Abstract
Recurrent chromosomal translocations involving the mixed lineage leukaemia (MLL) gene initiate aggressive forms of leukaemia, which are often refractory to conventional therapies1. Many MLL-fusion partners are members of the super elongation complex (SEC), a critical regulator of transcriptional elongation, suggesting that aberrant control of this process plays an important role in leukaemia induction2,3. Here we use a global proteomic strategy to demonstrate that MLL-fusions, as part of SEC2,3 and the polymerase associated factor (PAFc) complex4,5 are associated with the BET family of acetyl-lysine recognising, chromatin “adaptor” proteins. These data provided the basis for therapeutic intervention in MLL-fusion leukaemia, via the displacement of the BET family of proteins from chromatin. We show that a novel small molecule inhibitor of the BET family GSK1210151A (I-BET151) has profound efficacy against human and murine MLL-fusion leukaemic cell lines, through the induction of early cell cycle arrest and apoptosis. I-BET151 treatment in two human leukaemia cell lines with different MLL-fusions alters the expression of a common set of genes whose function may account for these phenotypic changes. The mode of action of I-BET151 is, at least in part, due to the inhibition of transcription at key genes (BCL2, C-MYC and CDK6) through the displacement of BRD3/4, PAFc and SEC components from chromatin. In vivo studies indicate that I-BET151 has significant therapeutic value, providing survival benefit in two distinct mouse models of murine MLL-AF9 and human MLL-AF4 leukaemia. Finally, the efficacy of I-BET151 against human leukaemia stem cells (LSC) is demonstrated, providing further evidence of its potent therapeutic potential. These findings establish the displacement of BET proteins from chromatin as a promising epigenetic therapy for these aggressive leukaemias.
Dysregulation of chromatin-modifiers is a recurrent and sentinel event in oncogenesis6. Therapeutic strategies which selectively alter the recruitment and/or catalytic activity of these enzymes at chromatin therefore hold great promise as targeted therapies6. In this regard the Bromodomain and Extra Terminal (BET) family of proteins (BRD2, BRD3, BRD4 and BRDT) provide an ideal ‘druggable’ target, since they share a common highly conserved tandem bromodomain at their amino-terminus. Selective bromodomain inhibitors that disrupt the binding of BET proteins to histones have recently been described7,8, however their true therapeutic scope remains untested.
To identify the nuclear complexes associated with ubiquitously expressed BETs (BRD2/3/4), we performed a systematic global proteomic survey. Specifically, this involved a tri-partite discovery approach (Fig. 1a). In the first approach, bead-immobilized analogues of I-BET7629 were incubated with HL60 nuclear extracts and bound proteins were analysed by quantitative mass spectrometry (Supplementary Table 1). This approach identified the BET isoforms, and a large number of co-purifying proteins (Supplementary Table 1 and 2), indicating that the BET isoforms reside in many distinct protein complexes. In the second approach, immunoprecipitation (IP) analyses with selective antibodies against BRD2/3/4 were performed (Supplementary Fig. 1 and Supplementary Table 3 and 4). This was complemented with additional IPs using selected antibodies against complex members (“baits”) selected from the sub-set of proteins that were identified in the first approach (Fig. 1b right panel, Supplementary Fig. 2 and Supplementary Table 3). In the third approach, bead-immobilized Histone H4(1-21; K5acK8acK12ac) acetylated peptides were used to purify protein complexes. These data were combined to highlight a list of complexes identified in all three methods (Fig. 1b left panel, Supplementary Fig. 3 and Supplementary Table 1). Finally, specificity of the I-BET762 and histone tail matrix was further assessed by competition experiments (Fig. 1c and Supplementary Fig. 4-5 and Supplementary Table 2). This strategy enabled the direct determination of the targets of the inhibitor, and the proteins associated with the target, with subunits of protein complexes exhibiting closely matching IC50 values10. Taken together these stringent and complementary approaches provide a high confidence global data set encompassing all known11-13 and several novel BET protein complexes (Fig 1b and Supplementary Fig. 3). Amongst the novel complexes, we observed a prominent enrichment and dose dependent inhibition of several components of the PAFc4,5 and SEC2,3 (Fig. 1b and Fig. 1c), which were confirmed by reciprocal IPs in HL60 cells (Fig. 1b). Moreover, reciprocal IPs in two MLL-Fusion leukaemia cell lines (MV4;11 and RS4;11) confirmed the relationship of SEC with BRD4 in different cellular contexts (Fig. 1d). Together these data indicate that BRD3/4 associate with the PAFc and SEC and may function to recruit these complexes to chromatin. Given that these complexes are crucial for malignant transformation by MLL fusions2-5 we tested the hypothesis that displacement of BET proteins from chromatin may have a therapeutic role in these leukaemias.
