Abstract
Group I introns are a distinct class of RNA self-splicing introns with an ancient origin. All known group I introns present in eukaryote nuclei interrupt functional ribosomal RNA genes located in ribosomal DNA loci. The discovery of the Tetrahymena intron more than 30 years ago has been essential to our understanding of group I intron catalysis, higher-order RNA structure, and RNA folding, but other intron models have provided information about the biological role. Nuclear group I introns appear widespread among eukaryotic microorganisms, and the plasmodial slime molds (myxomycetes) contain an abundance of self-splicing introns. Here, we summarize the main conclusions from previous work on the Tetrahymena intron on RNA self-splicing catalysis as well as more recent work on myxomycete intron biology. Group I introns in myxomycetes that represent different evolutionary stages, biological roles, and functional settings are discussed.
Keywords: Catalytic introns, Diderma, Didymium, Group I introns, Intron biology, Intron mobility, Physarum, RNA processing, Tetrahymena
Review
Introduction
Introns are genetic elements that interrupt functional RNA- or protein-coding genes, and are removed post-transcriptionally in a process termed splicing. Their ability to be spliced out at RNA level makes them almost invisible for the host and limits the phenotypic cost, and introns have often been labeled selfish elements or molecular parasites [1]. A major class of introns is represented by the self-splicing group I introns. These introns are widespread but sporadically distributed in nature, and they are present in the genomes of some bacteria, mitochondria, chloroplasts, bacteriophages, and eukaryotic viruses, and in the nuclei of eukaryotic microorganisms [2].
Group I introns in nuclear genomes are exclusively found within functional ribosomal RNA (rRNA) genes of a wide spectrum of eukaryotic microorganisms. Here they are frequently noted among red algae, chlorophyte algae, fungi, and myxomycetes, but only occasionally in ciliates [2-4]. Nuclear group I introns interrupt universally conserved sequences in the small subunit (SSU) and large subunit (LSU) rRNA genes. About 100 ribosomal DNA (rDNA) insertion sites have been noted to contain introns, 50 in the SSU and 50 in the LSU rRNA genes [3,4]. Interestingly, each insertion site appears to harbor at least one distinct family of group I introns with a separate evolutionary history [2,5,6], which probably reflects site-specific intron mobility (see below).
Despite the fact that thousands of nuclear group I introns have been annotated in sequence databases, only a very few have been submitted to molecular analyses and functional characterizations. The Tetrahymena LSU rRNA intron at insertion site L1925 has become the undisputedly most important group I intron model system for the study of RNA catalysis, RNA structure, and RNA folding [7]. Tth.L1925 (see [8] for nuclear group I intron and rDNA insertion site nomenclature) was discovered and reported more than 30 years ago within the extrachromosomal rDNA of the ciliate Tetrahymena thermophila [9]. This Tetrahymena intron has been thoroughly investigated and self-splicing was shown to occur by RNA catalysis based on a two-step transesterification reaction requiring a guanosine cofactor [7].
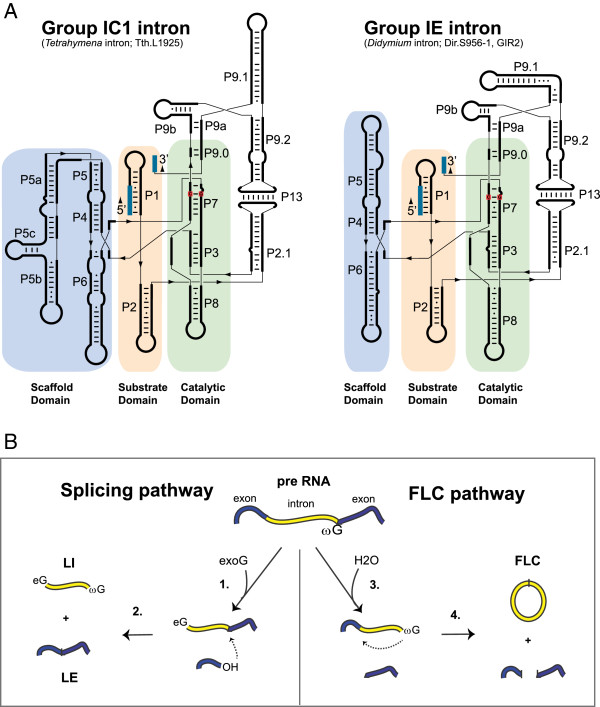
Further structural investigations have revealed a well-defined and highly conserved RNA core responsible for the catalysis, despite the fact that almost no universally conserved nucleotide residues are present among group I introns. Nuclear group I introns are mainly represented by two of the five subgroups, the group IC1 and group IE [6,10], and schematic drawings of secondary structures are shown in Figure 1A. The functional RNA part of the intron, the group I ribozyme core, consists of about nine paired segments (named P1 to P9), as well as one or more optional segments (for example, P10 and P13). These helices are further organized into three helical stacks referred to as the catalytic domain (P3 and P7, proximal P8 and P9), the substrate domain (P1 and proximal P2), and the scaffold domain (P4, P5 and P6) [11]. The domains are easily recognized in both the group IC1 and group IE introns, but with some notable differences. The group IC1 ribozyme, represented by the Tetrahymena intron, has a more complex structured scaffold domain than the Didymium group IE ribozyme (Figure 1A). Crystal structure analysis of the Tetrahymena ribozyme core has revealed a highly compact RNA architecture where the substrate domain is docked into a narrow cleft made by the catalytic domain wrapping around the scaffold domain [7,11,12]. The substrate specificity is in part dependent on a 4 to 6 nucleotide base pairing (P1) between the internal guide sequence and the 5′ exon, and the guanosine binding site (G site) is located in the P7 helix where a conserved G-C pair is the main component (Figure 1A).
Figure 1.
