Abstract
Administration of the mTORC1 inhibitor, rapamycin, to humans blocks the increase in skeletal muscle protein synthesis in response to resistance exercise or amino acid ingestion.
Objective
To determine whether rapamycin administration influences basal post-absorptive protein synthesis or breakdown in human skeletal muscle.
Materials/Methods
Six young (26±2 years) subjects were studied during two separate trials, in which each trial was divided into two consecutive 2h basal periods. The trials were identical except during one trial a single oral dose (16mg) of rapamycin was administered immediately prior to the second basal period. Muscle biopsies were obtained from the vastus lateralis at 0, 2, and 4h to examine protein synthesis, mTORC1 signaling, and markers of autophagy (LC3B-I and LC3B-II protein) associated with each 2h basal period.
Results
During the Control trial, muscle protein synthesis, whole body protein breakdown (phenylalanine Ra), mTORC1 signaling, and markers of autophagy were similar between both basal periods (p>0.05). During the Rapamycin trial, these variables were similar to the Control trial (p>0.05) and were unaltered by rapamycin administration (p>0.05). Thus, post-absorptive muscle protein metabolism and mTORC1 signaling were not affected by rapamycin administration.
Conclusions
Short-term rapamycin administration may only impair protein synthesis in human skeletal muscle when combined with a stimulus such as resistance exercise or increased amino acid availability.
Keywords: mTOR, Autophagy, FSR
1. Introduction
The preservation or growth of skeletal muscle mass represents an important treatment strategy for numerous debilitating clinical conditions [1-4]. Alterations in skeletal muscle mass are largely dependent upon the relationship between muscle protein synthesis and muscle protein breakdown, such that when one process is chronically favored over the other muscle mass is either gained or lost. Consequently, changes in skeletal muscle protein metabolism are commonly examined to understand the therapeutic potential of various treatment strategies (i.e., exercise and nutrition) aimed to preserve or increase skeletal muscle mass [5-8]. In addition, although not fully elucidated, key cellular mechanisms regulating changes in muscle protein metabolism are beginning to be uncovered [9,10], and consequently these findings provide important targets for intervention. While great strides have been made regarding the regulation of skeletal muscle protein metabolism in response to various interventions, the mechanism(s) involved in regulating basal skeletal muscle protein metabolism are less well defined. This research focus is important considering skeletal muscle mass is likely governed primarily by basal protein metabolism in numerous clinical conditions, in particular those that do not allow for physical activity.
Signaling through mTORC1 has become a focal point of numerous investigations examining the cellular mechanisms contributing to increased rates of muscle protein synthesis in response to various interventions [11]. The ability to better understand the regulatory role of mTORC1 signaling, with respect to increases in protein synthesis, has been accelerated with the use of rapamycin, a potent mTORC1 inhibitor [12]. For instance, several animal models have demonstrated that growth and/or increases in protein synthesis rate in response to anabolic stimuli (i.e., nutrition, mechanical stimulation) are blocked or attenuated with prior rapamycin administration, likely facilitated through inhibition of mTORC1 signaling [12-17]. Similarly, our laboratory has recently demonstrated that prior administration of rapamycin to humans blocks the increase in skeletal muscle protein synthesis rate in response to high intensity resistance exercise [18] or essential amino acid ingestion [19]. While it is well understood that rapamycin administration is able to block increases in protein synthesis in response to a stimulus, interestingly rapamycin administration has not been shown to effect basal rates of protein synthesis in animal models [14-16,20]. Whether rapamycin administration has an affect on basal skeletal muscle protein synthesis in humans has yet to be investigated.
In addition to its role in protein synthesis, mTORC1 signaling has also been shown to negatively regulate autophagy, an important process through which proteins, other macromolecules, and organelles are degraded in the cell [21]. Specifically, treatment of cells with rapamycin has been shown to stimulate autophagy in many cell types [22-24], including muscle cells [25], increasing the breakdown of protein and further highlighting a role for mTORC1 as a master switch between anabolic (e.g., protein synthesis) and catabolic (e.g., autophagy) cellular processes. Along these lines, we have recently observed that the ingestion of essential amino acids, a known stimulator of mTORC1 signaling and protein synthesis, was associated with reduced levels of autophagy markers (i.e., LC3B-II) in human skeletal muscle [26]. However, the relationship between mTORC1 signaling and autophagy in human skeletal muscle has yet to be mechanistically investigated, and consequently, it is unknown whether rapamycin administration impacts autophagy in human skeletal muscle.
To our knowledge, the mechanisms regulating human skeletal muscle protein metabolism in the basal state have yet to be the focus of any investigation. Thus, to provide a foundation from which to begin to examine these mechanisms, the purpose of this study was to determine whether rapamycin administration affects post-absorptive protein metabolism in human skeletal muscle. Specifically, we administered rapamycin under basal conditions and examined skeletal muscle protein synthesis rate, whole body protein breakdown, and downstream markers of mTORC1 signaling known to regulate both the synthesis (translation initiation) and breakdown (autophagy) of muscle proteins.
2. Methods
2.1. Subjects
Six healthy young male (n=3) and female (n=3) subjects (Table 1) volunteered for this investigation and were studied during two separate experimental trials. All subjects were healthy and considered recreationally active but not engaged in a regularly scheduled exercise-training program. Screening for all subjects was performed with clinical history, physical examination, and laboratory tests, including complete blood count with differential, liver and kidney function tests, coagulation profile, fasting blood glucose, oral glucose tolerance test, hepatitis B and C screening, HIV testing, thyroid-stimulating hormone, urinalysis, and drug screening. All subjects gave informed written consent prior to participation in the study, which was approved by the Institutional Review Board of the University of Texas Medical Branch (in compliance with the Declaration of Helsinki as revised in 1983).