Figure 1. A global proteomic survey identifies BET proteins as part of the PAFc and SEC.
(A). Proteomic strategy (B) Left: Cytoscape representation of the BET protein complex network (discussed in detail in supplementary figure 3). Bold circles indicate associations confirmed by the three orthogonal methods. Right: Heat map representing quantitative-MS data following co-IP of BETs, PAF and SEC complex members. (C) Differential proteomic analysis of the proteins interacting with I-BET and triple acetylated histone H4 tail. Left: Affinity matrices with immobilized I-BET762 or Histone H4(K5acK8acK12ac) peptide bind to the same set of BET complexes. Right: competitive inhibition of the binding of BET isoforms, and SEC and PAF complex components, to the I-BET762 matrix showing matching concentration dependence. (D) Brd4 and MLLT1 interact in HL60, MV4;11 and RS4;11 cells and binding to the I-BET762 matrix is blocked by excess I-BET151. (E) GSK1210151A (I-BET151). (F) I-BET151 binding to the acetyl-binding pocket of BRD4-BD1 (cyan) overlaid with H3K14-Acetyl peptide (green) (3jvk.pdb). A surface representation of the BRD4-BD1 is shown with key recognition and the specificity WPF shelf identified. (G) Ribbon representation of the BRD4-BD1 (cyan) crystal structure complexed with I-BET151 (shown in magenta stick format) overlaid with H3(12-19)K14ac peptide (green) taken from its complex with BRD4-BD1(3jvk.pdb). Secondary elements of the BRD4-BD1 structure have been highlighted. (H) Selectivity profile of IBET-151 showing average temperature shifts (Tm) using a fluorescent thermal shift assay. Numbering inside the spheres, e.g. 12 signifies both bromodomains 1 and 2 have been assessed. Overlaid is the selectivity profile generated using a proteomic approach (shown as boxes around proteins, discussed in Supplementary Fig. 5). Where the bromodomains have been profiled by both thermal shift and proteomic approaches the agreement is excellent. Proteins not assessed by either technique are shown in grey. (I) Comparison of I-BET762 and I-BET151 potency in ligand displacement assays, direct BIAcore binding and LPS stimulated IL6 cytokine production from human PBMCs or whole blood (WB).
To progress our studies with an optimised therapeutic agent we developed I-BET151(Fig 1e); a novel dimethylisoxazole template, previously undisclosed as a BET bromodomain inhibitor. It was identified and optimised to retain excellent BET target potency (Fig 1i) and selectivity (Fig 1h; Supplementary Fig. 5-10 and Supplementary Table 5) while enhancing the in vivo pharmacokinetics and terminal half-life to enable prolonged in vivo studies (Fig 4a and Supplementary Figure 20). We also generated a proteomic selectivity profiles comparing I-BET151 with I-BET762 (Fig. 1h, Supplementary Fig. 5 and Supplementary Table 6). We bead-immobilized a combination of differentially acetylated histone tail peptides (Supplementary Table 7), which captured a total of 27 bromodomain proteins from HL60 nuclear extracts. Competition with excess I-BET151 or I-BET762 blocked the capture of Brd2, Brd3, Brd4, and Brd9 but had no effect on the 23 other bromodomain proteins including MLL. The inhibition of Brd9 is likely to be indirect as this protein forms a complex with Brd4 (Supplementary Table 3). Finally, a high-resolution (1.5Å) crystal structure of I-BET151 bound to BRD4-bromodomain 1 (BD1) revealed binding to the acetylated-lysine (AcK) recognition pocket of the BET protein (Fig 1f, 1g and Supplementary Figure 10).
Figure 4. I-BET151 is efficacious in in vivo murine models and primary patients samples of MLL-fusion leukaemia.
