Abstract
MicroRNAs (miRNAs) are an abundant class of small non-coding RNAs (ncRNAs) that function to regulate gene expression at the post-transcriptional level. Although their functions were originally described during normal development, miRNAs have emerged as integral components of the oncogenic and tumor suppressor network, regulating nearly all cellular processes altered during tumor formation. In particular, mir-17-92, a miRNA polycistron also known as oncomir-1, is among the most potent oncogenic miRNAs. Genomic amplification and elevated expression of mir-17-92 were both found in several human B-cell lymphomas, and its enforced expression exhibits strong tumorigenic activity in multiple mouse tumor models. mir-17-92 carries out pleiotropic functions during both normal development and malignant transformation, as it acts to promote proliferation, inhibit differentiation, increase angiogenesis and sustain cell survival. Unlike most protein coding genes, mir-17-92 is a polycistronic miRNA cluster that contains multiple miRNA components, each of which has a potential to regulate hundreds of target mRNAs. This unique gene structure of mir-17-92 may underlie the molecular basis for its pleiotropic functions in a cell type and context dependent manner. Here we review the recent literature on the functional studies of mir-17-92, and highlight its potential impacts on the oncogene network. These findings on mir-17-92 indicate that miRNAs, together with protein coding genes, are integrated components of the molecular pathways that regulate tumor development and tumor maintenance.
Keywords: miRNAs, cancer, mir-17-92, oncomir-1
1-Introduction
1-1 miRNAs in post-transcriptional silencing
Data from comparative genomic studies have long implicated the functional importance of the non-protein coding regions of the genome, whose proportion increases as a function of genomic complexity (Mattick and Makunin, 2006). Extensive transcriptions have been observed from many of these regions, giving rise to numerous non-coding RNAs (ncRNAs) that regulate diverse developmental and physiological processes (Kapranov et al., 2007). We are only beginning to understand the realm of ncRNA funcitons, yet preliminary studies so far have revealed unexpected complexity and richness of their expression patterns, genomic structures and biological activities. In recent years, microRNAs (miRNAs) have emerged as a class of novel, small ncRNAs with potent capacity for gene regulation at the post-transcriptional level (Zamore and Haley, 2005, He and Hannon, 2004, Bartel, 2009, Ambros, 2004). First identified as novel ncRNAs essential for larval developmental timing in C. elegans (Wightman et al., 1993, Lee et al., 1993), thousands of such small RNAs have been and are still being discovered in almost all organisms, ranging from viruses and single-celled green algae Chlamydomonas (Zhao et al., 2007, Molnar et al., 2007) to complex mammalian species (Lee and Ambros, 2001, Lau et al., 2001, Lagos-Quintana et al., 2001). Upon maturation, miRNAs recognize specific mRNA targets through imperfect sequence complementarity, and largely dampen their expression at the post-transcriptional level. Given the ability of miRNAs to downregulate many mRNA targets, this level of gene regulation may provide robustness in a diverse range of biological processes, due to the simultaneous regulation of many components of the signaling network.
Genes encoding miRNAs are often transcribed into precursor transcripts (pri-miRNAs) that contain a stem loop hairpin structure(s) (Lee et al., 2002). Pri-miRNAs are sequentially processed by two RNaseIII enzymes, Drosha and Dicer, to yield mature miRNA duplexes ranging from 18 to 24 nucleotide in length (Lee et al., 2003, Hutvagner and Zamore, 2002). One strand of the mature duplex is then incorporated into the effector complex, RISC, to mediate the post-transcriptional silencing of specific mRNAs (Hutvagner and Zamore, 2002, Mourelatos et al., 2002). The recognition of the miRNA targets often involves imperfect base pairing (Wightman et al., 1993, Lee et al., 1993), which allows miRNA to potentially regulate a large number of protein coding genes (Lewis et al., 2003, Lewis et al., 2005, Krek et al., 2005, Grimson et al., 2007). Interestingly, multiple miRNAs can be produced within a single pri-miRNA transcript, each of which can act independently. This polycistronic structure of miRNA cluster genes sets them apart from most protein coding genes, and bestows them with a unique capacity and specificity for gene regulation in the complex molecular network for development and disease.
Despite the small size of the miRNA molecules, they exhibit enormous capacity for gene regulation, mostly due to the nature of imperfect base pairing required for target recognition (Lewis et al., 2003, Lewis et al., 2005, Krek et al., 2005, Grimson et al., 2007). Therefore, each miRNA is theoretically capable of regulating hundreds of mRNAs within a cell type and in a context dependent manner. Initially, a small number of miRNA targets were identified based on classic invertebrate genetics studies (Johnston and Hobert, 2003, Brennecke et al., 2003, Wightman et al., 1993, Lee et al., 1993). Examination of these validated target sites combined with biochemical studies revealed that the 5’ end of a miRNA, designated as the “seed” sequence, plays a critical role in target recognition and post-transcriptional repression (Lewis et al., 2005, Grimson et al., 2007, Doench and Sharp, 2004). The miRNA seed complementarity was employed to develop computational approaches for target prediction, and was later demonstrated experimentally as the major molecular basis for miRNA target recognition in cell culture systems (Selbach et al., 2008, Baek et al., 2008). Exceptions to this generalization still exist (Didiano and Hobert, 2006, Wightman et al., 1993). For example, in C. Elegans, a 6- to 8-base-pair perfect seed pairing is not always a reliable predictor for the lsy-6 miRNA interaction with its targets, because it has been shown that G.U base pairing is tolerated in the 'seed' region. It is likely that a variety of mechanisms may act together to regulate the specificity of miRNA:target interaction (Didiano and Hobert, 2006).