Secondary structures and processing pathways of nuclear group I intron RNAs. (A) Secondary structure diagrams of the group IC1 intron ribozyme in Tetrahymena (Tth.L1925) and the group IE intron ribozyme in Didymium (Dir.S956-1, GIR2). The paired segments (P1 to P9, P13) are noted. The three core domains and the conserved G-C pair at P7 are highlighted. The 5’ and 3’ exons are shown in blue. (B) The two main processing pathways, self-splicing and full-length intron circularization (FLC). The self-splicing pathway involves two transesterification reactions. The first reaction is initiated by a nucleophilic attack by the hydroxyl group of an exogenous guanosine cofactor (exoG) (1). The second transesterification reaction starts with a nucleophilic attack at the 3’ splice site (SS) (2), resulting in ligated exons (LEs) and linear intron (LI) RNA molecules. The FLC pathway involves hydrolysis and transesterification reactions. A hydrolytic cleavage at the 3’ SS (3) is followed by a nucleophilic attack at the 5’ SS by the terminal guanosine (ωG) (4) resulting in a full-length intron circle and non-ligated exons. The FLC pathway is independent of exoG. eG: exogenous guanosine factor covalently linked at the 5’ end of the free intron RNA; exoG: exogenous guanosine cofactor; FLC: full-length circularization; LE: ligated exon; LI; linear intron; SS: splice site.
A small fraction of the nuclear group I introns have the potential of being mobile elements since they harbor large homing endonuclease gene (HEG) insertions. The HEGs are located within the peripheral parts of the paired segments such as P1, P2, P6, P8 or P9, and expression of these protein-coding genes embedded in nucleolar rDNA utilizes unconventional strategies [13]. Interestingly, both sense and antisense HEG organizations relative to the group I ribozymes have been reported [6,14,15].
Several group I intron-based biotechnological applications have been suggested and realized. These involve the intron homing endonuclease (HE) as a highly specific and rare-cutting endonuclease, intron splicing as a therapeutic tool in fighting pathogens and engineered group I ribozymes as molecular tools in RNA reprogramming and RNA repair [16,17]. Ribozymes have features that can be modified and used in several applications involving gene regulation analysis and gene therapy [17,18]. These approaches are based on engineered trans-splice group I ribozymes, and most studies have been performed with the Tetrahymena ribozyme. One of the best characterized examples of RNA-based gene repair is the reprogramming of mutant p53 transcripts in human cancer cells [19,20]. The reprogramming includes an engineered ribozyme, which replaces a defective RNA sequence with a functional p53 homolog [21]. However, the low specificity and low efficiency are important limitations in further development of group I ribozymes in biotechnology [17].
Whereas the Tetrahymena intron has proven to be the undisputed prototype in the study of RNA catalysis and RNA structure, other nuclear group I introns have contributed to our understanding of intron functions beyond splicing. Recent studies have shown that the extrachromosomal nuclear rDNA of myxomycetes, eukaryotic microorganisms belonging to the Amoebozoa clade, contain an abundance of group I introns (Figure 2) [4,5,22]. Currently about 500 nuclear group I introns in myxomycetes have been described, and these introns are amazingly diverse in sequence, structure, organization and insertion sites. About 10% of the myxomycete rDNA introns harbor HEGs, and most group I introns tested are able to self-splice as naked RNA in vitro without any essential assistance from host factors.
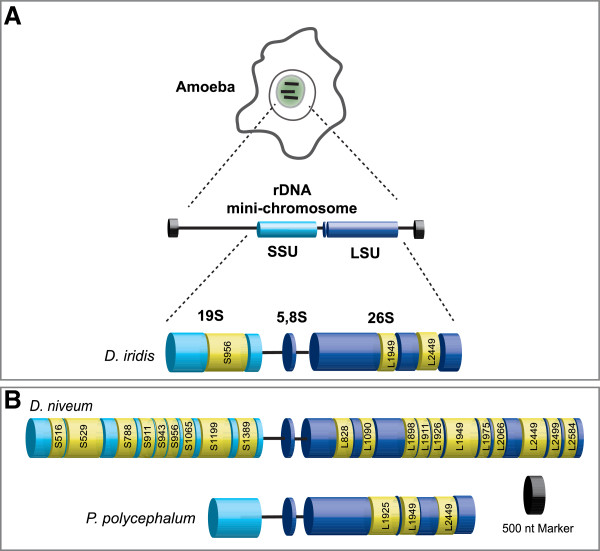
Figure 2.
Group I introns in extrachromosomal nuclear rDNA of myxomycetes. (A) The rDNA mini-chromosomes are located within the nucleolus of myxomycetes. The multicopy mini-chromosomes of Didymium iridis contain the SSU and LSU rRNA genes and have regular telomeres at the ends. The rRNA genes harbor three group I introns (yellow). The S956 intron in the SSU is self-splicing and contains HEG, while the two introns (L1949 and L2449) in the LSU are obligatory introns dependent on the host for splicing. In fact, two versions of D. iridis S956 have been described: the twin-ribozyme intron S956-1 in the Panama 2 isolate and S956-2 in the Costa Rica 8 isolate with antisense HEG orientation. (B) The location of the group I introns within the SSU and LSU are shown for the Diderma niveum Italian isolate and the Physarum polycephalum Carolina isolate. All species contain the obligatory introns L1949 and L2449. The 20 group I introns found in D. niveum have four main categories. P. polycephalum also contains a mobile intron at position L1925 encoding I-PpoI. For nomenclature of rDNA introns and insertion sites, see [8]. HEG: homing endonuclease gene; LSU rRNA: large subunit ribosomal RNA; nt: nucleotide; rDNA: ribosomal DNA; SSU rRNA: small subunit ribosomal RNA.
Three myxomycete species have been investigated in detail (Figure 2). Physarum polycephalum contains the intron (Ppo.L1925), which is cognate to that of the Tetrahymena intron; it harbors a HEG and is mobile in genetic crosses between intron-lacking and intron-containing strains [23,24]. The most complex organized nuclear group I intron known is the twin-ribozyme intron (Dir.S956-1) in Didymium iridis. Dir.S956-1 is mobile in genetic crosses and contains two distinct ribozymes with different functions in splicing and RNA processing, as well as a HEG [25,26]. A second variant of the Didymium intron (Dir.S956-2) harbors a HEG at the antisense orientation [15,27]. Finally, Diderma niveum has an extremely dense intron content with 20 or more group I introns present within the same rRNA primary transcript (Figure 2B) [22,28]. The myxomycetes P. polycephalum, D. iridis and D. niveum have all undergone whole genome sequencing analysis, including their rDNA mini-chromosomes and corresponding introns ([4,29]; our unpublished results).
Here we summarize the major hallmarks of nuclear group I intron catalysis and mobility based on key model introns in Tetrahymena, Physarum, and Didymium rDNAs. We then discuss the functional implications of different categories of introns and provide representative examples from Diderma. Finally, we present an example of a group I intron that recently has gained a new molecular function and biological role.