(A) Murine pharmacokinetic studies (mean ± SD (n = 4 per compound)) comparing the blood concentration of I-BET151 with I-BET762 and JQ1. (B) Kaplan-Meier curve of control and treated NOD-SCID mice transplanted with 1 × 107 MV4;11 cells. Green arrowhead = treatment commencement D21. (C) Haematoxylin and eosin (H&E) stained histological sections of the renal parenchyma of control and treated mice. Black arrows highlight leukaemic infiltration. (D) Representative FACS analysis from the peripheral blood of control or I-BET151 treated mice. (E) Kaplan-Meier curve of control and treated C57BL/6 mice transplanted with 2.5 × 106 syngeneic MLL-AF9 leukaemic cells. Green arrowhead = treatment commencement D9. (F) Photomicrograph of the spleen size from 5/8 control and 1/12 I-BET151 treated mice that died on day 12. (G) H&E stained histological sections of the liver parenchyma from control and IBET151 treated mice demonstrating reduced disease burden in the treated animal. (H) Peripheral blood white cell count, (I) liver weight and (J) spleen weights from all the control and treated mice at the time of necropsy. (K) Representative FACS analysis assessing apoptosis from a patient with MLL-AF6 leukaemia. (L) Clonogenic assays with human MLL-fusion LSC isolated by FACS sorting (CD34+/CD38-) and plated in the presence of DMSO or I-BET151. (M) Gene-expression changes in human MLL-fusion leukaemia cells following treatment with I-BET151 or DMSO. The log2 fold change in the expression level for all genes (expression level with IBET151 treatment/expression level with DMSO) is represented. (N) Schematic model proposing the mode of action for I-BET151 in MLL-fusion leukaemia.
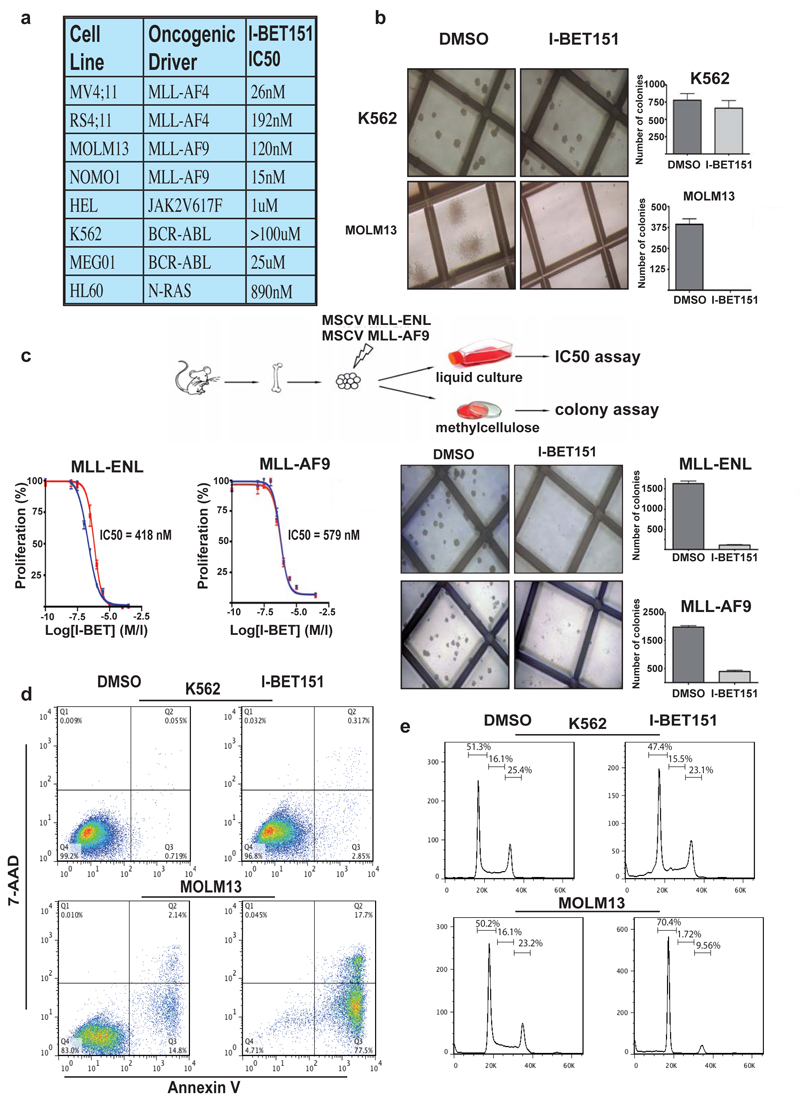
To assess the therapeutic efficacy and selectivity of I-BET151, we tested a panel of leukaemic cell lines harbouring a spectrum of distinct oncogenic drivers. These data demonstrated that I-BET151 has potent efficacy against cell lines harbouring different MLL-fusions (Fig. 2a and Supplementary Fig. 11). To extend these data we tested the clonogenic potential of human leukaemic cells grown in cytokine-supplemented methylcellulose containing DMSO (vehicle) or I-BET151. Consistent with the profound effects in liquid culture, the colony forming potential of MLL-fusion driven leukaemias (MOLM13) was completely ablated by I-BET151, whilst leukaemias driven by tyrosine kinase activation (K562) were unaffected (Fig. 2b). In addition to the data with human leukaemic cell lines, we also confirmed the potent efficacy of I-BET151 in both liquid culture and clonogenic assays using primary murine progenitors retrovirally transformed with either MLL-ENL or MLL-AF9 (Fig. 2c).