Table 1.
Subject characteristics by gender.
| Males (n=3) | Females (n=3) | |
|---|---|---|
| Age, year | 25±4 | 26±3 |
| Height, cm | 183±3 | 168±2 |
| Weight, kg | 84±3 | 61±5 |
| BMI, kg/m2 | 25±1 | 22±1 |
| LM, kg | 68±5 | 37±1 |
| Leg LM, kg; R+L | 25±2 | 15±1 |
| Fasting Plasma Glucose, mg/dl | 80±7 | 77±3 |
| 2h Plasma Glucose, mg/dl | 97±18 | 93±7 |
Values are means±SE. BMI, body mass index; LM, lean mass; R, right leg; L, left leg; 2h glucose, plasma glucose at 2 h of oral glucose tolerance test.
2.2. Study Design
Each subject completed an experimental trial (Fig. 1) on two separate occasions, separated by 2–4weeks, in a randomized, counterbalanced cross-over fashion. The experimental trial was designed to obtain measurements during two consecutive 2h basal post-absorptive periods, referred to as the “initial basal” period and “experimental basal” period, respectively. The two experimental trials completed by each subject were identical except during one trial subjects ingested 16mg (1mg tablets) of rapamycin (Rapamune/Sirolimus; Wyeth, Madison, NJ, USA) (RAP) at the start of the “experimental basal” period, whereas no rapamycin was ingested during the other trial (CON) (Fig. 1). The “initial basal” period was used to control for potential day-to-day variations in basal protein metabolism, whereas the “experimental basal” period was used to control for potential intraday variations in protein metabolism during a given trial.
Fig. 1.
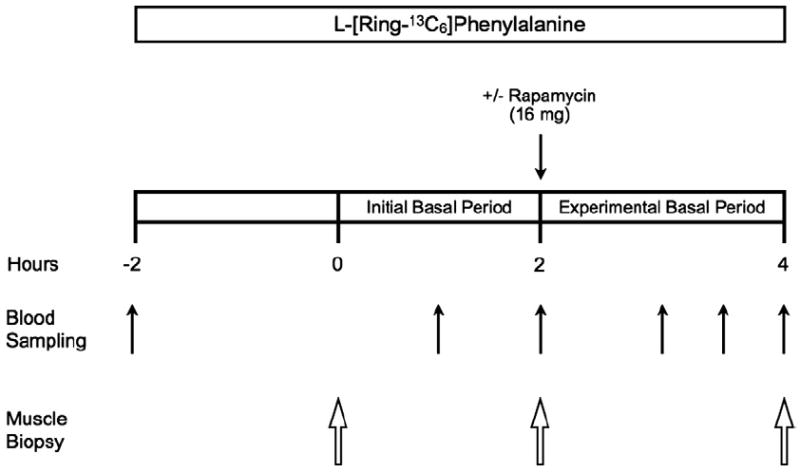
Schematic of the experimental trial. The experimental trial was designed to obtain measurements during two consecutive 2h basal post-absorptive periods, referred to as the initial basal period and experimental basal period, respectively. Each subject completed the experimental trial on two occasions (Control and Rapamycin) in a randomized, counterbalanced cross-over fashion. The experimental trial was identical for the Control and Rapamycin trials except that during one experimental trial subjects ingested 16mg of rapamycin (Rapamycin trial) at the start of the experimental basal period, whereas no rapamycin was ingested during the other experimental trial (Control trial).
On the evening prior to each experimental trial, subjects were admitted to the Institute for Translational Sciences-Clinical Research Center of the University of Texas Medical Branch and were fed a standardized meal at 1800h and a snack at 2200h. All subjects were studied during the same time of day (i.e., 0700–1300h) following an overnight fast under basal conditions and refrained from exercise for 24h prior to each experimental trial.
On the morning of each trial, an 18-gauge polyethylene catheter was inserted into a forearm vein for infusion of stable, isotopically labeled L-[ring-13C6]phenylalanine (Isotec, Sigma-Aldrich; St. Louis, MO). The phenylalanine tracer was dissolved in sterile 0.9% saline, passed through a 2μm filter prior to infusion, and infused at a constant rate of 0.05μmol·kg−1·min−1 throughout each trial, which was preceded by a priming dose of 2μmol·kg−1 [19]. During each trial a total of three muscle biopsies were obtained from the lateral portion of the vastus lateralis following local anesthesia (1% lidocaine) using a 5mm Bergström needle with suction [27]. The first biopsy was obtained 2h following the initiation of the tracer infusion, marking the beginning of the initial basal period. A second biopsy, marking the end of the initial basal period and the beginning of the experimental basal period, was obtained from the same incision 2h following the first biopsy, in which the biopsy needle was inclined at a different angle such that the second biopsy was taken approximately 5cm proximal to the first. Immediately following the second biopsy, subjects undergoing the RAP trial were administered 16mg of rapamycin. Subjects remained in their hospital beds for an additional 2h following the second biopsy and a third muscle biopsy was then obtained from the same incision as the first two biopsies to mark the end of the experimental basal period. During the third biopsy, the biopsy needle was inclined at a different angle so that the third biopsy was taken ~5cm proximal from the previous biopsy sampling site, as we have previously described [28,29]. A catheter was also placed in the antecubital vein of the opposite arm for blood sampling. Blood samples were obtained prior to initiating the tracer infusion and periodically during the experimental trial for measurement of tracer enrichment (Fig. 1).