The exact molecular basis underlying miRNA mediated gene silencing is not entirely clear. Biochemical studies using different reporter systems and endogenous miRNAs have yielded several distinct models, possibly reflecting the limitation of the in vitro reporter systems tested and/or the complex nature of miRNA mediated gene silencing (Liu et al., 2005, Kiriakidou et al., 2007, Eulalio et al., 2008, Chendrimada et al., 2007, Bhattacharyya et al., 2006). In all these models, miRNAs, upon maturation, are incorporated into the RISC (Hutvagner and Zamore, 2002), which mediates degradation of specific mRNAs (Eulalio et al., 2008, Giraldez et al., 2006) and/or represses their translation (Bagga et al., 2005, Chendrimada et al., 2007, Kiriakidou et al., 2007, Pillai et al., 2005). The key components of the RISC are the Argonaute (Ago) proteins, which, through their interactions with the P-body components, mediate the formation of P-bodies that contain miRNAs and their targets (Eulalio et al., 2008, Liu et al., 2005, Sen and Blau, 2005). Within the P-body, miRNA targets are sequestered from the translational machineries and are subjected to mRNA degradation, at least partially, through deadenylation (Giraldez et al., 2006). In several other studies, Ago proteins are suggested to mediate translational repression through various mechanisms (Filipowicz et al., 2008). Although there are debates about the exact mechanisms of the miRNA mediated repression, these findings implicated that both mRNA destabilization and translational repression contribute to miRNA mediated gene silencing. This is consistent with the recent findings using genome wide quantitative mass spectrometry to characterize the impact of miRNAs on the proteome in a cell culture system (Baek et al., 2008, Selbach et al., 2008). It is likely that multiple molecular pathways, either individually or collectively, mediate the biological effects of miRNAs in a context dependent manner.
1-2 miRNAs in cancer biology
miRNAs were initially studied as key regulators for normal development, as mutations, either in the founding members of the miRNA family or in the key components of the miRNA biogenesis pathway, give rise to pronounced developmental defects in nearly all model organisms. Even though all miRNAs share significant similarities in their gene structures, biogenesis and effector machineries, their expression patterns and biological functions vary tremendously. This unique mechanism of posttranscriptional gene silencing, along with transcription regulation, translation control and post-translation modification, are major gene regulatory mechanisms in diverse developmental and physiological processes.
Shortly after the miRNA identification in mammalian species, it was recognized that miRNAs may not solely regulate developmental processes. Initial studies indicated that genomic alterations and expression dysregulation of miRNAs are frequently associated with various human cancers (Calin et al., 2004a, Calin et al., 2004b, Iorio et al., 2005, Lu et al., 2005). These early findings prompted a number of subsequent studies to explore the functional connections between miRNAs and malignant transformation in a variety of cancer types. Two major approaches were undertaken. One approach aimed to identify candidate oncogenic and tumor suppressor miRNAs for the transformation of a specific cell type. This often involved performing expression studies to compare miRNA profiles between tumor and normal tissues. Under this rationale, mir-17-92 has been identified as a potential oncogene, due to its genomic amplification and elevated expression found in multiple hematopoietic malignancies, including diffuse large B-cell lymphomas (DLBCLs), mantle cell lymphomas and Burkitt’s lymphomas (Hayashita et al., 2005). Previous cancer genomic studies on DNA copy number alteration and insertional mutagenesis screens in mice also provide insights into candidate oncogenic and tumor suppressor miRNAs that were not fully recognized before. For example, the oncogenic potential for mir-155 and mir-106a-92 in lymphomagenesis were identified through this approach (Kluiver et al., 2005, Uren et al., 2008). The other rationale aims to explore the molecular crosstalk between miRNAs and well-characterized cancer pathways. From expression studies, miRNA functional screens, and computational analyses, a handful of candidate miRNAs have emerged as key components of the oncogenic and tumor suppressor network. For example, it is through extensive expression studies that miR-34 miRNAs were first identified as p53 transcriptional targets to mediate its tumor suppressor effects (Raver-Shapira et al., 2007, He et al., 2007, Chang et al., 2007); mir-372 and mir-373 were identified as potential oncogenes for testicular tumorigenesis out of a functional screen using a miRNA over-expression library (Voorhoeve et al., 2006); and finally bioinformatic prediction of let-7 miRNA targets lead us to explore its roles in post-transcriptional repression on key oncogenes Ras and HMGA2 (Johnson et al., 2005, Park et al., 2007, Mayr et al., 2007). Altogether, these lines of investigation lead to a surge of interest in the functional studies of miRNAs in the oncogenic and tumor suppressor network, a topic that has been largely unexplored until recently.