Group I ribozyme reactions
Group I intron RNAs catalyze transesterification and hydrolysis reactions, and the detailed mechanisms have been extensively reviewed [7,28,30]. These reactions involve two main processing pathways, splicing and full-length intron circularization (Figure 1B), which are parallel and mutually exclusive, and result in different end products [28]. The Tetrahymena intron has been the prototypical ribozyme in the study of the splicing pathway. Self-splicing depends on two consecutive transesterification reactions initiated by a nucleophilic attack of the 3′OH of an exogenous guanosine cofactor (exoG) at the 5′ splice site (SS) (Figure 1B). ExoG is specifically bound to the P7 catalytic core segment of the splicing ribozyme prior to the first splicing step. This reaction leaves exoG covalently attached to the 5′ end of the intron RNA as well as a free 5′ exon with an available 3′OH group. In the second transesterification reaction, exoG is replaced by the terminal guanosine (ωG) at P7, and the reaction is initiated when the 5′ exon attacks the 3′ SS, resulting in ligated exons and the released linear intron. In vitro studies of the Tetrahymena intron have shown that the linear intron RNA may undergo additional circularization reactions leaving a variety of truncated circles [31,32]. However, the biological significance of truncated intron circles, if any, is unclear.
The full-length intron circularization (FLC) pathway has been studied in detail for the Didymium group I intron Dir.S956-1 [33]. This pathway is common among self-splicing nuclear group I introns and is initiated by a hydrolytic cleavage at the 3′ SS [34]. The 3′OH of the ωG then attacks the 5′ SS resulting in a covalently linked full-length circular intron (Figure 1B). Interestingly, the FLC pathway produces fragmented RNA exons, which are unligated and expected to produce non-functional rRNAs.
Group I intron mobility at the DNA or RNA level
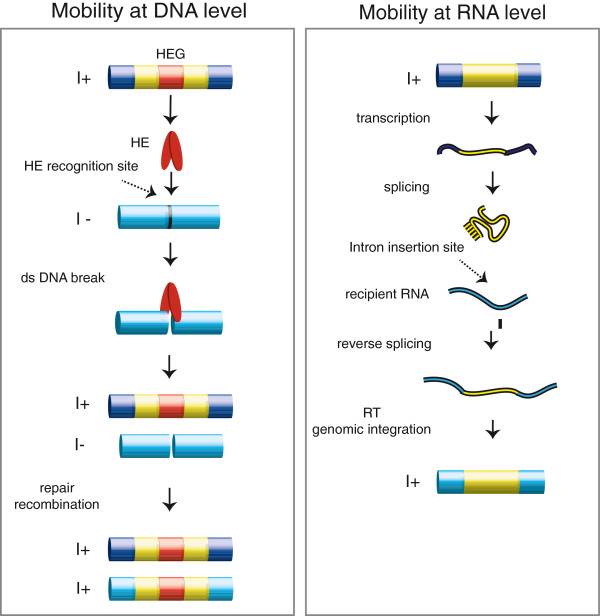
The spreading of group I introns may occur at either the DNA or RNA level (Figure 3). The most efficient mobility process is homing at the DNA level, which is initiated by a double-strand break performed by the intron-encoded HE close to, or at, the site of intron insertion at an intron-lacking allele [35]. HE-mediated group I intron homing involves a homology-dependent gene conversion event and results in the unidirectional spread of group I introns at the population level (Figure 3). Only a few nuclear HEs have been further characterized, and these include I-PpoI from P. polycephalum[36], I-DirI and I-DirII from D. iridis [27], and some isoschizomeric HEs from related Naegleria species [37]. Nuclear HEs all belong to the His-Cys family [38,39] and require specific recognition sequences of 15 to 20 bp spanning the intron insertion site.
Figure 3.
Group I intron mobility. Mobility at the DNA level (left) involves a dsDNA break by the homing endonuclease (HE) at the HE recognition site followed by recombination and repair. HE-dependent mobility is unidirectional and highly efficient. Mobility at the RNA level (right) involves reverse splicing into an intron insertion site in recipient RNA molecules followed by reverse transcription and genomic integration (see text for details). Exon sequences are indicated by blue cylinders (DNA) and lines (RNA), and introns by yellow cylinders (DNA) and lines (RNA). HE: homing endonuclease; HEG: homing endonuclease gene; I+: intron-containing allele; I–: intron-lacking allele; RT: reverse transcriptase; dsDNA: double stranded DNA.
Experimental evidence for homing in a biological setting has been collected from only two nuclear group I introns, both in the myxomycetes. The first system to be characterized was Ppo.L1925 in P. polycephalum, which encodes I-PpoI [23]. Here mobility was shown in mating experiments between intron-containing and intron-lacking amoeba cells. Similarly, homing was also detected in D. iridis for the Dir.S956-1 intron [40]. In addition, homing of nuclear group I introns has also been detected in yeast in artificial experimental settings using I-PpoI and the introns Ppo.L1925 and Tth.L1925 (Tetrahymena intron), which were integrated into all the approximately 150 genomic rDNA copies at chromosome XII in an elegant experiment [41-43].
It has been suggested that intron homing also occurs directly at the RNA level by reverse splicing. Here, an excised intron attacks the ligated exons at the intron-lacking cognate insertion site and integrates into the precursor RNA. Reverse splicing has been reported in vitro, in yeast and in Escherichia coli for both the Tetrahymena intron [44-46] and the Didymium intron [47]. Interestingly, in vitro integration of full-length circular intron RNA has also been noted, suggesting a biological role for the circularization pathway in propagation and intron spread [47]. Less frequently, reverse splicing may lead to intron spread at novel rRNA sites, and may explain the low frequency transposition features of nuclear group I introns observed in phylogenetic studies [5,22,48]. However, experimental evidence of the complete pathway including reverse transcription and genomic integration into rDNA is still lacking.
From parasitism to mutualism: lessons learned from the myxomycete group I introns
Recent studies of the rDNA mini-chromosome in myxomycetes have revealed more than 500 group I introns highly divergent in sequence, size and insertion site. Myxomycetes are eukaryotic microorganisms with a complex life cycle, which has several stages from haploid amoebae cells to a multi-nucleated plasmodium with synchronously dividing diploid nuclei [49]. The rDNA loci are exclusively located on extrachromosomal non-Mendelian mini-chromosomes (Figure 2A). For the myxomycete introns, we summarize results from P. polycephalum, D. iridis and D. niveum rDNAs.