2.3. Determination of tracer enrichment
Muscle samples were homogenized and separated into protein bound and intracellular free amino acids as previously described [30] for determination of mixed protein bound and muscle intracellular L-[ring-13C6]phenylalanine enrichment via gas chromatography–mass spectrometry (GCMS, 6890 Plus GC, 5973N MSD, 7683 autosampler, Agilent Technologies, Palo Alto, CA, USA). Mixed protein bound enrichment was determined by GCMS in triplicate following protein hydrolysis and amino acid extraction using the m+6/m+4 ratio and an external standard curve of known m+6/m+0 ratios [31,32]. Muscle intracellular enrichment was determined by GCMS in triplicate using the m+6/m+0 ratio. Blood L-[ring-13C6]phenylalanine enrichment was determined from deproteinized blood samples in duplicate using the m+6/m+0 ratio. All tracer enrichments were determined using tert-butyldimethylsilyl derivatives of the respective amino acid.
2.4. Calculations
The fractional synthesis rate (FSR) of mixed muscle protein was determined by examining the rate of L-[ring-13C6]phenylalanine incorporated into mixed muscle protein using the precursor product model:
where ΔEp is the increment in protein-bound L-[ring-13C6]phenylalanine enrichment between two muscle biopsies, t is the time between the two muscle biopsies, and EM(1) and EM(2) are the L-[ring-13C6]phenylalanine enrichments in the free intracellular pool in the two muscle biopsies. Data are expressed as percent per hour.
Whole body phenylalanine rate of appearance (Ra), an index of whole body protein breakdown, was calculated using the single pool model [33]:
where i is the infusion rate of L-[ring-13C6]phenylalanine and Ev is the mean L-[ring-13C6]phenylalanine enrichment in the blood during the respective basal period.
2.5. Immunoblot analysis
Immunoblot analysis was performed as previously detailed [30]. Briefly, frozen tissue was homogenized, centrifuged for 10min at 4°C, and the supernatant collected. Total protein concentrations were determined using the Bradford assay (Smartspec Plus, BioRad, Hercules, CA, USA). The supernatant was diluted (1:1) in a 2× sample buffer mixture containing 125 mM Tris, pH 6.8, 25% glycerol, 2.5% SDS, 2.5% β-mercaptoethanol and 0.002% bromophenol blue, then boiled for 3min at 100°C. Equal amounts of total protein (50μg) were loaded into each lane and the samples were separated by electrophoresis (150V for 60min) on a 7.5% or 15% polyacrylamide gel as determined by the size of the target protein (Criterion, BioRad). Each sample was loaded in duplicate and each gel contained an internal loading control and molecular weight ladder (Precision Plus, BioRad). Additionally, all samples from a given experimental trial were loaded onto the same gel and each gel contained samples from both the CON and RAP trials.
Following electrophoresis, protein was transferred to a polyvinylidene difluoride membrane (BioRad) at 50V for 60min. Blots were then blocked for 1h in 5% non fat dry milk and incubated in primary antibody overnight at 4°C (see Antibodies below). The next morning, blots were incubated in secondary antibody for 1h at room temperature. Blots were then incubated in a chemiluminescent solution (ECL plus, Amersham BioSciences, Piscataway, NJ, USA) for 5 min and optical density measurements were obtained with a phosphoimager (ChemiDoc, BioRad) and densitometric analysis was performed using Quantity One 4.5.2 software (BioRad). Membranes containing phospho-detected proteins were stripped of primary and secondary antibodies using restore western blot stripping buffer (Pierce Biotechnology, CA, USA) and were subsequently re-probed for total protein with the specific antibody of interest. All phospho and total density values were normalized to the internal loading control. Data for phospho-proteins are presented as phospho/total and adjusted to represent fold change from the first basal period of the same experimental trial. The antibody utilized for microtubule-associated protein 1 light chain 3 (LC3) B protein (see below) produces bands for both LC3B-I and LC3B-II. LC3B-I is conjugated to LC3B-II during stimulation of autophagy [34], and consequently the expression of LC3B-I and LC3B-II, as well as the ratio of LC3B-II/LC3B-I, was examined to gain insight into changes in autophagy [35,36]. Data for LC3B-I and LC3B-II are presented as total protein expression and adjusted to represent fold change from the initial basal period of the same trial. The LC3B-II/LC3B-I ratio was calculated by dividing the total protein expression value obtained for LC3B-II by that of LC3B-I and this ratio was adjusted to represent fold change from the initial basal period of the same trial.
2.6. Antibodies
All antibodies were purchased from Cell Signaling (Danvers, MA, USA) and utilized in the following dilutions: phospho-mTOR (Ser2448; 1:250), mTOR (1:1000), phospho-S6K1 (Thr389; 1:500), S6K1 (1:1000), phospho-4E-BP1 (Thr37/46; 1:1000), 4E-BP1 (1:1000), LC3B (1:1000). Anti-rabbit IgG horseradish peroxidase-conjugated secondary antibody was purchased from Amersham Bioscience (1:2000).
2.7. Statistical analysis
A two-way repeated measures ANOVA was used to test time by trial differences. A Tukey’s post hoc analysis was used when necessary to determine specific differences within an ANOVA. All data were analyzed using SigmaStat v.11.0 (Systat Software Inc, San Jose, CA, USA). Significance for all analyses was set to p<0.05. Data are presented as mean±SEM.