2 mir-17-92, a polycistronic miRNA oncogene
2-1 The unique gene structure of mir-17-92
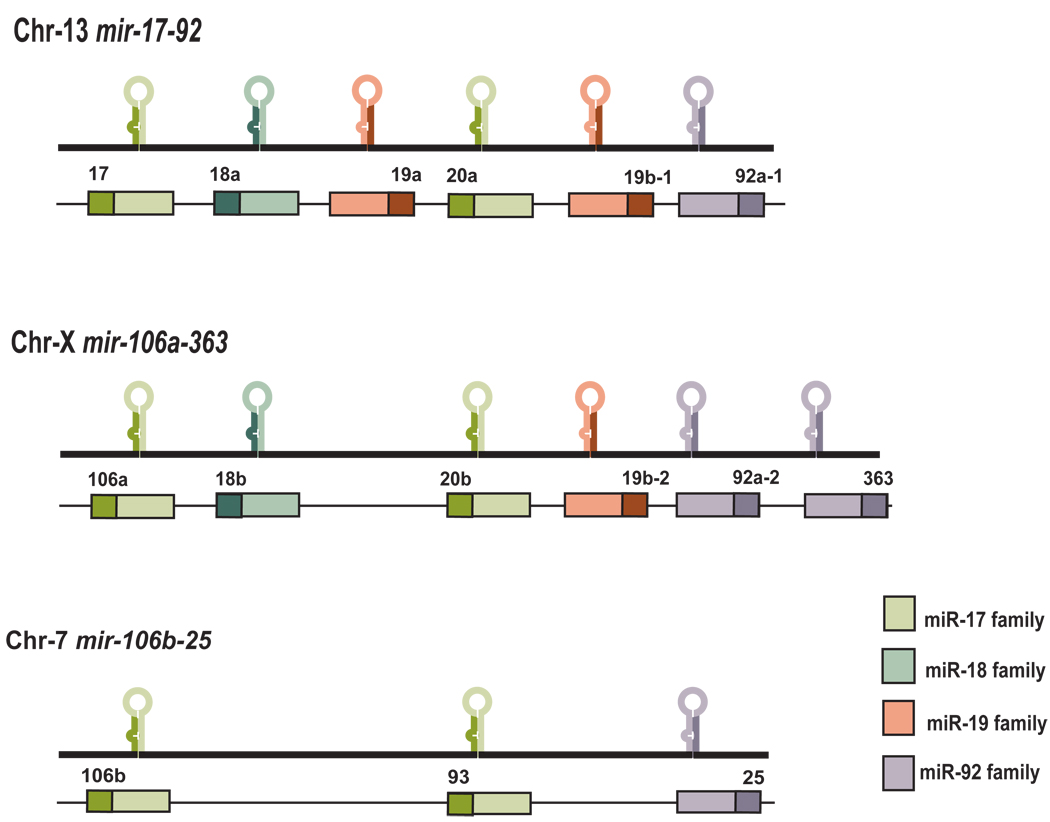
One of the best-characterized oncogenic miRNAs is mir-17-92, a polycistronic miRNA cluster also designated as oncomir-1 (He et al., 2005). The precursor transcript derived from the mir-17-92 gene contains six tandem stem-loop hairpin structures that ultimately yield six mature miRNAs, miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, miR-92-1 (Tanzer and Stadler, 2004). Recent high-throughput parallel sequencing efforts also discovered the rare species of miRNAs* within this cluster, which are the complementary miRNAs derived from the opposite arms of each mir-17-92 pre-miRNA. The six miRNAs encoded by mir-17-92 can be categorized into three separate miRNA families according to their seed sequences — the miR-17 family (including miR-17, miR-20 and miR-18), the miR-19 family (miR-19a and miR-19b) and the miR-92 family (Fig.1). It is worth noting that miR-18 exhibits a significant sequence homology with miR-17 and miR-20, despite one nucleotide difference within the “seed” regions. Ancient gene duplications have given rise to two mir-17-92 cluster paralogs in mammals — mir106a-363 and mir106b-25, each of which only contains homologous miRNAs to a subset of mir-17-92 components (Fig.1). Interestingly, not only the sequence of each miRNA component is highly conserved across species among all three paralogs, the organization of these miRNAs within mir-17-92 family also exhibits high level of conservation. The functional significance of this conservation is still unclear. It possibly reflects the evolutionary path through which gene duplications followed by subsequent loss of individual miRNA component(s) shaped the formation of this polycistronic miRNA family.
Fig1. Gene structure of human mir-17-92 and its homologues.
Primary transcript organization of the human mir-17-92 and its paralogs, mir106a-363 and mir106b-25 clusters. The miRNAs encoded by the 3 clusters can be categorized into four separate miRNA families according to their seed sequences, including the miR-17 family (miR-17, miR-20a, miR-20b, miR-106a, miR-106b and miR-93), the miR-18 family (miR-18a and miR-18b), the miR-19 family (miR-19a, miR-19b-1 and miR-19b-2) and the miR-92 family (miR-92a-1, miR-92a-2, miR-383 and miR-25).