The rDNA from D. niveum is highly unusual since at least 20 group I introns are present (Figure 2B). Recent deep-sequencing experiments of paired-end DNA libraries, performed on the SOLiD platform, have confirmed that all introns are present in all rDNA copies of D. niveum (our unpublished results). The high abundance of introns is a significant challenge to the host cell since the majority of group I introns have the ability to perform FLC and generate fragmented exons [22]. The myxomycete group I introns can be divided into four main categories based on splicing, mobility and pattern of occurrence.
Self-splicing HEG-containing introns
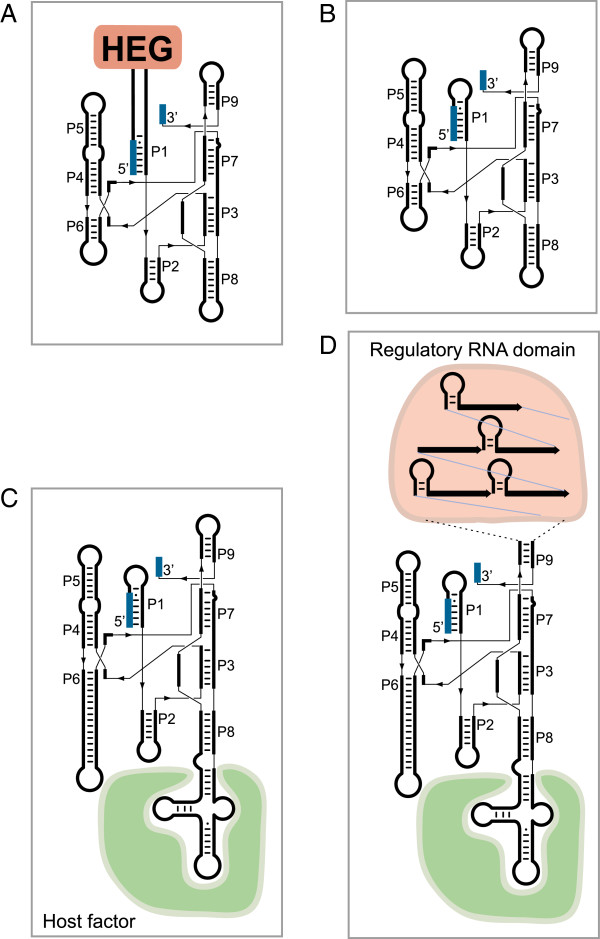
The first category consists of the mobile HEG-containing introns (Figure 4A). Of the representative introns presented in Figure 2, five belong to this category (two different S956 introns in Didymium, L1925 in Physarum, and S529 and S1199 in Diderma). HEGs have been noted in both sense and antisense orientations compared to the group I ribozyme and have been inserted at various locations (P1, P2, P6, P8 and P9). The HEG-containing introns belong to both the IC1 and IE subclass, are highly mobile in biological settings [23,40], and optional among strains and isolates of the same species [27]. Introns in this category are selfish parasitic introns.
Figure 4.
Schematic structural organization of the four main categories of group I introns observed in myxomycete rDNA. (A) Self-splicing and mobile introns with homing endonuclease genes (HEGs). (B) Self-splicing all-ribozyme introns without HEG. (C) Optional host-dependent introns with structural extensions available for host factors (green), here exemplified as associated with P8. (D) Obligatory host-dependent introns with large extension either associated with host factors and/or containing direct repeats (P9) or other motifs with the potential for being regulatory RNA domains (pink). HEG: homing endonuclease gene; rDNA: ribosomal DNA.
Self-splicing all-ribozyme introns
Most introns in Diderma belong to this category (as well as the Tetrahymena intron), and it has been suggested that the all-ribozyme group IC1 and IE introns are remnants of mobile introns after loss of HEGs (Figure 4B). This scenario is founded on the Goddard–Burt cyclic model based on intron invasion, degeneration and subsequent loss [50], which is well supported by several independent reports [51-53]. All-ribozyme introns are usually optional among strains and closely related species. Two of the Diderma introns deviate from the universal consensus features of group I introns, but still self-splice as naked RNA in vitro. S529 is inserted after a G-residue in rDNA (the U-residue is the consensus) and thus the U:G pair at the 5′ SS is replaced by a G:C pair [54]. Furthermore, the ωG in L2066 is replaced by ωA but still performs efficient and complete self-splicing in vitro[22].
Host-factor-dependent optional introns
Group I introns sometimes escape the Goddart–Burt cycle [28] and become dependent on host factors for splicing (Figure 4C). An example of an optional host-dependent intron in Diderma rDNA is S1389 (Figure 2B). This intron is commonly found among the myxomycete family Didymiaceae, but differs from most myxomycete group I introns as it does not self-splice as naked RNA in vitro [55]. One typical feature for the host-dependent splicing introns is extended peripheral loop regions (Figure 4C). In the case of S1389 there are significant size variations in the substrate domain (P1, P2) and the catalytic domain (P8, P9), but the scaffold domain is surprisingly uniform [55]. These structural and functional hallmarks resemble that of mitochondrial fungal group I introns dependent on splicing maturases [56-58]. However, unlike the fungal introns, no specific maturase has yet been identified or characterized for a nuclear group I intron.
Host-factor-dependent obligatory introns
Stable long-term relationships between a group I intron and its host have been noted in plant chloroplast genomes (trnL-intron) [59] and in hexacoral mitochondrial genomes (ND5-717 intron) [53]. A third example is found among nuclear group I introns in Physarales myxomycetes. L1949 and L2449 are present in LSU rDNA of all 60 species and isolates investigated of the Physarales families Didymiaceae and Physaraceae; they have been shown to be strictly vertically inherited [4,5,60-62]. None of the L1949 and L2449 introns tested self-splice in vitro as naked RNA and thus appear to be dependent on host factors for splicing. Further support for this notion is that a large subset of L1949 introns possesses a truncated catalytic core lacking the important P8 segment [5,60,61]. Another unusual feature of L1949 and L2449 introns is large sequence insertions at peripheral loop regions. No detectable protein-coding capacity can be found, but these large insertions sometimes contain complex direct-repeat motifs [4,62] (Figure 4D). The peripheral regions can, by duplications, increase in size over time and result in large introns [4,62].
