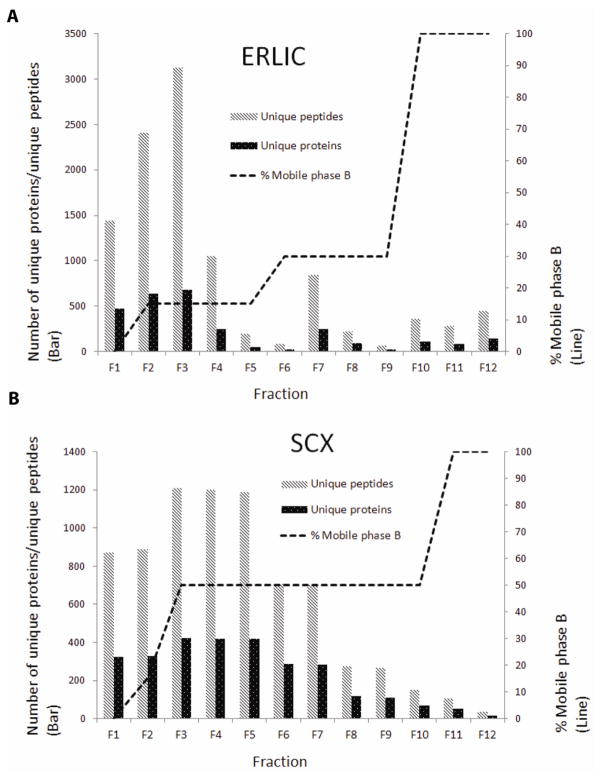
The nuclear proteomes have been studied in a variety of human organs and cells (Albrethsen et al., 2009; Andersen et al., 2002; Dreger et al., 2001; Henrich et al., 2007; Schenk et al., 2012; Tchapyjnikov et al., 2010; Torrente et al., 2011; Wilkie and Schirmer, 2006). Most of the studies used crude nuclear pellets without further purification or fractionation. It is well established that orthogonal protein or peptide fractionation prior to LC/MS/MS is a highly effective approach for identifying low-abundance proteins. We therefore evaluated the effectiveness of SCX- and ERLIC-based multidimensional peptide separation methods for expanding the nuclear proteome coverage relative to the coverage from analysis of unfractionated nuclear preparations. From 300 μg nuclear proteins prepared by combining Methods 2 and 3 (see Materials and Methods, Fig. 2A), average 2,497 non-redundant proteins and around 12,603 unique peptides were identified via ERLIC separation, whereas average 2623 non-redundant proteins and over 9,800 unique peptides were identified via SCX separation (Fig. 3A and 3B & Supplemental Tables 7, 8 & 9). Both separation methods were equally effective at dramatically improving the coverage of the nuclear proteomes, with each approach more than doubled the number of proteins identified over the unfractionated approach. Furthermore, a combined analysis involving both SCX and ERLIC fractionations almost tripled the number of proteins identified in comparison with the unfractionated analysis, bringing the total number of proteins to >3,000 (Fig. 3A and 3B). After multidimensional peptide separations, the predicted percentage of nuclear proteins decreased from ~64% to ~54–61% (Fig. 3C and 3D), suggesting that some low-abundance proteins identified only after SCX and ERLIC fractionations were mainly localized in the cytoplasm in SHSY-5Y cells. Even at a low nuclear protein purity of 50%, our combined SCX/ERLIC analysis detected >1,800 nuclear proteins, which constitute one of the largest datasets reported for a neuronal cell. To understand the details of the improved proteome coverage due to these two fractionation methods, we further analyzed the number of unique proteins and peptides identified in each ERLIC and SCX fraction (Fig. 4). Interestingly, the ERLIC method offered less peptide fractionation (Fig. 4A) compared with SCX (Fig. 4B), suggesting that the peptides derived from this nuclear preparation are homogeneous in polarity but heterogeneous in pI. All the experiments above were repeated at least twice and the results were reproducible (data not shown). Further optimization of these fractionation conditions (e.g. gradient) will likely lead to even deep coverage of the nuclear proteomes.
Fig. 3. Comparison of the proteome coverage among the unfractionated preparation and the ERLIC and SCX fractionated preparations.