Human mir-17-92 is located at 13q31.3, a region amplified in several hematopoietic malignancies and solid tumors, including Diffuse B-Cell Lymphomas (DLBCLs), follicular lymphomas, Burkitt’s lymphomas and lung carcinoma (Ota et al., 2004). The minimal 13q31 amplicon characterized contains the mir-17-92 precursor, whose elevated expression invariably correlates with the presence of the 13q31 amplicon (Ota et al., 2004, He et al., 2005). Over-expression of mir-17-92 has been observed in multiple tumor types (Volinia et al., 2006, Petrocca et al., 2008a, He et al., 2005), yet the amplitude and frequency of its over-expression vary depending on the specific tumor type. For example, the Golub groups have examined the miRNA expression profiles in 334 tumor samples across multiple major cancer types, and the mir-17-92 is predominantly over-expressed in the hematopoietic malignancies (Lu et al., 2005). Consistent with these findings, the murine mir-17-92 and mir106a-363 loci are common insertion sites for insertional mutagenesis screen in retrovirally induced erythroleukemia and T-cell leukemia further indicating the oncogenic potentials of this miRNA family (Uren et al., 2008, Cui et al., 2007, Wang et al., 2006, Landais et al., 2007)
2-2 mir-17-92 as a potential oncogene
One of the first functional evidence to support the oncogenic activity of mir-17-92 came from an in vivo mouse B-cell lymphoma model, in which enforced expression of mir17-19b, a truncated mir-17-92 lacking mir-92, collaborated with c-myc oncogene to accelerate B lymphomagenesis (He et al., 2005). In this model, c-myc is driven by the immunoglobulin heavy chain enhancer (Eμ) as a transgene. Elevated c-myc expression in the B-cell lineage promotes the formation of B-cell lymphomas with a relatively long latency. When mir17-19b is over-expressed in this model, it represses c-myc induced apoptosis, shortens the latency for tumor onset, and gives rise to highly invasive B-cell lymphomas. Interestingly, the over-expression of mir17-19b not only promotes the oncogenesis of c-myc expressing B-cells, but also alters the cell fate of the transformed B-cells. The majority of B-cell lymphomas derived from the Eμ-myc transgene are more mature B-cells. Yet B-cell lymphomas resulted from the collaboration between mir17-19b and c-myc are mostly of progenitor B-cell origin, implicating a possible role of mir-17-92 in B-cell development. This hypothesis was later validated by mouse genetic studies (Ventura et al., 2008). Deficiency of mir-17-92 indeed affects B-cell development, particularly in the pro-B to pre-B transition, due to enhanced apoptosis occurring in the pro-B cells during both fetal and adult B-cell development (Ventura et al., 2008).
Following this initial observation, the effects of mir-17-92 over-expression have been examined in multiple animal models, human cancers, and cell culture systems, for its abilities to regulate a number of cellular processes that favor malignant transformation. These studies altogether revealed the pleiotropic functions of mir-17-92, during both normal development and malignant transformation, to promote proliferation, inhibit differentiation, augment angiogenesis and sustain cell survival. It is worth noting that the exact functional readouts of mir-17-92 in a given biological system are largely dependent on the cell type as well as the developmental contexts. For example, mir-17-92 represses c-myc induced apoptosis in progenitor B-cells (Tagawa and Seto, 2005, He et al., 2005), and similarly, it inhibits cell death during pro-B to pre-B cell transition in normal development (Ventura et al., 2008). In contrast, mir-17-92 enhances the proliferation of normal lung epithelial progenitor cells and malignant lung cancer cells, while inhibiting their differentiation (Hayashita et al., 2005, Lu et al., 2005). Interestingly, mir-17-92 not only exhibits cell autonomous effects, but also performs cell non-autonomous functions. mir-17-92 in endothelial cells augments angiogenesis during normal development (Suarez et al., 2008), yet mir-17-92 up-regulation in cancer cells also promotes angiogenesis during tumor growth in a xenograft model (Dews et al., 2006).
Deletion of mir-17-92 results in multiple development defects, including lung hypoplasia and ventricular septal defect. In particular, both fetal and adult B-cell development is significantly impaired due to the enhanced pro-B apoptosis during pro-B to pre-B transition (Ventura et al., 2008). Other hematopoietic lineages, including erythrocytes, granulocytes, monocytes, and T-cells, seem to be unaffected by the deletion of mir-17-92 in the initial phenotypical characterization of the knockout mice. Interestingly, when Dicer was deleted specifically in B-cells, which abolished the entire miRNA biogenesis in the B-cell compartment, the resulting phenotype resembled that of mir-17-92 knockout mice to a certain degree. There was an accumulation of pro-B and a decrease of pre- and mature B cells (Koralov et al., 2008). Therefore, at least a portion of the Dicer phenotype could be attributed to the deficiency of mir-17-92. These observations are in line with findings from transgenic studies, where enforced expression of mir-17-92 in the lymphoid lineages promotes their proliferation and dampens the activation-induced cell death (Xiao et al., 2008). These mice develop lymphoproliferative disease and autoimmunity, suggesting that the pathological amplification of mir-17-92 can lead to an expansion of cells harboring this lesion, subsequently increasing the probability of oncogenic mutations that eventually lead to malignancy.
