Abstract
Recently, a novel mec gene conferring beta-lactam resistance in Staphylococcus aureus has been discovered. This gene, mecC, is situated on a SCCmec XI element that has to date been identified in clonal complexes 49, 130, 425, 599 and 1943. Some of the currently known isolates have been identified from animals. This, and observations of mecA alleles that do not confer beta-lactam resistance, indicate that mec genes might have a reservoir in Staphylococcus species from animals. Thus it is important also to screen wildlife isolates for mec genes. Here, we describe mecC-positive Staphylococcus aureus (ST130-MRSA-XI) and the lesions related to the infection in two diseased free-ranging European hedgehogs (Erinaceus europaeus). One was found dead in 2003 in central Sweden, and suffered from S. aureus septicaemia. The other one, found on the island of Gotland in the Baltic Sea in 2011, showed a severe dermatitis and was euthanised. ST130-MRSA-XI isolates were isolated from lesions from both hedgehogs and were essentially identical to previously described isolates from humans. Both isolates carried the complete SCCmec XI element. They lacked the lukF-PV/lukS-PV and lukM/lukF-P83 genes, but harboured a gene for an exfoliative toxin homologue previously described from Staphylococcus hyicus, Staphylococcus pseudintermedius and other S. aureus of the CC130 lineage. To the best of our knowledge, these are the first reported cases of CC130-MRSA-XI in hedgehogs. Given that one of the samples was taken as early as 2003, this was the earliest detection of this strain and of mecC in Sweden. This and several other recent observations suggest that CC130 might be a zoonotic lineage of S. aureus and that SCCmec XI/mecC may have originated from animal pathogens.
Introduction
Methicillin-resistant S. aureus (MRSA) has been known for just over 50 years, and it poses a serious problem for infection prevention and control and antibiotic treatment globally. In MRSA, resistance against almost all beta-lactam compounds in clinical use is caused by the expression of an alternate penicillin-binding protein (PBP2a) that is encoded by the mecA gene. It belongs to a family of genes that can be found in various staphylococci [1], 2. Staphylococcus sciuri and Staphylococcus vitulinus harbour mecA alleles that are not associated with beta-lactam resistance [2], [3], [4]. The alleles of mecA that are associated with resistance are situated on mobile genetic elements termed Staphylococcal Cassette Chromosome mec (SCCmec) elements [5] and can be found in S. aureus as well as in other staphylococcal species such as Staphylococcus epidermidis and Staphylococcus haemolyticus. Currently, ten different types of SCC elements harbouring mecA as well as a number of subtypes are known from S. aureus (see also http://www.sccmec.org/Pages/SCC_TypesEN.html).
A novel mec gene type was discovered in 2011 [6], [7]. It is localised on a novel SCCmec element designated as type XI [7]. Because of its highly divergent sequence, it cannot be detected by routinely used molecular assays designed to identify mecA [7], [8]. This gene, recently renamed mecC [1], has been found S. aureus belonging to clonal complexes (CCs, as defined by multilocus sequence typing) 49, 130, 425, 599 and 1943. They have been isolated from various animals including cattle, sheep, dogs, cats, a harbour seal (Phoca vitulina), a guinea pig, rabbits, rats, and a chaffinch (Fringilla coelebs) as well as from humans from Ireland, England, Scotland, Germany, Denmark, Sweden, Norway, France, Switzerland, Belgium and The Netherlands [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20]. In Sweden, where this study was performed, mecC was previously observed in four out of 537 S. aureus isolates recovered from nearly 9,000 cow milk samples [19]. Recently, mecC was also found in veterinary Staphylococcus xylosus [21].
These observations as well as the observation of mecA alleles in animal staphylococci that are not associated with beta-lactam resistance [2], [3] indicate that mec genes and their precursors might have a reservoir in animal strains of Staphylococcus species. Thus it is important to screen wildlife isolates for mec genes.
The aim of the present study was to characterize mecC-positive Staphylococcus aureus (ST130-MRSA-XI) recovered from two diseased free-ranging European hedgehogs (Erinaceus europaeus) from Sweden.