Obligatory introns could have gained new mutual functions that benefit the host, and one possibility is that peripheral insertions are further processed into long non-coding RNAs (lncRNAs). lncRNAs are known to regulate gene expression, translation, splicing and trafficking by acting as guides, scaffolds, decoys or enhancers [63,64], and are present in all eukaryote systems investigated. Interestingly, recent 454 pyrosequencing analysis on the D. iridis transcriptome revealed stable and differentially expressed L2449 intron RNA in four different life stages of myxomycetes (our unpublished results), which opens the possibility that intron RNA may have gained additional functions beyond splicing.
Group I intron ribozyme that evolved a new biological role
One notable example of group I introns that have evolved new biological roles are the twin-ribozyme introns, which are interrupting SSU rRNAs in the myxomycete Didymium, the amoebo-flagellates Naegleria and the amoeba Allovahlkampfia [26,60,65,66]. Twin-ribozyme introns have a highly complex structural organization that consists of a standard self-splicing ribozyme responsible for SSU rRNA exon ligation, intron excision and the generation of full-length intron RNA circles. Furthermore, the splicing ribozyme contains a large insertion in one of its peripheral helices. A schematic structural diagram of the best studied twin-ribozyme intron, Dir.S956-1 in D. iridis, is presented in Figure 5A. This Didymium intron carries a HEG inserted at P2 of the splicing ribozyme, and is mobile at the DNA level in genetic crosses between intron-containing and intron-lacking strains [26,40].
Figure 5.
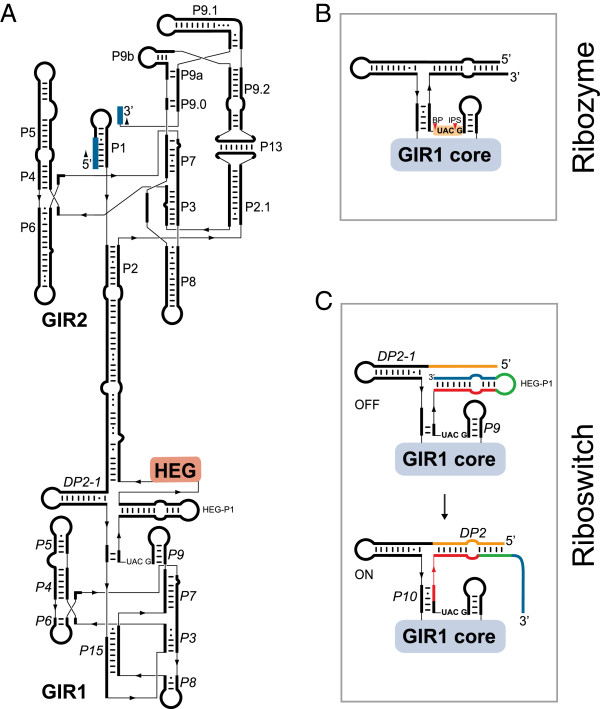
Structure diagrams of different stages of the GIR1 lariat capping ribozyme derived from a complex twin-ribozyme group I intron. (A) Twin-ribozyme intron (Dir.S956-1) from the D. iridis Panama 2 isolate. A standard group IE splicing ribozyme (GIR2; Figure 1B) contains an insertion in helix P2, which consists of a homing endonuclease gene (HEG) and the lariat capping group I-like ribozyme (GIR1). (B) The active GIR1 conformation performs a transesterification reaction at the junction between P9 and P10 resulting in a 3-nucleotide 2’,5’ lariat structure at the 5’ end of the HE messenger. (C) The regulatory domain of GIR1 resembles a complex riboswitch, which alternates between a catalytic inactive GIR1 (containing HEG-P1; the off state) and an active GIR1 conformation (containing DP2 and P10; the on state). This rearrangement involves replacement of RNA structures (color coded). BP: branch point; GIR1: group I-like ribozyme; GIR2: group I splicing ribozyme; HE: homing endonuclease; HEG: homing endonuclease gene; IPS: internal processing site.
Interestingly, a second ribozyme domain is located immediately upstream of the HEG. Detailed structural analysis has shown that the ribozyme is derived from a standard group I intron; it is named GIR1 (group I-like ribozyme 1) [67]. GIR1 lacks the 5′ and 3′ SSs, and has unique structural rearrangements in the catalytic core. GIR1 has an essential role in the expression of the intron HEG, which is transcribed by RNA polymerase I and embedded in the rDNA [14]. Recent reports have suggested dual functions for GIR1 in HEG expression: as a capping ribozyme [68] and as a riboswitch regulator [69,70].
Lariat capping ribozyme
The catalytic part of GIR1 has about 180 to 200 nucleotides, and when activated it catalyzes self-cleaving by branching [71]. This reaction is highly unusual for the group I ribozymes, but similar to that of group II ribozymes and the spliceosome [68]. GIR1 generates a 3-nucleotide lariat cap by joining the C residue at the internal processing site (IPS) and the U residue at the branch point (BP) by a 2′,5′ phosphodiester bond (Figure 5B) [68]. Similar lariat caps are generated by the Naegleria and Allovahlkampfia GIR1s [66,72].
On-off riboswitch
During the transcription and self-splicing of a twin-ribozyme intron, GIR1 has to be in an inactive conformation to avoid premature cleavage of the rRNA precursor [69]. Thus, the regulatory domain of GIR1 folds into an inactive off-state confirmation, which involves the HEG-P1 helix 3' of the BP (Figure 5C). The excised intron RNA then activates GIR1 into an on state by a conformational change in the regulatory domain [69]. HEG-P1 is replaced by two additional helical segments, DP2 and P10, which depend on base pairing of sequences 5′ and 3′ of the GIR1 core (Figure 5C). Related conformational changes, but differently organized, occur in the Naegleria and Allovahlkampfia GIR1s [66]. The on-off switch of GIR1 resembles that of many riboswitches [73], but currently no specific ligand has been identified.