2-3 The pleiotropic functions of mir-17-92
mir-17-92 is widely expressed in many different cell types. Its diverse functionality may result from the different target mRNAs subjected to mir-17-92 post-transcriptional silencing in different cell types and/or different developmental or physiological contexts. Yet the underlying molecular basis for its pleiotropic effects is still unclear. It is likely that the multiple miRNA components located within the mir-17-92 cluster may contribute to its functional complexity. Theoretically, the six mature miRNAs will act independently yet coordinately once the mir-17-92 pri-miRNA is cleaved by Drosha. This unique structural feature of the mir-17-92 cluster confers enormous potential and plasticity for gene regulation, as different target mRNAs may be targeted by a combination of mir-17-92 components, varying in the degree of binding affinity. Thus, multiple miRNA components within mir-17-92 can offer a wide dynamic range for miRNA-mRNA interactions, and consequently may result in diverse biological outputs in different cell types under different developmental and/or physiological contexts.
The exact biological activities of each mir-17-92 component have not been functionally dissected. However, mouse knockout studies have implicated that these mir-17-92 components may perform specific yet overlapping functions, arguing against the model where each miRNA component contributes equally to the overall biological functions of mir-17-92. Ventura et al. have knocked out the mir-17-92 cluster and its paralogous clusters, mir106a-363 and mir106b-25, both individually and in combination. Since these three miRNA clusters share extensive homology within a subset of their miRNA components, their different knockout phenotypes implicate the different functions of the mir-17-92 components.
It is very likely that mir-106a-363 is not expressed during embryonic development, so it is not surprising that deletion of mir106a-363 does not yield significant defects in embryonic development (Ventura et al., 2008). It is surprising, however, that deletion of the widely expressed mir106b-25 cluster alone or in combination with mir-106a-363 does not result in obvious phenotype, but the deficiency of mir-17-92 gives rise to defective embryonic development and postnatal death (Ventura et al., 2008). Double knockout mice with deficiency in both mir-17-92 and mir106b-25 yield quantitatively more severe phenotypes as well as a few novel defects, implicating the existence of a possible redundancy between mir-17-92 and mir-106b-25. mir-17-92 and mir106b-25 share extensive sequence homology within a subset of miRNA components, including miR-17/20/106b/93 and miR-92/25. However, miR-18 and miR-19 family miRNAs, while present in mir-17-92, are absent in mir106b-25. Despite the phenotypical differences between mir-17-92 knockout and mir106b-25 knockout, they exhibit similar embryonic expression patterns (Ventura et al., 2008). Thus, it is tempting to hypothesize that mir-17-92 knockout defects revealed the unique biological functions for miR-18 and miR-19 family miRNAs. Surprisingly, the double knockout mice for mir-17-92 and mir-106b-25, which presumably removed functional redundancy for miR-17, miR-20 and miR-92 during embryonic development, not only yield quantitatively different phenotypes with an increase in severity, but also give rise to a few qualitatively different defects. These findings raised an intriguing hypothesis that each of the miRNA components within mir-17-92 may perform specific functions, although a certain degree of functional overlap and cooperativity may exist among the different miRNA components.
We recently reported the functional dissection of the individual miRNA components of mir-17-92 by assaying their tumorigenic potential in vivo. Using the Eμ-myc mouse B-cell lymphoma model, we identified miR-19 as the key oncogenic component of mir-17-92. miR-19 is both necessary and sufficient for promoting c-myc induced lymphomagenesis through repression of apoptosis (Olive et. al., unpublished results). Therefore, mir-17-92 functions as a polycistronic miRNA cluster, whose miRNA components regulate gene expression either individually or in combination in a coordinated rather than an additive manner. However, more complex mechanism of coordination may also exist. Shan et al. indicate in a recent study that miR-17 overexpression in mice decreases cell proliferation, adhesion and migration, raising a possibility that components of mir-17-92 can both positively and negatively regulate the same cellular process to achieve homeostasis in vivo (Shan et al., 2009).