Materials and Methods
Animal data and post mortem examination
The two hedgehogs were submitted to the Swedish National Veterinary Institute (SVA) as part of the Swedish wildlife health surveillance program. Hedgehog V583/03 was an adult female found dead in a garden in June 2003 in Bankeryd (Jönköping county, Sweden, 57°51′N 14°07′E). Its body weight was 825 g. The carcass was moderately autolytic.
Hedgehog V5406/11 was an adult male with a body weight of 722 g, found alive in October 2011 on the island Gotland in the Baltic sea, off the East coast of Sweden (57°30′N 18°33′E), and taken care of in a wildlife rehabilitation centre. It had diffuse dermatitis, diarrhoea and difficulties moving, in particular in using the front legs. Due to its poor health it was finally euthanised. Three other hedgehogs with similar clinical signs had been observed in the same area and had died but were not submitted for necropsy.
Routine post mortem examination was performed and selected tissue samples were obtained fresh for bacteriological culture and fixed in 10% neutral buffered formalin for histopathology. The fixed samples were processed and embedded in paraffin wax, sectioned at 5 µm and stained with haematoxylin and eosin (H&E). Selected sections were Gram stained for visualisation of bacteria.
S. aureus isolates
Bacteriological culture was conducted on blood agar base (Difco/Becton Dickinson AB, Stockholm, Sweden) supplemented with 5% citrated equine blood and Bromcresol Purple Lactose Agar (SVA, Uppsala, Sweden). The plates were incubated at 37°C under aerobic conditions and examined after 24 h and 48 h. S. aureus was identified by morphological and physiological characteristics. Coagulase-positive S. aureus grew in pure culture from the brain and kidneys samples from hedgehog V583/03. An isolate from a kidney was used for further characterisation. Cultures of skin lesions from hedgehog V5406/11 also yielded S. aureus.
Susceptibility tests
Susceptibility tests were performed using the VITEK 2 automated microbiological identification system with the Gram-positive susceptibility panel AST-580 (bioMérieux, Nuertlingen, Germany). In addition, the Clearview ™ (Alere, Cologne, Germany) lateral flow test for the detection of PBP2a was performed according to the protocol provided by the manufacturer using culture material from a MRSA selective growth medium (chromID MRSA, bioMérieux). Minimum inhibitory concentration (MIC) determinations were performed using E-tests (bioMérieux; for methicillin and penicillin) or MIC Test Strips (bestbion dx, Cologne, Germany; for cefoxitin).
Array procedures
Isolates were characterised using the Alere Technologies StaphyType DNA microarray kit as described previously in detail [22]. The arrays were used to assign isolates to CCs and to identify SCCmec associated genes as well as a range of virulence and resistance determinants.
Furthermore, additional probes and primers were introduced to the Alere Technologies microarray and the corresponding master mix, respectively, to identify mecC and the blaZ allele associated with SCCmec XI [6], [7]. Primer and probe sequences are listed in Table 1.
Table 1. Novel primer and probe sequences and PCR conditions.
| Target | Forward primer | Reverse primer | Hybridisation probe | PCR conditions |
| mecC out frame | 5′-TGTTGTAGCAATGTTCACAC-3′ | 5′-CAAGCACTTAATATCAACGC-3′ | n.a. | 2 min at 95°C/30 cycles (30 sec at 95°C, 30 sec at 55°C, 2 min at 72°C)/5 min at 72°C |
| mecC | 5′-ATTAATTGGACCCACATAACC-3′ | n.a. | 5′-AAAGCCGTGTTTATCCATTGAACGAAG-3′, spotted on the microarray | n.a. |
| mecC | 5′-TATAGTTAAATGAAGATCTTTTCCG-3′ | n.a. | 5′-GGTTTTAAGGTATCCATTGCAAATACTTATGAC-3′, spotted on the microarray | n.a. |
| blaZ-M10/0065 | 5′-CAGCATTGCGCTACTTATAG-3′ | n.a. | 5′-AAGCGTTTTGCATATGCTTCCACATTA-3′, spotted on the microarray | n.a. |
| etD2 | 5′-TCAAGACACCACTAGAAGTC-3′ | 5′-CGTTTTCAGCTAATCGTGC-3′ | n.a. | 2 min at 94°C/35 cycles (30 sec at 94°C, 30 sec at 52°C, 30 sec at 72°C)/5 min at 72°C |
The identity of the mec gene was additionally confirmed using a previously described array that allows the known variants of mec from different staphylococcal species to be differentiated [2].