(A) and (B) Comparison of the numbers of unique proteins and peptides. Unfractionated (Blue bar), ERLIC (Red bar) and SCX (Green bar). (A) shows data from one of the representative experiment. ERLIC and SCX fractionations increased the number of unique proteins or peptides identified by more than 140% and 150%, respectively (* p≤ 0.05). (C) and (D) Annotated localizations of the proteins identified. Unfractionated (Blue bar), ERLIC (Red bar) and SCX (Green bar). (D) shows data from one of the representative experiment. Subcellular localization annotation: Nucleus (Blue); Cytoplasm (Red); Extracellular Space (Green); Plasma Membrane (Purple); Unknown (Orange).
Fig. 4.
Number of unique proteins and unique peptides identified in each (A) ERLIC and (B) SCX fraction.
3.4 Key neuronal transcription factors identified from in-depth nuclear proteomic analyses
Up until recently, conventional neuroproteomics techniques do not have the sensitivities for routine analysis of less abundant neuronal transcription factors, which play crucial roles in cells underlying neurological diseases. The new method described here can dramatically improve the coverage of the nuclear proteomes and enable the analysis of neuronal transcription factors (Supplemental Table 10). Here, we have identified more than 3,000 proteins in the nuclear extracts, which comprise one of the largest mammalian nuclear proteomics datasets published to date (Abdolzade-Bavil et al., 2004; Boisvert et al., 2010; Chaerkady et al., 2009; Shakib et al., 2005; Shiio et al., 2003; Tchapyjnikov et al., 2010; Torrente et al., 2011). Using an independent bioinformatics analysis by IPA, 215 proteins from the ERLIC separation and 303 proteins from the SCX separation were classified as “transcriptional regulators” (Supplemental Table 11), comparable to the ~300 transcription factors found in human brain tissues by a recent genetic survey (Vaquerizas et al., 2009). For example, zinc finger proteins are known to mediate specific protein–DNA interactions (Matthews and Sunde, 2002), over 30 zinc finger proteins were found in this study, some of which have been reported to be involved in transcriptional regulation (Blaiseau et al., 1997; Martinez-Pastor et al., 1996). One of the transcription factors detected in this study is CCAAT enhancer-binding protein (C/EBP) (Table 1). As a member of the basic leucine zipper DNA-binding protein family, it is enriched in neurons and up-regulated following brain injuries in animal models of neuronal regeneration (Cortes-Canteli et al., 2004; Nadeau et al., 2005). Its key functions include the regulation of neuronal cell growth, differentiation, learning, memory and apoptosis (Cortes-Canteli et al., 2002; Hatakeyama et al., 2006; Marshall et al., 2003; Menard et al., 2002). Additional transcription regulators found in this study included atrophin-1 (ATN1) and huntingtin (Table 1). ATN1 is localized in both the nuclei and cytoplasm of neurons in human CNS (Wood et al., 2000). Accumulations of ATN1 mutants have been associated with the development of dentatorubral-pallidoluysian atrophy neurodegeneration (Suzuki and Yazawa, 2011) and huntington’s disease (HD) (Costa Mdo et al., 2006; Schilling et al., 2001). Huntingtin protein polymorphism can lead to the incorporation of different number of glutamines in the protein; HD patients tend to have a large number of glutamines in huntingtin (Perutz, 1996). Overall, with the optimized multidimensional method, in-depth neuroproteomics analysis of neuronal cells and tissues appears quite feasible.
TABLE 1.
Select transcriptional regulators identified in this study
| Gene symbol | Protein name | Protein identification probability* | Number of unique peptides identified | Percentage sequence coverage | Identification approach |
|---|---|---|---|---|---|
| ATN1 | Atrophin-1 | 99.7% | 2 | 5% | SCX |
| CEBPZ | CCAAT/enhancer-binding protein zeta | 99.9% | 16 | 24% | ERLIC |
| CEBPG | CCAAT/enhancer-binding protein gamma | 99.7% | 2 | 48% | SCX |
| HTT | Huntingtin | 99.9% | 5 | 4% | ERLIC |
Protein identification probability calculated by Scaffold, using Protein Prophet method.
4.0 Conclusion
We have optimized a method to identify transcriptional regulators in a neuroblastoma cell line by a combination of improved nuclear protein isolation and multidimensional peptide fractionations. Using this method, we have identified many transcriptional regulators that were barely detectable in previous neuroproteomics analyses. Therefore, targeted nuclear proteomics analysis may provide an opportunity for a better understanding of neuronal cell functions and diseases. Because the approaches described in this study can be readily combined with different quantitative proteomics methodologies, discovering quantitative changes among the transcription regulators and their post-translational modifications underlying diverse neurological phenomena may soon become a reality.
Supplementary Material
Acknowledgments
The project described was supported in part by a grant P30NS046593 from the National Institute of Neurological Disorders And Stroke. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Neurological Disorders And Stroke or the National Institutes of Health.