Conclusions
Self-splicing, RNA structure and folding, and HE-dependent homing are fully described features of the group I introns in Tetrahymena and Physarum rDNA, but these studies represent only part of the story for nuclear group I introns. Additional studies have shown that there exist two main catalytic pathways for intron RNA: the intron splicing pathway and the intron FLC pathway. Intron homing is also represented by two distinct mechanisms: HE-dependent homing and the less efficient reverse-splicing-dependent homing. The latter mechanism sometimes results in intron insertion at non-allelic sites. The next important challenge is to understand the biological role of nuclear group I introns, and a first step has been achieved for the myxomycete protists, which appear to contain an abundance of diverse catalytic rDNA introns. Four main intron categories have been identified, from the true selfish HEG-containing and mobile group I introns, to introns that have become biochemically dependent on the host cell for splicing. Some introns appear obligatory for the host, and intron RNAs may evolve further to gain more regulatory functions. Finally, the lariat capping ribozyme (GIR1) is a unique example of a group I intron that has gained new catalytic properties and new biological roles in nuclear gene regulation.
Abbreviations
BP: branch point; dsDNA: double stranded DNA; eG: exogenous guanosine factor covalently linked at the 5’ end of the free intron RNA; exoG: exogenous guanosine cofactor; FLC: full-length circularization; GIR1: group I-like ribozyme; GIR2: group I splicing ribozyme; HE: homing endonuclease; HEG: homing endonuclease gene; I+: intron-containing allele; I–: intron-lacking allele; IPS: internal processing site; LE: ligated exon; LI: linear intron; LncRNA: long non-coding RNA; LSU rRNA: large subunit ribosomal RNA; Nt: nucleotide; RDNA: ribosomal DNA; RT: reverse transcriptase; SOLiD: sequencing by oligo ligation and detection; SS: splice site; SSU rRNA: small subunit ribosomal RNA.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AH and SDJ contributed equally to the writing for this article. Both authors read and approved the final manuscript.
Contributor Information
Annica Hedberg, Email: Annica.Hedberg@uit.no.
Steinar D Johansen, Email: Steinar.Johansen@uit.no.
Acknowledgments
We thank members of the RNA Research Unit for discussions and analysis, and especially Kari Haugli for myxomycete sampling and sequencing.
References
- Edgell DR, Chalamcharla VR, Belfort M. Learning to live together: mutualism between self-splicing introns and their hosts. BMC Biol. 2011;9:22. doi: 10.1186/1741-7007-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugen P, Simon DM, Bhattacharya D. The natural history of group I introns. Trends Genet. 2005;21:111–119. doi: 10.1016/j.tig.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D’Souza LM, Du Y. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinform. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandipati SC, Haugli K, Coucheron DH, Haskins EF, Johansen SD. Polyphyletic origin of the genus Physarum (Physarales, Myxomycetes) revealed by nuclear rDNA mini-chromosome analysis and group I intron synapomorphy. BMC Evol Biol. 2012;12:166. doi: 10.1186/1471-2148-12-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad EW, Einvik C, Rønning S, Haugli K, Johansen S. Twelve group I introns in the same pre-rRNA transcript of the myxomycete Fuligo septica: RNA processing and evolution. Mol Biol Evol. 2004;21:1283–1293. doi: 10.1093/molbev/msh126. [DOI] [PubMed] [Google Scholar]
- Haugen P, Reeb V, Lutzoni F, Bhattacharya D. The evolution of homing endonuclease genes and group I introns in nuclear rDNA. Mol Biol Evol. 2004;21:129–140. doi: 10.1093/molbev/msh005. [DOI] [PubMed] [Google Scholar]
- Golden BL. In: Ribozymes and RNA catalysis. Lilley DMJ, Eckstein F, editor. Cambridge: RSC Press; 2008. Group I introns: biochemical and crystallographic characterization of the active site structure; pp. 178–200. [Google Scholar]
- Johansen S, Haugen P. A new nomenclature of group I introns in ribosomal DNA. RNA. 2001;7:935–936. doi: 10.1017/S1355838201010500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Lu C, Wu Q-J, Wang Y, Sun Z-T, Deng J-C. GISSD: group I intron sequence and structure database. Nucleic Acids Res. 2008;36:31–37. doi: 10.1093/nar/gkn052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicens Q, Cech TR. Atomic level architecture of group I introns revealed. Trends Biochem Sci. 2006;31:41–51. doi: 10.1016/j.tibs.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Guo F, Gooding AR, Cech TR. Structure of the Tetrahymena ribozyme: base triple sandwich and metal ion at the active site. Mol Cell. 2004;16:351–362. doi: 10.1016/j.molcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Johansen SD, Haugen P, Nielsen H. Expression of protein-coding genes embedded in ribosomal DNA. Biol Chem. 2007;388:679–686. doi: 10.1515/BC.2007.089. [DOI] [PubMed] [Google Scholar]
- Vader A, Nielsen H, Johansen S. In vivo expression of the nucleolar group I intron-encoded I-DirI homing endonuclease involves the removal of a spliceosomal intron. EMBO J. 1999;18:1003–1013. doi: 10.1093/emboj/18.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen SD, Vader A, Sjøttem E, Nielsen H. In vivo expression of a group I intron HEG from the antisense strand of Didymium ribosomal DNA. RNA Biol. 2006;3:157–162. doi: 10.4161/rna.3.4.3958. [DOI] [PubMed] [Google Scholar]
- Johansen S, Einvik C, Elde M, Haugen P, Vader A, Haugli F. Group I introns in biotechnology: prospects of application of ribozymes and rare-cutting homing endonucleases. Biotechnol Ann Rev. 1997;3:111–150. [Google Scholar]
- Fiskaa T, Birgisdottir ÁB. RNA reprogramming and repair based on trans-splicing group I ribozymes. N Biotechnol. 2010;27:194–203. doi: 10.1016/j.nbt.2010.02.013. [DOI] [PubMed] [Google Scholar]