2-4 The transcription and biogenesis of mir-17-92
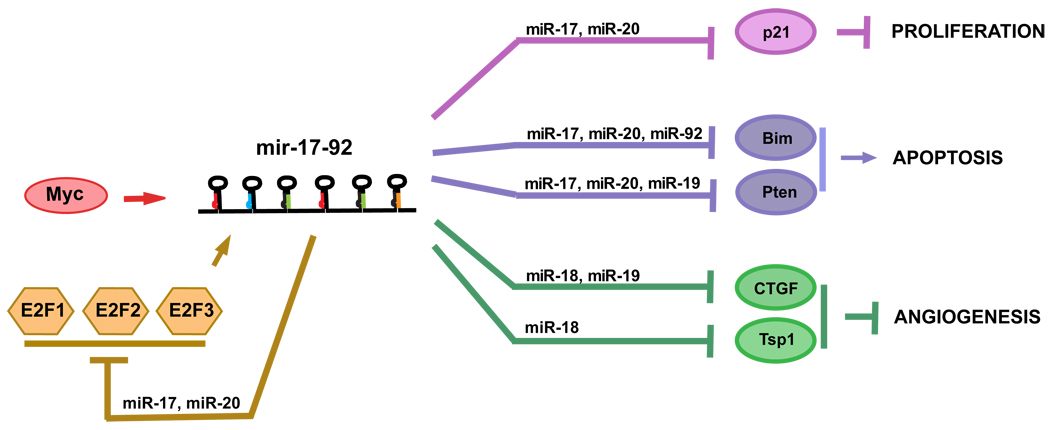
Given the functional specificity of different mir-17-92 components, it is likely that this miRNA polycistronic cluster is subjected to sophisticated transcriptional regulation. Due to the difficulty of defining the promoter region of ncRNAs in general, we are only beginning to reveal the transcriptional regulation for mir-17-92 through the combined approach of expression screens and bioinformatics approaches. O’Donnel et al. have documented that the transcription of mir-17-92 is directly activated by the c-myc oncogene (Fig.2) (O'Donnell et al., 2005), and not surprisingly, N-myc has also been shown to transcriptionally activate mir-17-92 (Schulte et al., 2008). These findings are consistent with the functions of mir-17-92 in promoting proliferation in a variety of cell types, including cells of lymphoid, epithelial, and neural origin. In addition to myc, E2F1 and E2F3 directly activate the transcription of mir-17-92, and chromatin immunoprecipitation demonstrates that E2F3 is the major E2F family member that binds to the mir-17-92 promoter region (Sylvestre et al., 2007, Woods et al., 2007). E2Fs are essential for the progression of the cell cycle, as they activate a large number of S phase genes, including thymidine kinase, DNA polymerase, and Cyclins E and A. As a result, cycling cells are likely to have elevated mir-17-92 due to the periodic burst of E2F activity during S phase, while quiescent cells may have reduced mir-17-92 levels. This is consistent with the high level of mir-17-92 expression in transformed cell lines, but not in the primary cell culture or fully differentiated cell types, suggesting mir-17-92 a key component of the complex regulatory signaling network for cell proliferation. Interestingly, recent studies also indicate mir-17-92 as a novel target for p53 mediated gene repression, most apparent in cells under hypoxia. Given the strong effect of mir-17-92 to promote cell survival, it is likely that repression of mir-17-92 by p53 gives rise to apoptosis under hypoxia treatment (Yan et al., 2009).
Fig2. The pleiotropic functions of mir-17-92 achieved by repressing specific targets.
Depending on both cell type and physiological context, mir-17-92 can promote proliferation, increase angiogenesis and sustain cell survival through the post-transcriptional repression of a number of target mRNAs.
mir-17-92 is not only regulated by a network of transcriptional machineries, its biogenesis is also subjected to intricate regulation through a largely unknown mechanism. We are only beginning to appreciate this novel regulation on the miRNA pathway, the best example of which is the repression of let-7 biogenesis by LIN-28 at both Drosha and Dicer cleavage (Heo et al., 2008, Newman et al., 2008, Viswanathan et al., 2008). It is recently identified that VEGF mediated up-regulation of mir-17-92 only elevates the expression level of three miRNA components of the entire cluster, miR-17, miR-18 and miR-20, to participate in the control of angiogenic phenotypes, suggesting a selective mir-17-92 biogenesis under given biological contexts (Suarez et al., 2008). Recent studies have revealed the RNA binding protein hnRNPA1 is specifically required for processing of miR-18 establishing a potential connection between the signaling transduction and specific miRNA biogenesis (Guil and Caceres, 2007). The specific processing of individual miRNAs within a polycistronic miRNA cluster leads to a new level of complexity in the regulation of miRNA expression and miRNA function in different cell types and under different contexts. It is also plausible that post-transcriptional silencing mediated by each miRNA component within mir-17-92 may also be regulated in a cell type and context dependent manner.
2-5 Target identification of mir-17-92
One of the least understood questions about mir-17-92 is the identity of its targets. Theoretically, each miRNA component within mir-17-92 has the potential to regulate hundreds of specific mRNAs, as miRNA target recognition largely depends on sequence complementarities at the 6 nucleotide seed regions (Lewis et al., 2003, Lewis et al., 2005). The post-transcriptional repression mediated by a specific miRNA often leads to moderate down-regulation of a large number of target mRNAs, which has been experimentally validated in two independent over-expression studies (Selbach et al., 2008, Baek et al., 2008). It is clear that mir-17-92 mediated gene silencing plays important roles in vivo given its knockout phenotypes. The current model in the field would predict that mir-17-92 gives rise to a moderate down-regulation of a large number of mRNAs in each cell type, which collectively mediates its biological functions. Yet it is still unclear if the physiological level of mir-17-92 indeed represses a large number of target mRNAs in a given cell type, and if mir-17-92 always leads to a moderate level of down-regulation of the key functional target mRNAs. The answer to these questions may be dependent on specific cellular context, and would require the disruption of the binding between mir-17-92 and its specific target mRNA(s) in vivo. Using morpholino oligos (Choi et al., 2007) and targeted deletions (Dorsett et al., 2008), several groups have started to examine the biological effects of a specific miRNA-mRNA interaction. Such an approach is likely to resolve the above questions and to dissect the contribution of each target mRNA to the biological functions of mir-17-92.