3. Results
3.1. Protein metabolism
Mixed muscle protein FSR was similar between trials during the initial basal period (p>0.05). Further, mixed muscle protein FSR was unchanged between the initial and experimental basal periods during both the CON and RAP trials (p>0.05) (Fig. 2), and was similar between trials during the experimental basal period (p>0.05). No differences in phenylalanine Ra (index of whole body protein breakdown) were observed between trials or between the initial and experimental basal periods during the CON and RAP trials (p>0.05) (Fig. 2).
Fig. 2.
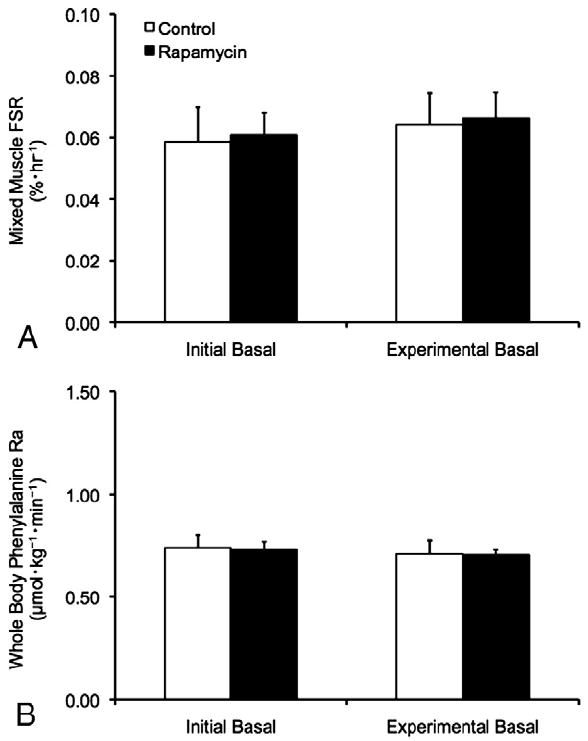
Mixed muscle protein fractional synthesis rate (FSR) in skeletal muscle (A) and whole body phenylalanine rate of appearance (Ra) (B) during the initial and experimental basal post-absorptive periods during the Control (CON) and Rapamycin (RAP) trials. To control for potential day-to-day variations in basal protein metabolism, both the CON and RAP trials included an initial basal period. However, during the RAP trial, subjects were administered 16mg of rapamycin at the start of the experimental basal period, whereas no rapamycin was ingested during the CON trial. Data are mean±SEM, n=6.
3.2. mTORC1 signaling
No differences in the phosphorylation status of mTOR, S6K1 or 4E-BP1 were observed between trials or between the initial and experimental basal periods during the CON and RAP trials (p>0.05) (Fig. 3).
Fig. 3.
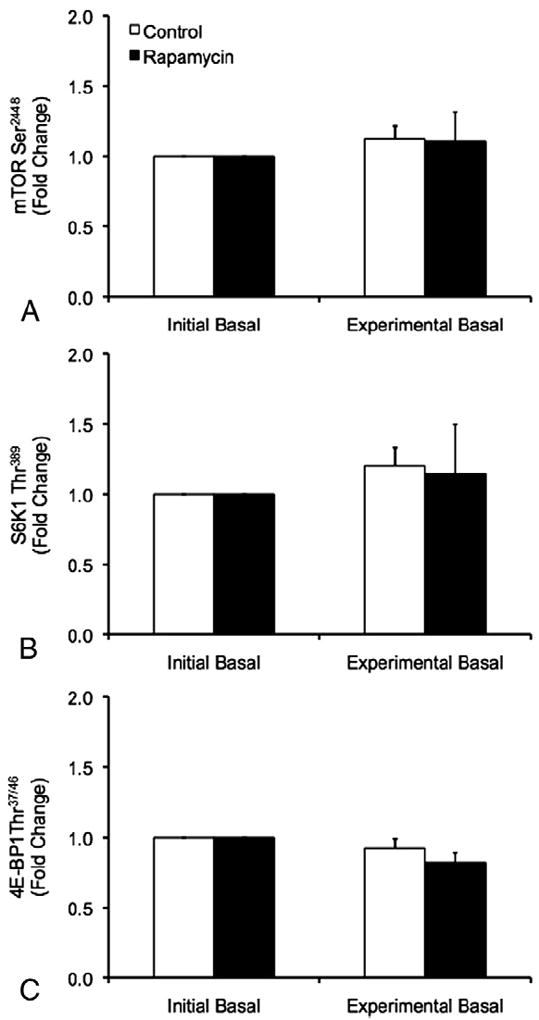
Phosphorylation of mTOR at Ser2448 (A), S6K1 at Thr389 (B), and 4E-BP1 at Thr37/46 (C) during the initial and experimental basal post-absorptive periods during the Control (CON) and Rapamycin (RAP) trials. To control for potential day-to-day variations in basal protein metabolism, both the CON and RAP trials included an initial basal period. However, during the RAP trial, subjects were administered 16mg of rapamycin at the start of the experimental basal period, whereas no rapamycin was ingested during the CON trial. Data are mean±SEM, n=6.
3.3. Autophagy
No differences between trials were observed for LC3B-I, however, a main effect of time was detected for LC3B-I such that LC3B-I protein was increased during the experimental basal period relative to the initial basal period independent of trial (p<0.05) (Fig. 4). No differences in LC3B-II protein or the LC3B-II/LC3B-I protein ratio were observed between trials or between the initial and experimental basal periods during the CON and RAP trials (p>0.05) (Fig. 4).
Fig. 4.