Figure 2. I-BET151 selectively and potently inhibits MLL-fusion leukaemic cell lines in vitro.
(A) Human leukaemia cell lines tested using I-BET151. (B) Clonogenic assays performed in the presence of DMSO or I-BET151. (C) Haematopoietic progenitors were isolated from mouse bone marrow and retrovirally transformed with MLL-ENL or MLL-AF9. These cells were used in both proliferation and clonogenic assays. (D) Apoptosis was assessed by FACS analysis following 72 hours incubation with DMSO or I-BET151. (E) Cell cycle progression was assessed by FACS analysis 24 hours after incubation with DMSO or I-BET151. Bar graphs are represented as the mean and error bars reflect standard deviation of results derived from triplicate experiments.
To investigate the mechanism of action for I-BET151, we performed flow cytometry (FACS) to assess apoptosis and cell cycle progression following I-BET151 treatment. Figure 2d-e and Supplementary Fig. 12 show a striking induction of apoptosis and a prominent G0/G1 arrest in two MLL-fusion cell lines driven by distinct MLL-fusions (MOLM13 and MV4;11 containing MLL-AF9 and MLL-AF4 respectively). In contrast, the cell cycle characteristics and apoptotic rate of K562 cells were largely unaffected at this time. These data suggest that I-BET151 alters the transcriptional programmes regulating apoptosis and cell-cycle progression in MLL-fusion leukaemias.
To identify the precise transcriptional pathways controlled by I-BET151, global gene-expression analysis was performed in MOLM13 and MV4;11 cells following treatment with I-BET151 or DMSO for 6 hours. This strategy allowed us to identify early I-BET151 responsive genes, prior to any discernable phenotypic alteration in cell cycle or apoptosis (Supplementary Fig. 12). As demonstrated previously7, we observed differential expression of a selective subset of genes (Fig. 3a), rather than global transcriptional dysregulation. Remarkably, the transcriptional programmes altered in the two MLL-fusion cell lines were highly correlated (Fig. 3b) and gene set enrichment analysis documented significant overlap with published MLL fusion signatures including MLL-fusion LSC14,15 (Supplementary Fig. 13). These data are consistent with the notion that MLL-fusions aberrantly co-opt the SEC and PAFc to regulate similar transcriptional programmes. Notably, the top 100 genes concomitantly decreased in both MOLM13 and MV4;11 (Fig. 3c) contained several previously reported direct MLL targets, such as BCL2, CDK6 and C-MYC, whose down-regulation was consistent with the phenotypic consequences of I-BET151.
Figure 3. Transcriptome and ChIP analyses provide mechanistic insights for the efficacy of I-BET151.
(A) Volcano plots for DMSO vs I-BET151 treated samples, showing the adjusted significance p-value (log10) vs. fold change (log2). (B) Correlation of log2 fold change between MV411/MOLM131 across all genes. Notably no genes show opposing expression changes. Lines represent the identity line (black solid), the line of best fit (black dotted), or log2 fold-change threshold values (green dotted). (C) Heatmap of top 100 genes down-regulated following treatment with IBET151. (D) BCL2 gene-expression (normalized to B2M expression) is shown. Expression level of BCL2 in DMSO was assigned a value of 1. (E) Immunoblotting demonstrating a decrease in BCL2 and an increase in cleaved PARP (*) after IBET151 treatment. (F) ChIP analysis at the TSS 3′-end of BCL2 is illustrated. Bar graphs are represented as the mean enrichment relative to input and error bars reflect standard deviation of results derived from biological triplicate experiments.