Using bioinformatic predictions, western analysis, and reporter assays, recent studies have identified a number of mir-17-92 targets, each of which is proposed to contribute to a specific functional readout of mir-17-92 (Fig.2). For example, the anti-apoptotic effects of mir-17-92 can be at least partially explained by its repression of Bim and PTEN, whose dysregulation may contribute to the increased apoptosis during pro-B to pre-B transition in mir-17-92 deficient animals and the aberrant cell survival in the lymphoid lineages of the mir-17-92 transgenic animals (Fig.2) (Xiao et al., 2008, Ventura et al., 2008, Koralov et al., 2008). It is interesting that E2F1 is also a target of miR-17 and miR-20 and that E2F2 and E2F3 are targets of miR-20 (O'Donnell et al., 2005). Since E2F directly activates transcription of mir-17-92 (Woods et al., 2007), these data suggest an auto-regulatory feedback loop between E2F factors and this miRNA cluster. This feedback possibly acts to maintain a homeostasis that dampens the apoptotic potential of E2F1 following a surge of proliferative signals. The proliferation effects of mir-17-92, on the other hand, can be partially attributed to its ability to repress the cyclin dependent kinase inhibitor CDKN1A (p21), a negative regulator of the G1-S checkpoint cell cycle progression. This repression is mostly achieved by miR-17 and the related miR-20 (Petrocca et al., 2008b, Ivanovska et al., 2008). Consistently, over-expression or inhibition of the miR-17 family members is sufficient to promote or delay the entry of cells into S phase, respectively (Ivanovska 2008). The angiogenic activity of mir-17-92 in tumor cells reflects its cell non-autonomous function. This effect of mir-17-92, at least in part, seems to be mediated by the direct repression on thrombospondin-1 (Tsp1) and connective tissue growth factor (CTGF) (Dews et al., 2006) (Fig.2). Altogether, these findings indicate the cell type specificity of mir-17-92 targets.
2-6 Remaining questions
Although a majority of the literature characterized the oncogenic potential for mir-17-92 from different angles, loss of heterogeneity at the 13q31.3 locus that harbors human mir-17-92 has been reported in a few tumor types (Zhang et al., 2007), including a small percentage of ovarian and breast cancers and melanomas. Enforced expression of miR-17 in breast cancer cell lines induces a reduction in proliferation of the cancer cells (Hossain et al., 2006), possibly through its repression on AIB1 (amplified in breast cancer 1), a transcriptional co-activator of E2F1 and of the estrogen receptor. These puzzling observations may reflect the functional complexity of mir-17-92 in a context dependent and cell type dependent manner, and more experimental investigations are needed to fully explore the functional complexity of mir-17-92.
3. Summary
Recent studies have revealed the unexpected abundance and complexity of ncRNAs transcribed in the mammalian genome, the functions of which are largely unexplored. The identification of ncRNA components in the cancer pathway reflects the necessity for complex gene regulation to achieve homeostasis and flexibility to maintain a normal developmental or physiological state. mir-17-92 is among the first miRNAs recognized as key components of the molecular network that impact tumorigenesis and tumor maintenance. Yet we are only beginning to understand its profound impacts and to appreciate its unique structural and functional relationship. The functional studies of mir-17-92 have served as a paradigm to investigate the interplay between protein coding genes and non-coding genes, and to understand the unique structural features of polycistronic miRNA clusters. In addition, miRNAs, and more broadly, ncRNAs, represent a novel class of molecules that offer great promise for developing new diagnostic markers and therapeutic treatments. Understanding the functions of mir-17-92 may serve as an entry point to explore the clinical applications of small RNA molecules that function in the oncogene and tumor suppressor network.
Acknowledgement
We thank Dr. Pengcheng Bu and Ms. Mona Foth for critical reading of this manuscript and for helpful discussions. L.H. is a fellow of the Searle Foundation and is supported by the K99 award from the NCI.