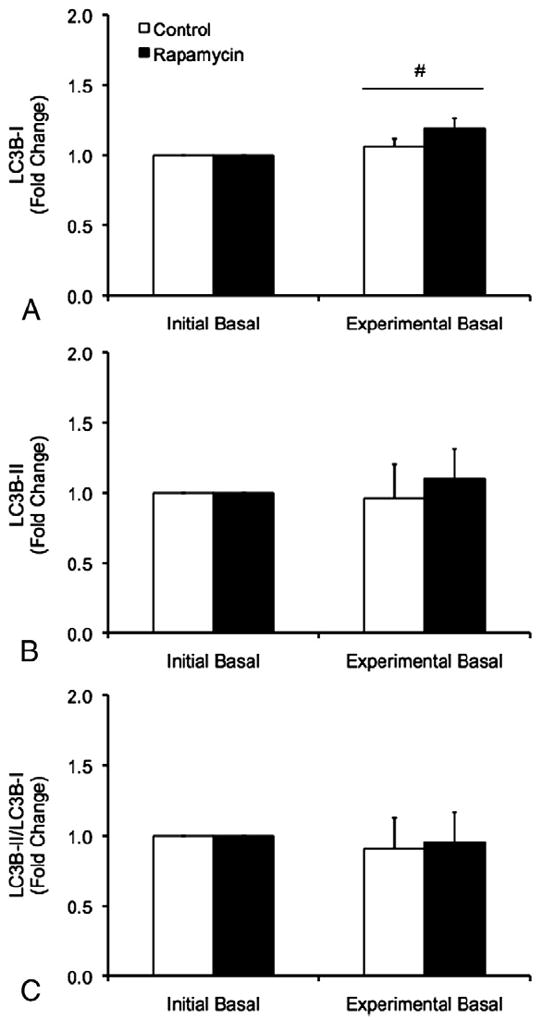
Protein expression of LC3B-I (A), LC3B-II (B), and the ratio of LC3B-II/LC3B-I protein (C) in skeletal muscle during the initial and experimental basal post-absorptive periods during the Control (CON) and Rapamycin (RAP) trials. To control for potential day-to-day variations in basal protein metabolism, both the CON and RAP trials included an initial basal period. However, during the RAP trial, subjects were administered 16mg of rapamycin at the start of the experimental basal period, whereas no rapamycin was ingested during the CON trial. Data are mean±SEM, n=6. #different from Initial Basal, main effect of time, p<0.05.
4. Discussion
This investigation was designed to determine whether administration of rapamycin affects basal post-absorptive protein metabolism in human skeletal muscle. The novel and important findings from the current investigation are that basal rates of protein synthesis are maintained in human skeletal muscle during the immediate hours following administration of rapamycin. In addition, rapamycin administration did not appear to affect basal mTORC1 signaling, the rate of whole body protein breakdown, or markers of autophagy in human skeletal muscle. These data suggest that short term rapamycin administration may only impair the increases in human skeletal muscle mTORC1 signaling and protein synthesis rate that occur as a result of a stimulus, such as that experienced in response to resistance exercise [18] or increased amino acid availability [19]. Furthermore, the results from this investigation provide a basis in which to begin to examine the mechanism(s) that regulate basal protein metabolism in human skeletal muscle.
In the current study, post-absorptive skeletal muscle protein synthesis rate was not influenced following rapamycin administration. The maintenance of basal rates of skeletal muscle protein synthesis following rapamycin administration is supported by previous research in animals [14-16,20], although a tendency for a reduced basal protein synthesis rate in rat skeletal muscle has been observed in one previous study [13]. Further, while the dose of rapamycin utilized in the current investigation (16mg, ~0.22mg·kg−1) was approximately one-third of the relative dose used in previous animal studies, the dose provided in the current study yields a blood rapamycin concentration [19] above what is necessary to achieve rapamycin-dependent effects in clinical practice [37-39]. Further, we have demonstrated that a dose similar to that utilized in the current study is capable of blocking the increase in protein synthesis in response to resistance exercise [18] or essential amino acid ingestion [19] in human skeletal muscle. Therefore, the results of the current study, in addition to those previously observed in animals, strongly suggest that post-absorptive rates of skeletal muscle protein synthesis are not affected in the immediate hours following rapamycin administration.
An additional aim of this investigation was to examine the effect of rapamycin administration on makers of autophagy (LC3B-I and LC3B-II) in human skeletal muscle to provide insight into whether rapamycin administration influences muscle protein breakdown in the basal state. This aim was premised on previous investigations demonstrating that treatment of cells with rapamycin is capable of stimulating autophagy in a variety of cell types [22-24], including C2C12 myotubes [25]. However, in the current investigation rapamycin administration had no effect on basal whole body protein breakdown rate, protein levels of LC3B-I and LC3B-II, or the ratio of LC3B-II/LC3B-I protein in skeletal muscle, collectively suggesting that rapamycin did not influence skeletal muscle autophagy or protein breakdown in the post-absorptive state. This finding is in agreement with a recent study demonstrating that autophagy in the skeletal muscle of mice under fasting conditions was largely resistant to rapamycin [40]. Collectively, these data support a hypothesis in which autophagy may be regulated through both mTORC1-dependent and mTORC1-independent processes, at least under catabolic states [41], which would include post-absorptive conditions. On the other hand, we have observed reductions in markers of autophagy in human skeletal muscle concomitant with increased mTORC1 signaling following essential amino acid ingestion [26], a known stimulus of mTORC1 signaling. The design of the current study however, focused solely on post-absorptive conditions, and therefore further research is necessary to define the mechanistic relationship between rapamycin, mTORC1 and autophagy in human skeletal muscle in response to an anabolic stimulus.