PCR and sequencing
A PCR for the detection of the SCCmec-associated phenol-soluble modulin, PSM-mec [23], was performed as previously described [24].
PCRs were also carried out to characterise the SCCmec element in detail in both isolates. The complete mecC gene was amplified and sequenced using GoTaq DNA polymerase and primers (Table 1) derived from the SCCmec XI sequence of an Irish CC130-MRSA-XI isolate (GenBank accession number FR823292.1, [7]). This strain, M10/0061, was also used as a positive control. The entire SCCmec XI element including the SCCmec XI-associated ccrA1B3 gene complex was amplified using previously described primers [7]. The sizes of the resulting amplimers obtained were compared to those of M10/0061, from which SCCmec XI has been fully sequenced.
The DNA sequence of a novel exfoliative toxin homologue, putatively named etD2, that was detected as part of the present study in an Irish CC130 genome sequence [7] as well as in two other CC130 genomes (GenBank accession number AEUQ01000009.1, bases 161312 to 162153 and AEUR01000016.1, bases 44379 to 45221 [25]) was submitted to GenBank under accession number HF563069. The presence of this gene in both isolates was investigated by PCR with primers as listed in Table 1. S. aureus reference strains COL (GenBank CP000046) and N315 (GenBank BA000018) were used as negative controls and M10/0061 [7] was used as a positive control. The PCR product had an expected length of ca. 130 bp.
Multilocus sequence typing and spa typing were performed on both isolates using previously described protocols [26], [27].The MLST allele sequences were analysed using the MLST website (http://saureus.mlst.net/). The spa spa types were assigned using SPATYPEMAPPER software (download at http://www.clondiag.com/fileadmin/Media/Downloads/SPATypeMapper_0_6.zip).
Results
Pathology
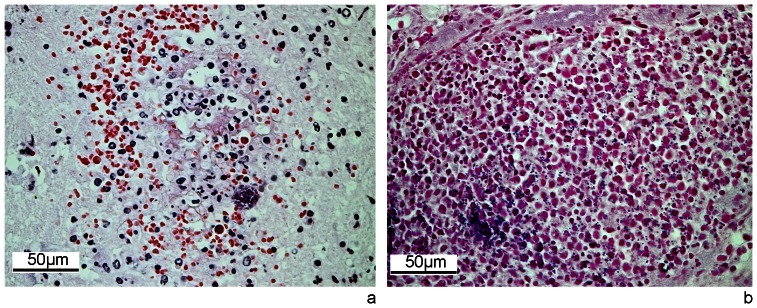
Hedgehog V583/03 was in poor physical condition, with depleted fat deposits and pregnant with nine embryonic vesicles. Gross changes included abundant red-brown liquid content in the stomach and intestines, enlarged spleen, pulmonary oedema and hyperaemia and severe hyperaemia of the meninges. Histologically, multiple foci of necrosis and inflammatory infiltration of neutrophils, macrophages and lymphocytes were observed in the cortex and medulla of the kidneys, in the spleen, lungs, meninges and brain parenchyma. In these foci there were numerous Gram-positive cocci, often forming aggregates (Figure 1a). Abundant Gram-positive cocci forming microcolonies were present in the lumen of small blood vessels in multiple organs. The microvasculature of the brain showed disruption of the wall with leakage of fibrin and focal haemorrhages (Figure 1b). In the lungs, eosinophilic bronchitis with nematodes in lumen of bronchi was observed.