List of abbreviations
- miRNA
microRNA
- ncRNAs
non-coding RNAs
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, Dell'Aquila ML, Alder H, Rassenti L, Kipps TJ, Bullrich F, Negrini M, Croce CM. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci U S A. 2004a;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004b;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, Pasquinelli AE, Shiekhattar R. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- Cui JW, Li YJ, Sarkar A, Brown J, Tan YH, Premyslova M, Michaud C, Iscove N, Wang GJ, Ben-David Y. Retroviral insertional activation of the Fli-3 locus in erythroleukemias encoding a cluster of microRNAs that convert Epo-induced differentiation to proliferation. Blood. 2007;110:2631–2640. doi: 10.1182/blood-2006-10-053850. [DOI] [PubMed] [Google Scholar]
- Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, Thomas-Tikhonenko A. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett Y, McBride KM, Jankovic M, Gazumyan A, Thai TH, Robbiani DF, Di Virgilio M, San-Martin BR, Heidkamp G, Schwickert TA, Eisenreich T, Rajewsky K, Nussenzweig MC. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol. 2008;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil S, Caceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Hossain A, Kuo MT, Saunders GF. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol Cell Biol. 2006;26:8191–8201. doi: 10.1128/MCB.00242-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, Kobayashi SV, Lim L, Burchard J, Jackson AL, Linsley PS, Cleary MA. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol. 2008;28:2167–2174. doi: 10.1128/MCB.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- Kiriakidou M, Tan GS, Lamprinaki S, De Planell-Saguer M, Nelson PT, Mourelatos Z. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell. 2007;129:1141–1151. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Kluiver J, Poppema S, de Jong D, Blokzijl T, Harms G, Jacobs S, Kroesen BJ, van den Berg A. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243–249. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, Jensen K, Cobb BS, Merkenschlager M, Rajewsky N, Rajewsky K. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Landais S, Landry S, Legault P, Rassart E. Oncogenic potential of the miR-106-363 cluster and its implication in human T-cell leukemia. Cancer Res. 2007;67:5699–5707. doi: 10.1158/0008-5472.CAN-06-4478. [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Liu J, Rivas FV, Wohlschlegel J, Yates JR, 3rd, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat Cell Biol. 2005;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(Spec No 1):R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature. 2007;447:1126–1129. doi: 10.1038/nature05903. [DOI] [PubMed] [Google Scholar]
- Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S, Yoshida Y, Seto M. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087–3095. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- Park SM, Shell S, Radjabi AR, Schickel R, Feig C, Boyerinas B, Dinulescu DM, Lengyel E, Peter ME. Let-7 prevents early cancer progression by suppressing expression of the embryonic gene HMGA2. Cell Cycle. 2007;6:2585–2590. doi: 10.4161/cc.6.21.4845. [DOI] [PubMed] [Google Scholar]
- Petrocca F, Vecchione A, Croce CM. Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 2008a;68:8191–8194. doi: 10.1158/0008-5472.CAN-08-1768. [DOI] [PubMed] [Google Scholar]
- Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M, Cavazzini L, Volinia S, Alder H, Ruco LP, Baldassarre G, Croce CM, Vecchione A. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008b;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Schulte JH, Horn S, Otto T, Samans B, Heukamp LC, Eilers UC, Krause M, Astrahantseff K, Klein-Hitpass L, Buettner R, Schramm A, Christiansen H, Eilers M, Eggert A, Berwanger B. MYCN regulates oncogenic MicroRNAs in neuroblastoma. Int J Cancer. 2008;122:699–704. doi: 10.1002/ijc.23153. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- Shan SW, Lee DY, Deng Z, Shatseva T, Jeyapalan Z, Du WW, Zhang Y, Xuan JW, Yee SP, Siragam V, Yang BB. MicroRNA MiR-17 retards tissue growth and represses fibronectin expression. Nat Cell Biol. 2009;11:1031–1038. doi: 10.1038/ncb1917. [DOI] [PubMed] [Google Scholar]
- Suarez Y, Fernandez-Hernando C, Yu J, Gerber SA, Harrison KD, Pober JS, Iruela-Arispe ML, Merkenschlager M, Sessa WC. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci U S A. 2008;105:14082–14087. doi: 10.1073/pnas.0804597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G, Chartrand P. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282:2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- Tagawa H, Seto M. A microRNA cluster as a target of genomic amplification in malignant lymphoma. Leukemia. 2005;19:2013–2016. doi: 10.1038/sj.leu.2403942. [DOI] [PubMed] [Google Scholar]
- Tanzer A, Stadler PF. Molecular evolution of a microRNA cluster. J Mol Biol. 2004;339:327–335. doi: 10.1016/j.jmb.2004.03.065. [DOI] [PubMed] [Google Scholar]
- Uren AG, Kool J, Matentzoglu K, de Ridder J, Mattison J, van Uitert M, Lagcher W, Sie D, Tanger E, Cox T, Reinders M, Hubbard TJ, Rogers J, Jonkers J, Wessels L, Adams DJ, van Lohuizen M, Berns A. Large-scale mutagenesis in p19(ARF)- and p53-deficient mice identifies cancer genes and their collaborative networks. Cell. 2008;133:727–741. doi: 10.1016/j.cell.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, Zlotorynski E, Yabuta N, De Vita G, Nojima H, Looijenga LH, Agami R. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- Wang CL, Wang BB, Bartha G, Li L, Channa N, Klinger M, Killeen N, Wabl M. Activation of an oncogenic microRNA cistron by provirus integration. Proc Natl Acad Sci U S A. 2006;103:18680–18684. doi: 10.1073/pnas.0609030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130–2134. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan HL, Xue G, Mei Q, Wang YZ, Ding FX, Liu MF, Lu MH, Tang Y, Yu HY, Sun SH. Repression of the miR-17-92 cluster by p53 has an important function in hypoxia-induced apoptosis. EMBO J. 2009;28:2719–2732. doi: 10.1038/emboj.2009.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- Zhao T, Li G, Mi S, Li S, Hannon GJ, Wang XJ, Qi Y. A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev. 2007;21:1190–1203. doi: 10.1101/gad.1543507. [DOI] [PMC free article] [PubMed] [Google Scholar]