Concomitant with the inability for rapamycin administration to affect basal protein synthesis rate and autophagy in human skeletal muscle, we did not observe any changes in the phosphorylation status of mTOR at Ser2448, a marker for mTORC1 activity [42], or well-described signaling components known to be influenced by mTORC1 (Fig. 3). In addition, in a subset of CON and RAP subjects we did not observe any affect of rapamycin administration on the phosphorylation status of mTOR at Ser2481 (data not shown). These findings are in agreement with previous studies in animal models in which basal levels of mTORC1 signaling appear to be largely unaffected by rapamycin treatment [15,16]. Further, these data suggest that the inability for rapamycin administration to affect basal protein metabolism in human skeletal muscle may be related to the inability for rapamycin to inhibit basal mTORC1 signaling, which could be related to mTORC1 translocation within the cell [43] as we have previously discussed [19]. Specifically, the ability to increase mTORC1 activation and downstream signaling in response to a stimulus appears to be facilitated through mTORC1 translocation [43], and thus it is interesting to speculate that the ability for rapamycin to impair mTORC1 signaling in response to a stimulus may be facilitated through inhibiting mTORC1 translocation. This reasoning could explain the inability for rapamycin administration to impair mTORC1 signaling during post-absorptive conditions, such as those examined in the current investigation, as it is likely that mTORC1 is already specifically localized within the cell (i.e., translocation is not necessary). Certainly, further work is needed to determine the precise role (if any) of mTORC1 signaling in the regulation of post-absorptive muscle protein turnover.
In the current study, human skeletal muscle basal protein metabolism was unaltered in the immediate hours following short-term rapamycin administration. While the timeframe analyzed in the current study encompassed peak circulating levels that occur during the first two hours following rapamycin ingestion [18,19], whether basal protein metabolism is affected at a later time course remains unknown. However, in a recent investigation Goodman et al. [17] observed that 14days of rapamycin treatment did not affect plantaris muscle cross sectional area in control mice (i.e., did not undergo any change in diet or activity), suggesting that the processes governing skeletal muscle mass under “maintenance” conditions are not substantially altered with chronic rapamycin treatment. The results of the current study would support a similar outcome in human skeletal muscle. In addition, while rapamycin administration does not appear to influence skeletal muscle protein metabolism in the basal post-absorptive state, rapamycin treatment has been shown to reduce basal protein synthesis rate in cardiomyocytes [44], indicating that the mechanisms regulating basal protein metabolism may be different across muscle tissues, and even could involve indirect affects on other signaling pathways involved in translation initiation (i.e., ERK signaling pathway) [18,45]. Nevertheless, further research is required to determine the influence of chronic rapamycin administration on skeletal muscle mass and the health of other human tissues, as well as the influence of rapamycin administration on the protein turnover of other clinically relevant protein pools (e.g., albumin and other liver derived blood proteins). This research would have practical importance given that rapamycin is not only utilized as a tool for research purposes, but also as a treatment for various clinical conditions [37-39,46].
In summary, we demonstrate that short-term administration of rapamycin, at a dose capable of blocking increases in muscle protein synthesis in response to an anabolic stimulus, does not affect basal post-absorptive protein synthesis rate in human skeletal muscle. Similarly, whole body protein breakdown rate and markers of autophagy in skeletal muscle were unaltered following rapamycin administration in the post-absorptive state, indicating that basal muscle protein breakdown also appears unaffected by rapamycin. The inability of rapamycin administration to alter basal skeletal muscle protein metabolism may be related to the inability of rapamycin to substantially influence basal mTORC1 signaling. We conclude that rapamycin administration may only impair protein synthesis in human skeletal muscle when combined with a stimulus such as resistance exercise or increased amino acid availability. Further, the results from this investigation can be used as a basis in which to begin to examine the mechanism(s) that regulate basal post-absorptive protein metabolism in human skeletal muscle.
Acknowledgments
We thank all the subjects for their participation. We thank Shelley Medina, Ming-Qian Zheng, and Junfang Hao for technical assistance.
Funding
NIH/National Institute of Arthritis and Musculoskeletal and Skin Diseases grant R01 AR049877; NIH/National Institute on Aging P30 AG024832; NIH T32-HD07539; 1UL1RR029876 from the NIH/National Center for Research Resources; and H133P110012 from NIDRR/National Institute on Disability and Rehabilitation Research.
Abbreviations
- CON
Control trial
- 4E-BP1
eukaryotic initiation factor 4E binding protein 1
- FSR
fractional synthesis rate
- LC3
microtubule-associated protein 1 light chain 3
- mTORC1
mammalian target of rapamycin complex 1
- Ra
rate of appearance
- RAP
Rapamycin trial
- S6K1
ribosomal S6 kinase 1
Footnotes
Author contributions
BBR, EV, and JMD designed the research; JMD, MJD, CSF, DMG, DKW, and KLT conducted research, collected data, and reviewed the manuscript; JMD, MJD, EV, and BBR analyzed data; JMD and BBR wrote the manuscript and had primary responsibility for final content. All authors read and approved the final draft of the manuscript.
Conflict of interest
The authors report no conflicts of interest.