Abbreviations
- 2DE
2D gel electrophoresis
- ACN
Acetonitrile
- ATN1
Atrophin-1
- C/EBP
CCAAT enhancer binding protein
- CID
Collision induced dissociation
- DMEM
Dulbecco’s Modified Eagle Medium
- DRPLA
Dentatorubral-pallidoluysian atrophy
- ERLIC
Electrostatic Repulsion-Hydrophilic Interaction Chromatographic
- FDR
False discovery rate
- HD
Huntington’s disease
- ICAT
Isotope-coded affinity tag
- IPA
Ingenuity Pathway Analysis
- SCX
Strong Cation Exchange
- TEAB
Triethylammonium bicarbonate
- TF
Transcription factor
Footnotes
The authors report no conflict of interest.
References
- Abdolzade-Bavil A, Hayes S, Goretzki L, Kroger M, Anders J, Hendriks R. Convenient and versatile subcellular extraction procedure, that facilitates classical protein expression profiling and functional protein analysis. Proteomics. 2004;4:1397–405. doi: 10.1002/pmic.200300710. [DOI] [PubMed] [Google Scholar]
- Albrethsen J, Knol JC, Jimenez CR. Unravelling the nuclear matrix proteome. J Proteomics. 2009;72:71–81. doi: 10.1016/j.jprot.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. Directed proteomic analysis of the human nucleolus. Curr Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- Blaiseau PL, Isnard AD, Surdin-Kerjan Y, Thomas D. Met31p and Met32p, two related zinc finger proteins, are involved in transcriptional regulation of yeast sulfur amino acid metabolism. Mol Cell Biol. 1997;17:3640–8. doi: 10.1128/mcb.17.7.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, Lam YW, Lamont D, Lamond AI. A quantitative proteomics analysis of subcellular proteome localization and changes induced by DNA damage. Mol Cell Proteomics. 2010;9:457–70. doi: 10.1074/mcp.M900429-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill DJ. Protein and antibody arrays and their medical applications. J Immunol Methods. 2001;250:81–91. doi: 10.1016/s0022-1759(01)00325-8. [DOI] [PubMed] [Google Scholar]
- Chaerkady R, Kerr CL, Marimuthu A, Kelkar DS, Kashyap MK, Gucek M, Gearhart JD, Pandey A. Temporal analysis of neural differentiation using quantitative proteomics. J Proteome Res. 2009;8:1315–26. doi: 10.1021/pr8006667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes-Canteli M, Pignatelli M, Santos A, Perez-Castillo A. CCAAT/enhancer-binding protein beta plays a regulatory role in differentiation and apoptosis of neuroblastoma cells. J Biol Chem. 2002;277:5460–7. doi: 10.1074/jbc.M108761200. [DOI] [PubMed] [Google Scholar]
- Cortes-Canteli M, Wagner M, Ansorge W, Perez-Castillo A. Microarray analysis supports a role for ccaat/enhancer-binding protein-beta in brain injury. J Biol Chem. 2004;279:14409–17. doi: 10.1074/jbc.M313253200. [DOI] [PubMed] [Google Scholar]
- do Costa MC, Teixeira-Castro A, Constante M, Magalhaes M, Magalhaes P, Cerqueira J, Vale J, Passao V, Barbosa C, Robalo C, Coutinho P, Barros J, Santos MM, Sequeiros J, Maciel P. Exclusion of mutations in the PRNP, JPH3, TBP, ATN1, CREBBP, POU3F2 and FTL genes as a cause of disease in Portuguese patients with a Huntington-like phenotype. J Hum Genet. 2006;51:645–51. doi: 10.1007/s10038-006-0001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–89. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreger M. Proteome analysis at the level of subcellular structures. Eur J Biochem. 2003;270:589–99. doi: 10.1046/j.1432-1033.2003.03426.x. [DOI] [PubMed] [Google Scholar]