BCL2 is a key anti-apoptotic gene implicated in the pathogenesis of MLL-fusion leukaemias16,17. Consistent with these data, I-BET151 reduced the expression of BCL2 in a third MLL-fusion cell line (NOMO1) but not in the un-responsive K562 cells (Fig 3d) and induction of apoptosis coincided with a marked reduction in BCL2 protein expression (Fig. 3e). Moreover, overexpression of BCL2 in the presence of IBET151 rescued the apoptotic phenotype (Supplementary Fig. 14). Chromatin Immuno-Precipitation (ChIP) analyses at the BCL2 locus showed that 6 hours of IBET151 treatment selectively decreased the recruitment of BRD3/4 and impaired recruitment of CDK9 and PAF1 (part of SEC and PAFc respectively) to the transcriptional start site (TSS). This correlated with reduced phosphorylation of POL-II on serine 2 of its carboxy-terminal domain (Pol-IIS2ph) (Fig. 3f). A similar pattern was observed at two other MLL target genes (MYC and CDK6) but not at housekeeping genes (B2M) whose expression was unaltered by I-BET151 (Supplementary Fig. 15). Together, these data suggest that the mechanism of efficacy for I-BET151 involves a selective abrogation of BRD3/4 recruitment to chromatin. The consequence of this is the inefficient phosphorylation/recruitment of Pol II. Further investigation is necessary to distinguish whether Pol-II recruitment and/or elongation is primarily affected by I-BET151
We next sought to establish the therapeutic potential of I-BET151 in vivo. We first characterised the pharmacokinetic properties of I-BET151 in several preclinical species (Supplementary Fig. 20) and also compared it to published inhibitors7,8 (Fig. 4a). We then assessed the efficacy of I-BET151 in two established models of MLL leukaemia. Our first model was a xenotransplant model of disseminated human MLL-AF4 leukaemia18. I-BET151 was delivered daily at 30mg/kg by IP injection from day 2118, and mice were humanely sacrificed if clinical disease dictated or if there was a sequential rise in peripheral blood (PB) disease. At the experimental end-point all the control mice had succumbed to fulminant or progressive disease whereas only 1/5 mice in the treated cohort had evidence of disease at low levels (Fig. 4b-d and Supplementary Figure 16). In our second syngeneic model of murine MLL-AF9 leukaemia, 2.5 × 106 leukaemic cells, established from serial transplantation, were injected into tertiary recipients. Despite the latency being reduced to < 15 days, we waited to initiate treatment from day 9 to test the efficacy of I-BET151 in the setting of overwhelming established disease (Fig. 4e), the scenario often encountered in clinical practice. Even here I-BET151 provided a clear and dramatic survival benefit (Fig. 4e-J and Supplementary Fig. 17). Taken together, these data demonstrate that I-BET151 provides excellent control of MLL leukaemia progression in two distinct and complementary murine models.
Finally, to demonstrate the applicability of our findings to human disease, we tested the efficacy of I-BET151 in leukaemia cells isolated from patients with various MLL-fusions. These data show that I-BET151 accelerates apoptosis (Fig. 4k and Supplementary Fig. 18), and abrogates clonogenic efficiency in bulk leukaemia (Supplementary Fig. 19) as well as isolated LSC (Fig. 4l). These effects are driven, at least in part, by downregulation of a similar transcription programme identified in MLL-fusion cell lines (Fig. 4m). Taken together these data provide compelling evidence of therapeutic potential and suggest that disease eradication is possible.
The paradigm for epigenetic drug discovery shown here highlights an emerging role for targeting aberrant transcriptional elongation in oncogenesis2-5 and provides the first example in epigenetic therapy where mechanistic insights have driven targeted drug discovery and application (Fig. 4n). Together, our results suggest that perturbing the interaction of BET proteins with chromatin using I-BET151, may be of great therapeutic value in human MLL fusion leukaemias. Using a complementary strategy and a different BET inhibitor, a separate study published in this issue concurs with this view19. Moreover, the extensive proteomic resource provided here, has identified other important disease-associated proteins binding to BET proteins, such as MMSET (WHSC1), which is implicated multiple myeloma20. This raises the possibility that BET inhibitors may have an even wider therapeutic scope in oncology and perhaps in other areas of unmet need within the clinical arena.
Methods Summary
Cell culture, gene expression, chromatin immunoprecipitation and FACS analysis were performed as previously described21. Proteomic profiling, and characterisation of inhibitor specificity, was performed using methodology previously described7,9,10. Detailed information about the reagents and methodology used in this study is available in full methods summary (supplementary information).