References
- 1.Matthews DE, Battezzati A. Regulation of protein metabolism during stress. Curr Opin Gen Surg. 1993:72–7. [PubMed] [Google Scholar]
- 2.Nair KS. Aging muscle. Am J Clin Nutr. 2005;81:953–63. doi: 10.1093/ajcn/81.5.953. [DOI] [PubMed] [Google Scholar]
- 3.Janssen I, Heymsfield SB, Ross R. Low relative skeletal muscle mass (sarcopenia) in older persons is associated with functional impairment and physical disability. J Am Geriatr Soc. 2002;50:889–96. doi: 10.1046/j.1532-5415.2002.50216.x. [DOI] [PubMed] [Google Scholar]
- 4.Burckart K, Beca S, Urban RJ, et al. Pathogenesis of muscle wasting in cancer cachexia: targeted anabolic and antic-atabolic therapies. Curr Opin Clin Nutr Metab Care. 2010;13:410–6. doi: 10.1097/MCO.0b013e328339fdd2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koopman R, Verdijk L, Manders RJ, et al. Co-ingestion of protein and leucine stimulates muscle protein synthesis rates to the same extent in young and elderly lean men. Am J Clin Nutr. 2006;84:623–32. doi: 10.1093/ajcn/84.3.623. [DOI] [PubMed] [Google Scholar]
- 6.Phillips SM, Tipton KD, Aarsland A, et al. Mixed muscle protein synthesis and breakdown after resistance exercise in humans. Am J Physiol. 1997;273:E99–107. doi: 10.1152/ajpendo.1997.273.1.E99. [DOI] [PubMed] [Google Scholar]
- 7.Short KR, Vittone JL, Bigelow ML, et al. Age and aerobic exercise training effects on whole body and muscle protein metabolism. Am J Physiol Endocrinol Metab. 2004;286:E92–101. doi: 10.1152/ajpendo.00366.2003. [DOI] [PubMed] [Google Scholar]
- 8.Fry CS, Drummond MJ, Glynn EL, et al. Aging impairs contraction-induced human skeletal muscle mTORC1 signaling and protein synthesis. Skelet Muscle. 2011;1:11. doi: 10.1186/2044-5040-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickinson JM, Rasmussen BB. Essential amino acid sensing, signaling, and transport in the regulation of human muscle protein metabolism. Curr Opin Clin Nutr Metab Care. 2011;14:83–8. doi: 10.1097/MCO.0b013e3283406f3e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suryawan A, Davis TA. Regulation of protein synthesis by amino acids in muscle of neonates. Front Biosci. 2011;16:1445–60. doi: 10.2741/3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drummond MJ, Dreyer HC, Fry CS, et al. Nutritional and contractile regulation of human skeletal muscle protein synthesis and mTORC1 signaling. J Appl Physiol. 2009;106:1374–84. doi: 10.1152/japplphysiol.91397.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bodine SC, Stitt TN, Gonzalez M, et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol. 2001;3:1014–9. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- 13.Anthony JC, Yoshizawa F, Anthony TG, et al. Leucine stimulates translation initiation in skeletal muscle of post-absorptive rats via a rapamycin-sensitive pathway. J Nutr. 2000;130:2413–9. doi: 10.1093/jn/130.10.2413. [DOI] [PubMed] [Google Scholar]
- 14.Kubica N, Bolster DR, Farrell PA, et al. Resistance exercise increases muscle protein synthesis and translation of eukaryotic initiation factor 2Bepsilon mRNA in a mammalian target of rapamycin-dependent manner. J Biol Chem. 2005;280:7570–80. doi: 10.1074/jbc.M413732200. [DOI] [PubMed] [Google Scholar]
- 15.Suryawan A, Jeyapalan AS, Orellana RA, et al. Leucine stimulates protein synthesis in skeletal muscle of neonatal pigs by enhancing mTORC1 activation. Am J Physiol Endocrinol Metab. 2008;295:E868–75. doi: 10.1152/ajpendo.90314.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimball SR, Jefferson LS, Nguyen HV, et al. Feeding stimulates protein synthesis in muscle and liver of neonatal pigs through an mTOR-dependent process. Am J Physiol Endocrinol Metab. 2000;279:E1080–7. doi: 10.1152/ajpendo.2000.279.5.E1080. [DOI] [PubMed] [Google Scholar]
- 17.Goodman CA, Frey JW, Mabrey DM, et al. The role of skeletal muscle mTOR in the regulation of mechanical load-induced growth. J Physiol. 2011;589:5485–501. doi: 10.1113/jphysiol.2011.218255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drummond MJ, Fry CS, Glynn EL, et al. Rapamycin administration in humans blocks the contraction-induced increase in skeletal muscle protein synthesis. J Physiol. 2009;587:1535–46. doi: 10.1113/jphysiol.2008.163816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dickinson JM, Fry CS, Drummond MJ, et al. Mammalian target of rapamycin complex 1 activation is required for the stimulation of human skeletal muscle protein synthesis by essential amino acids. J Nutr. 2011;141:856–62. doi: 10.3945/jn.111.139485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grzelkowska K, Dardevet D, Balage M, et al. Involvement of the rapamycin-sensitive pathway in the insulin regulation of muscle protein synthesis in streptozotocin-diabetic rats. J Endocrinol. 1999;160:137–45. doi: 10.1677/joe.0.1600137. [DOI] [PubMed] [Google Scholar]
- 21.Dunn WA., Jr Autophagy and related mechanisms of lyso-some-mediated protein degradation. Trends Cell Biol. 1994;4:139–43. doi: 10.1016/0962-8924(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 22.Nyfeler B, Bergman P, Triantafellow E, et al. Relieving autophagy and 4EBP1 from rapamycin resistance. Mol Cell Biol. 2011;31:2867–76. doi: 10.1128/MCB.05430-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi KC, Kim SH, Ha JY, et al. A novel mTOR activating protein protects dopamine neurons against oxidative stress by repressing autophagy related cell death. J Neurochem. 2010;112:366–76. doi: 10.1111/j.1471-4159.2009.06463.x. [DOI] [PubMed] [Google Scholar]