- Akashi H, Matsumoto S, Taira K. Gene discovery by ribozyme and siRNA libraries. Nat Rev Mol Cell Biol. 2005;6:413–422. doi: 10.1038/nrm1646. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Sullenger BA. Induction of wild-type p53 activity in human cancer cells by ribozymes that repair mutant p42 transcripts. Proc Natl Acad Sci USA. 2000;97:8490–8494. doi: 10.1073/pnas.150104097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin KS, Sullenger BA, Lee SW. Ribozyme-mediated induction of apoptosis in human cancer cells by targeted repair of mutant p53 RNA. Mol Ther. 2004;10:365–372. doi: 10.1016/j.ymthe.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Sullenger BA, Cech TR. Ribozyme-mediated repair of defective mRNA by targeted trans-splicing. Nature. 1994;371:619–622. doi: 10.1038/371619a0. [DOI] [PubMed] [Google Scholar]
- Andreassen M. The role of peripheral RNA structure domains in myxomycete group I ribozyme function. University of Tromsø; 2007. (PhD thesis). [Google Scholar]
- Muscarella DE, Vogt VM. A mobile group I intron in the nuclear rDNA of Physarum polycephalum. Cell. 1989;56:443–454. doi: 10.1016/0092-8674(89)90247-X. [DOI] [PubMed] [Google Scholar]
- Ruoff B, Johansen S, Vogt VM. Characterization of the self-splicing products of a mobile intron from the nuclear rDNA of Physarum polycephalum. Nucleic Acids Res. 1992;20:5899–5906. doi: 10.1093/nar/20.22.5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen S, Vogt VM. An intron in the nuclear ribosomal DNA of Didymium iridis codes for a group I ribozyme and a novel ribozyme that cooperate in self-splicing. Cell. 1994;76:725–734. doi: 10.1016/0092-8674(94)90511-8. [DOI] [PubMed] [Google Scholar]
- Decatur WA, Einvik C, Johansen S, Vogt VM. Two group I ribozymes with different functions in a nuclear rDNA intron. EMBO J. 1995;14:4558–4568. doi: 10.1002/j.1460-2075.1995.tb00135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugen P, Wikmark OG, Vader A, Coucheron D, Sjøttem E, Johansen SD. The recent transfer of a homing endonuclease gene. Nucleic Acids Res. 2005;33:2734–2741. doi: 10.1093/nar/gki564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Johansen SD. Group I introns: moving in new directions. RNA Biol. 2009;6:375–383. doi: 10.4161/rna.6.4.9334. [DOI] [PubMed] [Google Scholar]
- Glöckner G, Noegel AA. Comparative genomics in the Amoebozoa clade. Biol Rev. 2013;88:215–225. doi: 10.1111/j.1469-185X.2012.00248.x. [DOI] [PubMed] [Google Scholar]
- Hougland JL, Piccirilli JA, Forconi M, Lee J, Herschlag D. In: The RNA World. 3. Gesteland RF, Cech TR, Atkins JF, editor. New York: CSHL Press; 2006. How the group I intron works: a case study of RNA structure and function; pp. 133–205. [Google Scholar]
- Zaug AJ, Grabowski PJ, Cech TR. Autocatalytic cyclization of an excised intervening sequence RNA is a cleavage-ligation reaction. Nature. 1983;301:578–583. doi: 10.1038/301578a0. [DOI] [PubMed] [Google Scholar]
- Been MD, Cech TR. Selection of circularization sites in a group I IVS RNA requires multiple alignments of an internal template-like sequence. Cell. 1987;50:951–961. doi: 10.1016/0092-8674(87)90522-8. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Fiskaa T, Birgisdottir ÁB, Haugen P, Einvik C, Johansen S. The ability to form full-length intron RNA circles is a general property of nuclear group I introns. RNA. 2003;9:1464–1475. doi: 10.1261/rna.5290903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugen P, Andreassen M, Birgisdottir ÁB, Johansen S. Hydrolytic cleavage by a group I intron ribozyme is dependent on RNA structures not important for splicing. Eur J Biochem. 2004;271:1015–1024. doi: 10.1111/j.1432-1033.2004.04003.x. [DOI] [PubMed] [Google Scholar]
- Belfort M, Roberts RJ. Homing endonucleases: keeping the house in order. Nucleic Acids Res. 1997;25:3379–3388. doi: 10.1093/nar/25.17.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muscarella DE, Ellison EL, Ruoff BM, Vogt VM. Characterization of I-Ppo, an intron-encoded endonuclease that mediates homing of a group I intron in the ribosomal DNA of Physarum polycephalum. Mol Cell Biol. 1990;10:3386–3396. doi: 10.1128/mcb.10.7.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elde M, Willassen NP, Johansen S. Functional characterization of isoschizomeric His-Cys box homing endonucleases from Naegleria. Eur J Biochem. 2000;267:7257–7266. doi: 10.1046/j.1432-1327.2000.01862.x. [DOI] [PubMed] [Google Scholar]
- Johansen S, Embley TM, Willassen NP. A family of nuclear homing endonucleases. Nucleic Acids Res. 1993;21:4405. doi: 10.1093/nar/21.18.4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafez M, Hausner G. Homing endonucleases: DNA scissors on a mission. Genome. 2012;55:553–569. doi: 10.1139/g2012-049. [DOI] [PubMed] [Google Scholar]
- Johansen S, Elde M, Vader A, Haugen P, Haugli K, Haugli F. In vivo mobility of a group I twintron in nuclear ribosomal DNA of the myxomycete Didymium iridis. Mol Microbiol. 1997;24:737–745. doi: 10.1046/j.1365-2958.1997.3921743.x. [DOI] [PubMed] [Google Scholar]
- Muscarella DE, Vogt VM. A mobile group I intron from Physarum polycephalum can insert itself and induce point mutations in the nuclear ribosomal DNA of Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:1023–1033. doi: 10.1128/mcb.13.2.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Vogt VM. I-PpoI, the endonuclease encoded by the group I intron PpLSU3, is expressed from an RNA polymerase I transcript. Mol Cell Biol. 1998;18:5808–5817. doi: 10.1128/mcb.18.10.5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Vogt VM. Functional alpha-fragment of beta-galactosidase can be expressed from the mobile group I intron PpLSU3 embedded in yeast pre-ribosomal RNA derived from the chromosomal rDNA locus. Nucleic Acids Res. 2000;28:1428–1438. doi: 10.1093/nar/28.6.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman J, Woodson SA. Integration of the Tetrahymena group I intron into bacterial rRNA by reverse splicing in vivo. Proc Natl Acad Sci USA. 1998;95:2134–2139. doi: 10.1073/pnas.95.5.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman J, Woodson SA. Reverse splicing of the Tetrahymena IVS: evidence for multiple reaction sites in the 23S rRNA. RNA. 1995;1:478–490. [PMC free article] [PubMed] [Google Scholar]