Figure 1. Histological lesions in a hedgehog with MRSA septicaemia (V583/03).
a) H&E staining of a brain section showing disruption of the wall of microvasculature with leakage of fibrin and focal haemorrhage, focal infiltrate of inflammatory cells and aggregate of cocci. b) Gram staining of a kidney section showing a focus of necrosis and infiltrate of neutrophils, lymphocytes and macrophages in association to the presence of abundant dispersed or aggregated Gram positive cocci.
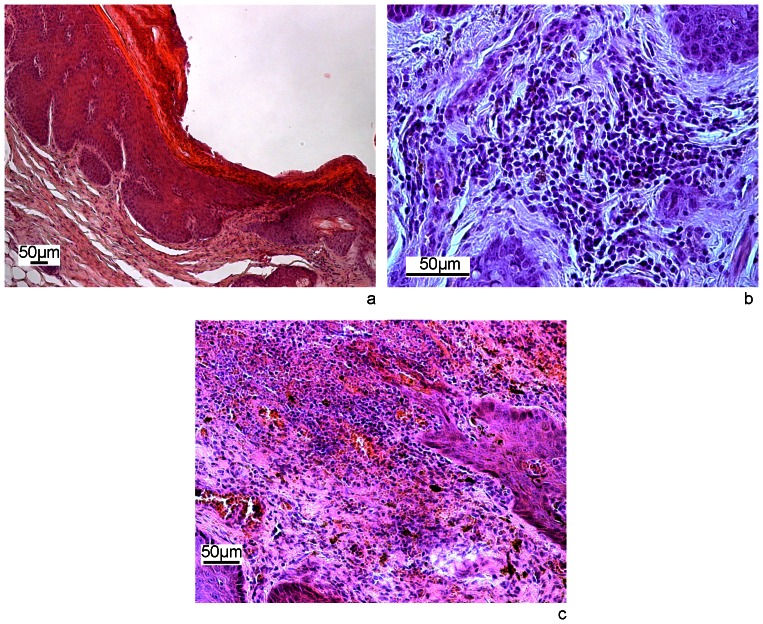
Hedgehog V5406/11 was in good physical condition but showed a diffuse dermatitis with thickening of the skin and prominent crusts affecting mostly skin areas free of spines. Histologically, skin lesions included ulcerations and formation of serocellular crusts. In the epidermis there was hyperkeratosis, parakeratosis, hyperpigmentation and irregular epidermal hyperplasia (Figure 2a) with formation of rete ridges uneven in length and shape. There was an interface dermatitis with infiltrate of macrophages, lymphocytes and few neutrophils (Figure 2b) and focal perivascular inflammation. Focal necroses and dense infiltration of neutrophils with few mononuclear cells were observed in the superficial dermis (Figure 2c). There was follicular lymphoid hyperplasia in the spleen and eosinophilic bronchitis with presence of nematodes in the lumen of bronchi in the lungs.
Figure 2. Skin lesions in a hedgehog with MRSA dermatitis (V5406/11).
a) H&E staining of an ulceration with necrosis of surface epithelium, thick serocellular crusts and irregular epidermal hyperplasia. b) H&E staining of an inflammatory infiltrate of lymphocytes, macrophages and few neutrophils in the superficial dermis. c) H&E staining of a superficial dermis subjacent to ulceration. Necrosis, cell debris, severe infiltrate of neutrophils and mononuclear cells and haemorrhages.
Susceptibility tests of S. aureus isolates
Phenotypically, both S. aureus isolates were resistant to penicillin and cefoxitin, but the oxacillin MICs as measured by VITEK were low (> = 0.5 µg/mL). However, they were identified by the Vitek software as MRSA based on cefoxitin resistance. Both isolates were susceptible to erythromycin, clindamycin, gentamicin, tobramycin, vancomycin, teicoplanin, tetracyclines, tigecycline, co-trimoxacole, levofloxacin, moxifloxacin, rifampicin, linezolid, mupirocin, fosfomycin and fusidic acid. The oxacillin MICs as determined by agar dilution (E-test) for V583/03 and V5406/11 were 4 µg/mL and 8 µg/mL, respectively, while the cefoxitin MICs were 12 µg/mL and 24 µg/mL, respectively. For both isolates, penicillin MICs were 16 µg/mL.