- Dreger M, Bengtsson L, Schoneberg T, Otto H, Hucho F. Nuclear envelope proteomics: novel integral membrane proteins of the inner nuclear membrane. Proc Natl Acad Sci USA. 2001;98:11943–8. doi: 10.1073/pnas.211201898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar MA, Hoelz DJ, Sandoval JA, Hickey RJ, Grosfeld JL, Malkas LH. Profiling of nuclear extract proteins from human neuroblastoma cell lines: the search for fingerprints. J Pediatr Surg. 2005;40:349–58. doi: 10.1016/j.jpedsurg.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Gauss C, Kalkum M, Lowe M, Lehrach H, Klose J. Analysis of the mouse proteome. (I) Brain proteins: separation by two-dimensional electrophoresis and identification by mass spectrometry and genetic variation. Electrophoresis. 1999;20:575–600. doi: 10.1002/(SICI)1522-2683(19990301)20:3<575::AID-ELPS575>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Hatakeyama D, Sadamoto H, Watanabe T, Wagatsuma A, Kobayashi S, Fujito Y, Yamashita M, Sakakibara M, Kemenes G, Ito E. Requirement of new protein synthesis of a transcription factor for memory consolidation: paradoxical changes in mRNA and protein levels of C/EBP. J Mol Biol. 2006;356:569–77. doi: 10.1016/j.jmb.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Henrich S, Cordwell SJ, Crossett B, Baker MS, Christopherson RI. The nuclear proteome and DNA-binding fraction of human Raji lymphoma cells. Biochim Biophys Acta. 2007;1774:413–32. doi: 10.1016/j.bbapap.2006.12.011. [DOI] [PubMed] [Google Scholar]
- Jung E, Hoogland C, Chiappe D, Sanchez JC, Hochstrasser DF. The establishment of a human liver nuclei two-dimensional electrophoresis reference map. Electrophoresis. 2000;21:3483–7. doi: 10.1002/1522-2683(20001001)21:16<3483::AID-ELPS3483>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–92. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Lin HW, Jain MR, Li H, Levison SW. Ciliary neurotrophic factor (CNTF) plus soluble CNTF receptor alpha increases cyclooxygenase-2 expression, PGE2 release and interferon-gamma-induced CD40 in murine microglia. J Neuroinflammation. 2009;6:7. doi: 10.1186/1742-2094-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, D’Mello V, Deng L, Hu J, Ricardo M, Pan S, Lu X, Wadsworth S, Siekierka J, Birge R, Li H. A multiplexed proteomics approach to differentiate neurite outgrowth patterns. J Neurosci Methods. 2006;158:22–9. doi: 10.1016/j.jneumeth.2006.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall J, Dolan BM, Garcia EP, Sathe S, Tang X, Mao Z, Blair LA. Calcium channel and NMDA receptor activities differentially regulate nuclear C/EBPbeta levels to control neuronal survival. Neuron. 2003;39:625–39. doi: 10.1016/s0896-6273(03)00496-3. [DOI] [PubMed] [Google Scholar]
- Martinez-Pastor MT, Marchler G, Schuller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE) EMBO J. 1996;15:2227–35. [PMC free article] [PubMed] [Google Scholar]
- Matthews JM, Sunde M. Zinc fingers--folds for many occasions. IUBMB Life. 2002;54:351–5. doi: 10.1080/15216540216035. [DOI] [PubMed] [Google Scholar]
- Menard C, Hein P, Paquin A, Savelson A, Yang XM, Lederfein D, Barnabe-Heider F, Mir AA, Sterneck E, Peterson AC, Johnson PF, Vinson C, Miller FD. An essential role for a MEK-C/EBP pathway during growth factor-regulated cortical neurogenesis. Neuron. 2002;36:597–610. doi: 10.1016/s0896-6273(02)01026-7. [DOI] [PubMed] [Google Scholar]
- Molina-Holgado F, Gaeta A, Francis PT, Williams RJ, Hider RC. Neuroprotective actions of deferiprone in cultured cortical neurones and SHSY-5Y cells. J Neurochem. 2008;105:2466–76. doi: 10.1111/j.1471-4159.2008.05332.x. [DOI] [PubMed] [Google Scholar]
- Nadeau S, Hein P, Fernandes KJ, Peterson AC, Miller FD. A transcriptional role for C/EBP beta in the neuronal response to axonal injury. Mol Cell Neurosci. 2005;29:525–35. doi: 10.1016/j.mcn.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–58. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Pandey A, Mann M. Proteomics to study genes and genomes. Nature. 2000;405:837–46. doi: 10.1038/35015709. [DOI] [PubMed] [Google Scholar]
- Perutz MF. Glutamine repeats and inherited neurodegenerative diseases: molecular aspects. Curr Opin Struct Biol. 1996;6:848–58. doi: 10.1016/s0959-440x(96)80016-9. [DOI] [PubMed] [Google Scholar]
- Salzano AM, Paron I, Pines A, Bachi A, Talamo F, Bivi N, Vascotto C, Damante G, Quadrifoglio F, Scaloni A, Tell G. Differential proteomic analysis of nuclear extracts from thyroid cell lines. J Chromatogr B Analyt Technol Biomed Life Sci. 2006;833:41–50. doi: 10.1016/j.jchromb.2005.12.025. [DOI] [PubMed] [Google Scholar]