- Roman J, Rubin MN, Woodson SA. Sequence specificity of in vitro reverse splicing of the Tetrahymena group I intron. RNA. 1999;5:1–13. doi: 10.1017/S1355838299981244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birgisdottir ÁB, Johansen S. Site-specific reverse splicing of a HEG-containing group I intron in ribosomal RNA. Nucleic Acids Res. 2005;33:2042–2051. doi: 10.1093/nar/gki341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D, Friedl T, Helms G. Vertical evolution and intragenic spread of lichen-fungal group I introns. J Mol Evol. 2002;55:74–84. doi: 10.1007/s00239-001-2305-x. [DOI] [PubMed] [Google Scholar]
- Burland TG, Solnica-Krezel L, Bailey J, Cunningham DB, Dove WF. Patterns of inheritance, development and the mitotic cycle in the protist Physarum polycephalum. Adv Microbiol Phys. 1993;35:1–69. doi: 10.1016/s0065-2911(08)60096-x. [DOI] [PubMed] [Google Scholar]
- Goddard MR, Burt A. Recurrent invasion and extinction of a selfish gene. Proc Natl Acad Sci USA. 1999;96:13880–13885. doi: 10.1073/pnas.96.24.13880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard MR, Leigh J, Roger AJ, Pemberton AJ. Invasion and persistence of a selfish gene in the Cnidaria. PLoS One. 2006;1:e3. doi: 10.1371/journal.pone.0000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikmark OG, Einvik C, De Jonckheere JF, Johansen SD. Short-term sequence evolution and vertical inheritance of the Naegleria twin-ribozyme group I intron. BMC Evol Biol. 2006;6:39. doi: 10.1186/1471-2148-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emblem Å, Karlsen BO, Evertsen J, Johansen SD. Mitogenome rearrangement in the cold-water scleractinian coral Lophelia pertusa (Cnidaria, Anthozoa) involves a long-term evolving group I intron. Mol Phylogenet Evol. 2011;61:495–503. doi: 10.1016/j.ympev.2011.07.012. [DOI] [PubMed] [Google Scholar]
- Birgisdottir ÁB. A functional study of catalytic RNAs from mobile group I introns. University of Tromsø; 2005. (PhD thesis). [Google Scholar]
- Wikmark OG, Haugen P, Lundblad EW, Haugli K, Johansen SD. The molecular evolution and structural organization of group I introns at position 1389 in nuclear small subunit rDNA of myxomycetes. J Euk Microbiol. 2007;54:49–56. doi: 10.1111/j.1550-7408.2006.00145.x. [DOI] [PubMed] [Google Scholar]
- Paukstelis PJ, Lambowitz AM. Identification and evolution of fungal mitochondrial tyrosyl-tRNA synthetases with group I intron splicing activity. Proc Natl Acad Sci USA. 2008;105:6010–6015. doi: 10.1073/pnas.0801722105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicens Q, Paukstelis PJ, Westhof E, Lambowitz AM, Cech TR. Toward predicting self-splicing and protein-facilitated splicing of group I introns. RNA. 2008;14:2013–2029. doi: 10.1261/rna.1027208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan CDS, Weeks KM. The Mrs1 splicing factor binds the bI3 group I intron at each of two tetraloop-receptor motifs. PLoS ONE. 2010;5:e8983. doi: 10.1371/journal.pone.0008983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhsel MG, Strickland R, Palmer JD. An ancient group I intron shared by eubacteria and chloroplasts. Science. 1990;250:1570–1573. doi: 10.1126/science.2125748. [DOI] [PubMed] [Google Scholar]
- Johansen S, Johansen T, Haugli F. Structure and evolution of myxomycete nuclear group I introns: a model for horizontal transfer by intron homing. Curr Genet. 1992;22:297–304. doi: 10.1007/BF00317925. [DOI] [PubMed] [Google Scholar]
- Vader A, Naess J, Haugli K, Haugli F, Johansen S. Nucleolar introns from Physarum flavicomum contain insertion elements that may explain how mobile group I introns gained their open reading frames. Nucleic Acids Res. 1994;22:4553–4559. doi: 10.1093/nar/22.22.4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikmark OG, Haugen P, Haugli K, Johansen SD. Obligatory group I introns with unusual features at positions 1949 and 2449 in nuclear LSU rDNA of Didymiaceae myxomycetes. Mol Phylogenet Evol. 2007;43:596–604. doi: 10.1016/j.ympev.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Ann Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einvik C, Decatur WA, Embley TM, Vogt VM, Johansen S. Naegleria nucleolar introns contain two group I ribozymes with different functions in RNA splicing and processing. RNA. 1997;3:710–720. [PMC free article] [PubMed] [Google Scholar]
- Tang Y. Molecular characteristics of the Naegleria GIR1 branching ribozyme. University of Tromsø; 2011. (PhD thesis). [Google Scholar]
- Beckert B, Nielsen H, Einvik C, Johansen SD, Westhof E, Masquida B. Molecular modelling of the GIR1 branching ribozyme gives new insight into evolution of structurally related ribozymes. EMBO J. 2008;27:667–678. doi: 10.1038/emboj.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Westhof E, Johansen S. An mRNA is capped by a 2′, 5′ lariat catalyzed by a group I-like ribozyme. Science. 2005;309:1584–1587. doi: 10.1126/science.1113645. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Einvik C, Lentz TE, Hedegaard MM, Johansen SD. A conformational switch in the DiGIR1 ribozyme involved in release and folding of the downstream I-DirI mRNA. RNA. 2009;15:958–967. doi: 10.1261/rna.669209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birgisdottir ÁB, Nielsen H, Beckert B, Masquida B, Johansen SD. Intermolecular interaction between a branching ribozyme and associated homing endonuclease mRNA. Biol Chem. 2011;392:491–499. doi: 10.1515/BC.2011.055. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Beckert B, Masquida B, Johansen SD. In: Ribozymes and RNA catalysis. Lilley DMJ, Eckstein F, editor. Cambridge: RSC Press; 2008. The GIR1 branching ribozyme; pp. 229–249. [Google Scholar]
- Tang Y, Nielsen H, Birgisdottir ÁB, Johansen SD. A natural fast-cleaving branching ribozyme from the amoeboflagellate Naegleria pringsheimi. RNA Biol. 2011;8:997–1004. doi: 10.4161/rna.8.6.16027. [DOI] [PubMed] [Google Scholar]
- Serganov A, Patel DJ. Molecular recognition and function of riboswitches. Curr Opin Struct Biol. 2012;22:279–286. doi: 10.1016/j.sbi.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]