Both isolates yielded positive signals in the Clearview™ PBP2a lateral flow test.
Molecular characterisation of S. aureus isolates
Both hedgehog isolates belonged to agr group III, capsule type 8 and ST130 (MLST alleles arcC-6, aroE-57, glpF-45, gmk-2, pta-7, tpi-58, yqil-52). Isolate V5406/11 belonged to spa type t843 (repeats: 04-82-17-25-17-25-25-16-17), whereas isolate V583/03 belonged to a similar, but yet not defined spa sequence with the repeats 04-17-25-16-17.
Both isolates tested positive for mecC and for a SCCmec XI-associated beta-lactamase gene blaZ by DNA microarray hybridisation as well as, for mecC, by PCR and sequencing. The mecC gene exhibited 99% DNA sequence identity (nucleotide change of A to C at nucleotide coordinate 1307 within mecC) and 100% amino acid sequence identity with mecC (GenBank FR821779.1, FR823292.1). In addition, the presence of ccrA1B3 and the entire SCCmec XI element, including the arsenic resistance operon, were confirmed in both isolates using PCR. Both isolates yielded amplicons of the expected size using all primer pairs. The gene encoding the SCCmec-associated phenol-soluble modulin PSM-mec [23] was absent from both isolates.
Apart from mecC and blaZ-SCCmec XI, no other genes associated with antibiotic resistance were identified.
Full array hybridisation profiles are provided as Table S1. In short, both isolates harboured the hlg-locus (hlgA, lukF/S) and the leukocidin homologue genes lukD/E. The Pantone-Valentine leukocidin and the animal-associated leukocidin homologue lukM/lukF-P83 were absent. The epidermal cell differentiation inhibitor gene edinB as well as an exfoliative toxin homologue putatively named “etD2” were detected. The hlb gene was intact and genes associated with beta-haemolysin converting phages (sea, sep, chp, sak and scn) were absent. Other enterotoxin genes and the toxic shock toxin gene were also not found. Protease genes aur, splA, splB, splE, sspA, sspB and sspP were detected. With regard to adhesion factors, cna and sasG were absent while bbp, clfA/B coa, ebh, ebpS, eno, fib, fnbA/B, map, sdrC/D and vwb were identified.
Hybridisation profiles of both hedgehog isolates were essentially indistinguishable from previously described isolates from Irish patients ([7], see Table S1).
Discussion
There are very few reports of S. aureus and MRSA in hedgehogs. In a study performed in the 1960s in New Zealand, a high prevalence of S. aureus (85% of animals tested) was found and a high rate of penicillin resistance (86.3% of isolates tested) was observed. Since these isolates were celbenin (oxacillin) susceptible, the resistance was attributed to a penicillinase [28]. Given the low MICs observed in mecC-positive strains as in the present study, its presence cannot safely be ruled out based on observed susceptibility to oxacillin. Thus, it cannot be concluded in retrospect whether these strains were negative for mecC, or not. In 2003, MRSA was isolated from a hedgehog suffering from rhinitis in the UK, but no typing data on this isolate were published [29].
To the best of our knowledge, this is the first report of MRSA carrying SCCmec XI in hedgehogs. Furthermore, isolate V583/03 recovered in 2003 is, to the best of our knowledge, the earliest known case of a mecC-positive S. aureus in Sweden. Several recent observations [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20] allow the speculation that CC130 might be a zoonotic strain, and that SCCmec XI/mecC may have originated in animal pathogens.