- Schenk LK, Bolger SJ, Luginbuhl K, Gonzales PA, Rinschen MM, Yu MJ, Hoffert JD, Pisitkun T, Knepper MA. Quantitative Proteomics Identifies Vasopressin-Responsive Nuclear Proteins in Collecting Duct Cells. J Am Soc Nephrol. 2012;23:1008–18. doi: 10.1681/ASN.2011070738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling G, Jinnah HA, Gonzales V, Coonfield ML, Kim Y, Wood JD, Price DL, Li XJ, Jenkins N, Copeland N, Moran T, Ross CA, Borchelt DR. Distinct behavioral and neuropathological abnormalities in transgenic mouse models of HD and DRPLA. Neurobiol Dis. 2001;8:405–18. doi: 10.1006/nbdi.2001.0385. [DOI] [PubMed] [Google Scholar]
- Shakib K, Norman JT, Fine LG, Brown LR, Godovac-Zimmermann J. Proteomics profiling of nuclear proteins for kidney fibroblasts suggests hypoxia, meiosis, and cancer may meet in the nucleus. Proteomics. 2005;5:2819–38. doi: 10.1002/pmic.200401108. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Jensen ON, Podtelejnikov AV, Sagliocco F, Wilm M, Vorm O, Mortensen P, Boucherie H, Mann M. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc Natl Acad Sci USA. 1996;93:14440–5. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiio Y, Eisenman RN, Yi EC, Donohoe S, Goodlett DR, Aebersold R. Quantitative proteomic analysis of chromatin-associated factors. J Am Soc Mass Spectrom. 2003;14:696–703. doi: 10.1016/S1044-0305(03)00204-6. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Yazawa I. Pathological accumulation of atrophin-1 in dentatorubralpallidoluysian atrophy. Int J Clin Exp Pathol. 2011;4:378–84. [PMC free article] [PubMed] [Google Scholar]
- Tchapyjnikov D, Li Y, Pisitkun T, Hoffert JD, Yu MJ, Knepper MA. Proteomic profiling of nuclei from native renal inner medullary collecting duct cells using LC-MS/MS. Physiol Genomics. 2010;40:167–83. doi: 10.1152/physiolgenomics.00148.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrente MP, Zee BM, Young NL, Baliban RC, LeRoy G, Floudas CA, Hake SB, Garcia BA. Proteomic interrogation of human chromatin. PLoS One. 2011;6:e24747. doi: 10.1371/journal.pone.0024747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L, Lamond AI. Nuclear functions in space and time: gene expression in a dynamic, constrained environment. FEBS Lett. 2008;582:1960–70. doi: 10.1016/j.febslet.2008.04.029. [DOI] [PubMed] [Google Scholar]
- Tyler WA, Jain MR, Cifelli SE, Li Q, Ku L, Feng Y, Li H, Wood TL. Proteomic identification of novel targets regulated by the mammalian target of rapamycin pathway during oligodendrocyte differentiation. Glia. 2011;59:1754–69. doi: 10.1002/glia.21221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10:252–63. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- Wilkie GS, Schirmer EC. Guilt by association: the nuclear envelope proteome and disease. Mol Cell Proteomics. 2006;5:1865–75. doi: 10.1074/mcp.R600003-MCP200. [DOI] [PubMed] [Google Scholar]
- Wood JD, Nucifora FC, Jr, Duan K, Zhang C, Wang J, Kim Y, Schilling G, Sacchi N, Liu JM, Ross CA. Atrophin-1, the dentato-rubral and pallido-luysian atrophy gene product, interacts with ETO/MTG8 in the nuclear matrix and represses transcription. J Cell Biol. 2000;150:939–48. doi: 10.1083/jcb.150.5.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan JX, Harry RA, Wait R, Welson SY, Emery PW, Preedy VR, Dunn MJ. Separation and identification of rat skeletal muscle proteins using two-dimensional gel electrophoresis and mass spectrometry. Proteomics. 2001;1:424–34. doi: 10.1002/1615-9861(200103)1:3<424::AID-PROT424>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Zhang C. Proteomic studies on the development of the central nervous system and beyond. Neurochem Res. 2010;35:1487–500. doi: 10.1007/s11064-010-0218-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.