- 24.Blommaart EF, Luiken JJ, Blommaart PJ, et al. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J Biol Chem. 1995;270:2320–6. doi: 10.1074/jbc.270.5.2320. [DOI] [PubMed] [Google Scholar]
- 25.Mordier S, Deval C, Bechet D, et al. Leucine limitation induces autophagy and activation of lysosome-dependent proteolysis in C2C12 myotubes through a mammalian target of rapamycin-independent signaling pathway. J Biol Chem. 2000;275:29900–6. doi: 10.1074/jbc.M003633200. [DOI] [PubMed] [Google Scholar]
- 26.Glynn EL, Fry CS, Drummond MJ, et al. Excess leucine intake enhances muscle anabolic signaling but not net protein anabolism in young men and women. J Nutr. 2010;140:1970–6. doi: 10.3945/jn.110.127647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bergstrom J. Muscle electrolytes in man. Scand J Med Sci Sports. 1962;68:1–110. [Google Scholar]
- 28.Dreyer HC, Drummond MJ, Pennings B, et al. Leucine-enriched essential amino acid and carbohydrate ingestion following resistance exercise enhances mTOR signaling and protein synthesis in human muscle. Am J Physiol Endocrinol Metab. 2008;294:E392–400. doi: 10.1152/ajpendo.00582.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Volpi E, Chinkes DL, Rasmussen BB. Sequential muscle biopsies during a 6-h tracer infusion do not affect human mixed muscle protein synthesis and muscle phenylalanine kinetics. Am J Physiol Endocrinol Metab. 2008;295:E959–63. doi: 10.1152/ajpendo.00671.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dreyer HC, Fujita S, Cadenas JG, et al. Resistance exercise increases AMPK activity and reduces 4E-BP1 phosphorylation and protein synthesis in human skeletal muscle. J Physiol. 2006;576:613–24. doi: 10.1113/jphysiol.2006.113175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calder AG, Anderson SE, Grant I, et al. The determination of low d5-phenylalanine enrichment (0.002-0.09 atom percent excess), after conversion to phenylethylamine, in relation to protein turnover studies by gas chromatography/electron ionization mass spectrometry. Rapid Commun Mass Spectrom. 1992;6:421–4. doi: 10.1002/rcm.1290060704. [DOI] [PubMed] [Google Scholar]
- 32.Patterson BW, Zhang XJ, Chen Y, et al. Measurement of very low stable isotope enrichments by gas chromatography/mass spectrometry: application to measurement of muscle protein synthesis. Metabolism. 1997;46:943–8. doi: 10.1016/s0026-0495(97)90084-6. [DOI] [PubMed] [Google Scholar]
- 33.Wolfe RR, Chinkes DL. Isotopic tracers in metabolic research: principles and practice of kinetic analysis. 2. Hoboken: John Wiley & Sons, Inc.; 2005. [Google Scholar]
- 34.Kabeya Y, Mizushima N, Ueno T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizushima N, Yoshimori T. How to interpret LC3 immuno-blotting. Autophagy. 2007;3:542–5. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 36.Klionsky DJ, Abeliovich H, Agostinis P, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–75. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paghdal KV, Schwartz RA. Sirolimus (rapamycin): from the soil of Easter Island to a bright future. J Am Acad Dermatol. 2007;57:1046–50. doi: 10.1016/j.jaad.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 38.Aranda-Dios A, Lage E, Sobrino JM, et al. Sirolimus experience in heart transplantation. Transplant Proc. 2006;38:2547–9. doi: 10.1016/j.transproceed.2006.08.075. [DOI] [PubMed] [Google Scholar]
- 39.Stallone G, Schena A, Infante B, et al. Sirolimus for Kaposi’s sarcoma in renal-transplant recipients. N Engl J Med. 2005;352:1317–23. doi: 10.1056/NEJMoa042831. [DOI] [PubMed] [Google Scholar]
- 40.Mammucari C, Milan G, Romanello V, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–71. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 41.Zhao J, Brault JJ, Schild A, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–83. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 42.Chiang GG, Abraham RT. Phosphorylation of mammalian target of rapamycin (mTOR) at Ser-2448 is mediated by p70S6 kinase. J Biol Chem. 2005;280:25485–90. doi: 10.1074/jbc.M501707200. [DOI] [PubMed] [Google Scholar]
- 43.Sancak Y, Peterson TR, Shaul YD, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang BP, Wang Y, Wang X, et al. Blocking eukaryotic initiation factor 4F complex formation does not inhibit the mTORC1-dependent activation of protein synthesis in cardiomyocytes. Am J Physiol Heart Circ Physiol. 2009;296:H505–14. doi: 10.1152/ajpheart.01105.2008. [DOI] [PubMed] [Google Scholar]
- 45.Carracedo A, Ma L, Teruya-Feldstein J, et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest. 2008;118:3065–74. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webster AC, Lee VW, Chapman JR, et al. Target of rapamycin inhibitors (sirolimus and everolimus) for primary immuno-suppression of kidney transplant recipients: a systematic review and meta-analysis of randomized trials. Transplantation. 2006;81:1234–48. doi: 10.1097/01.tp.0000219703.39149.85. [DOI] [PubMed] [Google Scholar]