No host-specific pathotypes for CC130-MRSA-XI can be distinguished, with the hedgehog and human isolates being virtually identical. The animal-specific leukocidin, lukM/lukF-P83, was absent from both, hedgehog and human isolates [7], although it has been observed in (methicillin susceptible) sheep and rat isolates of CC130 [7], [25]. The “etD2” exfoliative toxin homologue gene has been found in genome sequences of human and animal strains of CC130 [7], [25] as well as in the animal-specific staphylococcal species Staphylococcus hyicus (where it was named “shetb” ¸ GenBank AB036767.1) and Staphylococcus pseudintermedius (“exi” or “expb”; GenBank AB489850.1 and AB569087.1). A formation of skin blisters and exfoliation upon injection into neonatal mice has been observed [30] and a role in canine pyoderma has been suggested [31] as well as a role in exudative epidermitis of pigs [32]. These observations could suggest a wide host range for staphylococci harbouring “etD2”, but more clinical and experimental data are needed to assess the significance of that factor in the different host species.
In humans, CC130-MRSA-XI and other mecC-positive strains appear to be rare. In Germany, during the years 2006–2011, less than one out of 1000 MRSA isolates belonged to mecC-positive strains [11]. The rate in Denmark is higher, being 1.5% for the period of 2003–2011, and as much as 2.8% in 2011.Their rarity in humans and observations of cases with animal contacts [14] raise the question whether this MRSA strain evolved in animals rather than humans. It is also not yet clear whether domestic or wild animals are a reservoir to mecC-positive strains. If they emerged in wild animals such as chaffinches, squirrels, seals, or as in our study, in hedgehogs, the selective pressure(s) that may favour the evolution of MRSA in nature, i.e., under low-level exposure to beta-lactam compounds, need to be investigated. However, CC130 MRSA-XI also appears to be uncommon in wildlife, although systematic studies are needed. During Wildtech project activities in Sweden, 46 S. aureus isolates from a variety of wild mammal and bird species were identified and genotyped. The two isolates described herein were the only S. aureus isolates from hedgehogs, the only mecC-positive isolates and the only ones that were assigned to CC130.
The MRSA infection in the hedgehogs described in this report caused severe disease. One of the hedgehogs developed a lethal septicaemia with infection of multiple organs, and MRSA was isolated in abundant growth in pure culture from the two tissues analysed, kidney and brain. In the other case, the hedgehog developed a severe dermatitis. Bacterial shedding and environmental contamination for example through urine and skin purulent exudates is likely to have occurred in these cases. The role of colonised and infected hedgehogs in the epidemiology of S. aureus/CC130-MRSA-XI cannot currently be assessed due to lack of information and more studies are required to address this issue.
The presence of mecA variants in animal staphylococci [2], [3], [4] as well as of mecC in animal isolates of S. aureus and S. xylosus [21] indicates that bacterial populations from animals might serve as reservoir for precursors of resistance genes, that an emergence of antibiotic resistance in bacteria also takes place in animals and that the rise of drug-resistant pathogens thus can be regarded also as emerging zoonotic disease. Since, hedgehogs in particular often inhabit suburban gardens and parks, people and dogs often come into close contact with them. Therefore, it would be of interest to study the extent and frequency of MRSA infection in hedgehogs and other wildlife as well as the potential risk for humans and pets to contract the infection.
Supporting Information
Full array hybridisation and PCR results of the hedgehog isolates from this study and, for comparison, of CC130-MRSA-XI isolates from [7].
(PDF)
Acknowledgments
We acknowledge C. Bröjer (Uppsala), E. Prowse (Gotland), A. Ruppelt, S. Schubert, A. Gottwald (Dresden), J. Sachtschal and E. Mueller (Jena) for excellent technical assistance as well as Prof. Dr. E. Jacobs (Dresden) and E. Ermantraut (Jena) for their support.
Staphylococcus aureus isolates were collected within the framework of the Wildtech project (EU 7th Framework Program for Research and Technological Development, grant agreement no. 222633) in which wild animals are screened for various pathogens.
Funding Statement
These authors have no support or funding to report.