Supplementary Material
Acknowledgments
We thank SJ. Dawson, A. Bannister, S. Anand and all members of the Huntly and Kouzarides labs. We are grateful to Prof. H. Doehner, the NCRI AML trials biobank and A. Giles for the provision of patient samples. We acknowledge Prof. D. Huang for the BCL2 expression plasmid, L. Gordon for supplying FRET data and R. Woodward, C. Delves, E. Jones and P. Holmes for protein production. J. Witherington, N. Parr, S. Baddeley and J. Seal provided compound selectivity data. N. Deeks and L. Cutler for providing sample and pk data analysis. K. Smitheman and A. Wyce for help with the cellular analysis of the BET inhibitors. Dr P. Grandi for suggestions and discussion. S. Chan for biophysical assay data and members of the Epinova team for discussion and suggestions. We thank staff at the ESRF at Grenoble for beamline assistance. We thank T. Werner for assistance with Mass Spectrometry experiments and data analysis, and the members of the Cellzome Biochemistry, Mass Spectrometry, and IT teams for outstanding expertise and diligence. This work was supported by a Wellcome-Beit Intermediate Clinical Fellowship to M.A.D. The Huntly lab is funded by the Medical Research Council (UK), Leukaemia Lymphoma Research (UK), the Wellcome Trust, The Leukemia & Lymphoma Society of America, Cancer Research UK (CRUK) and the NIHR Cambridge Biomedical Research Centre. This work in the Kouzarides laboratory was funded by a program grant from Cancer Research UK (CRUK).
Footnotes
Author Contributions:
MAD, RKP, AD, GD, KL, PJ, BJPH and TK designed the research, interpreted data and wrote the manuscript. MAD, AD, GG, MB, WC, SR, CC, CH, MS, CH, EG, DL, SB, PES, KRA, and OM performed experiments and analysed data. All compound requests should be directed to KL. MAD, RKP and AD are joint first authors. MAD, BJPH and TK are equal corresponding authors.
References
- 1.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 2.Lin C, et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell. 2010;17:198–212. doi: 10.1016/j.ccr.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Milne TA, et al. Multiple interactions recruit MLL1 and MLL1 fusion proteins to the HOXA9 locus in leukemogenesis. Mol Cell. 2010;38:853–863. doi: 10.1016/j.molcel.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muntean AG, et al. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell. 2010;17:609–621. doi: 10.1016/j.ccr.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 7.Nicodeme E, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung CW, et al. Discovery and Characterization of Small Molecule Inhibitors of the BET Family Bromodomains. J Med Chem. 2011;54:3827–3838. doi: 10.1021/jm200108t. [DOI] [PubMed] [Google Scholar]
- 10.Bantscheff M, et al. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat Biotechnol. 2011;29:255–265. doi: 10.1038/nbt.1759. [DOI] [PubMed] [Google Scholar]
- 11.Jang MK, et al. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 12.Maruyama T, et al. A Mammalian bromodomain protein, brd4, interacts with replication factor C and inhibits progression to S phase. Mol Cell Biol. 2002;22:6509–6520. doi: 10.1128/MCB.22.18.6509-6520.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Z, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 14.Somervaille TC, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4:129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, et al. Conditional MLL-CBP targets GMP and models therapy-related myeloproliferative disease. EMBO J. 2005;24:368–381. doi: 10.1038/sj.emboj.7600521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson BW, et al. Abundant anti-apoptotic BCL-2 is a molecular target in leukaemias with t(4;11) translocation. Br J Haematol. 2008;141:827–839. doi: 10.1111/j.1365-2141.2008.07100.x. [DOI] [PubMed] [Google Scholar]
- 17.Wang QF, et al. MLL fusion proteins preferentially regulate a subset of wild type MLL target genes in the leukemic genome. Blood. 2011 doi: 10.1182/blood-2010-12-324699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Farrell AM, et al. SU11248 is a novel FLT3 tyrosine kinase inhibitor with potent activity in vitro and in vivo. Blood. 2003;101:3597–3605. doi: 10.1182/blood-2002-07-2307. [DOI] [PubMed] [Google Scholar]
- 19.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011 doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez-Garcia E, et al. The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood. 2011;117:211–220. doi: 10.1182/blood-2010-07-298349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dawson MA, et al. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461:819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.