References
- 1. Ito T, Hiramatsu K, Tomasz A, de Lencastre H, Perreten V, et al. (2012) Guidelines for Reporting Novel mecA Gene Homologues. Antimicrob Agents Chemother 56: 4997–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Monecke S, Muller E, Schwarz S, Hotzel H, Ehricht R (2012) Rapid microarray based identification of different mecA alleles in Staphylococci. Antimicrob Agents Chemother 56: 5547–5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Couto I, de Lencastre H, Severina E, Kloos W, Webster JA, et al. (1996) Ubiquitous presence of a mecA homologue in natural isolates of Staphylococcus sciuri . Microb Drug Resist 2: 377–391. [DOI] [PubMed] [Google Scholar]
- 4. Wu S, Piscitelli C, de Lencastre H, Tomasz A (1996) Tracking the evolutionary origin of the methicillin resistance gene: cloning and sequencing of a homologue of mecA from a methicillin susceptible strain of Staphylococcus sciuri . Microb Drug Resist 2: 435–441. [DOI] [PubMed] [Google Scholar]
- 5. IWG-SCC (2009) Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother 53: 4961–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Garcia-Alvarez L, Holden MT, Lindsay H, Webb CR, Brown DF, et al. (2011) Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis 11: 595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shore AC, Deasy EC, Slickers P, Brennan G, O'Connell B, et al. (2011) Detection of staphylococcal cassette chromosome mec type XI carrying highly divergent mecA, mecI, mecR1, blaZ, and ccr genes in human clinical isolates of clonal complex 130 methicillin-resistant Staphylococcus aureus . Antimicrob Agents Chemother 55: 3765–3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Laurent F, Chardon H, Haenni M, Bes M, Reverdy ME, et al. (2012) MRSA Harboring mecA Variant Gene mecC, France. Emerg Infect Dis 18: 1465–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Basset P, Prod'hom G, Senn L, Greub G, Blanc DS (2013) Very low prevalence of meticillin-resistant Staphylococcus aureus carrying the mecC gene in western Switzerland. Journal of Hospital Infection 83: 257–259. [DOI] [PubMed] [Google Scholar]
- 10.Bengtsson B, Börjesson S, Englund S, Unnerstad HE, Greko C, et al.. (2011) SVARM 2011, Swedish Veterinary Antimicrobial Resistance Monitoring. In: Department of Animal Health and Antimicrobial Strategies NVI, Uppsala. [Google Scholar]
- 11. Cuny C, Layer F, Strommenger B, Witte W (2011) Rare occurrence of methicillin-resistant Staphylococcus aureus CC130 with a novel mecA homologue in humans in Germany. PLoS One 6: e24360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elstrøm P, Jacobsen T, Larsen KW, Marstein L, Kilnes A, et al.. (2011) MRSA infections in humans in Norway 2011. In: NORM-VET N, editor. 2011 ed. Tromsø/Oslo. [Google Scholar]
- 13. Paterson GK, Larsen AR, Robb A, Edwards GE, Pennycott TW, et al. (2012) The newly described mecA homologue, mecALGA251, is present in methicillin-resistant Staphylococcus aureus isolates from a diverse range of host species. Journal of Antimicrobial Chemotherapy 67: 2809–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Petersen A, Stegger M, Heltberg O, Christensen J, Zeuthen A, et al. (2013) Epidemiology of methicillin-resistant Staphylococcus aureus carrying the novel mecC gene in Denmark corroborates a zoonotic reservoir with transmission to humans. Clin Microbiol Infect 19: E16–22. [DOI] [PubMed] [Google Scholar]
- 15. Robb A, Pennycott T, Duncan G, Foster G (2013) Staphylococcus aureus carrying divergent mecA homologue (mecALGA251) isolated from a free-ranging wild bird. Vet Microbiol 162: 300–301. [DOI] [PubMed] [Google Scholar]
- 16. Sabat AJ, Koksal M, Akkerboom V, Monecke S, Kriegeskorte A, et al. (2012) Detection of New Methicillin-Resistant Staphylococcus aureus Strains That Carry a Novel Genetic Homologue and Important Virulence Determinants. J Clin Microbiol 50: 3374–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stegger M, Andersen PS, Kearns A, Pichon B, Holmes MA, et al. (2012) Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecALGA251. Clin Microbiol Infect 18: 395–400. [DOI] [PubMed] [Google Scholar]
- 18. Unnerstad HE, Börjesson S, Bengtsson B (2012) MRSA pavisad hos svenska mjölkkor. Svensk Veterinärtidning 35–37. [Google Scholar]
- 19. Unnerstad HE, Bengtsson B, Af Rantzien MH, Borjesson S (2013) Methicillin-resistant Staphylococcus aureus containing mecC in Swedish dairy cows. Acta Vet Scand 55: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Walther B, Wieler LH, Vincze S, Antão EM, Brandenburg A, et al. (2012) MRSA Variant in Companion Animals. Emerg Infect Dis 18: 2017–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harrison EM, Paterson GK, Holden MTG, Morgan FJE, Larsen AR, et al. (2013) A Staphylococcus xylosus Isolate with a New mecC Allotype. Antimicrobial Agents and Chemotherapy 57: 1524–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Monecke S, Coombs G, Shore AC, Coleman DC, Akpaka P, et al. (2011) A Field Guide to Pandemic, Epidemic and Sporadic Clones of Methicillin-Resistant Staphylococcus aureus . PLoS One 6: e17936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Queck SY, Khan BA, Wang R, Bach TH, Kretschmer D, et al. (2009) Mobile genetic element-encoded cytolysin connects virulence to methicillin resistance in MRSA. PLoS Pathog 5: e1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Monecke S, Engelmann I, Archambault M, Coleman DC, Coombs GW, et al. (2012) Distribution of SCCmec-associated phenol-soluble modulin in staphylococci. Mol Cell Probes 26: 99–103. [DOI] [PubMed] [Google Scholar]
- 25. Le Maréchal C, Hernandez D, Schrenzel J, Even S, Berkova N, et al. (2011) Genome Sequences of Two Staphylococcus aureus Ovine Strains That Induce Severe (Strain O11) and Mild (Strain O46) Mastitis. Journal of Bacteriology 193: 2353–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Enright MC, Day NPJ, Davies CE, Peacock SJ, Spratt BG (2000) Multilocus Sequence Typing for Characterization of Methicillin-Resistant and Methicillin-Susceptible Clones of Staphylococcus aureus . J Clin Microbiol 38: 1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harmsen D, Claus H, Witte W, Rothganger J, Claus H, et al. (2003) Typing of Methicillin-Resistant Staphylococcus aureus in a University Hospital Setting by Using Novel Software for spa Repeat Determination and Database Management. J Clin Microbiol 41: 5442–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Smith JM (1965) Staphylococcus Aureus Strains Associated with the Hedgehog, Erinaceus Europaeus . J Hyg (Lond) 63: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Veterinary Laboratories Agency, Laboratory PR (2004) Wildlife diseases in the UK. In: Defra (Department of Environment FaRA, Penrith. [Google Scholar]
- 30. Futagawa-Saito K, Makino S, Sunaga F, Kato Y, Sakurai-Komada N, et al. (2009) Identification of first exfoliative toxin in Staphylococcus pseudintermedius . FEMS Microbiology Letters 301: 176–180. [DOI] [PubMed] [Google Scholar]
- 31. Iyori K, Hisatsune J, Kawakami T, Shibata S, Murayama N, et al. (2010) Identification of a novel Staphylococcus pseudintermedius exfoliative toxin gene and its prevalence in isolates from canines with pyoderma and healthy dogs. FEMS Microbiology Letters 312: 169–175. [DOI] [PubMed] [Google Scholar]
- 32. Watanabe T, Sato H, Hatakeyama Y, Matsuzawa T, Kawai M, et al. (2000) Cloning of the Gene Coding for Staphylococcus hyicus Exfoliative Toxin B and Its Expression in Escherichia coli . Journal of Bacteriology 182: 4101–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full array hybridisation and PCR results of the hedgehog isolates from this study and, for comparison, of CC130-MRSA-XI isolates from [7].
(PDF)